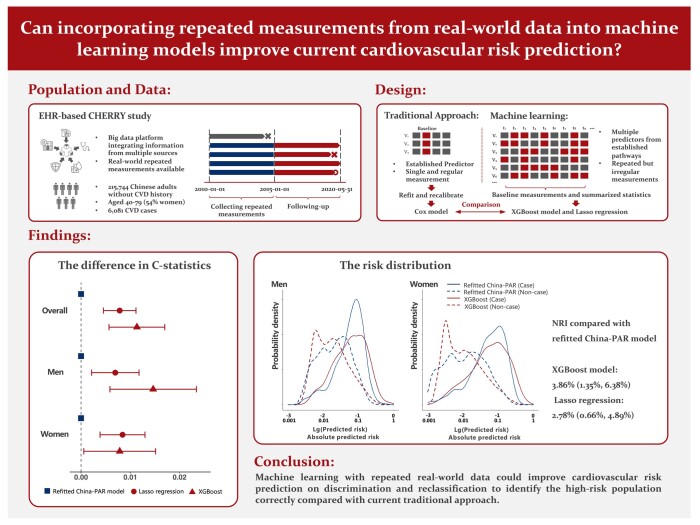
Abstract
Aims
Existing electronic health records (EHRs) often consist of abundant but irregular longitudinal measurements of risk factors. In this study, we aim to leverage such data to improve the risk prediction of atherosclerotic cardiovascular disease (ASCVD) by applying machine learning (ML) algorithms, which can allow automatic screening of the population.
Methods and results
A total of 215 744 Chinese adults aged between 40 and 79 without a history of cardiovascular disease were included (6081 cases) from an EHR-based longitudinal cohort study. To allow interpretability of the model, the predictors of demographic characteristics, medication treatment, and repeatedly measured records of lipids, glycaemia, obesity, blood pressure, and renal function were used. The primary outcome was ASCVD, defined as non-fatal acute myocardial infarction, coronary heart disease death, or fatal and non-fatal stroke. The eXtreme Gradient boosting (XGBoost) algorithm and Least Absolute Shrinkage and Selection Operator (LASSO) regression models were derived to predict the 5-year ASCVD risk. In the validation set, compared with the refitted Chinese guideline–recommended Cox model (i.e. the China-PAR), the XGBoost model had a significantly higher C-statistic of 0.792, (the differences in the C-statistics: 0.011, 0.006–0.017, P < 0.001), with similar results reported for LASSO regression (the differences in the C-statistics: 0.008, 0.005–0.011, P < 0.001). The XGBoost model demonstrated the best calibration performance (men: Dx = 0.598, P = 0.75; women: Dx = 1.867, P = 0.08). Moreover, the risk distribution of the ML algorithms differed from that of the conventional model. The net reclassification improvement rates of XGBoost and LASSO over the Cox model were 3.9% (1.4–6.4%) and 2.8% (0.7–4.9%), respectively.
Conclusion
Machine learning algorithms with irregular, repeated real-world data could improve cardiovascular risk prediction. They demonstrated significantly better performance for reclassification to identify the high-risk population correctly.
Keywords: Prediction, Preventive Cardiology, Risk
Graphical Abstract
Graphical Abstract.
See the editorial comment for this article ‘From data to wisdom: harnessing the power of multimodal approach for personalized atherosclerotic cardiovascular risk assessment’, by S. Al-Kindi and K. Nasir, https://doi.org/10.1093/ehjdh/ztad068.
Introduction
Safe and cost-effective treatments can reduce cardiovascular risk significantly. The magnitude of treatment benefit is directly related to the pre-treatment cardiovascular risk of individual patients. To accurately determine this risk, reliable risk prediction equations must be employed. The global cardiovascular guidelines recommend various risk assessment tools to tackle the heavy burden of cardiovascular disease (CVD), such as the Pooled Cohort Equation (PCE),1 the Systematic COronary Risk Evaluation (SCORE) model,2 and the Prediction for Atherosclerotic Cardiovascular Disease Risk in China (China-PAR) model.3 Although current risk prediction models using a number of traditional CVD risk factors have played an important role in CVD prevention, the predictive performance has yet to produce satisfactory results. For example, several external validation studies have demonstrated that the C-statistics of these models range only between 0.65 and 0.74,4,5 and may incorrectly estimate the absolute cardiovascular risk.4,6 It is known that traditional CVD risk prediction can be enhanced through additional information gained from either new predictors or repeated measurements. In addition to traditional predictors such as age, smoking, and systolic blood pressure (SBP), new predictors from various aetiological pathways [e.g. lipoprotein (a) and apolipoprotein,7,8 glucose metabolism,9,10 and renal function markers11] can also potentially improve prediction accuracy. However, implementing traditional prediction models using all these novel predictors for CVD primary prevention in the entire population is not realistic in real-world clinical practice. Secondly, recent evidence suggests that repeated measurements of CVD risk factors in traditional prediction models can improve performance,12–14 which may capture the longitudinal information of risk factors and help explain the cardiovascular residual risk.15 However, the current traditional modelling approaches have limitations in that they are able to consider only a limited number and type of repeated predictors,16 and they may overlook potential interactions among these predictors.17,18
In contrast, electronic health records (EHRs) can not only provide a wealth of information with repeated measurements on various predictors16 but also allow for automated screening if the risk prediction tool is embedded.19,20 However, the data structure in real-world EHR systems often differs from that in traditional cohort studies. Although new predictors may exist in subgroups of the population, the pattern of available risk factors is often irregular. For example, patients may undergo a series of repeated measurements, especially for traditional CVD risk factors, but the number of repeats varies among subjects. Moreover, it is also quite common that different predictors are measured between individuals, even from the same aetiological pathway. For example, someone having information on obesity will have his or her body mass index measured, whereas others will have their waist-hip ratio measured. Besides, risk factors are generally measured at different time points. Therefore, this EHR-based information remains challenging to be incorporated using conventional risk prediction models.
In this case, machine learning (ML) algorithms can be a valuable alternative to handle such complex data. While evidence shows that the benefits of ML algorithms over traditional models using the same predictors were limited, they can excel in accommodating multiple predictors and handling irregular measurements, making them suitable for leveraging the rich information present in EHRs effectively.21,22 While ML has been increasingly utilized to leverage information from repeated measurements in certain hospital-based scenarios,23,24 its application in primary care for cardiovascular risk assessment remains limited.25,26 Existing studies have demonstrated that ML can enhance risk prediction,22,27 but they have not fully utilized time-to-event information or comprehensively evaluated predictive performance. Developing fixed-term survival prediction models is crucial for CVD risk assessment, as they align with the recommended risk stratification cut-offs in clinical guidelines.1–3
Therefore, this study aims to investigate the improvement in CVD risk prediction by incorporating irregularly repeated real-world measurements of multiple predictors using ML models. The predictive performance is then compared against the guideline-recommended traditional Cox regression model.28
Methods
Study design
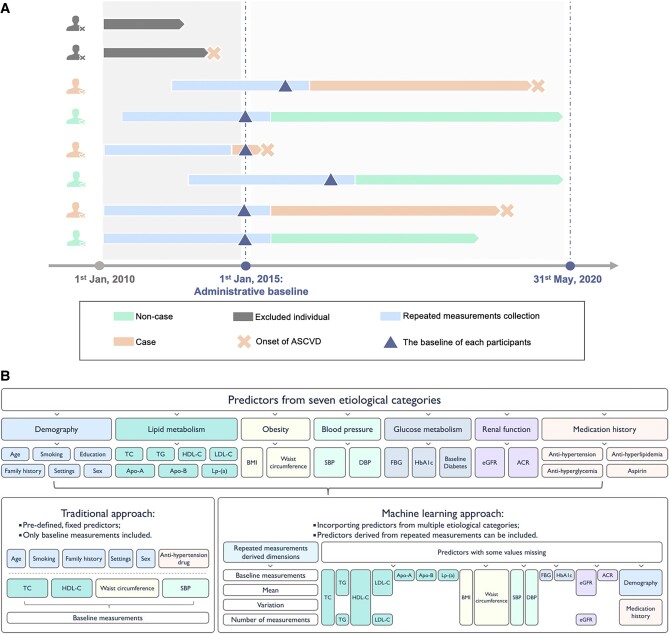
The concept of the study design is shown in Figure 1A. The population included in this study was taken from the Chinese Electronic Health Records Research in Yinzhou (CHERRY) study, which was an EHR-based cohort study in Yinzhou, Ningbo (a developed area in Eastern China). A detailed description of the CHERRY study has been published elsewhere.29 The inclusion criteria of this study population consist of (i) age ranging between 40 and 79 years at the entry time; (ii) Chinese residents registered in the health information system during the period between 1 January 2010 and 31 December 2016; and (iii) had been living in Yinzhou for at least 6 months. The exclusion criteria of this study are as follows: (i) had no records of serum lipid measurements. Since lipid-related predictors were causally related to atherosclerotic CVD (ASCVD), this may cause the model to be not applicable. (ii) had a history of CVD before being enrolled in the study. A flowchart of the inclusion and exclusion process is shown in Supplementary material online, Figure S1. Finally, 215 744 participants were included in the analysis set, among which a random sample of 80% (∼180 000) was separated as the training set to derive the models, and the remaining participants were left for the validation (shown in Supplementary material online, Figure S2). This study was approved by the Peking University Institutional Review Board (IRB00001052-16011).
Figure 1.
The study design and categories of predictors. (A) The cohort design of the study; (B) predictors of seven aetiological categories included in different approaches.
To maximize the number of repeated measurements collected, the baseline in this study was set as (i) the time when the participants registered in the system, (ii) the time when participants reached 40 years old, (iii) the time when the first serum lipid measurement was recorded, or (iv) 1 January 2015, whichever the latest. The repeated measurements were collected from the past 5 years before the baseline. Participants would be followed up to the time (i) when they experienced their first ASCVD event (further defined in the outcomes section), (ii) they were censored from following up, or (iii) 31 May 2020, whichever was the earliest.
Predictors
Seven common categories of cardiovascular risk factors (shown in Figure 1B) with 25 markers in total were pre-identified as the pool of predictors, including demography (age, sex, education levels, settings, smoke status, and family history), lipid metabolism {total cholesterol [TC], HDL cholesterol [HDL-C], LDL cholesterol [LDL-C], triglycerides [TG], apolipoprotein A [apo A], apolipoprotein B [apo B], and lipoprotein (a) [Lp-(a)]}, obesity (BMI and waist circumference), glucose metabolism [fasting blood glucose (FBG), diabetes at baseline, and haemoglobin A1c (HbA1c)], blood pressure [SBP and diastolic blood pressure (DBP)], renal function [estimated glomerular filtration rate (eGFR) and albumin creatinine ratio (ACR)], and medical treatments (antihypertension, antihyperglycaemic, antihyperlipidemic treatment, and aspirin). We selected these risk factors because they were universally incorporated into cardiovascular risk prediction,1,12,28,30–32 had possible causal relationships with ASCVD outcomes,8,9,33,34 or were closely associated with ASCVD from aetiological perspectives.7,10,11,35,36 Measurements of these predictors were collected from multiple sources in the regional health system, including census data, electronic medical records (EMRs), disease surveillance, chronic disease management system, and health check, which are summarized in Supplementary material online, Table S1. These records will be inherently linked to each other according to a unique and encoded identifier. Detailed data collection procedures of various data sources are described in Supplementary material online, Method S1. The exact definitions of each medical treatment are given in Supplementary material online, Table S2. Extreme outliers were removed according to pre-specified normal ranges of key predictors (shown in Supplementary material online, Table S3).
Considering the irregular nature of the predictors’ information available, we used a simple but effective approach to leverage these repeated measurements by summarized statistics.24,37 Standard deviation (SD), range, and the difference between the last and the first measurements were calculated as derived predictors since many studies have proposed that the variability of predictors is associated with CVD.17,18,38 The number of measurements was also counted and included in the pool of predictors.39 The mean values of the predictors were also summarized to represent the long-term average of these predictors. All the baseline and derived predictors included in this research are listed in Supplementary material online, Table S4.
Outcomes
The definition of the ASCVD was consistent with the one used in the China-PAR or PCE model, which was defined as the composite outcome of non-fatal or fatal stroke (ICD-10 code: I60, I61, I63, and I64), non-fatal myocardial infarction (I21 and I22), and coronary heart disease death (I20–I25).28 The outcomes in this study were collected from the following sources: disease surveillance, chronic disease management system, death registry, and EMR. Among these sources, the disease surveillance and death registry were recognized as the gold standard. The outcome used the first ASCVD events that occurred after the baseline and before 31 May 2020.
Risk prediction models
Since the China-PAR model was the Chinese guideline–recommended risk assessment tool in primary care, our study selected this model as the reference to be compared. The model was modified by two different approaches in this study to make the comparison fair: (i) the refitted China-PAR model was developed by directly replacing all the coefficients in the original model but preserving all the predefined terms (including all the interaction terms); (ii) the recalibrated China-PAR model was developed by replacing the baseline survivals and means of linear predictors in the original model without altering any predefined terms and their corresponding coefficients.
Two ML approaches were finally adopted in this study, which were eXtreme Gradient Boosting (XGBoost) algorithm and Least Absolute Shrinkage and Selection Operator (LASSO) regression. The choice of algorithms depends on various factors such as the nature of the data, the size of the dataset, the complexity of the problem, and the desired interpretability of the model.19 For large datasets with high dimensionality (many predictors), algorithms that can efficiently handle such data, such as Random Forest, Gradient Boosting, or Deep Learning models, may be suitable.40,41 Meanwhile, for CVD risk prediction, model interpretability is crucial. Simpler models such as regression-based models or decision trees are still preferred, as they can provide more transparent and easily interpretable results.19 Random Forest or Gradient Boosting can also offer feature importance rankings and handle missing values without imputation. Finally, the computational cost of training the model is a consideration, especially for large datasets. Linear models and tree-based models tend to be faster and easier to train than deep learning models. To control the potential overfitting, algorithms with built-in regularization, such as LASSO regression, were considered. After studying the performance and feasibility of the four aforementioned methods, XGBoost and LASSO regression were chosen because they performed better than the others and are relatively easy to be interpreted and implemented in the EHR system. These two algorithms utilize the information of predictors from different perspectives so this will also help to find the more suitable approach to leverage the repeated measurements.42,43 The importance of predictors was assessed according to the average reduction of information entropy in the XGBoost model and the absolute value of the β coefficients in LASSO regression, which reflected the information gains or the marginal effects of the predictors. Two ML classifiers were first trained in the 126 893 subjects with known outcome information at the end of the fifth year. Then, the two ML models were embedded into a Cox regression model to predict the absolute 5-year risk. Hyperparameter tuning was conducted by maximizing the area under the curve (AUC) in the five-fold cross-validation. Grid search were iterated 100 times to acquire the optimized hyperparameter sequence.44,45 The ranges of the hyperparameters are given in Supplementary material online, Table S5.
Statistical analysis
Continuous predictors were described using means and SDs, while categorical predictors were described using counts and percentages. The associations between predictors and ASCVD were given according to the hazard ratios of Cox proportional hazard regression adjusted for the variables from the China-PAR models. Proportions of missingness were described for each predictor. The predictors in the China-PAR models were multiple-imputed by chain equations (five imputation sets were created) to compare with the two ML models.12,46 The performance metrics were measured in each imputation set and then pooled according to Rubin’s rules.47 Machine learning models can handle the issue of missingness directly to preserve initial information. Details of data imputation are provided in Supplementary material online, Method S2. The performances of the models were evaluated in terms of the following perspectives: discrimination, calibration, and reclassification. The discrimination was assessed by using Harrell’s C-statistic. Calibration was used to measure the coordination between predicted risk and observed risk, which was evaluated using the Hosmer–Lemeshow and calibration plots.48 The C-statistics of different models were compared using the approach proposed by Kang et al.49 The risk distribution by different models was also illustrated. To pool the Hosmer–Lemeshow given by different imputation sets, a Dx statistic following the F distribution was generated based on the approach proposed by Rubin50 and Li et al.51 We provided the standard reclassification table. Net reclassification improvement (NRI) and integrated discrimination improvement (IDI) were calculated to quantify the reclassification benefits of the ML models over the refitted China-PAR model. The cut-offs of risk groups were selected according to the 2019 Guideline on the assessment and management of cardiovascular risk in China.3 A decision curve analysis (DCA) was also conducted to illustrate the clinical implications of the ML models. Sensitivity analyses were conducted as follows: (i) to further ascertain whether the possible improvement in risk prediction was driven by leveraging the information from the repeated measurements or by simply including more baseline predictors, a Cox regression model including all the baseline measurements of each predictor was constructed, and its performance was compared against the two ML models and the refitted China-PAR model; (ii) the performance of the recalibrated China-PAR was assessed and compared to evaluate how much improvement ML models can achieve compared with the per-guideline approach. All analyses were conducted using R version 4.0.4 with a statistically significant level of P < 0.05. The XGBoost model was constructed by using the xgboost package version 1.4.1.1, and the LASSO regression model was constructed with glmnet package version 4.1-1. The mice package version 3.13.0 was adopted for the multiple imputation by chain equations.
Results
Basic descriptions for participants and predictors
The characteristics of the 215 744 included participants are described in Table 1. Fifty-four per cent of the participants were women, and their mean age was ∼56.7 (SD = 9.6). The means of major risk factors for ASCVD: SBP, TC, and high-density lipoprotein cholesterol were 134.5 mmHg, 4.9, and 1.3 mg/dL, respectively. The average BMI was 23.3 kg/m2. Overall, 12.1% of them had diabetes at baseline. During a median of 5.4-year follow-up, 6081 individuals (2.82%) had ASCVD outcomes. The incidence rate of ASCVD was 6178 per million person-years. Only TC and anti-hyperglycaemia treatment significantly differed between derivation and internal validation datasets (shown in Supplementary material online, Table S6). The missing proportions of each predictor are shown in Supplementary material online, Table S7. The number of measurements and time intervals between each measurement of key predictors for each individual is given in Supplementary material online, Table S8. The mean number of measurements of TC, SBP, BMI, and fasting glucose were 3, 2, 1, and 3, respectively. The corresponding median time intervals between these measurements were 269, 136, 267, and 251 days.
Table 1.
Characteristicsa of the study population
| Overall (N = 215 744) | Men (n = 100 078) | Women (n = 115 666) | |
|---|---|---|---|
| Demography | |||
| Age, years | 56.70 (9.59) | 57.10 (9.75) | 56.35 (9.44) |
| Rural | 65 086 (30.34%) | 30 016 (30.15%) | 35 070 (30.51%) |
| Smokers (current or ever) | 57 961 (26.87%) | 53 861 (53.82%) | 4100 (3.54%) |
| Finished high school | 108 120 (50.11%) | 55 576 (55.53%) | 52 544 (45.43%) |
| Family history of ASCVD | 1318 (0.61%) | 701 (0.70%) | 617 (0.53%) |
| Blood pressure | |||
| SBP, mmHg | 134.45 (16.64) | 134.58 (16.37) | 134.32 (16.88) |
| DBP, mmHg | 82.63 (9.87) | 83.10 (9.90) | 82.18 (9.81) |
| Obesity | |||
| Waist circumference, cm | 81.76 (7.94) | 83.93 (7.61) | 79.90 (7.73) |
| BMI, kg/m2 | 23.31 (2.87) | 23.44 (2.71) | 23.21 (3.01) |
| Lipid metabolism | |||
| Total cholesterol, mmol/L | 4.90 (0.98) | 4.77 (0.97) | 5.01 (0.98) |
| HDL-C, mmol/L | 1.30 (0.34) | 1.25 (0.34) | 1.35 (0.33) |
| TG, mmol/L | 1.61 (1.09) | 1.66 (1.20) | 1.56 (0.99) |
| LDL-C, mmol/L | 2.84 (0.82) | 2.77 (0.81) | 2.90 (0.83) |
| Apo A, mmol/L | 1.22 (0.27) | 1.18 (0.27) | 1.26 (0.27) |
| Apo B, mmol/L | 0.95 (0.25) | 0.95 (0.25) | 0.95 (0.25) |
| Lp-(a), mmol/L | 4.87 (0.14) | 4.60 (0.14) | 5.12 (0.15) |
| Glucose metabolism | |||
| FBG, mmol/L | 5.67 (1.57) | 5.76 (1.72) | 5.60 (1.44) |
| HbA1c, % | 6.86 (1.90) | 6.99 (1.98) | 6.73 (1.82) |
| Diabetes mellitus | 26 090 (12.09%) | 12 364 (12.35%) | 13 726 (11.87%) |
| Renal function | |||
| eGFR, mL/min/1.73 m2 | 98.92 (15.30) | 97.71 (15.28) | 99.94 (15.25) |
| ACR, mg/g | 15.90 (45.36) | 16.32 (48.91) | 15.57 (42.39) |
| Medication | |||
| Anti-hypertension treatment | 75 857 (35.16%) | 35 590 (35.56%) | 40 267 (34.81%) |
| Anti-hyperlipidaemia treatment | 35 561 (16.48%) | 15 662 (15.65%) | 19 899 (17.20%) |
| Anti-hyperglycaemia treatment | 22 847 (10.59%) | 10 881 (10.87%) | 11 966 (10.35%) |
| Aspirin treatment | 19 064 (8.84%) | 9100 (9.09%) | 9964 (8.61%) |
| Outcome | |||
| ASCVD events | 6081 (2.82%) | 3272 (3.27%) | 2809 (2.43%) |
| Average follow-up time, years | 5.41 (1.36) | 5.41 (1.51) | 5.41 (1.22) |
| Incidence rate of ASCVD, per million person-years (95% CI) | 6178 (6177–6179) | 7242 (7241–7243) | 5245 (5244–5246) |
aCategorical variables are presented by counts and percentages; continuous variables are presented by means and SDs. All summarized statistics are given based on the complete sets of each predictor.
The discrimination of the models
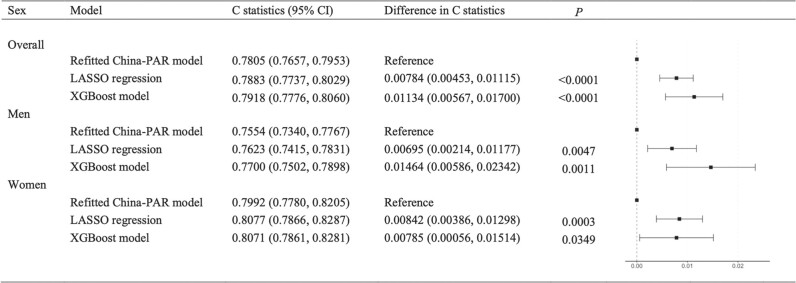
In the validation set, the C-statistics with the absolute differences compared with the refitted China-PAR model are given in Figure 2. The C-statistic of the XGBoost model was 0.7918 [95% confidence interval (CI): 0.7776–0.8060] and that of LASSO regression was 0.7883 (0.7737–0.8029). The two ML models performed better than the refitted China-PAR model in the parameter of discrimination (differences in the C-statistic for XGBoost: 0.01134, 0.00567–0.01700, P < 0.001; for LASSO: 0.00784, 0.00453–0.01115, P < 0.001). They also performed better than that of the refitted China-PAR model in the same parameter in both men and women, where the XGBoost model performed the best among men, while LASSO regression performed the best among women. The final hyperparameters in the final ML models are given in Supplementary material online, Table S9. The major structures of the final ML models are given in Supplementary material online, Figure S3 and Table S10.
Figure 2.
The difference in C-statistics scores compared with the refitted China-PAR model. The results are given based on the validation set of 31 544.
The calibration of the models
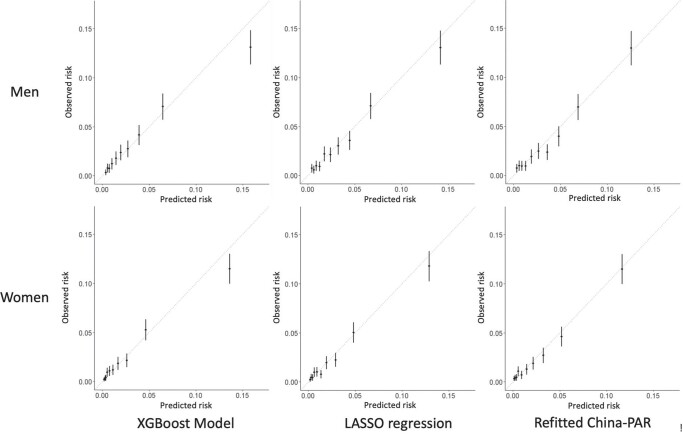
The XGBoost model showed better calibration than the refitted China-PAR model in both men and women (XGBoost: Dx = 0.598, P = 0.75 in men and Dx = 1.867, P = 0.08 in women; refitted China-PAR: Dx = 2.832 in men, P = 0.004 and Dx = 3.352 in women, P = 0.001), while LASSO regression was recalibrated well in men (Dx = 1.639, P = 0.11) but not in women (Dx = 1.950, P = 0.048). The calibration plots are shown in Figure 3. Although the XGBoost model slightly overestimated the risk in the highest risk group, the coordination of the predicted risks and Kaplan–Meier observed risks was much better than the LASSO model and refitted the China-PAR model, especially among low-risk deciles.
Figure 3.
Calibration plots of different models by sexa. The results are given based on the validation set of 31 544.
Clinical implications on outcomes
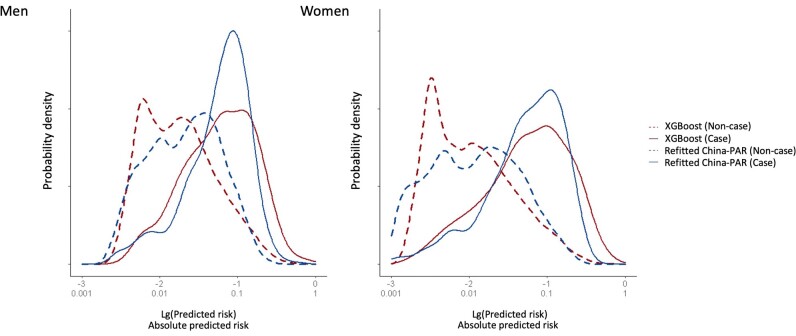
In the validation set, the reclassification table is shown in Table 2. By using the XGBoost model rather than the refitted China-PAR model, additional 3355 subjects out of total 24 247 non-case individuals were classified as lower risk. There were 667 subjects classified as higher risk. A net quantity of 2688 people (11.09%) was reclassified into the correct groups. Among 969 individuals who developed CVD during follow-up, XGBoost and the refitted China-PAR model selected a similar number of high-risk subjects, that is, 550 and 585. After taking the medium-risk group into account, China-PAR correctly selected 70 (7.22%) more case individuals. The overall NRI rate was 3.87% (1.35–6.38%). Similarly, the NRI rate for LASSO regression was 2.78% (0.66–4.89%). A direct comparison of the predicted risk against the refitted China-PAR model showed that the IDIs of the XGBoost model and LASSO regression were 0.0174 (0.0135–0.0212) and 0.0106 (0.0081–0.0131), respectively. The risk distributions predicted by the XGBoost and the refitted China-PAR models are illustrated in Figure 4. The risk predicted by XGBoost tended to centralize in the lower range in non-cases in both men and women, with a larger difference between the risks of cases and non-cases. The DCA demonstrated that all three models, namely XGBoost, LASSO, and the refitted China-PAR model, exhibited favourable performance by deviating from the curves of treating all or treating none within the common cardiovascular risk range of 0–20%. Moreover, the net benefit of the XGBoost model surpassed that of the refitted China-PAR model between the threshold range of 7.5 and 12.5%, while the net benefit of the LASSO regression model was superior within the range of 12.5–17.5% (see Supplementary material online, Figure S4).
Table 2.
Reclassification of machine models against the refitted China-PAR modela
| XGBoost | NRI (95% CI) | IDI (95% CI) | |||||
|---|---|---|---|---|---|---|---|
| <2.5% | 2.5–4.9% | ≥5% | Total | ||||
| Refitted China-PAR | |||||||
| Non-case | <2.5% | 14 142 | 270 | 74 | 14 486 | 0.0386 | 0.0174 |
| 2.5–4.9% | 2119 | 2506 | 323 | 4948 | (0.0135, 0.0638) | (0.0135, 0.0212) | |
| ≥5% | 53 | 1183 | 3577 | 4813 | |||
| Total | 16 314 | 3959 | 3974 | 24 247 | |||
| Case | <2.5% | 185 | 11 | 8 | 204 | ||
| 2.5–4.9% | 46 | 114 | 20 | 180 | |||
| ≥5% | 1 | 62 | 522 | 585 | |||
| Total | 232 | 187 | 550 | 969 | |||
| LASSO | |||||||
|---|---|---|---|---|---|---|---|
| <2.5% | 2.5–4.9% | ≥5% | Total | ||||
| Refitted China-PAR | |||||||
| Non-case | <2.5% | 14 147 | 324 | 15 | 14 486 | 0.0278 | 0.0106 |
| 2.5–4.9% | 1057 | 3543 | 348 | 4948 | (0.0066, 0.0489) | (0.0081, 0.0131) | |
| ≥5% | 3 | 826 | 3984 | 4813 | |||
| Total | 15 207 | 4693 | 4347 | 24 247 | |||
| Case | <2.5% | 188 | 14 | 2 | 204 | ||
| 2.5–4.9% | 22 | 132 | 26 | 180 | |||
| ≥5% | 0 | 41 | 544 | 585 | |||
| Total | 210 | 187 | 572 | 969 | |||
aThe results are given based on the subjects who were not censored (25 216) from the validation set of 31 544.
Figure 4.
Distribution of predicted risk given by the XGBoost model and the refitted China-PAR model in the validation set.
The importance of predictors
The associations between the predictors and ASCVD are presented in Supplementary material online, Table S11, which are adjusted by predictors in the China-PAR model. All predictors were included in the XGBoost model because it used the random subspace sampling technique, while the LASSO regression model selected only 78 of the total 101 predictors (i.e. baseline and summarized statistics of repeat information of 25 markers). The rank of importance is given in Supplementary material online, Figure S5. In general, age, anti-hypertension treatment history, glucose metabolism–related predictors, lipid metabolism–related predictors, blood pressure, eGFR, and a family history of ASCVD were most valued by both ML models. FBG ranked third and fifth in the rank of importance in XGBoost and LASSO regression, respectively. Novel lipid predictors such as apo B ranked eighth and tenth in the two ML models, while classic predictors such as TC ranked only 10th and 17th. The importance of smoking and predictors indicating obesity was relatively low (BMI: 16th in XGBoost and 19th in LASSO; waist circumference: 20th in XGBoost and 16th in LASSO; smoking: 18th in XGBoost and 14th in LASSO).
Sensitivity analysis
The Cox regression model with the baseline measurements of all predictors performed better than the refitted China-PAR model, while it was still worse than the XGBoost model in the whole validation set for the parameter of discrimination (the differences in the C–statistics: 0.00563, 0.00118–0.01009, P = 0.01, Supplementary material online, Table S12). Its discriminative performance was not significantly different from that of LASSO regression (0.00214, −0.00088–0.00515, P = 0.17, Supplementary material online, Table S12). The coordination of predicted and observed risk indicated by the calibration plot of the Cox model with all baseline measurements was insufficient, which was not even better than the refitted China-PAR model (Dx = 2.421, P = 0.01 in men and Dx = 2.216, P = 0.02 in women, Supplementary material online, Figure S6). Both ML models performed significantly better than the recalibrated China-PAR model in the two parameters of discrimination (all P < 0.001, Supplementary material online, Table S13) and calibration (recalibrated China-PAR: Dx = 2.421 in men and Dx = 2.216 in women, both P < 0.001, Supplementary material online, Figure S6).
Discussion
This study used two ML approaches (XGBoost and LASSO) to leverage the existing repeated measurements in EHR data to predict 5-year atherosclerotic cardiovascular risk. Both ML models outperformed the recalibrated and the refitted China-PAR model in the parameters of discrimination, calibration, and reclassification, which is the model recommended by the 2019 Guideline on the assessment and management of cardiovascular risk in China.3
Repeated measurements from EHRs offer valuable contributions to cardiovascular risk prediction. Notably, the QRISK3 model in the United Kingdom was derived from EHR data obtained from general practice computer systems, in which the SD of SBP was included as a predictor.12 This model stands as the first nationwide-used risk prediction model to incorporate predictors derived from repeated blood pressure measurements. Similarly, Paige et al.14 leveraged EHR data from the Health Improvement Network, a UK general practice electronic database, and applied a landmark model to utilize information from repeated measurements of smoking status, SBP, TC, and HDL-C, resulting in a significant improvement in C-statistics. Our study aligns with these findings, demonstrating that incorporating repeated measurements of multiple predictors from EHRs enhances predictive performance when compared with the Cox model which uses only baseline measurements. The temporal information present in repeated measurements is of great importance. It is reflected by the time intervals between measurements and trends or patterns observed over time. While the QResearch study and Paige et al. did not explicitly report the time intervals (or density) of measurements, Paige et al. did fit the temporal trend and dependency of repeated measurements using a multivariate linear mixed-effects model.12,14 In our study, we observed that the average time intervals between measurements of key predictors were generally <1 year (see Supplementary material online, Table S8), signifying the richness of information that can be harnessed from EHRs. The correlated predictors,22 irregularly missing records,52 and data with strong interaction in the EHR necessitate applying a novel modelling approach such as ML, which usually utilizes high-dimensional unstructured data to enhance predictive performance.53,54 However, it is worth noting that currently off-the-shelf ML algorithms lack a comprehensive approach to model secular trends and dependencies in irregularly structured data. This presents an area for further methodological investigation to effectively harness the temporal nature of the data for CVD risk prediction.
Although it is controversial whether ML can improve cardiovascular risk prediction using only baseline measurements of limited predictors,27,55 several pieces of evidence have demonstrated that predictive performance can be largely improved when predictors derived from repeated measurements are fed into ML models.25,26 For instance, Li et al. summarized the repeated measurements of blood lipid, blood pressure, and HbA1c from the EMRs of 101 110 people in a US regional healthcare system, into extremum, number of measurements, means, etc. Then, these longitudinal-derived predictors were used in the random forest ML model, causing large increments in the AUC (e.g. 0.823–0.902).25 Compared with those studies, our study demonstrated that: (i) by embedding XGBoost and LASSO regression algorithms into Cox regression to leverage time-to-event information, similar improvement could result in the parameter of discrimination when evaluated by using C-statistic; (ii) besides discrimination capability, our study comprehensively assessed the performance of the model from the perspectives of calibration and reclassification based on survival probabilities; (iii) it is feasible to conduct CVD risk prediction using rich but irregular existing EHR data for risk stratification without extra cost for screening new markers.
Under real-world scenarios, many predictors are not universally screened in the population. However, it is shown that these markers can predict cardiovascular risk. For example, the mean of FBG presents the long-term control of glucose metabolism, which is predictive for CVD independently.10 Apolipoprotein B and Lp-(a) are also useful biomarkers for ASCVD.7 Poor renal function (e.g. impaired eGFR) can result in hypertension, left ventricular hypertrophy, endothelial dysfunction, dyslipidaemia, and low-grade inflammation.56 In our study, these predictors were informative in predicting cardiovascular events as reflected by the importance of the predictors (see Supplementary material online, Figure S5) and the structures of the models (see Supplementary material online, Figure S3 and Table S10). Making the best use of these existing biomarkers in EHR data to enhance CVD risk prediction may change the current way of screening high-risk populations in clinical practice. Considering the irregular nature of the data, ML algorithms can be good alternatives. The ML models could accommodate residents with some unmeasured predictors flexibly. Including a predictor or its repeated measurement in the model does not necessitate requiring complete information on the whole population.
The absolute increment of C-statistic in our study was 0.0113. This gain in the parameter of discrimination was meaningful compared with the gains generated by adding established risk factors. For example, in the Emerging Risk Factors Collaboration study, incorporating C reaction protein or HDL-C into the traditional Cox model to predict ASCVD incidents will increase the C-statistics by 0.0039 or 0.0050, respectively.57 When SBP was removed from the Reynolds score in the Women’s Health Study, the change in the C-statistic was 0.01.58 C-statistic is an insensitive indicator that ranges from 0.5 to 1.0. The larger the C-statistic is, the more challenging it is to improve it.59 HDL-C can increase the C-statistics score only by 0.0013 in our cohort. As advised by Cook,58 the improvement in risk prediction provided by the two ML models was also evaluated using NRI and IDI in this study. The reclassification table of the XGBoost model indeed indicated significant net benefit. In the validation set with 25 216 subjects, according to the cut-offs defined by the current Chinese guideline, ∼4% more subjects will be allocated to proper risk groups and correspondingly receive more suitable recommendations on intervention. Assuming that statin therapy was recommended to the high-risk population, which helped to reduce the CVD risk by 20%,60 such assessments of individuals by the XGBoost and the refitted China-PAR models could assign 4529 (17.9%) and 5398 (21.4%) patients to initiate statin treatment and help prevent 110 and 117 CVD outcomes over 5 years, respectively. Correspondingly, for every 41 and 46 patients treated, 1 CVD outcome was prevented by using the XGBoost and the refitted China-PAR models. This is consistent with the calibration plot, in which the risk predicted by the XGBoost model was more coordinated to the observed risk than the refitted China-PAR model, especially among the low- or intermediate-risk groups. Such consistency indicates that the XGBoost model may gain benefit under the existing risk cut-off values. Considering the large numbers of the low-risk population, great benefits are likely to be achieved when this model is implemented for risk screening. In the DCA, the threshold probability defined the criteria for intervention in individuals. If the estimated risk exceeded the threshold probability, intervention would be recommended. The net benefit of the XGBoost model was higher than that of the refitted China-PAR model within the threshold probability range of 7.5–12.5%. This range aligns with the typical cut-off risk values recommended by the guidelines for initiating critically important interventions such as statin therapy.1,2 These results suggest the potential net benefit of implementing the XGBoost model based on the existing risk cut-offs. On the other hand, the risk predicted by LASSO regression may be more suitable for use in high-risk individuals, given its larger net benefit across a range of 12.5–17.5%. Finally, we note that this administrative data–based approach can enhance CVD primary prevention by offering a more accurate prediction without any extra cost for screening new markers.
In the present landscape, most risk prediction models have developed their own implementation tools, some of which are integrated into the health information system (e.g. QRISK in the United Kingdom and PREDICT in New Zealand),6,12 while others are offered independently through websites or applications (e.g. PCE, SCORE2, and the China-PAR model).1,28,30 Given the nature of utilizing comprehensive information from EHRs, we recommend implementing the ML model by embedding it within the healthcare information system. This approach can also facilitate automatic population screening, enhancing the sustainability of cardiovascular risk prediction. However, unlike the traditional Cox model, the implementation of an already derived ML model is not always straightforward. Our algorithm for 5-year prediction of CVD risk involved a two-stage process. The first stage utilized ML classification algorithms, while the second stage embedded the ML classifier into a Cox regression model to predict absolute 5-year risk. Therefore, we firmly believe that the baseline survival characteristics of local populations remain crucial for accurate absolute risk prediction. As a result, recalibration of the model may still be necessary when applying it to different populations, along with external validation to assess its performance in diverse settings.
Our study also has several limitations. First, although internally validated, the ML risk prediction models derived in our study were not externally and independently validated. Our study aims not to propose and generalize the ML models to other populations but to answer a methodological question by comparing the performance of two ML approaches to the locally refitted China-PAR models. The models’ relative performance was still valid since the performance was measured by the same scale from the same dataset. Secondly, only two ML methods were present in this study, considering the nature and sample size of the data, the complexity of the algorithm, and the desired model interpretability. Advanced ML methods such as neural networks can be adapted to use data in the future.61 Thirdly, our study is based on regional data, which may not fully represent the diversity of the Chinese population nationwide. Variations in genetic background, culture, socioeconomic levels, climate, geographic features, lifestyle, and dietary patterns among different ethnic groups within the Chinese population could influence the generalizability of our findings. Nevertheless, the primary objective of our study was to demonstrate the cardiovascular predictive value of repeated measurements using ML models. As such, the potential limitations arising from regional data may have a limited impact on the overall conclusions of this research. Additionally, we acknowledge that the analysis set, consisting of 215 744 Chinese participants, is a subset of the original CHERRY study, which included 1.05 million adults. Consequently, while our findings are informative, they may not fully represent the entire population. Nonetheless, this subset reflects the current clinical practice where lipid measurements are commonly requested, even when using traditional guideline-recommended models. Furthermore, it is important to note that the data source for our study primarily relied on EHRs, which are generally collected from individuals seeking medical care. This approach may lead to biased representations of certain health conditions or risk factors that are more likely to be captured in clinical settings. Novel risk factors, such as apolipoproteins or eGFR, may be particularly affected by this bias, as their availability could be associated with specific patient health conditions and outcomes. However, we mitigated this concern by leveraging ML algorithms, which effectively handled missing data and enabled us to capture valuable information for CVD risk prediction, including the association between the availability of specific markers and disease outcomes. Finally, although the use of summarized statistics to utilize repeated measurements is common, it is also important to model the time trend and consider the temporal dependence of the measurements from a single individual.16,24 Our study reinforces the importance of incorporating repeated measurements from EHRs in CVD risk prediction. The temporal aspect of repeated measurements adds valuable insights, but challenges remain in fully capturing this information using current ML algorithms. Future research efforts should focus on addressing these methodological limitations to unlock the full potential of EHR data for improved CVD risk assessment. While our study has several limitations, as listed above, we believe that our focus on assessing the cardiovascular predictive value of repeated measurements with ML models remains valuable and contributes to the current understanding of CVD risk assessment.
Conclusions
The irregularly repeated measurements in the EHR could be leveraged to improve the current 5-year ASCVD incident risk prediction by adopting XGBoost or LASSO regression algorithms. The XGBoost model displayed the best overall performance in the parameters of discrimination, calibration, and reclassification. A comprehensive consideration of the importance of predictors in both ML models showed that the average levels of blood glucose, renal function, and Apo B had relatively high predictive values. Real-world repeated measurements of risk factors have the potential to provide additive value for current ASCVD risk assessment.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health.
Supplementary Material
Acknowledgements
The authors thank the Yinzhou District Health Bureau for providing access to the administrative databases used in the study and all the CHERRY study investigators for their contributions.
Contributor Information
Chaiquan Li, Department of Epidemiology and Biostatistics, School of Public Health, Peking University Health Science Center, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China.
Xiaofei Liu, Department of Epidemiology and Biostatistics, School of Public Health, Peking University Health Science Center, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China.
Peng Shen, Yinzhou District Center for Disease Control and Prevention, No. 1221 Xueshi Road, Yinzhou District, 315199 Ningbo, China.
Yexiang Sun, Yinzhou District Center for Disease Control and Prevention, No. 1221 Xueshi Road, Yinzhou District, 315199 Ningbo, China.
Tianjing Zhou, Department of Epidemiology and Biostatistics, School of Public Health, Peking University Health Science Center, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China.
Weiye Chen, Department of Epidemiology and Biostatistics, School of Public Health, Peking University Health Science Center, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China.
Qi Chen, Yinzhou District Center for Disease Control and Prevention, No. 1221 Xueshi Road, Yinzhou District, 315199 Ningbo, China.
Hongbo Lin, Yinzhou District Center for Disease Control and Prevention, No. 1221 Xueshi Road, Yinzhou District, 315199 Ningbo, China.
Xun Tang, Department of Epidemiology and Biostatistics, School of Public Health, Peking University Health Science Center, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China; Key Laboratory of Epidemiology of Major Diseases, Peking University, Ministry of Education, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China.
Pei Gao, Department of Epidemiology and Biostatistics, School of Public Health, Peking University Health Science Center, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China; Key Laboratory of Epidemiology of Major Diseases, Peking University, Ministry of Education, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China; Center for Real-world Evidence Evaluation, Peking University Clinical Research Institute, No. 38 Xueyuan Road, Haidian District, 100191 Beijing, China.
Funding
This work was supported by the National Key Research and Development Program of China (grant number 2020YFC2003503) and the National Natural Science Foundation of China (NSFC) (grant number: 81973132).
Data availability
Data in this study were not publicly available due to administrative control. For placing any request, please contact the author directly.
References
- 1. Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, et al. 2019 ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2019;74:e177–e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Visseren FL, Mach F, Smulders YM, Carballo D, Koskinas KC, Bäck M, et al. 2021 ESC guidelines on cardiovascular disease prevention in clinical practice: developed by the task force for cardiovascular disease prevention in clinical practice with representatives of the European Society of Cardiology and 12 medical societies with the special contribution of the European Association of Preventive Cardiology (EAPC). Eur Heart J 2021;42:3227–3337.34458905 [Google Scholar]
- 3. Gu D. Guideline on the assessment and management of cardiovascular risk in China. Chin J Prev Med 2019;53:13–34. [DOI] [PubMed] [Google Scholar]
- 4. Kist JM, Vos RC, Mairuhu ATA, Struijs JN, van Peet PG, Vos HMM, et al. SCORE2 cardiovascular risk prediction models in an ethnic and socioeconomic diverse population in the Netherlands: an external validation study. EClinicalMedicine 2023;57:101862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muntner P, Colantonio LD, Cushman M, Goff DC, Howard G, Howard VJ, et al. Validation of the atherosclerotic cardiovascular disease Pooled Cohort risk equations. JAMA 2014;311:1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pylypchuk R, Wells S, Kerr A, Poppe K, Riddell T, Harwood M, et al. Cardiovascular disease risk prediction equations in 400 000 primary care patients in New Zealand: a derivation and validation study. Lancet 2018;391:1897–1907. [DOI] [PubMed] [Google Scholar]
- 7. Mehta A, Shapiro MD. Apolipoproteins in vascular biology and atherosclerotic disease. Nat Rev Cardiol 2022;19:168–179. [DOI] [PubMed] [Google Scholar]
- 8. Nordestgaard BG, Chapman MJ, Ray K, Borén Jan, Andreotti F, Watts GF, et al. Lipoprotein (a) as a cardiovascular risk factor: current status. Eur Heart J 2010;31:2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeung SLA, Luo S, Schooling CM. The impact of glycated hemoglobin (HbA1c) on cardiovascular disease risk: a Mendelian randomization study using UK Biobank. Diabetes Care 2018;41:1991–1997. [DOI] [PubMed] [Google Scholar]
- 10. Emergency Risk Factor Collaboration . Diabetes mellitus, fasting blood glucose concentration, and risk of vascular disease: a collaborative meta-analysis of 102 prospective studies. Lancet 2010;375:2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lim CC, Teo BW, Ong PG, Cheung CY, Lim SC, Chow KY, et al. Chronic kidney disease, cardiovascular disease and mortality: a prospective cohort study in a multi-ethnic Asian population. Eur J Prev Cardiol 2015;22:1018–1026. [DOI] [PubMed] [Google Scholar]
- 12. Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ 2017;357:j2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paige E, Barrett J, Pennells L, Sweeting M, Willeit P, Di Angelantonio E, et al. Use of repeated blood pressure and cholesterol measurements to improve cardiovascular disease risk prediction: an individual-participant-data meta-analysis. Am J Epidemiol 2017;186:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paige E, Barrett J, Stevens D, Keogh RH, Sweeting MJ, Nazareth I, et al. Landmark models for optimizing the use of repeated measurements of risk factors in electronic health records to predict future disease risk. Am J Epidemiol 2018;187:1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vanuzzo D. The epidemiological concept of residual risk. Intern Emerg Med 2011;6:45. [DOI] [PubMed] [Google Scholar]
- 16. Goldstein BA, Navar AM, Pencina MJ, Ioannidis J. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc 2017;24:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bangalore S, Fayyad R, Laskey R, DeMicco DA, Messerli FH, Waters DD. Body-weight fluctuations and outcomes in coronary disease. N Engl J Med 2017;376:1332–1340. [DOI] [PubMed] [Google Scholar]
- 18. Kim MK, Han K, Kim H-S, Park Y-M, Kwon H-S, Yoon K-H, et al. Cholesterol variability and the risk of mortality, myocardial infarction, and stroke: a nationwide population-based study. Eur Heart J 2017;38:3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med 2019;380:1347–1358. [DOI] [PubMed] [Google Scholar]
- 20. Forrest IS, Petrazzini BO, Duffy Á, Park JK, Marquez-Luna C, Jordan DM, et al. Machine learning-based marker for coronary artery disease: derivation and validation in two longitudinal cohorts. Lancet 2023;401:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goldstein BA, Navar AM, Carter RE. Moving beyond regression techniques in cardiovascular risk prediction: applying machine learning to address analytic challenges. Eur Heart J 2017;38:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rousset A, Dellamonica D, Menuet R, Lira Pineda A, Sabatine MS, Giugliano RP, et al. Can machine learning bring cardiovascular risk assessment to the next level? Eur Heart J Digit Health 2022;3:38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hyland SL, Faltys M, Hüser M, Lyu X, Gumbsch T, Esteban C, et al. Early prediction of circulatory failure in the intensive care unit using machine learning. Nat Med 2020;26:364–373. [DOI] [PubMed] [Google Scholar]
- 24. Goldstein BA, Pomann GM, Winkelmayer WC, Pencina MJ. A comparison of risk prediction methods using repeated observations: an application to electronic health records for hemodialysis. Stat Med 2017;36:2750–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q, Campan A, Ren A, Eid WE. Automating and improving cardiovascular disease prediction using machine learning and EMR data features from a regional healthcare system. Int J Med Inform 2022;163:104786. [DOI] [PubMed] [Google Scholar]
- 26. Zhao J, Feng Q, Wu P, Lupu RA, Wilke RA, Wells QS, et al. Learning from longitudinal data in electronic health record and genetic data to improve cardiovascular event prediction. Sci Rep 2019;9:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kakadiaris IA, Vrigkas M, Yen AA, Kuznetsova T, Budoff M, Naghavi M, et al. Machine learning outperforms ACC/AHA CVD risk calculator in MESA. J Am Heart Assoc 2018;7:e009476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang X, Li J, Hu D, Chen J, Li Y, Huang J, et al. Predicting the 10-year risks of atherosclerotic cardiovascular disease in Chinese population: the China-PAR project (Prediction for ASCVD Risk in China). Circulation 2016;134:1430–1440. [DOI] [PubMed] [Google Scholar]
- 29. Lin H, Tang X, Shen P, Zhang D, Wu J, Zhang J, et al. Using big data to improve cardiovascular care and outcomes in China: a protocol for the CHinese Electronic health Records Research in Yinzhou (CHERRY) study. BMJ Open 2018;8:e019698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. SCORE2 Working Group and ESC Cardiovascular Risk Collaboration . SCORE2 risk prediction algorithms: new models to estimate 10-year risk of cardiovascular disease in Europe. Eur Heart J 2021;42:2439–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. D’Agostino RB Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 2008;117:743–753. [DOI] [PubMed] [Google Scholar]
- 32. Kaptoge S, Pennells L, De Bacquer D, Cooney MT, Kavousi M, Stevens G, et al. World Health Organization cardiovascular disease risk charts: revised models to estimate risk in 21 global regions. Lancet Global Health 2019;7:e1332–e1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2019;73:e285–e350. [DOI] [PubMed] [Google Scholar]
- 34. Chan II, Kwok MK, Schooling CM. The total and direct effects of systolic and diastolic blood pressure on cardiovascular disease and longevity using Mendelian randomisation. Sci Rep 2021;11:21799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu K, Cedres LB, Stamler J, Nanas S, Berkson DM, Paul O, et al. Relationship of education to major risk factors and death from coronary heart disease, cardiovascular diseases and all causes, findings of three Chicago epidemiologic studies. Circulation 1982;66:1308–1314. [DOI] [PubMed] [Google Scholar]
- 36. Duran EK, Aday AW, Cook NR, Buring JE, Ridker PM, Pradhan AD. Triglyceride-rich lipoprotein cholesterol, small dense LDL cholesterol, and incident cardiovascular disease. J Am Coll Cardiol 2020;75:2122–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Plate JD, van de Leur RR, Leenen LPH, Hietbrink F, Peelen LM, Eijkemans MJC. Incorporating repeated measurements into prediction models in the critical care setting: a framework, systematic review and meta-analysis. BMC Med Res Methodol 2019;19:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stevens SL, Wood S, Koshiaris C, Law K, Glasziou P, Stevens RJ, et al. Blood pressure variability and cardiovascular disease: systematic review and meta-analysis. BMJ 2016;354:i4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goldstein BA, Bhavsar NA, Phelan M, Pencina MJ. Controlling for informed presence bias due to the number of health encounters in an electronic health record. Am J Epidemiol 2016;184:847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ambale-Venkatesh B, Yang X, Wu CO, Liu K, Hundley WG, McClelland R, et al. Cardiovascular event prediction by machine learning. Circ Res 2017;121:1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoogeveen RM, Pereira JPB, Nurmohamed NS, Zampoleri V, Bom MJ, Baragetti A, et al. Improved cardiovascular risk prediction using targeted plasma proteomics in primary prevention. Eur Heart J 2020;41:3998–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016, p785–794.
- 43. Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B (Methodol) 1996;58:267–288. [Google Scholar]
- 44. Al-Zaiti SS, Alghwiri AA, Hu X, Clermont G, Peace A, Macfarlane P, et al. A clinician’s guide to understanding and critically appraising machine learning studies: a checklist for Ruling Out Bias Using Standard Tools in Machine Learning (ROBUST-ML). Eur Heart J Digit Health 2022;3:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mathioudakis NN, Abusamaan MS, Shakarchi AF, Sokolinsky S, Fayzullin S, McGready J, et al. Development and validation of a machine learning model to predict near-term risk of iatrogenic hypoglycemia in hospitalized patients. JAMA Netw Open 2021;4:e2030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harel O, Mitchell EM, Perkins NJ, Cole SR, Tchetgen Tchetgen EJ, Sun B, et al. Multiple imputation for incomplete data in epidemiologic studies. Am J Epidemiol 2018;187:576–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rubin DB. Multiple imputation for nonresponse in surveys. Hoboken, NJ: Wiley-Interscience; 2004. [Google Scholar]
- 48. Alba AC, Agoritsas T, Walsh M, Hanna S, Iorio A, Devereaux PJ, et al. Discrimination and calibration of clinical prediction models: users’ guides to the medical literature. JAMA 2017;318:1377–1384. [DOI] [PubMed] [Google Scholar]
- 49. Kang L, Chen W, Petrick NA, Gallas BD. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med 2015;34:685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rubin DB. Multiple imputation for nonresponse in surveys. Hoboken, NJ: John Wiley & Sons; 1987. [Google Scholar]
- 51. Li K-H, Meng X-L, Raghunathan TE, Rubin DB. Significance levels from repeated p-values with multiply-imputed data. Stat Sin 1991;1:65–92. [Google Scholar]
- 52. An Y, Tang K, Wang J. Time-aware multi-type data fusion representation learning framework for risk prediction of cardiovascular diseases. IEEE/ACM Trans Comput Biol Bioinform 2021;19:3725–3734. [DOI] [PubMed] [Google Scholar]
- 53. Sun L, Pennells L, Kaptoge S, Nelson CP, Ritchie SC, Abraham G, et al. Polygenic risk scores in cardiovascular risk prediction: a cohort study and modelling analyses. PLoS Med 2021;18:e1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Al’Aref SJ, Anchouche K, Singh G, Slomka PJ, Kolli KK, Kumar A, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975–1986. [DOI] [PubMed] [Google Scholar]
- 55. Li Y, Sperrin M, Ashcroft DM, van Staa TP. Consistency of variety of machine learning and statistical models in predicting clinical risks of individual patients: longitudinal cohort study using cardiovascular disease as exemplar. BMJ 2020;371:m3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gansevoort RT, Correa-Rotter R, Hemmelgarn BR, Jafar TH, Heerspink HJL, Mann JF, et al. Chronic kidney disease and cardiovascular risk: epidemiology, mechanisms, and prevention. Lancet 2013;382:339–352. [DOI] [PubMed] [Google Scholar]
- 57. Emergency Risk Factor Collaboration . C-reactive protein, fibrinogen, and cardiovascular disease prediction. N Engl J Med 2012;367:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cook NR. Methods for evaluating novel biomarkers—a new paradigm. Int J Clin Pract 2010;64:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and updating. New York, NY: Springer-Verlag; 2009. [Google Scholar]
- 60. Collins R, Reith C, Emberson J, Armitage J, Baigent C, Blackwell L, et al. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet 2016;388:2532–2561. [DOI] [PubMed] [Google Scholar]
- 61. Barbieri S, Mehta S, Wu B, Bharat C, Poppe K, Jorm L, et al. Predicting cardiovascular risk from national administrative databases using a combined survival analysis and deep learning approach. Int J Epidemiol 2021;51:931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data in this study were not publicly available due to administrative control. For placing any request, please contact the author directly.