Abstract
C-reactive protein (CRP) tends to be elevated in individuals with psychiatric disorders. Recent findings have suggested a protective effect of the genetic liability to elevated CRP on schizophrenia risk and a causative effect on depression despite weak genetic correlations, but causal relationships with bipolar disorder were inclusive. We investigated the shared genetic underpinnings of psychiatric disorders and variation in CRP levels. Genome-wide association studies for CRP (n=575,531), bipolar disorder (n=413,466), depression (n=480,359), and schizophrenia (n=130,644) were used in causal mixture models to compare CRP with psychiatric disorders based on polygenicity, discoverability, and genome-wide genetic overlap. The conjunctional false discovery rate method was used to identify specific shared genetic loci. Shared variants were mapped to putative causal genes, which were tested for overrepresentation among gene ontology gene-sets. CRP was six to ten times less polygenic (n=1,400 vs 8,600–14,500 variants) and had a discoverability one to two orders of magnitude higher than psychiatric disorders. Most CRP-associated variants were overlapping with psychiatric disorders. We identified 401 genetic loci jointly associated with CRP and psychiatric disorders with mixed effect directions. Gene-set enrichment analyses identified predominantly CNS-related gene sets for CRP and each of depression and schizophrenia, and basic cellular processes for CRP and bipolar disorder. In conclusion, CRP has a markedly different genetic architecture to psychiatric disorders, but the majority of CRP associated variants are also implicated in psychiatric disorders. Shared genetic loci implicated CNS-related processes to a greater extent than immune processes, which may have implications for how we conceptualise causal relationships between CRP and psychiatric disorders.
Keywords: C-reactive protein, psychiatry, neuroimmunology, immunopsychiatry, schizophrenia, bipolar disorder, depression, chronic inflammation, pleiotropy, genetics
1. INTRODUCTION
Major psychiatric disorders, such as bipolar disorder (BIP), major depression (DEP), and schizophrenia (SCZ) are leading causes of morbidity and mortality worldwide (Whiteford et al., 2013). While biomedical research has advanced our understanding of many complex human disorders (Cífková et al., 2010; Foukakis et al., 2011), people with psychiatric disorders are yet to experience the same benefits. Strikingly, individuals with severe psychiatric disorders die on average 10–20 years earlier than the general population and recent evidence suggests this gap is not narrowing (Hjorthøj et al., 2017; Plana-Ripoll et al., 2019). There is therefore an urgent need for progress in mechanistic understanding and therapeutics.
Converging lines of evidence suggest that inflammation may play a causative role in psychiatric disorders (Goldsmith et al., 2016). Firstly, genome-wide association studies (GWASs) have implicated multiple immune-related loci and genes in DEP and SCZ (Howard et al., 2019; Pardiñas et al., 2018), including the major histocompatibility complex, which is the most significantly associated locus in SCZ, and the inflammation linked LIN28B gene in DEP. In vivo imaging studies have also indicated increased neuroinflammation in patients with DEP and SCZ compared to controls (Marques et al., 2019; Setiawan et al., 2018). Furthermore, alterations in the level of circulating inflammatory cytokines have been reported in the blood and cerebrospinal fluid of individuals with BIP, DEP, and SCZ in drug-naïve patients and persist during asymptomatic periods (Goldsmith et al., 2016; Leffa et al., 2019). The “inflammatory hypothesis” therefore represents a plausible avenue towards novel, targeted interventions that may provide a significant breakthrough in the treatment of major psychiatric disorders. Nonetheless, debate remains whether these associations are causal or not (Konsman, 2019).
Among inflammatory biomarkers associated with psychiatric disorders, C-reactive protein (CRP) is an acute phase protein synthesised by the liver and used as a biomarker for both acute and chronic inflammation (Pepys, 1995). Elevated CRP levels have consistently been reported in people with BIP, DEP, and SCZ and further increase during acute manic, depressive and psychotic episodes (Chang et al., 2020; Fernandes et al., 2016). Given that BIP, DEP, and SCZ are heritable, with SNP-based heritability estimated between 7–23% (Howard et al., 2019; Mullins et al., 2021; Trubetskoy et al., 2022), several recent studies have therefore applied statistical genetics tools to investigate causal relationships between CRP levels (hereafter CRP) and psychiatric disorders. These have revealed a potentially protective effect of genetically determined CRP on SCZ risk using mendelian randomisation (Prins et al., 2016; Said et al., 2022) and a putative risk-increasing effect of genetically determined CRP on DEP risk using latent causal variable models (Reay et al., 2022). However, there is currently no statistical genetic evidence of a causal relationship between CRP and BIP.
Despite these reports, there is limited understanding of the shared genetic architecture between CRP and psychiatric disorders. A recent large-scale GWAS of CRP reported nonsignificant genetic correlations with BIP, DEP, and SCZ (Said et al., 2022). Nevertheless, we have recently demonstrated extensive polygenic overlap between genetically uncorrelated traits (Bahrami et al., 2021; Hindley et al., 2021). Since genetic correlation summarises the proportion of shared variants and the distribution of effect sizes, minimal genetic correlation can result from a mixture of shared loci with concordant and discordant effects (Andreassen et al., 2023; Smeland et al., 2020), suggesting value in investigating the genetic overlap between CRP and psychiatric disorders beyond genetic correlation.
Furthermore, an improved understanding of specific genetic loci shared between CRP and psychiatric disorders may help to identify molecular mechanisms underlying their clinical associations and provide opportunities for improved disease classification and treatment. While two previous cross-GWAS analyses failed to identify any loci jointly associated between psychiatric disorders and CRP (Pouget et al., 2019; Tylee et al., 2018), a more recent study reported five genetic loci significantly associated with SCZ and CRP at genome-wide significance (Reay et al., 2022). However, this approach does not leverage the additional statistical power that can be derived from genetic overlap across traits (Smeland et al., 2019). In contrast, the conjunctional false discovery rate (FDR) method uses a Bayesian framework to directly compute the FDR of the joint association between two phenotypes beyond genome-wide significance without the same family-wise error rate penalty incurred by correcting for multiple testing (Andreassen et al., 2013; Smeland et al., 2019).
Given evidence of causal relationships between CRP and psychiatric disorders but minimal genetic correlation, we aimed to investigate the polygenic architecture of CRP and psychiatric disorders and estimate their total genetic overlap beyond genetic correlation to provide a more comprehensive understanding of their shared genetic architecture. We next aimed to leverage the putative polygenic overlap to identify loci jointly associated with each phenotype and characterise shared molecular mechanisms with functional annotation tools.
2. MATERIAL AND METHODS
2.1. Samples
2.1.1. UK Biobank
We obtained data on CRP and psychiatric disorders from 502,413 participants aged 40 to 69 years in the UK Biobank (Sudlow et al., 2015). Details on measurements and phenotype definitions are given in Supplementary materials 1.1.
2.1.2. GWAS summary statistics
We obtained GWAS summary statistics from meta-analyses on CRP (575,531 participants of from the Cohorts for Heart and Aging Research in Genomic Epidemiology Inflammation Working Group (Ligthart et al., 2018) and the UK Biobank) (Said et al., 2022), BIP (41,917 cases and 371,549 controls from the Psychiatric Genomics Consortium (PGC)) (Mullins et al., 2021), DEP (135,458 cases and 344,901 controls from the PGC and 23andMe, Inc) (Wray et al., 2018), and SCZ (53,386 cases and 77,258 controls from the PGC) (Trubetskoy et al., 2022). Most participants in the sub-studies were of European ancestry. Further details are described in the original studies (Mullins et al., 2021; Said et al., 2022; Trubetskoy et al., 2022; Wray et al., 2018) and Supplementary methods 1.2.
2.2. Data analysis
2.2.1. Genetic architecture
We applied univariate and bivariate Gaussian mixture models (Frei et al., 2019; Holland et al., 2020) incorporated in MiXeR v1.3 (https://github.com/precimed/mixer) to GWAS summary statistics from all phenotypes. The models were used to estimate polygenicity (number of ‘causal’ SNPs explaining 90% of SNP-heritability), discoverability (effect size of ‘causal’ SNPs), SNP-heritability (h2SNP), proportion of ‘causal’ SNPs shared between two phenotypes and unique to each phenotype, and genetic correlation between two phenotypes. Further details are provided in Supplementary methods 2.2. H2SNP estimates for psychiatric disorders were converted from the observed scale to the liability scale using population prevalence estimates as applied in the primary GWAS (SCZ = 0.01, BIP = 0.02, DEP = 0.30) (Mullins et al., 2021; Trubetskoy et al., 2022; Wray et al., 2018).
2.2.2. Locus discovery
The conditional false discovery rate (condFDR) is defined as the posterior probability that a SNP is null for the first phenotype given that the p-values for associations with both phenotypes are as small or smaller than their observed p-values. The conjunctional false discovery rate (conjFDR) is the maximum of two condFDR values (first phenotype conditioned on the second and vice versa). ConjFDR is defined as the posterior probability that a SNP is null for one or both phenotypes given that the p-values for associations with both phenotypes are as small or smaller than their observed p-values. Further details about the method and genomic loci definition are described in Supplementary materials 2.3 (Smeland et al., 2019).
2.2.3. Estimating local genetic correlations
We utilized LAVA to estimate pairwise local rg (Werme et al., 2022). LAVA allows for the estimation of local rg across 2495 semi-independent genetic regions, each of approximately equal size (~1Mb). To compute the local genetic covariance matrix for each region, LAVA utilizes the method of moments after estimating local h2SNP for each trait. Sample overlap was controlled using LDSR and significance testing was conducted using simulation-based p-values. To account for multiple testing, p values were adjusted using the false discovery rate (FDR) with a significance threshold set at FDR<0.05.
2.2.4. Colocalization analysis
We performed colocalization analysis using coloc (Giambartolomei et al., 2014). Briefly, coloc uses a Bayesian approach to estimate the posterior probability (PP) that a given genetic locus is not associated with either phenotype (H0), is uniquely associated with phenotype 1 (H1) or phenotype 2 (H2), harbours two independent causal variants for each phenotype (H3), or harbours a shared causal variant (H4). PP > 0.9 for H3 or H4 was considered strong evidence supporting each respective hypothesis. We subsequently used FUMA to identify blood and brain eQTLs from GTEx Consortium for all genetic loci with evidence of a shared causal SNP (GTEx Consortium et al., 2017).
2.2.5. Functional annotation and gene mapping
We functionally annotated all candidate SNPs using Functional Mapping and Annotation of GWAS (FUMA) and Open Targets variant to gene (V2G) tool to map lead SNPs to genes (Supplementary materials 2.4) (Ghoussaini et al., 2021). Mapped genes were defined as the top-ranking gene for all lead variants, prior to merging of physically approximate loci. This was because physically approximate loci with independent lead variants are likely to represent distinct causal genes.
We additionally estimated gene-level associations between the CRP gene and each psychiatric disorder using MAGMA as implemented in FUMA using standard settings. MAGMA computes the mean p-value for all SNPs residing within the borders of a given gene. 1000 genomes phase 3, European ancestry was used as the reference panel.
2.2.6. Gene-set enrichment analysis
We applied a hypergeometric test to investigate overrepresentation of mapped genes for each conjFDR pair-wise analysis within pre-defined gene-sets from the gene ontology resource using FUMA (Carbon et al., 2021). Mapped genes from all discovered shared loci were included as opposed to novel loci alone in order to maximise statistical power for the discovery of significantly enriched gene-sets. To test the effect of different gene mapping strategies on our gene-set analysis, we repeated our analysis using the nearest gene to each lead SNP as opposed to genes mapped using the V2G tool. We additionally tested for overrepresentation of mapped genes from loci with concordant and discordant effects on CRP and each psychiatric disorder separately. All mapped genes within areas of complex LD (i.e., MHC and 8p23) were excluded due to biased gene mapping in these regions. P-values were corrected for multiple comparisons using the Benjamini-Hochberg method.
3. RESULTS
3.1. The polygenic architecture of CRP and psychiatric disorders
Through application of univariate MiXeR, CRP was found to be substantially less polygenic (1,400 ‘causal’ variants accounting for 90% SNP-heritability, SD=200) than BIP (8,600 ‘causal’ variants, SD=200), SCZ (9,800 ‘causal’ variants, SD=300), and DEP (14,500 ‘causal’ variants, SD=700) (Figure 1). The mean effect size of ‘causal’ SNPs (i.e. discoverability) was one to two orders of magnitude higher for CRP (σ2β=1.5 x 10-4, SD=1.9 x 10-5) as compared to SCZ (σ2β=6.0 x 10-5, SD=1.6 x 10-6), BIP (σ2β=3.4 x 10-5, SD=9.0 x 10-7), and DEP (σ2β=7.4 x 10-6, SD=3.2 x 10-7). MiXeR-estimated h2SNP varied amongst CRP (h2SNP 0.14, SD=0.0027), SCZ (h2SNP=0.22, SD=0.0028), BIP (h2SNP=0.13, SD=0.0014), and DEP (h2SNP=0.011, SD=0.0015).
Figure 1: Polygenicity, genetic correlation, and genome-wide genetic overlap for C-reactive protein level (CRP, orange), bipolar disorder (BIP, blue), schizophrenia (SCZ, green), and depression (DEP, orange).
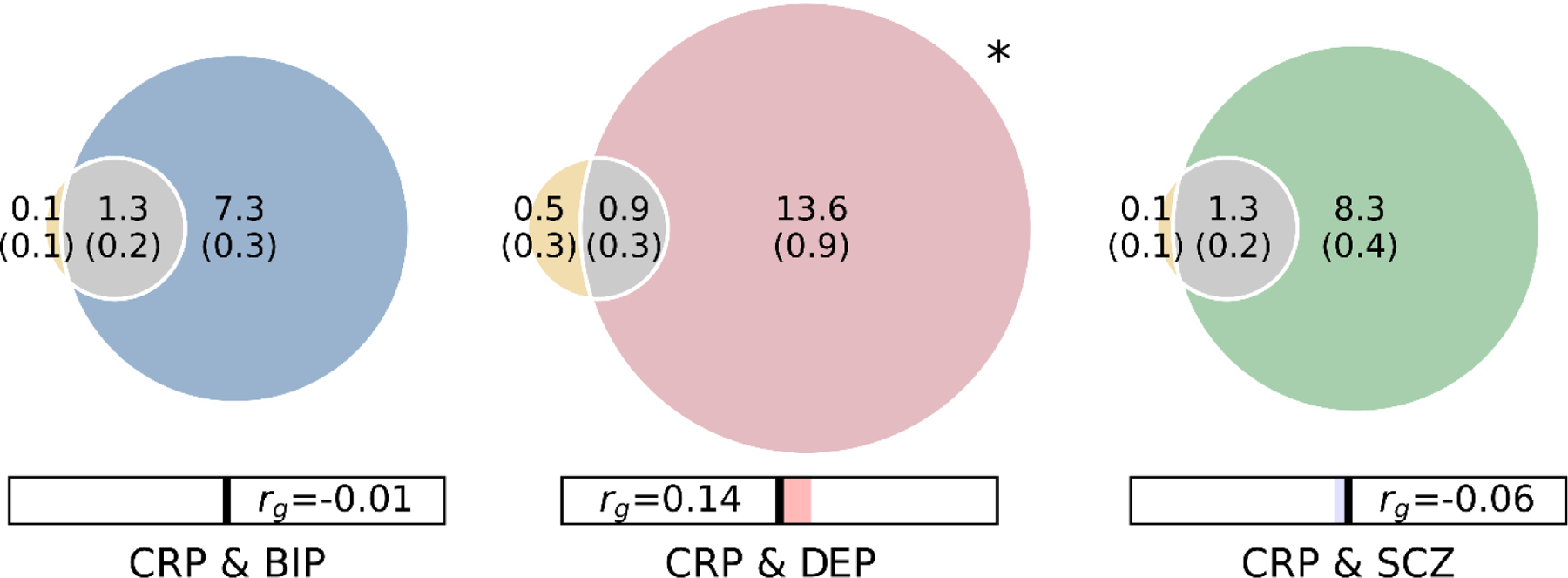
Polygenicity of each trait is represented by the size of each circle. Number of ‘causal’ variants for each component is presented in thousands, with standard deviations in brackets. Shared ‘causal’ variants are represented by grey shading. MiXeR-estimated genetic correlation (rg) is provided in the bar beneath the Venn diagrams, with blue shading representing negative correlations and red shading representing positive correlations. * Bivariate MiXeR model fit for CRP & DEP was sub-optimal as observed on the log likelihood plot (Supplementary figure 1). These findings should be interpreted with caution.
Bivariate MiXeR analysis estimated that 1,300 out of 1,400 causal CRP variants (93%) were shared with both BIP and SCZ. However, given the large differences in polygenicity between CRP and psychiatric disorders, this only represented 15% (1,300/8,600) and 14% (1,300/9,600) of BIP and SCZ ‘causal’ variants, respectively. There were fewer shared ‘causal’ variants estimated between DEP and CRP (900 ‘causal’ variants), but the absence of a clear minimum on the log-likelihood plot was indicative of unstable model fit suggesting that these results should be interpreted with caution (Supplementary figure 1). MiXeR-estimated genetic correlations (Figure 1, Table S1) were consistent with LDSR-based estimates (SCZ & CRP LDSR rg = −0.05, SE = 0.018, p = 0.002; BIP & CRP LDSR rg = −0.01, SE = 0.022, p = 0.675; DEP LDSR rg = 0.12, SE = 0.021, p = 4.60E-09). MiXeR-estimated proportion of shared variants with concordant effects was indicative of mixed effect directions, with 45% (SD = 1.2%) of shared variants for BIP and CRP predicted to have concordant effects, 82% (SD = 11.5%) for DEP and CRP, and 48% (SD = 0.8%) for SCZ and CRP.
3.2. Visualising cross-trait enrichment
Conditional QQ plots showed evidence of cross-trait enrichment between CRP and BIP, DEP, and SCZ respectively. This is illustrated by stepwise increases in enrichment of SNP-associations with each psychiatric disorder as the significance of association with CRP increased (Figure 2a). The same trend was observed when conditioning SNP-associations with CRP on psychiatric disorders (Figure 2b).
Figure 2: Cross-trait enrichment between C-reactive protein level (CRP) and bipolar disorder (BIP), depression (DEP) and schizophrenia (SCZ).
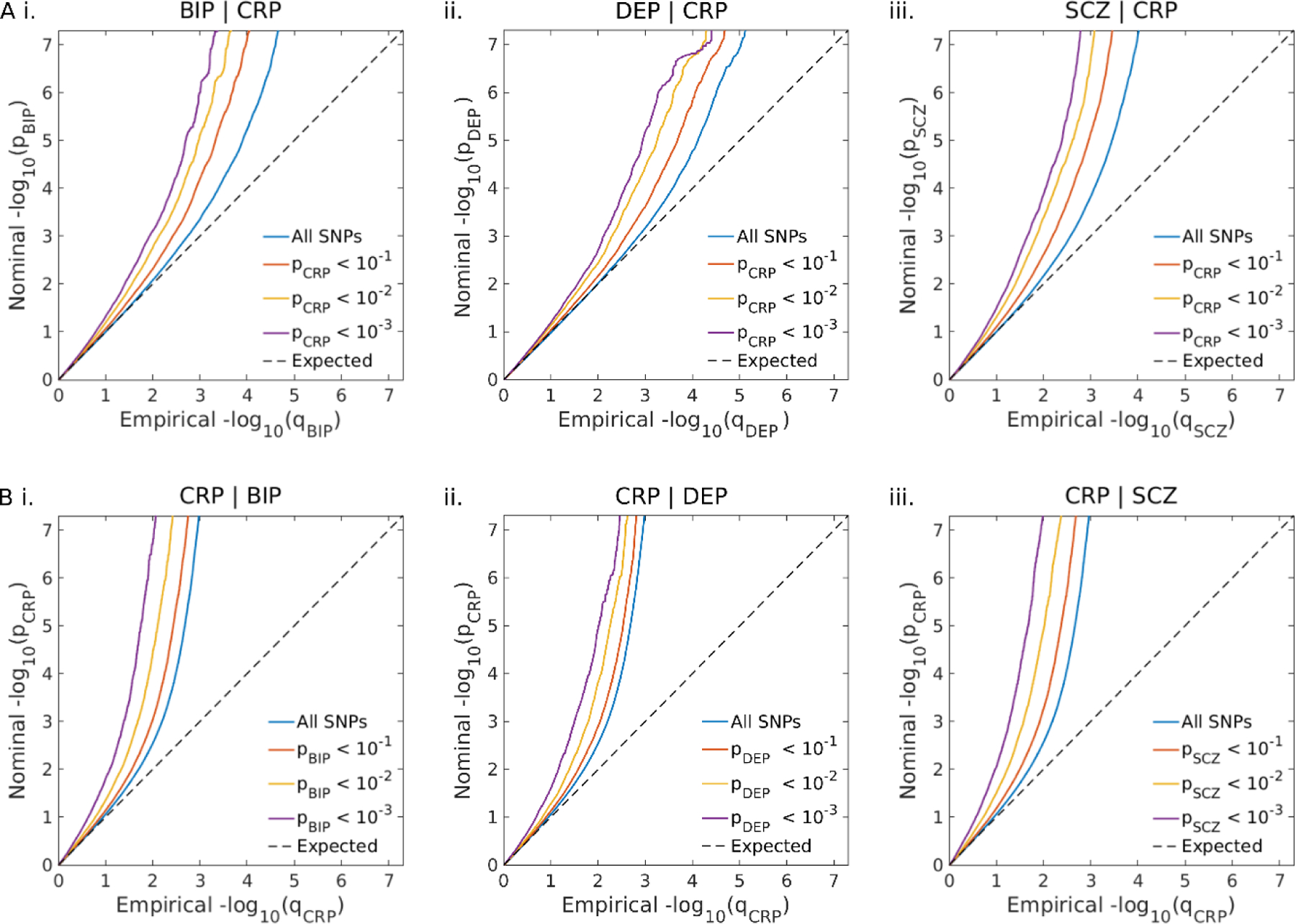
A. Conditional Q-Q plots of nominal versus empirical −log10p values in BIP (i.), DEP (ii.) and SCZ (iii.) as a function of the significance of their association with CRP at the level of p<0.1, p<0.01 and p<0.001, respectively. B. Conditional Q-Q plots of nominal versus empirical −log10p values in CRP as a function of the significance of their association with BIP (i.), DEP (ii.), and SCZ (iii.) at the level of p<0.1, p<0.01 and p<0.001, respectively.
3.3. Identifying loci jointly associated with CRP and psychiatric disorders
We identified 302 genetic loci jointly associated with CRP and SCZ at conjFDR<0.05, (Table S2), 111 loci jointly associated with BIP (Table S3), and 56 loci jointly associated with DEP (Table S4). When combining across all three analyses, these comprised a total of 401 unique genetic loci (Table S5). Effect directions of lead SNPs within shared loci were consistent with weak positive genetic correlations for CRP and DEP and weaker negative genetic correlations for CRP and each of BIP and SCZ. 66% (37/56) of lead SNPs shared between CRP and DEP had concordant effects on each phenotype, compared to 41% (46/111) and 42% (127/302) for BIP and SCZ, respectively.
3.4. Estimating local genetic correlations
Using LAVA, we identified 62 semi-independent genetic regions with significant local genetic correlations between CRP and SCZ, 37 between CRP and BIP, and 36 between CRP and DEP (Figure 4). Furthermore, the proportion of LAVA-identified positively correlated regions for SCZ and CRP (17/62, 28%), BIP and CRP (17/37, 48%), and DEP and CRP (24/36, 67%) were similar to the proportion of conjFDR lead SNPs with concordant effects (41%, 42% and 66%, respectively). Furthermore, 30 out of 62 locally correlated regions for SCZ and CRP were overlapping with conjFDR identified loci, compared to 11 out of 37 for BIP and CRP and four out of 36 for DEP and CRP.
Figure 4: Volcano plots of LAVA local genetic correlation coefficients (rho, y-axis) against −log10-p values for C-reactive protein level and each of schizophrenia (SCZ), bipolar disorder (BIP) and depression (DEP).
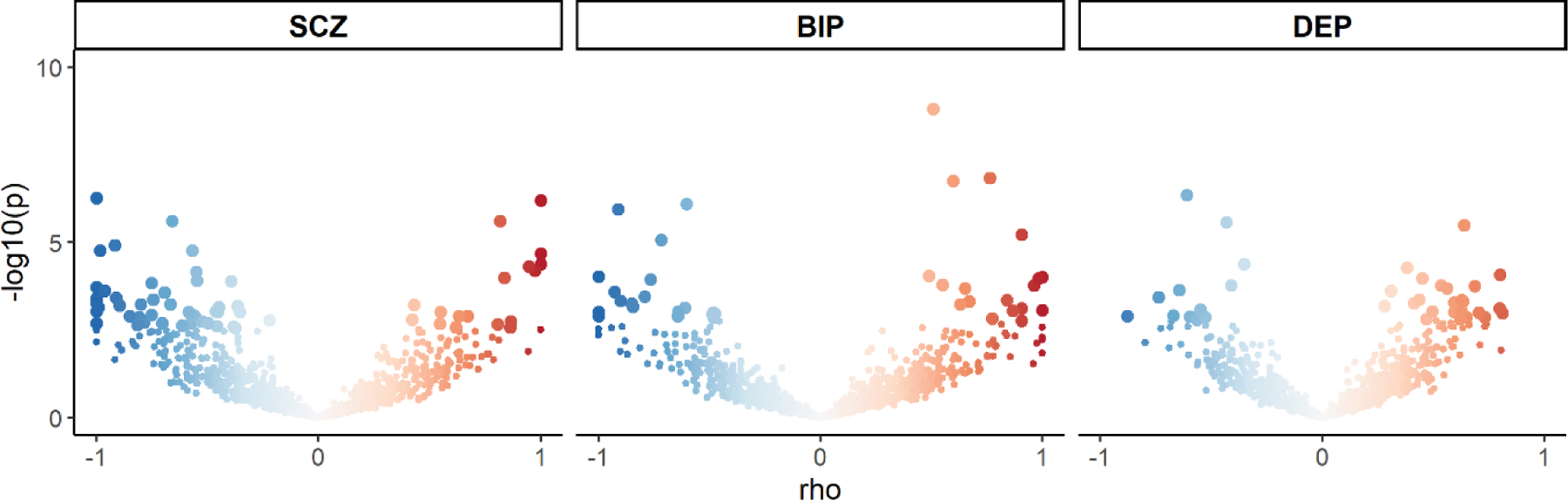
Each dot represents an individual locus. Larger dots represent significantly correlated loci after FDR-correction. The colour of the dots represents the strength of the local correlation from −1 (blue) to 1 (red).
3.5. Colocalization analyses
We used coloc to estimate the posterior probability that shared genetic loci as identified using conjFDR harboured a shared causal variant. The analysis was insufficiently powered to differentiate between these two scenarios for the majority of loci. For the 34 shared loci for SCZ and CRP with sufficient statistical power (11.2% of all loci), 17 loci displayed evidence of a shared causal variant (PP>0.9) and 17 loci displayed evidence of two independent causal variants (Table S6). For the 11 shared loci for BIP and CRP with sufficient statistical power (9.9% of all loci), five loci displayed evidence of a shared causal variant (PP>0.9) and six loci displayed evidence of independent causal variants (Table S7). Finally, for the 7 loci with sufficient statistical power for DEP and CRP (12.5% of all loci), four loci displayed evidence of a shared causal variant and three loci displayed evidence of independent causal variants (Table S8). We mapped blood and brain eQTLs from the GTEx to loci with evidence of a shared causal variant. Of 41 eQTLs for SCZ and CRP, 11 were differentially expressed in the blood and 15 in brain tissues, while 15 were differentially expressed in both blood and brain tissues. Similarly, for BIP and CRP there were four unique blood eQTLs, five unique brain tissue eQTLs, and 13 eQTLs in both blood and brain tissues. Finally for DEP and CRP there were three unique blood eQTLs, three unique brain tissue eQTLs, and five eQTLs in both blood and brain tissues. Among brain eQTLs there was a predominance of eQTLs in cerebellar tissues, basal ganglia, amygdala, and cortical tissues, in particular the anterior cingulate cortex (Table S9).
3.6. Mapped genes and gene-set enrichment analysis
Using open targets, we mapped a total of 355 genes to lead SNPs jointly associated with SCZ and CRP (Table S10), 128 genes for BIP and CRP (Table S11), and 57 genes for DEP and CRP (Table S12). MAGMA gene-level analysis revealed that the CRP gene was associated with SCZ, although this did not survive Bonferroni correction for multiple testing when considering all genes (z = 2.24, p = 0.0126). BIP (z = −0.83, p= 0.796) and DEP (z = −1.76, p = 0.961) were not associated with the CRP gene at the gene level.
Gene-set enrichment analysis of mapped genes identified 362 gene ontology biological processes which were overrepresented with mapped genes for SCZ and CRP, 68 cellular components, and 37 molecular functions (Table S13). Among both biological processes and cellular components there was a striking predominance of central nervous system (CNS)-related gene-sets, with seven of the top ten biological processes related to the CNS and eight out of the top ten cellular components implicated in neuronal structures. There were relatively few gene sets specifically related to immune system functioning. Besides basic nonspecific cellular processes which contribute to inflammatory processes such as “secretion”, only a few immune specific biological processes were identified and there were no significantly enriched immune related cellular components or molecular functions.
We next performed gene-set enrichment analysis on mapped genes from lead variants with concordant and discordant effects separately. A total of 120 gene-sets were enriched for concordant genes and 220 gene-sets for discordant genes, 57 of which were identified in both analyses (Table S14). When comparing gene-sets unique to each group there was a predominance of gene-sets related to metabolic processes (e.g. “regulation of lipid metabolic process”, padj = 1.60E-3), endocrine processes (e.g. “response to hormone”, padj = 9.43E-4) and transmembrane ion transport (e.g. “regulation of cation channel activity”, padj = 1.60E-3). In contrast, there was a predominance of gene-sets related to neurodevelopmental processes (e.g. “positive regulation of nervous system development”, padj = 5.77E-5), cell adhesion (e.g. “biological adhesion”, padj = 1.83E-3), and cell structure (“cytoskeletal organization”, padj = 1.83E-3) among gene-sets enriched for discordant mapped genes.
A similar pattern was observed for shared DEP and CRP genes (Table S115). Of a total 35 biological processes enriched with mapped genes, 16 were directly related to the CNS, including five of the top ten most significantly enriched gene sets. Comparatively, only two gene sets were directly related to the immune system. When performing gene-set enrichment analysis on mapped genes derived from concordant and discordant lead variants separately, 17 gene-sets were enriched for concordant mapped genes and a single gene-set, “nucleoplasm part” (padj = 0.0264), was enriched for discordant mapped genes (Table S16). Among gene-sets enriched for concordant mapped genes, there was a predominance of gene-sets related to development and neurodevelopment related gene-sets, in addition to “cognition” (padj = 0.0488) and “behaviour” (padj = 0.0490).
Gene sets jointly identified for BIP and CRP were predominantly non-specific cellular functions with no CNS related or immune related gene sets (Table S17). There was a single gene-set enriched with genes mapped to concordant lead SNPs (“nuclear membrane”, padj = 0.0467) and no gene-sets enriched with genes mapped to discordant lead SNPs (Table S18).
In our sensitivity analysis using nearest genes to lead SNPs as opposed to V2G-mapped genes, we identified 378 enriched gene-sets for SCZ and CRP, 32 for DEP and CRP and 39 for BIP and CRP. Of these, 218 were also identified in our primary analysis for SCZ and CRP (57.6%), 22 for DEP and CRP (68.8%), and 4 for BIP and CRP (10.3%).
3.7. Phenotypic associations
Using data from 468,446 participants in the UK Biobank and controlling for age, sex and BMI, we replicated previous associations between elevated CRP and diagnosis of SCZ (β=0.33, SE=0.035, p=1.95 x 10-21, n(cases)=974, n(controls)=460,405), BIP (β=0.14, SE=0.027, p=1.32 x 10-7, n(cases)=1,651, n(controls)= 459,728), and DEP (β=0.17, SE=0.007, p=2.05 x 10-18, n(cases)=26,730, n(controls)=434,649) (Supplementary material, Table S19).
4. DISCUSSION
In this cross-trait analysis, we found that the genetic architecture of CRP differs from psychiatric disorders in terms of lower polygenicity (i.e. number of ‘causal’ variants) and higher discoverability (i.e. the mean effect size of ‘causal’ variants). Despite weak to zero genetic correlations, the majority of CRP-associated ‘causal’ variants were predicted to be shared with both SCZ and BIP. Fewer ‘causal’ variants were predicted to be shared with DEP. Using conjFDR, a total of 401 unique genomic loci were jointly associated with CRP and one or more psychiatric disorder. Gene mapping and gene set enrichment analyses indicated that loci shared between CRP and each of SCZ and DEP were predominantly implicated in CNS-related gene sets, while CRP and BIP genes were enriched in gene sets related to nonspecific cellular functions. There was no evidence of an association between the CRP gene itself and SCZ, BIP or DEP.
A major finding of our study is that the genetic architecture of CRP differs from psychiatric disorders. The relatively low polygenicity and high discoverability of CRP aligns with the genetic architecture previously described for other biochemical traits and somatic disorders as compared to psychiatric disorders (Holland et al., 2020; Zhang et al., 2018). Differences in genetic architecture between CRP and psychiatric disorders may reflect differences in complexity. Biochemical traits like CRP seem to involve fewer molecular mechanisms linking common genetic variation to the phenotype, resulting in fewer ‘causal’ variants with larger effect sizes (Cai et al., 2020; Smoller et al., 2019). On the other hand, psychiatric disorders are likely to have more heterogenous biology and the pathways from ‘causal’ variant to the phenotype are more complex. This may explain the larger polygenicity and smaller average effect size in psychiatric disorders.
Differences in polygenicity contributed to the pattern of genome-wide genetic overlap observed using bivariate MiXeR. Given CRP’s low polygenicity compared to psychiatric disorders, only a minority of ‘causal’ variants for psychiatric disorders were predicted to influence CRP. As a result, CRP associated SNPs are unlikely to contribute substantial proportions of genetic liability for psychiatric disorders. It is, however, tempting to speculate that such a pattern of overlap may be consistent with sub-groups of patients for which inflammation plays a more prominent pathoetiological role. This is supported by findings that alterations in inflammatory markers in patients with BIP, DEP and SCZ are often reported in subsets of patients (Ormerod et al., 2022; North et al., 2022; Osimo et al., 2020; Sæther et al., 2022; Szabo et al., 2022; Tamouza et al., 2021).
On the other hand, large proportions of ‘causal’ variants for CRP were predicted to overlap with psychiatric disorders, despite weak genetic correlations. This indicates that the majority of common variants which influence CRP also influence risk for major psychiatric disorders, although the pattern of effect directions and effect sizes vary. If there is a balance of shared variants with concordant and discordant effects, as indicated by MiXeR and conjFDR findings, these cancel each other out resulting in minimal genetic correlation (Frei et al., 2019). This is intriguing given the evidence of a protective effect for genetically determined CRP (i.e. the genetic instrument used as a proxy for CRP in Mendelian randomization studies (Emdin et al., 2017)) on SCZ risk but increased CRP in patients with SCZ at the group level (Chang et al., 2020; Fernandes et al., 2016; Reay et al., 2022; Said et al., 2022). Mixed effect directions may therefore be indicative of SCZ subgroups, one which is characterised by a higher load of CRP lowering genetic variants which play a causative role in the disorder, and another with a higher load of CRP increasing variants, which may contribute to the higher CRP observed among SCZ patients.
A higher proportion of lead variants for DEP and CRP had concordant effects, consistent with MiXeR findings of weak positive genome-wide genetic correlation. Although MiXeR model fit was unstable for DEP and CRP, LDSR genetic correlation supported these findings. Since genetic correlation is a genome-wide measure and CRP is substantially less polygenic than DEP, even weak positive genome-wide genetic correlation may be indicative of a high degree of concordance among shared variants, demonstrating the advantages of estimating genetic overlap beyond genetic correlation. A high degree of concordance among shared variants indicates more closely aligned genetic effects which provide stronger evidence for the presence of shared molecular mechanisms with causal effects. This is particularly interesting in regard to the finding that genetically determined CRP may causatively increase risk of DEP (Reay et al., 2022; Said et al., 2022). Nevertheless, Pitharouli et al. found that the association between elevated CRP and DEP in UKB was largely driven by BMI, smoking and other environmental risk factors rather than shared genetic risk. Furthermore, the association between DEP PRS and CRP was mainly explained by BMI and smoking (Pitharouli et al., 2021). Similarly, Reay et al. found in multivariable Mendelian randomization analyses that genetically determined CRP conditioned on BMI was not associated with DEP. In contrast, the negative association between genetically determined CRP on SCZ remained significant after conditioning CRP on BMI as well as the CRP upstream signalling factor interleukin-6 (Reay et al., 2022).
One of the key advantages of our approach was a significant boost in genetic discovery compared to previous studies through the use of largest to date GWAS samples and the application of conjFDR (Reay et al., 2022; Tylee et al., 2018). The number of genetic loci identified by conjFDR is substantially smaller than the number of shared causal variants estimated by MiXeR because MiXeR applies mathematical modelling to predict the number of shared causal variants once fully powered GWAS for each input trait have been achieved (Frei et al., 2019). On the other hand, the number of loci identified by conjFDR is dependent on the statistical power of the input GWAS. The number of conjFDR discovered loci will therefore increase as GWAS sample sizes increase, whereas MiXeR estimates will only increase the precision of the estimate. LAVA also supported the presence of regions with shared genetic effects between CRP and the three mental disorders with a similar pattern of directional effects. There was, however, variable overlap between LAVA-identified regions and conjFDR-identified loci. These discrepancies may be explained by the fact that conjFDR functions on the level of individual SNPs, whereas LAVA requires consistent directional effects across a larger range of SNPs. This means that prioritised LAVA local genomic regions may not coincide with conjFDR identified loci given the different underlying assumptions of the two approaches (Smeland et al., 2019; Werme et al., 2022).
We additionally used coloc in an attempt to identify shared genetic loci with evidence of a shared causal variant as opposed to two independent causal variants. Unfortunately, only a minority of loci had sufficient statistical power to differentiate these two scenarios. This is to be expected to some degree since the conjFDR framework explicitly boosts statistical power to identify shared statistical associations beyond genome-wide significance, whereas coloc is dependent on the statistical power of the input GWAS (Giambartolomei et al., 2014; Smeland et al., 2019). Thus, the findings should be interpreted with caution. Still, within the sufficiently powered genetic loci the coloc results suggest that the shared loci with evidence of a shared causal variant were mapped to a mixture of unique blood eQTLs, unique brain eQTLs, and eQTLs in both blood and brain tissues. This may be consistent with a combination of pleiotropic mechanisms, brain specific mechanisms and immune mediated mechanisms.
Given the number of loci discovered, we focused on gene set enrichment analysis rather than discovery of individual genes which individually contribute small proportions of heritability. This revealed that there was a predominance of CNS structure and function gene sets jointly associated with CRP and each of SCZ and DEP. Further, by dividing mapped genes according to the direction of effect of lead variants we observed differences in the types of enriched gene-sets. In particular, neurodevelopmental gene-sets were predominantly enriched with discordant mapped genes for SCZ and CRP but concordant mapped genes for DEP and CRP. This suggests that genes influencing CRP level and liability for SCZ and DEP may both influence neurodevelopmental processes but with different directions of effect. Reay et al. have previously demonstrated that proteins whose expression is altered by genetically determined CRP are overrepresented in neuronal pathways, including axon guidance, neurogenesis, and dopaminergic synapse. In their study, overrepresentation of proteins associated with genetically determined CRP in the dopaminergic synapse pathway was driven by the CRP-associated genes AKT1, AKT2, and AKT3 (Reay et al., 2022). These genes encode a serine/threonine-protein kinase complex which is related to dopamine transmission (Li et al., 2016) and reduced in the prefrontal cortex of patients with SCZ (Emamian, 2012). As such, CRP might have CNS-effects related to risk of psychiatric disorders independently of inflammation. However, these findings could also be indicative of reverse causation, whereby genes which are implicated in primary CNS functions impact CRP level. This interpretation could be consistent with Pitharouli et al.’s findings in which the association between DEP PRS and CRP was no longer significant after controlling for behavioural phenotypes such as BMI and smoking (Pitharouli et al., 2021). Alternatively, for SCZ and CRP, the CNS-related gene sets may indicate that the protective effect of genetically determined CRP previously identified using mendelian randomization analyses (Reay et al., 2022) is a result of direct CNS effects of CRP-associated alleles rather than a causal effect of CRP. In contrast to the CNS specific gene set findings for the two mentioned psychiatric disorders and CRP, the BIP and CRP gene sets were predominantly related to basic cellular functions. The lack of positive findings for CRP in BIP using either mendelian randomization or latent causal variables and the lack of stability of our gene-set analyses when using different gene-mapping strategies may indicate that genetic overlap between BIP and CRP is capturing less specific molecular mechanisms than for either DEP or SCZ.
A limitation of the present study is that inflammation was only assessed by CRP, a non-specific marker of systemic inflammation, which has effects both in response to and resolution of inflammation (Del Giudice and Gangestad, 2018). We cannot exclude that some of the detected genetic overlap between the phenotypes could be explained by Berkson’s bias, e.g., that patients with psychiatric disorders and comorbid low-grade inflammation have a higher risk for contacting health services and thus be included in studies. Survival bias through lower inclusion of patients with comorbid psychiatric disorders and chronic inflammation due to premature death is another possible bias which could have had the opposite effect on our findings. GWAS are prone to confounding by population stratification that is previously demonstrated in data from the UK Biobank which maybe not be sufficiently accounted for by adjustments for principal components (Haworth et al., 2019). The lack of stability of CRP as a measure of chronic inflammation is likely to have increased noise in our analyses. Nonetheless, it is important to note that CRP is regarded as a relatively stable protein in the absence of infection displaying little diurnal (Meier-Ewert et al., 2001) and postprandial (Blackburn et al., 2006) variation. Further, the primary CRP GWAS excluded participants with CRP >4 SD from the mean, which should have reduced the impact of certain confounders like co-occurring infections and severe general inflammatory diseases. Finally, the GWAS samples used were also limited by their lack of ancestral diversity.
In conclusion, we have demonstrated that CRP has a distinct genetic architecture to psychiatric disorders, with lower polygenicity and higher discoverability limiting their maximum possible genetic overlap. Nonetheless, despite weak to zero genetic correlations, we identify overlapping genetic loci with a mixture of concordant and discordant effect directions, suggesting different subgroups of psychiatric disorders. Furthermore, we identify potential molecular mechanisms underlying the shared genetic architecture, with a predominance of CNS-related gene sets present for CRP and both of DEP and SCZ. Further studies are needed to investigate the longitudinal effects of gene sets shared between inflammatory markers and psychiatric disorders (e.g., through developmental phases and in interaction with environmental factors like stress and infections) on CNS function and structure (e.g., by cerebrospinal fluid analyses and magnetic resonance imaging in genotyped samples). Still, the current findings provide novel insights into the molecular pathways which may underlie altered immune marker levels in psychiatric disorders.
Supplementary Material
Figure 3: Chromosomal distribution of shared genetic loci between C-reactive protein level (CRP) and A. bipolar disorder (BIP, blue), B. depression (DEP, orange), and C. schizophrenia (SCZ, green).
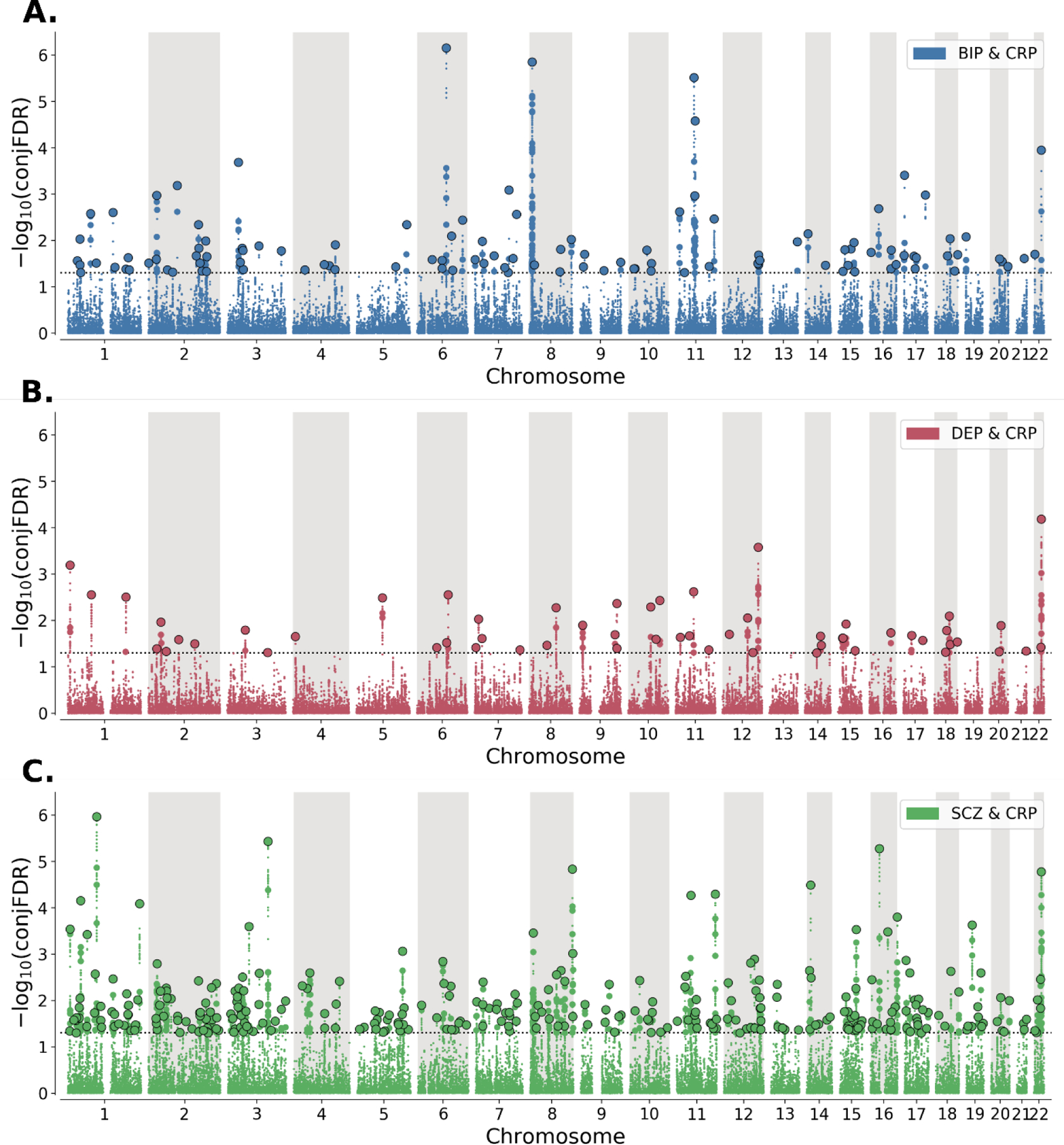
−log10conjFDR values (y-axis) are plotted against chromosomal position (x-axis). The threshold for statistical significance (conjFDR<0.05) is represented by the dashed line. Lead SNPs are represented by large black-outlined circles, independent significant SNPs are represented by medium-sized coloured circles, and all other SNPs are represented by small circles.
5. Acknowledgements
We would like to thank the research participants and employees of the Schizophrenia, Depression and Bipolar Disorder Working Groups of the Psychiatric Genomics Consortium, 23andMe, CHARGE, and UK Biobank for making this research possible. This research has been conducted using data from UK Biobank, a major biomedical database (www.ukbiobank.ac.uk). We gratefully acknowledge support from the American National Institutes of Health (NS057198, EB000790, 1R01MH124839), the Research Council of Norway (RCN 300309, 223273, 273291, 296030, 324252, 248980, 229129, 213837), the South-East Norway Regional Health Authority (2017–112, 2019–108), and KG Jebsen Stiftelsen (SKGJ-MED-008). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement Nos 847776, 964874, and 801133 (Marie Skadowska Curie grant agreement). This work was partly performed on the TSD (Services for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT). Computations were also performed on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway.
Footnotes
Declaration of Interests
O.A.A. has received speaker’s honorarium from Lundbeck, Janssen and Sunovion and is a consultant for Cortechs.ai. A.M.D. is a founder of and holds equity interest in CorTechs Labs and serves on its scientific advisory board. He is also a member of the Scientific Advisory Board of Healthlytix and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. Remaining authors have nothing to disclose.
7. References
- Andreassen OA, Hindley GFL, Frei O, Smeland OB, 2023. New insights from the last decade of research in psychiatric genetics: discoveries, challenges and clinical implications. World Psychiatry Off. J. World Psychiatr. Assoc. WPA 22, 4–24. 10.1002/wps.21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen OA, Thompson WK, Schork AJ, Ripke S, Mattingsdal M, Kelsoe JR, Kendler KS, O’Donovan MC, Rujescu D, Werge T, 2013. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet 9, e1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrami S, Hindley G, Winsvold BS, O’Connell KS, Frei O, Shadrin A, Cheng W, Bettella F, Rødevand L, Odegaard KJ, Fan CC, Pirinen MJ, Hautakangas HM, Headache HA-I, Dale AM, Djurovic S, Smeland OB, Andreassen OA, 2021. Dissecting the shared genetic basis of migraine and mental disorders using novel statistical tools. Brain 10.1093/brain/awab267 [DOI] [PMC free article] [PubMed]
- Blackburn P, Després J-P, Lamarche B, Tremblay A, Bergeron J, Lemieux I, Couillard C, 2006. Postprandial Variations of Plasma Inflammatory Markers in Abdominally Obese Men. Obesity 14, 1747–1754. 10.1038/oby.2006.201 [DOI] [PubMed] [Google Scholar]
- Cai N, Choi KW, Fried EI, 2020. Reviewing the genetics of heterogeneity in depression: operationalizations, manifestations and etiologies. Hum. Mol. Genet 29, R10–R18. 10.1093/hmg/ddaa115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, Hartline E, Fey P, Thomas PD, Albou LP, Ebert D, Kesling MJ, Mi H, Muruganujan A, Huang X, Mushayahama T, LaBonte SA, Siegele DA, Antonazzo G, Attrill H, Brown NH, Garapati P, Marygold SJ, Trovisco V, dos Santos G, Falls K, Tabone C, Zhou P, Goodman JL, Strelets VB, Thurmond J, Garmiri P, Ishtiaq R, Rodríguez-López M, Acencio ML, Kuiper M, Lægreid A, Logie C, Lovering RC, Kramarz B, Saverimuttu SCC, Pinheiro SM, Gunn H, Su R, Thurlow KE, Chibucos M, Giglio M, Nadendla S, Munro J, Jackson R, Duesbury MJ, Del-Toro N, Meldal BHM, Paneerselvam K, Perfetto L, Porras P, Orchard S, Shrivastava A, Chang HY, Finn RD, Mitchell AL, Rawlings ND, Richardson L, Sangrador-Vegas A, Blake JA, Christie KR, Dolan ME, Drabkin HJ, Hill DP, Ni L, Sitnikov DM, Harris MA, Oliver SG, Rutherford K, Wood V, Hayles J, Bähler J, Bolton ER, de Pons JL, Dwinell MR, Hayman GT, Kaldunski ML, Kwitek AE, Laulederkind SJF, Plasterer C, Tutaj MA, Vedi M, Wang SJ, D’Eustachio P, Matthews L, Balhoff JP, Aleksander SA, Alexander MJ, Cherry JM, Engel SR, Gondwe F, Karra K, Miyasato SR, Nash RS, Simison M, Skrzypek MS, Weng S, Wong ED, Feuermann M, Gaudet P, Morgat A, Bakker E, Berardini TZ, Reiser L, Subramaniam S, Huala E, Arighi CN, Auchincloss A, Axelsen K, Argoud-Puy G, Bateman A, Blatter MC, Boutet E, Bowler E, Breuza L, Bridge A, Britto R, Bye-A-Jee H, Casas CC, Coudert E, Denny P, Es-Treicher A, Famiglietti ML, Georghiou G, Gos AN, Gruaz-Gumowski N, Hatton-Ellis E, Hulo C, Ignatchenko A, Jungo F, Laiho K, Le Mercier P, Lieberherr D, Lock A, Lussi Y, MacDougall A, Ma-Grane M, Martin MJ, Masson P, Natale DA, Hyka-Nouspikel N, Orchard S, Pedruzzi I, Pourcel L, Poux S, Pundir S, Rivoire C, Speretta E, Sundaram S, Tyagi N, Warner K, Zaru R, Wu CH, Diehl AD, Chan JN, Grove C, Lee RYN, Muller HM, Raciti D, van Auken K, Sternberg PW, Berriman M, Paulini M, Howe K, Gao S, Wright A, Stein L, Howe DG, Toro S, Westerfield M, Jaiswal P, Cooper L, Elser J, 2021. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res 49, D325–D334. 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JPC, Mondelli V, Satyanarayanan SK, Chiang YJ, Chen HT, Su KP, Pariante CM, 2020. Cortisol, inflammatory biomarkers and neurotrophins in children and adolescents with attention deficit hyperactivity disorder (ADHD) in Taiwan. Brain. Behav. Immun 88. 10.1016/j.bbi.2020.05.017 [DOI] [PubMed] [Google Scholar]
- Cífková R, Škodová Z, Bruthans J, Holub J, Adámková V, Jozífová M, Galovcová M, Wohlfahrt P, Krajčoviechová A, Petržílková Z, Lánská V, 2010. Longitudinal trends in cardiovascular mortality and blood pressure levels, prevalence, awareness, treatment, and control of hypertension in the Czech population from 1985 to 2007/2008. J. Hypertens 28, 2196–2203. 10.1097/HJH.0b013e32833d4451 [DOI] [PubMed] [Google Scholar]
- Del Giudice M, Gangestad SW, 2018. Rethinking IL-6 and CRP: Why they are more than inflammatory biomarkers, and why it matters. Brain. Behav. Immun 70, 61–75. 10.1016/j.bbi.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Elkjaer Greenwood Ormerod MB, Ueland Thor, Frogner Werner MC, Hjell G, Rødevand L, Sæther LS, Lunding SH, Johansen IT, Ueland Torill, Lagerberg TV, Melle I, Djurovic S, Andreassen OA, Steen NE, 2022. Composite immune marker scores associated with severe mental disorders and illness course. Brain Behav. Immun. - Health 24, 100483. 10.1016/j.bbih.2022.100483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, 2012. AKT/GSK3 signaling pathway and schizophrenia. Front. Mol. Neurosci 5. 10.3389/fnmol.2012.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emdin CA, Khera AV, Kathiresan S, 2017. Mendelian Randomization [DOI] [PubMed]
- Fernandes BS, Steiner J, Molendijk ML, Dodd S, Nardin P, Gonçalves CA, Jacka F, Köhler CA, Karmakar C, Carvalho AF, Berk M, 2016. C-reactive protein concentrations across the mood spectrum in bipolar disorder: a systematic review and meta-analysis. Lancet Psychiatry 3, 1147–1156. 10.1016/S2215-0366(16)30370-4 [DOI] [PubMed] [Google Scholar]
- Foukakis T, Fornander T, Lekberg T, Hellborg H, Adolfsson J, Bergh J, 2011. Age-specific trends of survival in metastatic breast cancer: 26 years longitudinal data from a population-based cancer registry in Stockholm, Sweden. Breast Cancer Res. Treat 130, 553–560. 10.1007/s10549-011-1594-z [DOI] [PubMed] [Google Scholar]
- Frei O, Holland D, Smeland OB, Shadrin AA, Fan CC, Maeland S, O’Connell KS, Wang Y, Djurovic S, Thompson WK, Andreassen OA, Dale AM, 2019. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun 10, 1–11. 10.1038/s41467-019-10310-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M, Mountjoy E, Carmona M, Peat G, Schmidt EM, Hercules A, Fumis L, Miranda A, Carvalho-Silva D, Buniello A, Burdett T, Hayhurst J, Baker J, Ferrer J, Gonzalez-Uriarte A, Jupp S, Karim MA, Koscielny G, Machlitt-Northen S, Malangone C, Pendlington ZM, Roncaglia P, Suveges D, Wright D, Vrousgou O, Papa E, Parkinson H, MacArthur JAL, Todd JA, Barrett JC, Schwartzentruber J, Hulcoop DG, Ochoa D, McDonagh EM, Dunham I, 2021. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res 49, D1311–D1320. 10.1093/nar/gkaa840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V, 2014. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLOS Genet 10, e1004383. 10.1371/journal.pgen.1004383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith DR, Rapaport MH, Miller BJ, 2016. A meta-analysis of blood cytokine network alterations in psychiatric patients: Comparisons between schizophrenia, bipolar disorder and depression, in: Molecular Psychiatry. Nature Publishing Group, pp. 1696–1709. 10.1038/mp.2016.3 [DOI] [PMC free article] [PubMed]
- GTEx Consortium, Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, Mohammadi P, Park YoSon, Parsana P, Segrè AV, Strober BJ, Zappala Z, Cummings BB, Gelfand ET, Hadley K, Huang KH, Lek M, Li Xiao, Nedzel JL, Nguyen DY, Noble MS, Sullivan TJ, Tukiainen T, MacArthur DG, Getz G, Addington A, Guan P, Koester S, Little AR, Lockhart NC, Moore HM, Rao A, Struewing JP, Volpi S, Brigham LE, Hasz R, Hunter M, Johns C, Johnson M, Kopen G, Leinweber WF, Lonsdale JT, McDonald A, Mestichelli B, Myer K, Roe Bryan, Salvatore M, Shad S, Thomas JA, Walters G, Washington M, Wheeler J, Bridge J, Foster BA, Gillard BM, Karasik E, Kumar R, Miklos M, Moser MT, Jewell SD, Montroy RG, Rohrer DC, Valley D, Mash DC, Davis DA, Sobin L, Barcus ME, Branton PA, Abell NS, Balliu B, Delaneau O, Frésard L, Gamazon ER, Garrido-Martín D, Gewirtz ADH, Gliner G, Gloudemans MJ, Han B, He AZ, Hormozdiari F, Li Xin, Liu B, Kang EY, McDowell IC, Ongen H, Palowitch JJ, Peterson CB, Quon G, Ripke S, Saha A, Shabalin AA, Shimko TC, Sul JH, Teran NA, Tsang EK, Zhang H, Zhou Y-H, Bustamante CD, Cox NJ, Guigó R, Kellis M, McCarthy MI, Conrad DF, Eskin E, Li G, Nobel AB, Sabatti C, Stranger BE, Wen X, Wright FA, Ardlie KG, Dermitzakis ET, Lappalainen T, Aguet F, Ardlie KG, Cummings BB, Gelfand ET, Getz G, Hadley K, Handsaker RE, Huang KH, Kashin S, Karczewski KJ, Lek M, Li Xiao, MacArthur DG, Nedzel JL, Nguyen DT, Noble MS, Segrè AV, Trowbridge CA, Tukiainen T, Abell NS, Balliu B, Barshir R, Basha O, Battle A, Bogu GK, Brown A, Brown CD, Castel SE, Chen LS, Chiang C, Conrad DF, Cox NJ, Damani FN, Davis JR, Delaneau O, Dermitzakis ET, Engelhardt BE, Eskin E, Ferreira PG, Frésard L, Gamazon ER, Garrido-Martín D, Gewirtz ADH, Gliner G, Gloudemans MJ, Guigo R, Hall IM, Han B, He Y, Hormozdiari F, Howald C, Kyung Im H, Jo B, Yong Kang E, Kim Y, Kim-Hellmuth S, Lappalainen T, Li G, Li Xin, Liu B, Mangul S, McCarthy MI, McDowell IC, Mohammadi P, Monlong J, Montgomery SB, Muñoz-Aguirre M, Ndungu AW, Nicolae DL, Nobel AB, Oliva M, Ongen H, Palowitch JJ, Panousis N, Papasaikas P, Park YoSon, Parsana P, Payne AJ, Peterson CB, Quan J, Reverter F, Sabatti C, Saha A, Sammeth M, Scott AJ, Shabalin AA, Sodaei R, Stephens M, Stranger BE, Strober BJ, Sul JH, Tsang EK, Urbut S, van de Bunt M, Wang G, Wen X, Wright FA, Xi HS, Yeger-Lotem E, Zappala Z, Zaugg JB, Zhou Y-H, Akey JM, Bates D, Chan J, Chen LS, Claussnitzer M, Demanelis K, Diegel M, Doherty JA, Feinberg AP, Fernando MS, Halow J, Hansen KD, Haugen E, Hickey PF, Hou L, Jasmine F, Jian R, Jiang L, Johnson A, Kaul R, Kellis M, Kibriya MG, Lee K, Billy Li J, Li Q, Li Xiao, Lin J, Lin S, Linder S, Linke C, Liu Y, Maurano MT, Molinie B, Montgomery SB, Nelson J, Neri FJ, Oliva M, Park Yongjin, Pierce BL, Rinaldi NJ, Rizzardi LF, Sandstrom R, Skol A, Smith KS, Snyder MP, Stamatoyannopoulos J, Stranger BE, Tang H, Tsang EK, Wang L, Wang M, Van Wittenberghe N, Wu F, Zhang R, Nierras CR, Branton PA, Carithers LJ, Guan P, Moore HM, Rao A, Vaught JB, Gould SE, Lockart NC, Martin C, Struewing JP, Volpi S, Addington AM, Koester SE, Little AR, Brigham LE, Hasz R, Hunter M, Johns C, Johnson M, Kopen G, Leinweber WF, Lonsdale JT, McDonald A, Mestichelli B, Myer K, Roe Brian, Salvatore M, Shad S, Thomas JA, Walters G, Washington M, Wheeler J, Bridge J, Foster BA, Gillard BM, Karasik E, Kumar R, Miklos M, Moser MT, Jewell SD, Montroy RG, Rohrer DC, Valley DR, Davis DA, Mash DC, Undale AH, Smith AM, Tabor DE, Roche NV, McLean JA, Vatanian N, Robinson KL, Sobin L, Barcus ME, Valentino KM, Qi L, Hunter S, Hariharan P, Singh S, Um KS, Matose T, Tomaszewski MM, Barker LK, Mosavel M, Siminoff LA, Traino HM, Flicek P, Juettemann T, Ruffier M, Sheppard D, Taylor K, Trevanion SJ, Zerbino DR, Craft B, Goldman M, Haeussler M, Kent WJ, Lee CM, Paten B, Rosenbloom KR, Vivian J, Zhu J, 2017. Genetic effects on gene expression across human tissues. Nature 550, 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth S, Mitchell R, Corbin L, Wade KH, Dudding T, Budu-Aggrey A, Carslake D, Hemani G, Paternoster L, Smith GD, Davies N, Lawson DJ, Timpson J, N., 2019. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat. Commun 10, 333. 10.1038/s41467-018-08219-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindley G, Bahrami S, Steen NE, O’Connell KS, Frei O, Shadrin A, Bettella F, Rødevand L, Fan CC, Dale AM, Djurovic S, Smeland OB, Andreassen OA, 2021. Characterising the shared genetic determinants of bipolar disorder, schizophrenia and risk-taking. Transl. Psychiatry 11. 10.1038/S41398-021-01576-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorthøj C, Stürup AE, McGrath JJ, Nordentoft M, 2017. Years of potential life lost and life expectancy in schizophrenia: a systematic review and meta-analysis. Lancet Psychiatry 4, 295–301. 10.1016/S2215-0366(17)30078-0 [DOI] [PubMed] [Google Scholar]
- Holland D, Frei O, Desikan R, Fan C-C, Shadrin AA, Smeland OB, Sundar VS, Thompson P, Andreassen OA, Dale AM, 2020. Beyond SNP heritability: Polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLOS Genet 16, e1008612. 10.1371/journal.pgen.1008612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, Alloza C, Shen X, Barbu MC, Xu EY, Whalley HC, Marioni RE, Porteous DJ, Davies G, Deary IJ, Hemani G, Berger K, Teismann H, Rawal R, Arolt V, Baune BT, Dannlowski U, Domschke K, Tian C, Hinds DA, Trzaskowski M, Byrne EM, Ripke S, Smith DJ, Sullivan PF, Wray NR, Breen G, Lewis CM, McIntosh AM, 2019. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci 22, 343–352. 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konsman JP, 2019. Inflammation and depression: A nervous plea for psychiatry to not become immune to interpretation. Pharmaceuticals 12. 10.3390/ph12010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffa DT, Torres ILS, Rohde LA, 2019. A review on the role of inflammation in attention-deficit/hyperactivity disorder. NeuroImmunoModulation 25, 328–333. 10.1159/000489635 [DOI] [PubMed] [Google Scholar]
- Li Y-C, Yang S-S, Gao W-J, 2016. Disruption of Akt signaling decreases dopamine sensitivity in modulation of inhibitory synaptic transmission in rat prefrontal cortex. Neuropharmacology 108, 403–414. 10.1016/j.neuropharm.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligthart S, Vaez A, Võsa U, Stathopoulou MG, de Vries PS, Prins BP, Van der Most PJ, Tanaka T, Naderi E, Rose LM, Wu Y, Karlsson R, Barbalic M, Lin H, Pool R, Zhu G, Macé A, Sidore C, Trompet S, Mangino M, Sabater-Lleal M, Kemp JP, Abbasi A, Kacprowski T, Verweij N, Smith AV, Huang T, Marzi C, Feitosa MF, Lohman KK, Kleber ME, Milaneschi Y, Mueller C, Huq M, Vlachopoulou E, Lyytikäinen L-P, Oldmeadow C, Deelen J, Perola M, Zhao JH, Feenstra B, Amini M, Lahti J, Schraut KE, Fornage M, Suktitipat B, Chen W-M, Li X, Nutile T, Malerba G, Luan J, Bak T, Schork N, Del Greco MF, Thiering E, Mahajan A, Marioni RE, Mihailov E, Eriksson Joel, Ozel AB, Zhang W, Nethander M, Cheng Y-C, Aslibekyan S, Ang W, Gandin I, Yengo L, Portas L, Kooperberg C, Hofer E, Rajan KB, Schurmann C, den Hollander W, Ahluwalia Tarunveer S., Zhao J, Draisma HHM, Ford I, Timpson N, Teumer A, Huang H, Wahl S, Liu Y, Huang J, Uh H-W, Geller F, Joshi PK, Yanek LR, Trabetti E, Lehne B, Vozzi D, Verbanck M, Biino G, Saba Y, Meulenbelt I, O’Connell JR, Laakso M, Giulianini F, Magnusson PKE, Ballantyne CM, Hottenga JJ, Montgomery GW, Rivadineira F, Rueedi R, Steri M, Herzig K-H, Stott DJ, Menni C, Frånberg M, St. Pourcain B, Felix SB, Pers TH, Bakker SJL, Kraft P, Peters A, Vaidya D, Delgado G, Smit JH, Großmann V, Sinisalo J, Seppälä I, Williams SR, Holliday EG, Moed M, Langenberg C, Räikkönen K, Ding J, Campbell H, Sale MM, Chen Y-DI, James AL, Ruggiero D, Soranzo N, Hartman CA, Smith EN, Berenson GS, Fuchsberger C, Hernandez D, Tiesler CMT, Giedraitis V, Liewald D, Fischer K, Mellström D, Larsson A, Wang Y, Scott WR, Lorentzon M, Beilby J, Ryan KA, Pennell CE, Vuckovic D, Balkau B, Concas MP, Schmidt R, Mendes de Leon CF, Bottinger EP, Kloppenburg M, Paternoster L, Boehnke M, Musk AW, Willemsen G, Evans DM, Madden PAF, Kähönen M, Kutalik Z, Zoledziewska M, Karhunen V, Kritchevsky SB, Sattar N, Lachance G, Clarke R, Harris TB, Raitakari OT, Attia JR, van Heemst D, Kajantie E, Sorice R, Gambaro G, Scott RA, Hicks AA, Ferrucci L, Standl M, Lindgren CM, Starr JM, Karlsson M, Lind L, Li JZ, Chambers JC, Mori TA, de Geus EJCN, Heath AC, Martin NG, Auvinen J, Buckley BM, de Craen AJM, Waldenberger M, Strauch K, Meitinger T, Scott RJ, McEvoy M, Beekman M, Bombieri C, Ridker PM, Mohlke KL, Pedersen NL, Morrison AC, Boomsma DI, Whitfield JB, Strachan DP, Hofman A, Vollenweider P, Cucca F, Jarvelin M-R, Jukema JW, Spector TD, Hamsten A, Zeller T, Uitterlinden André G., Nauck M, Gudnason V, Qi L, Grallert H, Borecki IB, Rotter JI, März W, Wild PS, Lokki M-L, Boyle M, Salomaa V, Melbye M, Eriksson JG, Wilson JF, Penninx BWJH, Becker DM, Worrall BB, Gibson G, Krauss RM, Ciullo M, Zaza G, Wareham NJ, Oldehinkel AJ, Palmer LJ, Murray SS, Pramstaller PP, Bandinelli S, Heinrich J, Ingelsson E, Deary IJ, Mägi R, Vandenput L, van der Harst P, Desch KC, Kooner JS, Ohlsson C, Hayward C, Lehtimäki T, Shuldiner AR, Arnett DK, Beilin LJ, Robino A, Froguel P, Pirastu M, Jess T, Koenig W, Loos RJF, Evans DA, Schmidt H, Smith GD, Slagboom PE, Eiriksdottir G, Morris AP, Psaty BM, Tracy RP, Nolte IM, Boerwinkle E, Visvikis-Siest S, Reiner AP, Gross M, Bis JC, Franke L, Franco OH, Benjamin EJ, Chasman DI, Dupuis Josée, Snieder H, Dehghan A, Alizadeh BZ, Alizadeh BZ, Boezen HM, Franke L, van der Harst P, Navis G, Rots M, Snieder H, Swertz M, Wolffenbuttel BHR, Wijmenga C, Benjamin E, Chasman DI, Dehghan A, Ahluwalia Tarunveer Singh, Meigs J, Tracy R, Alizadeh BZ, Ligthart S, Bis J, Eiriksdottir G, Pankratz N, Gross M, Rainer A, Snieder H, Wilson JG, Psaty BM, Dupuis Josee, Prins B, Vaso U, Stathopoulou M, Franke L, Lehtimaki T, Koenig W, Jamshidi Y, Siest S, Abbasi A, Uitterlinden Andre G., Abdollahi M, Schnabel R, Schick UM, Nolte IM, Kraja A, Hsu Y-H, Tylee DS, Zwicker A, Uher R, Davey-Smith G, Morrison AC, Hicks A, van Duijn CM, Ward-Caviness C, Boerwinkle E, Rotter J, Rice K, Lange L, Perola M, de Geus E, Morris AP, Makela KM, Stacey D, Eriksson Johan, Frayling TM, Slagboom EP, 2018. Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders. Am. J. Hum. Genet 103, 691–706. 10.1016/j.ajhg.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques TR, Ashok AH, Pillinger T, Veronese M, Turkheimer FE, Dazzan P, Sommer IEC, Howes OD, 2019. Neuroinflammation in schizophrenia: meta-analysis of in vivo microglial imaging studies. Psychol. Med 49, 2186–2196. 10.1017/S0033291718003057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Ewert HK, Ridker PM, Rifai N, Price N, Dinges DF, Mullington JM, 2001. Absence of diurnal variation of C-reactive protein concentrations in healthy human subjects. Clin. Chem 47, 426–430. [PubMed] [Google Scholar]
- Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, Als TD, Bigdeli TB, Børte S, Bryois J, Charney AW, Drange OK, Gandal MJ, Hagenaars SP, Ikeda M, Kamitaki N, Kim M, Krebs K, Panagiotaropoulou G, Schilder BM, Sloofman LG, Steinberg S, Trubetskoy V, Winsvold BS, Won H-H, Abramova L, Adorjan K, Agerbo E, Al Eissa M, Albani D, Alliey-Rodriguez N, Anjorin A, Antilla V, Antoniou A, Awasthi S, Baek JH, Bækvad-Hansen M, Bass N, Bauer M, Beins EC, Bergen SE, Birner A, Bøcker Pedersen C, Bøen E, Boks MP, Bosch R, Brum M, Brumpton BM, Brunkhorst-Kanaan N, Budde M, Bybjerg-Grauholm J, Byerley W, Cairns M, Casas M, Cervantes P, Clarke T-K, Cruceanu C, Cuellar-Barboza A, Cunningham J, Curtis D, Czerski PM, Dale AM, Dalkner N, David FS, Degenhardt F, Djurovic S, Dobbyn AL, Douzenis A, Elvsåshagen T, Escott-Price V, Ferrier IN, Fiorentino A, Foroud TM, Forty L, Frank J, Frei O, Freimer NB, Frisén L, Gade K, Garnham J, Gelernter J, Giørtz Pedersen M, Gizer IR, Gordon SD, Gordon-Smith K, Greenwood TA, Grove J, Guzman-Parra J, Ha K, Haraldsson M, Hautzinger M, Heilbronner U, Hellgren D, Herms S, Hoffmann P, Holmans PA, Huckins L, Jamain S, Johnson JS, Kalman JL, Kamatani Y, Kennedy JL, Kittel-Schneider S, Knowles JA, Kogevinas M, Koromina M, Kranz TM, Kranzler HR, Kubo M, Kupka R, Kushner SA, Lavebratt C, Lawrence J, Leber M, Lee H-J, Lee PH, Levy SE, Lewis C, Liao C, Lucae S, Lundberg M, MacIntyre DJ, Magnusson SH, Maier W, Maihofer A, Malaspina D, Maratou E, Martinsson L, Mattheisen M, McCarroll SA, McGregor NW, McGuffin P, McKay JD, Medeiros H, Medland SE, Millischer V, Montgomery GW, Moran JL, Morris DW, Mühleisen TW, O’Brien N, O’Donovan C, Olde Loohuis LM, Oruc L, Papiol S, Pardiñas AF, Perry A, Pfennig A, Porichi E, Potash JB, Quested D, Raj T, Rapaport MH, DePaulo JR, Regeer EJ, Rice JP, Rivas F, Rivera M, Roth J, Roussos P, Ruderfer DM, Sánchez-Mora C, Schulte EC, Senner F, Sharp S, Shilling PD, Sigurdsson E, Sirignano L, Slaney C, Smeland OB, Smith DJ, Sobell JL, Søholm Hansen C, Soler Artigas M, Spijker AT, Stein DJ, Strauss JS, Świątkowska B, Terao C, Thorgeirsson TE, Toma C, Tooney P, Tsermpini E-E, Vawter MP, Vedder H, Walters JTR, Witt SH, Xi S, Xu W, Yang JMK, Young AH, Young H, Zandi PP, Zhou H, Zillich L, Adolfsson R, Agartz I, Alda M, Alfredsson L, Babadjanova G, Backlund L, Baune BT, Bellivier F, Bengesser S, Berrettini WH, Blackwood DHR, Boehnke M, Børglum AD, Breen G, Carr VJ, Catts S, Corvin A, Craddock N, Dannlowski U, Dikeos D, Esko T, Etain B, Ferentinos P, Frye M, Fullerton JM, Gawlik M, Gershon ES, Goes FS, Green MJ, Grigoroiu-Serbanescu M, Hauser J, Henskens F, Hillert J, Hong KS, Hougaard DM, Hultman CM, Hveem K, Iwata N, Jablensky AV, Jones I, Jones LA, Kahn RS, Kelsoe JR, Kirov G, Landén M, Leboyer M, Lewis CM, Li QS, Lissowska J, Lochner C, Loughland C, Martin NG, Mathews CA, Mayoral F, McElroy SL, McIntosh AM, McMahon FJ, Melle I, Michie P, Milani L, Mitchell PB, Morken G, Mors O, Mortensen PB, Mowry B, Müller-Myhsok B, Myers RM, Neale BM, Nievergelt CM, Nordentoft M, Nöthen MM, O’Donovan MC, Oedegaard KJ, Olsson T, Owen MJ, Paciga SA, Pantelis C, Pato C, Pato MT, Patrinos GP, Perlis RH, Posthuma D, Ramos-Quiroga JA, Reif A, Reininghaus EZ, Ribasés M, Rietschel M, Ripke S, Rouleau GA, Saito T, Schall U, Schalling M, Schofield PR, Schulze TG, Scott LJ, Scott RJ, Serretti A, Shannon Weickert C, Smoller JW, Stefansson H, Stefansson K, Stordal E, Streit F, Sullivan PF, Turecki G, Vaaler AE, Vieta E, Vincent JB, Waldman ID, Weickert TW, Werge T, Wray NR, Zwart J-A, Biernacka JM, Nurnberger JI, Cichon S, Edenberg HJ, Stahl EA, McQuillin A, Di Florio A, Ophoff RA, Andreassen OA, 2021. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet 53, 817–829. 10.1038/s41588-021-00857-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North HF, Weissleder C, Fullerton JM, Webster MJ, Weickert CS, 2022. Increased immune cell and altered microglia and neurogenesis transcripts in an Australian schizophrenia subgroup with elevated inflammation. Schizophr. Res 248, 208–218. 10.1016/j.schres.2022.08.025 [DOI] [PubMed] [Google Scholar]
- Osimo EF, Pillinger T, Rodriguez IM, Khandaker GM, Pariante CM, Howes OD, 2020. Inflammatory markers in depression: A meta-analysis of mean differences and variability in 5,166 patients and 5,083 controls. Brain. Behav. Immun 87, 901–909. 10.1016/j.bbi.2020.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, Legge SE, Bishop S, Cameron D, Hamshere ML, Han J, Hubbard L, Lynham A, Mantripragada K, Rees E, MacCabe JH, McCarroll SA, Baune BT, Breen G, Byrne EM, Dannlowski U, Eley TC, Hayward C, Martin NG, McIntosh AM, Plomin R, Porteous DJ, Wray NR, Caballero A, Geschwind DH, Huckins LM, Ruderfer DM, Santiago E, Sklar P, Stahl EA, Won H, Agerbo E, Als TD, Andreassen OA, Bækvad-Hansen M, Mortensen PB, Pedersen CB, Børglum AD, Bybjerg-Grauholm J, Djurovic S, Durmishi N, Pedersen MG, Golimbet V, Grove J, Hougaard DM, Mattheisen M, Molden E, Mors O, Nordentoft M, Pejovic-Milovancevic M, Sigurdsson E, Silagadze T, Hansen CS, Stefansson K, Stefansson H, Steinberg S, Tosato S, Werge T, Harold D, Sims R, Gerrish A, Chapman J, Abraham R, Hollingworth P, Pahwa J, Denning N, Thomas C, Taylor S, Powell J, Proitsi P, Lupton M, Lovestone S, Passmore P, Craig D, McGuinness B, Johnston J, Todd S, Maier W, Jessen F, Heun R, Schurmann B, Ramirez A, Becker T, Herold C, Lacour A, Drichel D, Nothen M, Goate A, Cruchaga C, Nowotny P, Morris JC, Mayo K, O’Donovan MC, Owen MJ, Williams J, Achilla E, Barr CL, Böttger TW, Cohen D, Curran S, Dempster E, Dima D, Sabes-Figuera R, Flanagan RJ, Frangou S, Frank J, Gasse C, Gaughran F, Giegling I, Hannon E, Hartmann AM, Heißerer B, Helthuis M, Horsdal HT, Ingimarsson O, Jollie K, Kennedy JL, Köhler O, Konte B, Lang M, Lewis C, MacCaba J, Malhotra AK, McCrone P, Meier SM, Mill J, Nöthen MM, Pedersen CB, Rietschel M, Rujescu D, Schwalber A, Sørensen HJ, Spencer B, Støvring H, Strohmaier J, Sullivan P, Vassos E, Verbelen M, Collier DA, Kirov G, Owen MJ, O’Donovan MC, Walters JTR, 2018. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389. 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepys MB, 1995. The acute phase response and C-reactive protein. Oxf. Textb. Med 2, 1527–1533. [Google Scholar]
- Pitharouli MC, Hagenaars SP, Glanville KP, Coleman JRI, Hotopf M, Lewis CM, Pariante CM, 2021. Elevated C-Reactive Protein in Patients With Depression, Independent of Genetic, Health, and Psychosocial Factors: Results From the UK Biobank. Am. J. Psychiatry 178, 522–529. 10.1176/appi.ajp.2020.20060947 [DOI] [PubMed] [Google Scholar]
- Plana-Ripoll O, Pedersen CB, Holtz Y, Benros ME, Dalsgaard S, De Jonge P, Fan CC, Degenhardt L, Ganna A, Greve AN, Gunn J, Iburg KM, Kessing LV, Lee BK, Lim CCW, Mors O, Nordentoft M, Prior A, Roest AM, Saha S, Schork A, Scott JG, Scott KM, Stedman T, Sørensen HJ, Werge T, Whiteford HA, Laursen TM, Agerbo E, Kessler RC, Mortensen PB, McGrath JJ, 2019. Exploring Comorbidity Within Mental Disorders among a Danish National Population. JAMA Psychiatry 76, 259–270. 10.1001/jamapsychiatry.2018.3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget JG, Han B, Wu Y, Mignot E, Ollila HM, Barker J, Spain S, Dand N, Trembath R, Martin J, Mayes MD, Bossini-Castillo L, López-Isac E, Jin Y, Santorico SA, Spritz RA, Hakonarson H, Polychronakos C, Raychaudhuri S, Knight J, 2019. Cross-disorder analysis of schizophrenia and 19 immune-mediated diseases identifies shared genetic risk. Hum. Mol. Genet 28, 3498–3513. 10.1093/hmg/ddz145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins BP, Abbasi A, Wong A, Vaez A, Nolte I, Franceschini N, Stuart PE, Guterriez Achury J, Mistry V, Bradfield JP, Valdes AM, Bras J, Shatunov A, Lu C, Han B, Raychaudhuri S, Bevan S, Mayes MD, Tsoi LC, Evangelou E, Nair RP, Grant SFA, Polychronakos C, Radstake TRD, van Heel DA, Dunstan ML, Wood NW, Al-Chalabi A, Dehghan A, Hakonarson H, Markus HS, Elder JT, Knight J, Arking DE, Spector TD, Koeleman BPC, van Duijn CM, Martin J, Morris AP, Weersma RK, Wijmenga C, Munroe PB, Perry JRB, Pouget JG, Jamshidi Y, Snieder H, Alizadeh BZ, 2016. Investigating the Causal Relationship of C-Reactive Protein with 32 Complex Somatic and Psychiatric Outcomes: A Large-Scale Cross-Consortium Mendelian Randomization Study. PLoS Med 13, e1001976. 10.1371/journal.pmed.1001976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay WR, Kiltschewskij DJ, Geaghan MP, Atkins JR, Carr VJ, Green MJ, Cairns MJ, 2022. Genetic estimates of correlation and causality between blood-based biomarkers and psychiatric disorders. Sci. Adv 8, eabj8969. 10.1126/sciadv.abj8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sæther LS, Ueland Thor, Haatveit B, Maglanoc LA, Szabo A, Djurovic S, Aukrust P, Roelfs D, Mohn C, Ormerod MBEG, Lagerberg TV, Steen NE, Melle I, Andreassen OA, Ueland Torill, 2022. Inflammation and cognition in severe mental illness: patterns of covariation and subgroups. Mol. Psychiatry 10.1038/s41380-022-01924-w [DOI] [PMC free article] [PubMed]
- Said S, Pazoki R, Karhunen V, Võsa U, Ligthart S, Bodinier B, Koskeridis F, Welsh P, Alizadeh BZ, Chasman DI, Sattar N, Chadeau-Hyam M, Evangelou E, Jarvelin M-R, Elliott P, Tzoulaki I, Dehghan A, 2022. Genetic analysis of over half a million people characterises C-reactive protein loci. Nat. Commun 13, 2198. 10.1038/s41467-022-29650-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan E, Attwells S, Wilson AA, Mizrahi R, Rusjan PM, Miler L, Xu C, Sharma S, Kish S, Houle S, Meyer JH, 2018. Association of translocator protein total distribution volume with duration of untreated major depressive disorder: a cross-sectional study. Lancet Psychiatry 5, 339–347. 10.1016/S2215-0366(18)30048-8 [DOI] [PubMed] [Google Scholar]
- Smeland OB, Frei O, Dale AM, Andreassen OA, 2020. The polygenic architecture of schizophrenia — rethinking pathogenesis and nosology. Nat. Rev. Neurol 16, 366–379. 10.1038/s41582-020-0364-0 [DOI] [PubMed] [Google Scholar]
- Smeland OB, Frei O, Shadrin A, O’Connell K, Fan C-C, Bahrami S, Holland D, Djurovic S, Thompson WK, Dale AM, 2019. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum. Genet 1–10. [DOI] [PubMed]
- Smoller JW, Andreassen OA, Edenberg HJ, Faraone SV, Glatt SJ, Kendler KS, 2019. Psychiatric genetics and the structure of psychopathology. Mol. Psychiatry 24, 409–420. 10.1038/s41380-017-0010-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R, 2015. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med 12, e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A, O’Connell KS, Ueland T, Sheikh MA, Agartz I, Andreou D, Aukrust P, Boye B, Bøen E, Drange OK, Elvsåshagen T, Engh JA, Hope S, Collier Høegh M, Joa I, Johnsen E, Kroken RA, Vik Lagerberg T, Lekva T, Malt UF, Melle I, Morken G, Nærland T, Steen VM, Sørensen K, Wedervang-Resell K, Auten Weibell M, Westlye LT, Steen NE, Andreassen O, Djurovic S, 2022. Increased circulating IL-18 levels in severe mental disorders indicate systemic inflammasome activation. Brain. Behav. Immun 99, 299–306. 10.1016/j.bbi.2021.10.017 [DOI] [PubMed] [Google Scholar]
- Tamouza R, Meyer U, Foiselle M, Richard J-R, Wu C-L, Boukouaci W, Le Corvoisier P, Barrau C, Lucas A, Perron H, Leboyer M, 2021. Identification of inflammatory subgroups of schizophrenia and bipolar disorder patients with HERV-W ENV antigenemia by unsupervised cluster analysis. Transl. Psychiatry 11, 377. 10.1038/s41398-021-01499-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, Bryois J, Chen C-Y, Dennison CA, Hall LS, Lam M, Watanabe K, Frei O, Ge T, Harwood JC, Koopmans F, Magnusson S, Richards AL, Sidorenko J, Wu Y, Zeng J, Grove J, Kim M, Li Z, Voloudakis G, Zhang W, Adams M, Agartz I, Atkinson EG, Agerbo E, Al Eissa M, Albus M, Alexander M, Alizadeh BZ, Alptekin K, Als TD, Amin F, Arolt V, Arrojo M, Athanasiu L, Azevedo MH, Bacanu SA, Bass NJ, Begemann M, Belliveau RA, Bene J, Benyamin B, Bergen SE, Blasi G, Bobes J, Bonassi S, Braun A, Bressan RA, Bromet EJ, Bruggeman R, Buckley PF, Buckner RL, Bybjerg-Grauholm J, Cahn W, Cairns MJ, Calkins ME, Carr VJ, Castle D, Catts SV, Chambert KD, Chan RCK, Chaumette B, Cheng W, Cheung EFC, Chong SA, Cohen D, Consoli A, Cordeiro Q, Costas J, Curtis C, Davidson M, Davis KL, de Haan L, Degenhardt F, DeLisi LE, Demontis D, Dickerson F, Dikeos D, Dinan T, Djurovic S, Duan J, Ducci G, Dudbridge F, Eriksson JG, Fañanás L, Faraone SV, Fiorentino A, Forstner A, Frank J, Freimer NB, Fromer M, Frustaci A, Gadelha A, Genovese G, Gershon ES, Giannitelli M, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, González Peñas J, González-Pinto A, Gopal S, Gratten J, Green MF, Greenwood TA, Guillin O, Gülöksüz S, Gur RE, Gur RC, Gutiérrez B, Hahn E, Hakonarson H, Haroutunian V, Hartmann AM, Harvey C, Hayward C, Henskens FA, Herms S, Hoffmann P, Howrigan DP, Ikeda M, Iyegbe C, Joa I, Julià A, Kähler AK, Kam-Thong T, Kamatani Y, Karachanak-Yankova S, Kebir O, Keller MC, Kelly BJ, Khrunin A, Kim S-W, Klovins J, Kondratiev N, Konte B, Kraft J, Kubo M, Kučinskas V, Kučinskiene ZA, Kusumawardhani A, Kuzelova-Ptackova H, Landi S, Lazzeroni LC, Lee PH, Legge SE, Lehrer DS, Lencer R, Lerer B, Li M, Lieberman J, Light GA, Limborska S, Liu C-M, Lönnqvist J, Loughland CM, Lubinski J, Luykx JJ, Lynham A, Macek M, Mackinnon A, Magnusson PKE, Maher BS, Maier W, Malaspina D, Mallet J, Marder SR, Marsal S, Martin AR, Martorell L, Mattheisen M, McCarley RW, McDonald C, McGrath JJ, Medeiros H, Meier S, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mitjans M, Molden E, Molina E, Molto MD, Mondelli V, Moreno C, Morley CP, Muntané G, Murphy KC, Myin-Germeys I, Nenadić I, Nestadt G, Nikitina-Zake L, Noto C, Nuechterlein KH, O’Brien NL, O’Neill FA, Oh S-Y, Olincy A, Ota VK, Pantelis C, Papadimitriou GN, Parellada M, Paunio T, Pellegrino R, Periyasamy S, Perkins DO, Pfuhlmann B, Pietiläinen O, Pimm J, Porteous D, Powell J, Quattrone D, Quested D, Radant AD, Rampino A, Rapaport MH, Rautanen A, Reichenberg A, Roe C, Roffman JL, Roth J, Rothermundt M, Rutten BPF, Saker-Delye S, Salomaa V, Sanjuan J, Santoro ML, Savitz A, Schall U, Scott RJ, Seidman LJ, Sharp SI, Shi J, Siever LJ, Sigurdsson E, Sim K, Skarabis N, Slominsky P, So H-C, Sobell JL, Söderman E, Stain HJ, Steen NE, Steixner-Kumar AA, Stögmann E, Stone WS, Straub RE, Streit F, Strengman E, Stroup TS, Subramaniam M, Sugar CA, Suvisaari J, Svrakic DM, Swerdlow NR, Szatkiewicz JP, Ta TMT, Takahashi A, Terao C, Thibaut F, Toncheva D, Tooney PA, Torretta S, Tosato S, Tura GB, Turetsky BI, Üçok A, Vaaler A, van Amelsvoort T, van Winkel R, Veijola J, Waddington J, Walter H, Waterreus A, Webb BT, Weiser M, Williams NM, Witt SH, Wormley BK, Wu JQ, Xu Z, Yolken R, Zai CC, Zhou W, Zhu F, Zimprich F, Atbaşoğlu EC, Ayub M, Benner C, Bertolino A, Black DW, Bray NJ, Breen G, Buccola NG, Byerley WF, Chen WJ, Cloninger CR, Crespo-Facorro B, Donohoe G, Freedman R, Galletly C, Gandal MJ, Gennarelli M, Hougaard DM, Hwu H-G, Jablensky AV, McCarroll SA, Moran JL, Mors O, Mortensen PB, Müller-Myhsok B, Neil AL, Nordentoft M, Pato MT, Petryshen TL, Pirinen M, Pulver AE, Schulze TG, Silverman JM, Smoller JW, Stahl EA, Tsuang DW, Vilella E, Wang S-H, Xu S, Indonesia Schizophrenia Consortium, PsychENCODE, Psychosis Endophenotypes International Consortium, SynGO Consortium, Adolfsson R, Arango C, Baune BT, Belangero SI, Børglum AD, Braff D, Bramon E, Buxbaum JD, Campion D, Cervilla JA, Cichon S, Collier DA, Corvin A, Curtis D, Forti MD, Domenici E, Ehrenreich H, Escott-Price V, Esko T, Fanous AH, Gareeva A, Gawlik M, Gejman PV, Gill M, Glatt SJ, Golimbet V, Hong KS, Hultman CM, Hyman SE, Iwata N, Jönsson EG, Kahn RS, Kennedy JL, Khusnutdinova E, Kirov G, Knowles JA, Krebs M-O, Laurent-Levinson C, Lee J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, Malhotra D, McIntosh A, McQuillin A, Menezes PR, Morgan VA, Morris DW, Mowry BJ, Murray RM, Nimgaonkar V, Nöthen MM, Ophoff RA, Paciga SA, Palotie A, Pato CN, Qin S, Rietschel M, Riley BP, Rivera M, Rujescu D, Saka MC, Sanders AR, Schwab SG, Serretti A, Sham PC, Shi Y, St Clair D, Stefánsson H, Stefansson K, Tsuang MT, van Os J, Vawter MP, Weinberger DR, Werge T, Wildenauer DB, Yu X, Yue W, Holmans PA, Pocklington AJ, Roussos P, Vassos E, Verhage M, Visscher PM, Yang J, Posthuma D, Andreassen OA, Kendler KS, Owen MJ, Wray NR, Daly MJ, Huang H, Neale BM, Sullivan PF, Ripke S, Walters JTR, O’Donovan MC, Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2022. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508. 10.1038/s41586-022-04434-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee DS, Sun J, Hess JL, Tahir MA, Sharma E, Malik R, Worrall BB, Levine AJ, Martinson JJ, Nejentsev S, Speed D, Fischer A, Mick E, Walker BR, Crawford A, Grant SFA, Polychronakos C, Bradfield JP, Sleiman PMA, Hakonarson H, Ellinghaus E, Elder JT, Tsoi LC, Trembath RC, Barker JN, Franke A, Dehghan A, Faraone SV, Glatt SJ, 2018. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am. J. Med. Genet. B Neuropsychiatr. Genet 177, 641–657. 10.1002/ajmg.b.32652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werme J, van der Sluis S, Posthuma D, de Leeuw CA, 2022. An integrated framework for local genetic correlation analysis. Nat. Genet 54, 274–282. 10.1038/s41588-022-01017-y [DOI] [PubMed] [Google Scholar]
- Whiteford HA, Degenhardt L, Rehm J, Baxter AJ, Ferrari AJ, Erskine HE, Charlson FJ, Norman RE, Flaxman AD, Johns N, Burstein R, Murray CJL, Vos T, 2013. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. The Lancet 382, 1575–1586. 10.1016/S0140-6736(13)61611-6 [DOI] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, 2018. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet 50, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi G, Park J-H, Chatterjee N, 2018. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet 50, 1318–1326. 10.1038/s41588-018-0193-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


