Abstract
Dysregulated Notch signaling is a common feature of cancer; however, its effects on tumor initiation and progression are highly variable, with Notch having either oncogenic or tumor-suppressive functions in various cancers. To better understand the mechanisms that regulate Notch function in cancer, we studied Notch signaling in a Drosophila tumor model, prostate cancer–derived cell lines, and tissue samples from patients with advanced prostate cancer. We demonstrated that increased activity of the Src-JNK pathway in tumors inactivated Notch signaling due to JNK pathway-mediated inhibition of the expression of the gene encoding the Notch S2 cleavage protease, Kuzbanian, which is critical for Notch activity. Consequently, inactive Notch accumulated in cells, where it was unable to transcribe genes encoding its target proteins, many of which have tumor-suppressive activities. These findings suggest that Src-JNK activity in tumors predicts Notch activity status and that suppressing Src-JNK signaling could restore Notch function in tumors, offering opportunities for diagnosis and targeted therapies for a subset of advanced prostate cancer patients.
Keywords: Notch, JNK, ADAM, Kuzbanian, Prostate Cancer, Drosophila
Introduction
First identified and characterized in Drosophila melanogaster (1–3), the Notch pathway is a highly evolutionarily conserved signaling mechanism with crucial roles in development and tissue homeostasis (4–7). In order to be activated, the Notch receptor undergoes three proteolytic cleavages at specific sites (schematically depicted in Fig 1A). The first cleavage site, known as S1, is targeted by the protease Furin while the protein transits from the endoplasmic reticulum (ER) to the plasma membrane. Upon interaction of the receptor with a transmembrane ligand on a neighboring cell, Notch is then cleaved at the S2 site by proteases of the ADAM (A disintegrin and metalloprotease) family, principally ADAM10/17 in mammals and Kuzbanian in Drosophila. This S2 cleavage leads to the detachment of the Notch extracellular domain (NECD) and the subsequent intracellular cleavage at the S3 site by the γ-secretase complex. The latter releases the Notch intracellular domain (NICD), which translocates to the cell nucleus where it interacts with CSL (Suppressor of Hairless in Drosophila) to regulate transcription of target genes. Although the resulting signaling outputs are diverse and context-dependent, they converge to control cell fate (6, 7).
Fig. 1. Change in Notch 1 cleavage correlates with changes in JNK signaling and LDH-A expression in prostate cancer cell lines.
(A) Schematic of the Notch 1 receptor depicting the 3 cleavage sites: S1, S2, and S3, with the resulting extracellular domain (ECD) and intracellular domain (ICD) fragments.
(B-J) Immunoblots of cell extracts from benign prostate (RWPE-1), early stage prostate cancer (LNCaP) and androgen resistant prostate cancer (PC-3) cell lines.
(B-D) Immunodetection of Notch 1 (NICD plus transmembrane fragment, 120 kDa), Notch1ICD (110 kDa) and GAPDH (36 kDa) as a loading control (B). Quantification of the normalized expression of the 120 kDa Notch1 fragment (C) and the 110 kDa Notch1ICD (D) in each cell line is shown as the ratio of the corresponding signal intensity over the corresponding GAPDH-associated signal intensity.
(E-F) Immunodetection of Notch 1 extracellular domain (Notch1ECD, 180 kDa) and α-tubulin (52 kDa) as a loading control (E). Quantification of the normalized expression of Notch1ECD in each cell line is shown as the ratio of the 180 kDa Notch1ECD-associated signal intensity scaled against the corresponding GAPDH-associated signal intensity (F).
(G-H) Immunodetection of phosphorylated JNK (pJNK, 46kDa), total JNK (46 kDa, 54 kDa) and GAPDH (36 kDa) as a loading control (G). Quantification of the relative levels of pJNK in each cell line is shown as the ratio of the 46kDa pJNK and the corresponding 46kDa total JNK band (H).
(I-J) Immunodetection of LDH-A (37 kDa) and total protein staining (I). Quantification of the normalized levels of LDH-A in each cell line is shown as the ratio of LDH-A-associated signal intensity over total protein staining intensity (J).
Unpaired t-tests were used to evaluate statistical significance, denoted as * for 0.01<p≤ 0.05, ** for 0.05<p≤ 0.01 and *** for 0.01<p≤0.001. Comparisons were performed between RWPE-1 and LNCaP cells and between RWPE-1 and PC-3 cells. For quantification of each immunoblot, N=3 biological replicates were used that were derived from independent cell lysates.
Dysregulations of Notch signaling, whether resulting from mutations in Notch itself or through other mechanisms, have been described in a large number of cancers and both hypo- and hyper-activation of the pathway have been associated with tumor formation (6–9). A truncated Notch protein, resulting from a chromosomal translocation and corresponding to NICD, is detected in increased abundance and contributes to the growth of cancers such as T-cell acute lymphoblastic leukemia (T-ALL), B-cell leukemias, lymphomas of mantle and splenic cells and breast cancer. An alternative scenario in squamous cell carcinomas, gliomas, neuroendocrine tumors, monocytic leukemias and others, indicates that Notch can function as a tumor suppressor. Both scenarios have been explored for prostate cancer with mixed results and possible therapeutic agents that target Notch function have yet to be shown to be fully effective (10, 11). Thus, it appears that Notch function is context-dependent in cancer, as it is in development. However, mechanistic details of a tumor suppressive role remain less explored (12).
In past studies of a glycolytic tumor model in Drosophila larvae, we showed that expression of an activated form of the PDGF/VEGF Receptor (Pvr) in the wing imaginal disc is sufficient to cause the formation of large tumors, in which three main signaling pathways, the phosphatidylinositol-3-kinase (PI3K), extracellular signal-regulated kinase (ERK) and c-Jun NH2-terminal kinase (JNK) pathways, are activated (13). Genetic analyses demonstrated that PI3K and ERK signaling together promote the activation of Sima (Hif1 homolog in flies), whereas Src promotes the phosphorylation and subsequent activation of JNK. An integration of these signals at multiple levels results in production of active JNK and lactate dehydrogenase (LDH), which converge in the establishment and maintenance of the Warburg effect, while decreasing oxidative phosphorylation (see fig. S2A for a redrawing of the schematic in (13)). While PI3K and ERK are major oncogenic pathways (14, 15), the JNK pathway has ambivalent roles in cancer and can be either oncogenic or tumor suppressive (16–18), in a manner similar to that of Notch. In prostate cancer, both of these effects have been proposed for JNK (19). As the Notch pathway is active and plays important roles in wing disc development, the Drosophila wing disc tumor model offers an ideal context to investigate the contribution of Notch in tumor progression and its potential relationship with JNK.
In the present study, we combine genetic manipulations in the Drosophila wing disc tumor model with observations in prostate cancer-derived cell lines and tissue samples from biopsies of patients with advanced prostate cancer. We show that elevated Src-JNK pathway activity in tumors inactivates Notch signaling, by decreasing the expression of the Notch S2 cleavage protease. Consequently, inactive Notch accumulates in the cell but is unable to transcribe genes encoding its target proteins, many of which share tumor-suppressive activities. Our findings suggest that Src-JNK activity in a tumor is predictive of Notch activity status and suggest that suppressing Src-JNK could restore Notch function in the tumor, offering new perspectives for diagnosis and targeted therapies for advanced prostate cancer.
Results
Changes in Notch 1 cleavage correlate with changes in JNK signaling and LDH-A expression in prostate cancer cell lines
To begin analyzing the role of Notch in prostate cancer, we performed Western blot analyses of prostate-derived cell lines. RWPE-1 are normal prostate immortalized cells, LNCaP are early androgen-sensitive prostate cancer cells and PC3 are advanced androgen independent prostate cancer cells (20).
We used two antibodies that identify the intracellular domain of Notch1. The first (clone D3B8) exclusively detects endogenous levels of Notch1ICD as a 110 kDa fragment resulting from the S3 cleavage of Notch1. The second (clone D1E11) identifies a slightly larger fragment of size 120 kDa that includes both NICD and the transmembrane domain, which results from the earlier S2 cleavage (Fig. 1A). We found that both antibodies give rise to similar trends. RWPE-1 showed higher amounts of Notch 1 intracellular fragments, while prostate cancer cell lines, in particular PC3, showed significantly lesser amounts of the protein fragments (Fig. 1B–D). We then used a third antibody raised against Notch1ECD that identifies the 180 kDa extracellular fragment resulting from S2 cleavage (Fig. 1A). The readily apparent 180 kDa band seen in RWPE-1 cells was detected in lower amounts in LNCaP cells and was further reduced in PC3 cells (Fig. 1E–F). Together, these data suggested a defect in Notch1 cleavage in prostate cancer cell lines, which should result in loss of Notch function in the tumor-derived cells.
We also analyzed the expression of Notch2, Notch3 and Notch4 in prostate cell lines. Similar to Notch1, we found that Notch3 and Notch4 intracellular fragments resulting from S2 cleavage were significantly less abundant in prostate cancer cell lines (fig. S1A,C–D). Such a decrease was also apparent for Notch2 in LNCaP cells, but not in PC3 cells (fig. S1A–B). While the reason for this difference in Notch 2 is unclear, overall, these results established that cleavage of other Notch isoforms is also altered in prostate cancer cells.
Similar to Notch, the JNK pathway can be oncogenic or tumor suppressive depending on contexts; both effects have been proposed in prostate cancer (18–19, 21–23). In mammals, JNK is encoded by three genes that are alternatively spliced to generate two protein isoforms (46 kDa and 54 kDa) differing in their C-terminal region (24). Western blot analysis of JNK in prostate-derived cell lines indicated that the isoform composition, as well as the total amount of JNK was not identical in RWPE-1, LNCaP and PC3 cells (Fig. 1G). We used an antibody that specifically binds to the phosphorylated, and thus activated, form of JNK (pJNK). We observed that the 46 kDa isoform was the predominant phosphorylated form in these cell lines (Fig. 1G) and that the ratio of the intensities associated with pJNK and total JNK for this 46kDa isoform was significantly higher in PC-3 cells than in RWPE-1 cells and LNCaP cells (Fig. 1H). These results showed that the JNK pathway is activated in advanced prostate cancer cells.
Active JNK contributes to metabolic reprogramming and, as a result, increased lactate dehydrogenase (LDH) expression is seen in Drosophila tumors (13). We therefore examined LDH-A expression in prostate cell lines, which is particularly relevant given that inhibition of LDH-A has a protective effect on the progression of prostate cancer (25). Consistent with this notion and with what is known in Drosophila, we found that expression of LDH-A was significantly increased in PC3 cells compared to RWPE-1 and LNCaP cells (Fig. 1I–J).
Activated JNK correlates with a loss of Notch activity in Drosophila tumors
The observation that changes in Notch 1 cleavage and JNK activity are correlated in prostate cancer cells in vitro prompted us to reexamine the Drosophila glycolytic tumor model (13) to investigate potential roles of Notch and mechanisms that link Notch and JNK signaling in these tumors.
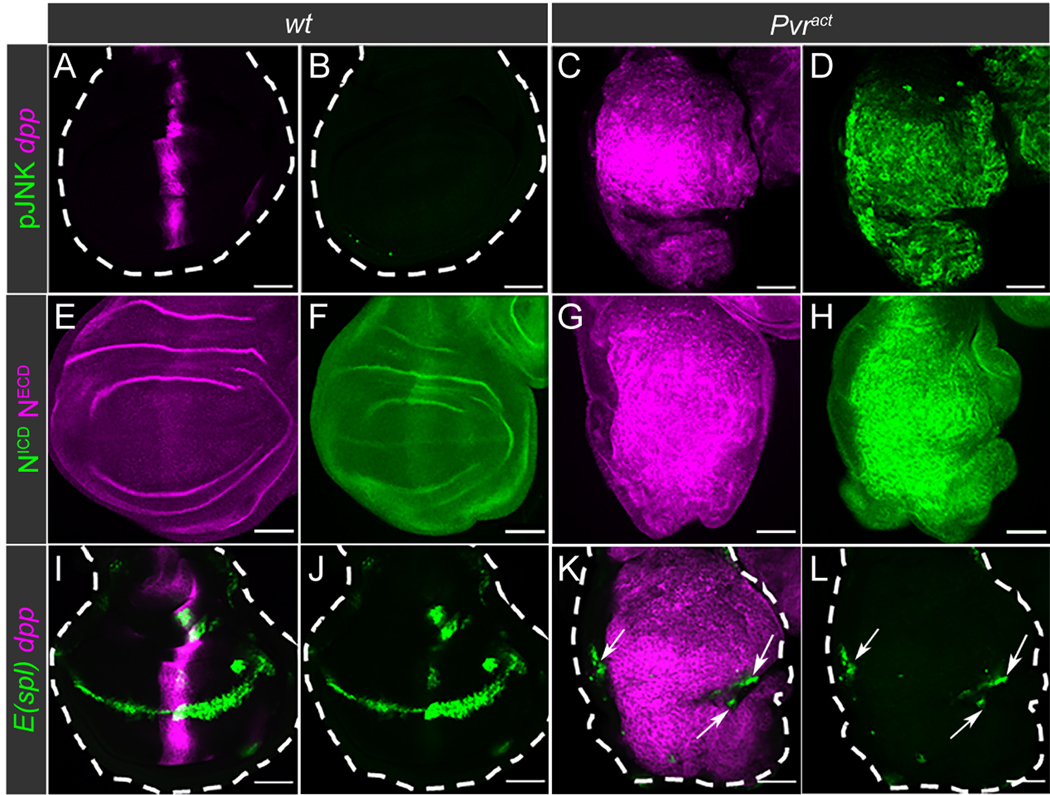
Large tumors can be induced by expressing Pvract along the antero-posterior (A/P) boundary of wing imaginal discs (13). In wild type, this A/P axis is a region defined as the narrow stripe of dpp-expressing cells (Fig. 2A). By immunostaining, pJNK was not detected in this region (Fig. 2B). When Pvract was expressed under the control of a dpp-enhancer (dpp-Gal4>UAS-Pvract,UAS-GFP), a large tumor formed that consists of dpp-expressing cells (Fig. 2C) and shows an increase in pJNK (Fig 2D). These results confirmed our previous observations (13) and further established that the JNK pathway is active in these Drosophila tumors. Using antibodies against the Drosophila NECD and NICD, we detected faint Notch protein expression in wild-type imaginal discs (Fig. 2E–F). As a strong contrast, using either of these antibodies, we detected an accumulation of Notch in the tumor (Fig. 2G–H). Given that increased Notch expression was detected using either antibody, we concluded that full length, uncleaved Notch accumulates in these tumors.
Fig. 2. Activation of JNK correlates with loss of Notch activity in Drosophila tumors.
(A-L) Immunostaining of Drosophila wing imaginal discs from wandering third instar larvae, with wild-type (wt) as control compared with Pvract. wt larvae express mCherry alone (magenta; dpp-GAL4>UAS-mCherry::CD8) and Pvract larvae express mCherry and Pvract (dpp-GAL4>UAS-mCherry::CD8,UAS-Pvract) along the A/P boundary of the wing discs.
(A-D) Activity of the JNK pathway, assessed by immunostaining of pJNK (green; B, D), in wt (A-B) and Pvract (C-D) wing discs. The dpp-expressing cells are identified by mCherry expression (magenta; A, C).
(E-H) Immunostaining of Notch with anti-NECD (magenta; E,G) and anti-NICD (green; F,H) antibodies, in wt (E-F) and Pvract (G-H) wing discs.
(I-L) Expression of the E(spl)m8-GFP reporter (green) in wt (I-J) and Pvract (K-L) wing discs. The dpp-expressing cells are identified by mCherry expression (magenta; I,K). Arrows in K-L point to E(spl)m8-GFP expression outside of the dpp-expressing region.
Each staining was repeated 3 times (N=3) and between 5 and 15 samples were analyzed per repeat.
Scale bars: 50μm.
To establish this further, we studied the expression of E(spl) (HES in humans (26)), a direct Notch transcriptional target, which is a reliable marker of Notch pathway activity (27, 28). In wild-type imaginal discs, E(spl)m8-GFP, a reporter for E(spl) expression (29), was observed along the dorso-ventral (D/V) boundary, indicating the normal site of Notch activation (Fig. 2I–J). Despite the large accumulation of Notch (Fig. 2G–H), expression of E(spl) was not detected by GFP fluorescence in tumor cells (Fig. 2K–L). The residual expression of E(spl) in the few D/V cells that do not express dpp, beyond the margins of the tumor (arrows in Fig. 2K–L), served as an internal control. In addition, the expressions of two other well defined targets of Notch signaling in the wing disc, Cut and Wingless (Wg), were affected in a way similar to that seen for E(spl). Both were expressed at the D/V boundary of the wild-type disc (fig. S2B–C and S2F–G), but neither of these Notch target proteins was expressed in Pvract tumors (fig. S2D–E and S2H–I). These results established that the accumulated Notch observed in Pvract tumors is inactive and unable to turn on the expression of downstream target genes. Consistently, RNAi-mediated depletion of Notch in the Pvract background did not prevent the formation of large tumors (fig. S2J–K and S3L). On the contrary, forced activation of Notch, achieved by overexpressing NICD (Nact) along the A/P boundary, caused the expression of E(Spl) to be much enhanced (fig. S2L–O). When Nact and Pvract were co-expressed in the same cells, the tumor recovered the ability to express E(spl) and was significantly reduced in size (fig. S2P–S and fig. S3L). Taken together, our data indicated that inactive Notch accumulates in Pvract tumors and forced activation of Notch decreases tumor size, in agreement with a tumor suppressive role for Notch in this tissue.
JNK controls Notch activity through negative regulation of Kuzbanian
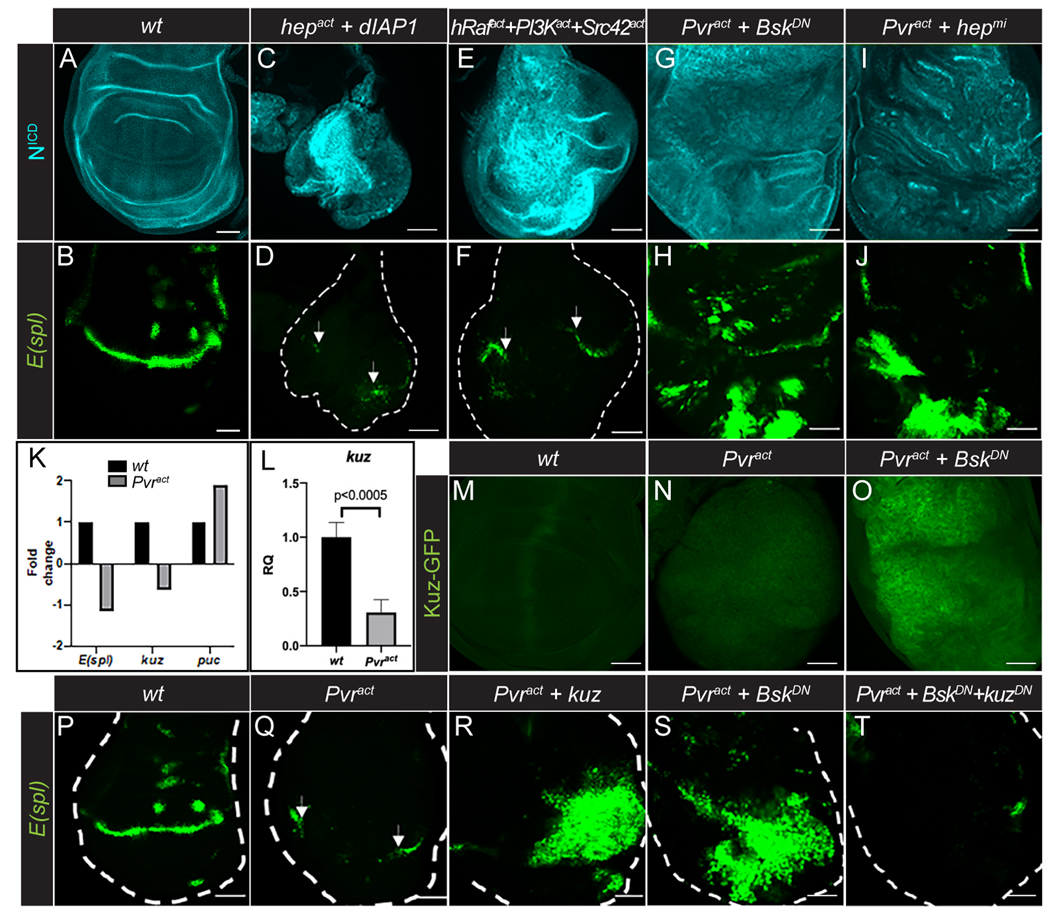
ERK, PI3K and JNK signaling pathways function downstream of Pvract in the wing disc tumors (13). To investigate whether these pathways contribute to the accumulation of inactive Notch in the tumor, we separately expressed the constitutively active forms, hRafact, PI3Kact, or JNKKact (hepact) and assessed NICD accumulation and E(spl) expression. hRafact and PI3Kact expression, either individually or in combination, gave rise to tumors, but they did not cause an obvious increase in NICD staining, nor a suppression of E(spl) expression (Fig. 3A–B; fig. S3A–H). In fact, these tumors showed ectopic E(spl) reporter expression (fig. S3D,F,H). Activation of the third pathway, JNK, cannot be achieved simply by overexpressing activated hep (JNKK), since it is established in Drosophila that this genotype causes cell death unless simultaneously protected by active Ras/Raf (17, 30, 31). Active Ras/Raf inhibits the cell death-promoting protein Hid (32, 33), which itself negatively regulates the caspase inhibitor Diap1 (34). Indeed, using cleaved caspase 3 staining, we found that Pvract tumors did not harbor high cell death, but they did so when combined with RasRNAi (fig. S3I–J). This increased cleaved caspase 3 staining was suppressed by inactivating JNK (BskDN) (fig. S3K). Therefore, we used the genetic combination hepact+Diap1 and this led to the formation of tumors with an accumulation of NICD and loss of E(spl) expression (Fig. 3C–D). This observation indicated that cells survive in this genetic background and linked inactive Notch accumulation to the JNK pathway. In this system, JNK can be activated by the Src kinase (13). Accordingly, we also found that the triple combination Srcact+PI3Kact+EGFRact (unlike the double combination PI3Kact+EGFRact) phenocopied Pvract in its accumulation of NICD and caused a loss of E(spl) reporter expression in a relatively large tumor (Fig. 3E,F). Thus, the activated JNK pathway was responsible for the accumulation of inactive Notch in the tumor.
Fig. 3. JNK, but not PI3K or Raf, controls Notch activity through negative regulation of Kuzbanian.
(A-J) NICD (cyan; A,C,E,G,I) and E(spl)m8-GFP reporter (green; B,D,F,H,J) expression in wild-type wing discs (A-B), wing discs with constitutive JNK activation (dpp-GAL4>UAS-hepact,UAS-dIAP1; C-D), constitutive ERK, PI3K and JNK activation (dpp-GAL4>UAS-hRafact,UAS-PI3Kact,UAS-Src42act; E-F), and combined Pvr activation and inhibition of JNK (dpp-GAL4>UAS-Pvract,UAS-BskDN; G-H, and dpp-GAL4>UAS-Pvract,UAS-hepmi; I-J) along the A/P axis. Arrows in D and F point to E(spl)m8-GFP expression outside of the dpp-expressing region. Note that E(Spl)GFP appears distorted and patchy due to extensive growth of the pouch in B, H and J.
(K) E(spl), kuz and puc transcript levels from RNA-Seq analysis of wild-type (black bars) and Pvract (grey bars) wing discs.
(L) Quantitative real-time PCR analysis of kuz levels in wild-type (black bar) and Pvract (grey bar) wing discs. Data represents mean ± SD (N=3).
(M-O) Kuz-GFP expression in wt (M), Pvract (N) and Pvract+BskDN (O) wing discs.
(P-T) E(spl)m8-GFP reporter expression in wt (P) and Pvract (Q) wing discs, wing discs with combined Pvr activation and overexpression of kuz (dpp-GAL4>UAS-Pvract,UAS-kuz; R), Pvract+BskDN (S) and Pvr activation combined with JNK and Kuz inhibition (dpp-GAL4>UAS-Pvract,UAS-BskDN,UAS-kuzDN; T). Arrows in Q point to E(spl)m8-GFP expression outside of the dpp-expressing region.
Each staining was repeated 3 times (N=3) and between 5 and 15 samples were analyzed per repeat. Scale bars: 50μm.
As loss of function counterparts to the above observations, a dominant negative form of JNK (bskDN) or a knockdown of JNKK (hepmicroRNA) in the context of the Pvract tumor reduced NICD accumulation and restored E(spl) reporter expression, while maintaining a large tumor (Fig. 3G–J and fig. S3L). These data confirmed that JNK is the principal regulator of Notch activity in Prvact tumors.
Results from immunostaining analysis of the whole mount tissue were fully consistent with those from an RNA-Seq analysis of the wing discs with or without tumors (13). The RNA level of E(spl) was reduced and that of the JNK pathway transcriptional target, puckered (puc), was significantly enhanced in Pvract tumors (Fig. 3K). The RNA-Seq data also revealed that expression of kuzbanian (kuz) (35, 36), the closest homolog of ADAM17 (37), which is implicated in the extracellular cleavage of NECD from the rest of the protein at the S2 site, was also significantly reduced in the tumor background (Fig. 3K). This result was confirmed by a qPCR analysis of the mutant tissue, which showed a significant decrease in kuz transcript levels (Fig. 3L). In addition, while a GFP-tagged Kuz protein (38) was undetectable in both wild-type and wing discs expressing Pvract (Fig. 3M–N), inactivation of JNK in Pvract+BskDN wing disc tumors caused high expression of Kuz-GFP (Fig. 3O), implying that JNK activity negatively regulates Kuz expression.
Next, we observed that E(spl) reporter expression was restored when the normal Kuz protein was co-expressed in the Pvract tumor cells (Fig. 3P–R). Furthermore, co-expression of BskDN and Pvract, which increased Kuz-GFP expression (Fig. 3O), also restored E(spl) reporter expression (Fig. 3S). This rescue of E(spl) by BskDN was blocked if a dominant negative Kuz protein was expressed in the same cells (Fig. 3T), providing further support that JNK inhibition restored kuz expression and thereby rescued Notch signaling. Altogether, our data indicated that activation of the JNK pathway downstream of Pvract is responsible for the accumulation of inactive Notch in the tumor, through negative regulation of the Notch S2 cleavage protease Kuzbanian.
JNK inhibits Kuzbanian expression and Notch activity in Drosophila blood cells
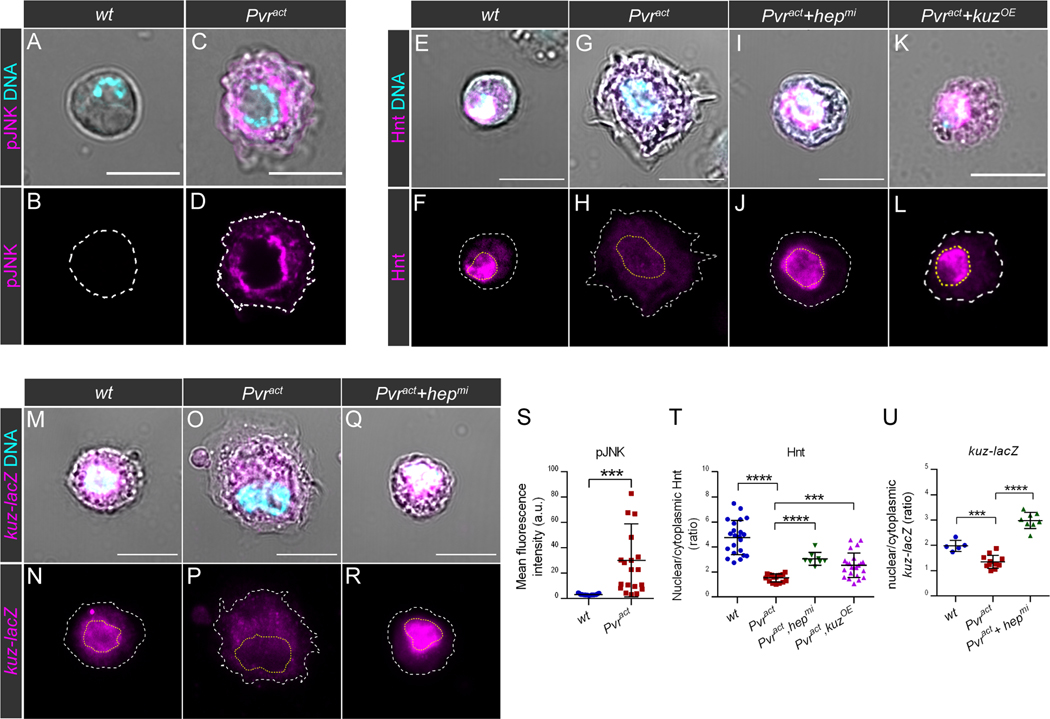
Our goal was to determine if human prostate cancer tissue showed similarities to the above model, but first, we asked if our observations could be extended to a second Drosophila tissue type of a different origin. The mesoderm-derived Drosophila blood cells, in particular the crystal cells (CCs) with platelet/granulocyte-like functions (39), are specified by a Runx protein, Lozenge, only in the presence of an active Notch signal (39). We found that when Pvract was expressed in these cells using a lozenge-GAL4 driver, the smooth CC plasma membrane structure exhibited morphological characteristics of invasive macrophages (called plasmatocytes in Drosophila), with membrane protrusions that resemble filopodial structures (Fig. 4A–D). We assessed the phosphorylation of JNK by immunostaining in these cells and found that pJNK was significantly increased in Pvract CCs compared to wild-type (Fig. 4A–D,S), indicating that the pathway was activated upon Pvract expression in blood cells, as was the case for the imaginal disc tumors.
Fig. 4. JNK inhibits Notch activity in Drosophila blood cells through negative control of Kuzbanian expression.
(A-R) Immunostaining of Drosophila circulating larval crystal cells (CCs) in which transgenes are expressed under the control of the lz-GAL4 driver. In all images, DNA (cyan) was labeled with DAPI and was used to outline the nucleus contours (yellow dashed lines). DIC images were used to outline the cell contours (white dashed lines). Scale bars: 10μm.
(A-D) Activity of the JNK pathway assessed by immunostaining of pJNK (magenta) in CCs of the following genotypes: lz-GAL4>UAS-GFP (wt; A-B) and lz-GAL4>UAS-GFP,UAS-pvract (Pvract; C-D). Images show representative examples from N=6 biological replicates per genotype.
(E-L) Immunostaining of Hnt (magenta) in CCs of the following genotypes: lz-GAL4>UAS-GFP (wt; E-F), lz-GAL4>UAS-GFP,UAS-pvract,UAS-mCherryRNAi (Pvract; G-H), lz-GAL4>UAS-GFP,UAS-pvract,UAS-hepmi (Pvract+hepmi; I-J) and lz-GAL4>UAS-GFP,UAS-pvract,UAS-kuz (Pvract+kuzOE; K-L). Images show representative examples from N=6 biological replicates per genotype.
(M-R) Immunostaining of β-galactosidase, as a readout for kuz-lacZ expression (magenta) in CCs of the following genotypes: lz-GAL4>UAS-GFP,kuz-lacZ (wt; M-N), lz-GAL4>UAS-GFP,UAS-pvract,kuz-lacZ (Pvract; O,P) and lz-GAL4>UAS-GFP,UAS-pvract,UAS-hepmi,kuz-lacZ (Pvract+hepmi; Q-R). Images show representative examples from N=4 biological replicates per genotype.
(S-U) Quantitative analysis of data represented in (A-R). Data points in scatter plots correspond to individual cells and lines indicate the mean and standard deviation. Unpaired t-tests were used to evaluate statistical significance, denoted as *** for p ≤ 0.001 and **** for p ≤ 0.0001.
(S) Quantification of the mean fluorescence intensity associated with pJNK immunostaining in wt CCs and Pvract CCs.
(T) Quantification of nuclear Hnt, shown as the ratio of the mean fluorescence intensity associated with Hnt in the nucleus over the mean fluorescence intensity associated with Hnt in the cytoplasm in wt, Pvract (N=16), Pvract+hepmi and Pvract+kuzOE CCs.
(U) Quantification of nuclear kuz-lacZ expression, shown as the ratio of the mean fluorescence intensity associated with β-galactosidase in the nucleus over the mean fluorescence intensity associated with β-galactosidase in the cytoplasm in wt, Pvract and Pvract+hepmi CCs.
Known direct Notch transcriptional targets in CCs include Hindsight (Hnt; human RREB1 (40)) and Cut (human Cux (41)). These proteins localized to nuclei in wild-type CCs (Fig. 4E–F and fig. S4A–B), but not in Pvract CCs (Fig. 4G–H,T and fig. S4C–D). These data are reminiscent of the loss of E(spl) reporter, Cut and Wg expression in wing disc tumors and suggest that Notch is also inactivated in Pvract CCs. To determine if loss of Notch target gene expression in CCs was also dependent on JNK activity, we assessed Hnt and Cut expression in CCs co-expressing Pvract and hepmi. This resulted in a suppression of ectopic cytoplasmic projections as well as a restoration of Hnt and Cut expression in the nucleus (Fig. 4I–J,T and fig. S4E–F). Therefore, JNK regulates Notch target gene expression in Pvract CCs. In epithelial tumors, our data indicated that this is through a negative regulation of Kuzbanian expression (Fig. 3K–T). Accordingly, forced expression of Kuz in Pvract CCs restored the expression of both Notch targets Hnt (Fig. 4K–L,T) and Cut (fig. S4G–L).
kuz-lacZ, a reporter for kuz expression, can be used to monitor kuz expression levels. We found that compared with the control (Fig. 4 M–N), kuz expression was significantly decreased in Pvract (Fig. 4O–P,U). Co-expression of hepmi with Pvract increased kuz-lacZ expression above that seen in control cells (Fig. 4Q–R,U), indicating that JNK also regulates Kuzbanian expression in CCs. Thus, JNK inhibits Notch pathway activity through the same mechanism in two distinct Drosophila cell types upon expression of the Pvract oncogene.
Low Notch activity correlates with high JNK activity in human prostate cancer tissues
To probe for possible consistency with tumor-derived tissue, we performed immuno-histochemical (IHC) staining on Formalin Fixed Paraffin Embedded (FFPE) sections from human prostate adenocarcinoma tissues obtained from the UCLA prostate cancer repository. Using appropriate antibodies, we found that Notch1ECD was faintly, if at all, detected in benign prostate tissue (Fig. 5A–B). By contrast, we observed an accumulation of Notch1ECD in prostate cancer tissues (Fig. 5C–E). The accumulation inside the cells suggests that this corresponds to uncleaved, and therefore inactive, Notch 1 receptor that saturates the cell surface and is detected in cytoplasmic vesicles (42, 43). Indeed, whereas the Notch1 target protein HES-7 (E(spl)m8 in Drosophila) was robustly expressed in the nuclei of benign prostate cells (Fig. 5F–G), its expression was strongly attenuated in the nuclei of prostate cancer cells (Fig. 5H–J). In agreement, an analysis based on the publicly available GEPIA tool (44), for curated RNAseq data obtained from a large collection of prostate adenocarcinomas, indicated that HES-7, in particular, and other Notch targets such as HES-1, HES-5 and HEY-1 to a lesser extent, are negatively regulated in prostate cancer tissue (44). Therefore, as in Drosophila wing disc and crystal cells, Notch signaling was found to be active in the normal prostate tissue, but became inhibited as Notch accumulated in an inactive form in advanced prostate cancer. We also observed an increase in pJNK-associated staining in advanced prostate cancer tissue when compared with benign tissue (Fig. 5K–O). Thus, the JNK pathway is active in tumors of patients with advanced prostate cancer but not in the benign tissue. Finally, qualitative analysis of LDH-A staining in benign prostate tissue and advanced prostate cancer biopsy samples suggested that these tumors are glycolytic (fig. S5). These observations correlated with those seen in Drosophila, leading to the conclusion that activation of JNK in advanced prostate cancer leads to inactivation of the Notch pathway through similar mechanisms in an evolutionarily conserved manner.
Fig. 5. Low Notch activity correlates with high JNK activity in human prostate cancer tissues.
(A-E) Immunohistochemical staining of Notch 1 with an antibody directed against the extracellular domain (NECD; brown staining) in paraffin sections of benign (A-B) and advanced prostate cancer (C-D) tissues, with quantification of the mean relative intensity associated with the staining (E), represented as the fold change between the average signal in benign prostate tissue (N=3, blue) and advanced prostate tumors (N=3, red).
(F-J) Immunohistochemical staining of HES7 (brown staining) in paraffin sections of benign (F-G) and advanced prostate cancer (H-I) tissue, with quantification of the mean relative intensity associated with the staining (J), represented as the fold change between the average signal in benign prostate tissue (N=3, blue) and advanced prostate tumors (N=3, red).
(K-O) Immunohistochemical staining of phosphorylated JNK (pJNK; brown staining) in paraffin sections of benign (K-L) and advanced prostate cancer (M-N) tissue, with quantification of the mean relative intensity associated with the staining (O), represented as the fold change between the average signal in benign prostate tissue (N=3, blue) and advanced prostate tumors (N=3, red).
Images in B, D, G, I, L and N are higher magnification of the regions framed with dashed lines in the corresponding A, C, F, H, K and M images. Scale bars: 100μm. Hematoxylin staining, enriched in nuclei, is in purple/blue.
Bar graphs in E, J and O represent the mean value and the standard deviation, based on a benign sample (blue) compared with three independent advanced tumor samples (red).
Discussion
Notch and JNK are two signaling pathways that play important roles in cancer and both have been alternately described as oncogenic or tumor suppressive in a context-dependent manner (8, 9, 18). As therapeutic strategies aimed at modulating the activity of these pathways are being developed (19, 45), it is crucial to be able to predict how they function in a particular tumor, in order to help make better informed treatment decisions.
In the present study, we took advantage of a well characterized Drosophila tumor model to uncover a mechanism by which activation of the JNK pathway can lead to inhibition of Notch signaling. In the imaginal wing disc epithelium, expression of a single oncogene, Pvract, causes the formation of large tumors in which the JNK pathway is activated. We demonstrated that, although the Notch protein accumulates in these tumors, it is largely inactive, as indicated by the lack of expression of Notch target genes. Using a genetic approach, we showed that decreasing JNK activity was sufficient to restore Notch activity in the tumor. We found that in this system, JNK inhibits the expression of the Notch S2 cleavage protease Kuzbanian, thereby preventing Notch activation. We showed that the same occurs in a population of Drosophila blood cells expressing the Pvract oncogene.
In parallel, we found a similar correlation between activation of JNK and accumulation of inactive Notch in prostate cancer cell lines and in prostate tissue samples derived from patients with advanced prostate cancer, suggesting that this mechanism is conserved through evolution. Accordingly, we found by Western-blot analysis that PC3 cells, derived from advanced prostate cancer, exhibit a defect in Notch cleavage at the S2 site and work by others has shown that, at a transcript level, PC3 cells express decreased amounts of the S2 protease ADAM17 (46).
The functional observations in the two different tissue types in Drosophila and the expression patterns in human prostate cancer cell lines and primary tumor samples, when taken together, support the model shown in Figure 6 (Fig. 6A–B). The model suggests that if cells with developmentally active Notch signaling, such as the luminal cells of the normal prostate, acquire an activating mutation in JNK or an upstream kinase such as Src, this would cause loss of a Notch S2 cleavage enzyme and result in accumulation of inactive, full-length Notch, which is unable to support the expression of its downstream targets. This model provides one mechanism through which Notch can act as a tumor suppressor. Many Notch target proteins are necessary to maintain tissue integrity and epithelial morphology (47). For example, some target proteins, such as HES and RREB, are essential in maintaining normal growth, whereas others, such as Cux, enable differentiation (40, 41). The combined results from this comparative analysis, in which functional relevance is established using genetic dissection in Drosophila and correlation to a human cancer, illustrates how context dependence can determine alternative ways to achieve an oncogenic vs a tumor suppressive mode (48–51). The most parsimonious interpretation of the model is that Notch can have a tumor suppressive role in prostate cancers that have activated JNK. These findings suggest that Src-JNK activation in a tumor can be predictive of the activity status of Notch.
Fig. 6. Model for control of Notch function by activated JNK in tumors.
Proposed model based on the observations made in Drosophila tumors, in human prostate cell lines and in patient-derived tissue samples:
(A) In normal/benign tissue in which the Notch pathway is homeostatically active, sequential proteolytic cleavages of the Notch receptor activate downstream target proteins that are important for controlled development. The stress-responsive JNK pathway is usually inactive.
(B) In advanced tumors, activating mutations in Src/JNK (or that of a kinase that activates Src/JNK) inhibit proteolytic cleavages of the Notch receptor, by negatively regulating the expression of the S2 cleavage protease (Kuzbanian in Drosophila), resulting in a loss of Notch target effector function and an accumulation of the full-length receptor.
Therefore, therapeutic strategies for advanced prostate cancer might benefit from identifying molecules that block JNK or its upstream components and those that facilitate controlled activation of Notch, rather than drugs that inhibit Notch function, as these are expected to work only for contexts where Notch functions as an oncogene. On the other hand, JNK activation could simultaneously have additional tumor suppressive effects, for example by inducing apoptosis, and therefore inhibiting all JNK activity may not always be beneficial for treatment. Further investigation is needed to elucidate the mechanism by which JNK regulates Notch1 proteolytic cleavage and activity in prostate cancer, as this could contribute to the refinement of drug design strategies to target downstream effectors and potentially maintain normal JNK functions.
Materials and Methods
Cell lines
Cell lines used in this study were acquired from the American Type Culture Collection (ATCC) and are the following: RWPE-1 (non-transformed human prostate epithelial cell line, ATCC CRL-11609), LNCaP (androgen receptor-positive prostate cancer cell line, ATCC CRL-1740), PC3 (androgen receptor-negative prostate cancer cell line, ATCC CRL-1435). Cells were cultured according to ATCC’s protocols. The cell identities were validated by a short tandem repeat-based authentication service (Laragen).
Immunoblotting of prostate cancer cell line samples
Protein lysates from prostate cancer cell lines were prepared by lysing the cells with RIPA buffer (Thermo Scientific ref. 89901) supplemented with protease inhibitors (cOmplete Protease Inhibitor Cocktail, Roche ref. 11836153001) and phosphatase inhibitors (Halt Phosphatase Inhibitor Cocktail, Thermo Scientific ref. 78428). For Western blotting, 20 μg of protein were loaded into each lane. Antibodies and their dilutions used are as follows: rabbit anti-Notch1ECD (1:1000; Millipore Sigma #07–218), mouse anti-pJNK (1:500; G-7, Santa Cruz Biotechnology #sc-6254), rabbit anti-JNK (1:500, Cell Signaling Technology #9252) mouse anti-LDH-A (1:2000; 5D2.1, Millipore MABC150), rabbit anti-cleaved Notch1 (Val1744) (1:1000; D3B8, Cell Signaling Technology #4147), rabbit anti-Notch1 (1:1000; D1E11, Cell Signaling Technology #3608), rabbit anti-Notch2 (1:1000; Cell Signaling Technology #5732), rabbit anti-Notch3 (1:1000; Cell Signaling Technology #5276), mouse anti-Notch4 (1:1000; Cell Signaling Technology #2423), mouse anti-alpha-tubulin (1:100; Developmental Study Hybridoma Bank 12G10), mouse anti-GAPDH (1:2000; Proteintech #60004–1-Ig), HRP-conjugated goat anti-rabbit IgG (1:10000; Invitrogen #31463) and HRP-conjugated goat anti-mouse IgG (1:10000; Invitrogen #31430). Chemiluminescent signals were obtained using ECL Prime Western Blotting Detection Reagent (Amersham ref. RPN2236) and iBright FL1500 Imaging System (Invitrogen ref. A44115), and with SuperSignal West Pico PLUS (Thermo Scientific ref. 34577) and West Atto (Thermo Scientific ref. 38554) substrates. Total protein was visualized by No-Stain Protein Labeling Reagent (Invitrogen ref. A44717). Quantification of the Western blots was performed using the ImageJ image analysis software.
Fly stocks and genetics
The Drosophila stocks used in this study are the following: w1118, dpp-GAL4blk−1 (BDSC#93385), lozenge-GAL4 (BDSC#6314) UAS-Pvract (λ-Pvr) (Norbert Perrimon), E(spl)m8-GFP (James W. Posakony), UAS-cRaf1gof (hRafact, BDSC#2074), UAS-dp110CAAX (PI3Kact, BDSC#25908), UAS-hepact (BDSC#9306), UAS-dIAP1 (BDSC#6657), UAS-src42act (BDSC#6410), UAS-BskDN (BDSC #9311), UAS-hepmicroRNA (hepmi, Guang-Chao Chen), kuz-GFP (François Schweisguth), UAS-kuz (BDSC#5816), UAS-kuzDN (BDSC#6578), kuz-lacZ (BDSC#10505), UAS-Nact (Yuh-Nung Jan), UAS-EGFRact (Trudi Schupbach). All fly stocks were raised on standard cornmeal medium at 25°C. Cell-type specific induction of transgene expression was performed using the binary GAL4/UAS system (52).
Immunostaining of Drosophila tissues
For immunofluorescent staining of Drosophila imaginal wing discs, wandering 3rd instar larvae were dissected in PBS, fixed for 30min in 4% paraformaldehyde, washed and permeabilized in 0.3% PBT (1x PBS, 0.1% Triton X-100). Tissues were incubated in 3% Bovine Serum Albumin (BSA)-0.1% PBT blocking solution for at least 30 minutes. Following overnight incubation with primary antibodies at 4°C, samples were washed with PBT three times for 15 minutes, incubated with secondary antibodies (Invitrogen, 1:500 in blocking solution) for 2h at room temperature, washed with PBT three times for 15 minutes and mounted in Vectashield® with DAPI (H-1200, Vector laboratories).
For staining of circulating hemocytes, wandering 3rd instar larvae were bled in PBS on 8-well glass slide in a humid chamber (one larva per well). Hemocytes were allowed to settle for 30min, before replacing the PBS with a drop of fixative solution (6% paraformaldehyde in PBS). After 30min fixation, hemocytes were washed and permeabilized in 0.3% PBT, incubated with blocking solution (3% BSA in PBT) for at least 30min and with primary antibodies overnight at 4°C. After washes in 0.3% PBT, hemocytes were incubated with secondary antibodies for 2h at room temperature, washed in PBT, stained with DAPI (1:1000 in PBS) for 10min and washed in PBS. Finally, PBS was replaced by a drop of Vectashield Vibrance® hardening antifade mounting medium (H-1700).
Primary antibodies were diluted in blocking solution as follow: mouse anti-NotchECD (1:50; C458.2H, DSHB), mouse anti-NotchICD (1:50; C17.9C6, DSHB), rabbit anti-pJNK (Thr183/Tyr185) (1:500; Cell Signaling #9251), mouse anti-Hnt (1:200; 1G9, DSHB), mouse anti-Cut (1:100; 2B10, DSHB) and mouse anti-Wg (1:5; 4D4, DSHB). Alexa-conjugated secondary antibodies (Invitrogen) were diluted 1:500 in blocking solution.
Quantitative Real-Time PCR Analysis
Total RNA was extracted from ~40 3rd instar imaginal wing discs using PureLink RNA mini kit (Ambion). The SuperScript III First-Strand synthesis SuperMix kit (Invitrogen) was used for first-strand cDNA synthesis. Relative quantitative PCR was performed by comparative CT method using Power SYBR Green PCR master mix kit (Applied Biosystems) and a StepOne Real-Time PCR detection thermal cycler (Applied Biosystems) using RT2 qPCR Primer Assay (SA Biosciences, QIAGEN), with primers specific for genes of interest and RpL10 (PPD10569B). The levels of RpL10 were used to normalize total cDNA input.
Prostate tissue samples
Formalin Fixed Paraffin-embedded tissue sections from localized prostate adenocarcinoma and adjacent benign prostate tissue were provided by the Translational Pathology Core Laboratory (TPCL) at UCLA. All samples were collected from patients with informed consent, and all related procedures were performed with the approval of the internal review and ethics boards.
Immunohistochemistry
For immunohistochemistry on human prostate tissue samples, paraffin-embedded tissue sections were incubated in a 60°C oven for 1hr and de-paraffinized in three changes of xylene, followed by 100% alcohol twice for 3 min each. Then the slides were transferred once in 95% and 70% alcohol, each for 3 min. A heat antigen retrieval step was used to unmask the antigenic epitopes. The remaining procedure was performed using HRP-DAB Cell and Tissue Staining Kits (CTS002 and CTS005, R&D Systems) according to the manufacturer’s protocol. Primary antibodies used for staining are: rabbit anti-NotchECD (1:500; Millipore Sigma #07–218), rabbit anti-pJNK (Thr183/Tyr185) (1: 500; Cell Signaling #9251), rabbit anti-HES7 (1:100; Invitrogen #PA5–73023) and mouse anti-LDH-A (1:1000; 5D2.1, Millipore MABC150). Biotinylated anti-mouse (CTS002, R&D Systems) and anti-rabbit (CTS005, R&D Systems) secondary antibodies are parts of the HRP-DAB Cell and Tissue staining kits.
Microscopy and image analysis
Fluorescent images were acquired with a Zeiss LSM 880 confocal microscope and ZEN 2018 acquisition software. Images were analyzed with the ImageJ software. All images shown are representative images of at least three independent experiments. Images of immunohistochemistry experiments were acquired with a fluorescence compound microscope with bright field imaging capability.
To quantify the ratio of the dpp positive area over the whole disc area in Drosophila imaginal discs, a reporter for dpp expression (UAS-GFP or UAS-mCherry) was used to delimitate the area of the dpp positive area, and nuclear staining with DAPI was used to delimitate the disc area. Statistical significance was evaluated using the software Graphpad Prism v6.0, after verifying the normality of values and equivalence of variances. Graphics display means with standard deviations and the statistical differences between control and test samples were assessed using unpaired t-tests.
To measure the staining intensity in IHC images, the Colour deconvolution function in the Fiji image analysis software was used. This function was created based on the method by Ruifrok and Johnston (53) to determine the relative contribution of each of the RGB color channels to the DAB staining. For NECD, pJNK and LDH-A stainings, the mean intensity in the deconvolved DAB images was measured in 40 to 60 square regions of identical size spanning the tissue sections in areas containing cells (avoiding regions devoid of tissue). For HES7, nuclei were segmented using hematoxylin staining and the mean intensity per nucleus was measured on the deconvolved DAB images. The integrated optical density (OD), referred to as “relative intensity”, was calculated as OD=log (maximum intensity/measured intensity), where the maximum intensity was determined for each image as the average mean intensity in lumen areas (devoid of cells). For each staining, three benign prostate tissue sample and three advanced prostate tumor samples (from different patients) were analyzed and the average relative intensity per sample was calculated. For each tumor sample, the relative intensity fold change compared to the average of the relative intensity of the three benign tissue samples, was calculated and plotted in Figure 5. In instances where the tissue sample contained both normal tubular tissue and prostate cancer tissue with disrupted tubules, the quantitative measures were applied to the morphologically affected parts.
Supplementary Material
Acknowledgments
We thank members of the Banerjee laboratory for support, intellectual input, and technical expertise. We thank UCLA’s Translational Pathology Core Laboratory. We thank Arunima Purkayastha (University of California, Los Angeles) for generating RNA seq data. We thank James Posakony (University of California, San Diego), Norbert Perrimon (Harvard University), Guang-Chao Chen (Institute of Biological Chemistry, Academia Sinica), François Schweisguth (Institut Pasteur), Yuh-Nung Jan (University of California, San Francisco) and Trudi Schüpbach (Princeton University) for kindly providing fly stocks, as well as the Vienna (VDRC) and Bloomington (BDSC) Drosophila stock centers.
Funding
MC was supported by a UCLA Broad Stem Cell Research Center training grant. LMG was supported by a Ruth L Kirschstein Institutional NRSA T32CA009056 and by NHLBI 3R01HL067395–16S1. We are grateful for the support provided to UB for most of this work by NIH R01 CA-217608 and partly by NIH R01 HL-067395.
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Data Materials Availability
All data needed to evaluate the conclusions are present in the paper or the Supplementary Materials.
References
- 1.Kidd S, Lockett TJ, Young MW, The Notch locus of Drosophila melanogaster. Cell. 34, 421–433 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Knust E, Campos-Ortega JA, The molecular genetics of early neurogenesis in Drosophila melanogaster. BioEssays. 11 (1989), pp. 95–100. [DOI] [PubMed] [Google Scholar]
- 3.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S, Nucleotide sequence from the neurogenic locus Notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 43, 567–581 (1985). [DOI] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Rand MD, Lake RJ, Notch signaling: Cell fate control and signal integration in development. Science (80-. ). 284 (1999), pp. 770–776. [DOI] [PubMed] [Google Scholar]
- 5.Bray S, Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Siebel C, Lendahl U, Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235–1294 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Zhou B, Lin W, Long Y, Yang Y, Zhang H, Wu K, Chu Q, Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 7, 1–33 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radtke F, Raj K, The role of Notch in tumorigenesis: Oncogene or tumour suppressor. Nat. Rev. Cancer. 3, 756–767 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Aster JC, Pear WS, Blacklow SC, The Varied Roles of Notch in Cancer. Annu. Rev. Pathol. Mech. Dis. 12 (2017), pp. 245–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng G, Ma L, Meng Q, Ju X, Jiang K, Jiang P, Yu Z, Notch signaling in the prostate: critical roles during development and in the hallmarks of prostate cancer biology. J. Cancer Res. Clin. Oncol. 142 (2016), pp. 531–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho FLF, Simons BW, Eberhart CG, Berman DM, Notch signaling in prostate cancer: A moving target. Prostate. 74, 933–945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nowell CS, Radtke F, Notch as a tumour suppressor. Nat. Rev. Cancer. 17 (2017), pp. 145–159. [DOI] [PubMed] [Google Scholar]
- 13.Wang CW, Purkayastha A, Jones KT, Thaker SK, Banerjee U, In vivo genetic dissection of tumor growth and the Warburg effect. Elife. 5 (2016), doi: 10.7554/eLife.18126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoxhaj G, Manning BD, The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 20, 74–88 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore AR, Rosenberg SC, McCormick F, Malek S, RAS-targeted therapies: is the undruggable drugged? Nat. Rev. Drug Discov. 19, 533–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner EF, Nebreda ÁR, Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 9 (2009), pp. 537–549. [DOI] [PubMed] [Google Scholar]
- 17.La Marca JE, Richardson HE, Two-Faced: Roles of JNK Signalling During Tumourigenesis in the Drosophila Model. Front. Cell Dev. Biol. 8 (2020), p. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tournier C, The 2 Faces of JNK Signaling in Cancer. Genes and Cancer. 4 (2013), pp. 397–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu R, Hu J, The role of JNK in prostate cancer progression and therapeutic strategies. Biomed. Pharmacother. 121 (2020), p. 109679. [DOI] [PubMed] [Google Scholar]
- 20.Sobel RE, Sadar MD, Cell lines used in prostate cancer research: A compendium of old and new lines - Part 1. J. Urol. 173 (2005), pp. 342–359. [DOI] [PubMed] [Google Scholar]
- 21.Yang YM, Bost F, Charbono W, Dean N, McKay R, Rhim JS, Depatie C, Mercola D, C-Jun NH2-terminal kinase mediates proliferation and tumor growth of human prostate carcinoma. Clin. Cancer Res. 9, 391–401 (2003). [PubMed] [Google Scholar]
- 22.Hübner A, Mulholland DJ, Standen CL, Karasarides M, Cavanagh-Kyros J, Barrett T, Chi H, Greiner DL, Tournier C, Sawyers CL, Flavell RA, Wu H, Davis RJ, JNK and PTEN cooperatively control the development of invasive adenocarcinoma of the prostate. Proc. Natl. Acad. Sci. U. S. A. 109, 12046–12051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parra E, Ferreira J, Modulation of the response of prostate cancer cell lines to cisplatin treatment using small interfering RNA. Oncol. Rep. 30, 1936–1942 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Davis RJ, Signal transduction by the JNK group of MAP kinases. Cell. 103 (2000), pp. 239–252. [DOI] [PubMed] [Google Scholar]
- 25.Xian ZY, Liu JM, Chen QK, Chen HZ, Ye CJ, Xue J, Yang HQ, Li JL, Liu XF, Kuang SJ, Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumor Biol. 36, 8093–8100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessho Y, Miyoshi G, Sakata R, Kageyama R, Hes7: A bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes to Cells. 6, 175–185 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Ligoxygakis P, Bray SJ, Apidianakis Y, Delidakis C, Ectopic expression of individual E(spl) genes has differential effects on different cell fate decisions and underscores the biphasic requirement for Notch activity in wing margin establishment in Drosophila. Development. 126, 2205–2214 (1999). [DOI] [PubMed] [Google Scholar]
- 28.De Celis JF, Tyler DM, De Celis J, Bray SJ, Notch signalling mediates segmentation of the Drosophila leg. Development. 125, 4617–4626 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Trylinski M, Mazouni K, Schweisguth F, Intra-lineage Fate Decisions Involve Activation of Notch Receptors Basal to the Midbody in Drosophila Sensory Organ Precursor Cells. Curr. Biol. 27, 2239–2247.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Morata G, Shlevkov E, Pérez-Garijo A, Mitogenic signaling from apoptotic cells in Drosophila. Dev. Growth Differ. 53, 168–176 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Shlevkov E, Morata G, A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death Differ. 19, 451–460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergmann A, Agapite J, McCall K, Steller H, The Drosophila gene hid is a direct molecular target of ras-dependent survival signaling. Cell. 95, 331–341 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Kurada P, White K, Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 95, 319–329 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Wang SL, Hawkins CJ, Yoo SJ, Müller HAJ, Hay BA, The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 98, 453–463 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Pan D, Rubin GM, Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 90, 271–280 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Lieber T, Kidd S, Young MW, Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16, 209–221 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bozkulak EC, Weinmaster G, Selective Use of ADAM10 and ADAM17 in Activation of Notch1 Signaling. Mol. Cell. Biol. 29, 5679–5695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, Schweisguth F, Rubinstein E, Tspanc8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol. 199, 481–496 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee U, Girard JR, Goins LM, Spratford CM, Drosophila as a genetic model for hematopoiesis. Genetics. 211, 367–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baechler BL, McKnight C, Pruchnicki PC, Biro NA, Reed BH, Hindsight/RREB-1 functions in both the specification and differentiation of stem cells in the adult midgut of Drosophila. Biol. Open. 5, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cubelos B, Sebastián-Serrano A, Kim S, Redondo JM, Walsh C, Nieto M, Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Dev. Neurobiol. 68, 917–925 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortini ME, Bilder D, Endocytic regulation of Notch signaling. Curr. Opin. Genet. Dev. 19, 323–328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinbuck MP, Winandy S, A review of Notch processing with new insights into ligand-independent notch signaling in T-cells. Front. Immunol. 9, 1230 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z, GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen F, Maillard I, Therapeutic Targeting of Notch Signaling: From Cancer to Inflammatory Disorders. Front. Cell Dev. Biol. 9, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCulloch DR, Harvey M, Herington AC, The expression of the ADAMs proteases in prostate cancer cell lines and their regulation by dihydrotestosterone. Mol. Cell. Endocrinol. 167, 11–21 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Li Y, Kong D, Sarkar FH, The Role of Notch Signaling Pathway in Epithelial-Mesenchymal Transition (EMT) During Development and Tumor Aggressiveness. Curr. Drug Targets. 11, 745–751 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobry C, Oh P, Mansour MR, Thomas Look A, Aifantis I, Notch signaling: Switching an oncogene to a tumor suppressor. Blood. 123 (2014), pp. 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lobry C, Oh P, Aifantis I, Oncogenic and tumor suppressor functions of Notch in cancer: It’s NOTCH what you think. J. Exp. Med. 208 (2011), pp. 1931–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Sha J, Yang G, Huang X, Bo J, Huang Y, Activation of Notch pathway is linked with epithelial-mesenchymal transition in prostate cancer cells. Cell Cycle. 16, 999–1007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marignol L, Rivera-Figueroa K, Lynch T, Hollywood D, Hypoxia, notch signalling, and prostate cancer. Nat. Rev. Urol. 10 (2013), pp. 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brand AH, Perrimon N, Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 118, 401–15 (1993). [DOI] [PubMed] [Google Scholar]
- 53.Ruifrok AC, Johnston DA, Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23, 291–299 (2001). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.