Abstract
The EUO gene (for early upstream open reading frame) of Chlamydia psittaci was previously found to be transcribed better at 1 than at 24 h postinfection. We found that the EUO gene encodes a minor protein that is expressed within 1 h of infection of host cells with C. psittaci 6BC but that protein quantity peaks during the logarithmic growth phase of reticulate bodies (RBs), declines late in the infection (after 20 h) when RBs reorganize into elementary bodies (EBs), and is absent in infectious EBs. EUO protein lacks homology to known proteins but does contain a putative helix-turn-helix motif. We found that recombinant EUO binds to DNA in vitro with a relatively broad specificity. Using the bp −200 to +67 promoter region of the cysteine-rich envelope protein (crp) operon as a model, we show that EUO protein preferentially binds to AT-rich sequences and protects crp DNA from DNase I from approximately bp −60 to −9. We also found that native EUO protein in extracts of RBs binds to the promoter region of the crp operon, demonstrating that the DNA binding property of EUO protein is not an artifact of recombinant methods. Although EUO protein appears to bind to the crp operon with high affinity in vitro (Kd of about 15 nM), it is not known whether the protein binds the crp DNA in vivo.
The bacterial genus Chlamydia is characterized by a distinct developmental cycle that includes the alternation between an extracellular infectious elementary body (EB) and an intracellular reticulate body (RB). Extracellular EBs are metabolically inert but commence synthesis of RNA and protein shortly after entry into host eukaryotic cells (38). The signals that trigger early gene activation and the identities of the initially activated genes are not known. Over a period of 6 to 10 h postinfection (p.i.), the small EB form reorganizes into a larger RB form, which undergoes cell division within a membrane-bound vacuole. By 18 to 20 h p.i., chlamydial development becomes asynchronous, with some RBs continuing to divide while others begin to reorganize into the EB form. The reorganization process includes a general step-down of gene expression and protein synthesis; however, some genes are activated at this late stage of the cycle, including the crp operon, which encodes two cysteine-rich envelope proteins (1, 2, 9, 15, 31), and two unlinked genes that encode histone-like proteins, Hc1 and Hc2 (18, 19, 35, 44). The cysteine-rich proteins are believed to contribute to the osmotic stability of EBs, which is absent in dividing RBs (16, 21–23, 33). The histone-like proteins are thought to be responsible for the condensation of DNA into a nucleoid present in EBs and may contribute to the down regulation of gene expression late in the developmental cycle (4, 5, 35–37). The presence of both dividing and reorganizing RBs within the same vacuole is a curiosity; it has been suggested that attachment of RBs to the vacuolar membrane permits continued procurement of host-supplied nutrients and log-phase growth whereas detachment from the membrane may trigger conversion to the EB form (20). For most strains of chlamydiae, the majority of RBs have reorganized into the EB form by 30 to 48 h p.i. and infectious EBs are released by lysis of the host cell.
Very little is known about the temporal regulation of gene expression late in the developmental cycle. The crp operon, the histone genes, and two other late-stage genes of unknown function have been shown to be dependent on the major chlamydial sigma factor (17) and thus are not regulated by a cascade of alternative sigma factors, as is the case for some other bacteria with distinct developmental morphologies, such as Bacillus subtilis (41). Even less is known about the early events that take place within the first few hours of infection. Wichlan and Hatch (45) used a radiolabeled RNA probe generated from Chlamydia psittaci 6BC isolated from host cells at 1 h p.i. to identify a clone from a chlamydial genomic library that is highly expressed early in the infection. Using RNA probes generated from host-free chlamydiae, these authors demonstrated that the gene carried by the clone was much more highly expressed at 1 than at 24 h p.i. compared to genes encoding the major outer membrane protein (MOMP) and the major sigma factor. The early clone was found to contain an open reading frame (ORF) of 182 codons and was designated EUO for early upstream ORF. Homologs of EUO have since been identified in the D and L2 serovars of Chlamydia trachomatis and in the GPIC strain of C. psittaci (8, 26, 29). Interestingly, EUO lies upstream of the crp operon, separated only by a gene homologous to bacterial glutamyl tRNA synthetases (8, 26).
Although the EUO gene appears to be transcribed, synthesis of EUO protein by chlamydiae has not yet been demonstrated, and the actual time course of EUO gene expression between 1 and 24 h p.i. is not known. The function of EUO also has not been firmly established. The predicted EUO peptide bears no convincing homology to known proteins; however, Kaul et al. (29) recently demonstrated that partially purified preparations of glutathione S-transferase-tagged recombinant EUO (rEUO) possess protease activity specific for the eukaryotic histone H1 and the C. trachomatis serovar L2 histone-like protein Hc1. The purpose of our study was to gain insight into the function of the EUO gene. We confirmed that EUO transcripts are most abundant early (1 to 2 h p.i.) in the developmental cycle relative to the MOMP gene transcripts and the 16S rRNA content of chlamydiae; however, transcripts can be detected until the end of the log phase of growth. We also found that the EUO ORF is translated into a protein in chlamydiae as early as 1 h p.i. However, EUO protein is most abundant, relative to MOMP, during the log phase of growth, declines at later times p.i. and is completely absent in EBs. We show that recombinant and native EUO bind to DNA in vitro, and we have characterized the binding characteristics of EUO, using the crp promoter region as a model.
MATERIALS AND METHODS
Preparation of rEUO.
For preparation of His tag rEUO, the entire EUO gene was amplified by PCR from pDGW-E47 (45) template DNA under conditions described by Fahr et al. (17); the forward primer was HE1 (5′ GCAGGAATTCATGGAATGCATACAACATG 3′ [the EcoRI site is underlined]), and the reverse primer (E2) was 5′ GCAGGAATTCGAAGACAAATTATGAACTAGTT 3′. The PCR fragment was cloned in frame into the EcoRI site of the pET-28a (+) vector (Novagen, Madison, Wis.) to obtain pLZ202F1, which was transformed into the nonexpressing Escherichia coli host XL1-Blue (Stratagene, La Jolla, Calif.) and confirmed to encode EUO by DNA sequencing. His tag rEUO was overexpressed as an insoluble inclusion body in E. coli BL21(DE3) (pLysE) (42), dissolved in 6 M urea, purified by nickel-agarose affinity chromatography following the instructions supplied by Novagen, and maintained as a stock solution in 0.2 M urea. The rEUO gene without a His tag was amplified by PCR amplification of pDGW-E47, using the forward primer E1 (5′ GCAGGAATTCGGGGAGGTTTGCTAACGTATGG 3′) and the reverse primer E2, and ligated into the EcoRI site of the pT7-5 vector (43) to obtain pLZ5F1. rEUO without a His tag was overexpressed in BL21(DE3) (pLysE) and fractionated by Laemmli (30) sodium dodecyl sulfate 7.5 to 15% gradient polyacrylamide gel electrophoresis (SDS–7.5 to 15% PAGE). A band containing a protein with a relative molecular mass of 27 kDa was excised, and the protein was eluted, concentrated, and renatured from 6 M guanidinium-HCl as previously described (12). The concentrations of the purified rEUO protein preparations were estimated by Coomassie brilliant blue staining of SDS-polyacrylamide gels, using bovine serum albumin as the standard.
Determination of the N-terminal amino acid sequence.
Gel-purified rEUO without a His tag was subjected to SDS-PAGE and transferred electrophoretically to Immobilon polyvinylidene difluoride membrane (Millipore Corp., Bedford, Mass.) in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11. The transferred protein band was visualized by a solution containing 0.1% Coomassie blue 250, 40% methanol, and 1% acetic acid and then destained by 50% methanol for 5 min. The band was cut out and sequenced by Edman degradation on an Applied Biosystems 475A protein sequencer.
Preparation of anti-rEUO antiserum.
A New Zealand White rabbit was prebled and immunized with approximately 500 μg of gel-purified rEUO without a His tag in 1 ml of adjuvant (RIBI Immunochem Research, Inc., Hamilton, Mont.). The rabbit was boosted at 28 days and bled at 42 days. His tag rEUO antiserum was prepared in rabbits by HTI Bio-Products (Ramona, Calif.), using 500 μg of purified rEUO. Both antisera were preadsorbed with E. coli BL21(DE3) (pLysE, pT7-5) lysate and titered by enzyme-linked immunoassay with a synthetic peptide corresponding to EUO amino acids 64-EIDLKDLEDYKRNRYSRKK-82 (45).
Detection of EUO protein and transcripts in C. psittaci.
L cells (109) were infected with 10 times the 50% infective dose of C. psittaci 6BC (10) in 1 liter of M199 (GIBCO BRL, Grand Island, N.Y.) plus 5% fetal calf serum. Infected cell samples ranging from 5 × 107 (late samples) to 2 × 108 cells (early samples) for EUO protein and from 2.5 × 106 to 1 × 107 cells for transcripts were pelleted at 1,000 × g at 1, 2, 4, 8, 12, 20, and 24 h p.i. For EUO protein detection, the infected cell pellets were lysed in an ultrasonic bath at 4°C in 5 ml of phosphate-buffered saline (GIBCO BRL) containing glass beads. The lysates were diluted to 15 ml in phosphate-buffered saline and layered on top of a cushion of Hypaque-76 (Sanofi Winthrop Pharmaceuticals, New York, N.Y.) diluted to a final concentration of 29%, and chlamydiae were pelleted for 30 min at 25,000 rpm in a Beckman SW28 rotor. EBs were harvested by lysis of host cells with 0.5% Nonidet P-40 (NP-40) at 48 h p.i. and purified on a three-step Hypaque gradient of 29, 34, and 40% at 25,000 rpm for 30 min. EBs, collected at the 40-34% interface, were treated for 2 min with 0.05% NP-40 to lyse any remaining RBs, diluted in phosphate-buffered saline, and pelleted at 10,000 × g for 20 min. All pellets were suspended in 100 μl of SDS-PAGE solubilization buffer (30) containing 5% β-mercaptoethanol and subjected to SDS–7.5 to 15% PAGE. Proteins were electrophoretically transferred to an Immobilon membrane, and chlamydial MOMP and EUO proteins were detected as described by Everett and Hatch (15) with 1:2,500 rabbit anti-MOMP (C. trachomatis serovar L2) and 1:1,000 rabbit anti-His tag rEUO. The sample volumes that were loaded on the gel were adjusted so that the amounts of MOMP at all time points were approximately the same. For EUO transcript detection between 1 and 24 h p.i., infected cells were pelleted at 10,000 × g for 2 min and RNA was prepared from the pellets with 1 ml of TRIzol reagent (GIBCO BRL), following the instructions of the manufacturer. RNA was extracted from Hypaque gradient-purified EBs by the same method. RNA was fractionated by electrophoresis on a formaldehyde-agarose gel, and EUO mRNA, MOMP mRNA, and 16S rRNA were detected on Northern blots as previously described (17). C. psittaci 6BC MOMP and rRNA probes were prepared with a random-primed DNA-labeling kit (Boehringer Mannheim, Indianapolis, Ind.), using 20 μCi of [α-32P]dCTP. The template for the MOMP probe was the ClaI-EcoRI fragment from pMp− (32); the template for 16S rRNA was the EcoRI-HindIII insert of pRRN1 (32). To obtain a probe with sufficient specific activity to detect EUO transcripts, pDGW-E47 (45) was amplified by PCR with 20 μCi of [α-32P]dCTP and primers RK3 (5′ CATAATCGTTATTCAAGAAG 3′) and SEQ2 (5′ TCAACAGGAGAGTCTCGGA 3′).
Gel mobility shift assays.
A protocol modified from that described by Carey (7) was used for both reverse (radiolabeled rEUO-unlabeled DNA fragment) and conventional (unlabeled EUO protein-radiolabeled DNA probe) gel mobility shift assays. 35S-labeled, gel-purified rEUO protein was prepared by growing E. coli BL21(DE3) (pLysE, pLZ5F1) in 10 ml of medium containing amino acids minus methionine and cysteine. Overexpression was carried out in the same medium for 1 h in the presence of 200 μg of rifampin per ml and 140 μCi of 70% [35S]methionine–30% [35S]cysteine (Pro-Mix; Amersham, Arlington Heights, Ill.). 35S-rEUO was gel purified by SDS-PAGE as described above. Crude extracts used in gel-shift assays were made from purified RBs obtained from 108 infected cells at 15 and 29 h p.i. Infected cells were lysed in an ultrasonic bath, and RBs were purified by centrifugation over a four-step Hypaque gradient of 24, 29, 34, and 40%. RBs at the 29-34% interface were lysed by 10 s of sonication in 250 μl of buffer H-500, consisting of 25 mM HEPES (pH 7.5), 10 mM EDTA, 5 mM dithiothreitol, 1 mM MgCl2, 500 mM NaCl, 25% (vol/vol) glycerol, and 0.5% NP-40. The lysate was spun in a microcentrifuge (9,000 × g) at 4°C for 10 min, and the supernatant fluid was used in the gel shift assays.
The DNA fragments used in the reverse gel shifts were prepared by digestion of pALR208 with XbaI and BamHI to produce a 267-bp fragment containing the C. psittaci 6BC crp operon from −200 to +67 (15) and 2.7 kb of pUC19spf′ vector DNA (13). pALR208 is a subclone of pALR207; it was constructed by PCR amplification of pCPM2 (14), using primers 12K1 (5′ CGGCTCTAGACTGGCTTGATCAAGTGGTTT 3′ [the XbaI site is underlined]) and 12K3 (5′ GCAAGGATCCCTTAATGTGCTTGATTCCTT 3′ [the BamHI site is underlined]), and ligation of the PCR fragment into pBluescript KS (+) (Stratagene).
In conventional gel mobility shifts, the XbaI-BamHI insert of pALR208 was PCR amplified by using 12K1 and 12K3 primer oligonucleotides, one of which was 5′ end-labeled with 125 μCi of [α-32P]ATP by T4 polynucleotide kinase as described previously (17). For competition experiments, single-stranded DNA (ssDNA) was made by PCR amplification of 10 ng of agarose gel-purified BamHI-XbaI fragment from pALR208 with either 12K1 or 12K3 as the primer. The single-strand nature of the probes was confirmed by their susceptibility to complete digestion with staphylococcal S1 nuclease. Sense-strand RNA was generated in vitro from BamHI-digested pALR207 template with phage T3 RNA polymerase (Promega) and the MEGAscript kit from Ambion Inc., Austin, Tex.
Binding reactions were carried out for 30 min at room temperature in a final volume of 20 μl of DNA binding buffer (25 mM Tris-HCl [pH 7.5], 70 mM KCl, 1 mM EDTA, 7% glycerol, 50 μg of bovine serum albumin per ml, 3 mM CaCl2, 7 mM MgCl2, 1 mM dithiothreitol) with either rEUO (10 ng) or RB extract (0.05 μl), 1 ng of radiolabeled crp operon DNA, and calf thymus DNA (Sigma Chemical Company, St. Louis, Mo.) at the concentrations indicated. In supershift experiments, His tag rEUO antiserum was added (final dilution, 1:1,600) after the initial 30-min incubation period and the reaction mixture was incubated for an additional 30 min at room temperature. Protein-bound DNA was separated from free probe by electrophoresis at 10 V/cm for 4 h in 0.5 × TBE (1×TBE is 0.89 M Tris base, 0.89 M boric acid, and 2 mM EDTA) on 5% native polyacrylamide gels prerun for 1 h before the samples were loaded.
DNase I protection assays.
DNA fragments were generated by PCR amplification of pALR208 with primers (12K1 for the top strand; 12K3 for the bottom strand) which were 5′ end-labeled with [α-32P]ATP (17). The binding reaction was carried out in a final volume of 50 μl of DNA binding buffer containing 10 ng of [32P]DNA (1.2 nM), various amounts of purified rEUO, and 10 μg of sonicated calf thymus DNA. After incubation for 30 min at room temperature, 2.25 U of DNase I (Pharmacia LKB Biotechnology, Piscataway, N.J.) was added and the mixture was incubated for 40 s at room temperature. The reaction was stopped with 100 μl of stop solution (200 mM NaCl, 10 mM EDTA, 1% SDS, 250 μg of tRNA per ml) and extracted with phenol-chloroform (1:1), and DNA was precipitated with ethanol. The samples were dissolved in loading buffer (deionized formamide containing 10 mM EDTA, 0.3% bromphenol blue, and 0.3% xylene cyanol), incubated for 2 min at 95°C, and electrophoresed on a 6% polyacrylamide-urea sequencing gel. Following electrophoresis, the gels were dried and bands were visualized by autoradiography. Protected regions were identified by comparison to DNA that had not been mixed with rEUO; the locations of the protected regions were determined by DNA sequence reactions generated with primers 12K1 and 12K3.
RESULTS
EUO is synthesized in C. psittaci.
The gene encoding EUO, the ORF identified and named by Wichlan and Hatch (45), was subcloned with the expression cloning vectors pET28a(+) (with a His tag at the N terminus) and pT7-5 (without a His tag) and overexpressed in E. coli. His tag rEUO was purified by nickel-agarose affinity chromatography, and rEUO without a tag was purified by elution of a 27-kDa band from an SDS-polyacrylamide gel (data not shown); antibodies against both recombinant proteins were raised in rabbits. The identity of rEUO was confirmed by comparing the amino acid sequence predicted from the ORF with the N-terminal amino acid sequence determined from gel-purified material (data not shown).
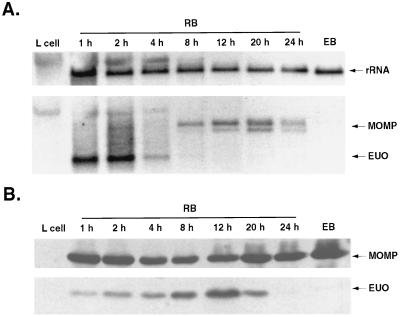
To determine the time of expression of the EUO gene, RNA was extracted from infected L cells and subjected to Northern blot analysis. In an attempt to analyze the same amount of total chlamydial RNA at all time points, the volume of material loaded was adjusted so that the amount of chlamydial 16S rRNA was approximately the same for all samples. Under these conditions of analysis, EUO transcripts were easily detected at 1 and 2 h p.i., barely detected 4 h p.i., and below detectable levels at later times in the developmental cycle (Fig. 1A). In other experiments (not shown), EUO transcripts could be detected through 24 h p.i. when 10- to 20-fold more total RNA was loaded on the gels. Thus, the EUO gene is transcribed during EB-to-RB conversion and during the phase of logarithmic RB cell division. However, the abundance of EUO transcripts, relative to the total RNA content of chlamydiae (reflected by 16S RNA content), is highest within the first few hours of infection. Wichlan and Hatch (45) previously demonstrated that transcription of the EUO gene by host-free chlamydiae isolated at 1 to 2 h p.i. is much more active than transcription of the MOMP. Using the less sensitive Northern blot method (Fig. 1A), which detects transcripts made by intracellular chlamydiae, we were unable to detect MOMP transcripts before 8 h p.i., confirming that EUO transcripts are more abundant than MOMP transcripts early in the infection cycle. Like the EUO transcripts, MOMP transcripts (40) were not detected by Northern blot analysis of Hypaque gradient-purified EBs harvested at 48 h p.i. (Fig. 1A).
FIG. 1.
Time course of EUO gene transcription and EUO protein synthesis in C. psittaci 6BC. (A) Northern blot. RNA was isolated with TRIzol reagent from uninfected L cells, whole infected L cells (RB) at the times indicated, and purified EBs harvested at 48 h p.i. The sample load was adjusted so that the amount of chlamydial rRNA in each lane was roughly the same. The blot shown was first reacted with a mixture of MOMP and EUO probes, exposed to film, and photographed and then stripped and reacted with the rRNA probe. The arrows at the right indicate signals to rRNA, MOMP (two transcripts at 12, 20, and 24 h p.i.), and EUO. At early times p.i. the amount of host RNA in the samples, particularly 18S rRNA, was sufficient to show cross-reaction with the chlamydial 16S rRNA. The length of the EUO transcript was estimated from unlabeled RNA standards (not shown) to be about 800 bases. (B) Immunoblot. C. psittaci RBs and EBs were harvested and partially purified at the times indicated, as described in Materials and Methods. The upper portion of the blot was reacted with C. trachomatis rabbit anti-MOMP, and the bottom portion was reacted with rabbit anti-rEUO protein.
To determine whether the EUO gene encodes a protein that is synthesized in C. psittaci, chlamydiae were harvested at various times p.i. and subjected to immunoblot analysis. The amount of sample subjected to electrophoresis was adjusted so that the immunoblot signals to MOMP antibody were approximately equal at all time points. MOMP has previously been demonstrated to be present at all times p.i., including in EBs (6, 24). Under these conditions of assay, EUO protein was detected in chlamydiae between 1 and 20 h p.i. but not at later times and not in isolated EBs (Fig. 1B). In another experiment (not shown), the amount of EUO peaked at 15 h p.i. These results indicate that the EUO gene encodes a protein that is made very early, when EBs are converting to RBs, but is also found and probably continues to be synthesized during the logarithmic growth phase of RBs (midcycle). The apparent paradox that EUO protein is most abundant at 15 h p.i. (relative to MOMP) whereas EUO transcripts are most abundant at 2 h p.i. (relative to 16S RNA) probably is a reflection of the stability of EUO mRNA versus the stability of EUO protein.
Although it is readily detected by immunoblotting, we have not detected an obvious 27-kDa band on Coomassie blue-stained gels when up to 50 μg of RB protein has been analyzed by SDS-PAGE (not shown). From these gels we estimate that EUO is present at 0.1 to 1% of the level of the most common chlamydial protein, MOMP.
EUO binds to DNA in vitro.
EUO bears no obvious homology to proteins currently in the Swiss protein database, and functional domains have not been identified with the Motifs (34) or Blocks (25) software programs. However, a potential helix-turn-helix DNA binding motif was identified by using the algorithm of Dodd and Egan (11) (Fig. 2). To determine whether EUO can bind to DNA, reverse gel mobility shift analysis was carried out. 35S-rEUO was partially purified and mixed with EcoRI-digested genomic C. psittaci DNA, and free EUO (which remains in the loading well) was separated from DNA-bound protein on a nondenaturing polyacrylamide-agarose gel. rEUO was found by this technique, to bind to many genomic DNA fragments, even in the presence of excess calf thymus DNA (Fig. 3A).
FIG. 2.
Predicted amino acid sequence and helix-turn-helix domain of EUO. A potential helix-turn-helix motif, as identified by the algorithm of Dodd and Egan (11), is underlined in the sequence. The helix-turn-helix motifs of EUO and selected other proteins, along with their Dodd and Egan standard deviation (SD) scores, are shown below the EUO sequence. A score above 2.5 is considered significant. Asterisks mark the most highly conserved amino acids in the motif.
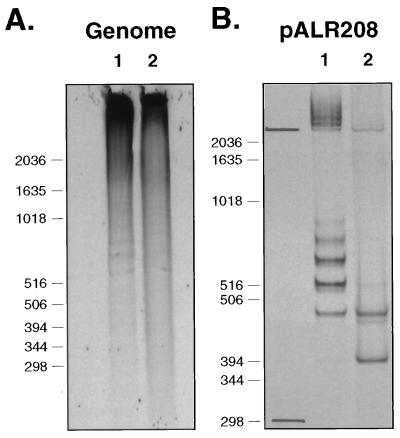
FIG. 3.
Reverse gel mobility shift assays showing binding of rEUO protein to C. psittaci genomic and crp operon DNA. 35S-rEUO (20 ng) was bound to unlabeled DNA, and the reaction mixtures were fractionated on a nondenaturing polyacrylamide gel, as described in Materials and Methods. (A) C. psittaci genomic DNA digested with EcoRI (200 ng/lane). Lanes: 1, genomic DNA with 35S-rEUO; 2, genomic DNA plus calf thymus DNA (4 μg) with 35S-rEUO. (B) pALR208 DNA (200 ng) digested with XbaI and BamHI to remove an insert containing the crp operon from bp −200 to +67. Lanes: 1, pALR208 with 35S-rEUO; 2, pALR208 plus 40 μg of calf thymus DNA with 35S-rEUO. The migration of the pALR208 vector (2.7 kbp) and insert fragment (267 bp) in the absence of rEUO is shown to the left of lane 1 in panel B. The migration of DNA standards (in basepairs) is shown to the left of each panel.
To simplify the characterization of the binding of rEUO to DNA, the binding of the protein to a defined sequence was analyzed. Several plasmid clones containing small chlamydial DNA inserts were examined, and rEUO bound to all of them to some degree (data not shown). One clone, pALR208, was used as a model for further investigations because of the high level of binding of rEUO to its insert DNA. pALR208 consists of a derivative pUC19 vector and an insert containing the C. psittaci crp operon sequence from bp −200 to +67 (15). The crp operon encodes two cysteine-rich envelope proteins that are made late in the developmental cycle, starting at 18 to 20 h p.i. (23, 33). In the absence of competing calf thymus DNA, rEUO bound to both vector and insert sequences (Fig. 3B, lane 2). The appearance of multiple shifted bands suggests that more than one molecule of rEUO can bind to a DNA fragment. In the presence of nonspecifically competitive calf thymus DNA, binding of rEUO to the 2.7-kbp vector DNA was almost eliminated. Binding to the smaller 267-bp fragment was also diminished, but two shifted bands—representing 2 and 1 molecules of rEUO bound per fragment—remained prominent. These observations suggest that, although the specificity of binding of rEUO to DNA is broad, some degree of specificity exists.
Specificity of binding of rEUO to crp operon DNA.
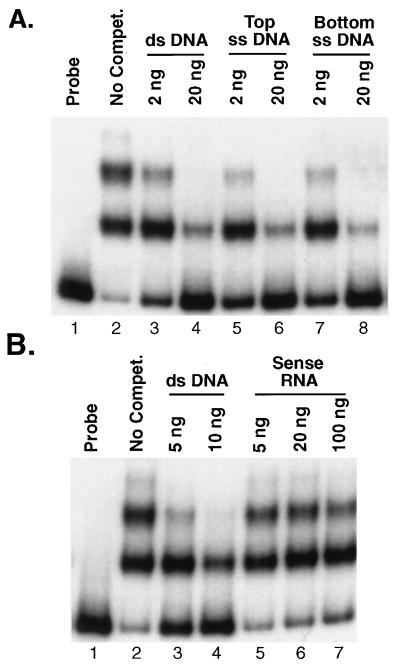
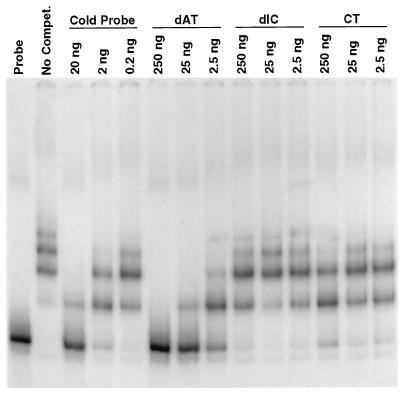
Unlabeled ssDNA and double-stranded DNA (dsDNA) probes were generated from the pALR208 template by PCR to examine the binding of rEUO by conventional gel shift assays. On a per-gram basis, ssDNA was found to compete as efficiently as unlabeled dsDNA for the labeled dsDNA probe (Fig. 4A). However, RNA generated in vitro by using phage T3 RNA polymerase competed poorly for binding even when it was present in 100-fold excess (Fig. 4B). Therefore, rEUO appears to be capable of binding to ds- and ssDNA and perhaps to RNA, but with considerably lower affinity. The specificity of binding of rEUO to insert DNA was further investigated by homopolymer competition experiments. We found that poly[d(A-T) · d(A-T)] but not poly[d(I-C) · d(I-C)] was an effective competitor, suggesting that the binding of rEUO to DNA is specific, with apparent preference for AT-rich sequences (Fig. 5). The affinity of rEUO for the fragment appears to be relatively strong, with a Kd of about 15 nM (data not shown).
FIG. 4.
Gel shift assays showing the effects of ssDNA and RNA on binding of rEUO to dsDNA. One nanogram of labeled crp dsDNA probe (bp − 200 to +67) and 10 ng of rEUO were used in all assays except in the free probe control, which lacked rEUO. Calf thymus DNA (100 ng) was included in all reactions. (A) Competition with crp operon ssDNA (bp −200 to +67). Lanes: 1, free probe; 2, probe plus rEUO, no competitor; 3, probe plus 2 ng of unlabeled dsDNA probe; 4, probe plus 20 ng of unlabeled dsDNA probe; 5, probe plus 2 ng of top (coding)-strand ssDNA; 6, probe plus 20 ng of top-strand ssDNA; 7, probe plus 2 ng of bottom-strand ssDNA; 8, probe plus 20 ng of bottom-strand ssDNA. (B) Competition with sense-strand RNA. Lanes: 1, free probe; 2, no competitor; 3, probe plus 5 ng of unlabeled dsDNA probe; 4, probe plus 10 ng of unlabeled dsDNA probe; 5, probe plus 5 ng of RNA; 6, probe plus 20 ng of RNA; 7, probe plus 100 ng of RNA.
FIG. 5.
Gel shift assay showing the effects of poly[d(A-T) · d(A-T)] (dAT), poly[d(I-C) · d(I-C)] (dIC), and calf thymus DNA (CT) on rEUO binding to crp DNA (bp −200 to +67). The amount of labeled probe was 1 ng and the amount of rEUO was 10 ng. The amount of competitor DNA in each reaction is indicated above the lanes.
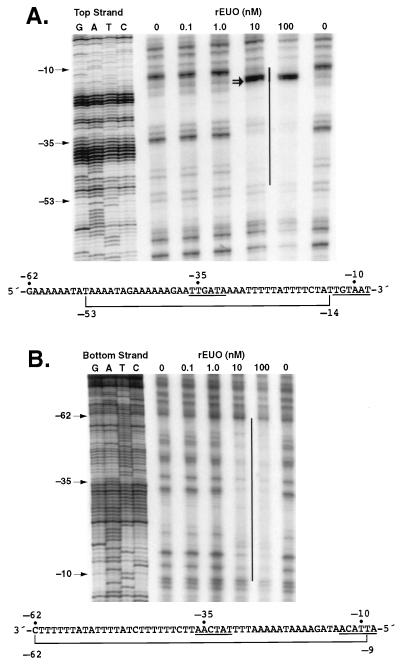
DNase I protection (footprint) analysis was carried out to determine the binding site of rEUO on the pALR208 DNA fragment. The top strand of the fragment was protected from about bp −53 to −14, and the bottom strand was protected from about bp −62 to −9 (Fig. 6). In addition, two prominent hypersensitive bands, corresponding to bp −16 and −17, were noted on the top strand. The protected region roughly spans the predicted −35 and −10 regions of the promoter. Although the protected region is extremely AT rich (89 to 90%, compared to a genomic A+T content of about 56%), it should be noted that rEUO did not protect other AT-rich sequences on the fragment (for example, 14 of 16 bp from −63 to −79 are AT).
FIG. 6.
Protection of crp DNA from DNase I by rEUO. (A) Top strand; (B) bottom strand. Protected regions are indicated by vertical lines. DNA sequencing ladders (in base pairs) prepared with the same primers used to generate the probes in the protection assay are shown at the left side of each panel. The double arrow indicates hypersensitive bands at positions −16 and −17. The DNA sequence (with the protected region bracketed) is shown at the bottom of each panel. The −35 and −10 promoter elements are underlined.
The protection experiment shown in Fig. 6 was carried out with rEUO without a His tag; an identical footprint was noted when rEUO with a His tag was used (data not shown). Indeed, we have found that all of the results reported here were independent of the nature of the rEUO used and the method of purification of the protein. This suggests that our observations are due to rEUO per se and not to a copurifying E. coli protein.
Native C. psittaci EUO protein binds to the crp operon promoter.
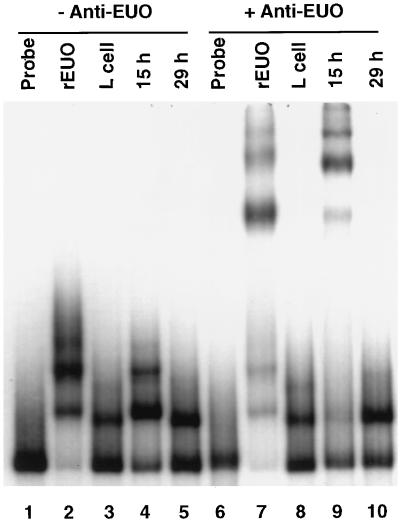
Gel shift experiments were performed with crude extracts of RBs purified from host cells at 15 (midcycle) and 29 (late cycle) h p.i. and bp −200 to +67 crp operon DNA to determine whether native EUO has binding activity. We noted several shifted bands in the 15-h p.i. preparation that resembled bands shifted by rEUO (Fig. 7A). These bands were not noted in the extract prepared at 29 h p.i., a time when EUO is not expected to be found in chlamydiae (Fig. 1B). A single shifted band with slightly greater mobility than the most rapidly migrating rEUO band was noted in the 29-h p.i. extract (Fig. 7A). The identity of the binding protein responsible for this band is not known. A comigrating band was noted in crude extracts of host L cells; however, the absence of the band in the 15-h p.i. preparation and the use of purified RBs for preparation of both extracts suggests that a chlamydial rather than a host protein is responsible for the 29-h band. The role of native EUO in the shifts noted in the 15-h p.i. preparation was confirmed by the addition of anti-EUO antibody, which resulted in the supershifting of these bands (Fig. 7B). These observations suggest that the binding of rEUO to the crp promoter region is a function of EUO protein structure and not an artifact of recombinant protein preparation.
FIG. 7.
Comparison of binding of native EUO and rEUO to crp operon promoter DNA. Gel shift assays were carried out with 1 ng of crp probe DNA (bp −200 to +67) as described in Materials and Methods. Lanes: 1, free probe; 2, probe plus 10 ng of rEUO; 3, probe plus crude L-cell extract; 4, probe plus 15-h p.i. RB extract; 5, probe plus 29-h p.i. RB extract; 6, free probe; 7, probe plus rEUO and anti-His tag rEUO; 8, probe plus L-cell extract and anti-rEUO; 9, probe plus 15-h p.i. RB extract and anti-rEUO; 10, probe plus 29-h p.i. RB extract and anti-rEUO.
Protease activity of rEUO.
Kaul et al. (29) have reported that glutathione S-transferase-tagged rEUO has protease activity specific for H1-like histones and chlamydial Hc1. We tested the effect of our His tag EUO preparations on extracts of C. psittaci 6BC EBs but could find no evidence for degradation of chlamydial 18-kDa Hc1 protein. Our preparations contain residual urea (necessary to maintain the solubility of rEUO), which may interfere with any enzymatic activity associated with rEUO.
DISCUSSION
We have determined that EUO is a quantitatively minor protein that is absent from C. psittaci EBs but is made very shortly after the EBs enter host cells. The amount of EUO protein peaks at midcycle and declines thereafter as the intracellular chlamydiae progress to the late stage of the developmental cycle, which is characterized by RB-to-EB conversion. We have also demonstrated that rEUO as well as native EUO protein can bind to DNA in vitro. The specificity of binding in vitro appears to be broad in that rEUO can bind to multiple EcoRI fragments of chlamydial genomic DNA as well as to pUC19 DNA. However, using pALR208 as a model, we found that rEUO protein binds preferentially to insert as opposed to vector DNA. The binding of rEUO to insert DNA appears to be specific in that poly[d(I-C) · d(I-C)] does not compete with binding to the insert, the apparent dissociation constant of the rEUO-insert complex is within the range of other DNA binding proteins, and rEUO protects DNA from DNase I at a specific location. The protected region roughly spans the putative −35 and −10 regions of the crp operon promoter; this region is extremely AT-rich, and binding is competed with poly[d(A-T) · d(A-T)], suggesting that EUO preferentially binds to AT-rich, sequences. However, the specificity of binding is likely more complicated than simple recognition of AT-rich sequences, since rEUO did not bind to other AT-rich stretches in the crp operon promoter region.
The binding properties of EUO are reminiscent of the properties of E. coli H-NS nucleoid protein, which also binds to AT-rich sequences and has a tendency to form multimeric complexes with DNA (reviewed in reference 3). AT-rich sequences can favor bends in DNA, and it is thought that H-NS specifically recognizes bent DNA. Additional studies are necessary to determine whether EUO also recognizes curved DNA. The most obvious difference between EUO and the E. coli nucleoid protein is quantity. H-NS is a major structural protein, present in thousands of copies per cell (3), whereas EUO is a minor protein, likely present in only a few copies per cell. Therefore, if the function of EUO in vivo is that of a DNA binding protein, it must bind to a limited number of sites at any given time. How these sites might be selected in vivo, in light of our observations that EUO appears to have a relatively broad specificity in vitro, is not known.
pALR208 was fortuitously chosen as a model clone to study EUO binding, and we do not know at present whether EUO binds to the crp operon in vivo. Nonetheless, some of our observations are intriguing and invite speculation. EUO binds with relatively high affinity to the crp promoter region, including over the −35 and −10 hexamers, and the time of synthesis and accumulation of EUO in chlamydial cells is just the reverse of that of the cysteine-rich proteins. The location of the gene encoding EUO close to the crp locus on the chlamydial genome also is interesting. A possible function of the quantitatively minor EUO protein, therefore, may be to repress the crp operon during the growth phases of the chlamydial developmental cycle. We emphasize, however, that we do not know the physiological function of EUO.
Two other chlamydial DNA binding proteins, the histone-like proteins Hc1 and Hc2, have been extensively characterized (4, 5, 18, 19, 29, 35–37, 44). The chlamydial histone-like proteins are unrelated to H-NS, but Hc1 homologs have been reported in Bordetella pertussis and Pseudomonas aeruginosa (27, 39). In contrast to EUO, Hc1 and Hc2 are major structural proteins. Their binding specificities are broad: Hc1, for example, can induce nucleoid formation when expressed in E. coli (5), and in vitro studies have demonstrated binding to a wide range of DNA fragments (4, 35–37). The pattern of recognition by the histone proteins has not been determined; however, Kaul et al. (28) have demonstrated a preference for binding of Hc1 to a specific locus on the 7.5-kb chlamydial plasmid.
The relationship between EUO and the histone-like proteins is unclear. Kaul et al. (29) have postulated that EUO is a protease that cleaves Hc1, possibly contributing to the activation of chlamydial gene expression shortly after the entry of EBs into host cells. The early synthesis of EUO is consistent with this idea; however, we found that peak levels of EUO protein are present in dividing RBs, long after chlamydial histone-like proteins dissociate from DNA and can be detected in extracts. Also, we have not detected protease activity in our rEUO preparations, although residual urea in our preparations and our use of His tag rather than glutathione S-transferase rEUO might explain our failure to detect enzymatic activity.
Many questions regarding the function of EUO need to be addressed. We are currently attempting to determine the chlamydial genomic sequences to which EUO preferentially binds and whether or not EUO binds to these sequences in vivo. We are also examining the role of EUO in the regulation of transcription of the crp operon.
ACKNOWLEDGMENT
This work was supported by U.S. Public Health Service research grant AI 19570.
REFERENCES
- 1.Allen J E, Cerrone M C, Beatty P R, Stephens R S. Cysteine-rich outer membrane proteins of Chlamydia trachomatis display compensatory sequence changes between biovariants. Mol Microbiol. 1990;4:1543–1550. doi: 10.1111/j.1365-2958.1990.tb02065.x. [DOI] [PubMed] [Google Scholar]
- 2.Allen J E, Stephens R S. Identification by sequence analysis of two-site posttranslational processing of the cysteine-rich outer membrane protein 2 of Chlamydia trachomatis serovar L2. J Bacteriol. 1989;171:285–291. doi: 10.1128/jb.171.1.285-291.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 4.Barry C E, III, Brickman T J, Hackstadt T. Hc1-mediated effects on DNA structure: a potential regulator of chlamydial development. Mol Microbiol. 1993;9:273–283. doi: 10.1111/j.1365-2958.1993.tb01689.x. [DOI] [PubMed] [Google Scholar]
- 5.Barry C E, III, Hayes S F, Hackstadt T. Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science. 1992;256:377–379. doi: 10.1126/science.256.5055.377. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 8.Chlamydia Genome Project (R. S. Stephens, S. Kalman, C. Fenner, and R. Davis). 1998. Chlamydia Genome Project, contig 5E. http://violet .berkeley.edu:4231.
- 9.Clarke I N, Ward M E, Lambden P R. Molecular cloning and sequence analysis of a developmentally regulated cysteine-rich outer membrane protein from Chlamydia trachomatis. Gene. 1988;71:307–314. doi: 10.1016/0378-1119(88)90047-9. [DOI] [PubMed] [Google Scholar]
- 10.Crenshaw R W, Fahr M J, Wichlan D G, Hatch T P. Developmental cycle-specific host-free RNA synthesis in Chlamydia spp. Infect Immun. 1990;58:3194–3201. doi: 10.1128/iai.58.10.3194-3201.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas A L, Saxena N K, Hatch T P. Enhancement of in vitro transcription by addition of cloned, overexpressed major sigma factor of Chlamydia psittaci 6BC. J Bacteriol. 1994;176:3033–3039. doi: 10.1128/jb.176.10.3033-3039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erickson J W, Gross C A. Identification of the ςE subunit of Escherichia coli RNA polymerase: a second alternative ς factor involved in high temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 14.Everett K D E, Andersen A A, Plaunt M, Hatch T P. Cloning and sequence analysis of the major outer membrane protein gene of Chlamydia psittaci 6BC. Infect Immun. 1991;59:2853–2855. doi: 10.1128/iai.59.8.2853-2855.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett K D E, Hatch T P. Sequence analysis and lipid modification of the cysteine-rich envelope proteins of Chlamydia psittaci 6BC. J Bacteriol. 1991;173:3821–3830. doi: 10.1128/jb.173.12.3821-3830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett K D E, Hatch T P. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol. 1995;177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahr M J, Douglas A L, Xia W, Hatch T P. Characterization of the late gene promoters of Chlamydia trachomatis. J Bacteriol. 1995;177:4252–4260. doi: 10.1128/jb.177.15.4252-4260.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackstadt T, Baehr W, Ying Y. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci USA. 1991;88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackstadt T, Brickman T J, Barry III C E, Sager J. Diversity in the Chlamydia trachomatis histone homologue Hc2. Gene. 1993;132:137–141. doi: 10.1016/0378-1119(93)90526-9. [DOI] [PubMed] [Google Scholar]
- 20.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt T, Todd W J, Caldwell H D. Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J Bacteriol. 1985;161:25–31. doi: 10.1128/jb.161.1.25-31.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatch T P, Allan I, Pearce J H. Structural and polypeptide differences between envelopes of infective and reproductive life cycle forms of Chlamydia spp. J Bacteriol. 1984;157:13–20. doi: 10.1128/jb.157.1.13-20.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatch T P, Miceli M, Sublett J E. Synthesis of disulfide-bonded outer membrane proteins during the developmental cycle of Chlamydia psittaci and Chlamydia trachomatis. J Bacteriol. 1986;165:379–385. doi: 10.1128/jb.165.2.379-385.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hatch T P, Vance D W, Jr, Al-Hossainy E. Identification of a major envelope protein in Chlamydia spp. J Bacteriol. 1981;146:426–429. doi: 10.1128/jb.146.1.426-429.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff S, Henikoff J G. Protein family classification based on searching a database of blocks. Genomics. 1994;19:97–107. doi: 10.1006/geno.1994.1018. [DOI] [PubMed] [Google Scholar]
- 26.Hsia R-C, Bavoil P M. Homologs of Escherichia coli recJ, gltX and a putative ‘early’ gene of avian Chlamydia psittaci are located upstream of the ‘late’ omp2 locus of Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Gene. 1997;176:163–169. doi: 10.1016/0378-1119(96)00240-5. [DOI] [PubMed] [Google Scholar]
- 27.Kato J, Misra T K, Chakrabarty A M. AlgR3, a protein resembling eukaryotic histone H1, regulates alginate synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1990;87:2887–2891. doi: 10.1073/pnas.87.8.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaul R, Allen M, Bradbury E M, Wenman W M. Sequence specific binding of chlamydial histone H1-like protein. Nucleic Acids Res. 1996;24:2981–2989. doi: 10.1093/nar/24.15.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaul R, Hoang A, Yau P, Bradbury E M, Wenman W M. The chlamydial EUO gene encodes a histone H1-specific protease. J Bacteriol. 1997;179:5928–5934. doi: 10.1128/jb.179.18.5928-5934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lambden P R, Everson J S, Ward M E, Clarke I N. Sulfur-rich proteins of Chlamydia trachomatis: developmentally regulated transcription of polycistronic mRNA from tandem promoters. Gene. 1990;87:105–112. doi: 10.1016/0378-1119(90)90500-q. [DOI] [PubMed] [Google Scholar]
- 32.Mathews S A, Douglas A, Sriprakash K S, Hatch T P. In vitro transcription in Chlamydia psittaci and Chlamydia trachomatis. Mol Microbiol. 1993;7:937–946. doi: 10.1111/j.1365-2958.1993.tb01185.x. [DOI] [PubMed] [Google Scholar]
- 33.Newhall W J, V. Biosynthesis and disulfide cross-linking of outer membrane components during the growth cycle of Chlamydia trachomatis. Infect Immun. 1987;55:162–168. doi: 10.1128/iai.55.1.162-168.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogiwara A, Uchiyama I, Takagi T, Kanehisa M. Construction and analysis of a profile library characterizing groups of structurally known proteins. Protein Sci. 1996;5:1991–1999. doi: 10.1002/pro.5560051005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perara E, Ganem D, Engel J N. A developmentally regulated chlamydial gene with apparent homology to eukaryotic histone H1. Proc Natl Acad Sci USA. 1992;89:2125–2129. doi: 10.1073/pnas.89.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen L B, Birkelund S, Christiansen G. Interaction of the Chlamydia trachomatis histone H1-like protein (Hc1) with DNA and RNA causes repression of transcription and translation in vitro. Mol Microbiol. 1994;11:1085–1098. doi: 10.1111/j.1365-2958.1994.tb00385.x. [DOI] [PubMed] [Google Scholar]
- 37.Petersen L B, Birkelund S, Christiansen G. Purification of recombinant Chlamydia trachomatis histone H1-like protein Hc2, and comparative functional analysis of Hc2 and Hc1. Mol Microbiol. 1996;20:295–311. doi: 10.1111/j.1365-2958.1996.tb02618.x. [DOI] [PubMed] [Google Scholar]
- 38.Plaunt M R, Hatch T P. Protein synthesis early in the developmental cycle of Chlamydia psittaci. Infect Immun. 1988;56:3021–3025. doi: 10.1128/iai.56.12.3021-3025.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scarlato V, Arico B, Goyard S, Ricci S, Manetti R, Prugnola A, Manetti R, Polverino-De-Laureto P A, Ullmann A, Rappuoli R. A novel chromatin-forming histone H1 homologue is encoded by a dispensable and growth-regulated gene in Bordetella pertussis. Mol Microbiol. 1995;15:871–881. doi: 10.1111/j.1365-2958.1995.tb02357.x. [DOI] [PubMed] [Google Scholar]
- 40.Stephens R S, Wagar E A, Edman U. Developmental regulation of tandem promoters for the major outer membrane protein of Chlamydia trachomatis. J Bacteriol. 1988;170:744–750. doi: 10.1128/jb.170.2.744-750.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stragier P, Losick R. Cascades of sigma factors revisited. Mol Microbiol. 1990;4:1801–1806. doi: 10.1111/j.1365-2958.1990.tb02028.x. [DOI] [PubMed] [Google Scholar]
- 42.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 43.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao S, Kaul R, Wenman W M. Identification and nucleotide sequence of a developmentally regulated gene encoding a eucaryotic histone H1-like protein from Chlamydia trachomatis. J Bacteriol. 1991;173:2818–2822. doi: 10.1128/jb.173.9.2818-2822.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wichlan D G, Hatch T P. Identification of an early-stage gene of Chlamydia psittaci 6BC. J Bacteriol. 1993;175:2936–2942. doi: 10.1128/jb.175.10.2936-2942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]