Abstract
Background
N4-acetylcytidine (ac4C) is a conserved and abundant mRNA modification that controls protein expression by affecting translation efficiency and mRNA stability. Whether the ac4C modification of mRNA regulates hepatocellular carcinoma (HCC) development or affects the immunotherapy of HCC is unknown.
Methods
By constructing an orthotopic transplantation mouse HCC model and isolating tumour-infiltrated immunocytes, we evaluated the ac4C modification intensity using flow cytometry. Remodelin hydrobromide (REM), an ac4C modification inhibitor, was systematically used to understand the extensive role of ac4C modification in immunocyte phenotypes. Single-cell RNA-seq was performed to comprehensively evaluate the changes in the tumour-infiltrating immunocytes and identify targeted cell clusters. RNA-seq and RIP-seq analyses were performed to elucidate the underlying molecular mechanisms. Tyramide Signal Amplification (TSA) analysis on the HCC tissue microarray was performed to explore the clinical relatedness of our findings.
Results
Ac4C modification promoted M1 macrophage infiltration and reduced myeloid-derived suppressor cell MDSCs infiltration in HCC. The inhibition of ac4C modification induces PDL1 expression by stabilising mRNA in the myeloid cells, thereby attenuating the CTL-mediated tumour cell-killing ability. High infiltration of ac4C+CD11b+ cells is positively related to a better prognosis in patients with HCC.
Conclusions
Ac4C modification of myeloid cells enhanced the HCC immunotherapy by suppressing PDL1 expression.
Subject terms: Cancer immunotherapy, Translational research
Background
Hepatocellular carcinoma (HCC) is one of the most prevalent and fatal cancers worldwide [1, 2]. In recent years, progress has been made in the systemic treatment of advanced HCC, which cannot be treated with resection or locoregional therapies, which provides survival benefits [3, 4]. As per the IMbrave150 trial, atezolizumab plus bevacizumab (atezo/beva), a combination therapy of immune checkpoint inhibitors and an anti-angiogenic agent, has been approved as a first-line systemic therapy option for advanced HCC [5, 6]. However, some HCC patients are insensitive to immunotherapy and show accelerated disease progression following treatment with it [7, 8].
HCC is a typical inflammation-related cancer in which the tumour microenvironment is a complex mixture of hepatic non-parenchymal resident cells, tumour cells, immune cells, and tumour-associated fibroblasts [9]. Like M2 polarised tumour-associated macrophages (TAM), regulatory inhibitory B cells, regulatory T cells (Tregs), or myeloid-derived suppressor cells (MDSCs), these negatively regulated cells interact to form a suppressive immune microenvironment, promote the progression of HCC, and weaken the effects of immunotherapy [3]. For instance, Treg cells and MDSCs in the liver can inhibit T cell function and are related to the poor prognosis of HCC by blunting immunotherapy [10, 11]. Deep investigations into the suppressive immune microenvironment of HCC are likely to provide new perspectives for HCC immunotherapy.
RNA modification is an important epigenetic regulatory mechanism that plays an essential role in the control of diverse biological functions by affecting RNA stability, decay, metabolism, and binding to RNA-binding proteins [12]. Up to now, several types of RNA modifications have been identified, including N6-methyladenosine (m6A), 5-methylcytosine (m5C), N7-methylguanosine (m7G), and N4-acetylcytidine (ac4C) [13]. For HCC, the overexpression of m6A writer methyltransferase-like 3 (METTL3) promotes HCC development and metastasis by degrading the suppressor of cytokine signaling-2 (SOCS2) mRNA [14]. Meanwhile, the m6A modification of snRNAs has been shown to affect the efficacy of immunotherapy in patients with HCC [15]. Deletion of METTL3 promotes sorafenib resistance in HCC by reducing FOXO3 mRNA stability [15]. These results suggest that RNA modifications, especially m6A, may be closely related to the effects of immunotherapy in patients with HCC. However, the relationship between ac4c modification and the suppressive tumour immune microenvironment remains unclear.
Ac4C modification in myeloid cells may be conducive to maintaining the effects of atezo/beva therapy. Once weakened, this may lead to enhanced inhibition of the suppressive immune microenvironment. To verify this hypothesis, we detected changes in the ac4C levels in various immune cell subsets during HCC immunotherapy. We used RNA-seq and RIP sequencing to explore the effects of RNA modifications on myeloid cells. Finally, combined with single-cell sequencing and in vitro and in vivo analyses, our team verified the vital role of ac4C modifications in HCC immunotherapy.
Methods
Cell isolation, culture and transfection
Bone marrow-derived macrophages were harvested from 6–8-week-old C57BL/6 mice. The macrophages were seeded in an incomplete medium. Following 6 h of incubation under 5% CO2 at 37 °C, the unattached cells were removed and the adherent cells were transferred to a complete 1640 medium containing 10% foetal bovine serum (FBS, Gibco, Australia), 100 IU/mL penicillin, and 100 μg/mL streptomycin. Hepa1–6 cells were cultured in a Dulbecco’s modification of Eagle’s medium supplemented with 10% FBS, 100 IU/mL penicillin, and 100 μg/mL streptomycin at 37 °C under a humidified 5% CO2 atmosphere. Hepa1–6 cells were purchased from American Type Culture Collection (ATCC) authenticated by STR profiling and tested for mycoplasma contamination before experiments. NAT10-overexpression plasmids (MR211470, Origene, USA) were transfected into cells using the jetPRIME® DNA transfection reagent (jetPRIME, PolyPlus-transfection, French) according to the manufacturer’s protocol.
Animal experiments and ethics statement
Six- to eight-week-old C57BL/6 and NOD-SCID-Il2rg (-/-) (NSG) mice were purchased from Hangzhou Medical College (Hangzhou, China). All the animal experiments were performed in accordance with the Institutional Animal Care and Use Committee of the Zhejiang Center of Laboratory Animals (Approval No: ZJCLA-IACUC-20050065). A 25 μL mixture of PBS and Matrigel (Corning, USA) (1:1) containing 5 × 105 Hepa1–6 cells were injected into the left liver lobe of the C57BL/6 mice to establish the HCC orthotopic transplantation mice model. For the inhibitor, drug, or immunotherapy experiments, the mice were intraperitoneally injected with remodelin hydrobromide (S7641, Selleck, USA) (10 mg/kg), an anti-PDL1 antibody (BP0101, Bio x Cell, China) (20 mg/kg), or apatinib (YN968D1, Invivochem, USA) (75 mg/kg) every 2 days.
Cell apoptosis assay
Flow cytometry using an Annexin V-FITC/PI detection kit (556547, BD, USA) was used to assess apoptosis in the Hepa1–6 cells. Briefly, the cells were plated in 6-well culture plates at a density of 2 × 106 per well and thereafter cultured with Remodelin hydrobromide for 24 h. Next, the cells were trypsinised, rinsed with PBS and re-suspended in 400 μL of 1× binding buffer. The cells were then stained with 5 μL annexin V-FITC and 5 μL PI in darkness for 20 min at room temperature (20–24 °C). After staining, the samples were immediately analysed using flow cytometry (BD, USA). The annexin V−/PI− cells were identified as viable, while the early-apoptotic cells were annexin V+/PI−, late-apoptotic cells were annexin V+/PI+, and cell debris were annexin V−/PI+.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8; Yeasen, China) was used to determine cell proliferation according to the manufacturer’s instructions. Briefly, 2 × 104 cells were seeded per well in a 96-well plate for 24 h and treated with remodelin hydrochloride. Ten microliters of the CCK-8 solution was added into each well with 100 μL of medium. The 96-well plates were incubated for 2 h at 37 °C, and the absorbencies were measured at 450 nm by a microplate reader at each timepoint.
RNA isolation, quantitative real-time PCR analysis
Total RNA from the peritoneal macrophages treated with inhibitors or drugs was extracted using a TRIzol reagent (Invitrogen, USA). Reverse transcription of RNA and RT-PCR was performed using the Prime Script TM Trimester Mix (Takara, Japan) and SYBR Premix Ex TaqTM11 (Yeasen, China) with a StepOnePlus RT-PCR system (Life Technologies, USA), according to the manufacturer’s instructions. GAPDH was used as an endogenous control. The primers for RT-PCR were synthesised by Tsingke Biotech (Beijing, China). The primer sequences used were as follows: Pdl1 forward primer, GCTCCAAAGGACTTGTACGTG; Pdl1 reverse primer, TGATCTGAAGGGCAGCATTTC. Gapdh forward primer-AGGTCGGTGTGAACGGATTTG; Gapdh reverse primer-TGTAGACCATGTAGTTGAGGTCA.
Mouse liver-derived lymphocyte isolation and cytometric analysis
Liver-derived lymphocytes were isolated from the mice using collagenase II/IV (Solarbio, China) and Lymphocyte Separation Medium (Dakewe Biotech, China). For the cytometric analysis, nonspecific binding of the cells was blocked using an Fc receptor block (clone 2.4G2, BD, USA). Following incubation for 10 min at 4 °C, the cells were stained with anti-surface antibodies for 30 min at 4 °C. After two washes with PBS, the cells were analysed using a BD FACSCanto II instrument. The data obtained were analysed using the FlowJo X software (TreeStar). The surface antigen-antibodies are as follows: CD11b (101254, BioLegend, USA), CD20 (152108, BioLegend, USA), CD45 (75-0451, Tonbo, USA), CD3e (100233, BioLegend, USA), CD8a (65-0081, Tonbo, USA), CD4 (100430, BioLegend, USA), Ly6G (127633, BioLegend, USA), Ly6C (20-5932, Tonbo, USA), PDL1 (124308, BioLegend, USA), FVS780 (565388, BD, USA), PD1 (109110, BioLegend, USA), CD107a (121606, BioLegend, USA), and KLRG1 (50-5893, Tonbo, USA). A FITC Conjugation Kit (ab188285, Abcam, UK) was used for the FITC-fluorescein conjugation of ac4C antibodies (ab252215, Abcam, UK).
RNA sequencing and data analysis
Macrophages harvested from 6–8-week-old C57BL/6 mice were incubated with or without Remodelin hydrobromide and then treated with or without IFN-γ. The cells were collected and lysed using the TRIzol reagent (Invitrogen, USA), and the total RNAs were extracted according to the manufacturer’s protocol. RNA-seq was performed at Novogene (Beijing, China) using a Hiseq6000 (Illumina, USA). LifeScope v2.5.1 was used to align the reads to the genome, generate raw counts corresponding to each known gene, and calculate the raw counts and reads per kilobase per million (RPKM) values.
RNA immunoprecipitation sequence and data analysis
The macrophages were harvested and lysed in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES, 0.5% NP-40, 1 mM DTT, 80 U/mL RNase inhibitor, and protease inhibitor cocktail) for 5 min on ice. The lysates were sonicated to fragment the chromatin and RNA and centrifuged, and the protein concentration in the supernatant was measured using a BCA assay (Beyotime, China). The proteins were incubated with 50 μL of Magnetic Dynabeads (Millipore, USA) pre-coated with 2 μg of anti-ac4C antibody (Abcam, UK) or IgG (Millipore, USA) for 4 h at 4 °C with rotation. Five per cent of the supernatant was used as the input. Following incubation, the beads were washed five times with an NT buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM MgCl2, 1% Triton X-100). Immunoprecipitated RNAs and input RNAs were extracted using the TRIzol reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The library was constructed by the Novogene Corporation (Beijing, China). Paired-end sequencing of the samples was performed using an Illumina platform (Illumina, CA, USA). Library quality was assessed using the Agilent Bioanalyzer 2100 system.
Mass cytometry by time-of-flight and data processing
HCC tissues were collected from the mice and lysed using a mouse tumour dissociation kit (130-096-730, Miltenyi, USA). In total, 3 × 106 cells were counted and stained with a live/dead dye. Subsequently, the cells were stained with a panel of 42 antibodies encompassing a broad range of immune subsets. Finally, CD45+ cells were analysed using a Helios mass cytometer (Fluidigm, USA). EQ four-element calibration beads were used to normalise the signal according to the manufacturer’s instructions. CD45+CD3+ T cells were gated for further analysis using X-shift algorithms and clusters were visualised using t-distributed stochastic neighbour embedding (t-SNE). The heavy metal isotopes and antibodies used for Cytometry by Time-Of-Flight (cyTOF) are listed in Supplementary Table 1.
Single-cell RNA-seq and data processing
Tumours extracted from the mice bearing HCC were excised and mechanically dissociated into individual cells using a Tumour Dissociation Kit (130-096-730, Miltenyi, USA). These individual cells were then collected using the 10 × Genomics Chromium Single Cell 5′ Solution. RNA-seq libraries were prepared according to the manufacturer’s protocol (10 × Genomics). These libraries were subjected to high-throughput sequencing using an Illumina HiSeq X Ten PE150 platform. Single-cell RNA-seq data from each experiment were processed using a cell ranger count (10 × Genomics CellRanger [v3.1.0]), with reference to the mouse genome GRCm38 (mm10). The digital gene expression matrices were analysed in R (v4.1.2) with the assistance of the Seurat (v4.0.0) package. DoubletFinder software was used to identify and eliminate any potential doublets based on the proximity of each cell to artificial doublets. Cells exhibiting the expression of more than 10,000 genes or fewer than 200 genes in UMI counts, along with those displaying over 10% of mitochondrial gene expression, were excluded from the analysis. Subsequently, the filtered data underwent log normalisation and scaling to mitigate any cell-to-cell variations attributable to the UMI counts and percentage of mitochondrial reads.
mRNA stability assay
The macrophages were transfected with Nat10-overexpression or empty plasmid for 24 h and thereafter incubated with actinomycin D (5 μg/mL) for 0, 6 or 10 h followed by RNA extraction. The half-life of the PDL1 mRNAs was analysed using qRT-PCR.
Tyramide signal amplification (TSA) multiplex IHC assay
The multiplex IHC assay was performed as follows: after heating, antigen retrieval, and rinsing, the slides were incubated with 3% hydrogen peroxide to remove endogenous peroxidase. Following multiplex staining, signal amplification, and fluorescence signal capture, the slides were placed in a microwave oven to remove the antibody complex. Subsequently, a second round of multiplex staining was performed. Finally, DAPI staining was completed, and image acquisition was performed using a Zeiss Axio Imager Z2 Microscope System. The StrataQuest software (version 7.0.1, TissueGnostics GmbH, Vienna, Austria) was applied to quantify cell density, nucleus area per cell, area per cell, and expression per cell. The antibodies are as follows: TCF1 (C63D9, CST, USA), CD11b (ab52478, Abcam, UK), PD1 (D4W2J, CST, USA), CD25 (ab231441, Abcam, USA), TIM3 (D5D5R, CST, USA), CD3 (ab16669, Abcam, USA), CD4 (ab133626, Abcam, USA), CD8 (ab237709, Abcam, USA), and ac4C (ab252215, Abcam, UK).
Statistics analysis
For the molecular and cellular studies, all the experiments were repeated in at least triplicates. For the animal experiments, a sample size of ≥six mice per group was used. Mice that died within 3 days after being injected the tumour cells were excluded. The animals were randomised according to their body weight before the administration of drug or control to avoid any bias and analysis was blinded. All the statistical tests, including the Mann–Whitney U test, Pearson’s χ2 test, Log-rank test, Spearman rank correlation coefficient, Student’s t-test, and analysis of variance (ANOVA) were performed using the GraphPad Prism 9.0 software (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was determined using unpaired two-tailed Student’s t-tests between two groups. One-way ANOVA was used to multiply the groups. Differences were considered statistically significant at p < 0.05.
Results
Inhibition of ac4C modification promotes myeloid cell and B cell infiltrations in the HCC tumour microenvironment
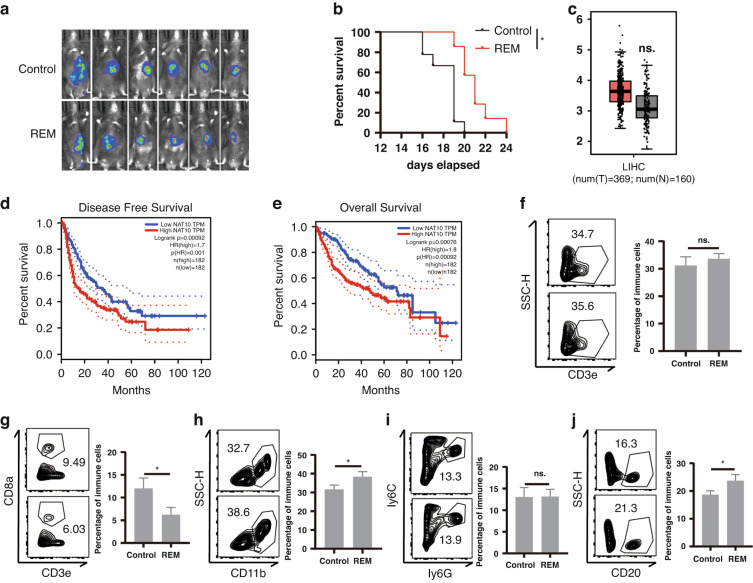
To investigate the role of ac4C modification in HCC development, we constructed a Hepa1–6-Luc orthotopic transplantation mouse HCC model (Luc-HCC) and evaluated the effect of remodelin hydrobromide (REM), a specific inhibitor of N-acetyltransferase 10 (NAT10) which catalyses the formation of ac4C [16, 17]. REM slightly slowed down the tumour progression and increased mouse survival (Fig. 1a, b). In vitro experiments showed that REM only delayed cell growth but had no prominent role in the apoptosis of the HCC cells (Supplementary Fig. 1a, b). Analysis of The Cancer Genome Atlas (TCGA) database indicated that the NAT10 expression was only slightly higher in the HCC tissues than in the adjacent tissues (Fig. 1c). However, the HCC patients with lower NAT10 expression had relatively longer disease-free survival and overall survival (Fig. 1d, e). Furthermore, we explored whether REM affects immune cell infiltration in the HCC tumour microenvironment (TME), and found that REM usage reduced CD8 T cell infiltration (Fig. 1g), increased myeloid cell (CD11b+, Fig. 1h) and B cell (CD20+, Fig. 1j) infiltrations, under the application of gating in flow cytometry (Supplementary Fig. 1c), but had no effect on the total T cell (Fig. 1f) and neutrophil infiltrations (ly6G+ly6C-, Fig. 1i). Next, we used NSG mice to construct a subcutaneous tumour model and found that REM inhibited tumour progression in the absence of most immune cells such as T, B, and natural killer (NK) cells (Supplementary Fig. 1e). Subsequently, through the analysis of single-cell data from the GEO database of humans [18], we found that the patients with high infiltration of NAT10+ myeloid cells had a better prognosis, whereas no such difference was observed in the T cells (Supplementary Fig. 1f, g). Collectively, these results indicate that the inhibition of ac4C modification promotes the infiltration of myeloid and B cells, and systemic usage prolongs the survival of mice with HCC.
Fig. 1. Inhibition of ac4C modification promotes myeloid cell and B cell infiltrations and accelerates HCC development.
a, b An orthotopic transplantation mice model with a Luc reporter-carrying Hepa1–6 HCC cell line was constructed, and REM was intraperitoneally injected every 2 days. In vivo, imaging was performed on the 14th day (a, n = 6, Control; n = 6, REM) as well as survival analysis (b). c Boxplot of NAT10 expression in HCC tumours and the adjacent tissues in the TCGA database. d, e Kaplan–Meier analysis from the TCGA database of overall survival (d, OS) and disease-free survival (e, DFS) for patients with a high or low expression of NAT10. f–j The Luc-Hepa1–6 model was constructed, and REM was intraperitoneally injected every 2 days. The percentage of CD3+ T cells in CD45+ T cells (f), CD3+ CD8+ T cells in CD45+ T cells (g), CD11b+ cells in CD45+ cells (h), Ly6G+Ly6C- cells in CD45+ cells (i), and CD20+ cells in CD45+ cells (j) were analysed using flow cytometry on the 14th day (n = 4, Control; n = 4, REM). The graph on the right shows the quantified results. *p < 0.05.
Ac4c modification in myeloid cells is required for effective immunotherapy
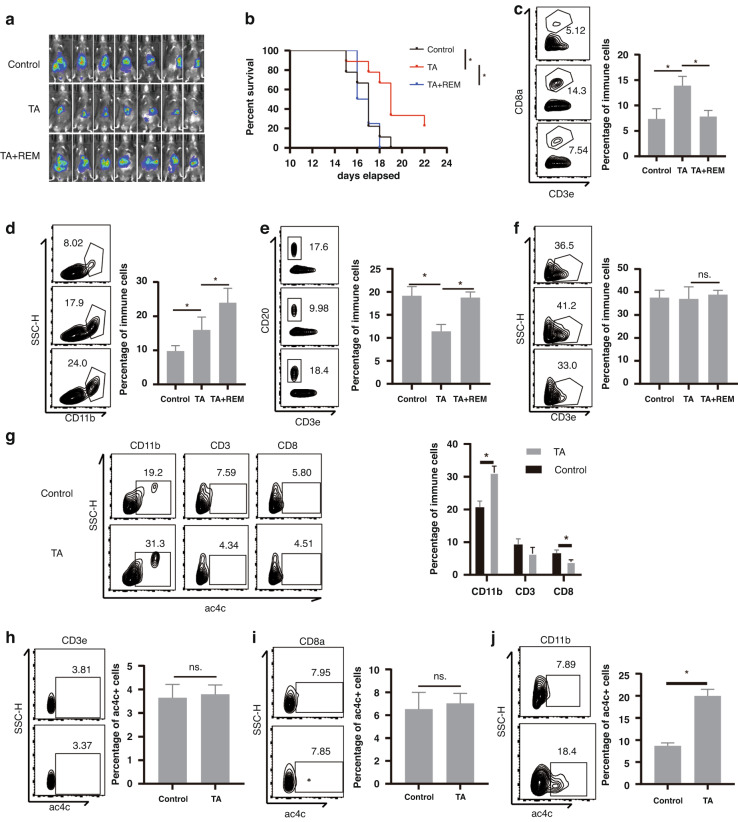
Since REM affects CD8 T cell infiltration in HCC, we investigated whether REM influences immunotherapy (TA, Tecentriq + Apatinib) in HCC. As expected, REM treatment reversed the effect of immunotherapy by promoting tumour development (Fig. 2a) and shortening survival (Fig. 2b). Flow cytometry analysis of the tumour-infiltrating immunocytes indicated that REM cancelled immunotherapy-induced CD8 T cell infiltration (Fig. 2c) and increased myeloid cell (Fig. 2d) and B cell infiltration (Fig. 2e) but had no prominent effect on T cell infiltration (Fig. 2f). We then evaluated the cellular composition of the ac4C+ cells and found that CD11b+ cells occupied the majority of the ac4C+ cells and immunotherapy induced this subset (Fig. 2g), whereas the ac4C+ cell ratio was reduced during tumour progression (Supplementary Fig. 1d). We also analysed the ac4C+ cell ratio in the respective cell types and obtained similar results (Fig. 2h–j). To verify our findings, we also conducted Cytometry by Time-Of-Flight (cyTOF) on the tumour-infiltrating lymphocytes (TILs) isolated from the three groups (C: control group, T: immunotherapy group, TR: immunotherapy plus REM group, n = 3, per group) (Supplementary Fig. 2a). Following the unsupervised cluster analysis using the two-dimensional t-distributed stochastic neighbour embedding (t-SNE) algorithm, we obtained 7 cell clusters with unique expression features, including B cells, dendritic cells (DCs), NK cells, CD4 T cells, CD8 T cells, neutrophils, and macrophages (Supplementary Fig. 2b, c). We observed significant changes in the immune cell subgroups among the three groups (Supplementary Fig. 2d, e). Among the 39 cell populations, we focused on the following five populations with significant differences: clusters C18 (KLRG1+ NKs), C20 (CD86+ myeloid cells), C24 (PDL1+ myeloid cells), C28 (Exhausted Th1 cells), and C35 (Effector T cells) (Supplementary Fig. 2f, g). Following the use of REM, the CD86+ myeloid cells and effector T cells were significantly reduced, with a significant increase in the PDL1+ myeloid cells. Next, we utilised a macrophage-purging agent, Clodronate Liposomes (CL), to eliminate the myeloid cells caused by REM. We discovered that in the case of immunotherapy plus REM, the efficacy of the immunotherapy was partially recovered and the survival of the mice was prolonged with the use of CL (Supplementary Fig. 2h, i). Simultaneously, cytotoxic T lymphocytes (CTLs) function also recovered (Supplementary Fig. 2j, k) and a reanalysis of published human HCC single-cell sequences [18] revealed that the infiltration of CD8+ T cells was positively correlated with the NAT10+ myeloid cells (Supplementary Fig. 2l). Overall, these results indicate that ac4c modification of myeloid cells increases CTLs infiltration, which is essential for effective immunotherapy.
Fig. 2. NAT10 expression in myeloid cells is required for effective immunotherapy.
a–f A Luc reporter-carrying Hepa1–6 HCC cell line orthotopic transplantation mice model was constructed and TA (Tecentriq + Apatinib) immunotherapy with or without REM treatment was injected every 2 days. In vivo imaging was performed on the 14th day (a, n = 7, Control; n = 7, TA; n = 7, TA+REM) as well as survival analysis (b). Tumour-infiltrated immune cells were isolated, and thereafter the percentage of CD8+ T cells in CD3+ T cells (c), percentage of CD11b+ cells in CD45+ cells (d), percentage of CD20+ cells in CD45+ cells (e), and percentage of CD3+ T cells in CD45+ cells (f) were detected using flow cytometry on the 7th day. The right panel shows statistical analysis. g–j A Luc reporter-carrying Hepa1–6 HCC cell line orthotopic transplantation mice model was constructed and T+A was intraperitoneally injected every 2 days. The percentage of CD11b+ cells, CD3+ T cells, and CD8+ T cells were detected by flow cytometry in the ac4C+ cells isolated from mouse splenocytes (g). Representative expression of ac4C in the CD3+ T cells (h), CD8+ T cells (i), and CD11b+ cells (j) were analysed by flow cytometry (n = 4, Control; n = 4, TA; n = 4, TA+REM). The graph on the right shows the statistical analysis. *p < 0.05.
Inhibition of ac4C modification promotes MDSC infiltration in the HCC TME
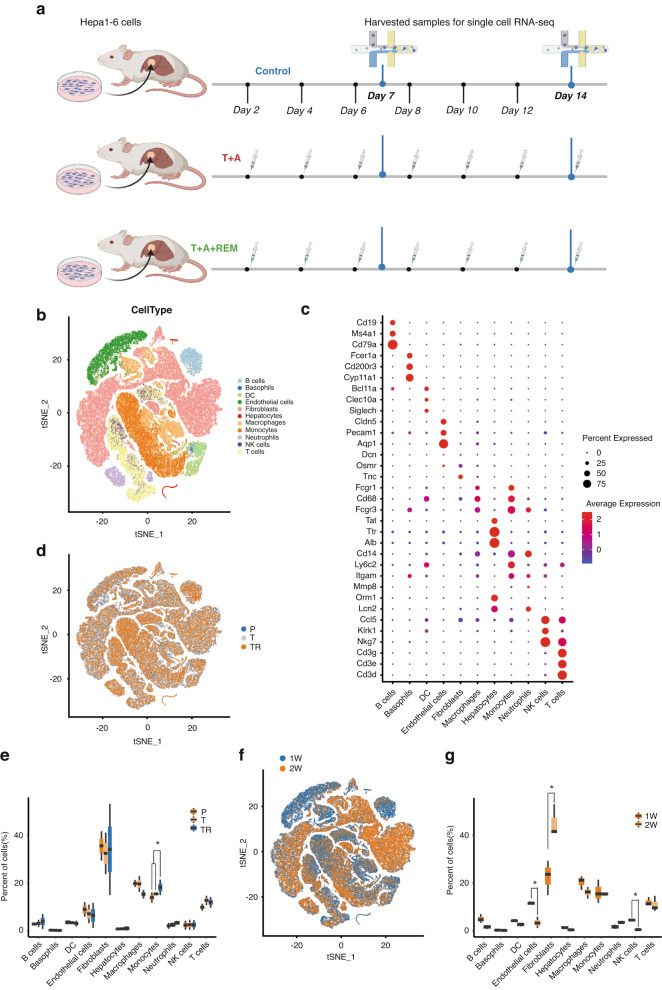
To resolve the patterns of TILs in HCC, we performed scRNA-seq on the TILs isolated from the three groups (P: control group, T: immunotherapy group, TR: immunotherapy plus REM group; n = 2 per group) at two timepoints (1 and 2 weeks) (Fig. 3a). Following an unsupervised cluster analysis using the two-dimensional t-distributed stochastic neighbour embedding (t-SNE) algorithm, we obtained 11 cell clusters with unique expression features, including B cells (Cd19, Ms4a1, Cd79a), basophils (Fcer1a, Cd200r3, Cyp11a1), dendritic cells (DCs) (Clec10a, Siglech), endothelial cells (Pecam1, Cldn5), fibroblasts (Tnc), myeloid cells (macrophages (Cd68, Fcgr1, Fcgr3), monocytes(Cd14, Ly6c2, Itgam)) neutrophils (Lcn2 Fcgr3), natural killer (NK) cells (Nkg7, Klrk1), T cells (Cd3d, Cd3e, Cd3g) and hepatocytes/tumour cells (Alb, Tat, Ttr) (Fig. 3b, c and Supplementary Fig. 3a, b). The cell cluster ratio analysis revealed that REM treatment induced monocyte infiltration but had no effect on T cell infiltration (Fig. 3d, e), compared to the immunotherapy group (T), which was consistent with our previous findings. Interestingly, the timepoint analysis indicated that the endothelial cell ratio was reduced, while the fibroblast ratio was prominently increased, compared to the 1-week group (Fig. 3f, g). Next, we validated the increase in fibroblasts during the progression of HCC using immunohistochemistry with Masson’s and Sirius Red staining (Supplementary Fig. 3c, d). The results suggested that the TME of HCC is reconstructed by fibroblasts in the 2nd week when the tumour cannot be controlled at an early stage.
Fig. 3. Inhibition of ac4C modification promotes MDSC infiltration in the TME of HCC.
a Schematic diagram of scRNA-seq experiments in the Hepa1–6 orthotopic transplantation model with TA immunotherapy combined with REM. b t-Distributed Stochastic Neighbor Embedding (t-SNE) plot of 10× genomics-based single cells showing 11 major cell types by manual annotation. c Bubble heatmap showing the expression levels of selected marker genes for each cell type. d t-SNE plot showing the origins of cells in the P (Control group), T (TA group), or TR (TA+REM group) groups. e Boxplot showing the proportion of major cell types from the samples with different treatments. f t-SNE plot showing the cells from the 1- or 2-week groups. g Boxplot showing the proportion of major cell types from the 1- or 2-week samples. B cell B lymphocyte, DC dendritic cell, NK cell natural killer cell, T cell T lymphocyte. *p < 0.05.
NAT10 induces M1 macrophages and reduces MDSCs infiltration
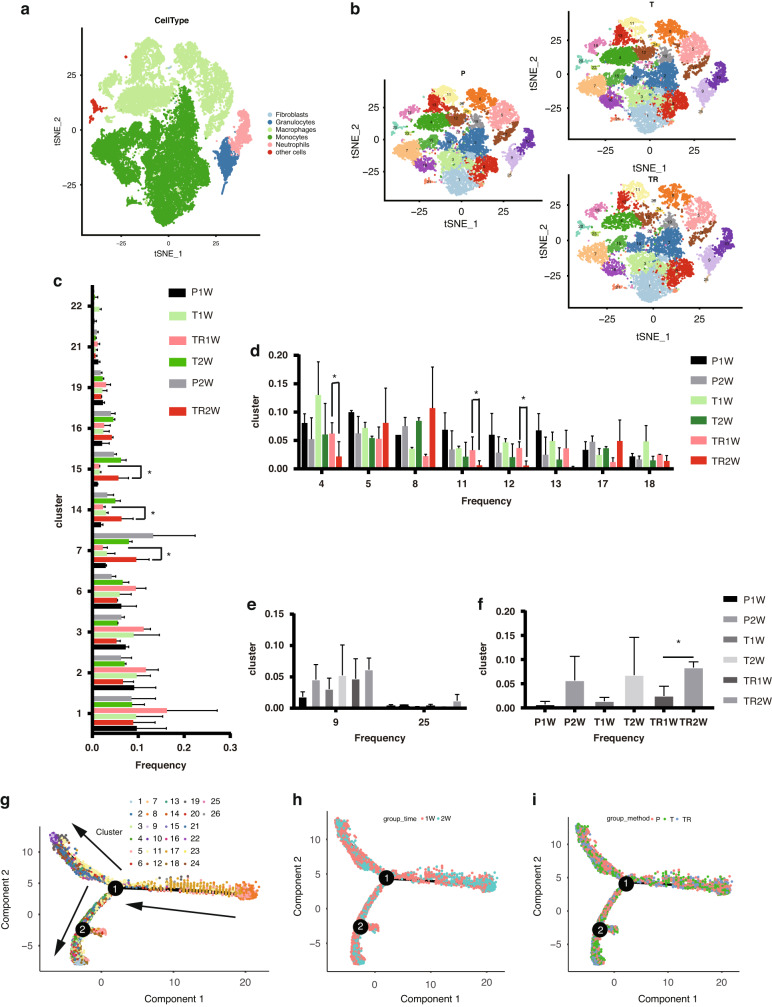
Given that MDSCs are the major target cells of REM, we re-clustered the total myeloid cells into six clusters, including fibroblast-like (expressed fibroblast marker) granulocytes, macrophages, neutrophils, and other cells (in a poorly differentiated state) (Fig. 4a, b and Supplementary Fig. 4a). The monocyte sub-cluster ratio analysis indicated that REM promoted the infiltration of clusters 7, 14, and 15 (Fig. 4c). KEGG analysis showed that cluster 7 (chil3+), cluster 14 (Cd74+), and cluster 15 (Ace+) were all enriched in the immunosuppressive pathways such as Fc gamma R-mediated phagocytosis (Supplementary Fig. 4b–g). Macrophage sub-cluster ratio analysis showed that REM reduced the infiltration of clusters 4, 11, and 12 (Fig. 4d). KEGG analysis revealed that clusters 4 (Folr2+), 11 (Fabp4+), and 12 (Marco+) were enriched in the inflammation-related pathways (data not shown) [19, 20]. These results indicated that REM induces MDSCs (clusters 7, 14, and 15) and reduces M1 macrophages (clusters 4, 11, and 12). Granulocyte and neutrophil cluster analyses showed that neutrophils (clusters 9 and 25) increased during the 2nd week (Fig. 4e, f). Finally, pseudotime analysis showed that the monocyte clusters (1, 2, 3, 14, 15) were less differentiated than cluster 7 and the macrophage clusters (5, 8, 17) (Fig. 4g). These results indicated that the TME can trigger the transformation of both poorly and highly differentiated myeloid cells into MDSCs. In addition, the TME in the early stage (1 week) had more poorly differentiated myeloid cells (Fig. 4h) than in the 2-week group. REM did not significantly affect the differentiation of MDSCs or macrophages (Fig. 4i). Moreover, we re-clustered the CD8+ T cells and found that clusters 1 and 7 were sharply reduced in the TR group compared to those in the T group (Supplementary Fig. 5a). Marker gene analysis showed that cluster 1 comprised IFNG+ effector cells (Supplementary Fig. 5b) and cluster 7 comprised TOX + TIGIT + exhausted T cells (Supplementary Fig. 5c), indicating that REM affects the effector function of CD8+ T cells.
Fig. 4. NAT10 induces M1 macrophages and reduces MDSCs.
a t-SNE plot showing the total clusters of myeloid cells. b t-SNE plot showing the total myeloid cells from the different treatments. c Boxplot showing the proportion of clusters (1, 2, 3, 6, 7, 14, 15, 16, 19, 21, 22) from the samples with different treatments. d Boxplot showing the proportion of clusters (4, 5, 8, 11, 12, 13, 17, 18) from the samples with different treatments. e Boxplot showing the proportion of granulocyte clusters (9, 25) from the samples with different treatments. f Boxplot showing the proportion of neutrophil clusters (9, 25) from the samples with different treatments. g–i Pseudotime trajectory analysis of all types of myeloid cells grouped by cluster (g), time (h) and treatment (i). *p < 0.05.
NAT10 inhibits PDL1 expression in myeloid cells and enhances the killing of CTL-mediated tumour cells
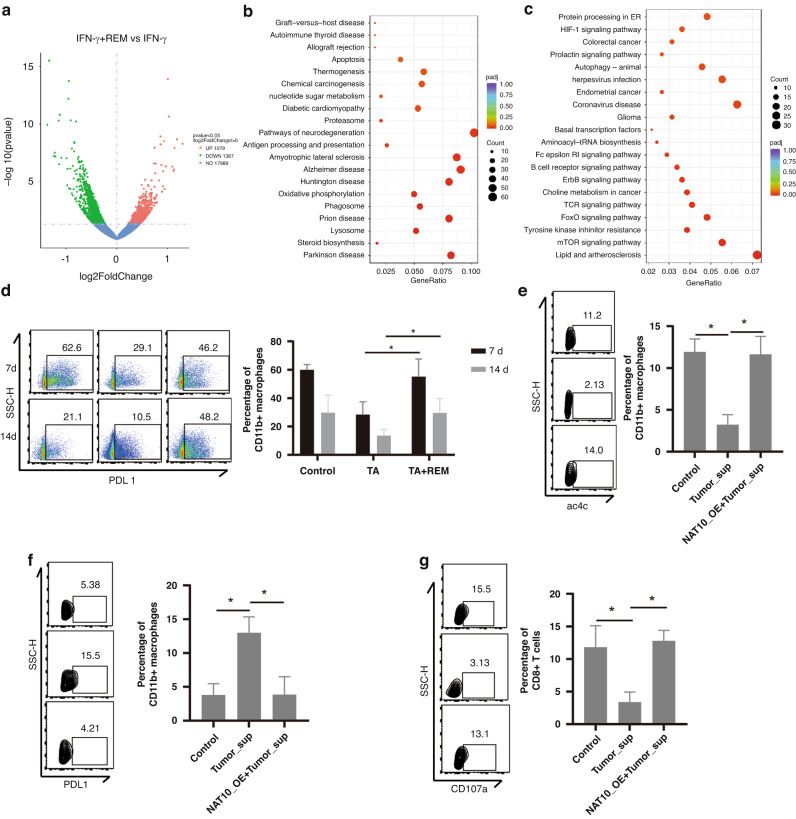
To explore the underlying mechanisms of REM-induced MDSCs differentiation, we performed an RNA-seq analysis of REM-treated bone marrow-derived macrophages (BMDM). In the REM-treated BMDM with IFN-γ stimulation, 1079 upregulated and 1367 downregulated genes were identified (Fig. 5a). KEGG analysis showed that the downregulated genes were mostly enriched in apoptosis, oxidative phosphorylation, antigen processing, and presentation (Fig. 5b), and the upregulated genes were mostly enriched in HIF-1 signalling, autophagy, and tyrosine kinase inhibitor resistance (Fig. 5c), indicating that REM treatment promotes BMDM survival, downregulates antigen processing and presentation, and promotes adaptation to low-oxygen conditions. In our study, REM treatment in vivo promoted PDL1 expression in the myeloid cells (CD45+CD11b+) based on the flow cytometry analysis conducted on days 7 and 14 (Fig. 5d). Subsequently, we assessed the expression of PD1 in the B, CD8+ T, and myeloid cells (Supplementary Fig. 6a–f). REM did not significantly alter the expression of PD1 in these cells under immunotherapy on days 7 and 14. To investigate whether this effect depends on the NAT10-mediated ac4C modifications, we transfected BMDM with a NAT10-overexpression plasmid (NAT10-OE) and found NAT10-OE cancelled tumour supernatant-induced ac4C downregulation in the BMDM (Fig. 5e) and PDL1 upregulation (Fig. 5f). As expected, NAT10-OE also reversed the tumour supernatant-induced BMDM-mediated CTL degranulation inhibition (Fig. 5g). Overall, these results indicated that NAT10 inhibition induces PDL1 expression in myeloid cells and attenuates CTL-mediated tumour cell death.
Fig. 5. NAT10 inhibits PDL1 expression in myeloid cells and enhances CTL-mediated tumour cell killing.
a Volcano analysis of differentially expressed genes in the IFN-γ+REM group compared to the IFN-γ group. Red or blue dots indicate upregulated or downregulated genes, respectively. b, c KEGG enrichment analysis of the downregulated (b) and upregulated (c) genes in the REM group compared to the control group. The Luc reporter-carrying Hepa1–6 HCC cell line orthotopic transplantation mice model was constructed and TA immunotherapy with or without REM was administered every 2 days to isolate the infiltrated immune cells. d Percentage of PDL1+ cells in the myeloid cells (CD45+CD11b+ cells) from the HCC orthotopic transplantation mice model treated with TA immunotherapy and/or REM were analysed using flow cytometry on the 7th and 14th day. The graph on the right shows the quantified results. e Representative expression of ac4C in the macrophages was analysed by flow cytometry. f Representative expression of PDL1 in the macrophages was analysed by flow cytometry. g Representative expression of CD107a+ in the CD8+ T cells was analysed by flow cytometry. The graph on the right shows the statistical analysis. *p < 0.05.
Ac4C modification promotes PDL1 mRNA degradation in myeloid cells
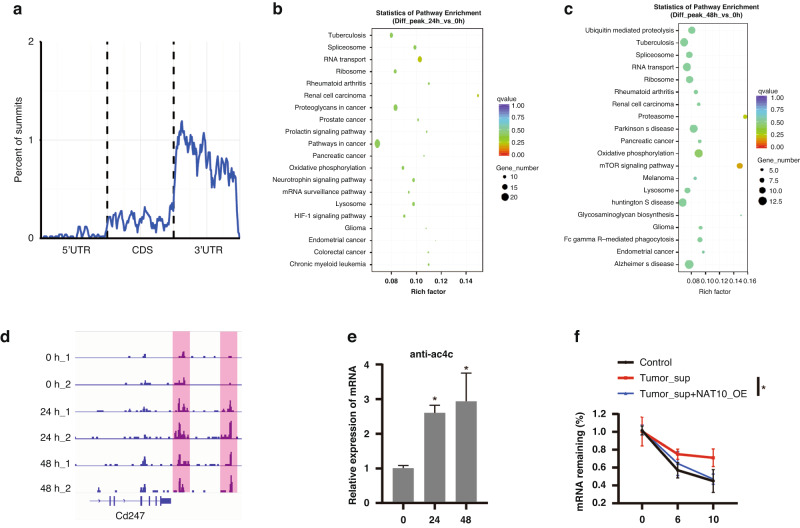
As NAT10 promotes the ac4C modification of mRNA [21], we suspect that ac4C modification occurs in the PDL1 mRNA of myeloid cells. We performed RIP-seq using an ac4C antibody in BMDM with IFN-γ incubation for 0, 24, and 48 h and found that the ac4C peaks were mostly distributed on the 3’ UTR of the gene body (Fig. 6a). Compared to the 0 h group, genes with ac4C peaks in the 24 h group were enriched in oxidative phosphorylation, lysosomes, and HIF-1 signalling (Fig. 6b), which was the same as the result of the 48 h group (Fig. 6c). These results indicate that metabolic pathway-related genes are extensively regulated by NAT10-mediated ac4C modifications. As expected, IFN-γ incubation promoted ac4C modification of the PDL1 mRNA (Fig. 6d); RIP-qPCR confirmed this finding (Fig. 6e). Given that ac4C modifications on PDL1 mRNA suppress PDL1 protein expression, we detected the effect of ac4C modification on the mRNA stability and found that it reversed the tumour supernatant-induced PDL1 mRNA stability (Fig. 6f). Collectively, these results indicate that the ac4C modification inhibits PDL1 expression by promoting mRNA degradation in the myeloid cells.
Fig. 6. Ac4C modification promotes PDL1 mRNA degradation in myeloid cells.
a The distribution of ac4C modification was detected by the ac4C antibody-mediated RIP-seq. b, c KEGG pathway enrichment analysis of the genes with significantly differential-ac4C modification in the 24 h (b) and 48 h (c) group treated with IFN-γ compared to the 0 h group. d The representative peaks of ac4C modified-PDL1 mRNA in the indicated groups. e The relative mRNA level of PDL1 was detected by RIP-qPCR with the ac4C antibody. f The relative PDL1 mRNA remaining ratio was shown in the control, tumour supernatant, and tumour supernatant with NAT10-OE groups by RIP-qPCR. KEGG Kyoto Encyclopedia of Genes and Genomes, RIP RNA Immunoprecipitation. *p < 0.05.
Ac4C+ myeloid cell infiltration results in a better prognosis in HCC patients
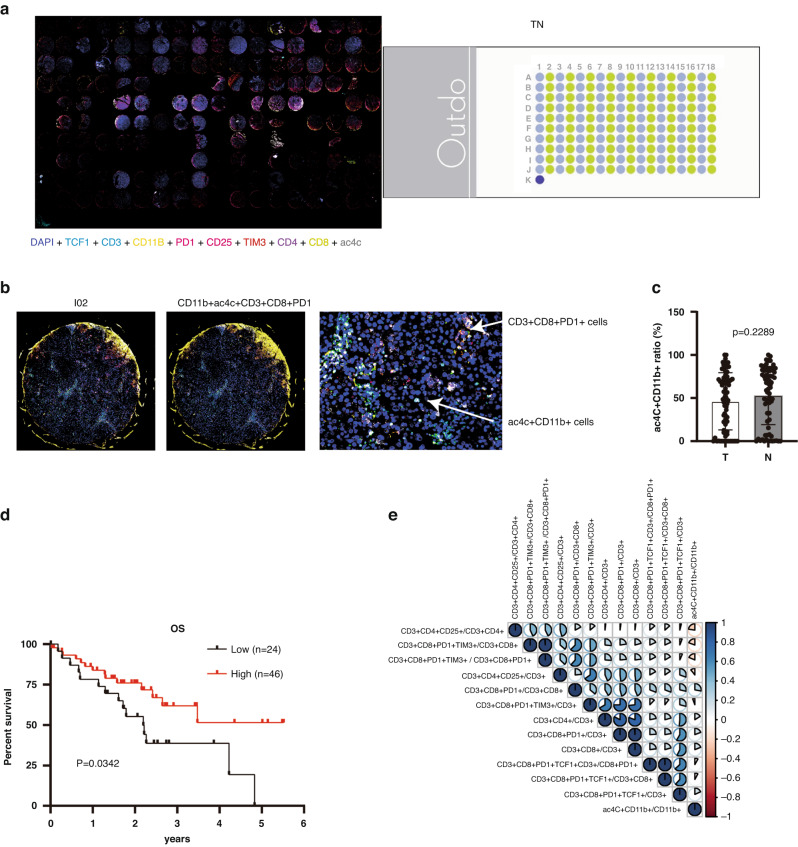
To connect our discovery with the clinic, we performed a TSA IHC analysis on HCC microarrays (Fig. 7a, b) and found that there was no significant difference in the ac4C+CD11b+ cell ratio between the tumour and adjacent tissues (Fig. 7c). However, high infiltration of the ac4C+CD11b+ cell subset indicated a better prognosis (Fig. 7d). Finally, we performed a correlation analysis between the ac4C+CD11b+ cell ratio and T subsets and found that the ac4C+CD11b+ cell ratio was positively correlated with activated CD8 T (PD1+CD3+CD8+) and negatively correlated with the Treg ratio (CD3+CD4+CD25+) and exhausted CD8 T (CD3+CD8+PD1+TIM3+) (Fig. 7e). In conclusion, these results show that ac4C+CD11b+ cells promote CTL infiltration and increase patient survival.
Fig. 7. Ac4C+CD11b+ infiltration relates to better prognosis in HCC patients.
a The images of multiple markers (DAPI, TCF1, CD11b, PD1, CD25, TIM3, CD3, CD4, CD8, and ac4C) were observed on the HCC microarray based on TSA IHC technology. b The images of ac4C+CD11b+ cells and CD3+CD8+PD1+ cells were shown in the I02 patient’s tissue. c The relative ac4c+CD11b+ cell ratio was calculated in both the tumour and adjacent tissues (p > 0.05). d The survival curve was created based on the HCC patients’ information (n = 70) and shows a better overall survival rate (p < 0.05) with high infiltration of ac4C+CD11b+ cells (n = 46). e The correlation analysis among ac4C+CD11b+ cells and T subsets was calculated. TSA tyramide signal amplification, IHC immunohistochemistry.
Discussion
Ineffective immunotherapy for HCC contributes to multiple factors, including tumour antigen evasion, suppressive metabolites, inhibitory cells, and dysfunctional CTLs [22]. Clarifying the mechanisms underlying immunotherapy resistance in HCC is essential to improve its therapeutic effect. In recent years, ac4C modification has received increasing attention in the field of cancer, and ac4C on mRNA is important for increasing and maintaining protein translation fidelity [23, 24]. In the present study, by systematically applying an ac4C modification inhibitor to HCC mouse models, we first explored the role of ac4C modification in HCC immunotherapy and found that it is essential in myeloid cells for the effective immunotherapy of HCC. Mechanistically, the ac4C modification of PDL1 mRNA promotes PDL1 degradation, hinders MDSCs differentiation, and strengthens CTL infiltration.
NAT10, a protein with histone acetylation activity, promotes hTERT transcription to enhance telomerase activity [25], and induces the ac4C modification of tRNA, rRNA, and mRNA [23]. NAT10-mediated mRNA N4-acetylcytidine modification promotes the progression of several cancers, including bladder, colon, and triple-negative breast cancers [26–28]. Consistently, we found that NAT10 is highly expressed in HCC tissues and is negatively associated with disease-free survival and overall survival of HCC patients. Importantly, we demonstrated that NAT10 is essential for the therapeutic effect of immunotherapy in HCC as it suppresses MDSCs infiltration and enhances CTL-mediated tumour cell-killing effects by regulating PDL1 expression. However, some questions remain to be addressed. First, the scRNA-seq analysis indicated that the inhibition of NAT10 by REM treatment not only decreased the number of effectors and exhausted CD8+ T cells in HCC, but also increased the number of naïve CD8+ T cells (cluster 9). Exhausted cells are cells under chronic antigen stimulation and originate from memory cell precursors [29], which indicates that REM reduces the transition from memory cells to exhausted cells [30]. Naïve CD8+ T cells accrued in the TME, implying that REM reduces the transition of naïve CD8+ T cells into effector T cells. Whether these processes are also involved in regulating the differentiation of other immune cells, such as antigen presentation cells (APCs) or Th1 cells, requires further investigation in future studies. However, based on these results, we speculated that ac4C modification also affects the autochthonous steady state of different T cell subsets in the TME of HCC. Second, from the ac4C antibody-mediated RIP-seq analysis, we found that metabolism, apoptosis, and inflammation pathways were enriched in the TA + REM group, which revealed that ac4C modification regulates extensive biological activities in monocytes. Thus, the activation of PDL1 signalling may be a regulatory factor for MDSCs to suppress CTL cytotoxicity and effector function. In future studies, multiomics detection, including proteomics, metabolomics, and translationomics, may contribute to clarifying the comprehensive role of ac4C modification in the biological function and differentiation state of monocytes. For further investigations, Lyz2-specific or CD4-specific NAT10 knockout mouse strains need to be constructed to clarify the detailed role of ac4C modifications in different immune cell subsets in the TME of HCC.
In conclusion, our group was the first to explore the potential effect of ac4C modification in HCC immunotherapy and revealed that ac4C modification in myeloid cells suppresses PDL1 expression and promotes the effectiveness of immunotherapy. This study highlighted the role of ac4C in HCC immunotherapy and deepened our understanding of immune cell differentiation in the TME during immunotherapy.
Supplementary information
Author contributions
QW, SW, and XX conceived and supervised the study. NX, JYZ, YYC, HC, ZSZ, ZXL, and XYW performed all the experiments. SW, RYS, DL, and NX analysed the data. NX and SW wrote the manuscript. SSZ and XX reviewed the manuscript.
Funding
The National Natural Science Foundation of China (Nos. 92159202 and 82273177). The Key Research and Development Plan of the Zhejiang Province (Nos. 2019C03050 and 2021C03118).
Data availability
The study data are available from the GEO database and are publicly available as of the date of publication (RNA-sequence (GSE220196), single-cell RNA-sequence (GSE244613) and ac4C-RIP-sequence (GSE227526)).
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All the individuals signed informed consent forms for the use of their clinical specimens in the present study according to the principles expressed in the Declaration of Helsinki and approved by the Institutional Research Ethics Committees of the Affiliated Hangzhou First People’s Hospital and the First Affiliated Hospital, Zhejiang University School of Medicine.
Consent for publication
All authors have agreed to publish this manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Nan Xu, Jianyong Zhuo, Yiyuan Chen, Renyi Su, Huan Chen.
Contributor Information
Xiao Xu, Email: zjxu@zju.edu.cn.
Shuai Wang, Email: wmmkao2@163.com.
Qiang Wei, Email: zjuwq@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02510-9.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Cao M, Ding C, Xia C, Li H, Sun D, He S, et al. Attributable deaths of liver cancer in China. Chin J Cancer Res. 2021;33:480–9. doi: 10.21147/j.issn.1000-9604.2021.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan AA, Liu ZK, Xu X. Recent advances in immunotherapy for hepatocellular carcinoma. Hepatob Pancreat Dis Int. 2021;20:511–20. doi: 10.1016/j.hbpd.2021.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345–62. doi: 10.1016/S0140-6736(22)01200-4. [DOI] [PubMed] [Google Scholar]
- 5.Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382:1894–905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 6.Chen CT, Feng YH, Yen CJ, Chen SC, Lin YT, Lu LC, et al. Prognosis and treatment pattern of advanced hepatocellular carcinoma after failure of first-line atezolizumab and bevacizumab treatment. Hepatol Int. 2022;16:1199–207. doi: 10.1007/s12072-022-10392-x. [DOI] [PubMed] [Google Scholar]
- 7.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–73. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 8.Xue M, Wu Y, Zhu B, Zou X, Fan W, Li J. Advanced hepatocellular carcinoma treated by transcatheter arterial chemoembolization with drug-eluting beads plus lenvatinib versus sorafenib, a propensity score matching retrospective study. Am J Cancer Res. 2021;11:6107–18. [PMC free article] [PubMed] [Google Scholar]
- 9.Lawal G, Xiao Y, Rahnemai-Azar AA, Tsilimigras DI, Kuang M, Bakopoulos A, et al. The immunology of hepatocellular carcinoma. Vaccines. 2021;9:1184. doi: 10.3390/vaccines9101184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586–93. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- 11.Veglia F, Perego M, Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–19. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22. doi: 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- 13.Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22:119–31. doi: 10.1038/s41576-020-00295-8. [DOI] [PubMed] [Google Scholar]
- 14.Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Zhang W, Shui Y, Li P, Tian Z, Duan S, et al. Implications of m6A-associated snRNAs in the prognosis and immunotherapeutic responses of hepatocellular carcinoma. Front Immunol. 2022;13:1001506. doi: 10.3389/fimmu.2022.1001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larrieu D, Rodriguez R, Britton S. [Chemical inhibition of NAT10 corrects defects of laminopathic cells] Med Sci. 2014;30:745–7. doi: 10.1051/medsci/20143008010. [DOI] [PubMed] [Google Scholar]
- 17.Ikeuchi Y, Kitahara K, Suzuki T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008;27:2194–203. doi: 10.1038/emboj.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng C, Zheng L, Yoo JK, Guo H, Zhang Y, Guo X, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–56.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 19.Nalio Ramos R, Missolo-Koussou Y, Gerber-Ferder Y, Bromley CP, Bugatti M, Nunez NG, et al. Tissue-resident FOLR2(+) macrophages associate with CD8(+) T cell infiltration in human breast cancer. Cell. 2022;185:1189–207.e1125. doi: 10.1016/j.cell.2022.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Xing Q, Feng Y, Sun H, Yang S, Sun T, Guo X, et al. Scavenger receptor MARCO contributes to macrophage phagocytosis and clearance of tumor cells. Exp Cell Res. 2021;408:112862. doi: 10.1016/j.yexcr.2021.112862. [DOI] [PubMed] [Google Scholar]
- 21.Arango D, Sturgill D, Yang R, Kanai T, Bauer P, Roy J, et al. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol Cell. 2022;82:2912. doi: 10.1016/j.molcel.2022.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Zhang W, Yang W, Zhou M, Liu F. Acquired resistance for immune checkpoint inhibitors in cancer immunotherapy: challenges and prospects. Aging. 2022;14:1048–64. doi: 10.18632/aging.203833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominissini D, Rechavi G. N(4)-acetylation of cytidine in mRNA by NAT10 regulates stability and translation. Cell. 2018;175:1725–7. doi: 10.1016/j.cell.2018.11.037. [DOI] [PubMed] [Google Scholar]
- 24.Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175:1872–86.e1824. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv J, Liu H, Wang Q, Tang Z, Hou L, Zhang B. Molecular cloning of a novel human gene encoding histone acetyltransferase-like protein involved in transcriptional activation of hTERT. Biochem Biophys Res Commun. 2003;311:506–13. doi: 10.1016/j.bbrc.2003.09.235. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Zhang M, Zhang Y, Xie Y, Zou J, Zhong J, et al. NAT10-mediated mRNA N4-acetylcytidine modification promotes bladder cancer progression. Clin Transl Med. 2022;12:e738. doi: 10.1002/ctm2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X, Wang Q, Zhou Y, Zhang D, Geng Y, Hu W, et al. N-acetyltransferase 10 promotes colon cancer progression by inhibiting ferroptosis through N4-acetylation and stabilization of ferroptosis suppressor protein 1 (FSP1) mRNA. Cancer Commun. 2022;42:1347–66. doi: 10.1002/cac2.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Zeng J, Wang J, Yang Z, Gao S, Liu H, et al. Revealing the potential markers of N(4)-acetylcytidine through acRIP-seq in triple-negative breast cancer. Genes. 2022;13:2400. doi: 10.3390/genes13122400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallies A, Zehn D, Utzschneider DT. Precursor exhausted T cells: key to successful immunotherapy? Nat Rev Immunol. 2020;20:128–36. doi: 10.1038/s41577-019-0223-7. [DOI] [PubMed] [Google Scholar]
- 30.Flecken T, Schmidt N, Hild S, Gostick E, Drognitz O, Zeiser R, et al. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology. 2014;59:1415–26. doi: 10.1002/hep.26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The study data are available from the GEO database and are publicly available as of the date of publication (RNA-sequence (GSE220196), single-cell RNA-sequence (GSE244613) and ac4C-RIP-sequence (GSE227526)).