Abstract
The hormone GDF15 is secreted in response to cellular stressors. Metformin elevates circulating levels of GDF15, an action important for the drug’s beneficial effects on body weight. Metformin can also inhibit mammalian respiratory complex I, leading to decreases in ATP:AMP ratio, activation of AMP Kinase (AMPK), and increased GDF15 production. We undertook studies using a range of mice with tissue-specific loss of Gdf15 (namely gut, liver and global deletion) to determine the relative contributions of two classical metformin target tissues, the gut and liver, to the elevation of GDF15 seen with metformin. In addition, we performed comparative studies with another pharmacological agent, the AMP kinase pan-activator, MK-8722. Deletion of Gdf15 from the intestinal epithelium significantly reduced the circulating GDF15 response to oral metformin, whereas deletion of Gdf15 from the liver had no effect. In contrast, deletion of Gdf15 from the liver, but not the gut, markedly reduced circulating GDF15 responses to MK-8722. Further, our data show that, while GDF15 restricts high-fat diet-induced weight gain, the intestinal production of GDF15 is not necessary for this effect. These findings add to the body of evidence implicating the intestinal epithelium in key aspects of the pharmacology of metformin action.
Subject terms: Obesity, Feeding behaviour, Hypothalamus, Endocrine system and metabolic diseases
Introduction
Metformin is the world’s most widely prescribed pharmacotherapy for Type 2 diabetes mellitus (T2DM). In addition to improving glycemic control, metformin causes modest, clinically-relevant weight loss1.
Metformin’s effects on energy balance have been linked to growth differentiation factor 15 (GDF15)2,3. A member of the TGFβ superfamily, GDF15 is a stress-responsive peptide hormone produced in peripheral tissues that acts centrally via its hindbrain-localized receptor, GFRAL-RET, to cause a reduction in food intake and body weight4,5. We and others have identified GDF15 as a mediator of metformin-induced weight loss in mice3,6,7.
Metformin’s actions continue to be the subject of intense scrutiny, with the drug reported to affect multiple biochemical, cellular, and endocrine processes across a variety of tissues8,9. For example, evidence has shown metformin to be an inhibitor of mammalian respiratory complex I8–10. Through its effect on ATP synthesis/oxidative phosphorylation, metformin causes activation of AMP kinase (AMPK), a cellular energy sensor known to increase glucose uptake and fatty acid oxidation10. AMPK has therefore been proposed as one of the mediators of the drug’s glucoregulatory effects11.
In addition, debate remains regarding the tissues and cell types where metformin exerts its actions. Most of the studies attempting to identify a specific biochemical effect of metformin, whether on AMP kinase12, mitochondrial glycerophosphate dehydrogenase13, or other biochemical processes11, have focused on the liver. Other evidence has suggested that, rather than the liver, the intestine might be the most important site of metformin action2. We and others have shown that metformin increases Gdf15 expression in the gastrointestinal (GI) tract and kidney, but not liver, of metformin-treated mice3,4. However, the relative contributions of different tissues to the metformin-stimulated rise in serum GDF15 levels remain uncertain.
Here, we employed a range of genetically-modified mouse models of tissue-specific Gdf15 deletion to identify the main sources of metformin-induced increases in circulating GDF15. We also performed comparative studies utilizing a systemic pan-AMPK activator compound, MK-87225, to assess whether this agent stimulates GDF15 production and secretion through targeting different tissues. Finally, as previous work has suggested that diet affects the metformin-mediated rise in circulatory GDF153,4,6, we performed studies in tissue-specific Gdf15-knockout animals using both a high-fat diet (HFD) and a high-fat high-sucrose (HFHS) diet.
Results
Gut-targeted Gdf15 ablation results in significant reductions of metformin-stimulated circulating GDF15, a response altered by antecedent diet
Gdf15flox/flox-Villin-Cre+ (Gdf15-gut-KO) mice were generated using Cre-Lox recombination under the control of the Villin promoter (Villin-Cre). Cre RNAscope in Gdf15-gut-KO mice showed the expected cytoplasmic RNA staining pattern in enterocytes (Supplementary Fig. A.1B). Additionally, a single spot was present in the nucleus of almost all cells, including lymphoid (Supplementary Fig. A.1B) and renal (Supplementary Fig. A.1E) cells, consistent with hybridization to the genomic Cre locus. To confirm and further investigate potential low level Cre expression in the kidney, DNase pre-treatment was used to remove genomic DNA prior to RNAscope. Nuclear hybridization was absent following DNase, while cytoplasmic mRNA signals were retained. No significant expression of Cre RNA was detected in the kidney following DNase treatment (Supplementary Fig. A.1C,F).
To validate loss of Gdf15 expression in the GI tract, we analyzed the colons and kidneys from sham- and metformin-treated wild-type and Gdf15-gut-KO mice via quantitative PCR (Supplementary Fig. A.2). Gdf15 expression was negligible in colon and kidney tissue from sham-treated, high-fat diet-fed wild-type and Gdf15-gut-KO mice (Supplementary Fig. A.2A,B,E,F). Metformin administration resulted in heightened Gdf15 expression in the colon and kidney of WT animals, whereas in Gdf15-gut-KO mice a strong signal was seen in renal tissue and was absent in the gut (Supplementary Fig. A.2C,D,G,H).
To assess the effect of gut-selective Gdf15 deletion on metformin-induced GDF15 production, as well as the impact of antecedent diet on this response, WT and Gdf15-gut-KO mice were fed either standard chow, a 60% HFD, or an HFHS diet for 4 weeks before receiving an acute dose of metformin.
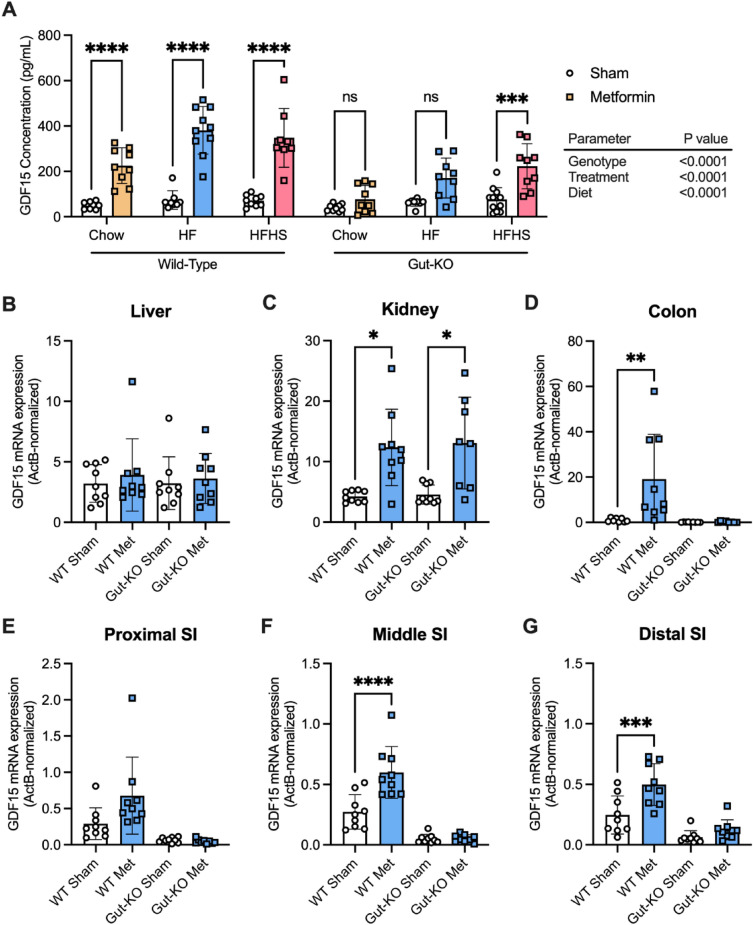
In WT mice, as expected, metformin significantly increased circulating GDF15 levels, with the absolute levels measured being higher in mice fed high-calorie (HF and HFHS) diets (Fig. 1A). In contrast, in mice lacking Gdf15 specifically in the gut, the circulating GDF15 response to metformin was significantly diminished. Specifically, on standard chow, the levels achieved in Gdf15-gut-KO animals were ~ 34% of those seen in WT animals, and on HFD, the metformin stimulated GDF15 rise in Gdf15-gut-KO mice was only ~ 40% of that of WT mice. Interestingly, the HFHS diet was the only dietary paradigm in which metformin treatment elicited a significant rise in circulating GDF15 in Gdf15-gut-KO mice, although, as with all previous dietary manipulations, there was a difference in the magnitude of the rise in GDF15 levels between WT and Gdf15-gut-KO animals.
Figure 1.
Gut-targeted Gdf15 ablation results in significant reductions of acute metformin-stimulated circulating GDF15, a response altered by antecedent diet. (A) Serum GDF15 levels (mean ± SEM) in 12-week-old wild-type and Gdf15-gut-KO mice fed either a standard chow (Chow), 60% high-fat (HF), or high-fat high-sucrose (HFHS) diet for 4 weeks followed by a single oral dose of water (sham) or 600 mg/kg metformin, n = 9/group, and ns = non-significant, ***p < 0.001, and ****p < 0.0001 as determined by three-way ANOVA. (B–G) Gdf15 mRNA expression (normalized to expression levels of actin B) in the (B) liver, (C) kidney, (D) colon, (E) proximal small intestine, (F) middle small intestine, and (G) distal small intestine from 60% high-fat diet-fed wild-type and Gdf15-gut-KO mice 6 h after receiving a single oral dose of water (Sham) or 600 mg/kg metformin (Met). n = 9/group, mean ± SEM, and *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 as determined by two-way ANOVA.
qPCR was performed on isolated kidney, liver, and sectioned small intestinal and colonic tissues from all three dietary groups. In WT animals fed an HFD, in keeping with previous results3, significant increases in Gdf15 mRNA were observed following metformin treatment in the kidney, colon, and small intestine. There was no such increase in the liver (Fig. 1B-G). The results of chow-fed (Supplementary Fig. A.3) and HFHS-fed (Supplementary Fig. A.4) tissue panels emulated the Gdf15 expression pattern in HFD-fed mice, and unchanged hepatic expression of Gdf15 post-metformin administration was displayed in both WT and Gdf15-gut-KO animals.
To further characterize the global and gut-specific Gdf15 knockout models, we measured circulating levels of GDF15 and GLP-1 following a lower dose (300 mg/kg) of metformin comparable to previous studies3,6. Compared to wild-type mice, there was no significant rise in GDF15 in Gdf15-gut-KO animals (Supplementary Fig. A.5A) while there was a significant rise in GLP-1 observed across all genotypes (Supplementary Fig. A.5B). Similarly, the response in PYY to an acute higher dose (600 mg/kg) of metformin was identical across genotypes (Supplementary Fig. A.5C).
Loss of liver Gdf15 does not impact the acute metformin-stimulated rise in circulatory GDF15
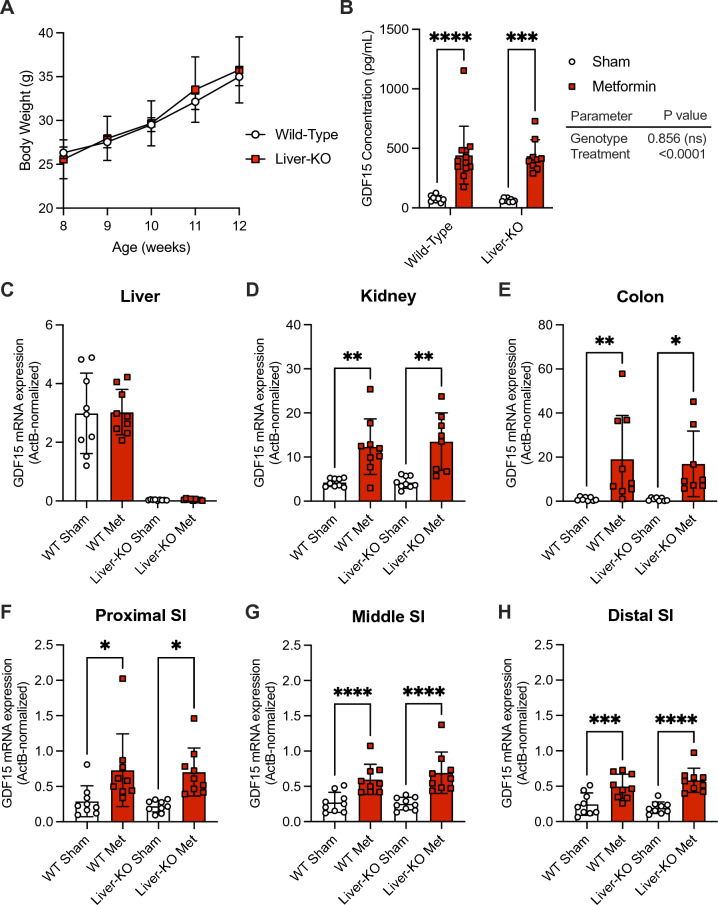
We used a Gdf15-liver-KO mouse model (Gdf15flox/flox-Albumin-Cre+) to explore the liver’s contribution to the GDF15 rise post-metformin. Gdf15-liver-KO mice displayed similar trends in body weight gain relative to WT mice when fed an HFD for 4 weeks (Fig. 2A). Loss of hepatic Gdf15 had no impact on response to acute metformin, with an identical rise in GDF15 seen in WT and Gdf15-liver-KO mice (Fig. 2B). Further, Gdf15 expression profiles in the kidney and the gut were identical in both genotypes (Fig. 2C-H).
Figure 2.
Loss of liver Gdf15 does not impact the acute metformin-stimulated rise in circulatory GDF15. (A) Body weight of 12-week-old wild-type (n = 17) and Gdf15-liver-KO (n = 10) mice on a 60% high-fat diet for 4 weeks beginning at 8 weeks of age, mean ± SEM. (B) Serum GDF15 levels (mean ± SEM) in 12-week-old wild-type and Gdf15-liver-KO mice fed a 60% high-fat diet for 4 weeks followed by a single oral dose of water (sham) or 600 mg/kg metformin, n = 9/group, and ns = non-significant, ***p < 0.001, and ****p < 0.0001 as determined by two-way ANOVA. (C–H) Gdf15 mRNA expression (normalized to expression levels of actin B) in the (C) liver, (D) kidney, (E) colon, (F) proximal small intestine, (G) middle small intestine, and (H) distal small intestine from 60% high-fat diet-fed wild-type and Gdf15-liver-KO mice 6 h after receiving a single oral dose of water (Sham) or 600 mg/kg metformin (Met). n = 9/group, mean ± SEM, and *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 as determined by two-way ANOVA.
AMPK activator treatment and GDF15
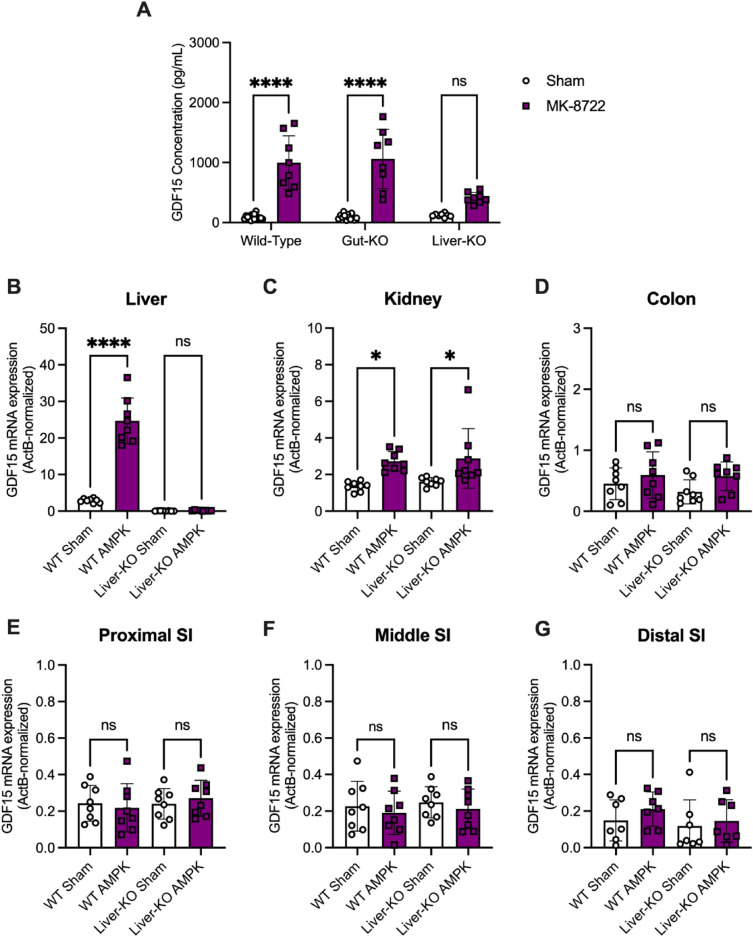
Having identified the gut as a major source of metformin-stimulated GDF15 production, we sought to assess whether other pharmacologic agents of GDF15 induction act through different organs. AMPK activation has been shown to elevate circulatory GDF15 levels through an increase in hepatic Gdf15 expression7, so we administered an AMPK activator, MK-8722, to our tissue-specific Gdf15-knockout mouse models (Fig. 3).
Figure 3.
The liver is a major contributor to the increase in circulating GDF15 in response to oral administration of an AMPK activator. (A) Serum GDF15 levels (mean ± SEM) in 20-week-old wild-type and Gdf15-liver-KO mice fed a 60% high-fat diet for 4 weeks followed by a single oral dose of water (sham) or 30 mg/kg MK-8722, n = 8/group, mean ± SEM, and ns = non-significant and ****p < 0.0001 as determined by two-way ANOVA. (B–G) Gdf15 mRNA expression (normalized to expression levels of actin B) in the (B) liver, (C) kidney, (D) colon, (E) proximal small intestine, (F) middle small intestine, and (G) distal small intestine from high-fat diet-fed wild-type and Gdf15-liver-KO mice 4 h after receiving a single oral dose of water (Sham) or 30 mg/kg MK-8722. n = 8/group, mean ± SEM, and ns = non-significant, *p < 0.05, and ****p < 0.0001 as determined by two-way ANOVA.
Although MK-8722 treatment increased circulating GDF15 ~ 10.7-fold in WT and Gdf15-gut-KO mice, Gdf15-liver-KO animals displayed a diminished (~ 3.4-fold) rise in GDF15 (Fig. 3A). In WT mice, significant increases in Gdf15 mRNA were observed following MK-8722 treatment in the liver (Fig. 3B) and kidney (Fig. 3C), while Gdf15 expression throughout the small intestine and colon remained unaffected by MK-8722 administration (Fig. 3D–G) independent of genotype. Together, these data identify the liver as the major contributor to the rise in serum GDF15 levels elicited by the AMPK activator, MK-8722, with the kidney also contributing to this response.
The gut does not contribute significantly to the protective effect of GDF15 on weight gain in response to a high fat diet
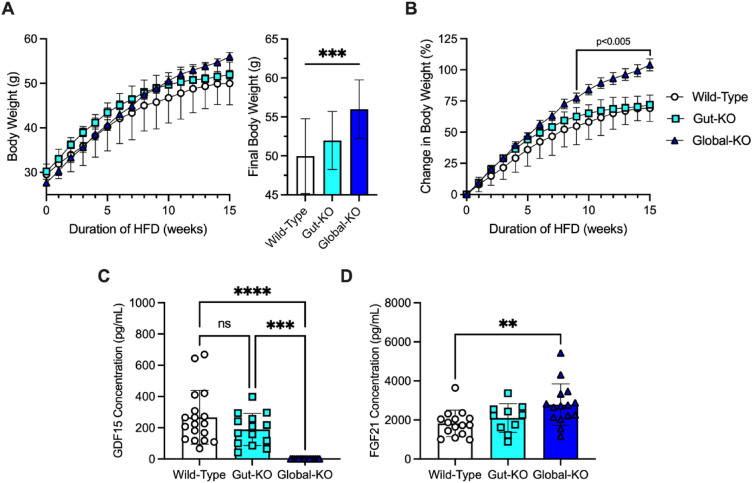
We next characterized the effects of gut-specific Gdf15 ablation on diet-induced body weight gain in WT, Gdf15-gut-KO, and Gdf15-global-KO animals fed a 60% HFD for 15 weeks (Fig. 4). Whilst global deletion of Gdf15 resulted in increased body weight gain, gut-specific GDF15 deficiency did not affect body weight trends on a long-term high-fat diet (BW at 15 weeks on a HFD WT vs Gdf15-gut-KO vs Gdf15-global-KO; 49.9 ± 4.8 g vs 51.9 ± 3.7 g vs 56.0 ± 3.8 g, respectively, p = 0.0003) (Fig. 4A,B).
Figure 4.
The gut does not contribute significantly to the protective effect of GDF15 on weight gain in response to a high fat diet. (A) Body weight of 23-week-old wild-type (n = 18), Gdf15-gut-KO (n = 15), and Gdf15-global-KO (n = 16) mice fed a 60% high-fat diet for 15 weeks beginning at 8 weeks of age, mean ± SEM. Inset, final body weight. (B) Percent body weight change of 23-week-old wild-type (n = 18), Gdf15-gut-KO (n = 15), and Gdf15-global-KO (n = 16) mice fed a 60% high-fat diet for 15 weeks beginning at 8 weeks of age, mean ± SEM. From 9 to 15 weeks on diet, global-KO body weight relative to wild-type mice p-value listed as determined by two-way ANOVA with multiple comparison adjustment by Tukey’s test. (C) Concentration of serum GDF15 (pg/mL) in 23-week-old wild-type (n = 18), Gdf15-gut-KO (n = 15), and Gdf15-global-KO (n = 16) mice fed a 60% high-fat diet for 15 weeks beginning at 8 weeks of age, mean ± SEM. ns = nonsignificant, ***p < 0.001, and ****p < 0.0001 as determined by two-way ANOVA with multiple comparison adjustment by Tukey’s test. (D) Concentration of serum FGF21 (pg/mL) in wild-type (n = 18), Gdf15-gut-KO (n = 15), and Gdf15-global-KO (n = 16) mice fed a 60% high-fat diet for 15 weeks beginning at 8 weeks of age, mean ± SEM. *p < 0.05 as determined by two-way ANOVA with multiple comparison adjustment by Tukey’s test.
Finally, chronic overfeeding is known to increase both circulating GDF15 and FGF21 in mice14. Wild-type mice fed a 60% fat diet had elevated levels of serum GDF15 compared to chow-fed mice (which exhibit basal levels of ~ 50–80 pg/mL (Fig. 1A)). Gut-selective Gdf15 ablation resulted in a slight, albeit nonsignificant decrease in circulating GDF15 levels relative to WT mice (Fig. 4C). Consistent with the findings of Patel et al.15, HFD-fed Gdf15-global-KO mice had significantly higher FGF21 levels than HFD-fed WT littermates (Fig. 4D).
Discussion
The reduction of body weight that occurs in response to metformin, though modest, contributes significantly to its beneficial impact on metabolic health1. Several3,6,16 but not all4 independent studies have reported that the rise in circulating GDF15 in response to metformin is necessary for its effects on energy balance and body weight. These studies have variously highlighted the gut3, the liver6, and the kidney3,16 as important sources of GDF15 production in response to metformin. Herein, we have studied mice in which Gdf15 expression has been tissue-specifically disrupted and conclude that, when metformin is acutely administered by oral gavage the gut is the main source of the GDF15 response, with the kidney, but not the liver, also making a significant contribution.
Once it has reached the gut, metformin enters enterocytes through organic cation transporters PMAT and OCT1. Bailey et al. reported that metformin concentrations in the jejunum are ~ 30- to ~ 300-fold higher than plasma concentrations17. After its absorption, metformin goes to the liver via the hepatic portal circulation. Hepatic metformin concentrations have been shown to be ~ two- to ~ fourfold greater than that of the plasma in rats18 and mice19, a finding reflected by similar measurements in humans20. Metformin is not metabolized and is instead rapidly taken up by the kidneys and excreted unchanged in the urine. The concentration of metformin reached in renal tubules has been approximated to be ~ eightfold greater than plasma concentrations20.
A growing body of evidence has implicated disruption of mitochondrial function as an important mechanism underlying the beneficial metabolic effects of metformin8–10. Bridges et al. recently reported the structural basis for metformin’s inhibition of mammalian respiratory Complex I10. Given that there is much evidence supporting mitochondrial dysfunction as a strong stimulus for increases in GDF15 expression21–23, this action of metformin seems to be a plausible mechanism through which it increases GDF1524.
Metformin has also been shown to activate AMP kinase by increasing AMP:ATP and ADP:ATP ratios12,25–27, although whether or not this effect is necessary for metformin to elicit its metabolic benefits remains under contention28–30. We found that a single, oral dose of an active AMP kinase activator elevated Gdf15 expression in the liver and kidney, but not the gut. The apparent absence of effect on gut-derived GDF15 levels, despite the fact that it must pass through those cells, may be seen as evidence that AMPK activation in enterocytes does not increase GDF15 levels. However, we have not looked specifically at intestinal AMPK activation after MK-8772 and are aware of previous reports from different model systems that intestinal AMPK activation can have a wider impact upon metabolism, for example, through altering hepatic glucose production31. We have not pursued longer-term studies of either metformin or MK-8772 and it may be the case that, with chronic administration, different patterns of tissue contribution would arise. Further, the majority of these studies used high doses of metformin and included only male mice, decisions based on previous studies as a way to robustly challenge the system under investigation. Again, other dosing regimens over longer time courses in both sexes may reveal different patterns of expression.
We also further characterized the impact of dietary status on the metformin-induced GDF15 response. High-fat feeding potentiates GDF15 responses to metformin, an effect further enhanced when high fat is combined with high sugar. While the underlying mechanism for these effects remains unclear, alterations in the gut microbiome following dietary manipulations32,33 and metformin treatment34–38 are well-defined, and may be a potential area for future exploration.
Animals lacking GDF15 have previously been reported to be more susceptible to weight gain from high-fat diets than WT mice15,39,40. Patel et al. recently compared mice lacking Gdf15 from liver vs. macrophages, and concluded that hepatocytes are the major source of high-fat diet-induced GDF1515. We report that mice lacking Gdf15 in the intestine are not more susceptible to gaining weight on a high-fat diet, suggesting that, under normal circumstances (i.e., in the absence of metformin) gut-derived GDF15 does not contribute significantly to the weight-restraining effects of GDF15 in the presence of high-fat feeding.
While the effects of metformin on GDF15 are important for the weight-lowering effects of the drug—and those effects contribute significantly to the ability of metformin to prevent the onset of T2DM in at-risk individuals—metformin has effects on lowering glucose and insulin that are independent of GDF15-GFRAL signaling3. Studies to elucidate the mechanism of metformin to reduce glucose and insulin levels have largely focused on the liver11,13. However, prior to the demonstration of the striking impact of metformin on enterocyte expression of Gdf153, there has been a long history of studies suggesting the gut as a key site of metformin action41,42.
Using insulin-resistant obese fa/fa rats, Pénicaud et al. identified the digestive tract as the tissue responsible for the majority of metformin-stimulated glucose utilization2. Patients taking metformin have robustly increased 18F-FDG uptake (a marker of glucose uptake and utilization), as observed by PET-CT, in the colon and small intestine43,44. A formulation which delivers metformin to the lower gastro-intestinal tract with much lower systemic exposure than the normal formulation is at least as efficacious as the latter in lowering glucose and HbA1c levels45,46.
Further evidence supporting the concept that metformin action in diabetes is not confined to the liver is provided by Gormsen et al. in which metformin is reported to actually increase endogenous glucose production in individuals with well-controlled T2DM and in normoglycemic controls47. Data from a series of detailed whole body glucose metabolism studies indicate that the main effect of metformin on fasting blood glucose is more likely to be through an extra-hepatic action not in skeletal muscle, and suggested to be in the gut47, with the increase in EGP the result of the lowering of plasma insulin and the rise in lactate production from anaerobic glycolysis in the “first responder” tissue. Recently Tobar et al. confirmed the effects of metformin on basolateral glucose uptake in the intestine and that this triggers an increase in hepatic glucose output in normoglycemic animals. In contrast, in hyperglycemic animals they observed a reduction of hepatic glucose output (at least as assessed by pyruvate tolerance testing) that they attribute to gut-liver crosstalk, the specific nature of which is yet to be precisely defined48.
Given the growing realization that the enterocyte may be a major site of action of metformin, further detailed exploration of its effects on Gdf15 expression in the gut may provide a window of opportunity for increasing understanding of the broader mechanism of action of this widely-used drug.
Methods
Animal studies
Animals
The research reported in this publication is solely the responsibility of the authors and does not necessarily represent the official views of the Medical Research Council. All mouse studies were performed at the University of Cambridge in accordance with UK Home Office Legislation regulated under the Animals (Scientific Procedures) Act 1986 Amendment, Regulations 2012, following ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB). The study is reported in accordance with ARRIVE guidelines. All methods were performed in accordance with the relevant guidelines and regulations. Mice were maintained in a 12 h:12 h light:dark cycle (lights on 07:00–19:00), temperature-controlled (22 °C) facility, with ad libitum access to food (RM3(E) Expanded Chow (Special Diets Services)) and water. Any mice bought from an outside supplier were acclimatized in a holding room for at least one week before study.
Mice carrying the GDF15 knockout-first “tm1a” allele [C57BL/6N-Gdf15tm1a(KOMP)Wtsi/H] were obtained through the IMPC, from the MRC Harwell Production Centre. A “conditional-ready Gdf15 tm1c” allele [C57BL/6N-Gdf15tm1c(KOMP)Wtsi/H] expressing mouse was generated in-house. Briefly, one-cell stage embryos (obtained from super-ovulated wild-type C57Bl/6N females fertilized in vitro with sperm from homozygous Gdf15Tm1a male mice) were injected into the pronucleus with 100 ng/μL StemMACS Flp Recombinase mRNA (Miltenyi Biotec) then transferred into the uteri of pseudo pregnant recipient females (F1 hybrids from C57Bl/6 J female × CBA/Ca male crosses). Mice from the transfer (F1 mice) were analyzed for the presence of the Gdf15 Tm1c and Tm1a alleles. F1 founders were then crossed twice with wild-type C657Bl/6N mice before establishing the Gdf15 Tm1c and Gdf15 Tm1a colonies.
The Gdf15 Tm1c mouse model contains loxP sites flanking exon 2 of Gdf15 gene. Gdf15-knockout mice can be derived from crossing mice carrying the Gdf15 Tm1c [C57BL/6N-Gdf15tm1c(KOMP)Wtsi/H] allele with either mice expressing Cre in the germline to create global knockout or with tissue-specific Cre-expressing mice to create mice lacking Gdf15 in specific tissues.
Gdf15-liver-deficient C57Bl/6N-Gdf15flox/flox-Albumin-Cre+ (referred to as Gdf15-liver-KO mice) were obtained from crossing mice carrying the Gdf15 Tm1c allele with transgenic mice expressing Cre under the control of the Albumin promoter. To establish Gdf15-liver-KO colonies, male Gdf15-liver-KO mice were obtained from Satish Patel and David Savage at the University of Cambridge Wellcome-MRC Institute of Metabolic Science-Metabolic Research Laboratories. Gdf15-gut-deficient C57Bl/6N-Gdf15flox/flox-Villin-Cre+ (referred to as Gdf15-gut-KO mice) were obtained from crossing mice carrying the Gdf15 Tm1c [C57BL/6N-Gdf15tm1c(KOMP)Wtsi/H] allele with Tg(Vil1-Cre)997Gum mice. Vil1-Cre mice express Cre recombinase in villus and crypt epithelial cells of the small and large intestines as previously described49. The Tg(Vil1-Cre)997Gum mice were obtained from The Jackson Laboratory (https://www.jax.org/strain/004586). C57Bl/6J and C57Bl/6N mice were obtained from Charles River. Sample sizes were determined based on homogeneity and consistency of characteristics in the selected models and were sufficient to detect statistically significant differences in body weight and serum parameters between groups. Experiments were performed with animals of a single sex in each study. Transgenic animals were randomised into the treatment groups based on body weight such that the mean body weights of each group were as close to each other as possible, but without using an excess number of animals. No samples or animals were excluded from analyses. Researchers were not blinded to group allocations.
Mouse study 1 (MS1)
Acute metformin and 4-week 60% high-fat diet, wild-type, Gdf15-liver-knockout, and Gdf15-gut-knockout mice
Experimental cohorts of male Gdf15-gut-KO and Gdf15-liver-KO mice were generated by GDF15flox/flox-Villin-Cre- x GDF15flox/flox-Villin-Cre+ and C57Bl/6N-Gdf15flox/flox-Albumin-Cre+ x C57Bl/6N-Gdf15flox/flox-Albumin-Cre- breeding pairs, respectively. Mice were approximately 8 weeks of age when transferred from a standard chow diet to a 60% high-fat diet (HFD; Research Diet Inc. no. 12492i) for 4 weeks before receiving a single dose by oral gavage of either 600 mg/kg metformin or a matched volume of vehicle (water). While fed an HFD, the body weight of each mouse was monitored weekly. GDF15flox/flox-Villin-Cre- (wild-type), Gdf15-gut-KO, and Gdf15-liver-KO animals were randomized into the treatment groups based on body weight such that the mean body weights of each group were as close to each other as possible. Oral gavaging was performed at 9:00 AM, GMT, after which mice were returned to ad libitum 60% HFD before being culled 6 h after treatment by cervical dislocation. Blood was collected into a Starstedt Serum Gel 1.1 mL Micro Tube, left for 30 min at room temperature, then spun for 10 min at 10,000 g at 4 °C before being frozen and stored at − 80 °C until assayed. Mouse GDF15 levels were measured using a Mouse GDF15 DuoSet ELISA (R&D Systems) which had been modified to run as an electrochemiluminescence assay on the Meso Scale Discovery assay platform. Tissues were fresh frozen on dry ice and kept at − 80 °C until the day of RNA extraction.
RNA extraction was carried out with approximately 100 mg of tissue in 1 ml Qiazol Lysis Reagent (Qiagen 79306 l) using Lysing Matrix D homogenisation tube and Fastprep 24 Homogenizer (MP Biomedicals) and Qiagen RNeasy Mini Kit (no. 74106) following manufacturers’ protocols. Five-hundred nanograms of RNA was used to generate cDNA using Promega M-MLV reverse transcriptase followed by TaqMan qPCR in triplicate for Gdf15. Samples were normalized to Actb. TaqMan Probes: Mm00442228 m1 GDF15, Mm02619580_g1 ActB, TaqMan; 2 × universal PCR Master mix (Applied Biosystems Thermo Fisher, 4318157); QuantStudio 7 Flex Real time PCR system (Applied Biosystems Life Technologies).
Mouse study 2 (MS2)
Acute metformin and standard chow diet, Gdf15-gut-knockout and wild-type mice
Experimental cohorts of male Gdf15-gut-KO were generated as in MS1. Mice were approximately 12 weeks of age before receiving a single dose by oral gavage of either 600 mg/kg metformin or a matched volume of vehicle (water). GDF15flox/flox-Villin-Cre- (wild-type) and Gdf15-gut-KO animals were randomized into treatment groups based on body weight such that the mean body weights of each group were as close to each other as possible. Oral gavaging was performed at 9:00 AM, GMT, and mice were culled 6 h after treatment by cervical dislocation. Blood was collected, processed, and analyzed for Mouse GDF15 as in MS1. Tissues were fresh frozen on dry ice and kept at − 80 °C until the day of RNA extraction.
Mouse study 3 (MS3)
Acute metformin and 4-week HFHS diet, Gdf15-gut-knockout and wild-type mice
Experimental cohorts of male Gdf15-gut-KO mice were generated as in MS1. Mice were approximately 8 weeks of age when transferred from a standard chow diet to a high-fat high-sucrose diet (HFHS; Research Diet Inc. no. 20022503) for 4 weeks before receiving a single dose by oral gavage of either 600 mg/kg metformin or a matched volume of vehicle (water). While fed an HFHS diet, the body weight of each mouse was monitored weekly. GDF15flox/flox-Villin-Cre- (wild-type) and Gdf15-gut-KO animals were randomised into treatment groups based on body weight such that the mean body weights of each group were as close to each other as possible. Oral gavaging was performed at 9:00 AM, GMT, and mice were returned to ad libitum HFHS diet after treatment administration. 6 h after treatment administration, animals were culled by cervical dislocation. Blood was collected, processed, and analyzed for Mouse GDF15 as in MS1. Tissues were fresh frozen on dry ice and kept at − 80 °C until the day of RNA extraction. RNA extraction was carried out and quantitative PCR was performed as in MS1.
Mouse study 4 (MS4)
AMPK activation and 4-week 60% high-fat diet, wild-type, Gdf15-liver-knockout, and Gdf15-gut-knockout mice
Experimental cohorts of male Gdf15-gut-KO and Gdf15-liver-KO mice were generated as in MS1. Mice were approximately 16 weeks of age when transferred from a standard chow diet to a 60% high-fat diet (HFD; Research Diet Inc. no. 12492i) for 4 weeks before receiving a single dose by oral gavage of either a pan-AMPK activator, MK-8722 (30 mg/kg), or a matched volume of vehicle (water). MK-8722 was purchased from MedChemExpress (MCE Cat. No. HY-111363). While fed on HFD, the body weight of each mouse was monitored weekly. GDF15flox/flox-Villin-Cre- (wild-type), Gdf15-gut-KO, and Gdf15-liver-KO animals were randomized into the treatment groups based on body weight such that the mean body weights of each group were as close to each other as possible. Oral gavaging was performed daily at 9:00 AM, GMT, after which mice were returned to ad libitum 60% HFD before being culled 4 h after treatment by cervical dislocation. Blood was collected, processed, and analyzed for Mouse GDF15 as in MS1. Tissues were fresh frozen on dry ice and kept at − 80 °C until the day of RNA extraction. RNA extraction was carried out and quantitative PCR was performed as in MS1.
Mouse study 5 (MS5)
Acute metformin and 4-week high-fat diet, wild-type, Gdf15-gut-knockout, and Gdf15-global-knockout mice
Experimental cohorts of male Gdf15-gut-KO and Gdf15-global-KO mice were generated as in MS2. Mice were between 13 and 16 weeks of age when transferred from a standard chow diet to a 60% high-fat diet (HFD; ResearchDiets no. 12492i) for 4 weeks before receiving a single dose by oral gavage of either 600 mg/kg metformin or a matched volume of vehicle (water). While fed an HFD, the body weight of each mouse was monitored weekly. GDF15flox/flox-Villin-Cre- (wild-type), Gdf15-gut-KO, and Gdf15-global-KO animals were randomized into treatment groups based on body weight such that the mean body weights of each group were as close to each other as possible. Oral gavaging was performed at 9:00 AM, GMT, and mice were returned to ad libitum HFD after treatment administration. 1 h after treatment administration, blood was collected in heparinized capillary tubes via the tail vein (approximately 50 μL) after which animals were returned to their cages for a 1-week washout period. Following blood collection, capillaries were immediately placed on ice and spun for 4 min at 10,000 RPM to isolate plasma before being frozen and stored at − 80 °C until assayed. After the wash-out period, animals received the same treatment (although metformin groups received a 300 mg/kg dose as opposed to 600 mg/kg) again via oral gavage. Oral gavaging was performed at 9:00 AM, GMT, and mice were returned to ad libitum HFD after treatment administration. 10 min after treatment administration, blood was collected in heparinized capillary tubes via the tail vein (approximately 50 μL) after which animals were returned to their cages for 4 h. Following blood collection, capillaries were immediately placed on ice and spun for 10 min at 2,000 g to isolate plasma before being frozen and stored at − 80 °C until assayed. 4 h after treatment administrations, animals were culled by cervical dislocation. Blood was collected, processed, and analyzed for Mouse GDF15 as in MS1. Mouse Total GLP-1 concentrations were measured using a V-PLEX GLP-1 Total Kit assay (Meso Scale Discovery). Mouse PYY concentrations were measured using a R-PLEX Mouse/Rat PYY Total Kit assay (Meso Scale Discovery). Tissues were fresh frozen on dry ice and kept at − 80 °C until the day of RNA extraction.
Mouse study 6 (MS6)
15-week high-fat diet, wild-type, Gdf15-gut-knockout, and Gdf15-global-knockout mice
Experimental cohorts of male Gdf15-gut-KO mice were generated as in MS1. C57BL/6N-Gdf15tm1a(KOMP)Wtsi/H mice (referred to as Gdf15-global-KO mice) were obtained from the MRC Harwell Institute, which distributes these mice on behalf of the European Mouse Mutant Archive (https://www.infrafrontier.eu/). Mice were approximately 8 weeks of age when transferred from a standard chow diet to a 60% high-fat diet (HFD; Research Diet Inc. no. 12492i) for 15 weeks. While fed an HFD, the body weight of each mouse was monitored weekly. Animals were single-housed after 13 weeks on high-fat diet. After 15 weeks on high-fat diet, mice were culled by cervical dislocation. Blood was collected, processed, and analyzed for Mouse GDF15 as in MS1. Mouse FGF21 levels were measured using a Mouse/Rat FGF21 Quantikine ELISA Kit (R&D Systems) which had been modified to run as an electrochemiluminescence assay on the Meso Scale Discovery assay platform. Tissues were fresh frozen on dry ice and kept at − 80 °C until the day of RNA extraction.
Experimental diets
Mice were given ad-libitum access to either standard laboratory chow (RM3(E) Expanded Chow (Special Diets Services)), a 60% high-fat diet (Research Diet Inc. no. 12492i), or a high-fat high-sucrose diet (ResearchDiets no. 20022503). Each diet’s composition is outlined in Tables 1, 2 and 3, respectively. The specific diet used was outlined in each mouse study (MS1-6).
Table 1.
Composition of chow diet used in mouse studies.
| Chow diet (CD) | Value | Source |
|---|---|---|
| AFE (kcal/g) | 3.63 | |
| Energy (% kcal) | ||
| Fat | 11.5 | Soybean oil |
| Protein | 27 | Amino acid blend |
| Carbohydrate | 61.5 | 7:1 maltodextrin:sucrose |
AFE Atwater free energy.
Table 2.
Composition of high-fat diet used in mouse studies.
| High-fat diet (HFD) | Value | Source |
|---|---|---|
| AFE (kcal/g) | 5.24 | |
| Energy (% kcal) | ||
| Fat | 60 | 10:1 lard:soybean oil |
| Protein | 20 | Casein (+ L-cystine) |
| Carbohydrate | 20 | 2:1 maltodextrin:sucrose |
AFE Atwater free energy.
Table 3.
Composition of HFHS diet used in mouse studies.
| High-fat high-sucrose diet | Value | Source |
|---|---|---|
| AFE (kcal/g) | 4.6 | |
| Energy (% kcal) | ||
| Fat | 41 | 6.4:1 lard:soybean oil |
| Protein | 20 | Casein (+ L-cystine) |
| Carbohydrate | 39 | 1:3 maltodextrin:sucrose |
AFE Atwater free energy.
In situ hybridization
Tissues were dissected and placed into 10% formalin for 48 h at room temperature, transferred to 70% ethanol and embedded into paraffin. Five-micrometre sections were cut using a Leica microtome, mounted onto Superfrost Plus slides (Thermo Fisher Scientific) and stained for hematoxylin and eosin. Detection of Cre mRNA was performed on formalin-fixed paraffin-embedded (FFPE) sections, obtained from 60% high-fat diet-fed GDF15flox/flox-Villin-Cre- (wild-type) and GDF15flox/flox-Villin-Cre+ (Gdf15-gut-KO) (MS1; Section 2.1.2), using Advanced Cell Diagnostics (ACD) RNAscope® 2.5 LS Reagent Kit-RED (no. 322150) and RNAscope® 2.5 LS Probe CRE-C2 (ACD, no. 312288-C2). Slides were processed as previously described3. Positive and negative controls were run in parallel each time. To prevent the Cre probe from binding to genomic DNA (where necessary), slides were treated with DNase I (QIAGEN, no. 79154) for 10 min at room temperature prior to probe incubation.
Detection of Mouse Gdf15 was performed on FFPE sections using Advanced Cell Diagnostics RNAscope® 2.5 LS Reagent Kit-RED (ACD, no. 322150) and RNAscope® LS 2.5 Probe Mm-Gdf15-O1 (ACD, no. 442948). Fast red detection of mouse Gdf15 was performed on the Bond RX using the Bond Polymer Refine Red Detection Kit (Leica Biosystems, no. DS9390) according to the ACD protocol. Slides were then counterstained with haematoxylin, removed from the Bond RX and were heated at 60 °C for 1 h, dipped in Xylene and mounted using EcoMount Mounting Medium (Biocare Medical, CA, USA. no. EM897L).
All slides were imaged using the Zeiss AxioScan.Z1 Slide Scanner at × 20 (standard) or × 40 magnification (RNAscope). For the RNAscope slides, three Z-stacks spanning a total of 2 μm were acquired and merged into a single extended depth of focus (EDF) image with maximum projection processing, and then sharpened using Unsharp Masking (ZEN Blue, Zeiss).
RNA isolation, cDNA synthesis, and qPCR
RNA extraction was carried out with approximately 100 mg of tissue in 1 ml Qiazol Lysis Reagent (Qiagen 79306l) using Lysing Matrix D homogenisation tube, Fastprep 24 Homogenizer (MP Biomedicals), and Qiagen RNeasy Mini Kit (no. 74106) following manufacturers’ protocols. RNA concentration and quality was determined by Nanodrop spectrophotometry. 400–500 ng of total RNA was then converted to cDNA using M-MLV Reverse Transcriptase with random primers (Promega). Quantitative RT–PCR was carried out with either TaqMan Universal PCR Master Mix or SYBR Green PCR master mix on the QuantStudio 7 Flex Real time PCR system (Applied Biosystems). All reactions were carried out in triplicate and Ct values were obtained. Relative differences in gene expression were normalized to the expression levels of the housekeeping gene Actin Beta (Actb) using the standard curve method. TaqMan probes used for these studies: mouse Gdf15 (Mm00442228_m1, ThermoFisher Scientific) and ActB (Mm02619580_g1, ThermoFisher Scientific).
Statistical analyses
GraphPad Prism (Version 9, GraphPad Software Inc., San Diego, CA) software was used for data visualization and statistical analyses. Unpaired Student’s t-test, two-way ANOVA, and three-way ANOVA with multiple comparison adjustment by Tukey’s or Sidak’s test were used for comparison of differences between experimental groups. Analysis of Covariance (ANCOVA) was performed using JASP (Version 0.16.3, JASP Team, Amsterdam, Netherlands) to determine metabolic rate with body weight as a covariate, genotype as a fixed factor, and energy expenditure as the dependent variable. The specific statistical method employed for individual data sets is listed in the figure legends.
Supplementary Information
Acknowledgements
The authors thank the Clinical Biochemistry Assay Lab, Disease Model Core (DMC), Histopathology Core and Imaging Core facilities at IMS-MRL, University of Cambridge, for experimental support for this study.
Author contributions
Study concept: J.W.R.K., A.P.C., and S.O. Design: All authors. Data acquisition: J.W.R.K., D.R., J.A.T., I.C. Data analysis and interpretation: J.W.R.K., A.P.C., and S.O. Drafting of the manuscript: J.W.R.K., A.P.C., and S.O. Critical revision of the manuscript for important intellectual content: All authors.
Funding
This work was supported by the Medical Research Council Metabolic Diseases Unit Disease Model Core (MC_UU_00014/5). J.W.R.K., A.P.C., I.C., D.R., J.A.T., and S.O. are supported by the MRC Metabolic Diseases Unit (MC_UU_00014/1). J.A.T. is also supported by a NIHR Clinical Lectureship (CL-2019-14-504).
Data availability
Data will be made available from the corresponding author upon request.
Competing interests
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: P.R. is an advisor and shareholder in Kallyope, Inc. S.O. undertakes remunerated consultancy work for Pfizer, AstraZeneca, Third Rock Ventures, Courage Therapeutics, Northsea Therapeutics, and LG Chem Life Sciences. I.C., S.O., and A.P.C. have filed a patent on therapeutic targeting of GDF15-GFRAL.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-51866-2.
References
- 1.Lachin JM, Christophi CA, Edelstein SL, Ehrmann DA, Hamman RF, Kahn SE, et al. Factors associated with diabetes onset during metformin versus placebo therapy in the Diabetes Prevention Program. Diabetes. 2007;56(4):1153–1159. doi: 10.2337/db06-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pénicaud L, Hitier Y, Ferré P, Girard J. Hypoglycaemic effect of metformin in genetically obese (fa/fa) rats results from an increased utilization of blood glucose by intestine. Biochem. J. 1989;262(3):881–885. doi: 10.1042/bj2620881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coll AP, Chen M, Taskar P, Rimmington D, Patel S, Tadross JA, et al. GDF15 mediates the effects of metformin on body weight and energy balance. Nature. 2020;578(7795):444–448. doi: 10.1038/s41586-019-1911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein AB, Nicolaisen TS, Johann K, Fritzen AM, Mathiesen CV, Gil C, et al. The GDF15-GFRAL pathway is dispensable for the effects of metformin on energy balance. Cell Rep. 2022;40(8):111258. doi: 10.1016/j.celrep.2022.111258. [DOI] [PubMed] [Google Scholar]
- 5.Myers RW, Guan H-P, Ehrhart J, Petrov A, Prahalada S, Tozzo E, et al. Systemic pan-AMPK activator MK-8722 improves glucose homeostasis but induces cardiac hypertrophy. Science. 2017;357(6350):507–511. doi: 10.1126/science.aah5582. [DOI] [PubMed] [Google Scholar]
- 6.Day EA, Ford RJ, Smith BK, Mohammadi-Shemirani P, Morrow MR, Gutgesell RM, et al. Metformin-induced increases in GDF15 are important for suppressing appetite and promoting weight loss. Nat. Metabol. 2019;1(12):1202–1208. doi: 10.1038/s42255-019-0146-4. [DOI] [PubMed] [Google Scholar]
- 7.Townsend LK, Weber AJ, Day EA, Shamshoum H, Shaw SJ, Perry CGR, et al. AMPK mediates energetic stress-induced liver GDF15. FASEB J. 2021;35(1):e21218. doi: 10.1096/fj.202000954R. [DOI] [PubMed] [Google Scholar]
- 8.El-Mir MY, Nogueira V, Fontaine E, Avéret N, Rigoulet M, Leverve X. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 2000;275(1):223–228. doi: 10.1074/jbc.275.1.223. [DOI] [PubMed] [Google Scholar]
- 9.Owen MR, Doran E, Halestrap AP. Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem. J. 2000;348(Pt 3):607–614. doi: 10.1042/bj3480607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bridges HR, Blaza JN, Yin Z, Chung I, Pollak MN, Hirst J. Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides. Science. 2023;379(6630):351–357. doi: 10.1126/science.ade3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaMoia TE, Shulman GI. Cellular and molecular mechanisms of metformin action. Endocr. Rev. 2020;42(1):77–96. doi: 10.1210/endrev/bnaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Investig. 2001;108(8):1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madiraju AK, Erion DM, Rahimi Y, Zhang X-M, Braddock DT, Albright RA, et al. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature. 2014;510(7506):542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel S, Alvarez-Guaita A, Melvin A, Rimmington D, Dattilo A, Miedzybrodzka EL, et al. GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metabol. 2019;29(3):707–718.e8. doi: 10.1016/j.cmet.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel S, Haider A, Alvarez-Guaita A, Bidault G, El-Sayed Moustafa JS, Guiu-Jurado E, et al. Combined genetic deletion of GDF15 and FGF21 has modest effects on body weight, hepatic steatosis and insulin resistance in high fat fed mice. Mol. Metabol. 2022;65:101589. doi: 10.1016/j.molmet.2022.101589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S-Y, Bruce K, Danaei Z, Li RJW, Barros DR, Kuah R, et al. Metformin triggers a kidney GDF15-dependent area postrema axis to regulate food intake and body weight. Cell Metabol. 2023 doi: 10.1016/j.cmet.2023.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Bailey CJ, Wilcock C, Scarpello JHB. Metformin and the intestine. Diabetologia. 2008;51(8):1552–1553. doi: 10.1007/s00125-008-1053-5. [DOI] [PubMed] [Google Scholar]
- 18.Wilcock C, Wyre ND, Bailey CJ. Subcellular distribution of metformin in rat liver. J. Pharm. Pharmacol. 1991;43(6):442–444. doi: 10.1111/j.2042-7158.1991.tb03507.x. [DOI] [PubMed] [Google Scholar]
- 19.Wilcock C, Bailey CJ. Accumulation of metformin by tissues of the normal and diabetic mouse. Xenobiotica. 1994;24(1):49–57. doi: 10.3109/00498259409043220. [DOI] [PubMed] [Google Scholar]
- 20.Gormsen LC, Sundelin EI, Jensen JB, Vendelbo MH, Jakobsen S, Munk OL, et al. In vivo imaging of human 11C-metformin in peripheral organs: Dosimetry, biodistribution, and kinetic analyses. J. Nucl. Med. 2016;57(12):1920–1926. doi: 10.2967/jnumed.116.177774. [DOI] [PubMed] [Google Scholar]
- 21.Montero R, Yubero D, Villarroya J, Henares D, Jou C, Rodríguez MA, et al. GDF-15 is elevated in children with mitochondrial diseases and is induced by mitochondrial dysfunction. PLoS One. 2016;11(2):e0148709. doi: 10.1371/journal.pone.0148709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulsen NS, Madsen KL, Hornsyld TM, Eisum A-SV, Fornander F, Buch AE, et al. Growth and differentiation factor 15 as a biomarker for mitochondrial myopathy. Mitochondrion. 2020;50:35–41. doi: 10.1016/j.mito.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Kang SG, Choi MJ, Jung S-B, Chung HK, Chang JY, Kim JT, et al. Differential roles of GDF15 and FGF21 in systemic metabolic adaptation to the mitochondrial integrated stress response. iScience. 2021;24(3):102181. doi: 10.1016/j.isci.2021.102181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang M, Darwish T, Larraufie P, Rimmington D, Cimino I, Goldspink DA, et al. Inhibition of mitochondrial function by metformin increases glucose uptake, glycolysis and GDF-15 release from intestinal cells. Sci. Rep. 2021;11(1):2529. doi: 10.1038/s41598-021-81349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawley SA, Gadalla AE, Olsen GS, Hardie DG. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes. 2002;51(8):2420–2425. doi: 10.2337/diabetes.51.8.2420. [DOI] [PubMed] [Google Scholar]
- 26.Meng S, Cao J, He Q, Xiong L, Chang E, Radovick S, et al. Metformin activates AMP-activated protein kinase by promoting formation of the αβγ heterotrimeric complex. J. Biol. Chem. 2015;290(6):3793–3802. doi: 10.1074/jbc.M114.604421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C-S, Li M, Ma T, Zong Y, Cui J, Feng J-W, et al. Metformin activates AMPK through the lysosomal pathway. Cell Metabol. 2016;24(4):521–522. doi: 10.1016/j.cmet.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Miller RA, Birnbaum MJ. An energetic tale of AMPK-independent effects of metformin. J. Clin. Investig. 2010;120(7):2267–2270. doi: 10.1172/JCI43661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foretz M, Hébrard S, Leclerc J, Zarrinpashneh E, Soty M, Mithieux G, et al. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. J. Clin. Investig. 2010;120(7):2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deschemin J-C, Foretz M, Viollet B, Vaulont S. AMPK is not required for the effect of metformin on the inhibition of BMP6-induced hepcidin gene expression in hepatocytes. Sci. Rep. 2017;7(1):12679. doi: 10.1038/s41598-017-12976-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duca FA, Côté CD, Rasmussen BA, Zadeh-Tahmasebi M, Rutter GA, Filippi BM, et al. Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 2015;21(5):506–511. doi: 10.1038/nm.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy EA, Velazquez KT, Herbert KM. Influence of high-fat diet on gut microbiota: A driving force for chronic disease risk. Curr. Opin. Clin. Nutr. Metabol. Care. 2015;18(5):515–520. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang Y, Khafipour E, Derakhshani H, Sarna LK, Woo CW, Siow YL, et al. Short-term high-fat diet induces obesity-enhancing changes in mouse gut microbiota that are partially reversed by cessation of the high-fat diet. Lipids. 2017;52(6):499–511. doi: 10.1007/s11745-017-4253-2. [DOI] [PubMed] [Google Scholar]
- 34.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 35.Sun L, Xie C, Wang G, Wu Y, Wu Q, Wang X, et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018;24(12):1919–1929. doi: 10.1038/s41591-018-0222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee H, Lee Y, Kim J, An J, Lee S, Kong H, et al. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes. 2018;9(2):155–165. doi: 10.1080/19490976.2017.1405209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauer PV, Duca FA, Waise TMZ, Rasmussen BA, Abraham MA, Dranse HJ, et al. Metformin alters upper small intestinal microbiota that impact a glucose-SGLT1-sensing glucoregulatory pathway. Cell Metabol. 2018;27(1):101–117.e5. doi: 10.1016/j.cmet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Lee H, Kim J, An J, Lee S, Choi D, Kong H, et al. Downregulation of IL-18 expression in the gut by metformin-induced gut microbiota modulation. Immune Netw. 2019;19(4):e28. doi: 10.4110/in.2019.19.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai VW-W, Macia L, Johnen H, Kuffner T, Manadhar R, Jørgensen SB, et al. TGF-b superfamily cytokine MIC-1/GDF15 is a physiological appetite and body weight regulator. PLoS One. 2013;8(2):e55174. doi: 10.1371/journal.pone.0055174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran T, Yang J, Gardner J, Xiong Y. GDF15 deficiency promotes high fat diet-induced obesity in mice. PLoS One. 2018;13(8):e0201584. doi: 10.1371/journal.pone.0201584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey CJ, Mynett KJ, Page T. Importance of the intestine as a site of metformin-stimulated glucose utilization. Br. J. Pharmacol. 1994;112(2):671–675. doi: 10.1111/j.1476-5381.1994.tb13128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCreight LJ, Bailey CJ, Pearson ER. Metformin and the gastrointestinal tract. Diabetologia. 2016;59:426–435. doi: 10.1007/s00125-015-3844-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gontier E, Fourme E, Wartski M, Blondet C, Bonardel G, Le Stanc E, et al. High and typical 18F-FDG bowel uptake in patients treated with metformin. Eur. J. Nucl. Med. Mol. Imaging. 2008;35(1):95–99. doi: 10.1007/s00259-007-0563-6. [DOI] [PubMed] [Google Scholar]
- 44.Oh J-R, Song H-C, Chong A, Ha J-M, Jeong S-Y, Min J-J, et al. Impact of medication discontinuation on increased intestinal FDG accumulation in diabetic patients treated with metformin. AJR Am. J. Roentgenol. 2010;195(6):1404–1410. doi: 10.2214/AJR.10.4663. [DOI] [PubMed] [Google Scholar]
- 45.Buse JB, DeFronzo RA, Rosenstock J, Kim T, Burns C, Skare S, et al. The primary glucose-lowering effect of metformin resides in the gut, not the circulation: Results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care. 2016;39(2):198–205. doi: 10.2337/dc15-0488. [DOI] [PubMed] [Google Scholar]
- 46.Henry RR, Frias JP, Walsh B, Skare S, Hemming J, Burns C, et al. Improved glycemic control with minimal systemic metformin exposure: Effects of metformin delayed-release (metformin DR) targeting the lower bowel over 16 weeks in a randomized trial in subjects with type 2 diabetes. PLoS One. 2018;13(9):e0203946. doi: 10.1371/journal.pone.0203946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gormsen LC, Søndergaard E, Christensen NL, Brøsen K, Jessen N, Nielsen S. Metformin increases endogenous glucose production in non-diabetic individuals and individuals with recent-onset type 2 diabetes. Diabetologia. 2019;62(7):1251–1256. doi: 10.1007/s00125-019-4872-7. [DOI] [PubMed] [Google Scholar]
- 48.Tobar N, Rocha GZ, Santos A, Guadagnini D, Assalin HB, Camargo JA, et al. Metformin acts in the gut and induces gut-liver crosstalk. Proc. Natl. Acad. Sci. 2023;120(4):e2211933120. doi: 10.1073/pnas.2211933120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002;277(36):33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available from the corresponding author upon request.