Abstract
Anaplasma marginale is an intraerythrocytic rickettsial pathogen of cattle in which infection persists for the life of the animal. Persistent A. marginale infection is characterized by repetitive rickettsemic cycles which we hypothesize reflect emergence of A. marginale antigenic variants. In this study, we determined whether variants of major surface protein 2 (MSP-2), a target of protective immunity encoded by a polymorphic multigene family, arise during persistent rickettsemia. By using a quantitative competitive PCR to identify rickettsemic cycles, msp-2 transcripts expressed in vivo were isolated from peak rickettsemia of sequential cycles. Cloning and sequencing of msp-2 cDNA revealed that genetic variants of MSP-2 emerge representing a minimum of four genetic variant types in each cycle during persistent infection. Two-color immunofluorescence using variant-specific antibody showed that emergence of MSP-2 variants resulted in expression of a minimum of three antigenic types of MSP-2 within one rickettsemic cycle. Therefore immune control of each cycle would require responses to an antigenically diverse A. marginale population. These findings demonstrate that polymorphic MSP-2 variants emerge during cyclic rickettsemia in persistent A. marginale infection and suggest that emergent variants play an important role in persistence.
Rickettsiae are obligate intracellular bacteria that invade and multiply within host cells and, depending on the host infected, cause anemia, leukopenia, thrombocytopenia, or vasculitis that can result in death (37). Animals that survive acute infection have an effective immune response that either eliminates the infection or reduces rickettsemia to low, microscopically undetectable levels which persist. How rickettsiae persist despite a controlling immune response is unknown. Several rickettsial pathogens which establish persistent infections in their hosts, including Anaplasma marginale (15, 25), Rickettsia tsutsugamushi (5, 8, 21, 29, 30), Ehrlichia canis (3), and Cowdria ruminantium (1, 27), show variation in outer membrane proteins. Major surface protein 2 (MSP-2) of A. marginale (22, 23, 26), the 56-kDa major outer membrane protein of R. tsutsugamushi (17, 20, 31), and the MAP-1 protein of C. ruminantium (32, 34), for example, are surface expressed and are highly immunogenic. While surface protein variation is clearly involved in strain-specific immunity, the role of outer membrane protein variation in persistence of rickettsial infections is unknown.
A. marginale is an intraerythrocytic rickettsia that infects cattle and can cause severe anemia, abortion, or death (14). Immunity to A. marginale is directed against outer membrane surface proteins (22, 23, 26), including MSP-2. MSP-2 is encoded by a large, polymorphic, multigene family that comprises at least 1% of the genome (25), providing the genetic capacity for antigenic variation. Variant msp-2 transcripts cloned and sequenced from acute rickettsemia encode unique MSP-2 polypeptides which are expressed in vivo (4). Similarly, A. marginale bearing MSP-2 antigenic variants during acute rickettsemia has been demonstrated in assays using copy-specific monoclonal antibodies reactive with MSP-2 on some but not all organisms (25). The development of a primary immune response, by either infection or MSP-2 immunization, dramatically reduces but does not clear A. marginale rickettsemia (24). However, these immune cattle are protected against high level rickettsemia and acute disease following subsequent homologous strain challenge. Thus, persistence occurs despite development of a protective immune response against the initial infecting organisms. We have hypothesized that persistence, characterized by cyclic rickettsemia, reflects continual emergence of new antigenic variants (4, 11). The goal of this study was to determine whether MSP-2 variants arise during rickettsemic cycles in persistent A. marginale infection. A competitive PCR was developed to quantitate the rickettsemia levels in persistent infection. We then examined each identified peak of cyclic rickettsemia to determine if MSP-2 variants emerge within sequential peaks, or if MSP-2 is invariant during persistent A. marginale infection.
MATERIALS AND METHODS
Sample collection.
Two Holstein steers (807 and 808) were experimentally infected with A. marginale Florida strain on October 3, 1994 (10-3-94 [this convention for expressing dates will be used hereafter]) by intravenous inoculation of 1 ml of whole blood from an acutely rickettsemic calf (2.4 × 109 infected erythrocytes/ml of blood). Beginning 3-6-96, peripheral blood was collected biweekly (in the morning) by venipuncture, and the following samples were stored: 10 ml of whole blood collected in acid citrate dextrose, mixed with TRIzol reagent (BRL) and frozen at −20°C for RNA extraction; 5 ml of whole blood collected in EDTA frozen at −20°C for DNA extraction; erythrocytes collected in heparin, washed three times in phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM sodium phosphate [pH 7.4]) and frozen at −20°C for immunoblots; 3 ml of serum frozen at −20°C; and unstained whole blood smears stored at −70°C for fluorescent antibody tests.
Identification of rickettsemic cycles in persistently infected cattle.
DNA was extracted from 100 μl of whole blood from persistently infected cattle, using a Puregene (Gentra) DNA extraction kit. DNA was then used in a competitive PCR to quantitate rickettsemia and identify cycles. Primers used for identification of rickettsemic cycles were based on A. marginale msp-5, a conserved single-copy gene (36). The external primers (5′ primer, GCATAGCCTCCGCGTCTTTC; 3′ primer, TCTGAGGGCCAAGGCGAGGA) amplify a 457-bp fragment. A 202-base msp-5 MIMIC (Clontech PCR MIMIC construction kit) with primer binding sites specific for A. marginale msp-5 was constructed. A constant amount of DNA extracted from persistently infected cattle was amplified in the presence of 10-fold dilutions of the msp-5 MIMIC. Amplifications were performed in 50-μl reaction volumes, using Boehringer Mannheim PCR Master. PCR products were evaluated densitometrically following electrophoresis on a 2% agarose gel. The initial molar amount of target msp-5 DNA (N0t) was extrapolated from a logarithmic plot of log [At/As] versus the log of the initial molar amount of mimic (log N0s), where At is the amount of amplified target, As is the amount of amplified standard, and N0s is the initial number of standard molecules. N0t is equal to N0s added in the reaction when an equimolar ratio of the two products is produced (i.e., where the log of At/As = log 1/1 = 0) (2). Rickettsemic cycles were identified by plotting the log of the target msp-5 concentration (in attomoles/microliter) versus the date of blood collection (Fig. 1). To ensure that the competitive PCR was consistent, standardized samples were evaluated. Blood from an acutely infected animal determined microscopically to have a rickettsemia level of 1.6 × 109 infected erythrocytes/ml of blood was diluted to 103 to 107 infected erythrocytes/ml of blood and run in the msp-5 competitive PCR. By determining the number of attomoles of msp-5 in samples with known numbers of infected erythrocytes, an approximation of the number of infected erythrocytes could be made for samples from persistently infected animals. Whole blood from an uninfected steer was also extracted and evaluated in the competitive PCR as a negative control.
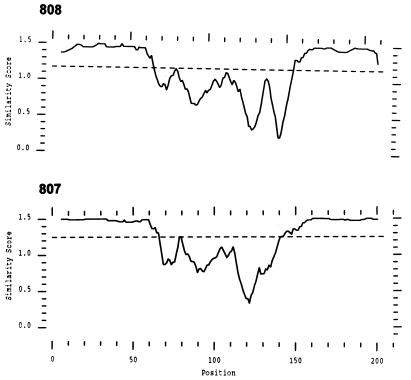
FIG. 1.
Detection of rickettsemic cycles by using competitive PCR. A. marginale organisms were quantitated in blood from two animals, 808 (A) and 807 (B), over an 18-week period.
Cloning and sequencing of msp-2 cDNA.
Total RNA was extracted using TRIzol (BRL) from whole blood taken at the peak of each rickettsemic cycle and reverse transcribed by using random hexamers. Primers were derived from conserved regions based on sequence comparisons of the existing full-length genomic clones, DF5 msp-2 and pCKR11.2 msp-2; a partial genomic clone, pCKR5.2 msp-2; and a cDNA clone from acute rickettsemia, AR5 msp-2 (4, 25). The 5′ and 3′ primers for msp-2 were TGGAGGAGCAAGGGTTGAAGT and TTGTGCTGGTATCGGTGGTAA, respectively. The amplified central 595-bp region of the 1.2-kb genes includes a polymorphic region (4, 25). PCR products were ligated into pCR 2.1 (Invitrogen) by using the TA cloning kit (Invitrogen). Competent Escherichia coli XL-1 Blue was transformed with the ligated vector and plated with 5 mM isopropyl-1-β-d-thiogalactopyranoside and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for blue/white screening. The same cDNA was also amplified with the msp-5-specific primers listed above as a control for nucleotide changes generated during amplification. Presence of msp-2 or msp-5 inserts in plasmids from transformed colonies was confirmed by restriction digests or PCR. Plasmid DNA was extracted, and clones were sequenced in both directions by using an ABI PRISM (Applied Biosystems) automated DNA sequencer. Sequence analysis using the Genetics Computer Group package from the University of Wisconsin, version 8.1, was performed on a VAX11/785 computer.
Generation of antibody against polymorphic MSP-2 variants.
To determine if different antigenic variants were expressed within one rickettsemic peak, genetic variants from peak 2 of animal 808 were used to generate specific antibodies. Two of the variant msp-2 sequences, designated Pk2-3 and Pk2-5, were subcloned in frame into the pET19b (Novagen) vector for expression of His-tagged fusion proteins. Pk2-3 and Pk2-5 were amplified by PCR with Taq DNA polymerase, using forward and reverse primers containing StuI and XhoI restriction sites, respectively. The vector was digested with NdeI, blunt ended with the Klenow fragment of DNA polymerase I, and subsequently digested with XhoI. The digested PCR fragments were then ligated into the pET19b plasmid between the NdeI and XhoI sites in the polylinker. Plasmids containing the inserts were sequenced to confirm reading frame and absence of nucleotide changes and used to transform competent E. coli BL21(DE3) cells. The clone generated from Pk2-3 was designated V3, and the clone from Pk2-5 was designated V5. V3 and V5 were then used for protein expression. Recombinant proteins were expressed and purified on Ni2+-charged columns under denaturing conditions as recommended by the manufacturer (Novagen) but modified by adding imidazole in the wash buffer (0.5 M NaCl, 20 mM Tris [pH 7.9], 80 mM imidazole) to minimize nonspecific binding of proteins to the column. Purified proteins were dialyzed against PBS for 48 h at 4°C. Rabbits were immunized twice subcutaneously at a 4-week interval with 100 μg of total protein emulsified in RIBI adjuvant (RIBI Immunochem Research, Inc.). Sera were obtained, and immunoglobulin G (IgG) was isolated by using a protein G-agarose column (Gibco BRL). The antibodies were cross-adsorbed: anti-V3 IgG was adsorbed with purified V5 protein, anti-V5 IgG was adsorbed with purified V3 protein, and as a negative control, anti-bovine interleukin-4 (IL-4) IgG was adsorbed with purified Babesia bigemina RAP-1 protein. Adsorption experiments were done by adding 20 μg of recombinant protein (V3, V5, or B. bigemina RAP-1) to 200 μg of purified IgG against V5, V3, or IL-4, respectively, and incubation for 4 h at room temperature followed by centrifugation for 5 min at 12,000 × g. Specific reactivity of the antibodies with respective variants was tested by immunoblotting. Purified proteins were electrophoresed on sodium dodecyl sulfate-containing polyacrylamide gels. Separated proteins were transferred electrophoretically to a nitrocellulose membrane and reacted with a 1:5,000 dilution of either the preadsorbed anti-V3, anti-V5, or anti-IL-4 IgG. Bound antibody was detected with peroxidase-labeled goat anti-rabbit IgG with enhanced chemiluminescence detection (25).
Identification of variant MSP-2 coexpressed in one peak during persistent rickettsemia.
Following confirmation of antibody specificity for the respective variants, IgG isolated from the anti-V3 serum was directly labeled with rhodamine, and IgG from anti-V5 or anti-IL-4 polyclonal sera was directly labeled with fluorescein (9). Solutions of 7 mg of IgG/ml were prepared by dilution with 0.1 M sodium carbonate. One milliliter of IgG solution was incubated for 8 h at 4°C with 50 μg of tetramethylrhodamine isothiocyanate or fluorescein isothiocyanate dissolved in dimethyl sulfoxide. The conjugated antibody was separated from unbound dye by gel filtration and subsequently used in direct immunofluorescence. Methanol-fixed blood smears from the second rickettsemic peak in animal 808 (5-10-96) were incubated first with rhodamine-conjugated anti-V3 for 30 min at 37°C. Slides were washed three times in PBS and then incubated with either fluorescein-conjugated anti-V5 or, as a negative control, anti-IL-4. Following three additional washes in PBS, slides were examined at a magnification of ×400, using an excitation wavelength of 450 to 490 nm to detect fluorescein-labeled organisms and a wavelength of 546 nm to detect rhodamine-labeled organisms.
RESULTS
Identification of rickettsemic cycles in persistently infected cattle.
Animals that survive acute A. marginale infection become persistently infected with low cyclic levels of rickettsemia (6, 7, 11). Rickettsemic cycles were identified in two persistently infected animals, 808 and 807, by quantitation using msp-5 competitive PCR. Since the msp-5 gene is present as a single genomic copy (36), each molecule of msp-5 represents a single organism. Major sequential cycles, defined as ≥106 rickettsiae per ml of blood, occurred at 6- to 8-week intervals during the 18-week study period (corresponding to 72 to 90 weeks or 504 to 630 days postinfection [Fig. 1]). Each major peak was followed by a rapid decline in rickettsemia of at least 102 per ml of blood. In Fig. 1, for animal 808 the highest major peak on 3-12-96 had 2.2 × 107 organisms per ml (−2.15 log attomol/μl of blood), and the low point on 5-31-96 had 4 × 103 organisms per ml of whole blood (−5.55 log amol/μl of blood). Rickettsemic cycles in animal 807 had similar fluctuations but reached a low point of 2 × 103 organisms per ml of blood (−5.82 log amol/μl of blood) on several sampling days (Fig. 1).
Since A. marginale organisms replicate within erythrocytes and are microscopically detectable in acute infection, rickettsemia levels have often been described as numbers of infected erythrocytes. To estimate the number of parasitized erythrocytes during persistent infection using competitive PCR, dilutions of known numbers of infected erythrocytes obtained from an experimental acute infection were used. In persistently infected animals, rickettsemia levels corresponded to approximately 107 infected erythrocytes per ml of blood at the highest peaks, 3-12-97 for animal 808 and 3-8-97 for animal 807 (Fig. 1). The low point of the rickettsemic cycles corresponded to approximately 103 infected erythrocytes per ml of blood on 5-31-96 in animal 808 and approximately 102.5 infected erythrocytes per ml of blood on 5-21-97 in animal 807. Samples from the uninfected animal were negative using competitive msp-5 PCR.
Variation in msp-2 transcripts during persistent rickettsemia.
To determine whether polymorphic msp-2 genes are expressed during persistent infection, total RNA was extracted from three sequential peaks of rickettsemia in animals 807 and 808, and msp-2 mRNA was amplified by reverse transcription-PCR (RT-PCR). The msp-2 gene-specific primers were selected from conserved regions that flank a central polymorphic region based on alignment of the full-length msp-2 genomic clones, pCKR11.2 msp-2 and DF5 msp-2; a partial genomic clone, pCKR5.2 msp-2; and a cDNA clone from acute rickettsemia, AR5 msp-2 (4, 25). RT-PCR products obtained from each peak of cyclic rickettsemia were cloned, and 10 msp-2 cDNA clones were randomly selected and sequenced. The msp-2 cDNA clones varied in size, ranging from 585 to 600 bp, compared to the 595 bp amplified in the pCKR11.2 msp-2 from acute rickettsemia. As described below, this size polymorphism reflects nucleotide deletions and insertions. Within each peak, the 10 cDNA clones represented a minimum of four unique sequence types. Sequence types are defined based on the variable region shown in Fig. 2, amino acids 185 to 280 (numbering is based on the predicted amino acid sequence of pCKR11.2 msp-2 [25]). To ensure that 10 clones were representative of msp-2 genetic types expressed within a given peak, an additional 10 msp-2 cDNA clones were generated from one peak (animal 808, peak 2), sequenced, and compared to the initial 10 clones from the same peak. This second set of clones had sequences identical to those of the original set of clones (data not shown) and therefore represent the same types.
FIG. 2.
Amino acid sequence alignment of the polymorphic region of types of clones from peak 2 of animal 808 and peak 3 of animal 807. Types of clones for 808 peak 2 are as follows: type I includes clones 9 and 10 (clone 10 has a gap at amino acid 218 relative to clone 9); type II includes clones 6 and 7; type III includes clone 5; type IV includes clone 8; and type V includes clones 1 to 4 (clone 4 has a single amino acid change relative to clones 1 to 3, amino acid 187 K→N). Types of clones for 807 peak 3 are as follows: type I includes clones 3 to 6 (clone 6 has an amino acid change relative to clones 3 to 5, amino acid 249 S→N); type II includes clone 7; type III includes clones 1 and 2; type IV includes clone 8; type V includes clone 9; and type IV includes clone 10. The central polymorphic region of each type is shown from amino acids 185 to 297 relative to the 11.2 msp-2 clone (25). Areas of amino acid substitutions, insertions, and deletions are indicated by a white background, areas of amino acid identity have a black background, and grey shading indicates conservative amino acid changes. The dots designate deletions.
All msp-2 cDNA clones obtained from both animals have the predicted open reading frame based on the previously sequenced full-length msp-2 clones (4, 25). As a control for sequence polymorphism generated by reverse transcriptase or Taq polymerase misincorporation, specific primers for the single-copy, genus-conserved msp-5 gene were also used in RT-PCR with the same total RNA as a template. Three nucleotide substitutions occurred following RT-PCR and sequencing of 10 457-bp msp-5 cDNA clones. Therefore, relatively few nucleotide changes were generated in the RT-PCR of the total A. marginale RNA; assuming that the three substitutions in a total of 4,570 msp-5 bases sequenced are errors and not true nucleotide changes, this is an error rate of 7 × 10−4, consistent with a predicted error rate of 2 × 10−4 for Taq polymerase (10). In addition, increased error rate of Taq polymerase due to msp-2-specific secondary structure (13) is not likely because comparison of several msp-2 cDNA clones following Taq-based subcloning did not result in nucleotide misincorporations relative to the original clones.
Comparison of all msp-2 cDNA clones representative of the different types obtained from animal 808 revealed a central region of polymorphism between nucleotides 555 and 882 (numbering is based on the full-length pCKR11.2 msp-2 clone [25]). Relative to the other cDNA types obtained from the same animal, the central polymorphic region had insertions, deletions, and substitutions that resulted in amino acid changes (Fig. 2). All msp-2 cDNA types obtained from animal 807 had similar central regions of polymorphism with nucleotide insertions, deletions, and substitutions. Interestingly, comparison of predicted amino acid sequences of all of the clones representative of the different types obtained from animal 808 revealed a distinct hypervariable region that spans amino acid positions 60 to 155 (Fig. 3). Similarly, clones from animal 807 have a prominent hypervariable region that spans amino acid positions 60 to 150 (Fig. 3). These positions represent amino acids 186 to 281 and 186 to 276, respectively, of the predicted amino acid sequence of pCKR11.2 msp-2 (25).
FIG. 3.
Plot similarity of amino acid sequences of all MSP-2 clones obtained from each of three peaks of cyclic rickettsemia in animals 808 and 807. Similarity score is plotted as a function of amino acid position. The dashed line that transects the plot at approximately 1.2 along the y axis indicates the average similarity score of all of the clones. Clones from animal 808 have a distinct hypervariable region that spans amino acid positions 60 to 155, and clones from animal 807 have a single hypervariable region that spans amino acid positions 60 to 150, corresponding to amino acids 186 to 281 and 186 to 276, respectively, of the pCKR11.2 MSP-2 (25).
Comparison of predicted amino acid sequences between peaks revealed a range of identities from 78 to 99% for animal 808 (Table 1). None of the clones representative of the different types obtained from one peak in animal 808 were identical to clones obtained from subsequent peaks. However, in animal 807, several clones from peak 1 recurred in peaks 2 and 3. Type I, represented by identical clones 1-1 and 1-2 from peak 1, showed 100% amino acid identity with clone 2-9 in peak 2 indicating the recurrence of this type. Similarly, type IV, represented by clones 1-5 and 1-6 from peak 1, recurred in peak 3 (clone 3-5 in peak 3), and type VII clone 1-7 from peak 1 showed 100% amino acid identity with peak 3 clones 3-6, 3-7, and 3-8.
TABLE 1.
Comparison of amino acid identity among MSP-2 clones obtained within a peak or from different peaks
| Comparison | % Identity
|
|
|---|---|---|
| Least | Most | |
| Between peaks | ||
| 808 | ||
| Peak 1-peak 2 | 78 | 99 |
| Peak 1-peak 3 | 75 | 96 |
| Peak 2-peak 3 | 80 | 97 |
| 807 | ||
| Peak 1-peak 2 | 78 | 100 |
| Peak 1-peak 3 | 83 | 100 |
| Peak 2-peak 3 | 79 | 95 |
| Within peaks | ||
| 808 | ||
| Peak 1 | 78 | 99 |
| Peak 2 | 83 | 100 |
| Peak 3 | 79 | 100 |
| 807 | ||
| Peak 1 | 83 | 100 |
| Peak 2 | 88 | 100 |
| Peak 3 | 81 | 100 |
Within each peak of cyclic rickettsemia, a minimum of four polymorphic sequence types were identified (Fig. 2). Amino acid sequence identity between clones obtained within the same peak or between any two peaks ranged from 78 to 100% (Table 1). The ranges of amino acid identities within a peak were similar for clones obtained from each of three peaks of cyclic rickettsemia from both animals, 807 and 808 (Table 1). In peaks 1 and 2 from both animals, comparison of clones representing different types revealed clusters around 80 and 90% identity (Fig. 4). As persistent infection progressed in both animals, however, types within peak 3 tended to diverge, with amino acid identities that clustered less and became more evenly distributed from 70 to 97% (Fig. 4).
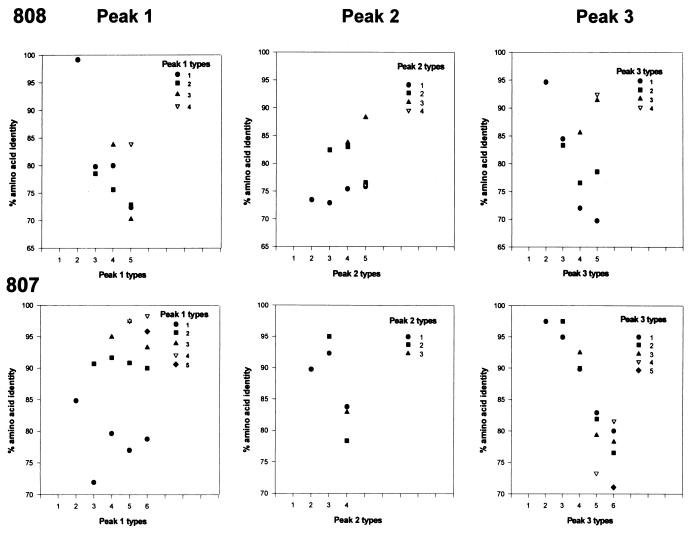
FIG. 4.
Pairwise comparison of amino acid identity of MSP-2 clone types obtained within peaks 1, 2, and 3 during persistent rickettsemia in animals 808 and 807. Types are defined based on the variable region shown in Fig. 2 and 3, amino acids 186 to 281. Each graph represents a single peak of cyclic rickettsemia. Within the peaks, each type (designated by a symbol) is compared to each of the other clone types obtained from the same peak (designated along the x axis) and plotted as the percent amino acid identity. Comparison of each type with itself is not shown. For example, from animal 808 peak 1, type 1 is compared with type 2 in the second column and the percent amino acid identity is indicated by a circle at 98.5%. Similarly, in the third column, types 1 and 2 are compared to type 3 and designated by a circle at 80% for type 1 and a square at 78% for type 2. In some cases the symbols overlap because the percent amino acid identities are similar; for example, in column 5, types 1 and 2 have similar percent amino acid identities (72.4 and 72.8%, respectively) compared to clone 5; therefore, the square that represents type 2 overlies the circle that represents type 1.
Expression of antigenically distinct MSP-2 during persistent rickettsemia.
To investigate whether antigenically distinct MSP-2 molecules were expressed during a single rickettsemic cycle, variant-specific antibodies were used in a two-color immunofluorescence assay. A. marginale-infected erythrocytes were collected at the same time as the blood used to generate MSP-2 clones V3 and V5 (5-10-96 in animal 808). Antibodies were generated against the V3 or V5 variants and cross-adsorbed with purified V5 or V3 protein to ensure variant specificity. Each antibody reacted specifically with its respective polypeptide in immunoblots (Fig. 5). Anti-V3 antibody reacted with the recombinant V3 polypeptide with an apparent molecular size of 27 kDa, and anti-V5 antibody reacted with the recombinant V5 polypeptide with an apparent molecular size of 29 kDa. The 2-kDa difference between the apparent molecular sizes of V3 and V5 reflected the predicted difference attributable to the nucleotide deletions within the polymorphic region of V3 relative to V5 (Fig. 2). The anti-V3 antibody did not react with the 29-kDa V5 polypeptide, nor did the anti-V5 antibody react with the 27-kDa V3 polypeptide. Therefore, these antibodies bound different variant-specific epitopes. Neither antiserum reacted with the negative control B. bigemina RAP-1 polypeptide (Fig. 5), nor did the control anti-IL-4 antibody react with the V3 and V5 polypeptides.
FIG. 5.
Binding of variant-specific antibody to respective MSP-2 polymorphic variants. Recombinant V3 (lanes 3 and 6), V5 (lanes 2 and 5), or the negative control RAP-1 (lanes 1 and 4) was purified, electrophoresed on a polyacrylamide gel, and transferred to nitrocellulose. The membranes were reacted with either anti-V3 (lanes 1 to 3) or anti-V5 (lanes 4 to 6) antibody followed by goat anti-rabbit IgG and developed by using enhanced chemiluminescence.
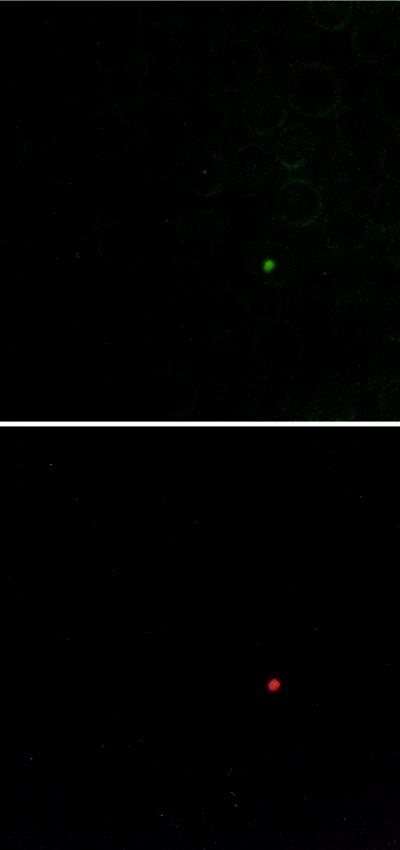
Immunofluorescence using the specific antibodies, anti-V3 and anti-V5, showed that polymorphic MSP-2 molecules can be coexpressed within a single A. marginale-infected erythrocyte obtained from a peak of cyclic rickettsemia (Fig. 6). These data confirm that A. marginale can contain epitopes encoded by different msp-2 transcripts. However, A. marginale that contained epitopes reactive with either the anti-V3 or anti-V5 antibodies were also identified (data not shown), indicating that a minimum of three antigenic types (V3 only, V5 only, and V3 plus V5) of MSP-2 are expressed within peak 2 of animal 808. The negative control anti-IL-4 antibodies did not bind organisms on the same blood smear.
FIG. 6.
Coexpression of MSP-2 variant types within a single A. marginale-infected erythrocyte. The smears used in the immunofluorescence were made from the same blood used to generate MSP-2 clones V3 and V5. The same microscopic field is shown in both panels. An excitation wavelength of 450 to 490 nm (top) or 546 nm (bottom) was used to detect A. marginale bound by fluorescein-labeled anti-V5 (top) or rhodamine-labeled anti-V3 (bottom) antibodies.
DISCUSSION
Recovery from acute A. marginale infection results in persistence characterized by repetitive cyclic fluctuations of rickettsemia (6, 7, 11). Sequential rickettsemic cycles during persistent infection were originally detected by using nucleic acid probe hybridization based on the multicopy msp-1b gene (6, 7, 11). Competitive msp-5 PCR has increased sensitivity over msp-1b probe hybridization, which is limited to approximately 103 infected erythrocytes per ml of blood (6), and allows quantitation of A. marginale based on the single-copy msp-5 gene (36). The rickettsemic cycles in this study occurred at intervals similar to those described previously (6, 7, 11), with major peaks of up to 107 infected erythrocytes per ml of blood occurring every 6 to 8 weeks. Each cycle terminated after the major peak by a dramatic reduction in rickettsemia, hypothesized to reflect a variant-specific immune response (11). Because the number of organisms could be determined in a sample containing a known number of infected erythrocytes, the number of organisms within a single infected erythrocyte could be ascertained. A single A. marginale organism invades and replicates within an erythrocyte, giving rise to a cluster of organisms within each infected erythrocyte (12, 28). The estimated average of two to four organisms per infected erythrocyte determined in this study is consistent with prior studies indicating that a single invading organism replicates one to three times within the erythrocyte (28).
Following identification of rickettsemic cycles by competitive PCR (Fig. 1), msp-2 cDNA clones were sequenced from each rickettsemic peak and shown to have an approximately 300-nucleotide area of polymorphism. The nucleotide changes resulted in amino acid substitutions, deletions, or insertions (Fig. 2). Interestingly, the genetic diversity of msp-2 variants within each peak was similar to that seen in acute infection, during which several classes of variant msp-2 genes are transcribed (4). Within each peak of cyclic rickettsemia, a minimum of four different msp-2 cDNA sequence types were isolated, and the translated amino acid sequences had the predicted open reading frames which were correspondingly polymorphic (Fig. 2). Clones were grouped into types defined by the polymorphic region between amino acids 186 and 281 (Fig. 2). This is a minimal estimate of sequence variation, as some of the sequences have known and may have unknown amino acid changes outside this region. The possibility of creating artificial variants during PCR amplification was considered. The large central region of polymorphism in the msp-2 clones is not likely due to Taq polymerase misincorporation because the number of nucleotide changes in the msp-2 clones is considerably greater than the deduced Taq misincorporation rate and greater than that seen in the msp-5 control, using the same RNA and identical amplification protocol. Because msp-5 is conserved within the genus Anaplasma (36), the low rate of nucleotide polymorphism observed in the msp-5 cDNA clones either reflects slight variation within the Florida strain or was generated in the reverse transcription or elongation during PCR. It has also been suggested that recombinant DNA molecules can form during PCR when two distinct gene sequences are coamplified (16). Polymorphic msp-2 cDNA clones generated in this study are not likely the result of recombination during PCR for several reasons: (i) all of the clones had the predicted msp-2 open reading frames, (ii) generation and sequencing of an additional 10 clones from peak 2 (5-10-96) of animal 808 yielded identical sequences, and (iii) frequency of homologous recombination is low (16) and would therefore not account for the high frequency of polymorphism observed in the msp-2 cDNA clones. Moreover, frequency of homologous recombination can be further reduced by an adequate DNA polymerase elongation time (16), a parameter that was taken into consideration in designing these experiments. Consequently, based on these data, we conclude that the observed MSP-2 polymorphism was generated in vivo, consistent with the msp-2 polymorphism in the genome (25).
The polymorphism of MSP-2 variants within each peak was prominent, with predicted amino acid sequence identities for clones ranging from approximately 80 to 100% (Table 1) and the identity between clones representing different types clustering around 80 and 90% (Fig. 4). Interestingly, the range of amino acid identities of clones representative of each type compared to other clones representative of different types obtained from within the same peak was similar to the range of amino acid identities of clones compared between any two peaks (Table 1). Comparison of AR5, an msp-2 cDNA clone transcribed during acute infection (4), with msp-2 cDNA clones from each peak in persistent infection showed blocks of identity between the acute and persistent infection types. However, none of the clones were 100% identical between acute and persistent infection. The occurrence of polymorphic msp-2 transcripts during rickettsemic cycles in persistent infection together with the polymorphism between AR5, an msp-2 clone from acute infection, and the msp-2 clones from persistent infection supports our hypothesis that polymorphic msp-2 variants emerge during rickettsemic cycles in persistent infection.
Even though multiple polymorphic msp-2 transcripts occurred within a peak, whether the whole population of emergent organisms expresses the same set of MSP-2 molecules was not known. Dual-staining immunofluorescence confirms that a minimum of three antigenically distinct populations are present in a single peak: rickettsia-expressed MSP-2 recognized only by the anti-V3 variant-specific antibody, MSP-2 recognized only by anti-V5 variant-specific antibody, or MSP-2 recognized by both antibodies. Expression of MSP-2 recognized by both antibodies (Fig. 6) may reflect two separate MSP-2 molecules expressed in the same A. marginale organism or expression of a hybrid MSP-2 including both V3 and V5 epitopes; less likely, native V3 MSP-2 is bound by anti-V5 antibody due to cross-reacting epitopes not represented in the recombinant proteins used in adsorption. Nonetheless, it is clear that complete clearance by immune responses to MSP-2 requires recognition of a combination of MSP-2 epitopes. During acute A. marginale infection, multiple MSP-2 antigenic variants are expressed (4), followed by development of a primary immune response which eliminates >99% of the rickettsiae but fails to completely clear the infection (11). Each rickettsemic cycle during persistent infection also attains a high-level peak rickettsemia that rapidly declines, similar to acute infection, and may result from recognition of most but not all variant MSP-2 by a new primary immune response.
Multiple msp-2 variant types emerge within each peak of cyclic rickettsemia. However, a few variant types recur in subsequent peaks; either these variant types evaded the existing immune response or their emergence is unrelated to immune pressure and persistence. Recurrent types were always accompanied by emergence of new variant types. Expression of recurrent types together with new types may contribute to the formation of new epitopes that were not previously recognized by the immune response. Because MSP-2 dimerizes on the surface of A. marginale (35), simultaneous expression of variants within a single organism could result in formation of conformationally unique epitopes. If a few organisms expressing a unique combination of variant MSP-2 are able to escape immune clearance in one rickettsemic cycle, then a new population of A. marginale expressing the combination of a recurrent type and a newly generated variant MSP-2 may emerge in the subsequent cycle. Another possibility is that other hypervariable regions occur 5′ or 3′ of the central polymorphic region and may result in expression of unique MSP-2 variants that are identical in the central polymorphic region examined in this study. Whether the expression of variant MSP-2 molecules results in a completely antigenically distinct population in each peak and how the immune system controls each rickettsemic cycle are unknown and appear to be critical for understanding the role of antigenic variation in persistent rickettsemia.
ACKNOWLEDGMENTS
We thank Curtis Brandt for excellent technical assistance and William P. Cheevers, Isidro Hötzel, and Carlos Suarez for helpful comments and discussion. We also acknowledge Jeff Abbott, Ignacio Echaide, Carla Robertson, and Bev Hunter for technical advice and support and Barbara von Beust for providing the anti-bovine IL-4 serum.
This work was supported by NIH/NIAID grant 5 K08 AI01371-02, USDA grant 95-372204-2348, the U.S.-Israel BARD program, and AVMF grant 96-09.
REFERENCES
- 1.Barbet A F, Semu S M, Chigagure N, Kelly P J, Jongejan F, Mahan S M. Size variation of the major immunodominant protein of Cowdria ruminantium. Clin Diagn Lab Immunol. 1994;1:744–746. doi: 10.1128/cdli.1.6.744-746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouboula M, Legoux P, Pességué B, Delpech B, Dumont X, Piechaczyk M, Casellas P, Shire D. Standardization of mRNA titration using a polymerase chain reaction method involving co-amplification with a multispecific internal control. J Biol Chem. 1992;267:21830–21838. [PubMed] [Google Scholar]
- 3.Brouqui P, Dumler J S, Raoult D, Walker D H. Antigenic characterization of ehrlichiae: protein immunoblotting of Ehrlichia canis, Ehrlichia sennetsu, and Ehrlichia risticii. J Clin Microbiol. 1992;30:1062–1066. doi: 10.1128/jcm.30.5.1062-1066.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eid G, French D M, Lundgren A M, Barbet A F, McElwain T F, Palmer G H. Expression of major surface protein 2 antigenic variants during acute Anaplasma marginale rickettsemia. Infect Immun. 1996;64:836–841. doi: 10.1128/iai.64.3.836-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisemann G H, Osterman J V. Identification of strain-specific and group-reactive antigenic determinants on the Karp, Gilliam, and Kato strains of Rickettsia tsutsugamushi. Am J Trop Med Hyg. 1997;34:1173–1178. doi: 10.4269/ajtmh.1985.34.1173. [DOI] [PubMed] [Google Scholar]
- 6.Eriks I S, Palmer G H, McGuire T C, Allred D R, Barbet A F. Detection and quantitation of Anaplasma marginale in carrier cattle by using a nucleic acid probe. J Clin Microbiol. 1989;27:279–284. doi: 10.1128/jcm.27.2.279-284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriks I S, Stiller D, Palmer G H. Impact of persistent Anaplasma marginale rickettsemia on tick infection and transmission. J Clin Microbiol. 1993;31:2091–2096. doi: 10.1128/jcm.31.8.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson B. Identification and partial characterization of Rickettsia tsutsugamushi major protein immunogens. Infect Immun. 1985;50:603–609. doi: 10.1128/iai.50.3.603-609.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 321–359. [Google Scholar]
- 10.Keohavong P, Thilly W G. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci USA. 1989;86:9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kieser S T, Eriks I S, Palmer G H. Cyclic rickettsemia during persistent Anaplasma marginale infection of cattle. Infect Immun. 1990;58:1117–1119. doi: 10.1128/iai.58.4.1117-1119.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krier J P, Gothe R, Ihler G M, Krampitz H E, Mernaugh G, Palmer G H. The hemotrophic bacteria: the families Bartonellaciae and Anaplasmataceae. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. New York, N.Y: Springer-Verlag; 1991. pp. 3994–4022. [Google Scholar]
- 13.Loewen P C, Switala J. Template secondary structure can increase the error frequency of the DNA polymerase from Thermus aquaticus. Gene. 1995;164:59–63. doi: 10.1016/0378-1119(95)00383-h. [DOI] [PubMed] [Google Scholar]
- 14.Losos G J. Anaplasmosis. In: Losos G J, editor. Infectious tropical diseases of domestic animals. Essex, United Kingdom: Longman Press; 1986. pp. 743–795. [Google Scholar]
- 15.McGuire T C, Palmer G H, Goff W L, Johnson M I, Davis W C. Common and isolate-restricted antigens of Anaplasma marginale detected with monoclonal antibodies. Infect Immun. 1984;45:697–700. doi: 10.1128/iai.45.3.697-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyerhans A, Vartanian J P, Wain-Hobson S. DNA recombination during PCR. Nucleic Acids Res. 1990;18:1687–1691. doi: 10.1093/nar/18.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata M, Yoshida Y, Osono M, Ohashi N, Oyanagi M, Urakami H, Tamura A, Nogami S, Tanaka H, Kawamura A J. Production and characterization of monoclonal strain-specific antibodies against prototype strains of Rickettsia tsutsugamushi. Microbiol Immunol. 1986;30:599–610. doi: 10.1111/j.1348-0421.1986.tb02987.x. [DOI] [PubMed] [Google Scholar]
- 18.Oaks E V, Stover C K, Rice R M. Molecular cloning and expression of Rickettsia tsutsugamushi genes for two major protein antigens in Escherichia coli. Infect Immun. 1987;55:1156–1162. doi: 10.1128/iai.55.5.1156-1162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oaks E V, Rice R M, Kelly D J, Stover C K. Antigenic and genetic relatedness of eight Rickettsia tsutsugamushi antigens. Infect Immun. 1989;57:3116–3122. doi: 10.1128/iai.57.10.3116-3122.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohashi N, Tamura A, Ohta M, Hayashi K. Purification and partial characterization of a type-specific antigen of Rickettsia tsutsugamushi. Infect Immun. 1989;57:1427–1431. doi: 10.1128/iai.57.5.1427-1431.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohashi N, Nashimoto H, Ikeda H, Tamura A. Diversity of immunodominant 56-kDa type-specific antigen (TSA) of Rickettsia tsutsugamushi. Sequence and comparative analyses of the genes encoding TSA homologues from four antigenic variants. J Biol Chem. 1992;267:12728–12735. [PubMed] [Google Scholar]
- 22.Palmer G H, McGuire T C. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J Immunol. 1984;133:1010–1015. [PubMed] [Google Scholar]
- 23.Palmer G H, Barbet A F, Davis W C, McGuire T C. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science. 1986;231:1299–1302. doi: 10.1126/science.3945825. [DOI] [PubMed] [Google Scholar]
- 24.Palmer G H, Oberle S M, Barbet A F, Goff W L, Davis W C, McGuire T C. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect Immun. 1988;56:1526–1531. doi: 10.1128/iai.56.6.1526-1531.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer G H, Eid G, Barbet A F, McGuire T C, McElwain T F. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect Immun. 1994;62:3808–3816. doi: 10.1128/iai.62.9.3808-3816.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer G H, McElwain T F. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet Parasitol. 1995;57:233–253. doi: 10.1016/0304-4017(94)03123-e. [DOI] [PubMed] [Google Scholar]
- 27.Reddy G R, Sulsona C R, Harrison R H, Mahan S M, Burridge M J, Barbet A F. Sequence heterogeneity of the major antigenic protein 1 genes from Cowdria ruminantium isolates from different geographical areas. Clin Diagn Lab Immunol. 1996;3:417–422. doi: 10.1128/cdli.3.4.417-422.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ristic M, Watrach A M. Anaplasmosis. VI. Studies and a hypothesis concerning the cycle of development of the causative agent. Am J Vet Res. 1963;24:267–277. [PubMed] [Google Scholar]
- 29.Seong S Y, Huh M S, Jang W J, Park S G, Kim J G, Woo S G, Choi M S, Kim I S, Chang W H. Induction of homologous immune response to Rickettsia tsutsugamushi Boryong with a partial 56-kilodalton recombinant antigen fused with the maltose-binding protein MBP-Bor56. Infect Immun. 1997;65:1541–1545. doi: 10.1128/iai.65.4.1541-1545.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stover C K, Marana D P, Carter J M, Roe B A, Mardis E, Oaks E V. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect Immun. 1990;58:2076–2084. doi: 10.1128/iai.58.7.2076-2084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura A, Ohashi N, Urakami H, Takahashi K, Oyanagi M. Analysis of polypeptide composition and antigenic components of Rickettsia tsutsugamushi by polyacrylamide gel electrophoresis and immunoblotting. Infect Immun. 1985;48:671–675. doi: 10.1128/iai.48.3.671-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Vliet A H, Jongejan F, van Kleef M, van der Zeijst B A. Molecular cloning, sequence analysis, and expression of the gene encoding the immunodominant 32-kilodalton protein of Cowdria ruminantium. Infect Immun. 1994;62:1451–1456. doi: 10.1128/iai.62.4.1451-1456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Vliet A H, van der Zeijst B A, Camus E, Mahan S M, Martinez D, Jongejan F. Use of a specific immunogenic region on the Cowdria ruminantium MAP1 protein in a serological assay. J Clin Microbiol. 1995;33:2405–2410. doi: 10.1128/jcm.33.9.2405-2410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vliet A H, van der Zeijst B A, Camus E, Mahan S M, Martinez D, Jongejan F. Recombinant expression and use in serology of a specific fragment from the Cowdria ruminantium MAP1 protein. Ann N Y Acad Sci. 1996;791:35–45. doi: 10.1111/j.1749-6632.1996.tb53509.x. [DOI] [PubMed] [Google Scholar]
- 35.Vidotto M C, McGuire T C, McElwain T F, Palmer G H, Knowles D P J. Intermolecular relationships of major surface proteins of Anaplasma marginale. Infect Immun. 1994;62:2940–2946. doi: 10.1128/iai.62.7.2940-2946.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visser E S, McGuire T C, Palmer G H, Davis W C, Shkap V, Pipano E, Knowles D P J. The Anaplasma marginale msp5 gene encodes a 19-kilodalton protein conserved in all recognized Anaplasma species. Infect Immun. 1992;60:5139–5144. doi: 10.1128/iai.60.12.5139-5144.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wanduragala L, Ristic M. Anaplasmosis. In: Woldehiwet Z, Ristic M, editors. Rickettsial and chlamydial diseases of domestic animals. Oxford, United Kingdom: Pergamon Press; 1993. pp. 65–87. [Google Scholar]