Abstract
One component of host defense at mucosal surfaces appears to be epithelium-derived peptides with antimicrobial activity called defensins. Human β-defensin 1 (hBD-1) represents the first member of the β-defensin family isolated from humans and has been implicated in the pathogenesis of cystic fibrosis. We describe in this report the isolation and characterization of a murine homolog of hBD-1 called mouse β-defensin 1 (mBD-1). The predicted amino acid sequence shows the hallmark features of other known epithelial β-defensins, including the ordered array of six cysteine residues. Analysis of a genomic clone of mBD-1 revealed two exons separated by a 15-kb intron. By use of fluorescence in situ hybridization, the mBD-1 gene was localized at the proximal portion of chromosome 8, the site where mouse α-defensins are found. Lysates from cells transfected with the mBD-1 cDNA showed antibacterial activity against gram-positive and gram-negative bacteria. mBD-1 transcripts were found in kidney, liver, and female reproductive organ tissues. In the airways, mBD-1 is expressed diffusely throughout the epithelial cells of the large proximal airways with less expression in the small distal airways and no expression in alveolar cells. The present study demonstrates that a β-defensin potentially homologous to human β-defensin 1 is present in the respiratory system and other mucosal surfaces in mice.
Antimicrobial peptides have been found in a wide array of animal species, ranging from insects to lower vertebrates and mammals (8). These peptides contribute to innate host defense against a number of bacterial and fungal pathogens (11). Mammalian defensins are small antimicrobial peptides (3.5 to 4.5 kDa) that are characterized by the presence of six cysteines which form three disulfide bonds whose ordered array defines the classification as α- or β-defensins. The α-defensins are found in granulocytes (8) and Paneth cells of the small intestine (14, 18) in various species, whereas β-defensins occur in leukocytes of cattle (28, 31) and fowl (7, 12). Further, β-defensins are expressed in epithelial cells, where they contribute to the host defense system of mucosal surfaces. Lingual and tracheal antimicrobial peptides are β-defensins expressed in bovine epithelial cells of the tongue and trachea (2, 4, 26). The first β-defensin isolated from humans, human β-defensin 1 (hBD-1), is found in salivary gland, airway, prostate, and placenta tissues, among other tissues (1, 10, 19, 32).
It has been suggested that the salt-sensitive defect of antimicrobial peptides may underlie the pathogenesis of cystic fibrosis (CF) lung disease (10, 29). Smith et al. (29) proposed that the defect in the CF gene product, the CF transmembrane conductance regulator, leads to elevated NaCl concentrations in the airway surface fluid, which inactivates the antimicrobial activity, leading to colonization of the airway with bacterial pathogens, which is the hallmark of the disease (9, 10, 15, 28, 29). Our data suggests that hBD-1 is one of the antimicrobial molecules disabled at the airway surface in CF (10). It would be of great importance to have a suitable animal model, such as a mouse or rat model, to study the role of β-defensins in pulmonary host defense, although little is known about defensin biology in rodents. Cryptins, which are expressed in the Paneth cells of the small intestine, are the only defensins identified in the mouse (5, 13, 22, 23); notably, mouse neutrophils lack defensins (6).
The aim of the present study was to evaluate defensin activity in epithelia of the mouse. We isolated a murine β-defensin, designated mouse β-defensin 1 (mBD-1), and analyzed its gene structure, antimicrobial function, and expression patterns.
MATERIALS AND METHODS
Cloning of mBD-1 cDNA and genomic sequence.
The hBD-1 cDNA sequence (10) was used to perform a BLAST search at the website of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov). A 444-bp mouse cDNA expressed sequence tag submitted by Marra et al. (accession no. AA065510) was found that showed 51% identity with hBD-1 at the amino acid level, including the six β-defensin-specific cysteines. Reverse transcriptase (RT)-PCR was used to clone the full-length cDNA sequence. Total RNA was isolated from a C57BL/6 mouse kidney by using Trizol (Gibco BRL) and further purified to poly(A)+ RNA by using oligo(dT) columns (Qiagen). Poly(A)+ RNA (approximately 100 ng) was reverse transcribed by using NotI-(dT)18 as the primer (First-Strand cDNA Synthesis Kit; Pharmacia Biotech), and 10% of the reaction product was used for a PCR. Primers specific to mBD-1 were designed as follows: forward primer (m-def 1), 5′-CGAAGCTTCACATCCTCTCTGCACTCTGG-3′ (nucleotides 4 to 24 of the GenBank sequence plus nine nucleotides at the 5′ end containing a restriction site for HindIII); reverse primer (m-def 2), 5′-CGACTAGTCCAGGCAGATGTTCTGG-3′ (nucleotides 433 to 444 of the GenBank sequence plus eight nucleotides at the 5′ end containing a restriction site for SpeI). The PCR products were analyzed on a 1.5% agarose gel, and a band of 440 bp was isolated and subcloned into Bluescript II SK− (Stratagene).
Cloning of the genomic sequence.
A mouse genomic library constructed in vector FIX II (Stratagene) was screened by standard procedures (25) with a probe generated by PCR using mBD-1 cDNA as the template and gene-specific primers (forward, 5′-TTTCACATCCTCTCTGCACT-3′ or 5′-TGCACTCTGGACCCTGGCT-3′; reverse, 5′-ACCTGGCTCCATCTGGGAGA-3′ or 5′-CCATCTGGGAGAAAAGAAAACA-3′). Positive clones were purified, and genomic DNA was isolated and subcloned into Bluescript II KS−. The genomic clones were analyzed by partial sequencing and digestion with restriction endonucleases.
Analysis of the chromosomal localization by fluorescence in situ hybridization (FISH).
Two gene-specific primers were used to amplify a sequence spanning a region including parts of exon 1 and the intron. The PCR product was used as a probe to screen a mouse genomic bacterial artificial chromosome library. The DNA of positive clones was labeled with digoxigenin dUTP by nick translation and hybridized to normal mouse metaphase chromosomes. The chromosome of interest was identified by analysis of the 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) banding pattern and finally by cohybridization of the mBD-1-specific probe with a probe specific for the telomeric region of chromosome 8 (GenomeSystems, Inc.).
Testing of antimicrobial activity.
For testing of antibacterial activity, we used lysates from cells transfected with the mBD-1 cDNA in a transfection vector. Staphylococcus aureus (clinical isolate provided by the Microbiology Laboratory, University of Pennsylvania Health System), Escherichia coli D31 (30), and Pseudomonas aeruginosa (clinical isolate provided by the Microbiology Laboratory, University of Pennsylvania Health System) were used as test organisms in both diffusion assays and liquid broth assays. To obtain the cell lysates, human SW 13 (ATCC CCL 105) adrenal adenocarcinoma cells grown in Dulbecco modified Eagle medium with 2.5% fetal calf serum in the absence of antibiotics were transfected with pcDNA3.1− (Invitrogen) containing the full-length mBD-1 cDNA by using calcium phosphate methods (Profection Mammalian Transfection Systems; Promega). As a control, we used cells transfected with the vector containing no insert. Transfected cells and mock-transfected control cells were harvested 48 h posttransfection, pelleted by centrifugation, resuspended in 500 μl of distilled water, and lysed by brief sonication.
Antibacterial liquid broth assays were performed as previously described for α-defensins (27). Bacteria (5 × 104 CFU) grown in Luria-Bertani broth were exposed at mid-log phase to 50 μl of the cell lysates for 2 h at 37°C. Cell viability was measured by plating several dilutions and counting colonies the following day. Antibacterial diffusion assays were performed as described by Lehrer et al. (16), with some modifications. Bacteria were grown to mid-log phase in a solution of yeast extract (5 g/liter) and tryptone (10 g/liter) buffered with 10 mM piperazine-N,N-bis(2-ethanesulfonic acid) (PIPES) at pH 7.5, and 5 × 106 CFU were diluted in 10 ml of a solution additionally containing 1% agarose. After being poured onto 150-mm-diameter plates and cooling, cell lysates (5 μl) were pipetted into wells formed with a 4-mm cork corer and incubated at 30°C overnight. To analyze the salt dependency of the activity of mBD-1 against E. coli D31, NaCl was added to the agarose solution to obtain final NaCl concentrations of 50, 100, 200, and 500 mM. The exact NaCl concentrations in the solutions were measured by Na- or Cl-sensitive electrodes (Microelectrodes). The pH dependency of mBD-1 activity was studied by adjusting the pH of the agarose solution to 9.0 or 5.5 by adding NaOH or HCl. As a positive control of antibacterial activity, 5 μl of magainin 1 (Magainin Pharmaceuticals, Inc.) at 1 mg/ml was applied to agarose plates. Further, liquid broth assays were performed by adding NaCl (final concentrations, 7, 50, 100, 200, and 500 mM) to the cell lysates.
RNase protection assays.
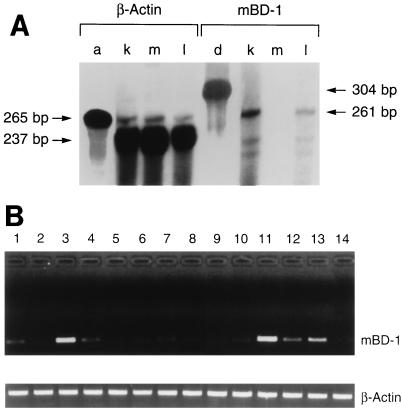
RNase protection assays were used to measure the level of mBD-1 transcripts in lung tissue. Total RNA from lung-trachea, skeletal muscle, and kidney tissues was isolated as described above. [32P]dCTP-labeled antisense riboprobes were prepared by in vitro transcription from the T7 RNA polymerase promoters of linearized Bluescript KS II− containing mBD-1 (304 bp) or β-actin cDNA (265 bp) (SP6/T7 Transcription Kit, Boehringer Mannheim). Hybridizations were performed in separate tubes by using 5 × 105 cpm of either an hBD-1 or a β-actin probe and 20 μg of total RNA. RNA-RNA hybrids were digested with RNases A and T1 to yield protected fragments of 261 and 237 bp for mBD-1 and β-actin, respectively (Lysate Ribonuclease Protection Kit; Amersham). Results were visualized by running the reaction products on a urea-polyacrylamide gel, followed by autoradiography.
RT-PCR.
A nested RT-PCR was used to detect low levels of mBD-1 mRNA in several tissues. Poly(A)+ RNA was isolated from mouse trachea, lung, tongue, esophagus, small bowel, large bowel, gall bladder, pancreas, skeletal muscle, heart, fallopian tube, ovary, vagina, and brain tissues and reverse transcribed as described above. The following primers were used in the first round of PCR: forward primer (m-def 5), 5′-TGGACCCTGGCTGCCACCACTATG-3′; reverse primer (m-def 6), 5′-GCTCATTCTTCAAACTACTGTCAG-3′. The products of this PCR were diluted 1:50 in distilled water, and an aliquot (2 μl) was used as the template for a nested PCR using the same reaction conditions as for the first round with the following primers: forward primer (m-def 7), 5′-ATGAAAACTCATTACTTTCTCCTGGTGATG-3′; reverse primer (m-def 8), 5′-CAATCCATCGCTCGCCTTTATGCTC-3′. The predicted size of the PCR product was 252 bp. RT was omitted from the negative control, whereas an RT-PCR with primers specific for mouse β-actin was used as a positive control. The PCR products were analyzed on a 1.5% agarose gel.
In situ hybridization.
Adult mouse nose, trachea, lung, kidney, liver, tongue, and heart tissues were harvested from CO2-euthanized animals, embedded in OCT (Tissue-TCK; Miles Inc.), and cryosectioned. Tissue sections 6 μm thick were mounted on slides and fixed in 4% paraformaldehyde in phosphate-buffered saline (pH 7.4) for 4 h at 4°C. Following dehydration through graded concentrations of ethanol, sections were desiccated overnight under vacuum and stored at −20°C. The following day, sections were treated with 10-μg/ml proteinase K for 30 min at 30°C, rinsed twice in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) for 30 s each time, fixed in 4% paraformaldehyde in phosphate-buffered saline, rinsed twice in 0.1 M triethanolamine (pH 8.0) for 4 min each time, incubated with 0.25% acetic anhydride in 0.1 M triethanolamine for 10 min at room temperature, and dehydrated through ethanol. Following drying under vacuum, prehybridization was performed for 4 h at 54°C in 10 mM Tris (pH 8.0)–50% formamide–2.5× Denhardt’s solution–0.6 M NaCl–1 mM EDTA–0.1% sodium dodecyl sulfate–500 μg of tRNA per ml–10 mM dithiothreitol (DTT). RNase control sections were treated with 200 μg of RNase A per ml for 1 h at 37°C before the prehybridization step. Sections were then hybridized with 35S-labeled antisense or sense probes at 5 × 106 cpm/ml for 16 h at 54°C in the prehybridization solution. Probes were synthesized by in vitro transcription from the T7 or T3 RNA polymerase promoter of full-length mBD-1 cDNA cloned in Bluescript KS II− (Promega In Vitro Transcription System). Following prehybridization, slides were washed in 4× SSC for 20 min at room temperature, treated with RNase (20 μg/ml) for 30 min at 37°C, and washed once with 2× SSC–1 mM DTT at room temperature for 10 min and three times in 0.5× SSC–1 mM DTT at 54°C. Slides were dehydrated with ethanol and air dried before dipping in photoemulsion (Kodak). Slides were developed and analyzed by bright- and dark-field microscopy using a Microphot-FXA Nikon microscope.
RESULTS
Cloning of the cDNA and genomic sequence for mBD-1 and localization of the mBD-1 gene.
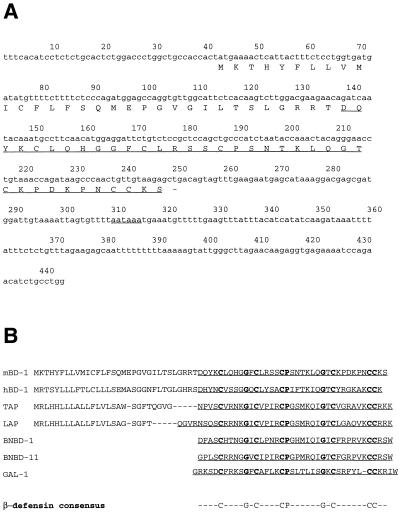
An expressed sequence tag homologous to the cDNA sequence of hBD-1 was obtained by performing a BLAST search at the NCBI website (Fig. 1A). This cDNA clone showed 62% nucleotide identity with the cDNA of hBD-1; the encoded peptide was 51% identical to the hBD-1 peptide. The murine clone was called mBD-1. The predicted peptide of mBD-1 showed the presence of the β-defensin-specific conserved amino acids, including the typical array of six cysteines (Fig. 1B). A cDNA clone for mBD-1 was isolated from kidney RNA by RT-PCR. A single product of the predicted size (440 bp) was obtained and cloned into Bluescript II SK−. The insert was sequenced and found to be identical to the database clone (Fig. 1A). The mBD-1 cDNA sequence consists of a 204-bp open reading frame encoding a peptide of 68 amino acids with a putative prepro sequence (Fig. 1A and B).
FIG. 1.
cDNA and peptide sequences of mBD-1. (A) cDNA and deduced amino acid sequences of mBD-1. The first underline indicates the putative mature peptide; the dash represents the termination codon; the second underline indicates the polyadenylation signal. (B) Comparison of the putative prepropeptide sequences of mBD-1, hBD-1, TAP, and LAP, as well as the peptide sequences of bovine neutrophil β-defensins 1 and 11 (BNBD-1 and BNBD-11, respectively) and gallinacin 1 (GAL-1). The bottom line presents the consensus sequence of β-defensins.
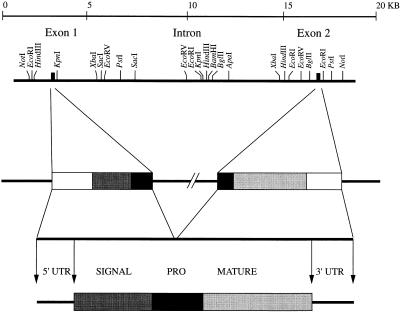
A mouse genomic library was screened with PCR-amplified sequences, and several positive clones were isolated and further analyzed by hybridization with radiolabeled probes corresponding to the 5′ and 3′ regions of the mBD-1 cDNA. Two clones contained the total sequence found in the cDNA. A restriction map of the mBD-1 gene is shown in Fig. 2. Comparison of the mBD-1 cDNA and the corresponding genomic clones indicated that the mBD-1 gene contains two exons separated by a 15-kb intron. A TATA box is located in the 5′ flanking region, and a polyadenylation signal is localized at position 309 of the cDNA. The exon-intron splice site sequences conform to the consensus rule (20) (data not shown). A sequence comparison analysis using a BLAST search at the NCBI identified no additional β-defensin-related mouse or human sequences, although a cDNA clone (accession no. X89820) obtained from a rat was found to be 94% identical to the mBD-1 cDNA.
FIG. 2.
Structure of the gene for mBD1. Restriction map of the gene for mBD-1 together with a schematic drawing of the gene, the cDNA, and the predicted structure of the prepropeptide. The gene is represented schematically with the following individual components: 5′ untranslated region (5′ UTR), open box; signal sequence, shaded box; interrupted prosequence (PRO), black box; mature peptide, shaded box; 3′ untranslated region (3′ UTR), open box.
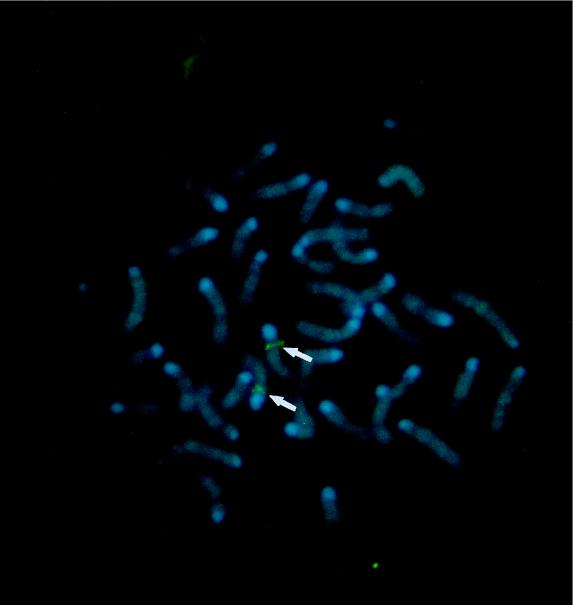
A bacterial artificial chromosome clone containing the mBD-1-encoding gene was labeled and used as a probe to localize the gene for mBD-1 by FISH analysis (GenomeSystems, Inc.). Initial experiments resulted in specific labeling of the proximal region of a medium-sized chromosome which was believed to be chromosome 8 on the basis of banding patterns. Further experiments using cohybridization of the mBD-1 probe with a probe specific for the telomeric region of chromosome 8 confirmed these results. Measurements of 10 metaphase chromosomes localized the mBD-1-specific hybridization signal to the proximal region of chromosome 8, an area that corresponds to band 8A4 (Fig. 3).
FIG. 3.
Chromosomal FISH of an mBD-1 probe to normal metaphase chromosomes derived from mouse embryo fibroblast cells. Chromosome 8 shows a positive green signal at its proximal portion (arrows), corresponding to band 8A4.
Analysis of antimicrobial activity.
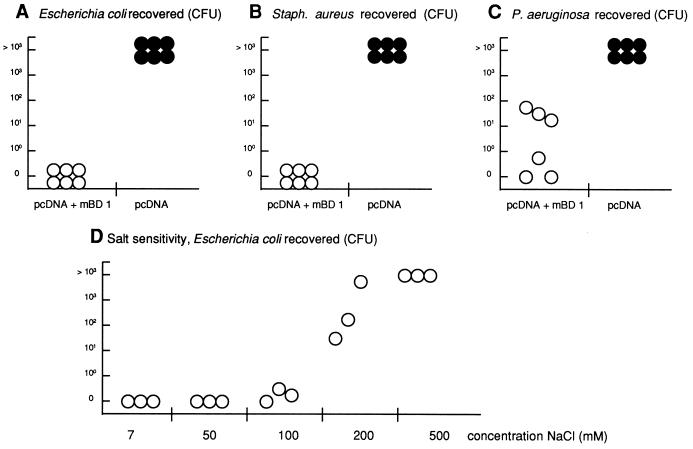
To test whether the cloned cDNA codes for an antimicrobial peptide, lysates from cells transfected with the mBD-1 cDNA were used in different assays and found to be active against all of the bacteria used, as well as in the diffusion assay (data not shown) and the liquid broth assay (Fig. 4A to C). Lysates from cells transfected with an empty vector failed to demonstrate killing activity, supporting the specificity of the assay (Fig. 4A to C). These results indicate that mBD-1 has antimicrobial activities against both gram-positive and gram-negative bacteria. Further, this activity is substantially diminished at high concentrations of NaCl (Fig. 4D) and at an acidic pH (data not shown).
FIG. 4.
Results of antibacterial activity assays. (A to C) Antibacterial liquid broth coincubation assay. E. coli (A), S. aureus (B), and P. aeruginosa (C) were added (103 to 104 CFU) to 50 μl of lysates from cells transfected with a vector containing the full-length cDNA of mBD-1 (empty circles) or with the vector alone (filled circles) and analyzed for microbial killing. Each circle represents results from an individual transfection experiment. (D) Salt sensitivity of the antimicrobial activity of mBD-1. Extracts from mBD-1-transfected cells were incubated with 5 × 104 CFU of E. coli D31 in 10 mM phosphate buffer (pH 7.4) and the indicated concentrations of NaCl. After incubation at 37°C for 1 h, serial dilutions were plated and colonies were counted the following day.
Expression of mBD-1 in mouse tissues.
RNA from various tissues was also evaluated for mBD-1 transcripts by RNase protection analysis. This sensitive assay detected mBD-1 mRNA from an extract of the whole lung (i.e., the lung and trachea) but at much lower levels than in the kidney. This assay failed to detect mBD-1 RNA in many other tissues, such as muscle (Fig. 5A). A β-actin probe was used as an internal control.
FIG. 5.
Analysis of mBD-1 expression by RNase protection analysis and RT-PCR (A) Measurement of mBD-1 expression by RNase protection analysis. Total RNA from the kidney (k), skeletal muscle (m), and whole lung (i.e., lung and trachea) (l) was investigated. Hybridization to a labeled riboprobe against β-actin (a) and mBD-1 (d) transcripts was performed a separate tube. (B) Detection of mBD-1 expression in various mouse tissues by nested RT-PCR. Poly(A)+ RNA was isolated from mouse tissues and reverse transcribed, and the cDNAs were amplified by using two pairs of mBD-1-specific primers. A single 250-bp band was amplified by using gene-specific primers. As a control, β-actin cDNA was amplified by using specific primers. Lanes: 1, trachea, 2, lung (lung parenchyma without cartilaginous airways); 3, tongue; 4, esophagus; 5, small bowel; 6, large bowel; 7, gall bladder; 8, pancreas; 9, skeletal muscle; 10, heart; 11, fallopian tube; 12, ovary; 13, vagina; 14, brain.
An RT-PCR using mRNA obtained from several mouse tissues was performed to evaluate low-level expression of mBD-1. Tongue, esophagus, fallopian tube, ovary, and vagina tissues showed expression of mBD-1, as did tracheal and, to a lesser extent, distal lung tissues (i.e., no cartilaginous airway tissue) (Fig. 5B). β-Actin was again used as a positive control. No bands were detected when RT was omitted (data not shown).
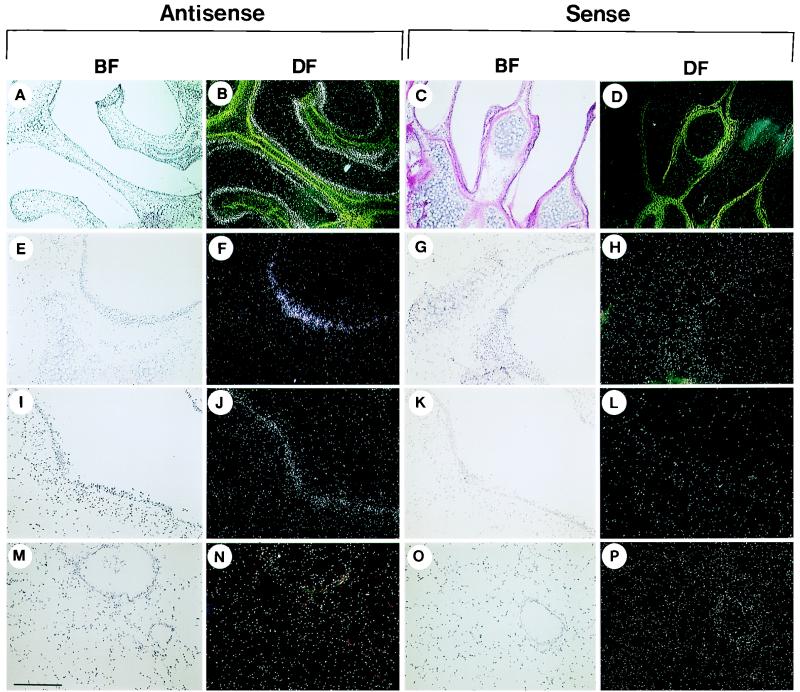
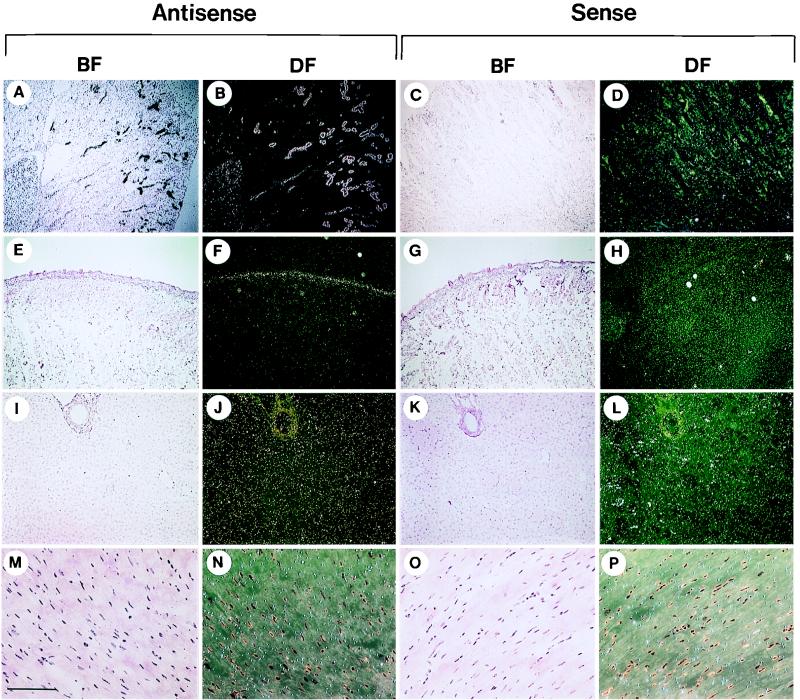
The tissue distribution of mBD-1 was analyzed at the cellular level by in situ hybridization. mBD-1 transcripts were detected in the epithelia of the nose (Fig. 6A and B), the large cartilaginous airways (Fig. 6E and F), and the larger bronchioles (Fig. 6I and J) when tissue sections were hybridized to the antisense probe. Hybridization signal appeared diffusely throughout virtually all of the surface epithelial cells of the conducting airway. The signal was substantially reduced or absent in the small bronchioles or lung parenchyma (Fig. 6M and N). Positive results were also seen in the distal tubules and collecting ducts of the kidney (Fig. 7A and B) and the epithelium covering the tongue (Fig. 7E and F) and as a diffuse signal in the liver (Fig. 7I-J). No signal over the background was detected in the heart muscle (Fig. 7M and N). Hybridization with sense mBD-1 riboprobes (Fig. 6 and 7) or digestion of the tissue with RNase before hybridization to the antisense probe (data not shown) resulted in no detectable signal, confirming the sensitivity of the assay.
FIG. 6.
Detection of transcripts encoding mBD-1 in the mouse respiratory tract. Antisense and sense probes were used to detect the tissue distribution of mBD-1 expression. Representative sections in dark and bright fields (DF and BF, respectively) from the nose (A to D), trachea (E to H), large bronchioles (I to L), and terminal bronchioles and lung parenchyma (M to P) are shown. Bars: A to D, 0.7 mm; E to P, 270 μm.
FIG. 7.
Detection of transcripts encoding mBD-1 in extrapulmonary organs. Antisense and sense probes were used to detect the tissue distribution of mBD-1 expression. Representative sections in dark and bright fields (DF and BF, respectively) from the kidney (A to D), tongue (E to H), liver (I to L), and heart muscle (M to P) are shown. Bars: A to H, 0.7 mm; I to L, 270 μm; M to P, 130 μm.
DISCUSSION
In the present report, we describe the isolation of a mouse cDNA with homology to hBD-1 whose expressed peptide in transfected cells has antimicrobial activity against various gram-positive and gram-negative organisms. We have named this antimicrobial peptide mBD-1.
The cDNA sequence was found during a BLAST search using the nucleotide sequence of hBD-1. Whereas the similarity of mBD-1 to hBD-1 is only 62% on the nucleotide level and 51% on the amino acid level, the amino acid sequence of the putative mBD-1 prepropeptide revealed the structural hallmarks of β-defensins. The amino-terminal prepro portion of the peptide contains several hydrophobic residues which are characteristic of β-defensins and are found in other β-defensin family members expressed on mucosal surfaces, such as the tracheal antimicrobial peptide (TAP) (2, 4), the lingual antimicrobial peptide (LAP) (26), or hBD-1 (1, 10). The putative mature peptide contains six cysteine residues spaced in a typical array and other conserved amino acids that may have important roles in determining the conformation and function of β-defensins, such as G10, P18, G25, and T26 (33). Several characteristic charged residues are present in the putative mature peptide.
Analysis of the mBD-1 gene revealed the presence of two exons surrounding a 15-kb intron. The structure of the mBD-1 gene is identical to those of other β-defensins, such as LAP (2) or hBD-1 (17), each of which contains two exons. The first exon contains the prepeptide and part of the propeptide, whereas the mature peptide and the remaining part of the propeptide are encoded by the second exon. These data indicate an evolutionary conservation of the members of the β-defensin group. Analysis of the cDNA and genomic clones of mBD-1 revealed the presence of typical regulatory elements, such as a TATA box and a polyadenylation signal; however, no binding sites for transcription factors that are involved in the inflammatory response could be found. This differs from the 5′ region of the TAP-encoding gene, which contains an NF-κB site upstream of the transcriptional start site (26). Expression of other mucosal β-defensins, such as LAP or TAP, is upregulated by inflammatory mediators and bacterial components (2, 3, 24, 26). Similar studies of mBD-1 are under way to evaluate the potential role of regulation in its biology in vivo.
The chromosomal localization of mBD-1 was analyzed by FISH and mapped to the proximal region of chromosome 8, an area which also contains the Defcr locus, where genes for mouse α-defensins are found (21). Further, this area is homologous to human chromosome 8p23, where human α- and β-defensins are located (17). These results indicate not only a close relationship of mBD-1 and cryptdins but, further, make evolutionary conservation of the α- and β-defensin groups likely. Thus, development of these two groups of antimicrobial peptides may have taken place during the development of mammalia before rodents and primates separated.
To further prove that the cloned cDNA sequence encodes an antimicrobial peptide, the activity of the expressed peptide against different bacteria was analyzed. Lysates of cells transfected with the mBD-1 cDNA revealed activity against gram-negative and gram-positive bacteria. Further, this activity was lost at high concentrations of salt and at an acidic pH. These antimicrobial activities are characteristic of defensins and are also shared by other known β-defensins (2, 4, 10, 26).
To compare the expression patterns of mBD-1 with those of other known epithelial β-defensins, an RNase protection assay, RT-PCR, and in situ hybridization were performed. Our data presented here indicate that mBD-1 is expressed in a pattern similar to that of hBD-1; the site of the most abundant expression is the distal collecting ducts of the kidney and other urogenital organs. This suggests that mBD-1 plays a role in preventing ascending infections such as pyelonephritis in the kidney and pelvic inflammatory disease in the female reproductive tract. Expression of mBD-1 is also found throughout the surface epithelia of the conducting airways. There appears to be a gradient of mBD-1 expression throughout the conducting airways, with the highest expression noted in proximal structures. This differs from hBD-1, which is expressed more uniformly throughout the conducting airways (10, 19). High expression of mBD-1 at sites within the oropharynx (e.g., the tongue) may also contribute to innate immunity in the lung by providing proximal defense against inhaled and aspirated organisms. The expression pattern of mBD-1 in airways is of specific importance in light of recent findings that implicate a salt-dependent defect of hBD-1 in the pathogenesis of CF (10, 29). This model specifically suggests that an elevated concentration of NaCl in the airway surface fluid of CF patients inactivates the antimicrobial activity of defensin and/or other antimicrobial molecules, possibly resulting in bacterial colonization and infection.
In summary, mBD-1 is a murine β-defensin diffusely expressed through the epithelia of multiple organs. It likely plays an important role in innate immunity at multiple mucosal sites.
ACKNOWLEDGMENTS
We thank the Microbiology Laboratory, University of Pennsylvania Health System, for making bacterial cultures available. The contribution of the Cell Morphology Core of the Institute of Human Gene Therapy was greatly appreciated.
This study was supported by the Cystic Fibrosis Foundation, NIDDK, and NHLBI of the NIH, as well as Genovo, Inc. R.B. received a fellowship from The Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Bensch K W, Raida M, Magert H-J, Schulz-Knappe P, Forssmann W G. hBD-1: a novel β-defensin from human plasma. FEBS Lett. 1995;368:331–335. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 2.Diamond G, Jones D E, Bevins C L. Airway epithelial cells are the site of expression of a mammalian antimicrobial peptide gene. Proc Natl Acad Sci USA. 1993;90:4596–4600. doi: 10.1073/pnas.90.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamond G, Russell J P, Bevins C L. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diamond G, Zasloff M, Eck H, Brasseur M, Maloy W L, Bevins C L. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenhauer P B, Harwig S S L, Lehrer R I. Cryptdins: antimicrobial defensins of the murine small intestine. Infect Immun. 1992;60:3556–3565. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenhauer P B, Lehrer R I. Mouse neutrophils lack defensins. Infect Immun. 1992;60:3446–3447. doi: 10.1128/iai.60.8.3446-3447.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans E W, Beach G G, Wunderlich J, Harmon B G. Isolation of antimicrobial peptides from avian heterophils. J Leukocyte Biol. 1994;56:661–665. doi: 10.1002/jlb.56.5.661. [DOI] [PubMed] [Google Scholar]
- 8.Ganz T, Lehrer R I. Defensins. Pharm Ther. 1995;66:191–205. doi: 10.1016/0163-7258(94)00076-f. [DOI] [PubMed] [Google Scholar]
- 9.Gilljam H, Ellin A, Strandvik B. Increased bronchial chloride concentration in cystic fibrosis. Scand J Clin Lab Invest. 1989;49:121–124. doi: 10.3109/00365518909105409. [DOI] [PubMed] [Google Scholar]
- 10.Goldman M J, Anderson G M, Stolzenberg E D, Kari U P, Zasloff M, Wilson J M. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 11.Hancock R E W. Peptide antibiotics. Lancet. 1997;349:412–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 12.Harwig S S L, Swiderek K M, Kokryakov V N, Tan L, Lee T D, Panyutich E A, Aleshina E M, Shamova O V, Lehrer R I. Gallinacins: cystine-rich antimicrobial peptides of chicken leukocytes. FEBS Lett. 1994;342:281–285. doi: 10.1016/0014-5793(94)80517-2. [DOI] [PubMed] [Google Scholar]
- 13.Huttner K M, Selsted M E, Ouellette A J. Structure and diversity of the murine cryptdin gene family. Genomics. 1994;19:448–453. doi: 10.1006/geno.1994.1093. [DOI] [PubMed] [Google Scholar]
- 14.Jones D E, Bevins C L. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 15.Joris L, Dab I, Quinton P M. Elemental composition of human airway surface fluid in healthy and diseased airways. Am Rev Respir Dis. 1993;148:1633–1637. doi: 10.1164/ajrccm/148.6_Pt_1.1633. [DOI] [PubMed] [Google Scholar]
- 16.Lehrer R I, Rosenman M, Harwig S S, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Heng H, Zhao C, Ganz T. Human α- and- β-defensins evolved from a common pre-mammalian ancestor peptide. J Invest Med. 1996;44:294A. [Google Scholar]
- 18.Mallow E B, Harris A, Salzman N, Russell J P, DeBerardinis R J, Ruchelli E, Bevins C L. Human enteric defensins. J Biol Chem. 1996;271:4038–4045. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
- 19.McCray P B, Bently L. Human airway epithelia express a β-defensin. Am J Respir Cell Mol Biol. 1997;16:343–349. doi: 10.1165/ajrcmb.16.3.9070620. [DOI] [PubMed] [Google Scholar]
- 20.Mount S. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouellette A J, Pravtcheva D, Ruddle F H, James M. Localization of the cryptdin locus on mouse chromosome 8. Genomics. 1989;5:233–239. doi: 10.1016/0888-7543(89)90051-7. [DOI] [PubMed] [Google Scholar]
- 22.Ouellette A J, Hsieh M M, Nosek M T, Cano-Gauci D F, Huttner K M, Buick R N, Selsted M E. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect Immun. 1994;62:5040–5047. doi: 10.1128/iai.62.11.5040-5047.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouellette A J, Miller S I, Henschen A H, Selsted M E. Purification and primary structure of murine cryptdin-1, a Paneth cell defensin. FEBS Lett. 1992;304:146–148. doi: 10.1016/0014-5793(92)80606-h. [DOI] [PubMed] [Google Scholar]
- 24.Russell J P, Diamond G, Tarver A P, Scanlin T F, Bevins C L. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor alpha. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Schonwetter B S, Stolzenberg E D, Zasloff M A. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- 27.Selsted M E, Szklarek D, Lehrer R I. Purification and antibacterial activity of antimicrobial peptides of rabbit granulocytes. Infect Immun. 1984;45:150–154. doi: 10.1128/iai.45.1.150-154.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selsted M E, Tang Y Q, Morris W L, McGuire P A, Novotny M J, Smith W, Henschen A H, Cullor J S. Purification, primary structures, and antibacterial activities of β-defensins, a new family of antimicrobial peptides from bovine neutrophils. J Biol Chem. 1993;268:6641–6648. [PubMed] [Google Scholar]
- 29.Smith J J, Travis S M, Greenberg E P, Welsh M J. Cystic fibrosis airway epithelia fail to kill bacteria because of abnormal airway surface fluid. Cell. 1996;85:229–236. doi: 10.1016/s0092-8674(00)81099-5. [DOI] [PubMed] [Google Scholar]
- 30.Steiner H, Hultmark D, Engstrom A, Bennich H, Boman H G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292:246–248. doi: 10.1038/292246a0. [DOI] [PubMed] [Google Scholar]
- 31.Tang Y Q, Selsted M E. Characterization of the disulfide motif in BNBD-12, an antimicrobial β-defensin peptide from bovine neutrophils. J Biol Chem. 1993;268:6649–6653. [PubMed] [Google Scholar]
- 32.Zhao C, Wang I, Lehrer R I. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 1996;396:319–322. doi: 10.1016/0014-5793(96)01123-4. [DOI] [PubMed] [Google Scholar]
- 33.Zimmerman G R, Legault P, Selsted M E, Pardi A. Solution structure of bovine β-defensin-12: the peptide fold of the β-defensins is identical to that of the classical defensins. Biochemistry. 1995;34:13663–13671. doi: 10.1021/bi00041a048. [DOI] [PubMed] [Google Scholar]