Abstract
Background
Recent research increasingly highlights a strong correlation between gut microbiota and the risk of gastrointestinal diseases. However, whether this relationship is causal or merely coincidental remains uncertain. To address this, a Mendelian randomization (MR) analysis was undertaken to explore the connections between gut microbiota and prevalent gastrointestinal diseases.
Methods
Genome-wide association study (GWAS) summary statistics for gut microbiota, encompassing a diverse range of 211 taxa (131 genera, 35 families, 20 orders, 16 classes, and 9 phyla), were sourced from the comprehensive MiBioGen study. Genetic associations with 22 gastrointestinal diseases were gathered from the UK Biobank, FinnGen study, and various extensive GWAS studies. MR analysis was meticulously conducted to assess the causal relationship between genetically predicted gut microbiota and these gastrointestinal diseases. To validate the reliability of our findings, sensitivity analyses and tests for heterogeneity were systematically performed.
Results
The MR analysis yielded significant evidence for 251 causal relationships between genetically predicted gut microbiota and the risk of gastrointestinal diseases. This included 98 associations with upper gastrointestinal diseases, 81 with lower gastrointestinal diseases, 54 with hepatobiliary diseases, and 18 with pancreatic diseases. Notably, these associations were particularly evident in taxa belonging to the genera Ruminococcus and Eubacterium. Further sensitivity analyses reinforced the robustness of these results.
Conclusions
The findings of this study indicate a potential genetic predisposition linking gut microbiota to gastrointestinal diseases. These insights pave the way for designing future clinical trials focusing on microbiome-related interventions, including the use of microbiome-dependent metabolites, to potentially treat or manage gastrointestinal diseases and their associated risk factors.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12967-024-04894-5.
Keywords: Gut microbiota, Gastrointestinal disease, Mendelian randomization, SNPs
Introduction
The human microbiome, a vast consortium of over 100 trillion microorganisms, coexists within the human body in a dynamic symbiotic or parasitic relationship. These microorganisms predominantly inhabit various niches such as the skin, respiratory, and gastrointestinal tracts [1, 2]. Extensive research has firmly established the crucial role of the gut microbiota in maintaining human health and modulating numerous physiological functions [3]. The gut microbiota is integral to processes like the breakdown and assimilation of nutrients, absorption of essential compounds, and synthesis of vital biological molecules like vitamins, providing the body with necessary energy and nutrients [4, 5]. Additionally, it plays a vital role in safeguarding the integrity of the intestinal barrier, protecting against pathogens, aiding immune system development, and regulating immune responses [6, 7]. Advancements in microbiota research have linked it to various diseases, notably gastrointestinal disorders like inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), gastric ulcers, and gastroesophageal reflux disease (GERD) [8]. Changes in the microbiota’s abundance, diversity, and composition are thought to weaken the intestinal barrier, leading to increased inflammation, immune dysregulation, and metabolic issues, thus exacerbating these diseases [9–11].
However, research on the gut microbiota is often based on observational studies, which are susceptible to the influence of confounding factors and reverse causality [12]. Observational studies may be limited in their ability to establish causality due to the potential presence of unmeasured or unknown confounders that can distort the observed associations [13]. Additionally, the bidirectional relationship between the gut microbiota and host health further complicates the interpretation of observational findings [14]. While these studies provide valuable insights into the associations between the gut microbiota and various diseases, they cannot definitively establish causation. Randomized controlled trials (RCTs) play a crucial role in controlling potential confounding factors and providing robust evidence to support the relationship between the gut microbiota and diseases [15, 16]. However, RCTs need a substantial sample size and complex data analysis methods. The exorbitant costs, time constraints, and ethical considerations pose significant obstacles to microbiota research, thereby impeding causal inference in this field [17, 18]. Therefore, it is crucial to choose appropriate research methods to explore the causal relationship between gut microbiota and gastrointestinal diseases.
Mendelian Randomization (MR) analysis is a powerful analytical tool that leverages genetic variants as instrumental variables (IVs) to investigate the causal relationship between an exposure and an outcome. Because genetic variations are randomly allocated during conception, MR studies are less prone to the typical confounding factors and reverse causality issues that often affect conventional observational research [19]. Furthermore, MR methods are not influenced by subjective factors such as self-reporting and memory distortion, thereby reducing the potential for information bias [20, 21]. Recently, the MiBioGen consortium released numerous microbiome abundance-associated loci, offering an unprecedented chance to explore the causality between the gut microbiota and diseases. Using genetic variations (single nucleotide polymorphisms, SNPs) closely associated with the gut microbiota as instrumental variables, MR analysis is conducted to simulate the effects of random allocation. This approach allowed us to assess the impact of instrumental variables on specific diseases, thereby providing more robust evidence for the causal relationship between the gut microbiota and diseases by mitigating confounding factors through the process of randomization [22]. Currently, MR analysis has been increasingly applied to investigate the causal relationship between gut microbiota and various diseases, including cancer, psychiatric disorders, metabolic disorders, and autoimmune diseases, providing novel insights into the underlying mechanisms of microbiota-mediated diseases [23–26]. Previous MR studies have delved into the causal connections between the gut microbiota and several gastrointestinal diseases. However, there has been a dearth of comprehensive investigations into the potential impact of gut microbiota.
In the present study, based on large-scale genome-wide association studies (GWAS), we conducted a two-sample Mendelian randomization analysis to evaluate the potential causal relationships between gut microbiota and 22 gastrointestinal diseases, including upper gastrointestinal diseases, lower gastrointestinal diseases, hepatobiliary diseases, and pancreatic diseases. Our study contributes to a more comprehensive understanding of the role of gut microbiota in gastrointestinal health. It provides valuable insights and guidance for clinical practice and public health decision-making. These novel insights and strategies may contribute to enhancing individualized treatment approaches for gastrointestinal diseases, offering new directions and strategies to advance the prevention and management of gastrointestinal disorders.
Methods
Study design
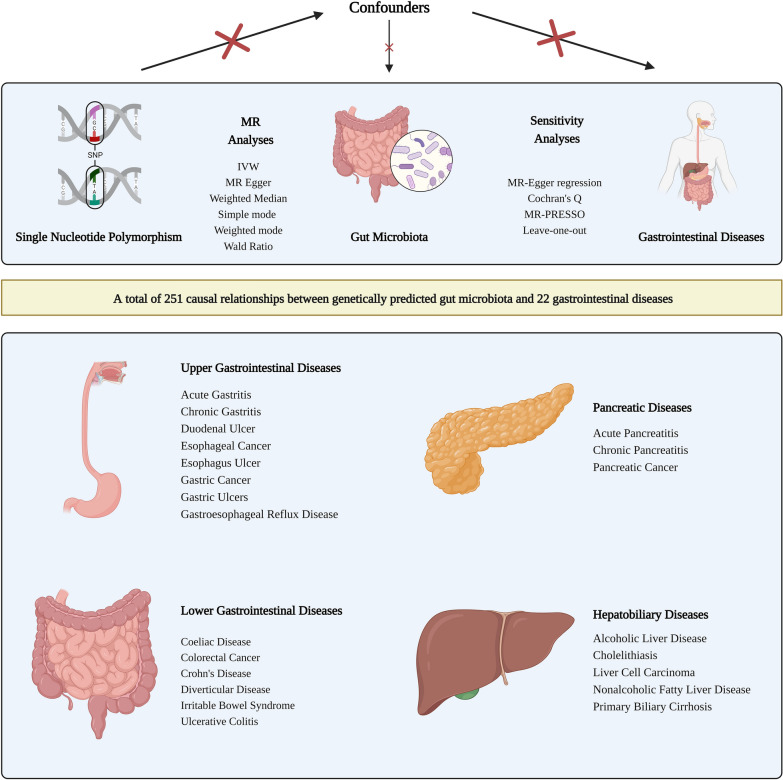
This study conducted a two sample MR analysis to investigate the potential causal relationship between gut microbiota and various gastrointestinal diseases. To ensure the validity of our findings, MR analysis needs to be grounded on three key assumptions: (1) genetic variants must exhibit a significant association with the exposure factor (gut microbiota); (2) genetic variants should not directly impact the outcome (gastrointestinal diseases); (3) genetic variants should have no causal relationship to any potential confounding factors [27]. The research process is depicted in Fig. 1.
Fig. 1.
The design and flowchart of MR analysis in our study. This schematic representation emphasizes the research question that we attempted to answer, the analysis workflow and the data used. Based on large-scale publicly available genome-wide association study (GWAS) summary statistics data, we conducted Mendelian randomization (MR) analysis to explore the causal relationship between the gut microbiota and 22 gastrointestinal diseases, including upper gastrointestinal diseases, lower gastrointestinal diseases, liver and gallbladder diseases, and pancreatic diseases. Sensitivity analysis was used to verify the robustness of the MR results. SNP, single nucleotide polymorphism; MR-PRESSO, Mendelian randomization pleiotropy residual sum and outlier; MR, Mendelian randomization; IVW, inverse variance weighted
Data collection for gut microbiota
The SNPs associated with the composition of the human gut microbiome were selected from the GWAS dataset of the MiBioGen Consortium. Researchers performed a large-scale and genome-wide meta-analysis of the associations between autosomal human genetic variants and the gut microbiome [22]. This study stands as the most comprehensive large-scale association analysis of human gut microbiota composition to date, shedding light on the intricate relationships between genetic variants and the human gut microbiome. Employing a standardized pipeline, microbiome trait loci (mbTL) mapping was conducted to pinpoint genetic loci influencing the relative abundance or presence (microbiome Binary Trait loci) of microbial taxa. In the initial investigation, the gut microbiota summary statistics were classified into 257 taxa across five taxonomic levels: phylum, class, order, family, and genus. Among these, a subset of 211 taxa (comprising 9 phyla, 16 classes, 20 orders, 35 families, and 131 genera) meeting the criteria for microbial quantitative trait locus (mbQTL) mapping analysis was incorporated into the present study. To ensure consistent genetic backgrounds, minimize confounding variables related to lifestyle and environment, and enhance the reliability and interpretability of the results, we exclusively utilized data from the European ancestry within the MiBioGen Consortium. We obtained 14,306 participants of European ancestry GWAS summary data that collected profiles of sequencing for the 16 S ribosomal RNA gene and genotyping information (Additional file 1).
Gastrointestinal disease data sources
The large-scale GWAS summary datasets for 22 gastrointestinal diseases were mainly collected from UK Biobank, FinnGen. Some studies of some large consortia were also included, such as the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC), PanScan consortium and UK primary biliary cirrhosis consortium. Gastrointestinal diseases can be classified into four categories based on the site of occurrence: (1) Upper gastrointestinal diseases: gastroesophageal reflux disease (GERD), esophageal cancer, esophagus ulcer, gastric ulcers, duodenal ulcer, chronic gastritis, acute gastritis, and gastric cancer. (2) Lower gastrointestinal diseases: irritable bowel syndrome, diverticular disease, Crohn’s disease, ulcerative colitis, coeliac disease and colorectal cancer. (3) Hepatobiliary diseases: alcoholic liver disease, nonalcoholic fatty liver disease (NAFLD), primary biliary cirrhosis, liver cell carcinoma and cholelithiasis. (4) Pancreatic diseases: acute pancreatitis, chronic pancreatitis and pancreatic cancer. To mitigate the impact of population structure and diminish the confounding effects of ethnic and genetic diversity in Mendelian randomization, we restricted the use of GWAS summary statistics to those derived from European participants. All datasets were freely accessed from the IEU Open GWAS project (https://gwas.mrcieu.ac.uk/). Since detailed information about participants was not collected, ethical approval was not requisite for this study. Detailed dataset information is provided in Additional file 2.
Instrumental variables selection
Bacterial taxa were analyzed at five levels (phylum, class, order, family, and genus). We selected SNPs that are closely associated with the gut microbiota as IVs. Initially, we screened the SNPs using a threshold of P < 5 × 10−8, resulting in only a small number of SNPs being included. Based on high-quality Mendelian randomization studies, we further selected SNPs using a threshold of P < 5 × 10−5. Furthermore, to reduce potential bias caused by allelic association, we also removed linkage disequilibrium (R2 > 0.001, kb = 10,000) to ensure the enhanced credibility of our results. To minimize the impact of weak instrument bias on causal inference, we used the following formula to calculate the F-statistic for each SNP: Fexposure = Beta2exposure / SE2exposure [28].
MR analysis
A Two sample MR analysis was used to estimate the potential casual relationships between the gut microbiota and gastrointestinal diseases. The inverse-variance weighted (IVW) method was selected as the main approach for data analysis. IVW combines effect estimates from individual genetic variants by weighting them inversely to their variances. By assigning higher weight to more precise estimates, IVW enhances the reliability of the overall causal effect estimate [29, 30]. In cases where pleiotropy is lacking and instrumental validity is assumed, the IVW method provides unbiased estimates of a causal effect if horizontal pleiotropy is balanced. In conjunction with the primary IVW method, supplementary analytical approaches such as MR Egger, Weighted Median, Simple Mode, and Weighted Mode were employed to provide a comprehensive assessment. IF only one SNP could be used as IVs, the causal relationship between exposure and outcome would be estimated by the Wald ratio [28, 31]. Furthermore, we also performed genetic risk scores (GRS) to obtain the combined estimate of the relationship between gut microbiota and gastrointestinal diseases, aiming to incorporate the genetic influences of chosen SNPs on the focal exposure using the genotyping data accessible at the individual level. GRS can integrate information from multiple genetic loci rather than focusing on individual genes. This facilitates a more comprehensive assessment of the complexity of genetic risk, enhancing accuracy and sensitivity in estimating individual genetic predisposition [32].
Pleiotropy and sensitivity analysis
MR Egger analysis was implemented for heterogeneity assessment and calculation of the Cochran’s Q value, crucial for evaluating the diversity among the genetic instruments used. Furthermore, MR-Egger regression was performed to assess potential horizontal pleiotropy effects in Mendelian randomization analysis. We also conducted Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) to rigorously test for horizontal pleiotropy. Additionally, leave-one-out analysis and funnel plots were used to examine the robustness and accuracy of the MR results. All analyses were conducted using the TwoSampleMR package (version 0.5.6) and MR-PRESSO package (version 1.0) in R software (version 4.0.5). The gtx package was used for GRS analysis. Heatmaps were generated using the R package ComplexHeatmap (version 2.6.2).
Results
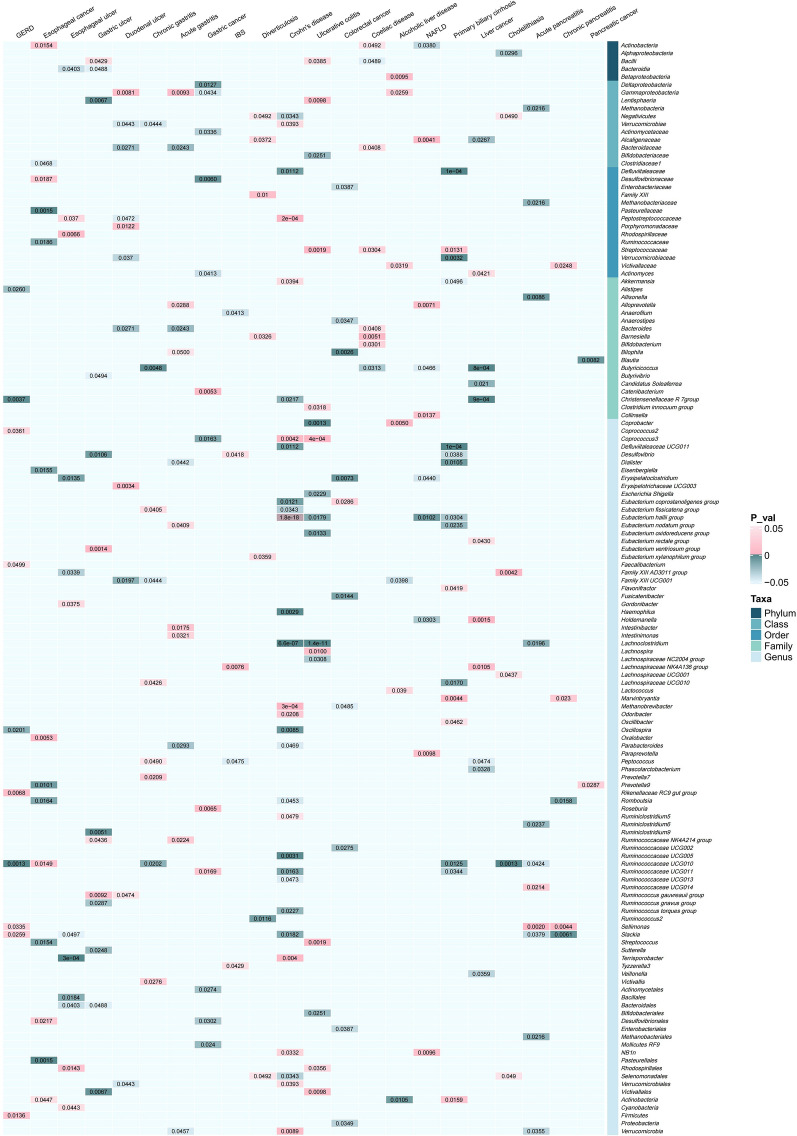
The F-statistic of all IVs is greater than 10, indicating that there is no weak instrumental bias in our analysis (Additional file 3). MR analysis suggested 251 causal relationships between genetically predicted gut microbiota and 22 gastrointestinal diseases (Fig. 2 and Additional file 4). In upper gastrointestinal diseases, there were 10, 14, 14, 13, 11, 9, 12 and 15 causal relationships in GERD, esophageal cancer, esophagus ulcer, gastric ulcer, duodenal ulcer, chronic gastritis, acute gastritis and gastric cancer, respectively. In lower gastrointestinal diseases, we found 5, 9, 32, 17, 10 and 8 causal relationships, including IBS, diverticulosis, Crohn’s disease, ulcerative colitis, colorectal cancer and coeliac disease, respectively. In hepatobiliary diseases, we identified 7, 13, 17, 11 and 6 causal relationships in alcoholic liver disease, NAFLD, primary biliary cirrhosis, liver cancer and cholelithiasis, respectively. Within the pancreatic diseases, the results suggested 11, 5 and 2 causal relationships in acute pancreatitis, chronic pancreatitis and pancreatic cancer, respectively. Furthermore, we observed 12 causal relationships in upper gastrointestinal diseases, 17 in lower gastrointestinal diseases, 15 in hepatobiliary diseases and 6 in pancreatic diseases according to P value corrected (Fig. 3). In addition, the results of the GRS analysis still obtained similar results to IVW method (Additional files 5 and 6).
Fig. 2.
Heatmap of correlation coefficients between gut microbiota abundance and gastrointestinal diseases. Pink blocks represent an increase in the abundance of gut microbiota, which may be associated with an increased risk of developing outcome diseases. Blue blocks demonstrate gut microbiota abundance was negatively correlated with outcome diseases (P value < 0.05). Other blocks represent that there is no causal relationship between gut microbiota and gastrointestinal diseases (P value > 0.05)
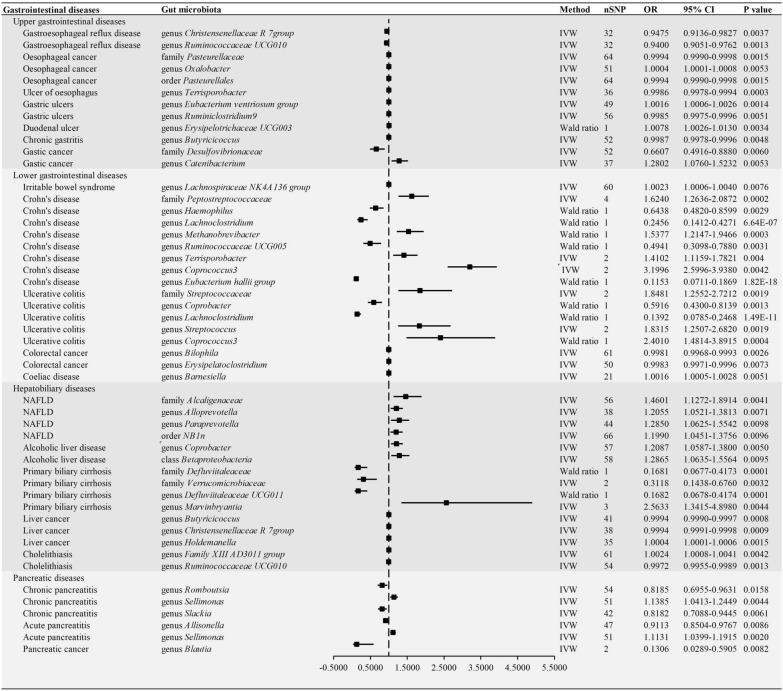
Fig. 3.
Forest plots of Mendelian randomization (MR) estimates between gut microbiota and gastrointestinal diseases according to P value corrected. MR results suggested 12 causal relationships in upper gastrointestinal diseases, 17 in lower gastrointestinal diseases, 15 in hepatobiliary diseases and 6 in pancreatic diseases according to P value corrected. NAFLD, nonalcoholic fatty liver disease; IVW, inverse variance weighted; CI, Confidence interval; OR, Odds ratios
Upper gastrointestinal diseases
Our results eindicated that genus Christensenellaceae R 7group (odds ratio [OR] = 0.9475, 95% confidence interval [CI] 0.9136–0.9827; P = 0.0037) and genus Ruminococcaceae UCG010 (OR = 0.9400, 95%CI 0.9051–0.9762; P = 0.0013) may be protective factors for GRED. Genus Pasteurellaceae (OR = 0.9994, 95%CI 0.9990–0.9998; P = 0.0015) and order Pasteurellales (OR = 0.9994, 95%CI 0.9990–0.9998; P = 0.0015) were associated with a reduced risk of esophageal cancer, while genus Oxalobacter (OR = 1.0004, 95%CI 1.0001–1.0008; P = 0.0053) is related with the risk of esophageal cancer. The MR results demonstrated that genetically predicted increases in genus Terrisporobacter (OR = 1.0016, 95%CI 1.0006–1.0026; P = 0.0014) were associated with an increased risk of esophagus ulcers and that genus Ruminiclostridium9 (OR = 0.9985, 95%CI 0.9975–0.9996; P = 0.0051) have causal relationships with gastric ulcers. Additionally, genus Erysipelotrichaceae UCG003 (OR = 1.0078, 95%CI 1.0026–1.0130; P = 0.0034) was associated with a higher risk of duodenal ulcers. The results suggest that genus Butyricicoccus (OR = 0.9987. 95%CI 0.9978–0.9996; P = 0.0048) may be related to chronic gastritis. Moreover, genetic predictions showed that high abundance of family Desulfovibrionaceae (OR = 0.6607, 95%CI 0.4916–0.8880; P = 0.0060) was associated with a reduced risk of gastric cancer, while genus Catenibacterium (OR = 1.2802, 95%CI 1.0760–1.5232; P = 0.0053) may be associated with an increased risk of gastric cancer.
Low gastrointestinal diseases
This Study have found that a high abundance of the genus Lachnospiraceae NK4A136 group (OR = 1.0023, 95%CI 1.0006–1.0040; P = 0.0076) may lead to an increased incidence of IBS. Family Peptostreptococcaceae, genus Haemophilus, genus Lachnoclostridium, genus Methanobrevibacter, genus Ruminococcaceae, genus Terrisporobacter, genus Coprococcus3, and genus Eubacterium hallii group with Crohn’s disease and genus Coprobacter, genus Lachnoclostridium, genus Streptococcus, and genus Coprococcus3 were causally associated with ulcerative colitis. Genus Lachnoclostridium (OR = 0.2456, 95%CI 0.1412–0.4271;, P = 6.64E-07 for Crohn’s disease and OR = 0.1392, 95%CI 0.0785–0.2468; P = 1.49E-11 for ulcerative colitis) was negatively correlated IBD, whereas a higher genetically predicted abundance of genus Coprococcus3 (OR = 3.1996, 95%CI 2.5996–3.9380; P = 0.0042 for Crohn’s disease and OR = 2.4010, 95%CI 1.4814–3.8915; P = 0.0004 for ulcerative colitis) was associated with an increased risk of Crohn’s disease and ulcerative colitis. In addition, genus Bilophila and genus Erysipelatoclostridium were associated with a reduced risk of colorectal cancer. Our study also demonstrated an inverse causal relationship between genetically predicted genus Barnesiella (OR = 1.0016, 95%CI 1.0005–1.0028; P = 0.0051) and the risk of coeliac disease.
Hepatobiliary diseases
We found that the higher genetically predicted abundance of family Alcaligenaceae (OR = 1.4601, 95%CI 1.1272–1.8914; P = 0.0041), genus Alloprevotella (OR = 1.2055, 95%CI 1.0521–1.3813; P = 0.0071), genus Paraprevotella (OR = 1.2850, 95%CI 1.0625–1.5542; P = 0.0098) and order NB1n (OR = 1.1990, 95%CI 1.0451–1.3756; P = 0.0096) were linked to an increased risk of NAFLD. The results showed some potential causal associations between the gut microbiota and cancers. Higher genetically predicted abundances of genus Coprobacter (OR = 1.2087, 95%CI 1.0587-1.3800; P = 0.0050) and class Betaproteobacteria (OR = 1.2865, 95%CI 1.0635–1.5564; P = 0.0095) were associated with an increased risk of alcoholic liver disease. In addition, family Defluviitaleaceae (OR = 0.1681, 95%CI 0.0677–0.4173; P = 0.0001), genus Defluviitaleaceae UCG011 (OR = 0.1682, 95%CI 0.0678–0.4174; P = 0.0001), family Verrucomicrobiaceae (OR = 0.3118, 95%CI 0.1438–0.6760; P = 0.0032), and genus Marvinbryantia (OR = 2.5633, 95%CI 1.3415–4.8980; P = 0.0001) had causal relationships with primary biliary cirrhosis. Furthermore, a high abundance of genus Butyricicoccus (OR = 0.9994, 95%CI 0.9990–0.9997; P = 0.0008) and genus Christensenellaceae R 7group (OR = 0.9994, 95%CI 0.9991–0.9998; P = 0.0009) are associated with a reduced risk of liver cancer. Additionally, the genus Family XIII AD3011 group (OR = 1.0024, 95%CI 1.0008–1.0041; P = 0.0042) and genus Ruminococcaceae UCG010 (OR = 0.9972, 95%CI 0.9955–0.9989; P = 0.0013) were significantly associated with cholelithiasis at the genetic prediction level.
Pancreatic diseases
In our MR analysis, genetically predicted genus Romboutsia (OR = 0.8185, 95% CI 0.6955–0.9631; P = 0.0158), genus Selemonas (OR = 1.1385, 95% CI 1.0413–1.2449; P = 0.0044), and genus Slackia (OR = 0.8182, 95% CI 0.7088–0.9445; P = 0.0061) were significantly correlated with chronic pancreatis. Genus Allisonella (OR = 0.9113, 95% CI 0.8504–0.9767; P = 0.0086) and genus Selemonas (OR = 1.1131, 95% CI 1.0399–1.1915; P = 0.0020) have a causal relationship with Acute pancreatis. Genus Selemonas may potentially serve as a risk factor for both acute and chronic pancreatitis. The genetic prediction level shows that genus Blautia (OR = 0.1306, 95% CI 0.0289–0.5905; P = 0.0082) may be a protective factor for Pancreative cancer.
Sensitivity analyses
Sensitivity analyses confirmed the robustness of the findings. MR Egger regression intercepts deviated from zero, indicating no evidence of heterogeneity was observed (all intercepts P > 0.05). The MR-PRESSO test also confirmed that the results did not identify horizontal pleiotropy and revealed that there are no obvious outliers for the instrumental variables in this study. Furthermore, most of the Cochrane Q statistic outcomes showed no significant heterogeneity (P > 0.05). Additionally, the vast majority of results from Cochrane Q statistics showed no significant heterogeneity (P > 0.05). Funnel plots exhibited a symmetrical distribution of effect points corresponding to causal associations, suggesting that the causal association is less likely to be impacted by potential bias. The leave–one–out sensitivity also confirmed the above conclusion (Additional files 7 and 8).
Discussion
In this study, based on the summary statistics of gut microbiota from the largest meta-analysis of GWAS conducted by the MiBioGen consortium, a two-sample MR analysis was conducted to evaluate the causal links between gut microbiota and 22 gastrointestinal diseases. The analysis revealed 251 genetically predicted causal relationships, highlighting the role of specific gut microbiota in influencing susceptibility to various gastrointestinal diseases. Besides, this finding emphasizes the complex relationship between gut microbiota and gastrointestinal health, potentially offering novel perspectives for public health interventions to mitigate the prevalence of these gastrointestinal disease risk factors.
Numerous studies have found a possible connection between the gut microbiota selected in our study and gastrointestinal diseases. Our research results suggested that some members of genus Ruminiclostridium and genus Ruminococcaceae (phylum Firmicutes) may act as protective agents against Crohn’ disease, gastric ulcers and GERD, which is consistent with previous reports [33–36]. Notably, Ruminococcaceae is one of the most abundant genera in the phylum Firmicutes, and it is also the most abundant bacterial group in the human genetically modified microbiome, encompassing numerous bacteria that produce short chain fatty acids (SCFAs), especially butyrate. In light of the evidence, SCFAs, produced by Ruminococcaceae, are widely believed to play a variety of important roles in maintaining gastrointestinal homeostasis, such as acting as special nutritional and energy components of the intestinal epithelium and enhancing gastrointestinal motor function [37–39]. SCFAs can significantly inhibit the production of pro-inflammatory cytokines, chemokines, and calprotectin in the intestinal tract, while downregulating myeloperoxidase, reactive oxygen species, and neutrophil extracellular traps formation. Furthermore, SCFAs help maintain the integrity of the intestinal epithelial barrier function by enhancing mucin secretion and other pathways. This explains the mechanism by which Ruminococcaceae may contribute to maintaining the integrity of the epithelial mucosa in disease of digestive tract such as GERD and Crohn’ disease [40–42]. Moreover, our research results suggested a negative causal relationship between the abundance of the genus Ruminococcaceae UCG010 and the risk of cholelithiasis. This relationship may be attributed to the butyrate produced by Ruminococcaceae, which is known to enhance bile salt hydrolase (BSH) activity. Enhanced BSH activity promotes the excretion of bile acids through feces. To replenish the bile acids lost, hepatocytes synthesize new bile acids from blood cholesterol, subsequently lowering blood cholesterol levels and potentially reducing the incidence of cholelithiasis [43, 44]. Beyond their role in producing SCFAs, Ruminococcaceae are among the limited groups within the intestinal microbiota capable of generating secondary bile acids (SBAs). These specific bacterial species can transform primary bile acids (PBAs) into SBAs when bile acids reach the colon [45]. The SBAs produced by The SBAs produced by Ruminococcaceae are believed to play a pivotal role in mitigating intestinal inflammation by modulating bile acid homeostasis. There is increasing evidence that SBAs can interact with the intestinal farnesoid X receptor (FXR) in various intestinal immune cells, including dendritic cells (DCs), natural killer cells (NKCs), and macrophages. This interaction leads to the suppression of pro-inflammatory cytokines such as IL-1 and TNF-α, thereby alleviating inflammatory responses in the intestinal mucosa [46, 47]. In addition to FXR, SBAs can activate Takeda G protein-coupled receptor 5 (TGR5) to promote the polarization of NKT cells towards NKT10 cells, which secrete the anti-inflammatory cytokine IL-10. Additionally, numerous studies have demonstrated that SBAs can regulate the proliferation and differentiation of intestinal stem cells and the self-renewal of intestinal epithelial cells, they can maintain homeostasis of the mechanical barrier of the intestinal mucosa by stimulating the TGR5. This provides additional insight into how Ruminococcaceae protects against IBD by influencing the pathway of SBAs [48]. However, it is worth noting that though studies have shown that Rumen microbiomes are mostly beneficial, different strains may have different impacts on human health Notably, our study is currently the first report to revealed that the genus Ruminococcus gauvreauii group may be a risk factor for gastric and duodenal ulcers. Xu et al. highlighted a positive correlation between the Ruminococcus gauvreauii group and systemic immune responses mediated by pro-inflammatory cytokines such as TNF-α and IL-6. This implies that the Ruminococcus gauvreauii group has the potential to initiate an inflammatory signaling cascade [49, 50]. However, the exact mechanism is not yet clear. Therefore, more epidemiological and basic research is needed in the future to expound the associations and mechanisms among them.
The outcomes of our investigation also identified that the genus Eubacterium hallii group (E. hallii) may reduce the risk of IBD and these findings seem to be consistent with results of a double-blind trial, which reported a significantly lower abundance of E. hallii in the inflamed areas of the intestine in IBD patients compared to healthy individuals [51]. A large cohort study also suggested a negative correlation between the E. hallii and inflammatory markers such as IL-2 and C-reactive protein in IBD and E. hallii negatively correlated with the intestinal inflammatory response [52]. These lend further support to the evidence of a negative correlation between E. hallii and intestinal inflammatory response. Besides producing butyrate, E. hallii also contributes to intestinal mucosal integrity by producing propionate, which activates the NLRP3 inflammasome in intestinal epithelial cells and enhances colonic regulatory T cell expansion, thereby mitigating inflammatory symptoms in IBD [53]. Moreover, propionate boosts the expansion of colonic regulatory T cells by modulating cell surface G-protein coupled receptors [54]. These contribute to alleviating inflammatory manifestations in the intestinal epithelium in IBD. Our results also indicated a negative causal relationship between E. hallii and the risk of NAFLD, which is consistent with previous research findings. In vivo experiments show that propionate, produced by E. hallii, reduces triglyceride accumulation by modulating the expression of genes related to lipogenesis, fatty acid uptake, and oxidation [55–57]. This mechanism contributes to the amelioration of hepatic steatosis, alleviating the progression of non-alcoholic fatty liver disease (NAFLD).
Recognizing its potential, Caelus Health is currently collaborating with the Danish firm Korhansson in developing oral formulations incorporating E. hallii as biotherapeutic agents. The objective is to mitigate insulin resistance and prevent the onset of Type 2 Diabetes in individuals diagnosed with metabolic syndrome (ClinicalTrials.gov, 2020), underscoring the significant value of E. hallii in the realm of biotherapeutic development [58]. E. hallii holds substantial potential for future applications. In the forthcoming phases of research and development, harnessing E. hallii as a key component in the creation of biotherapeutics appears particularly protecting the gastrointestinal mucosa, alleviating gastrointestinal inflammation, and mitigating hepatic steatosis. The intrinsic properties of E. hallii that contribute to metabolic health improvement could extend to beneficial effects on the gastrointestinal system.
Furthermore, the MR results from our study further indicated that gut microbiota producing SCFAs, such as genus Butyricicoccus, genus Christensenellaceae R 7group, and genus Lachnoclostridium, play a protective role in diseases like chronic gastritis, liver cancer, and Crohn’s disease. This further emphasizes the significant role of gut microbiota producing SCFAs in maintaining the integrity of the gastrointestinal mucosa. These findings not only provide new insights into understanding the impact of gut microbiota on various diseases but also deepen our understanding of the crucial role of short-chain fatty acids in regulating the immune system and participating in anti-inflammatory processes. Further research holds the promise of revealing the specific mechanisms through which these microorganisms contribute to the protection observed in chronic gastritis, liver cancer, and Crohn’s disease.
Currently, research suggests that administering metabolites derived from the gut microbiota formulations, such as SCFA, via enemas or oral intake, help preserve the integrity of the intestinal epithelium and the mucosal barrier [59]. However, it is important to acknowledge that supplementing short-chain fatty acids directly faces challenges in achieving and maintaining adequate concentrations within the gastrointestinal tract. Furthermore, the diffusion of these acids on the intestinal surface can be hindered [60]. In light of these challenges, utilizing specific members from the gut microbiota as a biological formulation offers distinct advantages. Firstly, these members of beneficial bacteria like Ruminococcaceae naturally inhabit the gastrointestinal tract, demonstrating inherent adaptability to the gut environment, which facilitates their establishment and maintenance [51]. Secondly, the metabolites produced by beneficial gut microbiota during their fermentation processes exhibit a more stable release profile, ensuring a sustained concentration that aligns with physiological requirements [45].
However, it is worth noting that a number of limitations need to be noted regarding the present study. Firstly, though using exclusively European ancestry helps mitigate potential confounding effects related to population stratification, it may limit the generalizability of results to broader populations. Additionally, a relatively small sample size may limit the extent to which further conclusions may be drawn. Future investigations using larger-scale GWAS are necessary to enhance the robustness and applicability of these findings. Secondly, the GWAS database we used is publicly available, so detailed information on participants cannot be obtained for further subgroup analysis. Thirdly, the analysis of gut microbiota in this study was confined to broader taxonomic levels, namely phylum, class, order, family, and genus. Given the nascent stage of gut microbiota research, the limited number of SNPs available for certain microbiota could potentially introduce biases in the results. Lastly, despite employing various methodologies to assess horizontal pleiotropy, and achieving consistent results across multiple analytical approaches, it cannot be guaranteed that potential horizontal pleiotropy has been entirely eliminated.
In summary, we performed two sample MR analysis to evaluate the potential relationship between the gut microbiota and 22 gastrointestinal diseases for the first time. Our findings suggest a potential causal relationship between specific gut microbiota and gastrointestinal diseases. These novel findings offer promising directions for the development of preventive and therapeutic strategies for gastrointestinal disorders. In the future, more comprehensive epidemiological and foundational research is needed to unravel the intricate mechanisms by which gut microbiota may influence the onset and progression of gastrointestinal diseases.
Supplementary information
Additional file 1: Information on GWAS samples of 22 gastrointestinal diseases used in this study.
Additional file 2: Detailed information on instrumental variables in the gut microbiota dataset.
Additional file 3: Detailed Information and F-statistics of Instrumental Variables Closely Associated with Gut Microbiota.
Additional file 4: The potential associations between gut microbiota and the risk of 22 gastrointestinal diseases.
Additional file 5: Genetic risk score (GRS) analysis for the associations of gut microbiota with gastrointestinal diseases.
Additional file 6: Genetic risk score GRSgut microbiota for gastrointestinal diseasess.
Additional file 7: Heterogeneity, level pleiotropy test, and the power of Mendelian randomization analysis.
Additional file 8: The scatter plot effect size, leave-one-out analyses, and funnel plot for gut microbiota on gastrointestinal diseases in MR analysis.
Acknowledgements
The authors express their gratitude to the participants and investigators of the UK Biobank and FinnGen study. The authors also appreciate the MiBioGen consortium for releasing the gut microbiota GWAS summary statistics.
Author contributions
XS and DX designed the study, analyzed and interpreted the data, and drafted the manuscript. KW, QL, YL and AL analyzed and interpreted the data. KW and DX concepted and designed the study and revised the manuscript. The authors read and approved the final manuscript.
Funding
This work was supported by the Nature Science Foundation of Hainan (823QN254, China).
Availability of data and materials
The datasets supporting the conclusions of this article are available in IEU open gwas project repository (https://gwas.mrcieu.ac.uk/).
Declarations
Ethics approval and consent to participate
Our analysis used publicly available GWAS summary statistics. No new data were collected, and no new ethical approval was required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kaiwen Wu and Qiang Luo have contributed equally to this study.
Contributor Information
Demeng Xia, Email: demengxia@163.com.
Xiaobin Sun, Email: xbsun1197@163.com.
References
- 1.Zmora N, Suez J, Elinav E. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 2.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017 doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69:1510–1519. doi: 10.1136/gutjnl-2019-320204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu H, Liu S, Jiang Y, Hu Y, Li Y, He L, Xing M, Li X, Wu L, Chen Z, et al. Are microplastics toxic? A review from eco-toxicity to effects on the gut microbiota. Metabolites. 2023 doi: 10.3390/metabo13060739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76:473–493. doi: 10.1007/s00018-018-2943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junca H, Pieper DH, Medina E. The emerging potential of microbiome transplantation on human health interventions. Comput Struct Biotechnol J. 2022;20:615–627. doi: 10.1016/j.csbj.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao L, Xia X, Shuai Y, Zhang H, Jin W, Zhang X, Zhang Y. Gut microbiota, a hidden protagonist of traditional Chinese medicine for acute ischemic stroke. Front Pharmacol. 2023;14:1164150. doi: 10.3389/fphar.2023.1164150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasso JM, Ammar RM, Tenchov R, Lemmel S, Kelber O, Grieswelle M, Zhou QA. Gut microbiome-brain alliance: a landscape view into mental and gastrointestinal health and disorders. ACS Chem Neurosci. 2023;14:1717–1763. doi: 10.1021/acschemneuro.3c00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Margolis KG, Cryan JF, Mayer EA. The microbiota-gut-brain axis: from motility to Mood. Gastroenterology. 2021;160:1486–1501. doi: 10.1053/j.gastro.2020.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimidi E, Christodoulides S, Scott SM, Whelan K. Mechanisms of action of probiotics and the gastrointestinal microbiota on gut motility and constipation. Adv Nutr. 2017;8:484–494. doi: 10.3945/an.116.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quaglio AEV, Grillo TG, De Oliveira ECS, Di Stasi LC, Sassaki LY. Gut microbiota, inflammatory bowel disease and colorectal cancer. World J Gastroenterol. 2022;28:4053–4060. doi: 10.3748/wjg.v28.i30.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Tong X, Zou Y, Lin X, Zhao H, Tian L, Jie Z, Wang Q, Zhang Z, Lu H, et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat Genet. 2022;54:52–61. doi: 10.1038/s41588-021-00968-y. [DOI] [PubMed] [Google Scholar]
- 13.Bowden J, Holmes MV. Meta-analysis and mendelian randomization: a review. Res Synth Methods. 2019;10:486–496. doi: 10.1002/jrsm.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M, Benayoun BA. The microbiome: an emerging key player in aging and longevity. Transl Med Aging. 2020;4:103–116. doi: 10.1016/j.tma.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wastyk HC, Fragiadakis GK, Perelman D, Dahan D, Merrill BD, Yu FB, Topf M, Gonzalez CG, Van Treuren W, Han S, et al. Gut-microbiota-targeted diets modulate human immune status. Cell. 2021;184:4137–4153e4114. doi: 10.1016/j.cell.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sergeev IN, Aljutaily T, Walton G, Huarte E. Effects of Synbiotic supplement on human gut microbiota, body composition and weight loss in obesity. Nutrients. 2020 doi: 10.3390/nu12010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabor EC, Kaizer AM, Hobbs BP. Randomized controlled trials. Chest. 2020;158:s79–s87. doi: 10.1016/j.chest.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes. 2017;8:238–252. doi: 10.1080/19490976.2017.1290757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318:1925–1926. doi: 10.1001/jama.2017.17219. [DOI] [PubMed] [Google Scholar]
- 20.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sleiman PM, Grant SF. Mendelian randomization in the era of genomewide association studies. Clin Chem. 2010;56:723–728. doi: 10.1373/clinchem.2009.141564. [DOI] [PubMed] [Google Scholar]
- 22.Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Q, Ni JJ, Han BX, Yan SS, Wei XT, Feng GJ, Zhang H, Zhang L, Li B, Pei YF. Causal relationship between gut microbiota and autoimmune diseases: a two-sample mendelian randomization study. Front Immunol. 2021;12:746998. doi: 10.3389/fimmu.2021.746998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni JJ, Xu Q, Yan SS, Han BX, Zhang H, Wei XT, Feng GJ, Zhao M, Pei YF, Zhang L. Gut microbiota and psychiatric disorders: a two-sample mendelian randomization study. Front Microbiol. 2021;12:737197. doi: 10.3389/fmicb.2021.737197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei Z, Yang B, Tang T, Xiao Z, Ye F, Li X, Wu S, Huang JG, Jiang S. Gut microbiota and risk of five common cancers: a univariable and multivariable mendelian randomization study. Cancer Med. 2023;12:10393–10405. doi: 10.1002/cam4.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo M, Sun M, Wang T, Zhang S, Song X, Liu X, Wei J, Chen Q, Zhong T, Qin J. Gut microbiota and type 1 diabetes: a two-sample bidirectional mendelian randomization study. Front Cell Infect Microbiol. 2023;13:1163898. doi: 10.3389/fcimb.2023.1163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burgess S, Thompson SG. Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol. 2011;40:755–764. doi: 10.1093/ije/dyr036. [DOI] [PubMed] [Google Scholar]
- 28.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 29.Bahls M, Leitzmann MF, Karch A, Teumer A, Dörr M, Felix SB, Meisinger C, Baumeister SE, Baurecht H. Physical activity, sedentary behavior and risk of coronary artery disease, myocardial infarction and ischemic stroke: a two-sample mendelian randomization study. Clin Res Cardiol. 2021;110:1564–1573. doi: 10.1007/s00392-021-01846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadik A, Dardani C, Pagoni P, Havdahl A, Stergiakouli E, Khandaker GM, Sullivan SA, Zammit S, Jones HJ, Davey Smith G, et al. Parental inflammatory bowel disease and autism in children. Nat Med. 2022;28:1406–1411. doi: 10.1038/s41591-022-01845-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:7. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin S, Wang T, Wenying C, Wu Y, Huang S, Zeng P. Maternal and fetal origins of offspring blood pressure: statistical analysis using genetic correlation and genetic risk score-based mendelian randomization. Int J Epidemiol. 2023;52:1360–1376. doi: 10.1093/ije/dyad034. [DOI] [PubMed] [Google Scholar]
- 33.Teofani A, Marafini I, Laudisi F, Pietrucci D, Salvatori S, Unida V, Biocca S, Monteleone G, Desideri A. Intestinal taxa abundance and diversity in inflammatory bowel Disease patients: an analysis including covariates and confounders. Nutrients. 2022 doi: 10.3390/nu14020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maukonen J, Kolho KL, Paasela M, Honkanen J, Klemetti P, Vaarala O, Saarela M. Altered fecal microbiota in paediatric inflammatory bowel disease. J Crohns Colitis. 2015;9:1088–1095. doi: 10.1093/ecco-jcc/jjv147. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Qi X, Han R, Mao T, Tian Z. Gut microbiota causally affects cholelithiasis: a two-sample mendelian randomization study. Front Cell Infect Microbiol. 2023;13:1253447. doi: 10.3389/fcimb.2023.1253447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinto S, Benincà E, Galazzo G, Jonkers D, Penders J, Bogaards JA. Heterogeneous associations of gut microbiota with Crohn’s disease activity. Gut Microbes. 2024;16:2292239. doi: 10.1080/19490976.2023.2292239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizrahi I, Wallace RJ, Moraïs S. The rumen microbiome: balancing food security and environmental impacts. Nat Rev Microbiol. 2021;19:553–566. doi: 10.1038/s41579-021-00543-6. [DOI] [PubMed] [Google Scholar]
- 38.Lillehoj H, Liu Y, Calsamiglia S, Fernandez-Miyakawa ME, Chi F, Cravens RL, Oh S, Gay CG. Phytochemicals as antibiotic alternatives to promote growth and enhance host health. Vet Res. 2018;49:76. doi: 10.1186/s13567-018-0562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 40.Chen G, Ran X, Li B, Li Y, He D, Huang B, Fu S, Liu J, Wang W. Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine. 2018;30:317–325. doi: 10.1016/j.ebiom.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Recharla N, Geesala R, Shi XZ. Gut microbial metabolite butyrate and its therapeutic role in inflammatory bowel disease: a literature review. Nutrients. 2023;15:2275. doi: 10.3390/nu15102275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Lin J, Zhang C, Gao H, Lu H, Gao X, Zhu R, Li Z, Li M, Liu Z. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes. 2021;13:1968257. doi: 10.1080/19490976.2021.1968257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Q, Zhang SS, Wang RR, Weng YJ, Cui X, Wei XT, Ni JJ, Ren HG, Zhang L, Pei YF. Mendelian randomization analysis reveals causal effects of the human gut microbiota on abdominal obesity. J Nutr. 2021;151:1401–1406. doi: 10.1093/jn/nxab025. [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Zhang H, Chen X, Chen Y, Menghebilige, Bao Q. Selection of potential probiotic lactobacilli for cholesterol-lowering properties and their effect on cholesterol metabolism in rats fed a high-lipid diet. J Dairy Sci. 2012;95:1645–1654. doi: 10.3168/jds.2011-4768. [DOI] [PubMed] [Google Scholar]
- 45.Li M, Yang L, Mu C, Sun Y, Gu Y, Chen D, Liu T, Cao H. Gut microbial metabolome in inflammatory bowel disease: from association to therapeutic perspectives. Comput Struct Biotechnol J. 2022;20:2402–2414. doi: 10.1016/j.csbj.2022.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang CX, Wang HY, Chen TX. Interactions between intestinal microflora/probiotics and the immune system. Biomed Res Int. 2019;2019:6764919. doi: 10.1155/2019/6764919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calzadilla N, Comiskey SM, Dudeja PK, Saksena S, Gill RK, Alrefai WA. Bile acids as inflammatory mediators and modulators of intestinal permeability. Front Immunol. 2022;13:1021924. doi: 10.3389/fimmu.2022.1021924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lajczak-McGinley NK, Porru E, Fallon CM, Smyth J, Curley C, McCarron PA, Tambuwala MM, Roda A, Keely SJ. The secondary bile acids, ursodeoxycholic acid and lithocholic acid, protect against intestinal inflammation by inhibition of epithelial apoptosis. Physiol Rep. 2020;8:e14456. doi: 10.14814/phy2.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu T, Ge Y, Du H, Li Q, Xu X, Yi H, Wu X, Kuang T, Fan G, Zhang Y. Berberis kansuensis extract alleviates type 2 diabetes in rats by regulating gut microbiota composition. J Ethnopharmacol. 2021;273:113995. doi: 10.1016/j.jep.2021.113995. [DOI] [PubMed] [Google Scholar]
- 50.Crost EH, Coletto E, Bell A, Juge N. Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol Rev. 2023;47:fuad01. doi: 10.1093/femsre/fuad014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, Paramsothy R, Walsh AJ, van den Bogaerde J, Samuel D, et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156:1440–1454e1442. doi: 10.1053/j.gastro.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Takahashi K, Nishida A, Fujimoto T, Fujii M, Shioya M, Imaeda H, Inatomi O, Bamba S, Sugimoto M, Andoh A. Reduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s disease. Digestion. 2016;93:59–65. doi: 10.1159/000441768. [DOI] [PubMed] [Google Scholar]
- 53.Li N, Wang M, Lyu Z, Shan K, Chen Z, Chen B, Chen Y, Hu X, Dou B, Zhang J, et al. Medicinal plant-based drug delivery system for inflammatory bowel disease. Front Pharmacol. 2023;14:1158945. doi: 10.3389/fphar.2023.1158945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding Y, Yanagi K, Cheng C, Alaniz RC, Lee K, Jayaraman A. Interactions between gut microbiota and non-alcoholic liver disease: the role of microbiota-derived metabolites. Pharmacol Res. 2019;141:521–529. doi: 10.1016/j.phrs.2019.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosh S, Yang X, Wang L, Zhang C, Zhao L. Active phase prebiotic feeding alters gut microbiota, induces weight-independent alleviation of hepatic steatosis and serum cholesterol in high-fat diet-fed mice. Comput Struct Biotechnol J. 2021;19:448–458. doi: 10.1016/j.csbj.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 57.Deng M, Qu F, Chen L, Liu C, Zhang M, Ren F, Guo H, Zhang H, Ge S, Wu C, Zhao L. SCFAs alleviated steatosis and inflammation in mice with NASH induced by MCD. J Endocrinol. 2020;245:425–437. doi: 10.1530/JOE-20-0018. [DOI] [PubMed] [Google Scholar]
- 58.Udayappan S, Manneras-Holm L, Chaplin-Scott A, Belzer C, Herrema H, Dallinga-Thie GM, Duncan SH, Stroes ESG, Groen AK, Flint HJ, et al. Oral treatment with Eubacterium hallii improves insulin sensitivity in db/db mice. NPJ Biofilms Microbiomes. 2016;2:16009. doi: 10.1038/npjbiofilms.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geirnaert A, Steyaert A, Eeckhaut V, Debruyne B, Arends JB, Van Immerseel F, Boon N, Van de Wiele T. Butyricicoccus pullicaecorum, a butyrate producer with probiotic potential, is intrinsically tolerant to stomach and small intestine conditions. Anaerobe. 2014;30:70–74. doi: 10.1016/j.anaerobe.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Cristofori F, Dargenio VN, Dargenio C, Miniello VL, Barone M, Francavilla R. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: a door to the body. Front Immunol. 2021;12:578386. doi: 10.3389/fimmu.2021.578386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Information on GWAS samples of 22 gastrointestinal diseases used in this study.
Additional file 2: Detailed information on instrumental variables in the gut microbiota dataset.
Additional file 3: Detailed Information and F-statistics of Instrumental Variables Closely Associated with Gut Microbiota.
Additional file 4: The potential associations between gut microbiota and the risk of 22 gastrointestinal diseases.
Additional file 5: Genetic risk score (GRS) analysis for the associations of gut microbiota with gastrointestinal diseases.
Additional file 6: Genetic risk score GRSgut microbiota for gastrointestinal diseasess.
Additional file 7: Heterogeneity, level pleiotropy test, and the power of Mendelian randomization analysis.
Additional file 8: The scatter plot effect size, leave-one-out analyses, and funnel plot for gut microbiota on gastrointestinal diseases in MR analysis.
Data Availability Statement
The datasets supporting the conclusions of this article are available in IEU open gwas project repository (https://gwas.mrcieu.ac.uk/).