Abstract
Background
X-linked nephrogenic diabetes insipidus (NDI) is a rare genetic renal disease caused by pathogenic variants in the AVPR2 gene. Single nucleotide variants and small insertions/deletions in AVPR2 are reliably detected by routine clinical sequencing. Nevertheless, structural variants involving AVPR2 are challenging to identify accurately by conventional genetic testing. Here, we report a novel deletion of AVPR2 in a Czech family identified for the first time by targeted long-read sequencing (T-LRS).
Methods
A male proband with X-linked NDI underwent clinical sequencing of the AVPR2 gene that failed and thus indicated possible whole-gene deletion. Therefore, PCR mapping and subsequent targeted long-read sequencing (T-LRS) using a Pacific Biosciences sequencer were applied to search for the suspected deletion. To validate the deletion breakpoints and prove variant segregation in the family with X-linked NDI, Sanger sequencing of the deletion junction was performed. Quantitative real-time PCR was further carried out to confirm the carrier status of heterozygous females.
Results
By T-LRS, a novel 7.5 kb deletion of AVPR2 causing X-linked NDI in the proband was precisely identified. Sanger sequencing of the deletion junction confirmed the variant breakpoints and detected the deletion in the probands´ mother, maternal aunt, and maternal cousin with X-linked NDI. The carrier status in heterozygous females was further validated by quantitative real-time PCR.
Conclusions
Identifying the 7.5 kb deletion gave a precise molecular diagnosis for the proband, enabled genetic counselling and genetic testing for the family, and further expanded the spectrum of structural variants causing X-linked NDI. Our results also show that T-LRS has significant potential for accurately identifying putative structural variants.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-01801-1.
Keywords: Long-read sequencing, PacBio, Breakpoint analysis, AVPR2 deletion, Nephrogenic diabetes insipidus
Background
X-linked nephrogenic diabetes insipidus (X-linked NDI, MIM: 300538) is a rare X-linked recessive disease characterized by the inability of the kidney to concentrate urine in response to the antidiuretic hormone arginine-vasopressin (AVP). The main clinical manifestation is polyuria with polydipsia, failure to thrive, feeding difficulty, and repeated vomiting. Nevertheless, without proper treatment, severe clinical symptoms may develop, such as intellectual disability [1, 2].
X-linked NDI is caused by pathogenic variants in the AVPR2 gene, which encodes the arginine-vasopressin V2 receptor [3]. AVPR2 is a G-protein coupled receptor that in response to AVP redistributes aquaporin-2 water channel (AQP2) in the renal collecting tubules to make the membrane permeable to water, thereby concentrating the urine [1, 2]. X-linked NDI accounts for approximately 90% of cases with congenital NDI. The remaining 10% of cases are caused by pathogenic variants in the AQP2 gene with autosomal recessive or dominant inheritance [1, 2] Most pathogenic variants in AVPR2 are single nucleotide variants [4, 5]. Therefore, the recommended first-tier genetic diagnostic test for individuals with suspected X-linked NDI is sequencing analysis of AVPR2 [1]. However, non-recurrent deletions of the entire AVPR2 causing X-linked NDI have also been observed [6]. The reported deletions varied in length but in most cases included not only AVPR2 but also a neighbouring gene of unknown clinical significance, ARHGAP4 [6]. Since these deletions, as well as other small structural variants (SVs), are challenging to identify by other clinical testing methods, such as chromosomal microarray or short-read sequencing [7], the next recommended step for patients with suspected large deletions is to perform gene-targeted deletion analysis of AVPR2 [1].
The most commonly used targeted methods to accurately detect causal large deletions involving AVPR2 are polymerase chain reaction mapping and Sanger sequencing [6, 8–10]. This approach is labour-intensive [8] and not always applicable. Large deletions and other SVs tend to occur in repetitive genomic regions that are challenging for Sanger and short-read sequencing [11]. However, it has been shown that such difficulties can be overcome by long-read sequencing (LRS) [12–15]. LRS on Pacific Biosciences (PacBio) platforms generates highly accurate long reads (HiFi reads) that can span SVs breakpoints and confidently identify SVs even in difficult-to-sequence regions [11]. Although targeted long-read sequencing (T-LRS) on the PacBio sequencer represents an effective way to accurately detect suspected SV [14], more clinical evidence is needed to confirm the clinical utility of T-LRS.
In this study, we report the first successful application of T-LRS on the PacBio sequencer to identify and fine-map a candidate causal structural variant in a male proband with X-linked NDI. In the proband diagnosed with X-linked NDI based on the clinical manifestations and the family history, genetic cause after clinical sequencing remained unknown, yet suspected. Therefore, we applied further genetic testing to search for suspected whole-gene deletion. Consequently, we precisely detected a novel 7.5 kb deletion of AVPR2 by T-LRS in the proband, and thus we were able to determine the genetic status of available at-risk relatives in the family and offer genetic testing for other family members that may prevent the birth of affected children or prevent primary manifestation. Our results further demonstrate the utility of T-LRS for accurately discriminating putative causal structural variants.
Materials and methods
Clinical description
The proband was born after an uncomplicated pregnancy at 39 + 1 weeks with a birth weight of 3660 g and birth length of 50 cm. Due to significant weight loss, early enteral intake was initiated. At the age of 23 hours, he developed hypernatremic dehydration (the maximum sodium level was 160 mmol/l), which was corrected by intravenous fluid administration (D5%W). The requirement for fluid was high, with a maximum of 300 ml/kg per day. At this time, Hydrochlorothiazide was administered temporarily. The child tolerated oral intake well and clinical symptoms were significantly reduced after targeted therapy. Thus, at 11 days of age, parenteral intake could be ended, and Hydrochlorothiazide was discontinued. Ultrasound showed a congenital duplex left kidney and normal sonography of the heart, liver, gallbladder, pancreas, and spleen. At the age of 19 days, the patient was readmitted to the Neonatology Department for hypernatremic dehydration and Hydrochlorothiazide therapy was restarted. Main symptoms at clinical manifestation were fever, irritability, polyuria, poor feeding and failure to thrive. The analysis of family history revealed the probands maternal uncle was clinically diagnosed with NDI after birth, and two more distant maternal male relatives were monitored and treated by a pediatric nephrologist during childhood and had discontinued treatment in adulthood. Based on the clinical manifestations in the male proband and pedigree analysis a diagnosis of the X-linked NDI was suspected. Other syndromes with polyuria such as Bartter syndrome were contemplated in the process of differential diagnosis, but their type of inheritance was not consistent with the pedigree analysis. The patient was referred for a nephrological follow-up.
Further treatment was based on free water intake, and low salt diet, a thiazide diuretic was given to lower urine output, as needed. The proband stayed in the care of the Department of Pediatrics, University Hospital Pilsen, Czech Republic, until referred to genetic testing by a pediatric nephrologist. Currently, at 8 years of age, the proband is treated with Hydrochlorothiazide, Verospiron, and KCl. His daily fluid intake is 4000–5000 ml.
DNA isolation
Genomic DNA was isolated from peripheral blood of the proband and his available family members using the Gentra Puregene Blood Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The concentration and purity of the DNA were assessed using a spectrophotometer DeNovix DS-11 FX (DeNovix, Wilmington, DE).
PCR and Sanger sequencing
The entire coding sequence with the flanking intronic regions of the AVPR2 gene (clinical sequencing) and the breakpoint junction (breakpoint sequencing) were amplified and sequenced in the proband using primers listed in Table S1 (Additional file 1). Breakpoint sequencing was also performed on available family members for segregation analysis. Genomic regions were amplified using AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific, Waltham, MA) in a final 10 μl PCR mixture containing 1 μl of H2O, 5 μl of AmpliTaq Gold 360 Master Mix, 2 μl of 10 μM forward and reverse primer mix and 2 μl of DNA (35 g/μl) according to the manufacturer’s protocols. PCR products were purified using Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA) and sequencing reactions were performed using the Gerbera Sequencing Kit v3.1 (SEQme, Dobříš, Czech Republic) according to the manufacturer’s protocols. Sequencing products were purified using CleanSEQ (Beckman Coulter, Brea, CA) and separated on the ABI PRISM 3130 Genetic Analyser (Thermo Fisher Scientific, Waltham, MA). Sequencing data were analysed using BioEdit sequence alignment editor v.7.0.5.3 [16].
PCR mapping
Several genomic regions flanking the deletion in the proband were amplified using AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific, Waltham, MA) in a final 10 μl PCR mixture containing 1 μl of H2O, 5 μl of AmpliTaq Gold 360 Master Mix, 2 μl of 10 μM forward and reverse primer mix and 2 μl of DNA (35 ng/μl) according to the manufacturer’s protocols. Primer sequences can be found in Table S1 (Additional file 1). PCR products were analysed by agarose gel electrophoresis (2%).
Targeted long-read sequencing and analysis
The selected target region in the proband was amplified by long-range PCR using the Phusion High-Fidelity PCR Kit (Thermo Fisher Scientific, Waltham, MA) in a final 50 μl PCR mixture containing 34 μl H2O, 10 μl of 5× Phusoin GC Buffer, 1 μl of 10 μM dNTPs, 2.5 μl of 10 μM forward and reverse primer mix, 2 μl of DNA (50 ng/μl) and 0.5 μl Phusion DNA Polymerase. Primer sequences can be found in Table S1 (Additional file 1). The PCR protocol was as follows: Initial denaturation at 98 °C for 30 s, 30 cycles of 98 °C for 5 s, 63.5 °C for 15 s and 72 °C for 35 s and final extension at 72 °C for 10 min. The amplified products (~ 3900 bp long) were verified by agarose gel electrophoresis (1%) and purified using 0.5× Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA). These amplicons were subjected to SMRTbell library preparation and sequenced on a Pacific Biosciences Sequel I system (Pacific Biosciences, Menlo Park, CA) as recommended by Pacific Biosciences. To generate highly accurate long reads (Hifi reads), Circular Consensus Sequence analysis was performed using SMRT Link v.10.1. HiFi reads were aligned to the reference human genome (hg19) using minimap2 v.2.1 [17] with default parameters and visualized in Integrative Genomics Viewer v.12.2.3 [18]. The flanking sequences of the breakpoints were manually evaluated for the presence of microhomology or repetitive elements.
Quantitative real-time PCR
The copy number of the AVPR2 gene in available healthy females was determined by quantitative real-time PCR on Rotor-Gene Q (Qiagen, Germantown, MD) in a final 25 μl PCR mixture containing 12.5 μl of ABsolute QPCR Mix, no ROX (Thermo Fisher Scientific, Waltham, MA), 1 μl of 10 μM forward primer and 1 μl of 10 μM reverse primer, 1.2 μl 20× EvaGreen Dye (Biotium, Freemont, CA), 2.5 μl of DNA (10 ng/μl) and 6.8 μl H2O. Primer sequences can be found in Table S1 (Additional file 1). Each sample was tested in three technical replicates with the following conditions: Initial denaturation at 95 °C for 15 min, 40 cycles of 95 °C for 15 s and 60 °C for 60 s and melting curve analysis at 60–90 °C/step 0.5 °C. qPCR data were analysed with double delta Ct analysis, and the copy number of AVPR2 was normalized to the copy number of the GAPDH reference gene in the same sample. The relative copy number of AVPR2 to an unrelated control female were compared and shown in the graph for each sample.
Results
Clinical genetic analysis
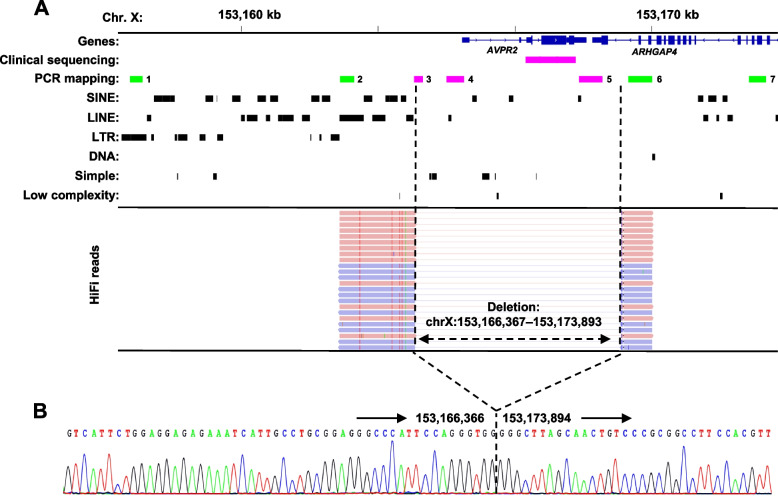
The proband with the clinical diagnosis of X-linked NDI underwent clinical sequence analysis of the AVPR2 gene. However, PCRs covering coding exons and flanking intronic regions of AVPR2 failed to amplify (Fig. 1A), which suggested a large deletion covering the entire AVPR2.
Fig. 1.
Identification of AVPR2 deletion. A Schematic presentation showing (from top to bottom) the position of genes, amplicons for clinical sequencing of AVPR2, amplicons for PCR mapping, and repetitive sequences in the RepeatMasker track. Amplified amplicons are coloured green, not amplified magenta. PacBio HiFi reads of long-range amplicons aligned to the human genome reference sequence (hg19) showing the unambiguous 7526 bp deletion (chrX:153,166,367–153,173,893) are shown at the bottom. Separate alignments from the same read are connected by a thin line. B Sanger sequencing of the breakpoint junction confirming the deletion breakpoints. The breakpoint junction is indicated by a dashed line
Identification of deletion
To refine and confirm the suspected deletion, we designed several primer pairs to produce amplicons surrounding AVPR2 within the region with previously published deletions [6]. Only PCR amplicons 1, 2, 6, and 7 were successfully produced (Fig. 1A), which indicates a deletion covering the entire AVPR2 and the last exon of adjacent ARHGAP4. To identify the size and location of the deletion at single nucleotide resolution in the region containing multiple repetitive elements, we applied T-LRS. Targeted amplicons overlapping the deletion were obtained by long-range PCR and subsequently sequenced on the PacBio sequencer. As a result, generated HiFi reads aligned to the human reference hg19 revealed a 7526 bp deletion of chrX:153,166,367–153,173,893 (Fig. 1A). The 5′ breakpoint was located near a repetitive element L1ME1 in the intergenic region between AVPR2 and adjacent L1CAM, while the 3′ breakpoint was delineated in intron 22 of ARHGAP4 with no repetitive elements. The breakpoint regions show no microhomologies.
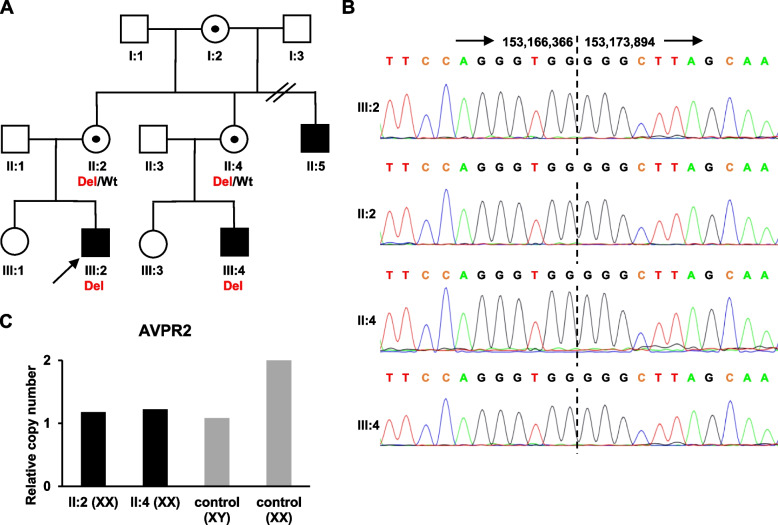
Confirmation of deletion and segregation analysis
The precise deletion breakpoints in the proband identified by T-LRS were validated by Sanger sequencing with PCR primers amplifying the breakpoint junction (Fig. 1B). Additionally, Sanger sequencing of the breakpoint junction in available at-risk family members (Fig. 2A) detected the identical deletion in a younger maternal cousin (III:4) also affected with X-linked NDI and in healthy females (II:2 and II:4) (Fig. 2B). To confirm the carrier status of the healthy females (II:2 and II:4), we performed copy number analysis of AVPR2 using quantitative real-time PCR (qPCR). The relative copy number of AVPR2 in all tested females was one copy that correlated with the hemizygous control male (Fig. 2C) and thus proved the heterozygous status of these females.
Fig. 2.
Family pedigree and segregation analysis. A Pedigree of the family with X-linked NDI and segregation of the 7.5 kb deletion (chrX:153,166,367–153,173,893). Available genotypes are shown below symbols: Del – deletion allele, Wt – wild-type allele. B Sanger sequencing of the breakpoint junction in the proband and available family members. The breakpoint junction is indicated by a dashed line. c qPCR data showing the relative copy number of AVPR2 in suspected female carriers (II:2 and II:4) and the control male to the copy number in the control female
Discussion
Timely and accurate diagnosis of X-linked NDI is beneficial for patients because untreated patients can develop severe symptoms. The diagnostic steps of X-linked NDI reflect the main clinical symptoms of the disease. However, early diagnosis of X-linked NDI can be clinically challenging as symptoms may not be specific [8]. Therefore, molecular genetic analysis of the AVPR2 gene is important to confirm the diagnosis at the molecular level. Identification of genetic cause of X-linked NDI in the proband also enables genetic testing for other family members to achieve the birth of unaffected child or to prevent primary manifestation. However, in some cases, widespread clinical sequencing of AVPR2 reveals difficult-to-confirm structural variants. Hence, there is a need to use better tools to provide a precise diagnosis. Here, we identified a novel pathogenic 7.5 kb deletion of AVPR2 in a family with X-linked NDI using T-LRS on the PacBio sequencer.
In our case, clinical sequence analysis of AVPR2 in the proband with clinically suspected X-linked NDI indicated a large deletion of the entire AVPR2. In contrast to previous studies using even thirty-two primer pairs around AVPR2 to characterize the large deletions [8], we performed only several PCRs. The approximately defined region was subsequently amplified and sequenced on the PacBio sequencer. As in previous studies using LRS [13, 15], T-LRS generated highly accurate sequence data even in the difficult-to-sequence regions (repetitive elements and homopolymer sequences) and precisely identified the deletion breakpoints. Detection of the deletion breakpoints was crucial for a complete diagnosis of the proband and for identifying related asymptomatic heterozygous female carriers who might otherwise stay undiagnosed, as routine clinical sequencing of AVPR2 gene would be falsely negative. Using precisely identified deletion breakpoints in the proband, we were able to design PCR primers to amplify the breakpoint junction. Sanger sequencing of the breakpoint junction in the proband demonstrated the accuracy of HiFi reads and segregation of the deletion in other family members based on X-linked recessive inheritance.
Since the deletion breakpoints were not mapped to be within repetitive elements and no sequence homology was found at the junction, proposed mechanisms for such deletion are non-homologous end joining or fork stalling and template switching [6, 19]. Both breakpoints lay in the previously defined regions with more breakpoints in which deletion is probably stimulated by the local genome architecture [6]. We report the first deletion within these two regions, which can contribute to elucidating the mechanisms of non-recurrent deletions causing X-linked NDI.
In addition to the entire AVPR2, the 7.5 kb deletion encompassed the last exon (exon 22) of the adjacent ARHGAP4 gene. This is consistent with previous results where all but one [20] of the previously published large deletions causing X-linked NDIs included a part or all of the ARHGAP4 gene [6, 8–10, 21–30]. Despite lacking the part of ARHGAP4, both patients in this family had no symptoms other than those associated with NDI [1] which supports the results of the majority of previous studies [6, 8, 21–24, 26–29]showing that, in such cases, disruptions of ARHGAP4 do not lead to a different clinical phenotype.
Conclusions
In summary, the identification of the novel 7.5 kb deletion enabled a precise molecular diagnosis for the proband, genetic counselling and genetic testing for at-risk family members, and further expanded the spectrum of SVs causing X-linked NDI. Our results also support a new strategic workflow for identifying pathogenic variants in rare disease cases [7]. According to this strategy, when a single-gene disease is suspected, targeted approaches are recommended. If Sanger sequencing data indicate the possibility of SV, LRS is then advised. We hope that the further application of T-LRS will help to identify and confirm the suspected causative SVs also in other genetic diseases.
Supplementary Information
Acknowledgements
The authors thank the family for their participation in the research study.
Authors’ contributions
L.S. designed the work, performed most of the experiments, analysed and interpreted the data. I.Š. supervised the study. I.Š. and E.S. collected the clinical data and samples from the family. L.S. and I.Š. wrote the manuscript. M.Č., M.H., T.Z., P.K., J.T., I.B., and J.S. contributed data, revised and edited the manuscript. All authors read and approved the final manuscript.
Funding
The authors acknowledge the National Center for Medical Genomics research infrastructure (LM2023067 funded by MEYS CR) for their support with obtaining scientific data presented in this paper. The study was also supported by the National Institute of Virology and Bacteriology (Programme EXCELES, ID Project No. LX22NP05103) funded by the European Union - Next Generation EU, and by the Charles University research programme Cooperatio – Pediatrics, No. 207040.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The ethics approval was obtained from the Ethics Committee of The University Hospital and the Faculty of Medicine, Charles University in Pilsen. Written informed consent was obtained from all subjects and/or their legal guardian(s). All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Written informed consent was obtained from the parents of the child for the publication of clinical details.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lukáš Strych, Email: Lukas.Strych@lfp.cuni.cz.
Ivan Šubrt, Email: subrti@fnplzen.cz.
References
- 1.Knoers N, Lemmink H. Hereditary nephrogenic diabetes insipidus. Seattle (WA): University of Washington, Seattle; 1993. [PubMed] [Google Scholar]
- 2.Bockenhauer D, Bichet DG. Pathophysiology, diagnosis and management of nephrogenic diabetes insipidus. Nat Rev Nephrol. 2015;11:576–588. doi: 10.1038/nrneph.2015.89. [DOI] [PubMed] [Google Scholar]
- 3.van den Ouweland AMW, Dreesen JCFM, Verdijk M, Knoers NVAM, Monnens LAH, Rocchi M, et al. Mutations in the vasopressin type 2 receptor gene (AVPR2) associated with nephrogenic diabetes insipidus. Nat Genet. 1992;2:99–102. doi: 10.1038/ng1092-99. [DOI] [PubMed] [Google Scholar]
- 4.Spanakis E, Milord E, Gragnoli C. AVPR2 variants and mutations in nephrogenic diabetes insipidus: review and missense mutation significance. J Cell Physiol. 2008;217:605–617. doi: 10.1002/jcp.21552. [DOI] [PubMed] [Google Scholar]
- 5.Bichet DG, Birnbaumer M, Lonergan M, Arthus MF, Rosenthal W, Goodyer P, et al. Nature and recurrence of AVPR2 mutations in X-linked nephrogenic diabetes insipidus. Am J Hum Genet. 1994;55:278–286. [PMC free article] [PubMed] [Google Scholar]
- 6.Anesi L, de Gemmis P, Galla D, Hladnik U. Two new large deletions of the AVPR2 gene causing nephrogenic diabetes insipidus and a review of previously published deletions. Nephrol Dial Transplant. 2012;27:3705–3712. doi: 10.1093/ndt/gfs359. [DOI] [PubMed] [Google Scholar]
- 7.Mitsuhashi S, Matsumoto N. Long-read sequencing for rare human genetic diseases. J Hum Genet. 2020;65:11–19. doi: 10.1038/s10038-019-0671-8. [DOI] [PubMed] [Google Scholar]
- 8.Leung MT, Sit JKK, Cheung HN, Iu YP, Chan WKY, Shek CC. Contiguous gene deletion in a Chinese family with X-linked nephrogenic diabetes insipidus: challenges in early diagnosis and implications for affected families. J Pediatr Endocrinol Metab. 2019;32:915–920. doi: 10.1515/jpem-2019-0028. [DOI] [PubMed] [Google Scholar]
- 9.Bai Y, Chen Y, Kong X. Contiguous 22.1-kb deletion embracing AVPR2 and ARHGAP4 genes at novel breakpoints leads to nephrogenic diabetes insipidus in a Chinese pedigree. BMC Nephrol. 2018;19:26. doi: 10.1186/s12882-018-0825-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Poke G, Gecz J, Gibson K. A novel contiguous gene deletion of AVPR2 and ARHGAP4 genes in male dizygotic twins with nephrogenic diabetes insipidus and intellectual disability. Am J Med Genet Part A. 2012;158A:2511–2518. doi: 10.1002/ajmg.a.35591. [DOI] [PubMed] [Google Scholar]
- 11.Marwaha S, Knowles JW, Ashley EA. A guide for the diagnosis of rare and undiagnosed disease: beyond the exome. Genome Med. 2022;14:23. doi: 10.1186/s13073-022-01026-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merker JD, Wenger AM, Sneddon T, Grove M, Zappala Z, Fresard L, et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med. 2018;20:159–163. doi: 10.1038/gim.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizuguchi T, Suzuki T, Abe C, Umemura A, Tokunaga K, Kawai Y, et al. A 12-kb structural variation in progressive myoclonic epilepsy was newly identified by long-read whole-genome sequencing. J Hum Genet. 2019;64:359–368. doi: 10.1038/s10038-019-0569-5. [DOI] [PubMed] [Google Scholar]
- 14.Reiner J, Pisani L, Qiao W, Singh R, Yang Y, Shi L, et al. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet–Biedl Syndrome 9 (BBS9) deletion. NPJ Genomic Med. 2018;3:3. doi: 10.1038/s41525-017-0042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizuguchi T, Okamoto N, Yanagihara K, Miyatake S, Uchiyama Y, Tsuchida N, et al. Pathogenic 12-kb copy-neutral inversion in syndromic intellectual disability identified by high-fidelity long-read sequencing. Genomics. 2021;113:1044–1053. doi: 10.1016/j.ygeno.2020.10.038. [DOI] [PubMed] [Google Scholar]
- 16.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 17.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel T, Lupski JR. Genomic disorders 20 years on-mechanisms for clinical manifestations. Clin Genet. 2018;93:439–449. doi: 10.1111/cge.13146. [DOI] [PubMed] [Google Scholar]
- 20.Tegay DH, Lane AH, Roohi J, Hatchwell E. Contiguous gene deletion involvingL1CAM andAVPR2 causes X-linked hydrocephalus with nephrogenic diabetes insipidus. Am J Med Genet Part A. 2007;143A:594–598. doi: 10.1002/ajmg.a.31536. [DOI] [PubMed] [Google Scholar]
- 21.Carroll P, Al-Mojalli H, Al-Abbad A, Al-Hassoun I, Al-Hamed M, Al-Amr R, et al. Novel mutations underlying nephrogenic diabetes insipidus in Arab families. Genet Med. 2006;8:443–447. doi: 10.1097/01.gim.0000223554.46981.7a. [DOI] [PubMed] [Google Scholar]
- 22.Demura M, Takeda Y, Yoneda T, Furukawa K, Usukura M, Itoh Y, et al. Two novel types of contiguous gene deletion of the AVPR2 and ARHGAP4 genes in unrelated Japanese kindreds with nephrogenic diabetes insipidus. Hum Mutat. 2002;19:23–29. doi: 10.1002/humu.10011. [DOI] [PubMed] [Google Scholar]
- 23.Dong Y, Sheng H, Chen X, Yin J, Su Q. Deletion of the V2 vasopressin receptor gene in two Chinese patients with nephrogenic diabetes insipidus. BMC Genet. 2006;7:53. doi: 10.1186/1471-2156-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto M, Imai K, Hirata K, Kashiwagi R, Morinishi Y, Kitazawa K, et al. Immunological profile in a family with nephrogenic diabetes insipidus with a novel 11 kb deletion in AVPR2 and ARHGAP4 genes. BMC Med Genet. 2008;9:42. doi: 10.1186/1471-2350-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knops NBB, Bos KK, Kerstjens M, van Dael K, Vos YJ. Nephrogenic diabetes insipidus in a patient with L1 syndrome: a new report of a contiguous gene deletion syndrome including L1CAM and AVPR2. Am J Med Genet Part A. 2008;146A:1853–1858. doi: 10.1002/ajmg.a.32386. [DOI] [PubMed] [Google Scholar]
- 26.Kotnik P, Battelino T, Debeljak M, Podkrajšek KT, Waldhauser F, Frøkiaer J, et al. Correlation between AVPR2 mutations and urinary AQP2 excretion in patients with nephrogenic diabetes insipidus. J Pediatr Endocrinol Metab. 2007;20 [DOI] [PubMed]
- 27.Schöneberg T, Pasel K, von Baehr V, Schulz A, Volk H-D, Gudermann T, et al. Compound deletion of the rhoGAP C1 and V2 vasopressin receptor genes in a patient with nephrogenic diabetes insipidus. Hum Mutat. 1999;14:163–174. doi: 10.1002/(SICI)1098-1004(1999)14:2<163::AID-HUMU8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 28.Schulz A, Sangkuhl K, Lennert T, Wigger M, Price DA, Nuuja A, et al. Aminoglycoside pretreatment partially restores the function of truncated V 2 vasopressin receptors found in patients with nephrogenic diabetes insipidus. J Clin Endocrinol Metab. 2002;87:5247–5257. doi: 10.1210/jc.2002-020286. [DOI] [PubMed] [Google Scholar]
- 29.Cho SY, Law CY, Ng KL, Lam CW. Novel large deletion in AVPR2 gene causing copy number variation in a patient with X-linked nephrogenic diabetes insipidus. Clin Chim Acta. 2016;455:84–86. doi: 10.1016/j.cca.2016.01.032. [DOI] [PubMed] [Google Scholar]
- 30.Broides A, Ault BH, Arthus M-F, Bichet DG, Ellen CM. Severe combined immunodeficiency associated with nephrogenic diabetes insipidus and a deletion in the Xq28 region. Clin Immunol. 2006;120:147–155. doi: 10.1016/j.clim.2006.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.