ABSTRACT
Both influenza A virus genome transcription (vRNA→mRNA) and replication (vRNA→cRNA→vRNA), catalyzed by the influenza RNA polymerase (FluPol), are dynamically regulated across the virus life cycle. It has been reported that the last amino acid I121 of the viral NS2 protein plays a critical role in promoting viral genome replication in influenza mini-replicon systems. Here, we performed a 20 natural amino acid substitution screening at residue NS2-I121 in the context of virus infection. We found that the hydrophobicity of the residue 121 is essential for virus survival. Interestingly, through serial passage of the rescued mutant viruses, we further identified adaptive mutations PA-K19E and PB1-S713N on FluPol which could effectively compensate for the replication-promoting defect caused by NS2-I121 mutation in the both mini-replicon and virus infection systems. Structural analysis of different functional states of FluPol indicates that PA-K19E and PB1-S713N could stabilize the replicase conformation of FluPol. By using a cell-based NanoBiT complementary reporter assay, we further demonstrate that both wild-type NS2 and PA-K19E/PB1-S713N could enhance FluPol dimerization, which is necessary for genome replication. These results reveal the critical role NS2 plays in promoting viral genome replication by coordinating with FluPol.
IMPORTANCE
The intrinsic mechanisms of influenza RNA polymerase (FluPol) in catalyzing viral genome transcription and replication have been largely resolved. However, the mechanisms of how transcription and replication are dynamically regulated remain elusive. We recently reported that the last amino acid of the viral NS2 protein plays a critical role in promoting viral genome replication in an influenza mini-replicon system. Here, we conducted a 20 amino acid substitution screening at the last residue 121 in virus rescue and serial passage. Our results demonstrate that the replication-promoting function of NS2 is important for virus survival and efficient multiplication. We further show evidence that NS2 and NS2-I121 adaptive mutations PA-K19E/PB1-S713N regulate virus genome replication by promoting FluPol dimerization. This work highlights the coordination between NS2 and FluPol in fulfilling efficient genome replication. It further advances our understanding of the regulation of viral RNA synthesis for influenza A virus.
KEYWORDS: influenza virus, virus genome transcription and replication, NS2, FluPol, adaptive mutations
INTRODUCTION
Influenza A viruses (IAV) can cause acute and highly contagious respiratory infections leading to annual seasonal epidemics worldwide. Avian IAV, in particular, have great potential to induce global pandemics (1, 2). The genome of IAV consists of eight single-stranded negative-sense RNA segments (3). Each viral RNA (vRNA) segment is encapsidated by influenza viral RNA-dependent RNA polymerase (FluPol) and viral nucleoprotein (NP), forming a rod-shaped ribonucleoprotein complex (vRNP) (3). FluPol, a heterotrimeric complex composed of polymerase basic 1 (PB1), polymerase basic 2 (PB2), and polymerase acidic (PA), is responsible for transcribing and replicating the viral genome in the nucleus of infected cells (4).
Upon the entry of the virus into cells, vRNPs are released and rapidly transported into the cell nucleus where they are primarily transcribed into viral mRNAs (5). These mRNAs are then nuclear exported to cytoplasm for new viral protein production. Once new viral proteins are produced and transported into the nucleus, secondary transcription (vRNA→mRNA) and genome replication (vRNA→cRNA→vRNA) occur (5). The synthesis of the three viral RNA species shows very distinct synthetic dynamics during the virus life cycle (6, 7). The accumulation of viral mRNA peaks and then drops as infection proceeds, whereas the accumulation of vRNA keeps rising during the entire course of infection, and the accumulation of cRNA remains at low levels throughout (8). FluPol transcribes and replicates the viral genome with very different initiation strategies. The initiation of transcription is primer dependent, using capped RNAs as primers that are snatched by FluPol from host pre-mRNAs (9, 10). The viral genome replication is a two-step procedure involving the synthesis of intermediate complementary RNA, which then serves as a template to synthesize progeny vRNA. The initiations of both replication steps are primer independent, whereas the cRNA synthesis is terminal initiated on the vRNA template, while the vRNA synthesis uses internal initiation and realign strategy on the cRNA template (11).
Crystal structure studies have advanced significantly in resolving nearly the complete FluPol of influenza A, B, C, and D viruses in recent years (10, 12–14). They all showed very similar overall architecture with a central core region consisting of PB1, PB2-N, and PA-C, as well as several peripheral flexible domains consisting of PA-N with an endonuclease domain and the C-terminal two-thirds of PB2 (PB2-C) with the key cap-binding, 627, and nuclear localization signal (NLS) domains (13, 15). Interestingly, structural studies have demonstrated that these peripheral domains could be repacked flexibly around the core region to form different functional states depending on which kind of viral RNA promoters or protein ligands are bound (4, 16). The two most representative conformations are the transcriptase conformation, which was resolved by the presence of capped RNA, and the replicase conformation, which was resolved by the presence of vRNA or cRNA promoter (4). The transition from transcriptase to replicase conformation involves the collapse of the PB2 cap-binding domain downward, migrating closer to the PB1 core, the ascending of the PB2-627 domain, and the in situ rotation of PA-N in aiding its binding to PB2-NLS (13, 15, 17, 18). Furthermore, structural studies have revealed that during viral genome replication, FluPol is dimerized, including symmetric dimerization for trans-activation of cRNA-to-vRNA synthesis and asymmetric dimerization for encapsidation of newly synthesized RNA (19, 20).
In contrast to the largely resolved intrinsic mechanisms of FluPol in catalyzing viral transcription and replication (19–21), the mechanisms of how transcription and replication are dynamically regulated remain elusive. It has been reported that, as viral infection proceeds, the accumulation of viral NP proteins or virus-derived small viral RNAs could facilitate in promoting replication and inhibiting transcription (22–25). In addition, nuclear export of vRNP mediated by viral non-structural protein 2 (NS2, also named nuclear export protein) and matrix protein 1 (M1) could also affect viral RNA synthesis during the late stage of viral infection (26–29). Furthermore, it has been reported that NS2 is able to regulate both transcription and replication in a manner that is independent of its nuclear export function (8). Interestingly, it has been proposed that the inefficient splicing of the eight (NS) segments, which leads to the early expression of NS1 and the slow accumulation of NS2, acts as a “molecular timer” to coordinate the IAV life cycle (30). Recently, we reported that the stoichiometric changes in viral NS1 and NS2 proteins, generated from unspliced and spliced mRNA transcripts of the NS segment, are involved in the fine regulation of influenza virus RNA synthesis dynamics (31). Particularly, we pinpointed that the last amino acid I121 of NS2 plays an essential role in promoting viral genome replication in an influenza mini-replicon system.
In this report, we investigated the biological significance of NS2 on its specific role in promoting replication in the context of virus infection. We performed a 20 amino acid screening at residue 121 of NS2 for virus rescue, followed by serial passage of the rescued mutant viruses. We identified two adaptive mutations, PA-K19E and PB1-S713N, on FluPol which could compensate the defect of NS2-I121 mutation in promoting viral genome replication both in the mini-replicon and infection systems. The structural analysis indicated that the two adaptive mutations could contribute to stabilizing the FluPol in its replicase conformation. With a cell-based NanoBiT complementary reporter assay, we found that NS2 and the two adaptive mutations could facilitate the FluPol dimerization.
RESULTS
The property of the last C-terminal amino acid of NS2 is crucial for influenza virus survival
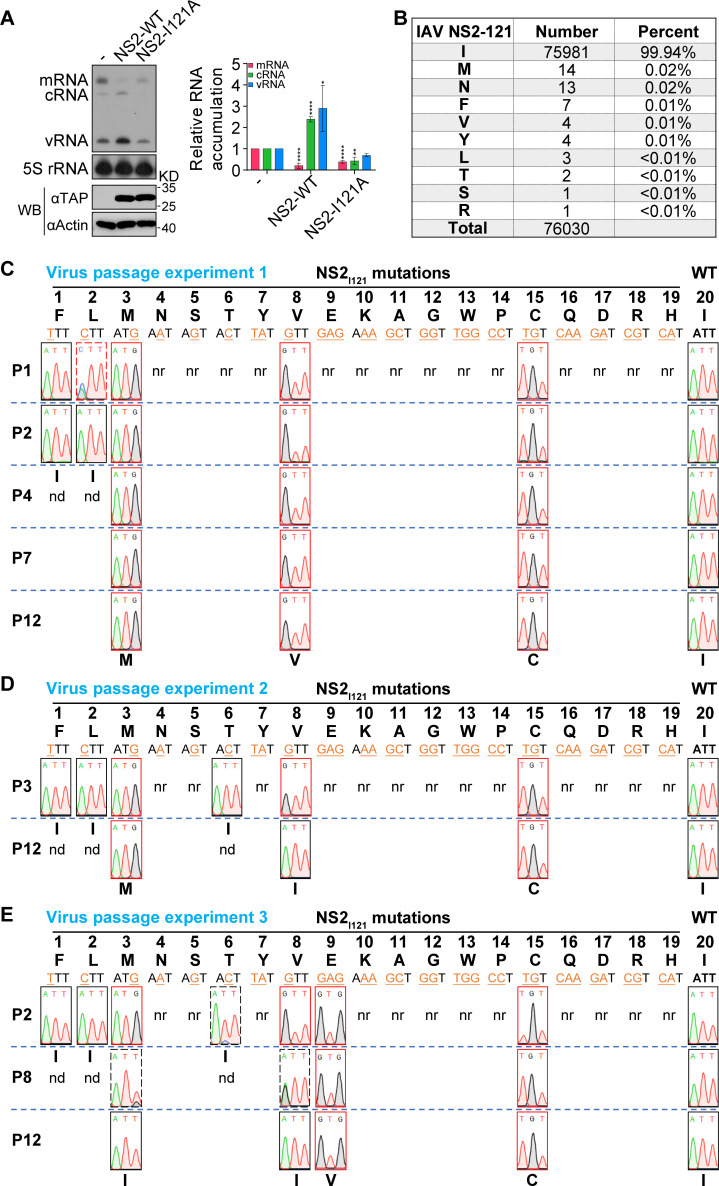
We have recently reported that the point mutation of the last amino acid I121 of NS2 to alanine completely abolishes NS2 function in promoting viral genome replication in the mini-replicon system as shown in Fig. 1A. The analysis of a total of 76,030 sequences of IAV NS2 from the NCBI Influenza Virus Resource database on 17 July 2022 displays that the last amino acid of IAV NS2 is isoleucine (Ile, I) with 99.94% conservation. A residual percentage (<0.02%) of other amino acids is present (Fig. 1B).
Fig 1.
The property of the last C-terminal amino acid of NS2 is crucial for influenza virus survival. (A) HEK 293T cells were co-transfected with plasmids expressing PB2, PB1, PA, NP, and NA vRNA, together with either NS2 wild-type (WT) or I121A mutant. The steady-state levels of RNAs and proteins were determined by primer extension analysis (top) and western blotting (bottom) at 24 h p.t. The graph shows the relative mean intensity of viral RNA normalized to 5S rRNA. The data represent the mean ± SD from three biological replicates. Asterisks represent a significant difference from the control group (by one-way ANOVA with Dunnett’s post hoc test) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (B) The percentage of a specific amino acid at residue 121 of NS2 was calculated based on the NS2 sequences of influenza A virus obtained from the NCBI Influenza Virus Resource database. (C–E) The virus rescue and passage of NS2-I121 mutants in three independent experiments. NS2 WT or I121 mutant viruses were rescued in HEK 293T cells mixed with MDCK cells by transfecting eight pHW2000 plasmids and then passaged in MDCK cells. Amino acids and corresponding nucleotide codons are shown. The mutated nucleotides are shown in orange and underlined. Virion RNA was isolated, amplified by RT-PCR, and sequenced. The sequence traces of residue 121 of NS2 from viruses are shown. P1–P12 represent generations of virus passage. nr indicates those not rescued, and nd indicates those not detected further.
To further investigate this property that the last amino acid of NS2 plays in regulating virus infection, we conducted virus rescue experiments with the I121 residue mutated into other alternative 19 amino acids in the eight-plasmid influenza A/WSN/33 (H1N1) (WSN) reverse genetic system (32). As shown in Fig. 1C, we were able to rescue five mutant viruses with substitutions of phenylalanine (Phe, F), leucine (Leu, L), methionine (Met, M), valine (Val, V), and cysteine (Cys, C). The amino acids F, L, M, V, and wild-type (WT) I are all hydrophobic amino acids. Besides, C is now often grouped among the hydrophobic amino acids because its slightly polar S-H side chain is too mild to interact with water (33). Therefore, we concluded that the hydrophobic property of the residue 121 of NS2 plays a critical role for virus survival. Next, we further passaged the rescued mutant viruses in MDCK cells. It was found that the mutants NS2-I121F and I121L reverted to wild-type residue isoleucine in P1 and P2, respectively. In contrast, the mutants NS2-I121M, I121V, or I121C could maintain the mutation until P12 (Fig. 1C).
Considering the randomness of mutagenesis caused by the viral error-prone FluPol in the process of virus rescue and serial passage, we conducted two other independent experiments (Fig. 1D and E). In general, the results of virus rescue were essentially the same as the first experiment except that I121T was rescuable in the two repeated experiments, while I121E was rescuable only in the third experiment. In the passage experiments, we found that I121T reproducibly reversed into wild type, and the I121E reversed into 121V in early passage. Taken together, these data strongly indicate that the hydrophobicity of the last amino acid of NS2 is crucial for virus survival.
Adaptive mutations occurred on FluPol during serial passage of NS2-I121 mutant viruses
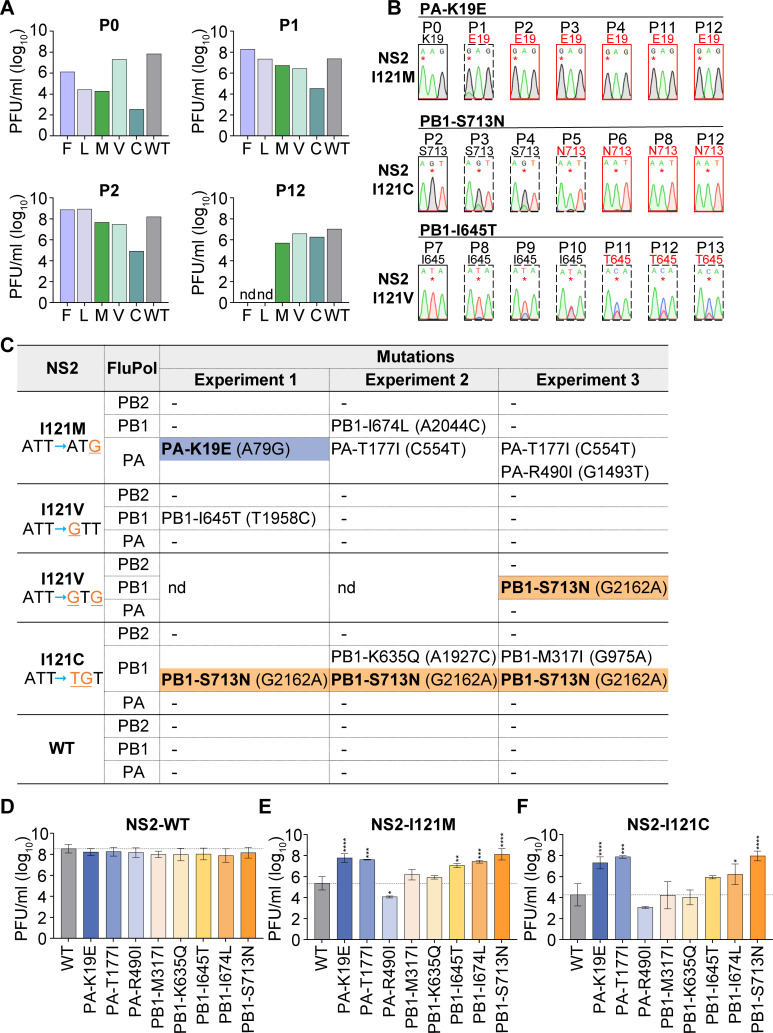
For those mutant viruses (NS2-I121M, NS2-I121C, and NS2-I121V) which could maintain the mutation in our serial passage, we also monitored their virus titers during passaging. As shown in Fig. 2A, in P0, the titer of NS2-I121V virus was close to that of WT virus, while the titers of NS2-I121M and NS2-I121C viruses were significantly lower (about 4 log reduction for NS2-I121M and 6 log reduction for NS2-I121C) than that of the WT virus. Interestingly, in P1, we observed significantly increased virus titers (about ~2 log increase) for both NS2-I121M and NS2-I121C viruses. The titer of NS2-I121C virus could further increase to nearly wild-type level at P12. We, therefore, speculated that these mutants may have acquired adaptive mutations that could enhance the viral replication capacity. Since we have found that the last amino acid I121 of NS2 plays a critical role in promoting viral genome replication, we, therefore, focused on searching for adaptive mutations in the three subunits of FluPol (PB2, PB1, and PA).
Fig 2.
Adaptive mutations occurred on the viral polymerase during serial passage of NS2-I121 mutant viruses. (A) Infectious viral titers from different passages (P0, P1, P2, and P12) were determined by plaque assay. (B) Amino acid mutations were identified in the viral polymerase during serial passage of NS2-I121 mutant viruses. Virion RNA was isolated, amplified by RT-PCR, and sequenced. The sequence traces of the nucleotide change (*) are shown. The dashed boxes indicate that the nucleotide mutation is in a mixed state, while the red boxes represent stabilized nucleotides. (C) The amino acid mutations which occurred on viral polymerase from three independent passages of NS2-I121 mutant viruses are summarized. PA-K19E, blue background, and PB1-S713N, orange background, indicate that the mutations are stabilized, whereas the other mutations are in a mixed state until P12. nd indicates those not detected further. (D–F) The wild-type or mutant polymerase in the context of the NS2-WT (D), I121M (E), and I121C (F) background was rescued, and the infectious viral titers were determined by plaque assay. The data represent the mean ± SD from two to five biological replicates. Asterisks represent a significant difference from the control group (by one-way ANOVA with Dunnett’s post hoc test) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Interestingly, we found that, in the passage of NS2-I121M mutant virus, PA-K19E appeared immediately in P1 and became stabilized (Fig. 2B). Meanwhile, in the passage of NS2-I121C virus, PB1-S713N mutation appeared in P3 and became stabilized in P6. In contrast, in the passage of NS2-I121V virus, PB1-I645T appeared in P8 and maintained the mixed state until P13. In order to examine the reproducibility of these mutations, we further conducted two other independent passage experiments (Fig. 2C). Surprisingly, we found that the PB1-S713N mutation reproducibly appeared in the passage of NS2-I121C and NS2-I121V viruses. Meanwhile, we also observed other mutations, including PA-T177I, PA-R490I, PB1-M317I, PB1-K635Q, and PB1-I674L. However, these mutations maintained a mixed state throughout the passaging.
By using reverse genetics, we examined the effects of these mutations on virus growth in the context of either wild-type NS2 or NS2-I121 mutant viruses. The PFU titers of viruses were determined by plaque assay on MDCK cells. As shown in Fig. 2D, the titers of these polymerase mutants were comparable to wild-type virus in the wild-type NS2 background. Interestingly, in the context of the NS2-I121 (NS2-I121M or I121C) mutant virus, which showed significant titer reduction of about 2.5–4 log compared to wild-type virus, PA-K19E, PA-T177I, or PB1-S713N remarkably improved the titers of both NS2-I121M and I121C mutant viruses (Fig. 2E and F). The other mutations appeared to increase the virus titers mildly except in the case of PA-R490I which showed decreased virus titers. These results indicate that mutations occurred in FluPol can compensate for the replication-promoting defect of NS2-I121 mutation in the context of viral infection.
PA-K19E and PB1-S713N promote viral genome replication in the RNP/NS reconstitution system
Given that PA-K19E and PB1-S713N exhibit the properties of fast adaption/stabilization and multiple reproducible appearance/stabilization, respectively, we next focused on an in-depth mechanistic study of the effect PA-K19E and PB1-S713N have in compensating the defect of NS2-I121 mutation. We first examined the effects of the two-point mutations, either alone or in combination with our previously reported RNP/NS reconstitution assay (31), in which the four-protein expression plasmids expressing PB2, PB1, PA, and NP were co-transfected with various individually modified RNAs expressing pPOLI-NS plasmids which introduced mutations expressing either inactivated NS1 and NS2, wild-type NS1 and NS2, or wild-type NS1 and NS2-I121C, respectively (Fig. 3). Protein expression of wild-type or mutant PA and PB1 was confirmed by western blot (Fig. 3D and E).
Fig 3.
PA-K19E and PB1-S713N promote viral genome replication in the RNP/NS reconstitution system. (A–C) HEK 293T cells were co-transfected with plasmids expressing PB2, PB1 (either wild type or mutant), PA (either wild type or mutant), NP, and different NS vRNA templates. The steady-state levels of RNAs were determined by primer extension analysis at 24 h p.t. LE, long exposure. The graph shows the relative mean intensity of viral RNA normalized to 5S rRNA. The data represent the mean ± SD from three biological replicates. Asterisks represent a significant difference from the control group (by one-way ANOVA with Dunnett’s post hoc test) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (D and E) Expression levels of wild type or mutant of PA (D) and PB1 (E) proteins in HEK 293T cells.
It can be seen that, in the absence of NS1 and NS2 expressions, the introduction of PA-K19E or PB1-S713N mutation alone slightly inhibited transcription and promoted replication (Fig. 3A). As expected, these effects were more obvious when both NS1 and NS2 were expressed (Fig. 3B). Interestingly, the combination of PA-K19E and PB1-S713N led to further increased replication and reduced transcription (Fig. 3A and B). We further examined the effect of these mutations in the presence of wild-type NS1 and NS2-I121C mutant (Fig. 3C). The combination of the two mutations showed the similar effect as the presence of wild-type NS2, suggesting that the two mutations make the polymerase to synthesize vRNA more efficiently.
PA-K19E and PB1-S713N are able to restore the replication defect of NS2-I121 mutant viruses in the context of virus infection
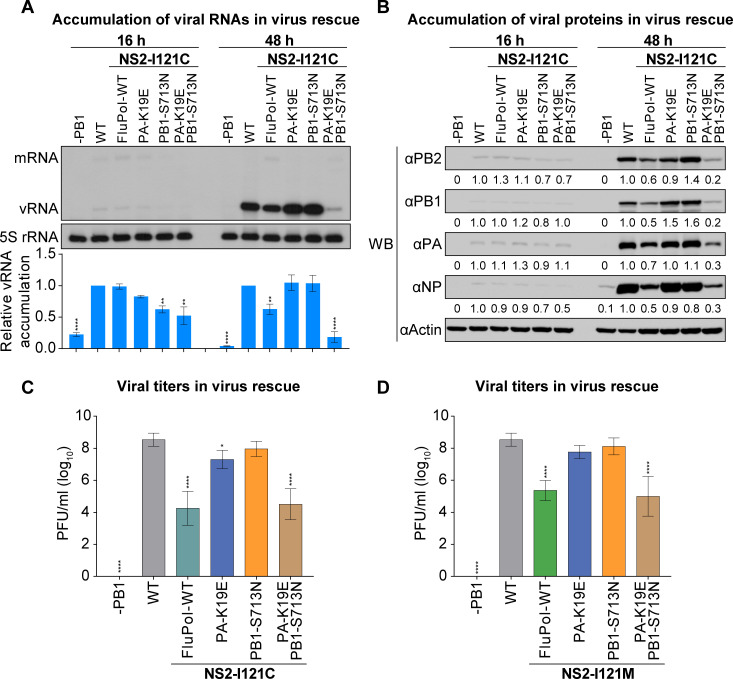
We determined the effects of PA-K19E and PB1-S713N mutations, alone or in combination, in the context of virus infection. HEK 293T cells mixed with MDCK cells were co-transfected with the pHW2000 eight-plasmid rescue system of influenza virus A/WSN/1933 (H1N1/WSN), and the plasmids containing indicated amino acid mutation in PA, PB1, and NS2 were used to replace respective wild-type plasmids (Fig. 4A and B). Accumulation of viral RNAs and proteins at post-transfection 16 and 48 h (p.t.) were measured by primer extension and western blot analyses (Fig. 4A and B).
Fig 4.
PA-K19E and PB1-S713N are able to restore the replication defect of NS2-I121 mutant viruses in the context of virus infection. (A and B) HEK 293T cells mixed with MDCK cells were co-transfected with indicated eight pHW2000 plasmids (either wild type or mutant). The steady-state levels of RNAs (A) and proteins (B) were determined by primer extension analysis and western blotting (WB) at 16 and 48 h p.t. The graph in panel A shows the relative mean intensity of viral RNA normalized to 5S rRNA. The data represent the mean ± SD from three biological replicates. Asterisks represent a significant difference from the control group (by one-way ANOVA with Dunnett’s post hoc test) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (C and D) HEK 293T cells mixed with MDCK cells were co-transfected with indicated eight pHW2000 plasmids (either wild type or mutant). The virus supernatants were harvested at 48 h p.t., and the infectious viral titers were determined by plaque assay. The data represent the mean ± SD from three biological replicates. Asterisks represent a significant difference from the control group (by one-way ANOVA with Dunnett’s post hoc test) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
The results showed that there was little difference in viral RNA and protein accumulation among different virus-rescued groups at the early stage of transfection in general (16 h p.t.). However, at 48 h p.t., the accumulation levels of viral RNAs and proteins were significantly lower in the NS2-I121C mutant virus than those in the wild-type virus. Interestingly, this reduction was reversed when the PA-K19E or PB1-S713N point mutations were individually introduced into the viral polymerase in the context of NS2-I121C mutant virus. However, the combination of PA-K19E and PB1-S713N mutations led to significantly reduced viral RNA and protein accumulations in the context of virus infection. At the same time, we also measured virus titers for all mutant viruses. As expected, remaining consistent with viral RNA and protein levels, mutations in either PA-K19E or PB1-S713N increased viral titers by more than 1,000-fold in the context of NS2-I121C mutant virus, while the combination of PA-K19E and PB1-S713N mutations did not improve the virus titer (Fig. 4C). We further confirmed these effects of PA-K19E and PB1-S713N in the context of NS2-I121M virus (Fig. 4D).
In terms of the detrimental effects of the PA-K19E/PB1-S713N double mutations on RNA/protein accumulation and virus titer, it can be expected that double mutations were too drastic in promoting viral genome replication which would break the optimal dynamics of transcription and replication. Taken together, these results clearly demonstrate that PA-K19E and PB1-S713N could compensate the defect of NS2-I121 mutation in the context of virus infection. It further confirms that the replication-promoting function of NS2 is essential for efficient virus production.
PA-K19E and PB1-S713N could contribute in stabilizing the replicase conformation of FluPol
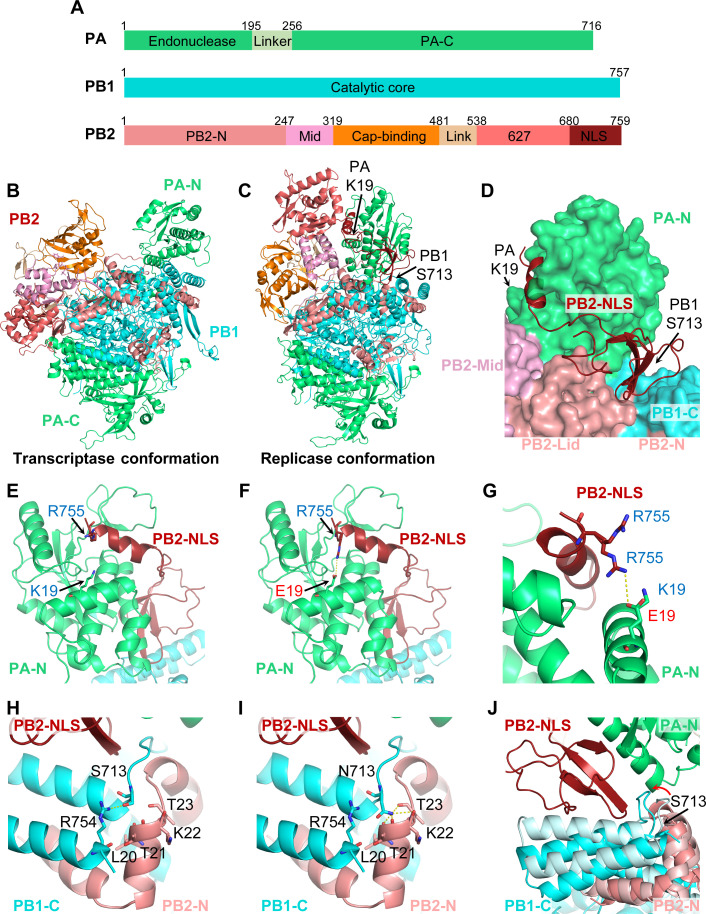
Substantial structural studies have demonstrated that FluPol, composed by PB2, PB1, and PA (Fig. 5A), can switch between different functional conformations by rearrangement of peripheral domains around the core region (4, 15, 16, 18). To further investigate the molecular mechanism by which PA-K19E and PB1-S713N promote viral genome replication, we analyzed the structural models of the WSN viral polymerase. Using SWISS-MODEL homology modeling, we generated transcriptase and replicase conformations of WSN polymerase, respectively (Fig. 5B and C). To our surprise, both the K19 of PA and the S713 of PB1 amino acids are located around the PB2-NLS domain in the replicase conformation (Fig. 5C and D). Specifically, the K19 of PA is spatially close to the R755 of PB2 (Fig. 5E). When the K19 of PA is mutated to E19, it can directly form hydrogen bond with the R755 of PB2 (Fig. 5F and G). We, therefore, propose that PA-K19E may stabilize the replicase conformation by enhancing the interaction between PA-N and the PB2-NLS domain.
Fig 5.
PA-K19E and PB1-S713N could contribute in stabilizing the replicase conformation of the viral polymerase. (A) Schematic of the three subunits of influenza polymerase. (B and C) Ribbon representation of structural models of the WSN viral polymerase in the transcriptase (B) and replicase (C) conformations created by homology modeling. (D) Surface diagram showing the specific site of PA-K19 and PB1-S713 in the replicase conformation. (E–G) Close-up of PA-K19 (E) or PA-E19 (F) and superposition of PA-K19 and PA-E19 (G) in the replicase conformation. The PB2-R755 of PB2 NLS domain is shown. Potential interactions between the PA-E19 and PB2-R755 are represented by broken lines. (H–I) Close-up of PB1-S713 (H) and PB1-N713 (I) in the replicase conformation. Potential interactions between residues are represented by broken lines. (J) Superposition of the transcriptase (lighter color) and replicase [colored as in (A)] conformations. Red arrow highlights the difference between the two conformations in the loop where PB1-S713 is located.
On the other hand, structural modeling also revealed that the S713 of PB1 is located close to the last two helices of the C terminus of PB1 (PB1-C), which is bound by PB2-N. In the replicase conformation, the S713 of PB1 interacts with the side chain of R754 of PB1 (Fig. 5H). However, when the S713 is mutated to N713, the side chain of N713 extends downward to make multivalent hydrogen bonds with the main chain carbonyl oxygens of L20, T21, K22, and T23 of PB2-N (Fig. 5I). Furthermore, superposition of the transcriptase and replicase conformations showed that the loop where the PB1 S713 is located (so-called PB1-713-loop) has an angular rotation change when moving from transcriptase conformation to replicase conformation which would bring the PB1-713-loop closer to the PB2-NLS domain (Fig. 5J). PB1-S713N substitution probably facilitates this PB1-713-loop turn toward PB2-NLS, thus facilitating the inlay of PB2-NLS into the groove formed by PB1-C/PB2-N/PA-N which would keep FluPol in the replicase conformation.
Taken together, the structural analysis results suggest that both the PA-K19E and PB1-S713N mutations could sterically contribute to the stabilization of the replicase conformation of FluPol, which may be the underlying mechanism in compensating the role of NS2 in promoting replication.
NS2 and PA-K19E/PB1-S713N facilitate FluPol dimerization
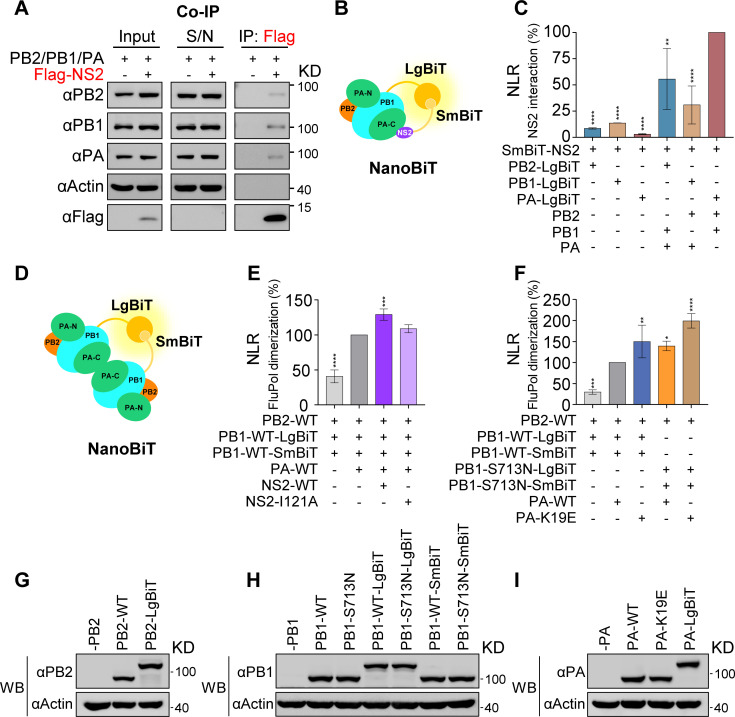
It has been reported that NS2 interacts with the subunits PB2 and PB1 of the viral polymerase but not with the PA subunit (34, 35). To better understand how NS2 regulates transcription and replication, the interaction between NS2 and the polymerase was further investigated. We first performed Co-immunoprecipitation (Co-IP) to examine the interaction between Flag-tagged NS2 and FluPol (Fig. 6A). HEK 293T cells were co-transfected with pCAGGS plasmids expressing the PB2, PB1, PA proteins, together with Flag-NS2 or empty vector, and harvested at 24 h p.t., followed by co-IP analysis with anti-Flag M2 magnetic beads. The results showed that Flag-NS2 could co-precipitate all three subunits (PB2, PB1, and PA), indicating that NS2 can bind to the intact FluPol. However, we found that the amounts of FluPol binding to NS2 were extremely low, as large amounts of the FluPol proteins were retained in the precipitate supernatant, suggesting the interaction between NS2 and FluPol is transient or very weak.
Fig 6.
NS2 and PA-K19E/PB1-S713N can facilitate viral polymerase dimerization. (A) Interaction between NS2 and the viral polymerase complex assessed by co-immunoprecipitation. HEK 293T cells were co-transfected with pCAGGS plasmids expressing PB2, PB1, PA, and either Flag-tagged NS2 or empty vector. Co-immunoprecipitation was performed by using anti-Flag magnetic beads at 48 h p.t. Immunoprecipitated proteins were detected by western blot (WB). S/N indicates supernatant. (B) Schematic of the NanoBiT luciferase complementation-based assay for the interaction between NS2 and polymerase. The large BiT (LgBiT) subunit and the small BiT (SmBiT) subunit interact to reconstitute an active Nano luciferase enzyme when viral polymerase and NS2 protein are associated. (C) Interaction between NS2 and the polymerase subunits or complex assessed by NanoBiT luciferase complementation assay. HEK-293T cells were co-transfected with the pCAGGS plasmids expressing SmBiT-tagged NS2 and one LgBiT-tagged subunit alone or combined with the other two non-tagged subunits. After 24 h p.t., the Nano luciferase enzymatic activities displayed as nomalized luminescence ratio (NLR) were measured. (D) Schematic of the NanoBiT luciferase complementation-based polymerase dimerization assay. The active Nano luciferase enzymatic activities were reconstituted when two polymerase complexes are associated. (E) Effect of NS2 wild-type or I121A mutant on polymerase dimerization. HEK 293T cells were co-transfected with the pCAGGS plasmids expressing PB1-LgBiT, PB1-SmBiT, PB2, PA, and either NS2-WT or I121A mutant. Omit of PA used as a control. After 24 h p.t., the Nano luciferase enzymatic activities were measured. (F) Effects on polymerase dimerization exerted by PA-K19E and PB1-S713N mutations alone or in combination. HEK 293T cells were co-transfected with the pCAGGS plasmids expressing PB2, PB1-LgBiT (either wild type or mutant), PB1-SmBiT (either wild type or mutant), and PA (either wild type or mutant). Omit of PA was used as a control. After 24 h p.t, the Nano luciferase enzymatic activities were measured. The results are expressed as percentages and are shown as mean ± SD of three (C) or four (E and F) independent experiments. Asterisks represent a significant difference from the control group (by one-way ANOVA with Dunnett’s post hoc test) (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (G–I) Expression levels of PB2-related (G), PB1-related (H), and PA-related (I) plasmids in HEK 293T cells.
To further verify the interaction between NS2 and FluPol, we developed a cell-based complementary reporter assay based on Nano luciferase binary technology (NanoBiT), which allows quantitative investigation of protein interactions within living cells under physiological conditions (36). The large BiT (LgBiT) subunit was fused to the C terminus of PB2, PB1, and PA, respectively, and the small BiT (SmBiT) subunit was fused to the N terminus of NS2. When SmBiT-NS2 was expressed with the three individually expressed PB2-LgBiT, PB1-LgBiT, and PA-LgBiT, the results of NanoBiT assay showed that SmBiT-NS2 interacts weakly with all three subunits, with the interaction intensities of PB1 and PB2 being stronger than that of PA, which is consistent with previous reports (Fig. 6B and C) (34, 35).
Interestingly, when SmBiT-NS2 was co-expressed with the combinations of all three subunits in which only one C-terminal LgBiT-tagged subunit was combined with the other two non-tagged subunits as indicated in Fig. 6C, the NanoBiT assay showed that interaction signals were significantly enhanced in comparison with the signals in individually expressed samples. These results further confirm that NS2 interacts much stronger with the intact FluPol complex than with the individual subunits. Interestingly, we also noticed that within the FluPol complex, SmBiT-NS2 showed the strongest binding to PA-LgBiT, followed by PB2-LgBiT, with the weakest binding being to PB1-LgBiT. This suggests that NS2 might be spatially closer to the C terminus of PA in the polymerase complex.
It is known that the newly synthesized polymerase could form both symmetric and asymmetric dimers necessary for vRNA synthesis (19, 20). This prompted us to explore whether NS2 is involved in the polymerase dimerization. With the developed cell-based NanoBiT technology in hand, we examined the dimerization level of FluPol in the cellular context (Fig. 6D). Interestingly, the results showed that wild-type NS2 can increase the dimerization extent of the polymerase, and NS2-I121A mutant can reduce the promotion effect of wild-type NS2 (Fig. 6E). Inspired by this, we then examined whether PA-K19E and PB1-S713N could also enhance FluPol dimerization. As expected, the results showed that FluPol dimerization was enhanced by the introduction of either PA-K19E or PB1-S713N mutation, and it can be further enhanced by the combination of the two mutations (Fig. 6F). The protein-expressing plasmids used for the NanoBiT assay were confirmed by western blot (Fig. 6G-I). It should be pointed out that neither PA-K19 nor PB1-S713 localizes to any of the interfaces of the symmetric and asymmetric FluPol dimers. Taken together, we concluded that wild-type NS2 promotes viral genome replication by enhancing polymerase dimerization though interacting with the intact polymerase complex. The two adaptive mutations PA-K19E and PB1-S713N could compensate for the defect of NS2-I121A in FluPol dimerization.
DISCUSSION
The viral genome transcription and replication of influenza virus are tightly controlled in the viral life cycle to ensure the most efficient virus multiplication. We recently reported that the stoichiometric changes of viral NS1 and NS2 protein expressions are involved in the fine regulation of viral genome transcription and replication (31). Specifically, we discovered that the last amino acid I121 of NS2 plays a critical role in promoting virus genome replication. To follow this up, we performed 20 amino acid screening at residue NS2-I121 for virus rescue. We found that the hydrophobicity of this residue is very important for virus survival. Interestingly, we also identified PA-K19E and PB1-S713N adaptive mutations in the passage of the NS2-I121 mutant viruses. Structural analysis revealed that PA-K19E and PB1-S713N could be beneficial in stabilizing the viral polymerase into the replicase conformation. With the self-developed NanoBiT assay, we further found evidence that both NS2 and NS2-I121mutant adaptive mutations (PA-K19E/PB1-S713N) can enhance FluPol dimerization in cells. Together, we report that NS2 promotes virus genome replication by enhancing FluPol dimerization. Therefore, we reveal that the replication-promotion function of NS2 is essential for virus survival and efficient virus multiplication.
Influenza NS2 protein is translated from the spliced mRNA of the NS segment, which accounts for 10%–15% of total unspliced NS transcripts and appears relatively late during infection (6, 37). Interestingly, the inefficient splicing of the NS segment has been proposed to act as a “molecular timer” to ensure efficient virus multiplication (30). NS2 was renamed as the nuclear export protein when it was found that NS2 plays a critical role in the nuclear export of vRNP (26). Later on, a number of studies reported that low levels of NS2 could enhance viral polymerase activities independent of its nuclear export function (34, 38), which has been implicated in facilitating the adaption of avian influenza viruses into human cells (34, 35). On the other hand, it has been reported that NS2 is capable of differentially inhibiting viral genome transcription but promoting replication (8, 24). Furthermore, we recently reported that the effect of NS2 on viral RNA transcription and replication is dose sensitive, and the late-expressed NS2 at high concentration could inhibit viral genome transcription and promote viral genome replication in an uncoupled manner (31). In this study, we further demonstrated that wild-type NS2 and NS2-I121 mutant adaptive mutations PA-K19E and PB1-S713N can promote viral genome replication by enhancing FluPol dimerization. However, how NS2 manipulates viral RNA polymerase activities in a concentration-sensitive manner and how the high levels of NS2 inhibit viral genome transcription independent of its function in promoting replication at the same time remain elusive. Further structural studies are needed to elucidate these underlying mechanisms.
Using the cell-based NanoBiT complementary reporter assay, we show that NS2 interacts weakly with all three individually expressed FluPol subunit in cells. Among them, the interaction between NS2 and PA was the weakest, which is highly consistent with the literature (34, 35). Interestingly, the interaction signals were significantly enhanced when the three subunits were co-expressed, indicating that NS2 binds primarily to the intact FluPol complex. Moreover, SmBiT-NS2 showed the strongest binding signal to PA-LgBiT, followed by PB2-LgBiT, with the weakest binding to PB1-LgBiT in the context of the intact FluPol. This suggests that the N terminus of NS2 might be spatially closer to the C terminus of PA in the FluPol-NS2 complex. Interestingly, the structure of the symmetric FluPol dimer shows that the PA-C locates in the interface of the dimer and directly mediates the dimerization (19). We speculate that NS2 probably directly interacts with PA-C to manipulate the FluPol symmetric dimerization. However, considering the recent reports that NS2 cooperates with host ANP32 protein to regulate avian influenza virus polymerase activity (35) and that ANP32A protein is involved in mediating asymmetric polymerase dimerization (20), the involvement of NS2 protein in mediating asymmetric polymerase dimerization could not be ruled out at present.
The structural studies have revealed that the conformational transition of FluPol from transcriptase conformation to replicase conformation requires in situ rotation of PA-N and significant translocation of PB2-C around the core of FluPol (4, 15). Furthermore, in a recently reported asymmetrical dimerization conformation, one of the FluPol dimers that functions in encapsidating the nascent RNA exhibited a PB2-C flip upward to pack onto the PA-C domain (20). Therefore, it is conceivable that the high flexibility of PB2-C drives the switching of FluPol among multiple states to fulfill the different functions. According to this, it makes more sense that the adaptive mutations to NS2-I121 mutant viruses were detected in less flexible PA-N and PB1-C but not in PB2. It can be speculated that, by cooperating the translocation of PB2-C, adaptive mutations in specific regions of PA and PB1 can regulate viral genome transcription or replication more appropriately. Indeed, our structural analysis indicates that both PA-K19E and PB1-S713N could exquisitely stabilize the replicase conformation by enhancing the binding of PB2-NLS to PA-N/PB1-C.
A number of specific mutations in FluPol have been identified to affect an exact function of the polymerase. For instance, point mutation PB2-F363A affects PB2 cap-binding capacity, and PA-D108A destroys PA endonuclease activity; both mutations result in the loss of transcription but not replication (39–42). On the other hand, point mutations PA-C95A and PA-T157A have been found to be able to preferentially inhibit viral genome replication with unknown mechanisms (40). Moreover, it has been shown that amino acid mutations introduced in the polymerase dimerization interface could also specifically destroy viral genome replication (19, 20). Recently, Günl et al. reported that the ubiquitin-mediated charge neutralization at PB1 K578 disrupts the interaction to an unstructured loop in the PB2 N-terminus that is required to coordinate polymerase dimerization (43). On the contrary, we identified here that PA-K19E and PB1-S713N mutations could significantly promote viral genome replication by enhancing polymerase dimerization, probably through stabilizing the replicase conformation. To the best of our knowledge, this is the first observation that specific amino acid mutations in a FluPol subunit can upregulate viral genome replication by fine-tuning the conformational changes of the viral polymerase.
In summary, this study confirms the importance of the replication-promotion function of NS2 for influenza virus survival and efficient multiplication. Moreover, it reveals that NS2 could regulate viral genome transcription and replication by participating in the modulation of FluPol conformational states. These findings not only provide further insights into the dynamic regulation of viral genome transcription and replication in the context of the virus infection but also provide basis for future structural studies in elucidating the molecular mechanisms of such regulations.
MATERIALS AND METHODS
Cells
Human embryonic kidney HEK 293T (ATCC, CRL-3216) and Madin-Darby canine kidney (MDCK) (ATCC, CCL-34) cells were purchased from the American Type Culture Collection (ATCC) and were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin. Cells were cultured in humidified incubators at 37°C with 5% CO2.
Antibodies
The following antibodies were used: the primary antibodies rabbit anti-influenza virus PB2 (prepared in our laboratory), rabbit anti-influenza virus PB1 (catalog number GTX637313; Genetex), rabbit anti-influenza virus PA (catalog number GTX125932; Genetex), mouse anti-influenza virus NP (catalog number MAB8251; Sigma-Aldrich), mouse anti-β-actin (catalog number A5441; Sigma-Aldrich), mouse anti-Flag M2 (catalog number F3165; Sigma-Aldrich), rabbit anti-tandem affinity purification (TAP) tag (catalog number sc-25768; Santa Cruz), and the secondary antibodies goat anti-rabbit IgG (catalog number BE0101-100; Easybio) and anti-mouse IgG (catalog number BE0102-100; Easybio).
Plasmids
The pHW2000 eight-plasmid rescue system of influenza virus A/WSN/1933 (H1N1/WSN) was described previously (32). The protein expression (PB2, PB1, PA, and NP) plasmids of the RNP reconstitution system of the WSN virus were described previously (44). The NS2 expression plasmids were constructed in either pcDNA or pCAGGS plasmids with the tags fused at the N terminus. pPOLI-WSN/NS plasmids used in the WSN RNP/NS reconstitution system were described previously (31). The LgBiT subunit or SmBiT subunit fused to the C terminus of PB2, PB1 (either wild type or mutant), and PA was constructed in pCAGGS plasmids according to reference (36). Site-directed mutagenesis to introduce mutations into the reverse genetics or expression plasmids was performed by using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs).
Generation and characterization of recombinant viruses
Approximately 106 HEK 293T cells mixed with MDCK cells (at a ratio of 2:1) were co-transfected with 0.5 µg of each plasmid (either wild type or mutant) of the influenza virus A/WSN/33 (H1N1) reverse genetic system using Lipofectamine 2000 (Invitrogen) and Opti-MEM (Gibco) according to the manufacturer’s instructions. After 24 h p.t., the medium was replaced with DMEM containing 0.5% FBS and 0.5 mg/mL tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Sigma-Aldrich), and the virus supernatant was harvested 48 h after the medium was changed. The concentration of infectious viral particles (PFU per milliliter) was determined by a plaque assay in MDCK cells. For viral passage, MDCK cells were infected with NS2-I121 mutant viruses at the MOI of 0.01–0.001 in DMEM supplemented with 0.5% FBS and 0.5 mg/mL TPCK-treated trypsin for 48 h. For sequencing of the viral polymerase and NS segments, viral RNA was extracted with TRI LS reagent (Sigma-Aldrich) and reverse transcribed using the SuperScript III first-strand synthesis system (Invitrogen) with a universal influenza A virus reverse transcription (RT) primer (45). The RT products were then amplified by PCR using primers (PB2 forward primer, 5′-AGCGAAAGCAGGTCAATTATATTCAATATGG-3′; PB1 forward primer, 5′-AGCGAAAGCAGGCAAACCATTTGAATGGATG-3′; PA forward primer, 5′-AGCGAAAGCAGGTACTGATTCAAAATGGAAG-3′; NS forward primer, 5’- AGCAAAAGCAGGGTGACAAAGACATAATG-3′; and universal reverse primer, 5′-AGTAGAAACAAGG-3′). The successfully amplified PCR products were purified and sequenced by Sanger sequencing (Beijing Tsingke Biotech, China).
RNP reconstitution and primer extension analyses
Approximately 106 HEK 293T cells were co-transfected with 0.5 µg each of pcDNA plasmids expressing PB2, PB1 (either wild type or mutant), PA (either wild type or mutant), NP, and 0.5 µg of the vRNA expression plasmids using Lipofectamine 2000 and Opti-MEM according to the manufacturer’s instructions. Where required, 1 µg of the pcDNA-TAP-NS2 (either wild type or mutant) was co-transfected. For all RNP reconstitution assays, a certain concentration of the empty vector was co-transfected to maintain equal concentrations of the total plasmid transfected. For primer extension analysis, cells were harvested at the indicated time points, and total RNA was extracted using TRIzol reagent (Invitrogen). The steady-state levels of three viral RNA species (mRNA, cRNA, and vRNA) were analyzed by primer extension analysis with 32P-labeled specific primers for negative- or positive-sense RNA of NA (8) and NS (37). The primer used for the detection of the internal control 5S rRNA was described previously (8). Primer extension products were analyzed on a 6% PAGE gel containing 7 M urea and detected by autoradiography.
Immunoprecipitation assay for examining the interaction between NS2 and FluPol
Approximately 106 HEK 293T cells were co-transfected with 1 µg of each of the pCAGGS plasmids expressing the PB2, PB1, PA, together with 1 µg of Flag-tagged NS2 or empty vector, using Lipofectamine 2000 and Opti-MEM according to the manufacturer’s instructions. After 48 h, cell lysates were prepared with 500 µL of lysis buffer [25 mM HEPES, 200 mM NaCl, 10% glycerol, 1 mM TCEP, 0.5% Igepal CA-630, 1 mM PMSF, and 1× EDTA-free protease inhibitor cocktail tablet (Roche)] at 4°C for 30 min. After centrifugation at 12,000 g for 15 min to remove cell debris, cell lysates were incubated at 4°C for 6 h with 10 µL/well of anti-Flag M2 magnetic beads (Sigma), in a 1-mL volume adjusted with washing buffer (10 mM HEPES, 150 mM NaCl, 10% glycerol, 0.1% Igepal CA-630, and 1 mM PMSF). The magnetic beads were then washed three times with washing buffer. The proteins were eluted using 1× Laemmli buffer. Immunoprecipitated proteins were detected by western blot.
NanoBiT luciferase complementation assay
To solidify the interaction between NS2 and polymerase, we utilized the SmBiT-tagged NS2 expression plasmid (pCAGGS-SmBiT-NS2) with the LgBiT-tagged polymerase subunit alone (pCAGGS-PB2-LgBiT, pCAGGS-PB1-LgBiT, or pCAGGS-PA-LgBiT) or combined with the other two non-tagged subunits, respectively. All of the above plasmids were co-transfected in HEK 293T cells for 30 ng and incubated for 24 h. To test the dimerization level of the polymerase, HEK 293T cells were co-transfected with 30 ng of each of the pCAGGS-PB1-LgBiT (either wild type or mutant) and pCAGGS-PB1-SmBiT (either wild type or mutant) plasmids together with 30 ng of each the pCAGGS plasmids expressing the PB2 and PA (either wild type or mutant). To determine background signal, the PA plasmid was omitted, and the total amount of plasmid was adjusted to 120 ng using the empty pCAGGS plasmid. After 24 h of incubation at 37°C, the Nano luciferase enzymatic activities were measured using the Nano-Glo Live Cell Assay System (Promega) and a GLOMAX 96 Microplate luminometer (Promega). To perform the NanoBiT luciferase complementation assay in the NS2 context, 30 ng of each of the pCAGGS-PB1-LgBiT, pCAGGS-PB1-SmBiT, pCAGGS-PB2, and pCAGGS-PA was co-transfected with 30 ng of pcDNA-TAP-NS2 (either wild type or mutant) plasmids. Likewise, the PA or NS2 plasmid was left out to test background signal. Total amount of plasmid was adjusted to 150 ng using the empty pCAGGS or pcDNA plasmid.
Western blot analysis
Cells were lysed using CytoBuster protein extraction reagent (Novagen). The lysates were separated by SDS-PAGE and immunoblotted with the indicated primary antibodies, with anti-β-actin antibody as an internal control. Protein expression levels were visualized with an enhanced chemiluminescence detection kit (NCM Biotech) according to the manufacturer’s instructions.
Homology modeling of WSN polymerase structures
Both models of WSN polymerase in the transcriptase and replicase conformations were generated by using SWISS-MODEL homology modeling with the default parameters. The crystal structure of the heterotrimeric influenza A polymerase (PDB: 6RR7 and 6QNW) was served as templates for the transcription and replication conformations, respectively. The structural figures were generated using PyMOL (version 2.4.0).
Statistics
GraphPad Prism software, version 8, was used for statistical analysis. One-way analysis of variance with Dunnett’s correction was used for one-variable comparisons.
ACKNOWLEDGMENTS
We thank Ervin Fodor (University of Oxford) for helpful discussions.
This work was supported by grants from the National Key R&D Program of China (2022YFF1203200 and 2021YFC2300700 to T.D.), the National Natural Science Foundation of China (81871669 and 32070173 to T.D.), and Beijing Natural Science Foundation (M22031 to T.D.).
Contributor Information
Tao Deng, Email: dengt@im.ac.cn.
Kanta Subbarao, The Peter Doherty Institute for Infection and Immunity, Melbourne, Australia.
REFERENCES
- 1. Petrova VN, Russell CA. 2018. The evolution of seasonal influenza viruses. Nat Rev Microbiol 16:47–60. doi: 10.1038/nrmicro.2017.146 [DOI] [PubMed] [Google Scholar]
- 2. Wang D, Zhu W, Yang L, Shu Y. 2021. The epidemiology, virology, and pathogenicity of human infections with avian influenza viruses. Cold Spring Harb Perspect Med 11:a038620. doi: 10.1101/cshperspect.a038620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fodor E, Te Velthuis AJW. 2020. Structure and function of the influenza virus transcription and replication machinery. Cold Spring Harb Perspect Med 10:a038398. doi: 10.1101/cshperspect.a038398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wandzik JM, Kouba T, Cusack S. 2021. Structure and function of influenza polymerase. Cold Spring Harb Perspect Med 11:a038372. doi: 10.1101/cshperspect.a038372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eisfeld AJ, Neumann G, Kawaoka Y. 2015. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13:28–41. doi: 10.1038/nrmicro3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hatada E, Hasegawa M, Mukaigawa J, Shimizu K, Fukuda R. 1989. Control of influenza virus gene expression: quantitative analysis of each viral RNA species in infected cells. J Biochem 105:537–546. doi: 10.1093/oxfordjournals.jbchem.a122702 [DOI] [PubMed] [Google Scholar]
- 7. Phan T, Fay EJ, Lee Z, Aron S, Hu W-S, Langlois RA. 2021. Segment-specific kinetics of mRNA, cRNA and vRNA accumulation during influenza infection. J Virol 95:e02102-20. doi: 10.1128/JVI.02102-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robb NC, Smith M, Vreede FT, Fodor E. 2009. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J Gen Virol 90:1398–1407. doi: 10.1099/vir.0.009639-0 [DOI] [PubMed] [Google Scholar]
- 9. Walker AP, Fodor E. 2019. Interplay between influenza virus and the host RNA polymerase II transcriptional machinery. Trends Microbiol 27:398–407. doi: 10.1016/j.tim.2018.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crépin T, Hart D, Lunardi T, Nanao M, Ruigrok RWH, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. doi: 10.1038/nature14009 [DOI] [PubMed] [Google Scholar]
- 11. Deng T, Vreede FT, Brownlee GG. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol 80:2337–2348. doi: 10.1128/JVI.80.5.2337-2348.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008 [DOI] [PubMed] [Google Scholar]
- 13. Hengrung N, El Omari K, Serna Martin I, Vreede FT, Cusack S, Rambo RP, Vonrhein C, Bricogne G, Stuart DI, Grimes JM, Fodor E. 2015. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527:114–117. doi: 10.1038/nature15525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, Wang P, Yan J, Zhang B, Qi J, Deng T, Gao GF, Shi Y. 2019. Structural insight into RNA synthesis by influenza D polymerase. Nat Microbiol 4:1750–1759. doi: 10.1038/s41564-019-0487-5 [DOI] [PubMed] [Google Scholar]
- 15. Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, Hart DJ, Beck M, Cusack S. 2016. Influenza polymerase can adopt an alternative configuration involving a radical repacking of PB2 domains. Mol Cell 61:125–137. doi: 10.1016/j.molcel.2015.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu Z, Fodor E, Keown JR. 2023. A structural understanding of influenza virus genome replication. Trends Microbiol 31:308–319. doi: 10.1016/j.tim.2022.09.015 [DOI] [PubMed] [Google Scholar]
- 17. Pflug A, Lukarska M, Resa-Infante P, Reich S, Cusack S. 2017. Structural insights into RNA synthesis by the influenza virus transcription-replication machine. Virus Res 234:103–117. doi: 10.1016/j.virusres.2017.01.013 [DOI] [PubMed] [Google Scholar]
- 18. Li H, Wu Y, Li M, Guo L, Gao Y, Wang Q, Zhang J, Lai Z, Zhang X, Zhu L, Lan P, Rao Z, Liu Y, Liang H. 2023. An intermediate state allows influenza polymerase to switch smoothly between transcription and replication cycles. Nat Struct Mol Biol 30:1183–1192. doi: 10.1038/s41594-023-01043-2 [DOI] [PubMed] [Google Scholar]
- 19. Fan H, Walker AP, Carrique L, Keown JR, Serna Martin I, Karia D, Sharps J, Hengrung N, Pardon E, Steyaert J, Grimes JM, Fodor E. 2019. Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature 573:287–290. doi: 10.1038/s41586-019-1530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carrique L, Fan H, Walker AP, Keown JR, Sharps J, Staller E, Barclay WS, Fodor E, Grimes JM. 2020. Host ANP32A mediates the assembly of the influenza virus replicase. Nature 587:638–643. doi: 10.1038/s41586-020-2927-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wandzik JM, Kouba T, Karuppasamy M, Pflug A, Drncova P, Provaznik J, Azevedo N, Cusack S. 2020. A structure-based model for the complete transcription cycle of influenza polymerase. Cell 181:877–893. doi: 10.1016/j.cell.2020.03.061 [DOI] [PubMed] [Google Scholar]
- 22. Vreede FT, Jung TE, Brownlee GG. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J Virol 78:9568–9572. doi: 10.1128/JVI.78.17.9568-9572.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vreede FT, Brownlee GG. 2007. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J Virol 81:2196–2204. doi: 10.1128/JVI.02187-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, García-Sastre A, tenOever BR. 2010. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A 107:11525–11530. doi: 10.1073/pnas.1001984107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez JT, Zlatev I, Aggarwal S, Subramanian S, Sachidanandam R, Kim B, Manoharan M, tenOever BR. 2012. A small-RNA enhancer of viral polymerase activity. J Virol 86:13475–13485. doi: 10.1128/JVI.02295-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O’Neill RE, Talon J, Palese P. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J 17:288–296. doi: 10.1093/emboj/17.1.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hankins RW, Nagata K, Kato A, Ishihama A. 1990. Mechanism of influenza virus transcription inhibition by matrix (M1) protein. Res Virol 141:305–314. doi: 10.1016/0923-2516(90)90002-z [DOI] [PubMed] [Google Scholar]
- 28. Watanabe K, Handa H, Mizumoto K, Nagata K. 1996. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol 70:241–247. doi: 10.1128/JVI.70.1.241-247.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heldt FS, Frensing T, Reichl U. 2012. Modeling the intracellular dynamics of influenza virus replication to understand the control of viral RNA synthesis. J Virol 86:7806–7817. doi: 10.1128/JVI.00080-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chua MA, Schmid S, Perez JT, Langlois RA, Tenoever BR. 2013. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep 3:23–29. doi: 10.1016/j.celrep.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang L, Wang Y, Shao Y, Guo J, Gao GF, Deng T. 2023. Fine regulation of influenza virus RNA transcription and replication by stoichiometric changes in viral NS1 and NS2 proteins. J Virol 97:e0033723. doi: 10.1128/jvi.00337-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nagano N, Ota M, Nishikawa K. 1999. Strong hydrophobic nature of cysteine residues in proteins. FEBS Lett 458:69–71. doi: 10.1016/s0014-5793(99)01122-9 [DOI] [PubMed] [Google Scholar]
- 34. Mänz B, Brunotte L, Reuther P, Schwemmle M. 2012. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat Commun 3:802. doi: 10.1038/ncomms1804 [DOI] [PubMed] [Google Scholar]
- 35. Sun L, Kong H, Yu M, Zhang Z, Zhang H, Na L, Qu Y, Zhang Y, Chen H, Wang X. 2023. The SUMO-interacting motif in NS2 promotes adaptation of avian influenza virus to mammals. Sci Adv 9:eadg5175. doi: 10.1126/sciadv.adg5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, Wood MG, Eggers CT, Encell LP, Wood KV. 2016. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol 11:400–408. doi: 10.1021/acschembio.5b00753 [DOI] [PubMed] [Google Scholar]
- 37. Robb NC, Jackson D, Vreede FT, Fodor E. 2010. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J Gen Virol 91:2331–2340. doi: 10.1099/vir.0.022004-0 [DOI] [PubMed] [Google Scholar]
- 38. Reuther P, Giese S, Götz V, Kilb N, Mänz B, Brunotte L, Schwemmle M. 2014. Adaptive mutations in the nuclear export protein of human-derived H5N1 strains facilitate a polymerase activity-enhancing conformation. J Virol 88:263–271. doi: 10.1128/JVI.01495-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fechter P, Mingay L, Sharps J, Chambers A, Fodor E, Brownlee GG. 2003. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J Biol Chem 278:20381–20388. doi: 10.1074/jbc.M300130200 [DOI] [PubMed] [Google Scholar]
- 40. Hara K, Schmidt FI, Crow M, Brownlee GG. 2006. Amino acid residues in the N-terminal region of the PA subunit of influenza A virus RNA polymerase play a critical role in protein stability, endonuclease activity, cap binding, and virion RNA promoter binding. J Virol 80:7789–7798. doi: 10.1128/JVI.00600-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RWH, Ortin J, Hart DJ, Cusack S. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15:500–506. doi: 10.1038/nsmb.1421 [DOI] [PubMed] [Google Scholar]
- 42. Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, Fodor E, Rao Z, Liu Y. 2009. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature 458:909–913. doi: 10.1038/nature07720 [DOI] [PubMed] [Google Scholar]
- 43. Günl F, Krischuns T, Schreiber JA, Henschel L, Wahrenburg M, Drexler HCA, Leidel SA, Cojocaru V, Seebohm G, Mellmann A, Schwemmle M, Ludwig S, Brunotte L. 2023. The ubiquitination landscape of the influenza A virus polymerase. Nat Commun 14:787. doi: 10.1038/s41467-023-36389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol 76:8989–9001. doi: 10.1128/jvi.76.18.8989-9001.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pan M, Zhang W, Xiao Y, Lai Y, Cao M, Wang J, Deng T. 2022. The hierarchical sequence requirements of the H1 subtype-specific noncoding regions of influenza A virus. Microbiol Spectr 10:e0315322. doi: 10.1128/spectrum.03153-22 [DOI] [PMC free article] [PubMed] [Google Scholar]