ABSTRACT
Porcine epidemic diarrhea virus (PEDV) is an enteric coronavirus that causes high mortality in piglets, thus posing a serious threat to the world pig industry. Porcine epidemic diarrhea (PED) is related to the imbalance of sodium absorption by small intestinal epithelial cells; however, the etiology of sodium imbalanced diarrhea caused by PEDV remains unclear. Herein, we first proved that PEDV can cause a significant decrease in Na+/H+ exchanger 3 (NHE3) expression on the cell membrane, in a viral dose-dependent manner. Further study showed that the PEDV nucleocapsid (N) protein participates in the regulation of NHE3 activity through interacting with Ezrin. Flame atomic absorption spectroscopy results indicated a serious imbalance in Na+ concentration inside and outside cells following overexpression of PEDV N. Meanwhile, molecular docking technology identified that the small molecule drug Pemetrexed acts on the PEDV N-Ezrin interaction region. It was confirmed that Pemetrexed can alleviate the imbalanced Na+ concentration in IPEC-J2 cells and the diarrhea symptoms of Rongchang pigs caused by PEDV infection. Overall, our data suggest that the interaction between PEDV N and Ezrin reduces the level of phosphorylated Ezrin, resulting in a decrease in the amount of NHE3 protein on the cell membrane. This leads to an imbalance of intracellular and extracellular Na+, which causes diarrhea symptoms in piglets. Pemetrexed is effective in relieving diarrhea caused by PEDV. Our results provide a reference to screen for anti-PEDV targets and to develop drugs to prevent PED.
IMPORTANCE
Porcine epidemic diarrhea (PED) has caused significant economic losses to the pig industry since its initial outbreak, and the pathogenic mechanism of porcine epidemic diarrhea virus (PEDV) is still under investigation. Herein, we found that the PEDV nucleocapsid protein interacts with Ezrin to regulate Na+/H+ exchanger 3 activity. In addition, we screened out Pemetrexed, a small molecule drug, which can effectively alleviate pig diarrhea caused by PEDV. These results provide support for further exploration of the pathogenesis of PEDV and the development of drugs to prevent PED.
KEYWORDS: Pemetrexed, porcine epidemic diarrhea virus, nucleocapsid protein, Ezrin, piglet diarrhea
INTRODUCTION
Porcine epidemic diarrhea (PED) is an infectious disease causing high mortality in suckling piglets. The main clinical symptoms are acute diarrhea, vomiting, and dehydration (1). Porcine epidemic diarrhea virus (PEDV) mainly attacks the jejunum of piglets, resulting in intestinal villus atrophy and intestinal epithelial cell damage, ultimately causing vomiting and diarrhea, which lead to dehydration and death of the piglets (2). PEDV has a large and highly variable genome, making it difficult to study its pathogenesis. At the same time, the virus has evolved multiple immune escape mechanisms, leading to poor effectiveness of commercially available vaccines to prevent and control the disease (3). Therefore, there is an urgent need to develop new treatments for PED, and to clarify the diarrheal pathogenesis of PEDV, which will aid prevention and treatment.
The PEDV genome is about 28 kb in length, encoding non-structural proteins (pp1a, pp1ab), structural proteins [spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N)], and protein 3 open reading frame (ORF3) (4, 5). The PEDV S protein plays an important role in the binding of the virus to receptors, fusion with the cell membranes, and cell entry. PEDV E and PEDV M proteins are important in virion assembly and budding. The PEDV N gene is 1,326 bp, encoding a protein of about 55 kDa, which is the basic structural protein produced by the virus in the host cell replication process. PEDV N has multiple potential phosphorylation sites, and as a phosphorylated protein, it plays a role in the transcription and replication of the viral genome (6).
Na+/H+ exchanger 3 (NHE3) is a major transmembrane transporter protein mediating sodium-hydrogen ion exchange, thus playing a key role in sodium and fluid uptake (7). NHE3 circulates between the plasma membrane and circulating endosomes, and is stimulated by the activity of key proteins, such as Ezrin, after reprocessing by the endoplasmic reticulum and Golgi apparatus. Ezrin is a junction protein between the cytoskeleton and the cell membrane that directly stimulates NHE3 translocation in epithelial cells (8, 9). Phosphorylated Ezrin functions to connect the cell membrane and actin filaments in the cytoplasm (10, 11), thus stimulating the transportation of NHE3 to the cytoplasmic membrane for fixation and maintenance of acid-base homeostasis in the intestine (12). Phosphorylated Ezrin transported to the cell membrane has been shown to attach NHE3 to the actin backbone in microvilli, thereby anchoring NHE3 and allowing it to perform Na+/H+ transport function in microvilli at the tip of epithelial cells, which is essential to maintain the intestinal electrolyte balance (13). In addition, several studies have demonstrated that reduced NHE3 expression in the small intestine causes a general decrease in paracellular water flux and diffusion drive, leading to reduced sodium absorption and hydrogen ion retention in the intestine, ultimately triggering diarrhea (14). Selective deletion of intestinal NHE3 leads to disruption of intestinal structural integrity, watery diarrhea, swelling of the intestine, and even death. Therefore, NHE3 is necessary to maintain the normal intestinal physiology (15, 16). Previously, we found that NHE3 plays an important role in piglet diarrhea caused by porcine alpha coronavirus (transmissible gastroenteritis virus, TGEV) and PEDV. However, the mechanisms regulating NHE3 activity in PEDV-infected host cells remain unknown.
In this study, we found that PEDV N and Ezrin are co-localized in IPEC-J2 cells, and regulating PEDV N expression could significantly affect the level of phosphorylated Ezrin. Furthermore, the small molecule lead drug Pemetrexed was screened out using molecular docking technology and shown to target the interaction region of PEDV N and Ezrin. Pemetrexed was proven to be effective to alleviate the symptoms of diarrhea caused by PEDV in piglets via in vitro and in vivo tests. These results provide new insights for the clarification of sodium imbalance diarrhea caused by PEDV, and provide a theoretical basis for the treatment of PEDV.
RESULTS
PEDV dose-dependent inhibition of membrane surface protein NHE3
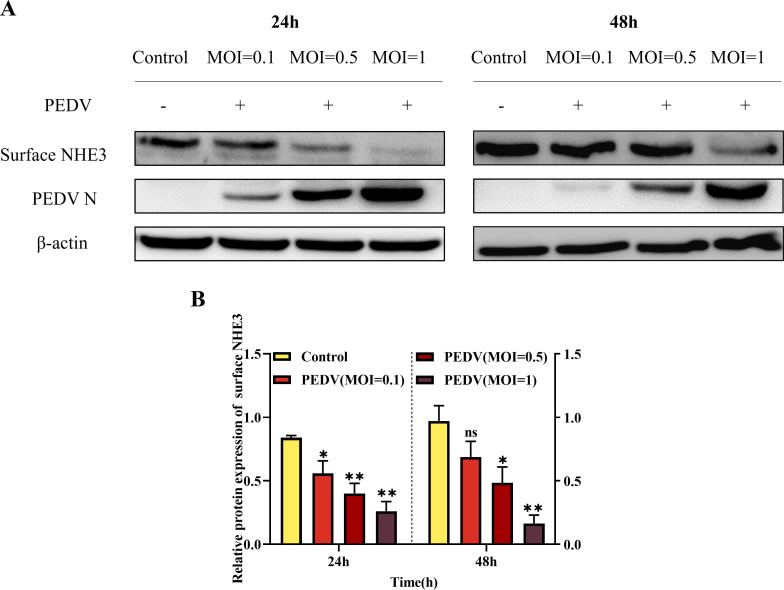
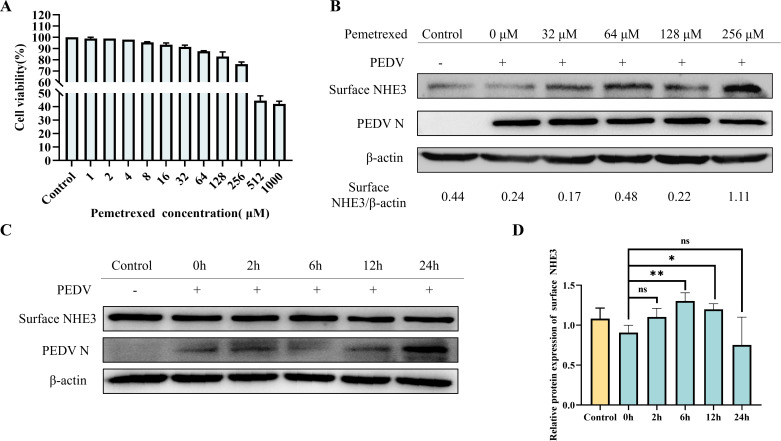
PEDV infection has previously been shown to cause a significant decrease in NHE3 expression and activity on the cell membrane, leading to diarrhea in piglets (17). Therefore, we further explored the relationship between PEDV and NHE3. The results showed that with increasing PEDV multiplicity of infection (MOI), the levels of surface NHE3 protein decreased significantly at 24 h and 48 h (Fig. 1). Thus, the decrease of membrane surface protein NHE3 expression caused by PEDV infection is related to the virus dose.
Fig 1.
Changes in surface NHE3 protein levels after PEDV infection at different MOIs. (A) Western blotting results for surface NHE3 protein expression after PEDV infection at different MOIs. (B) Quantitative analysis of surface NHE3 levels after PEDV infection at different MOIs.
PEDV N protein interacts with Ezrin
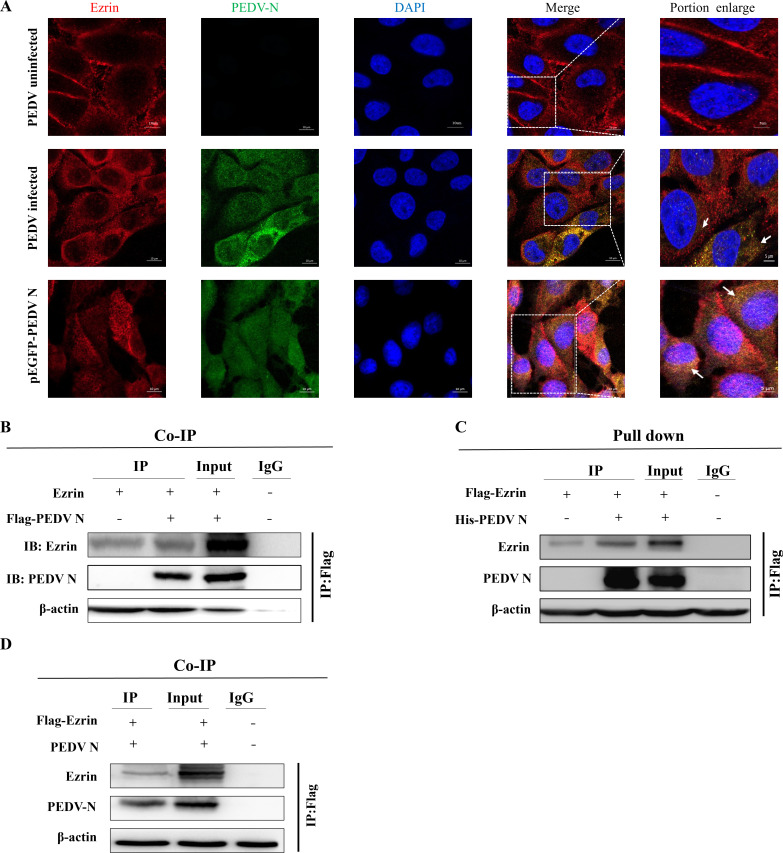
Immunofluorescence assays showed that PEDV N and Ezrin were spatially co-localized in the cytoplasm after PEDV infection of IPEC-J2 cells, and the co-localization region between PEDV N and Ezrin increased significantly after PEDV N overexpression (Fig. 2A). Subsequently, we used co-immunoprecipitation (co-IP) and FLAG pull down assays to confirm the relationship between these two proteins. The co-IP results showed the presence of specific PEDV N and Ezrin bands in the PEDV-infected group, and the reverse co-IP results also showed that there was an interaction between Ezrin and PEDV N (Fig. 2B and D). In addition, the FLAG pull down assay using human embryonic kidney cells 293 (HEK-293T) cells also showed that specific bands for PEDV N and Ezrin appeared after PEDV infection in the IP group (Fig. 2C). The above results confirmed the interaction between PEDV N and Ezrin.
Fig 2.
Determination of the interaction between PEDV N and Ezrin. (A) After treatment of IPEC-J2 cells with PEDV or PEDV N overexpression plasmids, yellow overlapping regions between PEDV N and Ezrin were observed. (B) Co-IP assay demonstrating the mutual interaction between PEDV N and Ezrin. (C) FLAG pull down assay demonstrating the interaction between PEDV N and Ezrin. (D) Reverse co-IP assay demonstrating then interaction between Ezrin and PEDV N.
PEDV N participates in the regulation of NHE3 activity by interacting with Ezrin
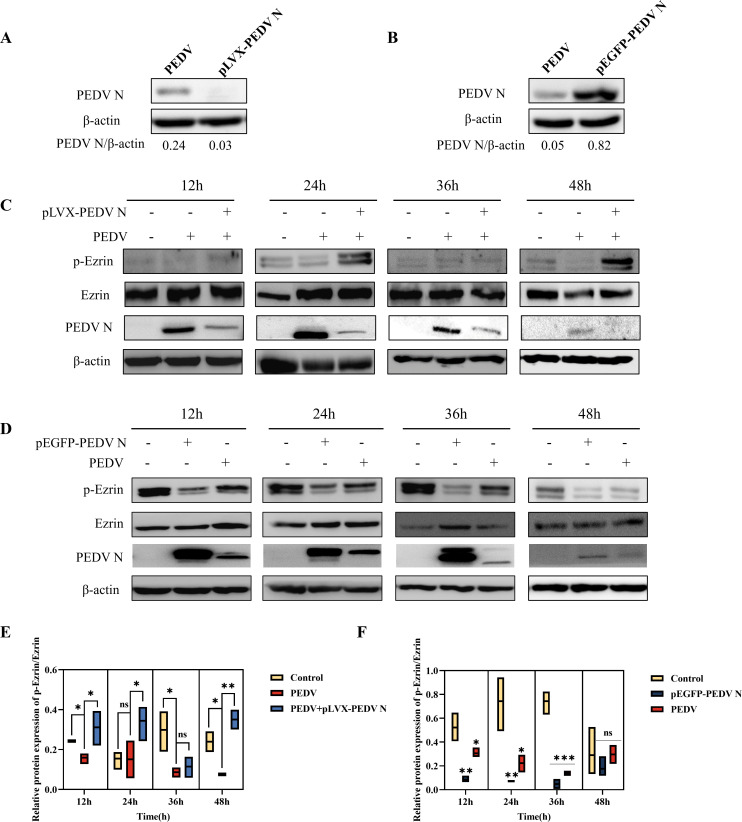
To further explore the changes in the levels of phosphorylated Ezrin after PEDV N regulation, recombinant interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N were constructed and their effects on PEDV N expression were detected using western blotting. The results showed that PEDV N protein levels in IPEC-J2 cells were downregulated and upregulated, respectively, indicating that the two recombinant plasmids could support subsequent experiments (Fig. 3A and B). Phosphorylated (p)-Ezrin/Ezrin levels were significantly downregulated after PEDV infection compared with those in the control group (Fig. 3C through F). Compared with that in the PEDV-infected group, the decreasing trend of p-Ezrin/Ezrin was significantly reversed after downregulation of PEDV N protein expression (Fig. 3C and E), and p-Ezrin/Ezrin levels were significantly reduced at 12–36 h after PEDV N overexpression (Fig. 3D and F). These results suggested that PEDV N can significantly reduce the level of phosphorylated Ezrin, and that the two are negatively correlated.
Fig 3.
Detection of the levels of PEDV N, p-Ezrin, and Ezrin after transfection of interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N in IPEC-J2 cells. (A) Western blotting results of PEDV N levels after transfection of interference plasmid pLVX-PEDV N. (B) Western blotting results of PEDV N levels after transfection of overexpression plasmid pEGFP-PEDV N. (C) Western blotting results of p-Ezrin, Ezrin, and PEDV N levels after PEDV N interference. (D) Western blotting results of p-Ezrin, Ezrin, and PEDV N levels after PEDV N overexpression. (E) Quantitative analysis of p-Ezrin/Ezrin levels after PEDV N interference. (F) Quantitative analysis of p-Ezrin/Ezrin levels after PEDV N overexpression.
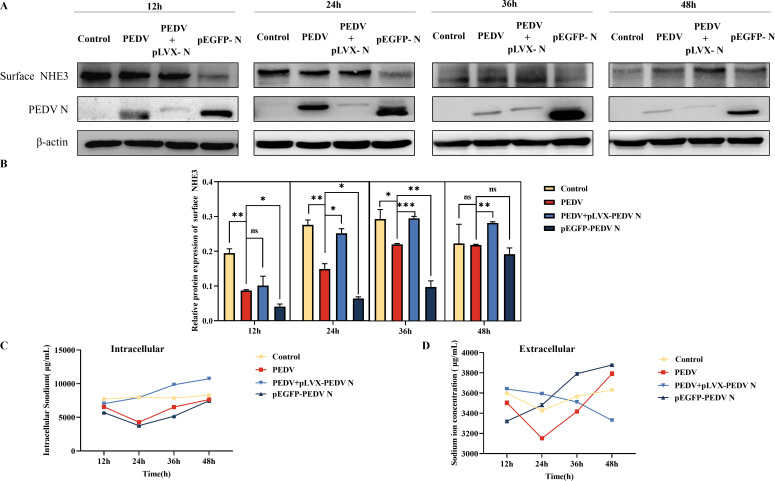
We confirmed the regulatory effect of p-Ezrin on NHE3 by detecting the effect of regulating PEDV N protein on NHE3 expression and activity. The western blotting results showed that, compared with that in the PEDV infected group, the level of surface NHE3 increased after interfering with PEDV N expression, and the level of surface NHE3 was reduced after overexpression of PEDV N, especially within 24–36 h (Fig. 4A and B). The results of flame atomic absorption spectroscopy showed that the intracellular and extracellular Na+ concentrations in the control group were relatively stable over 12–48 h. The intracellular Na+ concentrations in PEDV group and pEGFP-PEDV N group were lower than those in the control group over 12–48 h, but were higher than that in the control group in the late stages of infection. After interfering with the PEDV N protein expression, the intracellular Na+ concentration gradually increased, while the extracellular Na+ concentration gradually decreased (Fig. 4C and D). These results suggested that regulation of PEDV N can affect intracellular and extracellular Na+ exchange by altering surface NHE3 protein levels.
Fig 4.
Detection of surface NHE3 protein levels, and extracellular and intracellular Na+ concentrations after transfection of interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N in IPEC-J2 cells. (A) Western blotting results of surface NHE3 protein levels after transfection of the interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N. (B) Quantitative analysis of surface NHE3 levels after transfection of the interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N. (C) Changes in the intracellular Na+ concentration after transfection of the interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N. (D) Changes in the extracellular Na+ concentration after transfection of the interference plasmid pLVX-PEDV N and overexpression plasmid pEGFP-PEDV N.
In summary, the level of phosphorylated Ezrin increased after interfering with PEDV N, which in turn increased the expression of surface NHE3, and promoted the restoration of intracellular and extracellular Na+ exchange activity. In contrast, overexpression of PEDV N reduced the level of phosphorylated Ezrin, thereby inhibiting the expression and activity of surface NHE3 protein, ultimately leading to impaired Na+/H+ transport in small intestinal epithelial cells, which would result in piglet diarrhea.
Screening the specific inhibitor Pemetrexed for the interaction between PEDV N protein and Ezrin
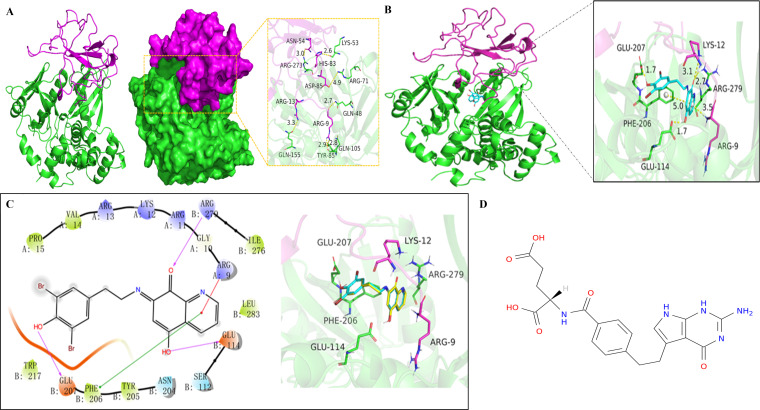
We modeled Ezrin and PEDV N proteins based on their amino acid sequences and then performed rigid docking using HDOCK and Maestro 11.9 to select the best conformation based on the output model score. APExBIO compound entity libraries of FDA (Food and Drug Administration)-approved drugs were screened as small molecule libraries and molecular docking was performed using Autodock Vina. Pymol visualization and interaction analysis showed that PEDV N protein binding sites (including amino acid residues such as Arg-9, Arg-13, Asn-54, His-83, and Asp-85) and Ezrin protein binding sites (including amino acid residues such as Gln-48, Gln-105, Tyr-85, Gln-155, Arg-273, Lys-53, and Arg-71) can form salt bridges, hydrogen bonds, hydrophobic interactions, and other interactions (Fig. 5A). These interactions might improve the stability of the PEDV N and Ezrin protein complex. In addition, we found that the Ezrin inhibitor NSC668394 could form strong cationic π and π π conjugation interactions with key residues Arg-9 (from PEDV N) and Phe-206 (from Ezrin), and strong hydrogen bond interactions with Arg-279, Glu-114, and Glu-207 (all from Ezrin) (Fig. 5B). The hydrophobic benzene ring and nitrogen heterocyclics of NSC668394 form good hydrophobic interactions with Pro-15 (from PEDV N), Val-14 (from PEDV N), Trp-217 (from Ezrin), and Leu-283 (from Ezrin). These interactions might play an important role in stabilizing NSC668394 with the PEDV N-Ezrin complex. We paired NSC668394 with the PEDV N-Ezrin protein complex and found that it had good overlap with ligands in the PEDV-N-Ezrin protein complex, indicating that the screening method was feasible (Fig. 5C). Combining the energy scoring and evaluation methods of key residues at the active site, we finally screened out Pemetrexed, with the molecular formula C20H21N5O6 (Fig. 5D).
Fig 5.
Screening out the drug Pemetrexed acting on the interaction region of the PEDV N and Ezrin using molecular docking technology. (A) Overall three-dimensional structure and interaction site of PEDV N (purple) and Ezrin (green), with yellow dashed lines representing hydrogen bonds or salt bridges. (B) Overall three-dimensional structure of the PEDV N-Ezrin complex and a close-up view of the active site where the NSC668394 ligand (blue rod) binds to the PEDV N-Ezrin complex. (C) Interaction diagram of NSC668394 ligand (yellow) with the PEDV N-Ezrin complex, with protein residues represented by circles (green, hydrophobic residues; purple, polar residues). (D) The chemical structure of Pemetrexed.
Determining the optimal treatment concentration and pretreatment time for Pemetrexed
The 50% lethal dose of Pemetrexed toward IPEC-J2 cells was 512 µM according to the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 6A). To verify the effect of Pemetrexed, we examined the effect of different concentrations of Pemetrexed on surface NHE3 protein levels. The results showed that increasing the Pemetrexed concentration increased surface NHE3 levels under PEDV infection (Fig. 6B). Combining the results of the cytotoxicity test and the effect on membrane NHE3 protein expression levels, we selected an optimal concentration of Pemetrexed that maintained a cell survival rate greater than 50% and significantly increased membrane NHE3 expression, which was 256 µM. We then examined the effect of 256 µM Pemetrexed on surface NHE3 protein expression after different pretreatment times, which showed that the optimal pretreatment time point for Pemetrexed was 6 h (Fig. 6C and D). According to the above results, the concentration of Pemetrexed used in the follow-up tests was 256 µM and the treatment time was 6 h.
Fig 6.
Screening of Pemetrexed for optimal treatment concentration and pretreatment time. (A) Statistical plot of cell viability after treatment with different concentrations of Pemetrexed. (B) Western blotting results of surface NHE3 protein levels after treatment with different concentrations of Pemetrexed in the presence of PEDV infection. (C) Western blotting results of surface NHE3 protein levels at different time points after Pemetrexed treatment in the presence of PEDV infection. (D) Quantitative analysis of surface NHE3 levels at different time points after Pemetrexed treatment in the presence of PEDV infection.
Pemetrexed blocks the interaction between PEDV N and Ezrin
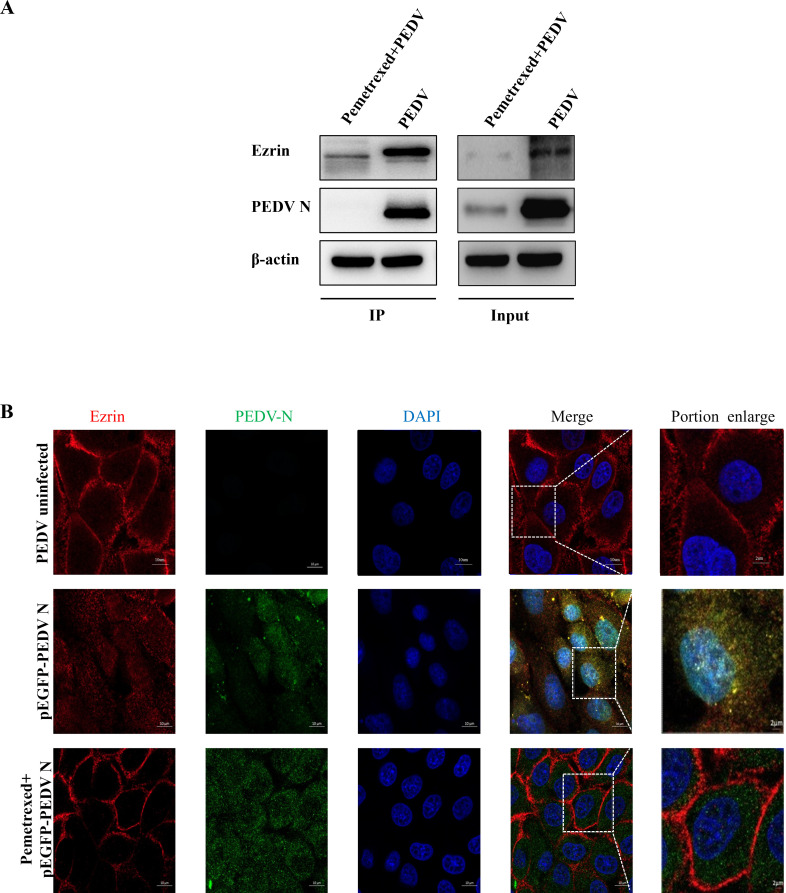
We tested whether PEDV N continued to interact with Ezrin after Pemetrexed treatment using co-IP and immunological co-localization assays. The co-IP results showed that there were no specific PEDV N bands in the PEDV-infected group after Pemetrexed pretreatment (Fig. 7A). The immunofluorescence co-localization results also showed that there was no PEDV N-Ezrin co-localization region around the nucleus after Pemetrexed pretreatment (Fig. 7B). The results suggested that Pemetrexed could effectively block the interaction between PEDV N and Ezrin.
Fig 7.
Effect of Pemetrexed on the interaction between PEDV N and Ezrin. (A) Co-IP results showing that Pemetrexed pretreatment blocked the interaction between PEDV N and Ezrin. (B) Immunofluorescence analysis showed that the co-localization region (yellow) between PEDV N and Ezrin was significantly reduced after pretreatment with Pemetrexed.
Pemetrexed pretreatment significantly increased the level of NHE3, decreased PEDV titers, and restored the balance of intracellular and extracellular Na+ concentrations
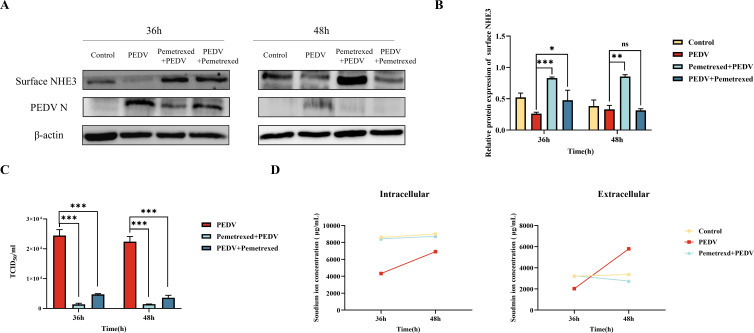
A pretreatment infection group (Pemetrexed + PEDV group) and a post-infection administration group (PEDV + Pemetrexed group) were set up, respectively, and it was found that the level of surface NHE3 was significantly increased by Pemetrexed, to a level higher than that in the control group, and the effect was more significant after Pemetrexed pretreatment (Fig. 8A and B). The PEDV titers in the Pemetrexed + PEDV and PEDV + Pemetrexed groups were significantly lower than those in the PEDV group, and the PEDV titers in the Pemetrexed + PEDV group were lower than those in the PEDV + Pemetrexed group (Fig. 8C). The flame atomic absorption spectrometry data showed that the changes in the intracellular and extracellular Na+ concentrations in the control group were relatively stable, while the extracellular Na+ concentration increased rapidly after PEDV infection, and was significantly higher than that in the control group at 48 h. PEDV inhibited Na+ entry into cells, and the intracellular concentrations were much lower than that in the control group. After Pemetrexed pretreatment, the fluctuation of intracellular and extracellular Na+ concentrations decreased, the extracellular Na+ concentration decreased slowly, and the intracellular Na+ concentration increased gradually, to a level close to that in the control group (Fig. 8D).
Fig 8.
Effects of Pemetrexed on surface NHE3 protein levels, PEDV titer, intracellular and extracellular Na+ concentrations under PEDV infection conditions. (A) Western blotting of surface NHE3 protein levels after Pemetrexed treatment. (B) Quantitative analysis of surface NHE3 levels after Pemetrexed treatment. (C) TCID50 (50% tissue culture infective dose) assay to detect the effect of Pemetrexed treatment on PEDV titers. (D) Changes in intracellular and extracellular Na+ concentrations after Pemetrexed pretreatment.
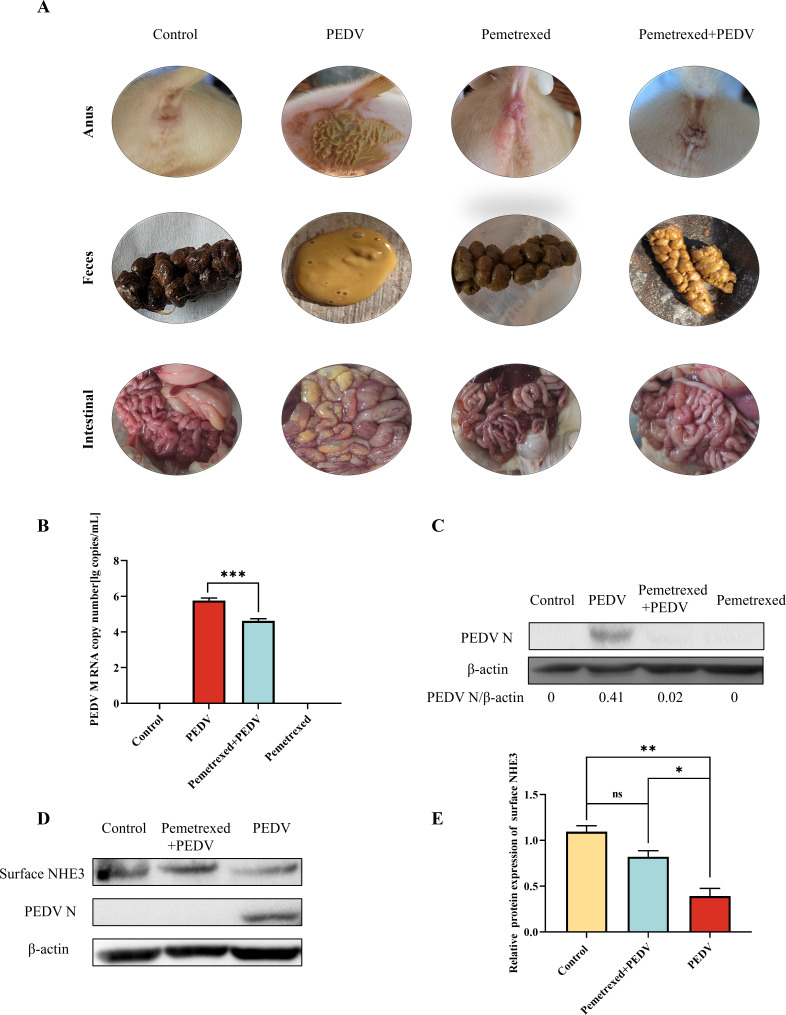
Pemetrexed is effective in preventing diarrhea in piglets
To determine the clinical effect of Pemetrexed, we conducted a clinical trial of Pemetrexed as a prophylactic drug for PEDV using Rongchang piglets as animal models. Piglets in the PEDV group all showed obvious diarrhea symptoms. None of the piglets in the control group, the Pemetrexed group, and the Pemetrexed + PEDV group showed diarrhea symptoms. After preventive administration of Pemetrexed, the feces of piglets were dry and the diarrhea symptoms of piglets were effectively relieved. The anatomical results showed that compared with that of the control group, the gastrointestinal tract of piglets pretreated with Pemetrexed showed no significant pathological changes (Fig. 9A). The expression of the PEDV membrane (M) gene was detected using quantitative real-time reverse transcription PCR (RT-qPCR) in small intestine tissues of piglets in the different treatment groups as a proxy measurement of PEDV infection. The results showed that the PEDV M gene expression in the intestinal tissues of piglets in the Pemetrexed + PEDV group was significantly lower than that in the PEDV infected group (Fig. 9B). Western blotting results showed that the level of PEDV N in the Pemetrexed + PEDV group was significantly lower than that in the PEDV group (Fig. 9C) and the levels of surface NHE3 protein in the Pemetrexed + PEDV group was significantly higher than that in the PEDV group (Fig. 9D and E).
Fig 9.
Animal experiments to test the clinical effect of Pemetrexed. (A) Clinical efficacy of Pemetrexed in preventing diarrhea in piglets caused by PEDV infection. (B) Expression of PEDV M mRNA in the small intestine tissues of piglets. (C) Western blotting of PEDV N protein levels in the small intestine tissue of piglets. (D) Western blotting of surface NHE3 and PEDV N protein levels in the small intestine tissues of piglets. (E) Quantitative analysis of surface NHE3 levels in the small intestine tissues of piglets.
DISCUSSION
Since the outbreak of the novel coronavirus in 2019, researchers have paid more attention to the study of other coronaviruses. PEDV is a member of the α-coronavirus genus, which mainly causes severe diarrhea and vomiting in piglets aged 3–10 days (18). PEDV-induced diarrhea is associated with reduced mobility and expression of NHE3 in the organism (17). However, the exact mechanism is unknown. An important biological role of NHE3 is to maintain the concentration gradient of Na+ on the cell membrane (19). Reduced expression of NHE3 results in reduced absorption of Na+ by the jejunum, leading to disruption of the normal structure of the intestinal microvilli, which increases diarrhea. NHE3 is a highly regulated ion exchange protein whose regulation depends on its C-terminal cytoplasmic domain, which acts as a scaffold to bind a variety of regulatory proteins (including Ezrin), thereby exerting its biological activity (20).
Ezrin is a backbone protein localized on the apical membrane of epithelial cells (such as the uterus, small intestine, and renal tubules), where it maintains the apical microvilli of polarized epithelial cells, and interacts with a variety of signaling pathway-related proteins (such as CD44, receptor tyrosine kinase) (21, 22). A correlation has been identified between NHE3 and Ezrin, linking NHE3 to the actin cytoskeleton in microvilli, which is necessary to regulate the endocytosis rate and stimulates changes in the exocytosis rate. At the brush borders of intestinal and renal epithelial cells, the Na+/H+ exchange regulatory factor 1 (NHERF1) and Ezrin binding domains are essential to regulate the transport of ion transporters and the function of receptors on the cell membrane. Ezrin and NHERF1 anchor or retain NHE3 in microvilli through multiple protein interaction domains (23). NHE3’s function is regulated by assembling it into large complexes. Ezrin can regulate NHE3 activity through direct or indirect binding. On the one hand, p-Ezrin can interact directly with NHE3 by binding to its C-terminus of NHE3 or through the FERM domain (a tetrahymena shamrock-shaped protein structural domain) (24), which determines the expression of surface NHE3. Thus, reducing the direct binding of NHE3 to Ezrin could decrease NHE3 activity at the base of the ileum. On the other hand, p-Ezrin indirectly binds to the C-terminus of the membrane protein NHE3 via NHERF1 or NHERF2 (25, 26), relying on Na+-K+-ATPase to provide energy, such that Na+ and H+ are alternately transported in equal proportions, and the dynamic balance of Na+ inside and outside the cell is maintained (27).
PEDV N protein is a highly functionally conserved structural protein (28), comprising three domains: The N-terminal domain, linkage region, and C-terminal domain (29). PEDV N recognizes and binds to viral genomic RNA to eventually form the helical nucleocapsid (30). PEDV N interacts with tumor suppressor p53 to induce cell cycle arrest in S-phase and promote viral replication (31). PEDV inhibits histone deacetylase 1 expression to promote replication by binding its nucleocapsid protein to the host transcription factor sp1 (32).
The present study found that PEDV N interacts with Ezrin, and identified an inverse relationship between PEDV N and Ezrin phosphorylation. We further found that PEDV N significantly affected the amount and activity of NHE3, demonstrating that PEDV N is involved in regulating NHE3 activity by influencing Ezrin phosphorylation levels. Finally, we used virtual screening to identify the small molecule Pemetrexed as an inhibitor of the PEDV N-Ezrin interaction. Pemetrexed is often used as a standard drug for the treatment of rectal and pancreatic cancer. The results of co-IP and fluorescence co-localization showed that Pemetrexed pretreatment could block the interaction between PEDV N and Ezrin. In vitro experiments showed that Pemetrexed could increase the expression and activity of NHE3, and in vivo experiments showed that Pemetrexed pretreatment could effectively prevent diarrhea in piglets caused by PEDV infection. These results increase our understanding of the pathogenesis of coronavirus N protein and provide new ideas for screening novel therapeutic drugs for animal infectious diseases.
MATERIALS AND METHODS
Cells and virus
Porcine small intestinal epithelial cells (IPEC-J2) and African green monkey kidney cells (Vero) were purchased from Shanghai Suer Biotechnology Co., Ltd (Shanghai, China). HEK-293T and the PEDV-LJX strain were preserved in our laboratory. All cell assays were performed at MOI = 0.1. All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (Biological Industries, Beit Haemek, Israel) and 1% double antibiotics (streptomycin + penicillin; Gibco).
Extraction of total proteins
Samples in lysis buffer (Beyotime, Shanghai, China) were lysed on ice for 15 min, and vortexed every 5 min to fully lyse the cells. The supernatant was collected by centrifugation at 12,000 × g for 15 min at 4°C, and 1/5 vol of 6 × Protein Loading Buffer (TransGen Biotech, Beijing, China) was added. The proteins were denatured by boiling at 100°C for 10 min and then frozen at −80°C.
Extraction of membrane proteins
Membrane extraction reagent A (Beyotime) containing 1% phenylmethylsulfonyl fluoride (PMSF) was added to the cells, frozen and thawed in liquid nitrogen, and centrifuged at 4°C and 700 × g for 10 min. The supernatant was pipetted into a new centrifuge tube, centrifuged at 4°C and 14,000 × g for 30 min, the precipitate was retained, then 200 µL of membrane protein extraction reagent B containing 1% PMSF (Beyotime) was added. The samples were incubated on ice for 15 min, and centrifuged at 4°C and 14,000 × g for 5 min. The supernatant was collected as the membrane protein solution.
Western blotting
Equal amounts of protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then the proteins were transferred to polyvinylidene fluoride membranes (Merck Millipore, Billerica, MA, USA). The membranes were blocked with Tris-buffered saline-Tween 20 (TBST) with 5% skim milk at room temperature for 1 h, and then washed with TBST. Rabbit-derived anti-p-Ezrin polyclonal antibodies (Abcam, Cambridge, UK), rabbit-derived anti-Ezrin polyclonal antibodies (Proteintech, Rosemont, IL, USA), mouse-derived anti-NHE3 polyclonal antibodies (Proteintech), mouse-derived anti-PEDV N polyclonal antibodies (kindly gifted by Guangliang Liu, Research Fellow, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences), and mouse-derived anti-β-actin polyclonal antibody (Proteintech) were diluted in TBST and incubated with the membranes overnight at 4°C. The next day, goat anti-rabbit or goat anti-mouse antibodies (Proteintech) labeled with horseradish peroxidase were incubated with the membranes at room temperature for 1 h. The immunoreactive protein bands were visualized using the FX5 imaging system (VILBER, Collégien, France), and the grayscale values of the bands were analyzed using Image J software (National Institutes of Health, Bethesda, MD, USA).
Immunofluorescence co-localization
IPEC-J2 cells were inoculated into 24-well plates pre-lined with cell climbing tablets cell crawlers (Solarbio, Beijing, China), and transfected with plasmids when the cells’ adhesive growth reached 70%.
Twenty-four hours after transfection of the plasmid, the cells were then infected with PEDV. After 24 h of PEDV infection, the cells were treated with 4% paraformaldehyde (Solarbio) for 25 min and permeabilized with 0.03% Triton X-100 (Solarbio) for 10 min. The cells were then blocked using 0.3% bovine serum albumin (Beyotime) for 1 h and incubated with anti-Ezrin antibodies (1:150) and anti-PEDV N antibodies (1:1) at 4°C for 14 h. The cells were washed using phosphate-buffered saline-Tween 20 three times and incubated with fluorescent secondary antibodies (cy3 mouse secondary antibody 1:500; AlexaFluor 647 rabbit secondary antibody 1:500; Proteintech) in the dark for 1 h. The nuclei were stained with 4′,6-diamidino-2-phenylindole (Beyotime) in the dark. Finally, the cells were observed using a laser confocal microscope and photographed. The images were processed and analyzed using ZEN software (Carl Zeiss, Oberkochen, Germany).
co-IP
The PEDV N overexpression vector with a Flag tag was transfected into IPEC-J2 cells and samples were collected after 48 h. Cell samples were lysed using 1 mL of lysis buffer (Beyotime) containing 1% PMSF, centrifuged at 10,000–14,000 × g for 3–5 min at 4°C, and the supernatant was collected. Anti-Flag Agarose Gel magnetic beads (Beyotime) were added to the sample and Mouse IgG Agarose Gel magnetic beads (Beyotime) were added to the negative control for immunoprecipitation. Then, the samples were placed on a rotary mixer and incubated for 12 h at 4°C. After incubation, the samples were centrifuged at 6,000 × g for 30 s at 4°C and the supernatant was discarded. Then, 0.5 mL of lysis buffer containing inhibitor lysate was added, placed in an ice bath for 5 min, centrifuged at 4°C 6,000 × g for 30 s, and the supernatant was discarded. Then, 2 × SDS PAGE Sample Loading Buffer (Beyotime) was added and heated at 95°C for 5 min, and the supernatant was removed and subjected to western blotting.
Flag pull down assay
When HEK-293T cells reached 80% confluence, His-pEGFP-PEDV N, Flag-pEGFP-Ezrin, and empty plasmid pEGFP were transfected into the cells using TransIntro EL Transfection Reagent separately. Samples were collected after 48 h, and a portion of each sample was collected as the Input group. After adding Anti-Flag Agarose Gel and Mouse IgG Agarose Gel beads to the samples, HEK-293T cell samples transfected with His-pEGFP-PEDV N and Flag-pEGFP-Ezrin were mixed on the rotary mixer, and then treated in the same way as in the co-IP assay.
Construction and transfection of overexpression and shRNA interference vectors
The overexpression vector and shRNA vectors of PEDV N protein (Table 1) were synthesized by Genecreat Biotechnology Co., Ltd. (Wuhan, China). TransIntro EL Transfection Reagent (TransGen Biotech, Beijing, China) was used to transfect the vectors into IPEC-J2 cells, IPEC-J2 cells were infected with PEDV when the plasmid-transfected cells showed stable fluorescence (usually 24 h after transfection), and samples were collected at different time points.
TABLE 1.
shRNA vector sequence
| shRNA vector name | Sequence |
|---|---|
| PEDV N-shRNA | GCAACTGGCACTTCTACTACCCTCGAGGGTAGTAGAAGTGCCAGTTGCTTTTT |
Flame atomic absorption spectrometry
The Na+ standard solution was configured in advance. The acetylene valve was opened, and the instrument and computer software were switched on in turn according to the operating instructions of the instrument and computer software. The Na element lamp was selected, the parameters on the computer software were set, and the working wavelength of Na was measured. After selecting the peak, energy balancing was carried out. When the energy value reached 100%, zero point calibration was performed. After cleaning the igniter with ultrapure water and then with 5% KNO3 media solution, each sample was finally measured (including the dilutions of the standard Na solutions with mass concentrations of 0.05 µg/mL, 0.10 µg/mL, 0.20 µg/mL, 0.30 µg/mL, and 0.40 µg/mL, respectively), and the values were recorded.
Molecular docking and screening of blocking drugs
Acquisition of the structure of the binding target protein
Both the PEDV N and Ezrin protein structures were constructed using the HDOCK online server based on their amino acid sequences [http://hdock.phys.hust.edu.cn/, homologous templates: 2GEC (37.7%), 4RM8 (95.6%)]. HDOCK protein docking software was used to perform molecular docking between PEDV N and Ezrin, and the lowest energy structural conformation was selected as the receptor template protein through the scoring function, followed by compound screening. The processing and optimization of the virtual screening was carried out using the Glide module in Schrödinger Maestro software (Schrödinger, New York, NY, USA), which selects the prediction site of the protein as the center of a 20 Å box. First, the ligand was re-docked to confirm the feasibility of the selected docking method. The data set was then screened using standard-precision (SP) docking, which is suitable to screen a large number of compounds. Finally, the extra-precision docking template was used to screen the high scores of the primary ligands determined using the SP method.
Reliability validation of binding target protein structures
To verify the validity of the docking method, we selected the Ezrin inhibitor NSC668394 as a positive control. NSC668394 molecules can form strong cation-π and π-π conjugate interactions with key residues, and can also form strong hydrogen bond interactions. To determine the appropriate docking protocol for screening potential active compounds, NSC668394 was attached to the receptor binding site between Ezrin and PEDV N, and its binding conformation overlapped well with the ligands in the complex mentioned above, indicating that the screening method was effective and feasible.
Virtual screening of blocking drugs
The FAD-approved APExBIO compound entity library (https://www.apexbio.cn/discoveryprobetm-clinical-fda-approved-drug-library) was selected as an alternative library, and all compounds were protonated and minimized using the LigPre module in the Maestro11.9 platform. The docking data were filtered according to the energy score, the first 155 compounds were screened, and the target drug Pemetrexed was finally obtained by combining the energy score and the evaluation of key residues in the active site.
MTT test
Pemetrexed was dissolved in dimethyl sulfoxide (DMSO). IPEC-J2 cells were inoculated in 96-well plates. When the cell growth reached about 85% confluence, the medium was discarded, DMEM semi-culture medium was added to dilute the drug to different concentrations, and the culture medium was changed 90 min later. After a further 24 h, the MTT reagent (Beyotime) was added to each well, incubation was continued for 4 h, formazan lysate was then added, and incubation continued for another 4 h. The absorbance of each well was measured at OD570. The maximum non-toxic dose of the drug to cells was calculated according to the cell survival rate (%), calculated as (the OD of the experimental group − the OD of the blank group) / (the OD of the control group − the OD of the blank group) × 100.
TCID50
Vero cells were seeded in 96-well plates at a density of 4 × 104/0.1 mL, and when they reached 90% confluence, the cell supernatant was collected at different time points after PEDV infection and diluted 10-fold with DMEM semi-culture medium supplemented with 10 µg/mL trypsin at a total of six dilutions of 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6, while a blank control group was established and placed at 37°C, 5% CO2 incubator for 72 h. The cytopathic conditions in each well were observed every 12 h and the viral titer was calculated using the Reed-Muench method (33).
Animal experiments
Twelve 7-day-old lactating Rongchang piglets (PEDV and TGEV-negative) were randomly divided into a control group, PEDV group, Pemetrexed group, and Pemetrexed + PEDV group, with three piglets in each group. Piglets in the Pemetrexed group and Pemetrexed + PEDV group were fed 0.5 mg/kg Pemetrexed (MedChemExpress, Monmouth Junction, NJ, USA) 6 h in advance. Pemetrexed was diluted with 10% DMSO, 40% polyethylene glycol (PEG300), 5% Tween 80, and saturated saline. After 6 h, piglets in the PEDV group and Pemetrexed + PEDV group were fed 15 mL of PEDV-LJX solution (derived from Vero cell propagation) with a titer of 1.35 × 106 TCID50/mL, and piglets in the control group and Pemetrexed group were fed 15 mL of normal saline. The clinical symptoms of piglets infected with PEDV were observed, and photographs were daily. When the piglets in the PEDV group showed obvious diarrheal symptoms, the piglets were humanely sacrificed and clinically dissected, and the intestinal lesions were observed and recorded. The jejunal tissues were frozen at −80°C for the extraction of tissue protein and total RNA.
Absolute fluorescence quantitative PCR
The PEDV M fluorescent PCR primers were designed and synthesized by Sangon BiotechShanghai Co., Ltd. (Shanghai, China). cDNA was obtained by reverse transcription of PEDV-LJX solution (derived from Vero cell propagation), and was then gradient diluted 10-fold to 10−7 as a positive qPCR standard template. The primers are shown in Table 2. After qPCR, the linear relationship between the Ct value and copy number was analyzed using Bio-Rad CFX Manager software (Bio-Rad, Hercules, CA, USA), and the standard curve was generated using the random matrix method. The Ct values of cDNAs from different intestinal tissues were substituted into the standard curve to obtain the copy number of the M gene of the PEDV-LJX strain.
TABLE 2.
Primer sequences
| Primer names | Primer sequences (5´−3´) | Product size (bp) |
|---|---|---|
| PEDV M-F | AGGTTGCTACTGGCGTACAG | 157 |
| PEDV M-R | GAGTAGTCGCCGTGTTTGGA |
Statistical analysis
The experimental results were statistically analyzed using GraphPad Prism 8.0 (GraphPad Inc., La Jolla, CA, USA) software. All data are expressed as the mean ± SD or standard error of the mean of three independent experiments. In addition, one-way analysis of variance and t-tests were used to detect statistical differences between the groups. A P-value less than 0.05 was considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001).
ACKNOWLEDGMENTS
We would like to thank Guangliang Liu (Research Fellow, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences) for providing the mouse-derived anti-PEDV N polyclonal antibodies.
Contributor Information
Xingcui Zhang, Email: zhangxc923@swu.edu.cn.
Zhenhui Song, Email: szh7678@126.com.
Tom Gallagher, Loyola University Chicago - Health Sciences Campus, Maywood, Illinois, USA.
ETHICS APPROVAL
All animal experiments were approved by Southwestern University’s Institutional Animal Care and Use Committee (Animal Protocol Approval Number: IACUC-20230509–02) and followed the National Institutes of Health’s Animal Performance Guidelines.
REFERENCES
- 1. Li Z, Luo Y, Huang Z, Zhao C, Chen H, El-Ashram S, Huang J, Su L, Zhang W, Ma G, Liang Y, Guo J, Huang S, Zhao Y. 2023. An ultrasensitive electrochemical sensor for detecting porcine epidemic diarrhea virus based on a Prussian blue-reduced graphene oxide modified glassy carbon electrode. Anal Biochem 662:115013. doi: 10.1016/j.ab.2022.115013 [DOI] [PubMed] [Google Scholar]
- 2. Chen Y-M, Burrough E. 2022. The effects of swine coronaviruses on ER stress, autophagy, apoptosis, and alterations in cell morphology. Pathogens 11:940. doi: 10.3390/pathogens11080940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang Y, Huang H, Li D, Zhao C, Li S, Qin P, Li Y, Yang X, Du W, Li W, Li Y. 2023. Identification of niclosamide as a novel antiviral agent against porcine epidemic diarrhea virus infection by targeting viral internalization. Virol Sin 38:296–308. doi: 10.1016/j.virs.2023.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yao X, Qiao W-T, Zhang Y-Q, Lu W-H, Wang Z-W, Li H-X, Li J-L. 2023. A new PEDV strain CH/HLJJS/2022 can challenge current detection methods and vaccines. Virol J 20:13. doi: 10.1186/s12985-023-01961-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhuang H, Sun L, Wang X, Xiao M, Zeng L, Wang H, Yang H, Lin F, Wang C, Qin L, Wang C. 2022. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus strains circulating in China from 2020 to 2021. BMC Vet Res 18:392. doi: 10.1186/s12917-022-03481-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li Z, Zeng W, Ye S, Lv J, Nie A, Zhang B, Sun Y, Han H, He Q. 2018. Cellular hnRNP A1 interacts with nucleocapsid protein of porcine epidemic diarrhea virus and impairs viral replication. Viruses 10:127. doi: 10.3390/v10030127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anbazhagan AN, Priyamvada S, Kumar A, Jayawardena D, Borthakur A, Gill RK, Alrefai WA, Dudeja PK, Saksena S. 2022. Downregulation of NHE-3 (SLC9A3) expression by microRNAs in intestinal epithelial cells. Am J Physiol Cell Physiol 323:C1720–C1727. doi: 10.1152/ajpcell.00294.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cha B, Tse M, Yun C, Kovbasnjuk O, Mohan S, Hubbard A, Arpin M, Donowitz M. 2006. The NHE3 juxtamembrane cytoplasmic domain directly binds Ezrin: dual role in NHE3 trafficking and mobility in the brush border. Mol Biol Cell 17:2661–2673. doi: 10.1091/mbc.e05-09-0843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yun CH, Lamprecht G, Forster DV, Sidor A. 1998. NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein Ezrin. J Biol Chem 273:25856–25863. doi: 10.1074/jbc.273.40.25856 [DOI] [PubMed] [Google Scholar]
- 10. Senju Y, Hibino E. 2023. Moesin-Ezrin-radixin-like protein merlin: its conserved and distinct functions from those of ERM proteins. Biochim Biophys Acta Biomembr 1865:184076. doi: 10.1016/j.bbamem.2022.184076 [DOI] [PubMed] [Google Scholar]
- 11. Barik GK, Sahay O, Paul D, Santra MK. 2022. Ezrin gone rogue in cancer progression and metastasis: an enticing therapeutic target. Biochim Biophys Acta Rev Cancer 1877:188753. doi: 10.1016/j.bbcan.2022.188753 [DOI] [PubMed] [Google Scholar]
- 12. Nwia SM, Li XC, Leite A de O, Hassan R, Zhuo JL. 2022. The Na+/H+ exchanger 3 in the intestines and the proximal tubule of the kidney: localization, physiological function, and key roles in angiotensin II-induced hypertension. Front Physiol 13:861659. doi: 10.3389/fphys.2022.861659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayashi H, Tamura A, Krishnan D, Tsukita S, Suzuki Y, Kocinsky HS, Aronson PS, Orlowski J, Grinstein S, Alexander RT. 2013. Ezrin is required for the functional regulation of the epithelial sodium proton exchanger, NHE3. PLoS One 8:e55623. doi: 10.1371/journal.pone.0055623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kovesdy CP, Adebiyi A, Rosenbaum D, Jacobs JW, Quarles LD. 2021. Novel treatments from inhibition of the intestinal sodium-hydrogen exchanger 3. Int J Nephrol Renovasc Dis 14:411–420. doi: 10.2147/IJNRD.S334024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xue J, Dominguez Rieg JA, Thomas L, White JR, Rieg T. 2022. Intestine-specific NHE3 deletion in adulthood causes microbial dysbiosis. Front Cell Infect Microbiol 12:896309. doi: 10.3389/fcimb.2022.896309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Woo AL, Gildea LA, Tack LM, Miller ML, Spicer Z, Millhorn DE, Finkelman FD, Hassett DJ, Shull GE. 2002. In vivo evidence for interferon-gamma-mediated homeostatic mechanisms in small intestine of the NHE3 Na+/H+ exchanger knockout model of congenital diarrhea. J Biol Chem 277:49036–49046. doi: 10.1074/jbc.M205288200 [DOI] [PubMed] [Google Scholar]
- 17. Song Z, Yan T, Ran L, Niu Z, Zhang Y, Kan Z, Xu S, Zhang S, Zhang J, Zou H, Lei C. 2021. Reduced activity of intestinal surface Na+/H+ exchanger NHE3 is a key factor for induction of diarrhea after PEDV infection in neonatal piglets. Virology 563:64–73. doi: 10.1016/j.virol.2021.08.011 [DOI] [PubMed] [Google Scholar]
- 18. Wang F, Zhang Q, Zhang F, Zhang E, Li M, Ma S, Guo J, Yang Z, Zhu J. 2023. Adenovirus vector-mediated single chain variable fragments target the nucleocapsid protein of porcine epidemic diarrhea virus and protect against viral infection in piglets. Front Immunol 14:1058327. doi: 10.3389/fimmu.2023.1058327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernardazzi C, Sheikh IA, Xu H, Ghishan FK. 2022. The physiological function and potential role of the ubiquitous Na+/H+exchanger Isoform 8 (NHE8): an overview data. Int J Mol Sci 23:10857. doi: 10.3390/ijms231810857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang J, Sarker R, Singh V, Sarker P, Yin J, Chen T-E, Chaerkady R, Li X, Tse CM, Donowitz M. 2015. The NHERF2 sequence adjacent and upstream of the ERM-binding domain affects NHERF2-Ezrin binding and dexamethasone stimulated NHE3 activity. Biochem J 470:77–90. doi: 10.1042/BJ20150238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Centonze M, Saponaro C, Mangia A. 2018. NHERF1 between promises and hopes: overview on cancer and prospective openings. Transl Oncol 11:374–390. doi: 10.1016/j.tranon.2018.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demacopulo B, Lema BE, Cabrini RL, Kreimann EL. 2016. Similar expression pattern of NHERF1 and EZRIN in papillary but not in solid areas of human serous ovarian carcinomas. Acta Histochem 118:797–805. doi: 10.1016/j.acthis.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 23. Chen Y, Wu S, Qi L, Dai W, Tian Y, Kong J. 2020. Altered absorptive function in the gall bladder during cholesterol gallstone formation is associated with abnormal NHE3 complex formation. J Physiol Biochem 76:427–435. doi: 10.1007/s13105-020-00751-3 [DOI] [PubMed] [Google Scholar]
- 24. He P, Zhao L, Zhu L, Weinman EJ, De Giorgio R, Koval M, Srinivasan S, Yun CC. 2015. Restoration of Na+/H+ exchanger NHE3-containing macrocomplexes ameliorates diabetes-associated fluid loss. J Clin Invest 125:3519–3531. doi: 10.1172/JCI79552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J, Singh V, Cha B, Chen T-E, Sarker R, Murtazina R, Jin S, Zachos NC, Patterson GH, Tse CM, Kovbasnjuk O, Li X, Donowitz M. 2013. NHERF2 protein mobility rate is determined by a unique C-terminal domain that is also necessary for its regulation of NHE3 protein in OK cells. J Biol Chem 288:16960–16974. doi: 10.1074/jbc.M113.470799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang X, Liu Y, Zhang X-Y, Liu X, Liu X, Wu X, Jose PA, Duan S, Xu F-J, Yang Z. 2022. Intestinal gastrin/CCKBR (cholecystokinin B receptor) ameliorates salt-sensitive hypertension by inhibiting intestinal Na+/H+ exchanger 3 activity through a PKC (protein kinase C)-mediated NHERF1 and NHERF2 pathway. Hypertension 79:1668–1679. doi: 10.1161/HYPERTENSIONAHA.121.18791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khundmiri SJ, Ecelbarger CM, Amponsem J, Ji H, Sandberg K, Lee DL. 2022. PPAR-α knockout leads to elevated blood pressure response to angiotensin II infusion associated with an increase in renal α-1 Na+/K+ ATPase protein expression and activity. Life Sci 296:120444. doi: 10.1016/j.lfs.2022.120444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dong S, Kong N, Wang C, Li Y, Sun D, Qin W, Zhai H, Zhai X, Yang X, Ye C, Ye M, Liu C, Yu L, Zheng H, Tong W, Yu H, Zhang W, Tong G, Shan T. 2022. FUBP3 degrades the porcine epidemic diarrhea virus nucleocapsid protein and induces the production of type I interferon. J Virol 96:e0061822. doi: 10.1128/jvi.00618-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morse M, Sefcikova J, Rouzina I, Beuning PJ, Williams MC. 2023. Structural domains of SARS-CoV-2 nucleocapsid protein coordinate to compact long nucleic acid substrates. Nucleic Acids Res 51:290–303. doi: 10.1093/nar/gkac1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sungsuwan S, Kadkanklai S, Mhuantong W, Jongkaewwattana A, Jaru-Ampornpan P. 2022. Zinc-finger antiviral protein-mediated inhibition of porcine epidemic diarrhea virus growth is antagonized by the coronaviral nucleocapsid protein. Front Microbiol 13:975632. doi: 10.3389/fmicb.2022.975632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Su M, Shi D, Xing X, Qi S, Yang D, Zhang J, Han Y, Zhu Q, Sun H, Wang X, Wu H, Wang M, Wei S, Li C, Guo D, Feng L, Sun D. 2021. Coronavirus porcine epidemic diarrhea virus nucleocapsid protein interacts with p53 to induce cell cycle arrest in S-phase and promotes viral replication. J Virol 95:e0018721. doi: 10.1128/JVI.00187-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu J, Mao J, Han X, Shi F, Gao Q, Wang T, Zhang Z, Shan Y, Fang W, Li X. 2021. Porcine epidemic diarrhea virus inhibits HDAC1 expression to facilitate its replication via binding of its nucleocapsid protein to host transcription factor Sp1. J Virol 95:e0085321. doi: 10.1128/JVI.00853-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]