Abstract
Background:
Metabolic differences have been reported between individuals with and without Major Depressive Disorder (MDD), but their consistency and causal relevance has been unclear.
Methods:
We conducted a metabolome-wide association study of MDD with 249 metabolomic measures available in UK Biobank (N = 29, 757). We then applied 2-sample bidirectional Mendelian Randomisation (MR) and colocalization analysis to identify potentially causal relationships between each metabolite and MDD.
Results:
One hundred and ninety-one metabolites tested were significantly associated with MDD (PFDR < 0.05), which reduced to 129 after adjustment for likely confounders. Lower abundance of Omega-3 fatty acid measures and a higher Omega-6: Omega-3 ratio showed potentially causal effects on liability to MDD. There was no evidence of a causal effect of MDD on metabolite levels. Furthermore, genetic signals associated with Docosahexaenoic Acid colocalized with loci associated with MDD within the FADS gene cluster. Post-hoc MR of gene-transcript abundance within the FADS cluster demonstrated a causal association with MDD. In contrast, colocalization analysis did not suggest a single causal variant for both transcript abundance and MDD liability, but the likely existence of two variants in LD with one another.
Conclusion:
Our findings suggest that decreased Docosahexaenoic Acid and increased Omega-6: Omega-3 fatty acids ratio may be causally related to MDD. These findings provide further support for the causal involvement of fatty acids in MDD.
Keywords: Metabolome, Depression, Mendelian Randomisation, Colocalization, Multi-Omics, Fatty Acids
Introduction
Major Depressive Disorder (MDD) is a debilitating condition estimated to affect 322 million people, leading to a global total of 50 million years lived with disability (1). This is due to both the high prevalence of MDD and its frequent comorbidity with other conditions (2-4), particularly cardiovascular disease (5). Most antidepressants target monoamine related pathways (6) but are ineffective in 40% of cases (7). Better understanding of the molecular mechanisms of MDD may aid in the development of more effective treatments.
Twin studies have estimated the heritability of MDD as ~37% (8) and investigating this genetic component of MDD may help to identify its pathophysiological basis. Recently, genome wide association studies (GWAS) have been increasingly successful in identifying genetic variants associated with MDD (9-11). Despite this success, the high polygenicity of MDD presents significant challenges to researchers who wish to identify actionable mechanisms lying between genetic variants and clinical phenotype (12). One means of translating genetic findings into biomarkers and biological mechanisms is through the incorporation of molecular phenotype data, such as metabolomics. These data may help to clarify the downstream functional impact of identified risk single nucleotide variant (SNVs) on molecular traits. This information can then be utilized to stratify risk, provide therapeutic targets or enable lifestyle interventions (13).
There have been several recent studies investigating the links between genetic risk to MDD and molecular phenotypes, including DNA methylation (14,15), mRNA levels (16,17) and protein markers (18,19). Metabolomics considers the role of circulating metabolites (20), including lipids, amino acids and other small molecules that have been implicated in a range of disorders and some hypothesis-driven studies of MDD (21,22). The metabolome has not been characterized to the same extent as other molecular phenotypes (23), but the recent availability of reproducible high-throughput metabolomics makes studies feasible for many thousands of individuals (24). Studying the association between metabolite levels and MDD may offer potential biomarkers and interventional targets for MDD, and provides a means to investigate MDD’s comorbidity with cardiometabolic disorders (2,3). The majority of previous metabolic studies of MDD have concentrated on small numbers of metabolites (25), often used sample sizes of less than 100 (25-29), and have implemented a variety of methodological approaches all leading to somewhat heterogeneous findings (30). However, larger and more standardized studies are beginning to emerge; a recent meta-analysis of 230 metabolite levels in 15, 428 individuals (5, 283 Cases and 10, 145 Controls) found evidence of a distinct metabolomic profile associated with MDD (31).
The UK Biobank recently released a large metabolomic dataset (31), in which we conducted a comprehensive and agnostic examination of metabolite associations with MDD. Association analysis between metabolites and MDD is limited because of the susceptibility of these analyses to confounding and reverse causation. We therefore also assessed potentially causal associations between MDD, and metabolite levels using two-sample Mendelian Randomisation (MR). Finally, we sought to identify potentially causal gene targets using MR and colocalization analysis of expression quantitative trait loci (eQTL) (Figure S1).
Methods and Materials
Study Population
The UK Biobank (UKB) is a large prospective study (N = 502, 492) of participants recruited from around the United Kingdom between the ages of 40 and 69 (33). Baseline data collection, including extensive phenotyping and genotyping for a range of health outcomes, occurred between 2006 and 2010. UKB data acquisition was approved by the North West NHS Research Ethics Committee (11/NW/0382). The analysis and data acquisition for the present study were conducted under application #4844. Written informed consent was obtained for all participants.
Metabolic biomarkers
Two hundred and forty-nine metabolite biomarkers were quantified in non-fasting baseline EDTA plasma samples (N = 118, 021) (33), collected at baseline assessment between April 2007 and December 2010 (34), and measured between June 2019 and April 2020 (31). Measurement was performed using the high-throughput nuclear magnetic resonance (NMR) spectroscopy profiling platform, developed by Nightingale Health Ltd, Finland. Detailed methodology of the Nightingale NMR pipeline has been described elsewhere (35-37). These biomarkers include detailed cholesterol measures, fatty acid compositions and low weight metabolites such as amino acids and ketones (Table S1). Most biomarkers were measured in absolute concentration (mmol/L), excluding apolipoproteins (A&B; g/L), diameter measures (Diameter of VLDL/LDL/HDL particles; nm) and the degree of unsaturation measure (degree; the total number of pi bonds and rings within a molecule). Prior to analysis, the levels of each metabolite biomarker were transformed to a standard normal distribution using rank-based inverse normalisation. Therefore, all reported beta coefficients from regression analyses represent the standardized change in ranked metabolite levels between MDD cases and controls.
Assessment of Major Depressive Disorder status
The MDD phenotype used in this study was derived from the online Thoughts and Feelings Questionnaire, fully completed by 126,077 participants in July 2017 (38). The questionnaire included the Composite International Diagnostic Interview Short Form (CIDI-SF), from which MDD lifetime diagnoses were derived (38) (SI: Methods & Materials). This sample (referred to as the mental-health questionnaire (MHQ) sample) comprised 37, 430 cases and 88, 647 controls of which 29,757 (8, 840 Cases and 20, 917 controls) also had metabolite levels derived from their blood samples (Table 1).
Table 1: Demographics of MDD Cases and Controls.
The demographics of MDD cases and controls are displayed (rows: MDD cases and MDD controls). Age, BMI, and SES are displayed as the mean values within the cohorts. % Ethnicity shows the ratio of White, Mixed and Other populations (Supplementary Information: Materials and Methods: Covariates).
| MDD controls | MDD cases | |
|---|---|---|
| N | 20917 | 8840 |
| Age | 56.67 (SD = 7.67) | 54.12 (SD = 7.55) |
| BMI | 26.48 (SD = 4.18) | 27.26 (SD = 5.12) |
| % Female | 49.95 | 68.82 |
| SES | −1.92 | −1.37 |
| % Never Smoked | 59.72 | 52.58 |
| % Ethnicity | 97:0.45:2.2 | 97:0.7:2 |
| % Attended University/College | 46.36 | 45.67 |
Metabolome-wide association study
Linear regression models were used to test the association of each metabolite level with the MDD phenotype (N = 29, 757), referred to as a metabolome-wide association study (MetWAS). In the basic MetWAS, only age, sex, assessment centre and NMR spectrometer were included as covariates. MetWAS analysis was also conducted with adjustment for socioeconomic status (SES), smoking status, ethnicity, educational attainment and body mass index (BMI) (SI: Methods and Materials) due to the strong and potentially confounding association between these factors with both MDD and metabolite levels (39-42). Significance was defined by FDR-corrected P values < 0.05.
Metabolite GWAS
For each metabolite a genome-wide association study (GWAS) was performed on unrelated individuals of European ancestry (N = 88, 329) using PLINK 2.0 (43). Unrelated individuals were identified as previously reported (44). In brief, related individuals were initially excluded based on a shared relatedness of up to the third-degree using kinship coefficients (> 0.044) calculated using the KING toolset. One member of each group of related individuals was then subsequently added back in, by creating a genomic relationship matrix (GRM) and selecting individuals with a genetic relatedness of less than 0.025 with any other participant. Association testing was performed on the imputed genotypes using an additive model, adjusted for age, sex, genotyping array, assessment centre, spectrometer and 10 genetic principal components (PC1-10). Variants with minor allele frequency (MAF) < 0.001, minor allele count (MAC) < 20 and an INFO score < 0.1 were removed. Insertion-deletions (indels) were included in the analysis. To account for multiple comparisons, significance was defined as the genome-wide threshold divided by the number of PCs which account for 99% of the variance within the metabolomic data (5e−8/42 = 1.19e−9), as described previously (45,46). An F statistic for the strength of the association between the sentinel SNVs and metabolite levels was calculated using the method: F = ((N-k-1) / k) × (r 2 / (1-r 2)), where N = sample size, k = number of SNVs and r 2 = variance explained in metabolite levels by the genetic instruments. The r 2 statistic was calculated as follows: r 2 = 2 × MAF × (1-MAF) × beta 2, where beta = effect size of the SNV and MAF = the SNV effect allele frequency. SNV heritability for each metabolite and the genetic correlation of the metabolite with MDD was performed using linkage disequilibrium (LD) score regression as implemented in the LDSC package (available at https://github.com/bulik/ldsc) (47,48). Pre-computed LD scores based on the 1000 Genomes project data for European ancestry (49) were used as the reference panel for regression weights in the analysis (available at https://data.broadinstitute.org/alkesgroup/LDSCORE).
Metabolite bidirectional Mendelian Randomisation analysis
Bidirectional MR was performed using the ‘TwoSampleMR’ R package, version 0.5.6 (50,51) to assess causality between the metabolite (exposure) and MDD (outcome) for all metabolites, and vice versa. The MR analysis used SNV effect sizes from the UKB metabolite GWAS conducted in this study and the MDD meta-analysis GWAS, performed by the Psychiatric Genetics Consortium (9) with UKB participants removed using three MR methods (Inverse variance weighted, MR Egger, and Weighted median). Genetic instruments were identified by clumping genome-wide significant SNVs (P < 1.19x10−9 and MAF > 0.005) using the ‘clump_data’ function in the R package TwoSampleMR (r2 = 0.001, within a 10Mb window). Metabolites with < 15 genetic instruments after harmonisation with MDD summary statistics were omitted from the MR analysis.
Colocalization
Colocalization analysis was performed between MDD and metabolite genetic signals that showed potentially causal associations using MR, with a window of 1Mb. Lead SNVs were identified using the default clumping method (r2 = 0.1) on the ‘SNP2GENE’ tool within FUMA (52). To avoid running multiple colocalization tests of overlapping regions, lead SNVs were ordered by significance, and iteratively filtered so that each lead SNV was > 1Mb away from any other lead SNV. For each of the final lead SNVs, all SNVs within a 1Mb window from the metabolite GWAS and the MDD GWAS were extracted from the respective summary statistics. Colocalization analysis was then performed using the R coloc package (v5.1.1) (53). The prior probabilities were set to default values: P1 (SNV is associated with the metabolite) and P2 (SNV is associated with MDD) were both set at 1e−4, and P12 (SNV is associated with both MDD and the metabolite) was set at 1e−5. An arbitrary posterior probability (PP) > 0.8 was set for evidence of each of the four hypotheses: PP.H0 - no causal variant; PP.H1 and PP.H2 - causal variant for one of the traits; PP.H3 – distinct causal variant for each trait, PP.H4 - shared causal variant for both traits.
MR and colocalization with expression Quantitative Trait Loci Data
To probe possible mechanistic pathways between exposure (metabolite) and outcome (MDD), colocalized MDD and metabolite genetic signals were investigated to identify if they localised to any causal genes within the colocalized region. For genes lying within the same LD window as co-localized MDD-metabolite genetic signals, we first identified any cis- expression quantitative trait loci (cis-eQTLs) from whole blood gene expression data (N = 948) from the Genotype Tissue Expression (GTEx) Project (47). Secondly, MR and complementary colocalization analysis was performed to assess the causality of gene expression levels (mRNA) with MDD. The MR analysis was performed using a Wald Ratio Test (55) using a single instrumental SNV, defined as the most significant eQTL present in the dataset. For this post-hoc analysis, significance was defined as P < 0.05. The instrumental SNVs were also used as lead SNVs in the colocalization analysis. For each lead SNV, MDD GWAS (9) and GTEx (54) summary statistics for variants within ±1Mb were extracted as inputs for the colocalization analysis.
Results
MetWAS MDD association analysis
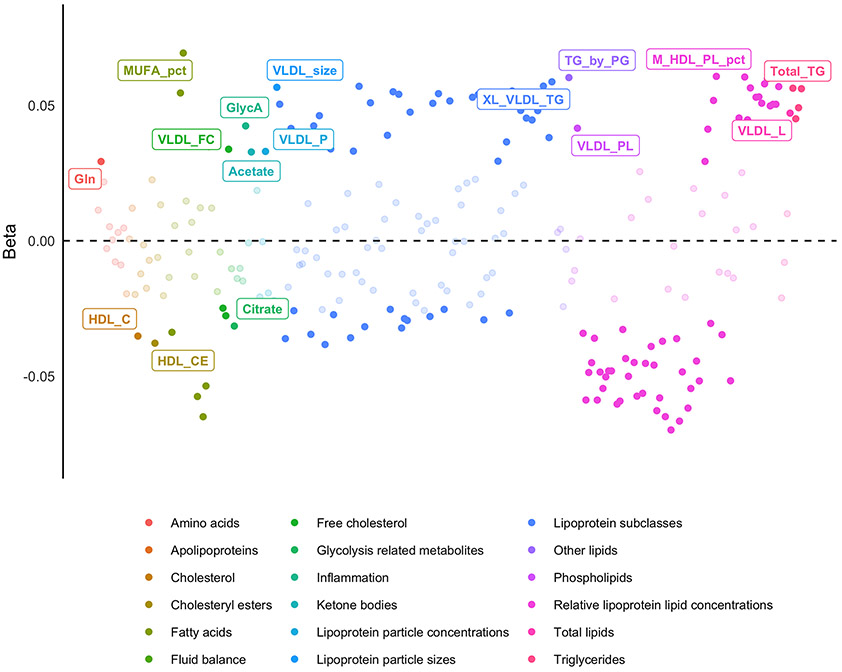
The demographics of the MHQ sample (metabolomic and MDD data available; n = 29, 757), alongside other subsets of UKB are presented in Table 1. The basic MetWAS, without adjustment for common confounders, found that 191 (~76%) metabolites were significantly associated with MDD (PFDR < 0.05) (Table S2: Figure S2). The most significant positive metabolite-MDD association was for Monosaturated Fatty Acids to Total Fatty Acids (%) (MFA-TFA), (β = 0.177, PFDR 1.52x10−43), and the most significant negative association was for Polyunsaturated Fatty Acids to Monounsaturated Fatty Acids ratio (PFA-MFAR: β= −0.176, PFDR = 1.85 x10−43). After adjustment for possible confounders, 133 (~ 53 %) metabolites were significantly associated with MDD, 129 of which (~52 %) were shared in the unadjusted model (Figure 1) (Table S3). The most significant positive metabolite-MDD association remained MFA-TFA (β = 0.069, PFDR = 2.22x10−7), whilst the most significant negative metabolite-MDD association changed to Free Cholesterol to Total Lipids in Large LDL percentage (FC-TL-LDL, %) (B = −0.070, PFDR = 2.82x10−7). Across most metabolites (N = 224, Unadjusted β range: −0.176 to 0.177), inclusion of confounders into the analysis attenuated the absolute effect sizes by an average of 61% (Adjusted β range: −0.070 to 0.069) (Figure S3).
Figure 1: Adjusted MetWAS.
Standardized effect sizes for Metabolite ~ MDD + covariates. Significant associations (PFDR < 0.05) are shaded in, and the most significant association for each metabolite group are labelled.
Metabolite GWAS
Manhattan plots for all 249 GWAS can be found in the Supplementary Folder 1. All metabolites had multiple independent (r2 = 0.001, within a 10Mb window) genome wide significant SNV associations (P < 1.19 x 10−9), all with F-statistics > 10 (Table S4). The number of significant independent SNVs ranged from 4 (Phenylalanine and Acetoacetate) to 60 (Cholesteryl Esters in Large HDL) (Table S5). SNV-heritability of metabolites varied from 2.41% (Histidine) to 19.9% (Triglycerides to Phosphoglycerides ratio). One hundred and forty-four metabolites (57%) showed a significant genetic correlation with MDD (Table S6), with absolute correlations ranging from (−) 0.073 (Omega-3 Fatty Acids to Total Fatty Acids (%)) to (+) 0.180 (Phospholipids to Total Lipids in Large HDL percentage). Of the metabolites which showed a significant genetic correlation with MDD, 119 (~ 83 %) also had significant associations with the MDD phenotype in both the unadjusted and adjusted MetWAS analysis.
Mendelian Randomisation
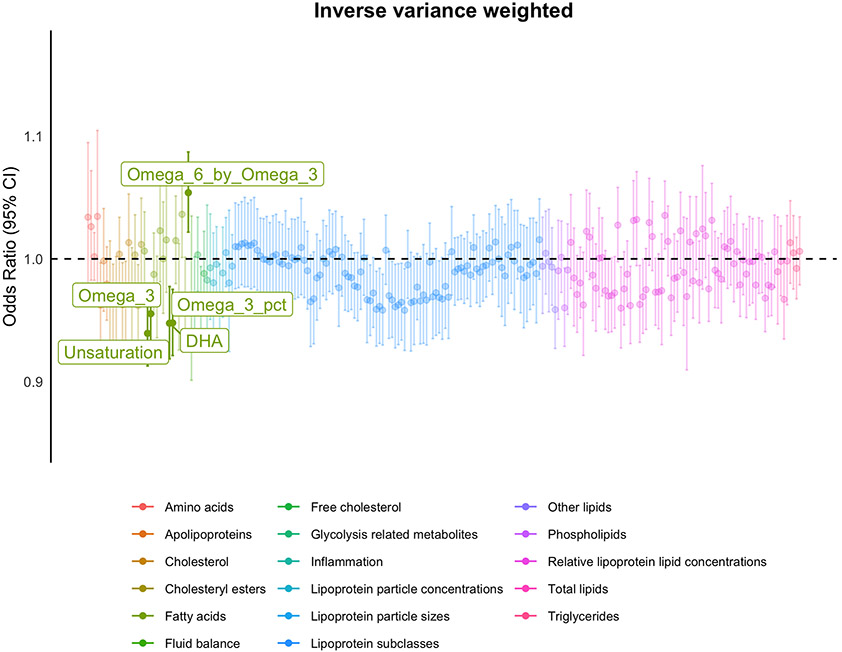
In the metabolite (exposure) to MDD (outcome) MR analysis, 21 metabolites were excluded due to having too few (< 15) genetic instruments (Figure S4). MR analysis on the remaining 228 metabolites (Figure 2; Figure S5) found five with significant evidence for a causal relationship with MDD (Table 2; Table S7), all relating to long-chain polyunsaturated fatty acids (LC-PUFAs). Specifically, Omega-3 fatty acid levels (Omega-3 Fatty Acids, Omega-3 to Total Fatty Acids (%), Docosahexaenoic Acid) and the Degree of Unsaturation had a negative causal effect on MDD, whilst Omega-6: Omega-3 ratio had a positive causal effect on lifetime MDD (Figure S6). These metabolites also had strong correlations with the other metabolites that significantly associated with MDD in the MetWAS phenotypic analysis, ranging from Omega-3 Fatty Acids (significantly correlated with 79% of associated metabolites) to the Degree of Unsaturation (significantly correlated with 100% of the other MDD-associated metabolites) (Figure S7). Of the five putatively causal metabolites, only the Degree of Unsaturation showed consistency in previous observational analyses, holding a significant association and genetic correlation with MDD (Figure S8). In the MR leave-one-SNV-out analysis, three of the significant metabolites-to-MDD MR results; Degree of Unsaturation, Omega-3 Fatty Acids and Docosahexaenoic Acid were dependent on rs174528, a single SNV in the FADS gene cluster on chromosome 11 (Table S8, Figure S9).
Figure 2: Metabolite to MDD Mendelian Randomisation (MR) analysis.
The odds ratios of metabolite on lifetime MDD status given by the inverse variance weighted MR method, coloured by metabolite group. Significant MR findings (PFDR < 0.05) are filled in and labelled. DHA; Docosahexaenoic Acid, Unsaturation; Degree of Unsaturation, Omega_3_pct; Omega-3 to Total Fatty Acids (%), Omega-3; Omega-3 Fatty Acids, Omega_6_by_Omega_3; Omega-6: Omega-3 Ratio.
Table 2: Significant Metabolite to MDD MR results.
All three MR methods for the five metabolites which showed a significant causal relationship with MDD in any MR method. Colocalization analysis was performed for each metabolite for multiple lead SNVs, the top colocalization result for each metabolite is reported in this table under PP.H4. The full table of colocalization results is found in Table S10. (Egger = MR Egger, WM = Weighted Median, IVW= Inverse Variance Weighted).
| Metabolite | Method | N (SNPS) |
Beta | SE | P | PFDR | PP.H4 |
|---|---|---|---|---|---|---|---|
|
Degree of
Unsaturation |
Egger | 25 | −0.063 | 0.021 | 6.20x10−3 | 0.89 | 0.026 |
| WM | 25 | −0.069 | 0.016 | 1.20x10−5 | 0.0015 | ||
| IVW | 25 | −0.063 | 0.015 | 1.80x10−5 | 0.0041 | ||
|
Omega-3
Fatty Acids |
Egger | 23 | −0.044 | 0.021 | 5.10x10−2 | 0.89 | 0.13 |
| WM | 23 | −0.068 | 0.016 | 1.30x10−5 | 0.0015 | ||
| IVW | 23 | −0.046 | 0.013 | 3.30x10−4 | 0.025 | ||
|
Docosahexaenoic
Acid |
Egger | 24 | −0.066 | 0.025 | 1.60x10−2 | 0.89 | 0.89 |
| WM | 24 | −0.078 | 0.019 | 4.50x10−5 | 0.0034 | ||
| IVW | 24 | −0.054 | 0.016 | 6.80x10−4 | 0.039 | ||
|
Omega-3
Fatty Acids To Total Fatty Acids (%) |
Egger | 23 | −0.057 | 0.021 | 1.30x10−2 | 0.89 | 0.16 |
| WM | 23 | −0.063 | 0.018 | 4.80x10−4 | 0.022 | ||
| IVW | 23 | −0.054 | 0.014 | 2.20x10−4 | 0.025 | ||
|
Omega-6
Fatty Acids to Omega-3 Fatty Acids ratio |
Egger | 18 | 0.062 | 0.026 | 3.10x10−2 | 0.89 | 0.15 |
| WM | 18 | 0.067 | 0.019 | 4.70x10−4 | 0.022 | ||
| IVW | 18 | 0.052 | 0.016 | 8.80x10−4 | 0.04 |
In the MDD (exposure) to metabolite (outcome) MR analysis, all metabolite measures were included. In this analysis, there was no significant evidence that MDD was significantly causal to a change in any of the metabolite levels tested (β range: −0.28 to 0.26, PFDR range: 0.91 to 0.99) (Table S9: Figure S10).
Colocalization
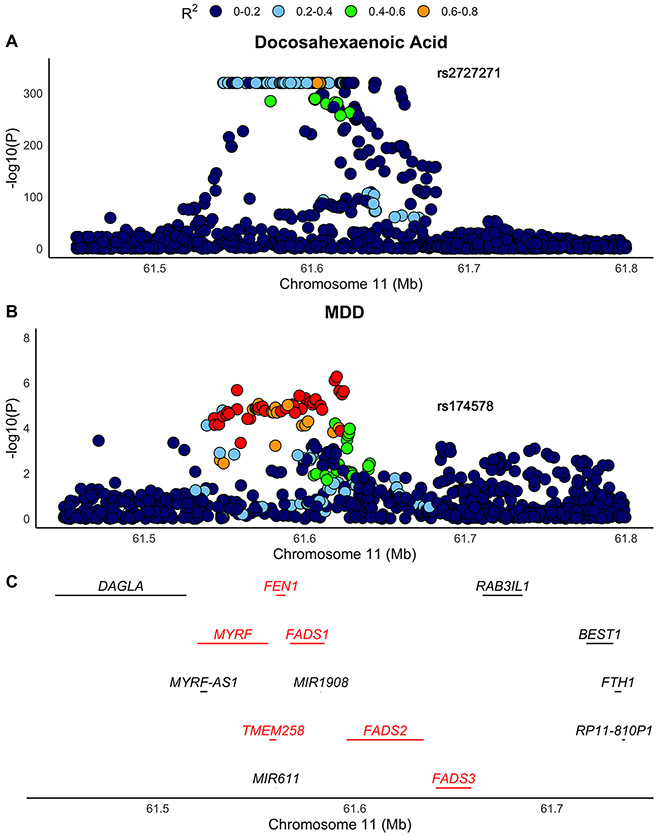
The number of lead SNVs identified across the genome for the 5 metabolites with MR PFDR < 0.05 ranged from 20 (Omega-3 Fatty Acids to Total Fatty Acids (%)) to 30 (Docosahexaenoic Acid) (Table S10). Docosahexaenoic Acid had strong evidence for colocalization (PP.H4 = 0.89) with MDD at SNV rs2727271 (Table 2), which is an intronic variant for the Fatty Acid Desaturase II (FADS2) gene (Figure 3). The rest of the metabolites showed weak evidence of colocalization with MDD (PP.H4 < 0.2) (Table S11; Figure S11).
Figure 3: Colocalization of Docosahexaenoic Acid with MDD.
A) The association of SNVs in the FADS cluster in the Docosahexaenoic Acid GWAS. B) The association of SNVs in the FADS cluster in the MDD GWAS (Without UKB). C) The genes present in the FADS cluster. Genes colored red are those which were present in the GTEx data and consequently tested for a causal relationship with MDD in the post-hoc analysis.
MR and colocalization with expression Quantitative Trait Loci Data
Transcriptomic data were used in downstream MR analysis with MDD for the genes Myelin Regulatory Factor (MYRF), Flap endonuclease 1 (FEN1), Transmembrane protein 258 (TMEM258) and Fatty Acid Desaturase 1-3 (FADS1/2/3) (Figure 3D), as these were both within DHA-MDD colocalized region on chromosome 11 and had eQTL data available from GTEx. mRNA-MDD MR analysis found the mRNA levels of MYRF, TMEM258, FADS1, FADS2 and FADS3 to have significant positive causal effect on lifetime MDD status (OR range: 1.01 to 1.12, P = 7.58x10−7 to 0.011) (Table 3: Figure S12). In contrast, mRNA levels of FEN1 were not significantly causal to MDD status (OR = 0.98, P = 0.62). Colocalization analysis for the FADS cluster eQTLs and MDD found minimal evidence of shared causal variant for both gene expression and MDD. Supporting evidence was strongest for the existence two independent causal variants for each trait at the same locus (PP.H3 > 0.7) (Table S12).
Table 3: Expression quantitative trait loci to MDD Wald Ratio MR Results.
These results indicate a positive causal effect of FADS1, FADS2, FADS3, MYRF and TMEM258 on lifetime MDD status. OR: Odds Ratio, CI: Confidence Intervals.
| Gene | SNV | OR | LOW CI (95%) |
HIGH CI (95%) |
MR_P |
|---|---|---|---|---|---|
| FADS1 | rs174567 | 1.08 | 1.05 | 1.10 | 7.58x10−7 |
| FADS2 | rs968567 | 1.01 | 1.01 | 1.02 | 6.33x10−5 |
| FADS3 | rs2524288 | 1.05 | 1.01 | 1.10 | 0.010995 |
| FEN1 | rs10897119 | 0.98 | 0.89 | 1.07 | 0.617075 |
| MYRF | rs198462 | 1.03 | 1.01 | 1.05 | 0.000607 |
| TMEM258 | rs7943728 | 1.12 | 1.06 | 1.19 | 5.78x10−5 |
Discussion
This study reports a large-scale (N = 29, 757) association and causal analysis of blood metabolites with MDD. We found that 191 metabolites were significantly associated with lifetime MDD status, 129 of which withstood further adjustment for common confounders (39). These findings provide greater confidence that the significant metabolite-MDD associations observed in this study are not due solely to the effects of confounding. Furthermore, we found that 144 metabolites showed a significant genetic correlation with MDD, suggesting a shared genetic architecture between the traits. Bidirectional MR analyses found evidence for potentially causal relationships between five metabolite measures relating to long-chain polyunsaturated fatty acids (LC-PUFAs) and MDD. Complementary colocalization analyses between these metabolites and MDD, found that an Omega-3 Fatty Acid, Docosahexaenoic Acid (DHA) colocalized with MDD within the FADS region. Subsequent MR analysis of eQTL data of genes in the FADS cluster found evidence of causality between MYRF, FADS1, FADS2, FADS3 and TMEM258 mRNA abundance and MDD. However, complementary colocalization analysis found evidence that this locus contains separate independent causal variants for mRNA abundance and MDD.
The MR analysis provides evidence that Omega-3 LC-PUFAs may have a negative causal effect on lifetime MDD, whilst the ratio of Omega-6: Omega-3 LC-PUFAs may have a positive causal effect on lifetime MDD. Specifically, colocalization analysis provided further support for a shared causal variant between MDD and Docosahexaenoic Acid. Research on Omega-3 and Omega-6 levels and MDD has been lengthy and complex (25,56-61). Although many cross-sectional and prospective studies have observed a reduction in Omega-3 fatty acids, and an increase in the Omega-6: Omega-3 ratio in those with MDD (30,57,62), it is important to note both the considerable methodological heterogeneity and contradictory evidence (30,60,63). Clinical trial studies testing the efficacy of Omega-3 supplements in the prevention and treatment of MDD also show inconsistent findings (64-67). Of note, our findings contradict results from a two-sample MR study in 2019, which found no evidence of a significant causal effect of Omega-3 levels on MDD (68). However, this study leveraged summary statistics from smaller GWAS, and therefore may have been underpowered to detect causal effects. The enzymatic oxygenation of Omega-3 and Omega-6 fatty acids and their derived eicosanoids have opposing anti-inflammatory and pro-inflammatory effects respectively (Figure S13). Importantly, humans do not show endogenous biosynthesis of active Omega-3 and Omega-6 fatty acids (EPA and ARA respectively) and the levels of each are reliant on the intake of their precursors LA and alpha-Linoleic acid (ALA) from the diet (69). Over the last century, the ratio of Omega-6: Omega-3 has increased dramatically in many western diets (20-30:1 compared to 1:1) (70), resulting in an imbalance in pro-inflammatory and pro-resolving mediators that may contribute to MDD pathology (71). Furthermore, there is a FADS haplotype associated with more efficient LC-PUFA synthesis from the diet (72). Whilst this is evolutionarily advantageous in environments with limited LC-PUFA availability, it may heighten the imbalance between Omega-6 and Omega-3 levels resulting from modern western diets (72). Overall, this indicates the possibility that disproportionate dietary levels of Omega-6: Omega-3 may be contributing to MDD through increased inflammatory processes, and that these may be exacerbated by certain genetic haplotypes.
The post-hoc MR analysis found evidence of a causal association between MYRF, FADS1, FADS2, FADS3 and TMEM258 mRNA levels and MDD. MYRF and TMEM258 each have plausible mechanisms for their association with MDD, due to links to abnormal myelination and intestinal stress respectively (73-75). The FADS enzymes, however, are directly linked to the metabolism of both Omega-3 and Omega-6 LC-PUFAs (76,77), and have been shown to be central regulators of the ratio of their respective anti-inflammatory and proinflammatory lipid mediators (78). Therefore, this suggests FADS enzymes act as key regulatory molecules in the activation and resolution of inflammatory processes, which may be important in the pathophysiology of MDD. A recent GWAS of Bipolar Disorder of Japanese populations also found a genetic association with FADS intron variants (79), indicating that this association may not be specific to MDD, but rather shared across different psychiatric disorders. Although this aligns with evidence that MDD and Bipolar Disorder share genetic architecture (80), whether the disorders share the putative causal effect of FADS found in this study would require additional analysis dissecting the causal relationship of FADS cluster on Bipolar Disorder.
Despite these putative causal mechanisms, complementary colocalization analysis between all eQTLs and MDD found evidence that each trait had distinct causal variants in the locus. This indicates that the instrumental variable in MR exhibits horizontal pleiotropy on the outcome (a direct violation of the exchangeability assumption for MR), thus weakening the evidence for causality (81,82). Therefore, it cannot be determined with high confidence that the transcript abundance of the genes have a direct causal relationship with MDD, but rather may be commonly seen alongside the presentation of MDD. Despite these putative causal mechanisms, complementary colocalization analysis between all eQTLs and MDD found evidence that each trait had distinct, rather than shared, causal variants at the locus. This indicates that the instrumental variable for FADS gene cluster gene expression abundance may be in LD with the MDD causal variant in the region, suggesting independent genetic effects on both traits at the same locus (81,82). We cannot confidently conclude that transcript abundance of FADS region genes has a direct causal relationship with MDD.
This study overcomes various limitations that existed in previous metabolomic analyses of MDD. Firstly, this study analysed the association and causal relationship between MDD and 249 metabolites in a hypothesis-free manner, and therefore reduces the possibility of confirmation bias, which has been criticised in previous studies with an a priori focus on LC-PUFAs (25). Secondly, previous studies often had small sample sizes, each with distinct study methodology and often suffering from self-report bias (30). This study utilises a large sample size from which metabolite levels were biologically measured in the blood using NMR spectroscopy in a standardized, well-documented procedure (35,37). Our findings are also strengthened by the integration of eQTL data from GTEx v8 (54), which interrogated the role of the gene-products from the colocalized FADS region.
This study is limited as it tests the association of MDD with plasma metabolite levels, despite MDD primarily being a brain-based disorder. However, blood samples are minimally invasive to collect, which is essential for enabling large-scale high-powered studies and affords them the most promise for future clinical applications. Another limitation to this analysis is the high LD within the FADS region (72), meaning that a direct violation of the exclusion-restriction assumption in MR may exist, generating false-positive results (83). However, the complementary use of MR and colocalization analysis in this study acts as a sensitivity analysis (81). Another limitation is the focus of this analysis on European ancestry, especially as the frequencies of the FADS haplotypes differs geographically. The divergent FADS haplotypes are more commonly seen in non-European populations (84), and are associated with an increased expression of FADS1, and a more efficient LC-PUFA synthesis (72). Consequently, the mechanistic effects hypothesised in this study may be greater in non-European ancestral groups which are not currently captured in this analysis. Additionally, although this is a large-scale metabolomic analysis, it is not wholly comprehensive for the human metabolome (85). Indeed, the recent human metabolome database (86) detailed 18,557 metabolites which had been quantified and estimated the presence of thousands more. Finally, as this study focused MDD prevalence, it may be biased by cases of chronic MDD within the cohort (87). Future work should therefore investigate whether Omega-3 Fatty Acid levels and the Omega 6: Omega 3 ratio associate with incident MDD.
In summary, this study interrogated the links between the metabolome and MDD and indicated a protective role of Docosahexaenoic Acid against MDD. Furthermore, this study also suggests a role of an increased ratio of Omega-6: Omega-3 fatty acids in the pathophysiology of MDD.
Supplementary Material
Acknowledgements
This work is supported by two Wellcome Trust grants to AMM (Investigator Award in Science: 220857/Z/20/Z and Strategic Award 104036/Z/14/Z). Data acquisition and analyses were conducted using the UK Biobank Resource under approved project #4844. Participation of the UK Biobank subjects is gratefully appreciated. We also thank UK Biobank team for collecting and preparing data for analyses. Funding from the Biotechnology and Biological Sciences Research Council (BBSRC) and Medical Research Council (MRC) is gratefully acknowledged. The PGC has received major funding from the US National Institute of Mental Health (5 U01MH109528-03). We thank the research participants and employees of 23andMe for supporting this study.
ED was supported by the United Kingdom Research and Innovation (grant EP/S02431X/1), UKRI Centre for Doctoral Training in Biomedical AI at the University of Edinburgh, School of Informatics. For the purpose of open access, the author has applied a creative commons attribution (CC BY) licence to any author accepted manuscript version arising.
DAG is supported by funding from the Wellcome Trust 4-year PhD in Translational Neuroscience–training the next generation of basic neuroscientists to embrace clinical research [108890/Z/15/Z]. AMM is also supported by a UK Research and Innovation award (MR/W014386/1) and by European Union Horizon 2020 funding (Grant agreement 847776.)
Footnotes
Disclosures
Required financial disclosures and conflict of interest statements for each author.
Ella Davyson reports no relevant disclosures.
Xueyi Shen reports no relevant disclosures.
Danni Gadd is a scientific consultant for Optima partners.
Elena Bernabeu reports no relevant disclosures.
Robert Hillary has received consultant fees from Illumina and is a scientific consultant for Optima Partners.
Daniel McCartney reports no relevant disclosures.
Mark Adams reports no relevant disclosures.
Riccardo Marioni has received speaker fees from Illumina, is an advisor to the Epigenetic Clock Development Foundation, and a scientific consultant for Optima Partners.
Andrew McIntosh reports no relevant disclosures.
Figure S1 and Figure S13 were created with licensed Biorender.com.
References
- 1.GBD 2017 Dis Injury Incidence Pr, James SLG, Abate D, Abate KH, Abay SM, Abbafati C, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. The Lancet. 2018. Nov 10;392(10159):1789–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oladeji BD, Gureje O. The Comorbidity between Depression and Diabetes. Curr Psychiatry Rep. 2013. Aug 11;15(9):390. [DOI] [PubMed] [Google Scholar]
- 3.Nemeroff CB, Goldschmidt-Clermont PJ. Heartache and heartbreak—the link between depression and cardiovascular disease. Nat Rev Cardiol. 2012. Sep;9(9):526–39. [DOI] [PubMed] [Google Scholar]
- 4.Cizza G, Ravn P, Chrousos GP, Gold PW. Depression: a major, unrecognized risk factor for osteoporosis? Trends Endocrinol Metab. 2001. Jul 1;12(5):198–203. [DOI] [PubMed] [Google Scholar]
- 5.Glassman AH. Depression and cardiovascular comorbidity. Dialogues Clin Neurosci. 2007. Mar 31;9(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robinson E. Psychopharmacology: From serendipitous discoveries to rationale design, but what next? Brain Neurosci Adv. 2018. Jan 1;2:2398212818812629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souery D, Serretti A, Calati R, Oswald P, Massat I, Konstantinidis A, et al. Switching antidepressant class does not improve response or remission in treatment-resistant depression. J Clin Psychopharmacol. 2011. Aug;31(4):512–6. [DOI] [PubMed] [Google Scholar]
- 8.Sullivan PF, Neale MC, Kendler KS. Genetic Epidemiology of Major Depression: Review and Meta-Analysis. Am J Psychiatry. 2000. Oct;157(10):1552–62. [DOI] [PubMed] [Google Scholar]
- 9.Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019. Mar;22(3):343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018. May;50(5):668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levey DF, Stein MB, Wendt FR, Pathak GA, Zhou H, Aslan M, et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci. 2021. Jul;24(7):954–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McIntosh AM, Sullivan PF, Lewis CM. Uncovering the Genetic Architecture of Major Depression. Neuron. 2019. Apr 3;102(1):91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyama M. Multi-omics study for interpretation of genome-wide association study. J Hum Genet. 2021. Jan;66(1):3–10. [DOI] [PubMed] [Google Scholar]
- 14.Barbu MC, Shen X, Walker RM, Howard DM, Evans KL, Whalley HC, et al. Epigenetic prediction of major depressive disorder. Mol Psychiatry. 2020. Jun 10;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, Caramaschi D, Adams MJ, Walker RM, Min JL, Kwong A, et al. DNA methylome-wide association study of genetic risk for depression implicates antigen processing and immune responses. Genome Med. 2022. Mar 31;14(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pain O, Glanville KP, Hagenaars S, Selzam S, Fürtjes A, Coleman JRI, et al. Imputed gene expression risk scores: a functionally informed component of polygenic risk. Hum Mol Genet. 2021. Apr 15;30(8):727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabbri C, Pain O, Hagenaars SP, Lewis CM, Serretti A. Transcriptome-wide association study of treatment-resistant depression and depression subtypes for drug repurposing. Neuropsychopharmacology. 2021. Sep;46(10):1821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Haeringen M, Milaneschi Y, Lamers F, Penninx BWJH, Jansen R. Dissection of depression heterogeneity using proteomic clusters. Psychol Med. 2022. Jan 18;1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wingo TS, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM, et al. Brain proteome-wide association study implicates novel proteins in depression pathogenesis. Nat Neurosci. 2021. Jun;24(6):810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idle JR, Gonzalez FJ. Metabolomics. Cell Metab. 2007. Nov 7;6(5):348–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillbrand M, Waite BM, Miller DS, Spitz RT, Lingswiler VM. Serum Cholesterol Concentrations and Mood States in Violent Psychiatric Patients: An Experience Sampling Study. J Behav Med. 2000. Dec 1;23(6):519–29. [DOI] [PubMed] [Google Scholar]
- 22.Holthoff VA, Beuthien-Baumann B, Zündorf G, Triemer A, Lüdecke S, Winiecki P, et al. Changes in brain metabolism associated with remission in unipolar major depression. Acta Psychiatr Scand. 2004;110(3):184–94. [DOI] [PubMed] [Google Scholar]
- 23.Humer E, Probst T, Pieh C. Metabolomics in Psychiatric Disorders: What We Learn from Animal Models. Metabolites. 2020. Feb;10(2):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ala-Korpela M, Kangas AJ, Soininen P. Quantitative high-throughput metabolomics: a new era in epidemiology and genetics. Genome Med. 2012. Apr 30;4(4):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mocking RJT, Assies J, Ruhé HG, Schene AH. Focus on fatty acids in the neurometabolic pathophysiology of psychiatric disorders. J Inherit Metab Dis. 2018. Jul 1;41(4):597–611. [DOI] [PubMed] [Google Scholar]
- 26.Kawamura N, Shinoda K, Sato H, Sasaki K, Suzuki M, Yamaki K, et al. Plasma metabolome analysis of patients with major depressive disorder. Psychiatry Clin Neurosci. 2018;72(5):349–61. [DOI] [PubMed] [Google Scholar]
- 27.Adams PB, Lawson S, Sanigorski A, Sinclair AJ. Arachidonic acid to eicosapentaenoic acid ratio in blood correlates positively with clinical symptoms of depression. Lipids. 1996;31(1Part2):S157–61. [DOI] [PubMed] [Google Scholar]
- 28.McNamara RK, Liu Y. Reduced expression of fatty acid biosynthesis genes in the prefrontal cortex of patients with major depressive disorder. J Affect Disord. 2011. Mar 1;129(1):359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaddurah-Daouk R, Yuan P, Boyle SH, Matson W, Wang Z, Zeng ZB, et al. Cerebrospinal Fluid Metabolome in Mood Disorders-Remission State has a Unique Metabolic Profile. Sci Rep. 2012. Sep 19;2(1):667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appleton KM, Voyias PD, Sallis HM, Dawson S, Ness AR, Churchill R, et al. Omega-3 fatty acids for depression in adults. Cochrane Database Syst Rev [Internet]. 2021. [cited 2022 Apr 29];(11). Available from: https://www.cochranelibrary.com/cdsr/doi/10.1002/14651858.CD004692.pub5/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bot M, Milaneschi Y, Al-Shehri T, Amin N, Garmaeva S, Onderwater GLJ, et al. Metabolomics Profile in Depression: A Pooled Analysis of 230 Metabolic Markers in 5283 Cases With Depression and 10,145 Controls. Biol Psychiatry. 2020. Mar 1;87(5):409–18. [DOI] [PubMed] [Google Scholar]
- 32.Julkunen H, Cichońska A, Slagboom PE, Würtz P, Nightingale Health UK Biobank Initiative. Metabolic biomarker profiling for identification of susceptibility to severe pneumonia and COVID-19 in the general population. eLife. 2021. May 4;10:e63033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLOS Med. 2015. Mar 31;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elliott P, Peakman TC, on behalf of UK Biobank. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol. 2008. Apr 1;37(2):234–44. [DOI] [PubMed] [Google Scholar]
- 35.Würtz P, Kangas AJ, Soininen P, Lawlor DA, Davey Smith G, Ala-Korpela M. Quantitative Serum Nuclear Magnetic Resonance Metabolomics in Large-Scale Epidemiology: A Primer on -Omic Technologies. Am J Epidemiol. 2017. Nov 1;186(9):1084–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzoulaki I, Ebbels TMD, Valdes A, Elliott P, Ioannidis JPA. Design and Analysis of Metabolomics Studies in Epidemiologic Research: A Primer on -Omic Technologies. Am J Epidemiol. 2014. Jul 15;180(2):129–39. [DOI] [PubMed] [Google Scholar]
- 37.Soininen P, Kangas AJ, Würtz P, Suna T, Ala-Korpela M. Quantitative Serum Nuclear Magnetic Resonance Metabolomics in Cardiovascular Epidemiology and Genetics. Circ Cardiovasc Genet. 2015. Feb;8(1):192–206. [DOI] [PubMed] [Google Scholar]
- 38.Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, et al. Mental health in UK Biobank – development, implementation and results from an online questionnaire completed by 157 366 participants: a reanalysis. BJPsych Open. 2020. Feb 6;6(2):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avila C, Holloway AC, Hahn MK, Morrison KM, Restivo M, Anglin R, et al. An Overview of Links Between Obesity and Mental Health. Curr Obes Rep. 2015. Sep 1;4(3):303–10. [DOI] [PubMed] [Google Scholar]
- 40.Han TS, Lean ME. A clinical perspective of obesity, metabolic syndrome and cardiovascular disease. JRSM Cardiovasc Dis. 2016. Feb 25;5:2048004016633371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hebebrand J, Peters T, Schijven D, Hebebrand M, Grasemann C, Winkler TW, et al. The role of genetic variation of human metabolism for BMI, mental traits and mental disorders. Mol Metab. 2018. Jun 1;12:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerstenberg G, Aoshima T, Fukasawa T, Yoshida K, Takahashi H, Higuchi H, et al. Effects of the CYP 2D6 Genotype and Cigarette Smoking on the Steady-State Plasma Concentrations of Fluvoxamine and Its Major Metabolite Fluvoxamino Acid in Japanese Depressed Patients. Ther Drug Monit. 2003. Aug;25(4):463–8. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007. Sep 1;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard DM, Adams MJ, Shirali M, Clarke TK, Marioni RE, Davies G, et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat Commun. 2018. Apr 16;9(1):1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallois A, Mefford J, Ko A, Vaysse A, Julienne H, Ala-Korpela M, et al. A comprehensive study of metabolite genetics reveals strong pleiotropy and heterogeneity across time and context. Nat Commun. 2019. Oct 21;10(1):4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin X, Chan LS, Bose D, Jackson AU, VandeHaar P, Locke AE, et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat Commun. 2022. Mar 28;13(1):1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015. Mar;47(3):291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ReproGen Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015. Nov;47(11):1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McVean GA, Altshuler (Co-Chair) DM, Durbin (Co-Chair) RM, Abecasis GR, Bentley DR, Chakravarti A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012. Nov;491(7422):56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemani G, Tilling K, Smith GD. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLOS Genet. 2017. Nov 17;13(11):e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Loos R, editor. eLife. 2018. May 30;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017. Dec;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace C, Rotival M, Cooper JD, Rice CM, Yang JHM, McNeill M, et al. Statistical colocalization of monocyte gene expression and genetic risk variants for type 1 diabetes. Hum Mol Genet. 2012. Jun 15;21(12):2815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.THE GTEX CONSORTIUM, Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015. May 8;348(6235):648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017. Oct 1;26(5):2333–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hibbeln JR. Fish consumption and major depression. The Lancet. 1998. Apr 18;351(9110):1213. [DOI] [PubMed] [Google Scholar]
- 57.Grosso G, Micek A, Marventano S, Castellano S, Mistretta A, Pajak A, et al. Dietary n-3 PUFA, fish consumption and depression: A systematic review and meta-analysis of observational studies. J Affect Disord. 2016. Nov 15;205:269–81. [DOI] [PubMed] [Google Scholar]
- 58.Liao Y, Xie B, Zhang H, He Q, Guo L, Subramanieapillai M, et al. Efficacy of omega-3 PUFAs in depression: A meta-analysis. Transl Psychiatry. 2019. Aug 5;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berger ME, Smesny S, Kim SW, Davey CG, Rice S, Sarnyai Z, et al. Omega-6 to omega-3 polyunsaturated fatty acid ratio and subsequent mood disorders in young people with at-risk mental states: a 7-year longitudinal study. Transl Psychiatry. 2017. Aug;7(8):e1220–e1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wani AL, Bhat SA, Ara A. Omega-3 fatty acids and the treatment of depression: a review of scientific evidence. Integr Med Res. 2015. Sep;4(3):132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grosso G, Galvano F, Marventano S, Malaguarnera M, Bucolo C, Drago F, et al. Omega-3 Fatty Acids and Depression: Scientific Evidence and Biological Mechanisms. Oxid Med Cell Longev. 2014. Mar 18;2014:e313570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li F, Liu X, Zhang D. Fish consumption and risk of depression: a meta-analysis. J Epidemiol Community Health. 2016. Mar;70(3):299–304. [DOI] [PubMed] [Google Scholar]
- 63.Ellis FR, Sanders TA. Long chain polyunsaturated fatty acids in endogenous depression. J Neurol Neurosurg Psychiatry. 1977. Feb 1;40(2):168–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thesing CS, Milaneschi Y, Bot M, Brouwer IA, Owens M, Hegerl U, et al. Supplementation-induced increase in circulating omega-3 serum levels is not associated with a reduction in depressive symptoms: Results from the MooDFOOD depression prevention trial. Depress Anxiety. 2020. Nov;37(11):1079–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bot M, Brouwer IA, Roca M, Kohls E, Penninx BWJH, Watkins E, et al. Effect of Multinutrient Supplementation and Food-Related Behavioral Activation Therapy on Prevention of Major Depressive Disorder Among Overweight or Obese Adults With Subsyndromal Depressive Symptoms: The MooDFOOD Randomized Clinical Trial. JAMA. 2019. Mar 5;321(9):858–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu MC, Tung CY, Chen HE. Omega-3 polyunsaturated fatty acid supplementation in prevention and treatment of maternal depression: Putative mechanism and recommendation. J Affect Disord. 2018. Oct 1;238:47–61. [DOI] [PubMed] [Google Scholar]
- 67.Okereke OI, Vyas CM, Mischoulon D, Chang G, Cook NR, Weinberg A, et al. Effect of Long-term Supplementation With Marine Omega-3 Fatty Acids vs Placebo on Risk of Depression or Clinically Relevant Depressive Symptoms and on Change in Mood Scores: A Randomized Clinical Trial. JAMA. 2021. Dec 21;326(23):2385–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milaneschi Y, Peyrot WJ, Nivard MG, Mbarek H, Boomsma DI, W. JH Penninx B. A role for vitamin D and omega-3 fatty acids in major depression? An exploration using genomics. Transl Psychiatry. 2019. Sep 5;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saini RK, Keum YS. Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance — A review. Life Sci. 2018. Jun 15;203:255–67. [DOI] [PubMed] [Google Scholar]
- 70.Simopoulos AP. Evolutionary aspects of omega-3 fatty acids in the food supply. Prostaglandins Leukot Essent Fatty Acids. 1999. May 1;60(5):421–9. [DOI] [PubMed] [Google Scholar]
- 71.Simopoulos AP. The Importance of the Omega-6/Omega-3 Fatty Acid Ratio in Cardiovascular Disease and Other Chronic Diseases. Exp Biol Med. 2008. Jun;233(6):674–88. [DOI] [PubMed] [Google Scholar]
- 72.Ameur A, Enroth S, Johansson Å, Zaboli G, Igl W, Johansson ACV, et al. Genetic Adaptation of Fatty-Acid Metabolism: A Human-Specific Haplotype Increasing the Biosynthesis of Long-Chain Omega-3 and Omega-6 Fatty Acids. Am J Hum Genet. 2012. May 4;90(5):809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, et al. Myelin Gene Regulatory Factor Is a Critical Transcriptional Regulator Required for CNS Myelination. Cell. 2009. Jul 10;138(1):172–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poggi G, Boretius S, Möbius W, Moschny N, Baudewig J, Ruhwedel T, et al. Cortical network dysfunction caused by a subtle defect of myelination. Glia. 2016;64(11):2025–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Graham DB, Lefkovith A, Deelen P, de Klein N, Varma M, Boroughs A, et al. TMEM258 Is a Component of the Oligosaccharyltransferase Complex Controlling ER Stress and Intestinal Inflammation. Cell Rep. 2016. Dec 13;17(11):2955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marquardt A, Stöhr H, White K, Weber BHF. cDNA Cloning, Genomic Structure, and Chromosomal Localization of Three Members of the Human Fatty Acid Desaturase Family. Genomics. 2000. Jun 1;66(2):175–83. [DOI] [PubMed] [Google Scholar]
- 77.Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism. 2010. Jul 1;59(7):993–9. [DOI] [PubMed] [Google Scholar]
- 78.Gromovsky AD, Schugar RC, Brown AL, Helsley RN, Burrows AC, Ferguson D, et al. Δ-5 Fatty Acid Desaturase FADS1 Impacts Metabolic Disease by Balancing Proinflammatory and Proresolving Lipid Mediators. Arterioscler Thromb Vasc Biol. 2018. Jan;38(1):218–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. 2018. Mar;23(3):639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coleman JRI, Gaspar HA, Bryois J, Breen G. The genetics of the mood disorder spectrum: genome-wide association analyses of over 185,000 cases and 439,000 controls. Biol Psychiatry. 2020. Jul 15;88(2):169–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zuber V, Grinberg NF, Gill D, Manipur I, Slob EAW, Patel A, et al. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am J Hum Genet. 2022. Apr;S0002929722001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018. Aug 1;27(R2):R195–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44(4):313–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mathias RA, Sergeant S, Ruczinski I, Torgerson DG, Hugenschmidt CE, Kubala M, et al. The impact of FADS genetic variants on ω6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet. 2011. May 20;12(1):50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pearson H. Meet the human metabolome. Nature. 2007. Mar 1;446(7131):8–8. [DOI] [PubMed] [Google Scholar]
- 86.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018. Jan 4;46(Database issue):D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bromet EJ. Cross-National Comparisons: Problems in Interpretation When Studies Are Based on Prevalent Cases. Schizophr Bull. 2008. Mar 1;34(2):256–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.