ABSTRACT
SARS-CoV-2 variants with undetermined properties have emerged intermittently throughout the COVID-19 pandemic. Some variants possess unique phenotypes and mutations which allow further characterization of viral evolution and Spike functions. Around 1,100 cases of the B.1.640.1 variant were reported in Africa and Europe between 2021 and 2022, before the expansion of Omicron. Here, we analyzed the biological properties of a B.1.640.1 isolate and its Spike. Compared to the ancestral Spike, B.1.640.1 carried 14 amino acid substitutions and deletions. B.1.640.1 escaped binding by some anti-N-terminal domain and anti-receptor-binding domain monoclonal antibodies, and neutralization by sera from convalescent and vaccinated individuals. In cell lines, infection generated large syncytia and a high cytopathic effect. In primary airway cells, B.1.640.1 replicated less than Omicron BA.1 and triggered more syncytia and cell death than other variants. The B.1.640.1 Spike was highly fusogenic when expressed alone. This was mediated by two poorly characterized and infrequent mutations located in the Spike S2 domain, T859N and D936H. Altogether, our results highlight the cytopathy of a hyper-fusogenic SARS-CoV-2 variant, supplanted upon the emergence of Omicron BA.1. (This study has been registered at ClinicalTrials.gov under registration no. NCT04750720.)
IMPORTANCE
Our results highlight the plasticity of SARS-CoV-2 Spike to generate highly fusogenic and cytopathic strains with the causative mutations being uncharacterized in previous variants. We describe mechanisms regulating the formation of syncytia and the subsequent consequences in a primary culture model, which are poorly understood.
KEYWORDS: SARS-CoV-2, syncytia, fusion, cytopathy, hNECs
INTRODUCTION
Over the timespan of the COVID-19 pandemic, SARS-CoV-2 has been subjected to selection pressures, leading to emerging variants carrying their own repertoire of mutations and to temporal waves of epidemiological resurgence (1, 2). The most successful variants evolved to evade the immune response (3–8) displaying differing abilities to form syncytia in cell culture systems (9, 10). During the period of co-circulation, the disease severity of Omicron (BA.1) was reduced in comparison to Delta (11–13). Proposed explanations for this include background immunity, different tissue tropisms, with BA.1 preferentially replicating in the upper respiratory tract, and reduced cell-cell fusogenicity of BA.1 Spike due to mutations (14–17). Therefore, the mutations and subsequent mechanisms surrounding SARS-CoV-2 pathogenicity and Omicron’s attenuation are still debated.
SARS-CoV-2 fuses with the cell plasma membrane to transfer its genome into the cytoplasm and instigate replication. This process is initiated through the binding of the Spike to its receptor angiotensin-converting enzyme 2 (ACE2) (18). Spike comprises two subunits, S1 and S2, separated by a polybasi cleavage site cleaved during viral production in the trans-Golgi network. Certain mutations in Spike, such as P681H/R, allow for this process to occur more readily, subsequently improving viral fusion (19, 20). During entry, Spike is cleaved at the S2′ site by host proteases, such as TMPRSS2, at the cell surface (21, 22) or cathepsins in endosomes (23, 24). This allows the conformational changes necessary to project the fusion peptide (FP) into the host membrane, leading to membrane fusion. Thus, a series of proteolytic events regulate SARS-CoV-2 entry and tropism prior to replication of the viral RNA.
The later stages of the SARS-CoV-2 replication cycle occur in the endoplasmic reticulum (ER) and Golgi network. Here, the host protein COPI binds to the Spike cytoplasmic tail and traffics it to the packaging site of SARS-CoV-2 virions (25). However, suboptimal Spike binding to COPI results in leakage to the plasma membrane. Consequently, Spike at the cell surface may interact with ACE2 on neighboring cells leading to cell-cell fusion and syncytia formation (21, 26). The relationship between ACE2 affinity of Spike and fusogenicity is debated and not necessarily direct. For example, Alpha Spike displays higher affinity for ACE2 than Delta yet lower fusogenicity (9). Furthermore, Omicron sublineage BA.2.86 exhibits very high ACE2 affinity yet lower fusion than the D614G (27–29). Therefore, Spike fusogenicity is influenced by other factors aside receptor affinity.
Histological studies of lung tissue from severe COVID-19 patients describe the presence of abnormal pneumocytes and large, multinucleated syncytia (30, 31). Syncytia are also observed in the lungs of long-term COVID patients who eventually succumb to the disease up to 300 days after testing negative, yet their role in pathogenesis is unknown (32). Syncytia formation by SARS-CoV-2 has also been demonstrated in various human cell lines and primary cell cultures, including primary airway epithelia and induced pluripotent stem cell-derived cardiomyocytes (9, 33). Spike mutations can impact fusogenicity. Notably, D215G and P681R/H increase fusion, with the latter promoting S1/S2 cleavage. Conversely, N856K and N969K in Omicron decrease fusogenicity (16, 24). Nevertheless, the role of syncytia in SARS-CoV-2 replication and their impact on disease severity has yet to be fully explored.
Minor SARS-CoV-2 variants harboring uncharacterized mutations represent opportunities for understanding certain viral processes. Variant B.1.640.1 was first identified in the Central African Republic in January 2021 and later circulated in France in October 2021 (34). As of November 2023, 1,107 sequences of B.1.640.1 are available on the GISAID database (35), 895 from France, with the most recent dating to February 2022. Here, we isolated a B.1.640.1 strain and investigated the humoral immune response, replication, fusogenicity, and cytopathy of this variant.
MATERIALS AND METHODS
Plasmids
Human codon-optimized SARS-CoV-2 Spikes (Alpha, Delta, BA.1, BA.5, B.1.640.1, and B.1.640.2) were produced in silico (GeneArt, Thermo Fisher Scientific). Spike sequences were then cloned into a phCMV backbone (GenBank: AJ318514) using Gateway cloning (Thermo Fisher Scientific). Individual mutations (D614G, T859N, and D936H) were produced through Q5 site-directed mutagenesis (NEB) in indicated backbones. Spike chimeras were generated through Gibson assembly of PCR generated fragments (NEB). Primers used for site-directed mutagenesis and Gibson assembly are described in Table 1. pQCXIP-Empty plasmid was used as previously described (36). All plasmids were sequenced by the Eurofins Genomics TubeSeq service with the primers listed in Table 1.
TABLE 1.
List of primers used throughout this study
| Experiment | Primer name | Primer sequence 5′−3′ |
|---|---|---|
| Site-directed mutagenesis of S2 mutations | N859T Fwd | CAACGGACTGACCGTGCTGCCTC |
| N859T Rev | AACTTCTGGGCGCAAATCAG | |
| T859N Fwd | TAACGGACTGAACGTCCTGCCAC | |
| T859N Rev | AACTTCTGGGCGCAAATCAGATC | |
| H936D Fwd | CAAGATTCAGGACAGCCTGAGCA | |
| H936D Rev | CCGATGGCGCTGTTGAAC | |
| D936H Fwd | CAAGATTCAGCACAGCCTGAGTAG | |
| D936H Rev | CCGATGGCGCTGTTGAAC | |
| Spike chimera generation | Spike NTDa Fwd | AAGGGCATCTACCAGACCAGC |
| Spike NTD Rev | TTGCTGGTCTGGTAGATGCCC | |
| Spike (−) NTD Fwd | AAGGGCATCTACCAGACCAGC | |
| Spike (−) NTD Rev | CACGTGGATGGCGTGGAAC | |
| Spike RBDa Fwd | AAGGGCATCTACCAGACCAGC | |
| Spike RBD Rev | GGAGCATGCAGCAGTTCGAA | |
| Spike (−) RBD Fwd | GGTGCTGAGCTTCGAACTG | |
| Spike (−) RBD Rev | TTGCTGGTCTGGTAGATGCCC | |
| Spike S1 Fwd | CTCCATAGAAGACACCGGGACC | |
| Spike S1 Rev | TAGGCAATGATGCTCTGGCT | |
| Spike S2 Fwd | GTGGCCAGCCAGAGCATCA | |
| Spike S2 Rev | GTTCTCAGGATCGGTCGACC | |
| TMPRSS2 siRNA RT-qPCR | b-Tubulin Fwd | CTTCGGCCAGATCTTCAGAC |
| b-Tubulin Rev | AGAGAGTGGGTCAGCTGGAA | |
| TMPRSS2 Fwd | GGGGATACAAGCTGGGGTTC | |
| TMPRSS2 Rev | GATTAGCCGTCTGCCCTCAT | |
| Sequencing primers | phCMV-Fwd | CTCTTTCCTACAGCTCCTGG |
| Pan-CoV-2-S-1 | CTACCCCGACAAGGTGTTCC | |
| Pan-CoV-2-S-2 | CTGATGGACCTGGAAGGCAAG | |
| Pan-CoV-2-S-3 | CAACTGCGTGGCCGACTAC | |
| Pan-CoV-2-S-4 | GCGTGAACTTCAACTTCAACGG | |
| Pan-CoV-2-S-5 | CCACCAACTTCACCATCAGCG | |
| Pan-CoV-2-S-6 | GTGCTGTACGAGAACCAGAAG |
NTD, N-terminal domain; RBD, receptor-binding domain.
Cells
HEK293T, Caco2/TC7, VeroE6, and A549 cells and their derivatives were purchased from the ATCC or generously provided by fellow staff at Institut Pasteur. Cells were cultured in DMEM with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. GFP-split cells, expressing either GFP subunits 1–10 or GFP subunit 11, were generated through transduction of HEK293T or VeroE6 cells with respective pQCXIP-derived plasmids. Transduced cells were cultured with 1 µg/mL puromycin (InvivoGen). A549-ACE2 cells were generated through transduction of human ACE2 and cultured with 10 µg/mL blasticidin (21). All cells used in this study were tested negative for mycoplasma.
Cohorts
To study antibody neutralization of SARS-CoV-2 variants following infection or vaccination, a cohort from the prospective, monocentric, longitudinal, interventional cohort clinical study (ABCOVID) from Orléans was used. This study was commenced in August 2020 with the objective of studying the kinetics of COVID-19 antibodies in patients with confirmed SARS-CoV-2 infection (ClinicalTrials.gov identifier NCT04750720). A substudy aimed to describe the kinetic of neutralizing antibodies after vaccination. The cohort is described in previous publications (3, 8, 37). To exclude individuals infected before vaccination, anti-N antibodies were measured upon sera collection. This study was approved by the Ile-de-France IV ethical committee. Written informed consent from the participants was collected, and a questionnaire covering sociodemographic characteristics was completed.
Origin of the B.1.640.1 strain
A nasopharyngeal swab, collected from a patient tested positive for SARS-CoV-2 on November 2021, was sent to Hôpital Henri Mondor sequencing platform in the context of a nationwide survey. Briefly, private and public diagnostic laboratories in France participated in the national SARS-CoV-2 genomic surveillance by providing a random subsampling of positive SARS-CoV-2 samples to national sequencing platforms weekly (38). After sequencing, the leftover sample was used for viral isolation. The virus was isolated and amplified by one or two passages on VeroE6 cells as previously described (37).
Virus sequencing
Viral RNA was extracted from the nasopharyngeal swab in viral transport medium. Sequencing was performed with the Illumina COVIDSeq Test (Illumina, San Diego, California), using 98 target multiplex amplifications along the full SARS-CoV-2 genome. The libraries were sequenced with NextSeq 500/550 High Output Kit v2.5 (75 Cycles) on a NextSeq 500 device (Illumina). The sequences were demultiplexed and assembled as full-length genomes using the DRAGEN COVIDSeq Test Pipeline on a local DRAGEN server (Illumina). The sample was identified as B.1.640.1 (hCoV-19/France/GES-HMN-21112100277/2021) before being submitted to the GISAID database (35), with the following ID: EPI_ISL_6470307. The sequence obtained by metagenomic sequencing (random hexamer cDNA generation and Nextera XT library preparation) after amplification was identical, and no contamination was detected.
Other SARS-CoV-2 isolates
The D614G isolate was obtained through the European Virus Archive goes Global (Evag) platform and received from the National Reference Center for Respiratory Viruses at Institut Pasteur. The Delta and BA.1 isolates were previously described (8). Strains were amplified and titrated on VeroE6 cells. Viral stock titers were calculated through TCID50 measurements. Viral stocks were diluted 1:10 in DMEM complete media and then serially diluted 10-fold for a total of eight dilutions (10−1–10−8 dilution). Titrated virus was added to a 96-well plate containing 1.2 × 104 VeroE6 cells per well. This was replicated six times per viral stock. Five days post-infection, cells were assessed for cytopathic effect using a light microscope. The lowest dilutions showing cytopathic effect were used to calculate the TCID50 of the viral stock. Manipulations of SARS-CoV-2 isolates were performed in a BSL-3 laboratory, under the guidelines of the risk prevention service of Institut Pasteur.
GFP-split fusion assay and video microscopy
To assess the fusion of the respective Spike constructs, HEK293T-GFP1-10 cells (6 × 104 cells per well) were transfected in suspension at 37°C using a shaking incubating at 750 rpm for 30 min. The transfection mix was prepared using Lipofectamine 2000 (Thermo Fisher Scientific) with 50 ng DNA in a 1:10 ratio of phCMV-SARS-CoV-2-Spike and pQCXIP-Empty (control), respectively, before being added to the cells. Following transfection, cells were washed and resuspended in DMEM 10% FBS. Transfected HEK293T cells were then co-cultured with either 1.5 × 104 VeroE6-GFP11 cells or 1.5 × 104 Caco2-GFP11 cells per well in a µClear black 96-well plate for 18 hours. Spike expression was assessed by staining with 1 µg/mL anti-S2 mAb (mAb10) targeting an unknown yet conserved epitope within the S2 subunit (8). For TMPRSS2 knockdown (KD) experiments, Caco-2-GFP11 cells were transfected with either control siRNA targeting luciferase (5′-CGUACGCGGAAUACUUCGA-3′) or siGENOME Humam TMPRSS2 SMARTpool (#7113—Horizon Discovery) at 50 nM using Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 48 hours. For protease inhibition assays, 10 µM camostat (CliniSciences, HY-13512), 10 µM marimastat (Abcam), or 10 µM E64d (Enzo Life Sciences) was added to Caco-2-GFP11 cells prior to co-culturing with HEK293T-GFP1-10 cells transfected with Spike. Hoechst 33342 was added to the media at a 1:10,000 dilution. At 18 hours post-transfection, images were acquired using the Opera Phenix High-Content Screening System (PerkinElmer). A total of 20 images were acquired per well. Analysis was performed using Harmony High-Content Imaging and Analysis Software (PerkinElmer, HH17000012, v.5.0), including nuclei count and GFP area. For video microscopy, as described above, the cells were incubated (37°C, 5% CO2) inside the Mica Microhub system using the internal incubator (Leica microsystems). Videos were analyzed using the complementary software (Leica microsystems).
A549-ACE2 viral infection, immunofluorescence, and video microscopy
At 6 hours prior to infection, 3 × 104 A549-ACE2 cells were plated in a µClear black 96-well plate. Cells were then infected with MOI 0.1, 0.01, and 0.001 of SARS-CoV-2 virus. Cells were fixed using 4% PFA at 24-, 48-, and 72-hour timepoints, and supernatant was collected for RT-qPCR and LDH analysis. Cells were then stained using 1 µg/mL of anti-Spike mAb [mAb102, (37)] and 0.05% saponin in PBS, 1% BSA, and 0.05% sodium azide for 30 min at room temperature. For the secondary antibody, cells were washed twice with PBS and stained as per the primary antibody with 1:600 goat anti-human IgG-FC alexa-647 antibody for 30 min, then washed twice with PBS. Hoechst 33342 (Invitrogen) was added to the final PBS wash. Images and analyses were then performed as described above using the Opera Phenix High-Content Screening System. For video microscopy, A549-ACE2 cells were seeded in a µ-Dish 35-mm Quad (Ibidi) 6 hours prior to infection. Media was then replaced with DMEM 10% FBS with SARS-CoV-2 virus at an MOI of 0.1. Propidium iodide (PI) and Hoechst dyes were also added to the media. The microscopy was carried out using the BioStation video microscope, with six fields acquired per chamber. Images were acquired every 10 min over 36 hours. Video analysis and editing and nuclei counting for syncytia quantification were performed using ImageJ software (Fiji).
LDH activity assay
Cell supernatants were collected and stored in LDH storage buffer + Triton 10% as per the manufacturer’s instructions (Promega). Supernatants were inactivated for 2 hours at room temperature prior to usage outside the BSL-3. A volume of 50 µL of diluted supernatant was then added to 50 µL LDH substrate mix as described by Promega. LDH activity was recorded using the VICTOR3 multilabel plate reader (Perkin-Elmer).
Human nasal epithelium culture, infection, and imaging
MucilAir, reconstructed human nasal epithelial cells (hNECs) that had been differentiated for 4 weeks prior to obtention, was cultured in 700 µL MucilAir media on the basal side of the air/liquid interface (ALI) cultures and monitored for healthy cilia movements. One hour prior to infection, mucus was removed from the apical side of the culture by washing the apical side with warm 200 µL MucilAir media. Cells were then infected with equal virus titers in 100 µL MucilAir media for 2 hours. Viral input was removed and stored at −80°C. Cells were then washed for 10 min at 37°C in warm PBS and then for 20 min in 200 µL MucilAir media for the day 0 recording. Washing with warm media was repeated every 24 hours for 96 hours. Every wash was subsequently centrifuged at 1,500 rpm to remove cell debris and frozen at −80°C. After 96 hours, cells were fixed on the apical and basal sides with 4% PFA for 15 min. For imaging, fixed cells were stained intracellularly with rabbit anti-ZO1 (40-2200; Invitrogen), anti-SARS-CoV-2 N AlexaFluor-488 Dylight, as described in reference (3), and rabbit anti-cleaved caspase-3 (D175; Cell Signaling Technology) and imaged using the LSM-700 confocal microscope (Zeiss).
Flow cytometry
For surface staining, both primary and secondary antibodies were diluted with MACS buffer. Further information on the monoclonal antibodies (mAbs) used in this study is listed in Table 2. Serum samples were diluted 1:300, and primary monoclonal antibodies were used at 1 µg/mL. Soluble-ACE2-human-IgG-FC was diluted to 10 µg/mL and serially diluted threefold six times. Cells were mixed with 50 µL of primary antibody and incubated in the dark at 4°C. Cells were then washed in 100µL PBS after staining. Goat anti-human FC AlexaFluor-647 was used as the secondary antibody at a dilution of 1:400. Cells were fixed in 4% PFA and acquired on an Attune NxT Flow Cytometer (Thermo Fisher). Data were analyzed using FlowJo software (BDBioSciences).
TABLE 2.
List of antibodies used throughout this study
| Antibody | Target | Concentration (μg/mL) | Source |
|---|---|---|---|
| 1 | NTDa | 1 | Humoral Immunology Unit—Institut Pasteur (39) |
| 10 | S2 | 1 | |
| 18 | NTD | 1 | |
| 20 | NTD | 1 | |
| 21 | NTD | 1 | |
| 32 | NTD | 1 | |
| 40 | NTD | 1 | |
| 45 | NTD | 1 | |
| 53 | NTD | 1 | |
| 69 | NTD | 1 | |
| 71 | NTD | 1 | |
| 83 | NTD | 1 | |
| 105 | NTD | 1 | |
| 114 | NTD | 1 | |
| 118 | NTD | 1 | |
| 127 | NTD | 1 | |
| 129 | RBDa | 1 | |
| Sotrovimab (VIR-7831) | RBD | 1 | CHR Orleans |
| Cilgavimab (AZD1061) | RBD | 1 | |
| Tixagevimab (AZD8895) | RBD | 1 | |
| Imdevimab (REGN10897) | RBD | 1 | |
| Bebtelovimab (LY-CoV1404) | RBD | 1 | (40) |
| Anti-IgG AF647 | Human-IgG Fc | 3.3 | Thermo Fisher Scienfitic |
NTD, N-terminal domain; RBD, receptor-binding domain.
S-fuse neutralization assay
U2OS-ACE2 GFP1-10 or GFP11, termed S-fuse cells, were mixed (1:1), and 8 × 103 cells per well were plated in a mClear black 96-well plate. Heat-inactivated serially diluted sera were incubated with the indicated SARS-CoV-2 viruses for 15 min at a starting dilution of 1:30 before being added to the cells. Therapeutic mAbs were serially diluted starting at a concentration of 10 µg/mL prior to 15 min incubation with virus. At 18 hours post-infection (hpi), cells were fixed in 4% PFA and stained with Hoechst 33342 (Invitrogen). Cell images and GFP and nuclei quantification were performed using the Opera Phenix system as described above. Percentage neutralization was calculated with the following formula: {100*(1-(x with “serum” - x with “non infected”) - (x with “no serum” - x with “non infected”))} with x = number of syncytia. Percentage neutralization was then used to calculate the ED50 of each serum.
RT-qPCR, RNA extraction, and reverse transcription
For quantification of viral RNA release, cell supernatants were collected and diluted 1:4 in H2O and then heat inactivated at 80°C for 20 min. An amount of 10 µM SARS-CoV-2 E-gene Forward (5′-ACAGGTACCTTAATAGTTAATAGCGT-3′) and Reverse (5′-ATATTGCAGCAGTACGCACACA-3′) primers was used with Luna Universal One-step RT-qPCR Kit (New England Biolabs) and added to 1 µL supernatant ( 5µL total) in a 384-well plate. A standard curve was produced through a 1:10 serial dilution of EURM-019 ssRNA SARS-CoV-2 fragments for reference (European Commission). To evaluate Caco-2 TMPRSS2 KD, 5 × 105 cells were lysed in RLT buffer (QIAGEN) with 10 µL of β-mercaptoethanol. RNA extraction and reverse transcription were performed using the RNeasy Plus Mini Kit (QIAGEN) and Superscript II (Thermo Fisher Scientific), respectively, according to manufacturer’s instructions. RT-qPCR was carried out using iTaq universal SYBR green supermix (BioRad) and the primers listed in Table 1. All RT-qPCR experiments were performed with a QuantStudio 6 Flex Real-Time PCR machine (Thermo Fisher).
Scanning electron microscopy
MucilAir samples were fixed with 4% PFA for 1 hour at RT to inactivate SARS-CoV-2 and stored in PBS at 4°C prior to preparation. Samples were then washed in 0.1 M cacodylate buffer and several times in water and processed by alternating incubations in 1% osmium tetroxide and 0.1 M thiocarbohydrazide (OTOTO). Samples were then dehydrated by incubating with increasing concentrations of ethanol. Samples were critical point dried and mounted on a stub for analysis. Analysis was performed by field emission scanning electron microscopy with a Jeol JSM6700F microscope operating at 3 kV.
PyMOL
The SARS-CoV-2 Spike structure (PDB: 6VXX) was imported in PyMOL. Mutations and residue proximities were calculated and annotated with the mutagenesis and measurement tools.
Western blotting
HEK293T cells were transfected with Spike for 24 hours. Cells were lysed by resuspending in TXNE buffer [1% triton X-100, 50 mM Tris-HCL (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1× Roche complete protease inhibitors] for 30 min on ice. Also, 15 µg of protein was loaded per condition. PageRuler Plus-prestained protein ladder (Thermo Fisher Scientific) was used as a reference. Membranes were blocked in PBS—5% BSA. Antibodies were diluted in PBS—1% BSA, 0.05% Tween, and 0.1% sodium azide. Primary antibodies consisted of mouse anti-S2 (GeneTex, GTX632604, 1:2000), mouse anti-actin (Cell Signaling, 8H10D10; 1:2000). Anti-mouse IgG conjugated to DyLight-680 (Thermo Fisher Scientific, SA5-35521) was used as the secondary antibody. Membranes were revealed using a Licor imager and analyzed using Image Studio Lite v5.2.5 software.
Statistical analysis
Calculations were performed using Excel 365 (Microsoft). Figures and statistical analyses were conducted using GraphPad Prism 9. Statistical significance between different groups was calculated using the tests indicated in each figure legend. No statistical methods were used to predetermine sample size.
RESULTS
B.1.640.1 epidemiology
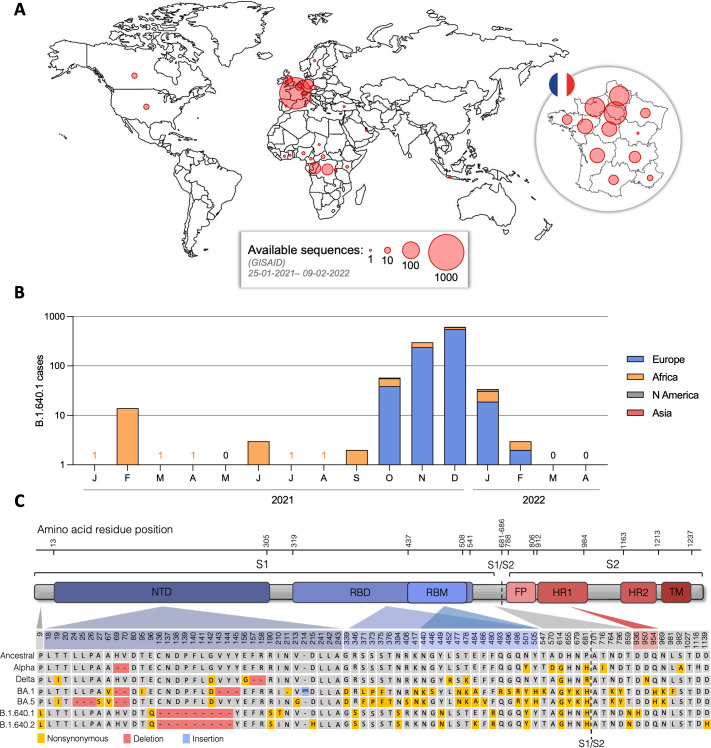
Analysis of the available B.1.640.1 sequences on the GISAID database suggests B.1.640.1 was first detected in the Central African Republic in January 2021 before spreading to neighboring central African countries (Fig. 1A and B). The first detected sequence in Europe was followed 9 months later, with the majority of B.1.640.1 cases seen in central and northern France. Cases of B.1.640.1 rapidly declined in January 2022, and the variant was eventually supplanted in March 2022, culminating in 1,107 total recorded sequences.
Fig 1.
Epidemiology and Spike mutations of SARS-CoV-2 B.1.640.1. (A) Total number of B.1.640.1 variant cases by country as displayed by red circles. Also shown is total case number of B.1.640.1 variant by region of France. Data on sample number and location were obtained from GISAID EpiCoV database. Total case number = 1,107. (B) Total case number of B.1.640.1 variant per month during its circulation between January 2021 and April 2022. Data are grouped by continent. Data on sample collection date and sample location were obtained from GISAID EpiCoV database. (C) Schematic view of B.1.640.1 and B.1.640.2 Spike sequences and their respective amino acid mutations compared to the ancestral Wuhan Spike sequence (NC_045512.2) and Alpha, Delta, BA1, and BA5 variants.
B.1.640.1 displays NTD antibody-binding escape and reduced neutralization sensitivity to Delta
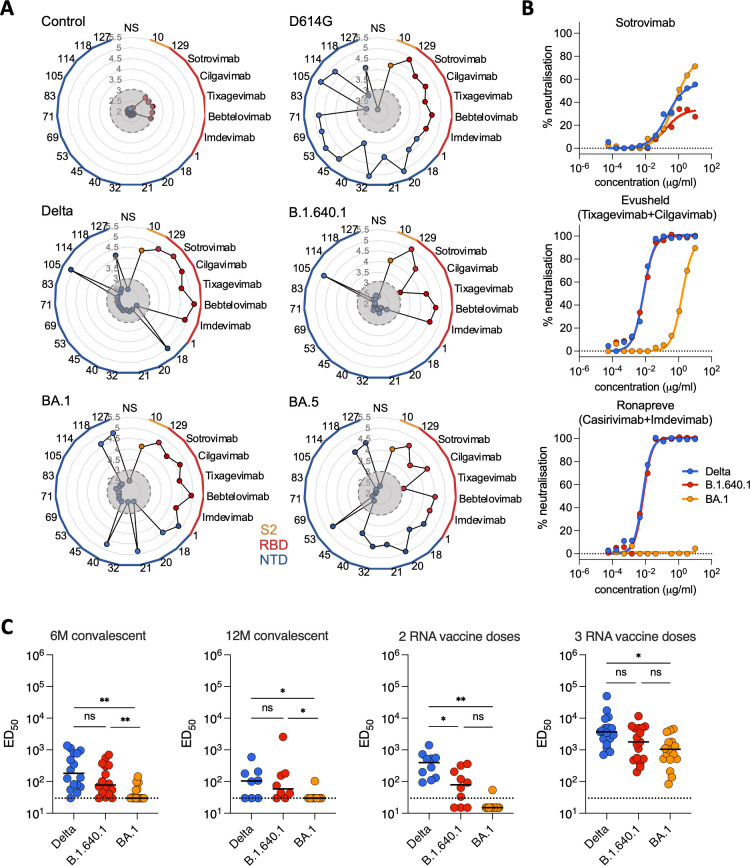
The N-terminal domain (NTD) and receptor-binding domain (RBD) of Spike are continuously subjected to immune selection pressures (2, 41). B.1.640.1 carries several unique mutations in these regions, including an unusually large NTD deletion of nine amino acids (Fig. 1C). To investigate their contribution to immune evasion, HEK293T cells expressing Spikes from D614G, Delta, B.1.640.1, BA.1, and BA.5 were incubated with a set of anti-NTD monoclonal antibodies or anti-RBD therapeutic mAbs (Table 2). The therapeutic mAbs incubated at 1 µg/mL bound to all variants with the exceptions of cilgavimab to B.1.640.1 and tixagevimab to BA.5 (Fig. 2A). Conversely, D614G and BA.5 showed a high level of binding to the anti-NTD mAbs, with 13/15 and 9/15 mAbs binding, respectively. B.1.640.1 displayed the greatest reduction in anti-NTD binding with only 1 mAb retaining activity (Fig. 2A).
Fig 2.
Binding and neutralization of B.1.640.1 by monoclonal antibodies and sera from convalescent and vaccinated individuals. (A) Radial plots showing binding of a panel of mAbs that target the S2, RBD, and NTD of Spike, as depicted by color. HEK293T cells were transfected with Spike or control plasmids 24 hours before staining with mAbs. Binding is quantified by mean fluorescence intensity (MFI) of each mAb, with the y-axis showing log10(MFI). Gray circles indicate the limit of detection [MFI = log10 (3)]. N = 3. (B) Neutralization activity of therapeutic mAbs. Dose-response analysis of the neutralization of Delta, B.1.640.1, and BA.1 virus by sotrovimab and the combinations of tixagevimab + cilgavimab and casirivimab + imdevimab. Data points are a mean of two independent repeated experiments. (C) Neutralization activity of sera from convalescent individuals, 6 and 12 months after infection, and 1 month post second and third Pfizer RNA vaccine doses. Dotted line represents the limit of detection (ED50 = 30). Solid black bars represent median values. Data points are a mean of two independent repeated experiments. Mann-Whitney tests were performed to compare the respective variants, *P < 0.05, **P < 0.001, ns = not significant.
Next, we used therapeutic mAbs and a longitudinal cohort of sera from convalescent and vaccinated individuals established in Orléans to test the sensitivity of B.1.640.1 to neutralizing antibodies. We isolated a B.1.640.1 strain from a swab of an infected individual. Comparisons were drawn against BA.1 and to Delta which circulated at the same time as B.1.640.1. To measure sera neutralization, we calculated the 50% effective dilution (ED50) of each serum sample to each variant. B.1.640.1 virus showed similar neutralization to the therapeutic mAb cocktails Evusheld (tixagevimab + cilgavimab) and Ronapreve (casirivimab + imdevimab) to Delta but was less sensitive to sotrovimab neutralization compared to both Delta and BA.1, with no virus being 100% neutralized at 10 µg/mL (Fig. 2B), consistent with previous studies (8). Against sera, B.1.640.1 exhibited a consistent reduction in neutralization compared to Delta with median ED50 values reduced by 2.3-fold and 1.7-fold, at month 6 (M6) and month 12 (M12), respectively (Fig. 2C). As expected, BA.1 showed the greatest reduction in neutralization, with median ED50 values reducing 2.6-fold and 2.0-fold at M6 and M12, respectively, compared to B.1.640.1 (Fig. 2C).
Circulation of B.1.640.1 ceased in early 2022 at the time of Omicron emergence and concomitantly to the second and third RNA vaccine rollout in France. To investigate the sensitivity of B.1.640.1 to Pfizer vaccination, sera from individuals that received their second or third doses of vaccine were used to test viral neutralization. Here, Delta showed a fivefold higher EC50 compared to B.1.640.1, whereas BA.1 was minimally neutralized after two vaccine doses (Fig. 2C). After three vaccine doses, B.1.640.1 displayed a 2.1-fold decrease and 1.7-fold increase in median EC50 over Delta and BA.1, respectively.
In all, B.1.640.1 Spike showed anti-NTD mAb-binding escape, with the consequences of this on the humoral response needing further investigation. Neutralization of the variant was consistently reduced compared to Delta, yet higher than BA.1. B.1.640.1 was efficiently neutralized after three doses of Pfizer vaccine.
B.1.640.1 Spike displays a high fusogenic phenotype
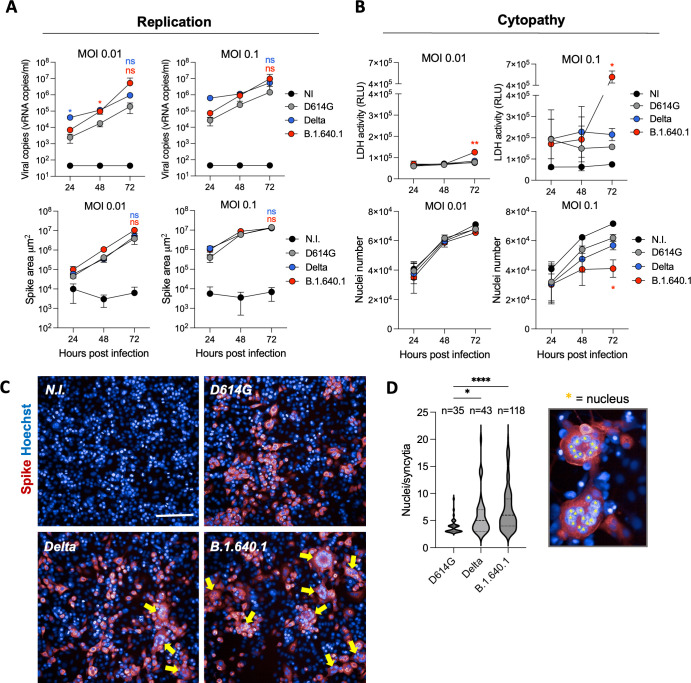
Viral fitness is influenced by factors such as immune evasion, tissue tropism, and replication capacity (17). To investigate the replication kinetics of B.1.640.1, compared to D614G, Delta, and BA.1, we infected A549 human lung cell line expressing human ACE2 (referred to herein as A549-ACE2 cells) with a range of MOIs. Viral release was measured by RT-qPCR, and the frequency of infected cells was quantified by Spike immunostaining. At 72 hours post-infection (hpi), there was no significant difference in viral RNA release of B.1.640.1 or Delta compared to D614G at MOI 0.01 and 0.1 (Fig. 3A). In accordance, no significant difference in the total area of Spike staining was observed between variants (Fig. 3A). BA.1 replicated poorly in this cell line after 72 hpi.
Fig 3.
Cytopathic effects of B.1.640.1 infection on A549-ACE2 cells. (A) (Top) Replication kinetics of D614G, Delta, and B.1.640.1 in A549-ACE2 cells shown by quantification of the viral E protein gene in the cell supernatant by RT-qPCR at the respective timepoints. (Bottom) Area of Spike-positive A549-ACE2-infected cells from respective variants over 72 hours. Cells were stained with anti-Spike mAb (mAb102) before staining with an Alexa-Fluor 647 conjugated secondary to allow quantification of infection. N = 4. (B) (Top) Quantification of LDH release in A549-ACE2-infected cell supernatant through a luciferase-based assay. N = 4. (Bottom) Nuclei count following infection of A549-ACE2 cells at the indicated timepoints with the respective variants. Nuclei were stained with Hoechst dye prior to quantification. (C) Confocal immunofluorescence of A549-ACE2 cells infected with MOI 0.1 D614G, Delta, and B.1.640.1 cells and control cells 48 hpi. Yellow arrows indicate the presence of syncytia. Scale bar = 200 µm. (D) Violin plot of number of nuclei per syncytia 48 hpi with D614G, Delta, and B.1.640.1 infection (MOI = 0.1). Results were taken from two independent experiments, with 12 fields analyzed per variant. Nuclei counting was performed manually using ImageJ software. Ordinary one-way ANOVA tests were performed with Tukey’s multiple comparison test to compare D614G to respective variants, *P < 0.05, ****P < 0.00001, ns = not significant. For A and B, two-way ANOVA tests were performed with Geisser-Greenhouse correction to compare Delta and B.1.640.1 to D614G, *P < 0.05. Error bars represent SD.
Microscopy analysis of B.1.640.1-infected cells revealed striking levels of cell-cell fusion at 48 hpi compared to other variants (Fig. 3C). Syncytia size and number were quantified through manual nuclei counting. Here, B.1.640.1-induced syncytia were significantly larger than D614G (3.91 versus 6.98 nuclei/syncytia on average) and larger than Delta (5.81 nuclei/syncytia). In the 12 fields analyzed per variant, B.1.640.1 infection produced 118 syncytia versus 35 and 43 for D614G and Delta, respectively. To further investigate this phenomenon, we infected A549-ACE2 cells and monitored the cells using video microscopy for over 48 hours. Video microscopy analysis showed rapid fusion and syncytial formation in B.1.640.1-infected cells commencing 24 hpi, ultimately leading to large syncytia and cell death as detected by PI dye (Fig. S1A; Movie S1). D614G also induced syncytial formation but to a far lesser extent.
To confirm the high cytopathicity of B.1.640.1 observed by video microscopy, lactate dehydrogenase (LDH) activity was measured on cell supernatants to assess the level of plasma membrane damage in the infected cells. LDH activity peaked significantly at 72 hpi following B.1.640.1 infection at MOI 0.1 (5.2 × 105 RLU and 1.6 × 105 RLU for B.1.640.1 and D614G, respectively) indicating a surge in cytotoxicity (Fig. 3B). Infection of B.1.640.1 at MOI 0.1 also showed a significant reduction in nuclei number compared to Delta and D614G, with 2%–6%, 17%–33%, and 38%–51%, fewer nuclei at 24, 48, and 72 hours, respectively (Fig. 3B).
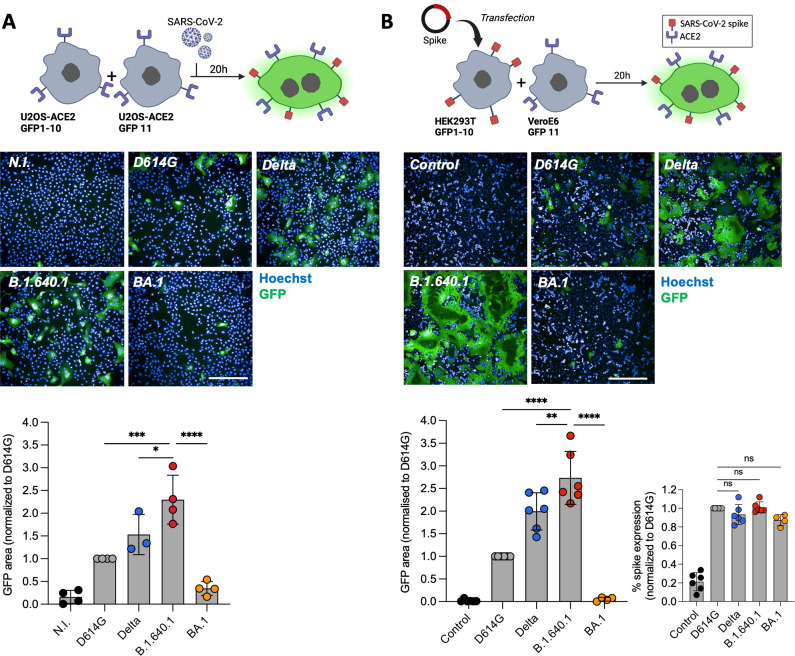
We then quantified cell-cell fusion using a GFP-split S-fuse assay in U2OS-ACE2 cells (Fig. 4A), as described previously (9, 21, 37). B.1.640.1 infection at MOI 0.01 induced a 2.3-fold and 1.4-fold increase in cell-cell fusion compared to D614G and Delta, respectively (Fig. 4A). Next, we investigated the fusogenicity of the variant Spike proteins independently of viral replication. We transfected Spikes into HEK293T cells expressing GFP1–10 subunits and incubated them with VeroE6 cells expressing GFP11 subunit for 18 hours (Fig. 4B). Spike expression on transfected HEK293T cells was similar across variants, as measured by anti-S2 mAb staining (Fig. 4B). As seen during infection, B.1.640.1 Spike displayed the greatest fusogenicity compared to other variants, with a 2.6-fold and a 1.3-fold increase over D614G and Delta, respectively (Fig. 4B). To further confirm the increased fusogenicity of B.1.640.1 Spike, we transfected HEK293T GFP-Split cells with variant Spikes and ACE2 and monitored cell fusion by video microscopy for over 18 hours (Fig. S1B; Movie S2). B.1.640.1 Spike demonstrated a more rapid fusion kinetic across 18 hours of cell co-incubation.
Fig 4.
B.1.640.1 virus and Spike display high fusogenicity in cell lines. (A) (Top) Schematic of S-fuse assay utilizing U2OS S-fuse GFP-split cells to quantify viral fusion following infection (MOI = 0.01). (Middle) Confocal microscopy images of cells infected with respective variants at MOI 0.01. Hoechst dye stains the nuclei. (Bottom) Quantification of GFP area was performed 20 hpi, and data are normalized to D614G fusion. (B) (Top) Schematic of HEK293T-GFP1-10 and VeroE6-GFP11 co-culture system. HEK293T-GFP1-10 cells were transfected with Spike or control plasmids before co-culture with VeroE6-GFP11 cells. (Middle) Confocal microscopy images of co-cultured cells 20 hpi. Hoechst dye stains the nuclei. (Bottom) Quantification of GFP area 20 hours post-transfection and surface Spike expression by staining of HEK293T-GFP1-10 cells with an anti-S2 mAb. For A and B, ordinary one-way ANOVA tests were performed with Tukey’s multiple comparison test to compare D614G to respective variants, *P < 0.05, **P < 0.001, ***P < 0.0001, ****P < 0.00001, ns = not significant. Error bars represent SD. Scale bars = 400 µm.
Overall, B.1.640.1 showed a highly fusogenic phenotype and an increase cytopathy while having similar replication kinetics to the Delta in A549-ACE2 cells. In addition, B.1.640.1 Spike alone displayed higher fusogenicity than D614G and Delta while being similarly expressed.
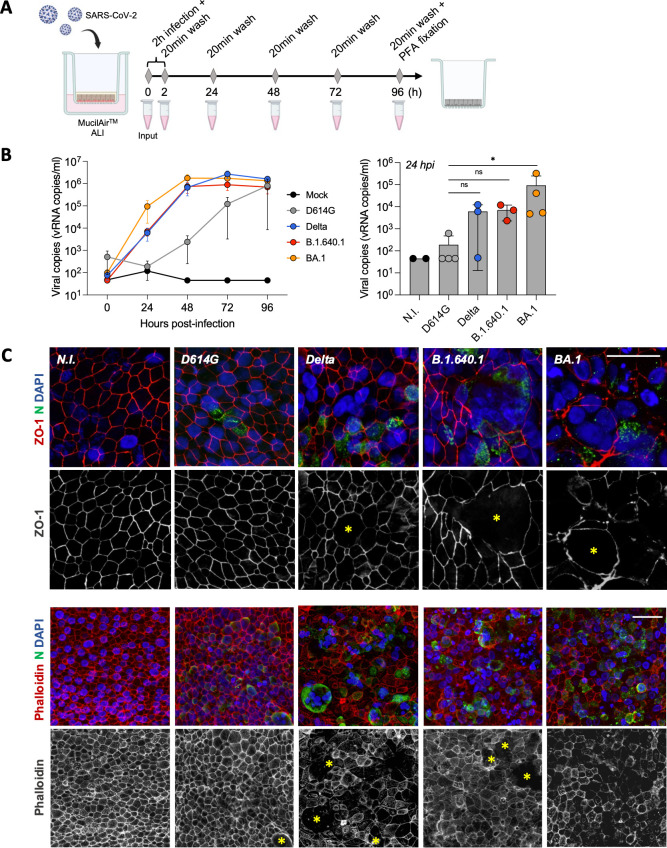
B.1.640.1-infected human nasal epithelial cells form large syncytia
To investigate our findings in a physiologically relevant model, we infected MucilAir primary hNECs in an ALI culture with D614G, Delta, B.1.640.1, and BA.1 variants for 96 hours (Fig. 5A). This culture system is an effective tool to study SARS-CoV-2 infection (10, 14, 42). As previously described (5), BA.1 exhibited a replication advantage compared to Delta and D614G, as quantified by viral RNA release (Fig. 5B). Compared to BA.1, B.1.640.1 replication was delayed, with a 13-fold reduction in average RNA release after 24 hours of infection. B.1.640.1 replication kinetics closely resembled Delta after 48 hours of infection, with a 2.7-fold increase in peak Delta replication at 72 hpi (Fig. 5B). We next sought to examine if B.1.640.1 induced syncytia in this primary cell model. The cell cultures were fixed and stained 96 hpi when all variants had comparable viral RNA release. Due to the absence of fusion reporter systems, we assessed fusion through staining of Zonula Occludens 1 (ZO-1), a tight junction protein found at cell-cell borders, and phalloidin, binding to F-actin. ZO-1 and phalloidin staining revealed the presence of large, infected nucleoprotein-positive syncytia in the B.1.640.1 and Delta conditions (Fig. 5C; Fig. S2A). D614G induced smaller and rarer syncytia, however, replicated to a far lesser extent. Conversely, BA.1 infection resulted in considerable deconstruction of the epithelium and presence of small syncytia. However, its significantly higher replication in the ALI culture system may impact comparisons to the other variants. Delta and B.1.640.1 displayed similar replication kinetics, and both induced the formation of syncytia, with B.1.640.1 inducing large-rounded syncytia, confirmed through ZO-1 staining (Fig. 5C; Fig. S2A). Ultrastructural SEM analysis also revealed extensive loss of cilia on the surface in Delta- and B.1.640.1-infected hNECs (Fig. S3A). B.1.640.1 thus efficiently induces cell-cell fusion in primary hNEC ALI cultures with no replicative advantage compared to Delta.
Fig 5.
Apical replication and syncytia formation in hNEC air-liquid interface culture. (A) Schematic of hNEC ALI culture infection with SARS-CoV-2 variants over 96 hours followed by cell fixation. (B) Viral RNA release from apical side of hNECs as measured by RT-qPCR targeting the SARS-CoV-2 E gene (n = 3/4; left). Apical viral RNA release measured at 24 hpi; bars represent mean values (right). Dotted lines and error bars represent SD. Mann-Whitney tests were performed to compare the respective variants to D614G, *P < 0.05. (C) Confocal immunofluorescence of hNECs 96 hpi with respective variants displaying syncytia formation (yellow asterisks) through ZO-1, phalloidin, SARS-CoV-2 nucleoprotein, and DAPI staining. Upper scale bar = 20 µm. Lower scale bar = 40 µm.
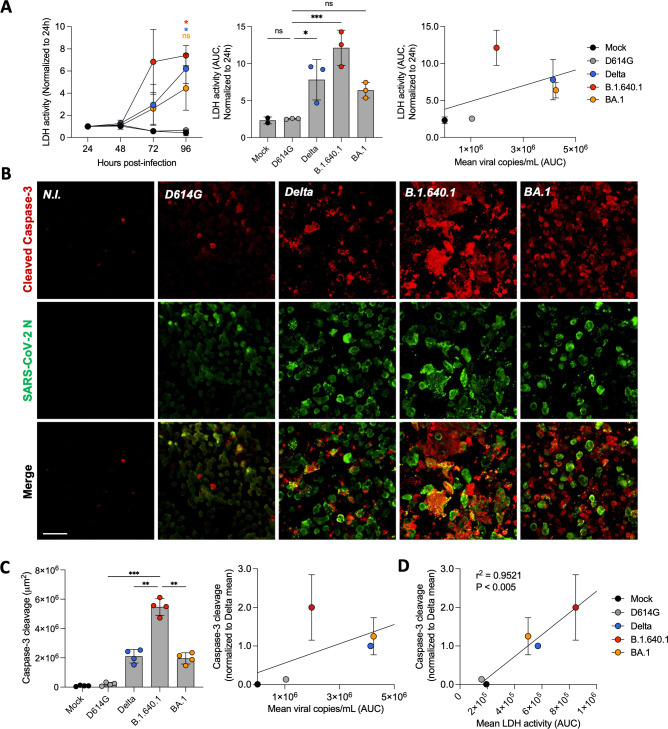
Elevated caspase-3 cleavage and LDH release during B.1.640.1 hNEC infection
Next, we investigated the cytopathy of B.1.640.1 in the primary cell model. We first quantified LDH release in the supernatant of infected cells. LDH release was detected at 72 hours and was significantly increased at 96 hpi for Delta and B.1.640.1, most notably for B.1.640.1 (Fig. 6A). BA.1 induced detectable but non-significant LDH, while D614G did not induce detectable LDH release 96 hpi. As SARS-CoV-2 replication results in cytopathy (42–44), we normalized the LDH release to the levels of replication. Once normalized, B.1.640.1 infection increased LDH activity by 35% compared to BA.1 following linear regression, suggesting B.1.640.1 is more cytopathic in primary nasal ALI cultures (Fig. 6A). To further investigate the mechanism of cell death, we examined the levels of cleaved caspase-3 induced by each variant. Caspase-3 undergoes cleavage following activation of the intrinsic or extrinsic cell death pathways; thus, its detection is a reliable marker for cell demise (45–47). Caspase-3 activation, assessed by immunostaining, was increased significantly by infection with B.1.640.1 compared to D614G, Delta, and BA.1 (Fig. 6B and C). A similar but non-significant trend was seen in the replicate infection (Fig. S2B). After normalizing for replication, B.1.640.1 infection resulted in a twofold increase in caspase-3 activation. Caspase-3 cleavage correlated significantly (P < 0.005) with LDH release across all variants (Fig. 6D). In accordance with the cell line observations, B.1.640.1 displays elevated markers of cytopathy in hNEC ALI cultures.
Fig 6.
Cytopathic effects of hNEC infection with B.1.640.1. (A) LDH release from apical side of hNEC ALI culture over the time course of infection with respective SARS-CoV-2 variants (n = 3/4; left). Area under the curve (AUC) representation of LDH activity, bars represent mean values (middle). Linear regression analysis of LDH release (AUC) compared to viral copies/mL (AUC) from 96 hours of infection with respective SARS-CoV-2 variants (right). Asterisk colours represent respective variants. (B) Immunofluorescence of hNECs stained for cleavage products of caspase-3 and SARS-CoV-2 nucleoprotein. Shown is one field of each variant. Scale bar = 40 µm. (C) Quantification of total area of cleavage products of caspase-3. Each data point represents one randomly assigned field from a single biological repeat (left). An ordinary one-way ANOVA test was performed with Tukey’s multiple comparison test to compare D614G to respective variants, *P < 0.05, ***P < 0.0001, ns = not significant. Linear regression analysis of caspase-3 cleavage from two biological repeats (total number of fields = 8) normalized to Delta compared to mean viral copies/mL (AUC) 96 hpi (right). (D) Linear regression analysis of caspase-3 cleavage normalized to Delta compared to LDH activity (AUC) over 96 hours of infection. Error bars represent SD.
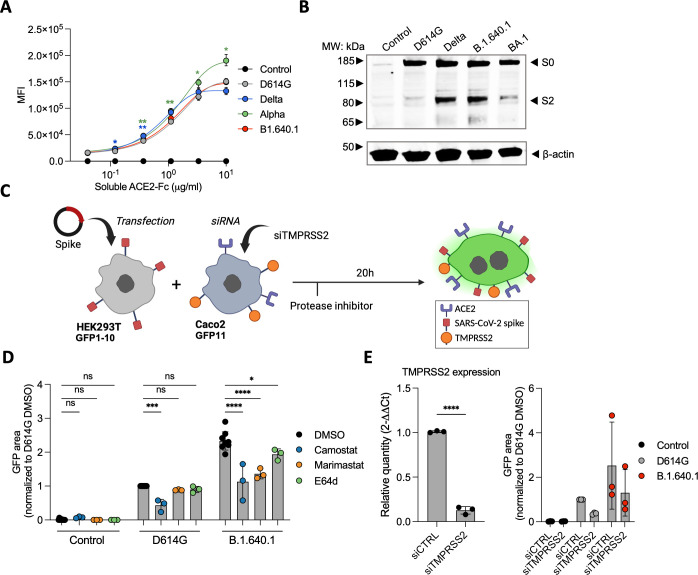
B.1.640.1 Spike is preferentially cleaved at polybasic cleavage site and by matrix metalloproteinases
We next explored the different mechanisms which could explain the increased fusogenicity of B.1.640.1 Spike. We first checked the affinity of Spike to ACE2 (Fig. 7A). Using a soluble ACE2-Fc-binding assay, B.1.640.1 Spike affinity was similar to that of D614G, suggesting that the increase in fusion is not due to differences in ACE2 affinity, as shown previously (9, 27, 29). Next, we investigated if B.1.640.1’s increased fusion can be explained by increased S1/S2 cleavage. Western blot analysis revealed greater S1/S2 cleavage of B.1.640.1 Spike compared to D614G while exhibiting a similar profile to Delta (Fig. 7B). This is consistent with the P681H/R mutation increasing S1/S2 cleavage (48).
Fig 7.
The impact of host cell proteases on B.1.640.1 Spike fusion. (A) Staining of HEK293T cells transfected with Spike or control plasmids and stained with a serial dilution of soluble human ACE2-Fc followed by staining with anti-human-Fc secondary antibody. A two-way ANOVA test was performed with Geisser-Greenhouse correction to compare D614G to the respective variants, *P < 0.05, **P < 0.001. (B) S1/S2 cleavage of respective variant Spike proteins measured by western blot. HEK293T cells transfected with respective Spikes or control plasmids and cell lysates were analyzed 24 hours later (n = 2). (C) Schematic of HEK293T-GFP1-10 cells transfected with D614G Spike, B.1.640.1 Spike, or control plasmids and co-cultured with Caco-2-GFP11 in the presence of protease inhibitors. (D) Fusion of D614G and B.1.640.1 Spikes, quantified by GFP area, in the presence 10 mM camostat, marimastat, E64d, or a DMSO control. (E) (Left) Total RNA was extracted from Caco2 cells treated with TMPRSS2 siRNA or siCTRL, and RT-qPCR was performed. Data were normalized to β-Tubulin levels. Relative mRNA expression normalized to siCtrl condition (2−ΔΔCT) was plotted. (Right) Fusion of D614G and B.1.640.1, quantified by GFP, in the presence of TMPRSS2 knockdown or wild-type Caco-2 cells. For (D) and (E), ordinary one-way ANOVA tests were performed with Tukey’s multiple comparison test to compare D614G to respective variants, *P < 0.05, ***P < 0.0001, ****P < 0.00001. Error bars represent SD.
Next, we examined the processing of Spike by other cellular proteases. Serine proteases, such as TMPRSS2, and matrix metalloproteinases (MMPs) cleave Spike at the S2′ site to release the fusion peptide (21, 22). To study the impact of these proteases on fusion, we treated Caco-2-GFP11 cells with camostat, a TMPRSS2 inhibitor, marimastat, an MMP inhibitor, and E64d, a cysteine protease inhibitor (Fig. 7C). We also performed siRNA knockdown of TMPRSS2 confirmed by RT-qPCR (Fig. 7E). Both inhibitor-treated and siRNA-treated Caco-2 cells were incubated with HEK293T-GFP1-10 cells expressing Spike. Both camostat treatment and TMPRSS2 knockdown resulted in a 2.4-fold and 2.0-fold reduction in D614G and B.1.640.1 fusion, respectively (Fig. 7D). Nevertheless, B.1.640.1 Spike maintained a twofold increase in fusion over D614G in TMPRSS2 inhibition or TMPRSS2-knockdown conditions (Fig. 7D and E). This suggests that the increase in B.1.640.1 Spike fusion compared to D614G is independent of TMPRSS2 processing. E64d had no effect on D614G fusion and a minor effect on B.1.640.1, suggesting cysteine proteases play a minor role in the cell-cell fusion process. Conversely, marimastat significantly inhibited B.1.640.1 Spike fusion but not D614G fusion, suggesting preferential MMP-mediated Spike processing and S1/S2 cleavage contribute to the increase in B.1.640.1 fusion.
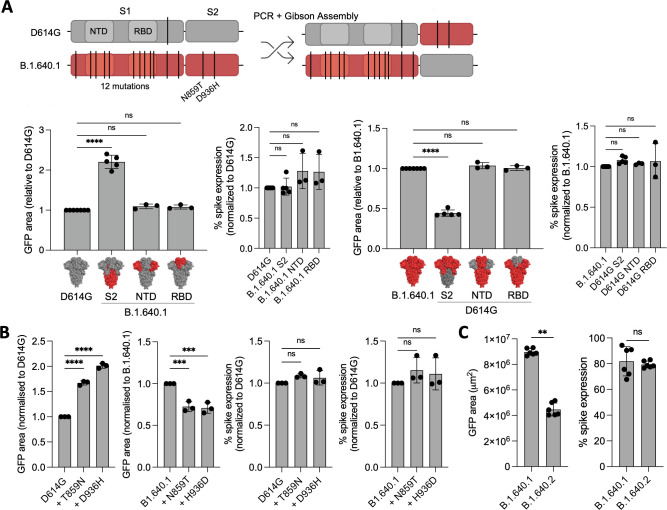
S2 domain mutations T859N and D936H promote the high fusogenicity of B.1.640.1
Mutations in Spike can alter the fusion capacity of SARS-CoV-2 variants. To investigate this, we generated chimeric Spike plasmids by swapping the NTD, RBD, or S2 domains of D614G and B.1.640.1 Spikes (Fig. 8A). We first confirmed that the chimeras were expressed at the surface at the same level (Fig. 8A). Next, using the HEK293T/VeroE6 GFP-split system, we observed that B.1.640.1 S2 approximately doubled D614G fusion, while D614G S2 halved B.1.640.1 fusion, while the NTD and RBD regions had no significant impact on fusion (Fig. 8A). Interestingly, only two mutations reside in the B.1.640.1 S2 domain, T859N and D936H. Using site-directed mutagenesis, we introduced these mutations and their respective reversions individually into the D614G and B.1.640.1 Spike backbones. Concurrent with the Spike chimeras, both mutations significantly increased D614G fusion, while the reversion mutations significantly reduced B.1.640.1 fusion (Fig. 8B) while being expressed similarly (Fig. 8B). Altogether, these findings reveal that the increased fusogenic property of B.1.640.1 is also linked to the two S2 subunit mutations T859N and D936H.
Fig 8.
B.1.640.1 Spike S2 mutations increase fusogenicity. (A) (Top) Schematic of D614G and B.1.640.1 chimeric Spike production through Gibson’s assembly. The NTD, RBD, and S2 domains of both Spikes were swapped to generate chimeric Spikes. For simplicity, only S1/S2 chimeras are shown. (Bottom) Fusion of D614G and B.1.640.1 Spike chimeras in HEK293T-GFP1-10 and VeroE6-GFP11 co-culture system. HEK293T-GFP1-10 cells were transfected with Spikes or control plasmids and then co-cultured with VeroE6-GFP11. Fusion was quantified by GFP area 18 hours post-transfection. Spike expression was assessed by surface staining of transfected HEK293T-GFP1-10 cells with an anti-S2 mAb. (B) D614G and B.1.640.1 Spikes were mutated to incorporate or revert B.1.640.1 S2-specific mutations, respectively. HEK293T-GFP1-10 cells were transfected with Spikes or control plasmids before co-culture with VeroE6-GFP11 cells. Fusion was assessed by GFP area 18 hours post-transfection. Spike expression was assessed by surface staining of transfected HEK293T-GFP1-10 cells with an anti-S2 mAb. (C) HEK293T-GFP1-10 cells were transfected with B.1.640.1 or B.1.640.2 Spikes or control plasmids before co-culture with VeroE6-GFP11 cells. Fusion was assessed by GFP area 18 hours post-transfection. Spike expression was assessed by surface staining of transfected HEK293T-GFP1-10 cells with an anti-S2 mAb. Mann-Whitney tests were performed to compare GFP area and spike expression, **P < 0.001. For (A) and (B), ordinary one-way ANOVA tests were performed with Tukey’s multiple comparison test to compare respective variants, **P < 0.001, ***P < 0.0001, ****P < 0.00001, ns = not significant. Error bars represent SD.
B.1.640.2 Spike is less fusogenic than B.1.640.1
The sister variant of B.1.640.1, B.1.640.2, circulated in France over a similar timespan. The two variants differ by five mutations in the Spike protein (Fig. 1A). Notably, the D936H HR1 mutation is not present in B.1.640.2. Consistent with this, fusion of B.1.640.2 Spike was reduced by twofold in comparison to B.1.640.1 (Fig. 8C) while being expressed similarly (Fig. 8C). This is a greater reduction than the single-point mutation alone, suggesting that other mutations within B.1.640.2 reduce its fusogenic potential.
DISCUSSION
We describe the phenotypic properties of the SARS-CoV-2 B.1.640.1 variant which transiently circulated in Europe and Africa in 2021–2022. Analysis of a panel of mAbs identified a loss of anti-NTD binding to this variant. The role of the NTD in neutralizing and non-neutralizing antibody (nnAb) functions is of current interest (39, 49, 50). A region termed the NTD antigenic supersite is a favorable target for potently neutralizing antibodies (2, 51). The supersite comprises three loops, N1 (residues 14–26), N3 (residues 141–156), and N5 (residues 246–260). Deletion or mutation of these residues and proximal residues, such as C136Y, G142D, and Del144, greatly reduces neutralization by mAbs by changing loop conformations and antibody epitopes (51). The epitopes of the NTD mAbs we tested are unknown. It is likely that Del136-144 in B.1.640.1 NTD results in alteration to the antibody supersite and thus loss of antibody recognition, but this requires further structural analysis. B.1.640.1 Spike demonstrates a plasticity of the NTD to undergo large levels of mutation while maintaining the protein functions. Supersite deletions are seen during persistent infections of immunocompromised individuals (52, 53). Additionally, the NTD is a region of interest for non-neutralizing antibody functions such as antibody-dependent cell-cytotoxicity and complement-dependent cytotoxicity (39, 50, 54). Future studies would be of interest to reveal if the B.1.640.1 NTD and Del136-144 arose as a mechanism of nnAb evasion.
Our mAb-binding analysis revealed the loss of cilgavimab (AZD1061) binding to B.1.640.1, whereas other anti-RBD mAbs maintained their binding capacities. A similar loss of binding was seen in recent Omicron subvariants BA.2.75.2 and BQ.1.1, all containing Spike mutation R346T, now known to cause the loss of binding (3, 55, 56). As such, cilgavimab is no longer recommended for therapeutic use. B.1.640.1 carries an R346S substitution, demonstrating that convergent mutations with clinical relevance have circulated in older variants.
Emergence and dominance of variants of concern are facilitated by many viral and host factors. The success of the initial Omicron variant, BA.1, was, in part, due to its considerable evasion of neutralizing antibodies (8, 57–60), among other factors. While B.1.640.1 virus showed a slight yet consistent reduction in neutralization by sera from convalescent or vaccinated individuals compared to Delta, it was more sensitive than BA.1. Therefore, while B.1.640.1 was able to circulate throughout France, its disappearance coincided with the rollout of the third SARS-CoV-2 vaccine dose and the emergence of BA.1.
We report the ability of B.1.640.1 to induce large syncytia in our culture systems. Syncytia are observed in histological studies of COVID-19 patients’ lungs; however, their role in pathogenesis is still debated, and their presence during mild disease remains unclear (31, 32, 61). We found B.1.640.1 elevated LDH release and caspase-3 cleavage beyond that of the other variants. The replication of SARS-CoV-2 is sufficient to cause cytopathic effects in airway epithelial cells (42, 44, 62). To account for this and the differences in viral replication between the variants, we normalized measures of cytopathy to replication. Here, B.1.640.1 caused greater LDH release and caspase-3 cleavage than the other variants. In accordance with the previous literature, SARS-CoV-2 Spike-induced syncytia formation in HeLa cells causes activation of caspase-3 (45). Furthermore, cell-cell fusion induced by the FAST fusion protein of reoviruses enhances viral pathogenicity (63). Thus, it would be of interest to investigate the impact of the differing fusogenic phenotypes of SARS-CoV-2 variants on pathogenicity. Together, our results suggest that the highly fusogenic property of B.1.640.1 is responsible for its increased cytopathy.
The role of cell-cell fusion in SARS-CoV-2 transmission remains unclear. Syncytia formation facilitates cell-to-cell transmission of virus and provides a mechanism of evasion from the host immune response (64). Additionally, syncytia may allow for dissemination of virus within the host. Syncytia from multiple cellular origins containing high levels of infection are detected in the bronchoalveolar fluid of COVID-19 patients (65). On the contrary, increased cytopathy relating to syncytia formation may also be detrimental to SARS-CoV-2 replication at timepoints beyond those we have studied. Therefore, further understanding into how syncytia formation impacts viral transmission within and between hosts is warranted.
We show that B.1.640.1 Spike displays increased S1/S2 cleavage as well as increased protease activation by cell surface MMPs compared to D614G. P681H, present in B.1.640.1, increases S1/S2 cleavage and Spike fusogenicity (48). Additionally, this mutation reduces the dependence of S2′ processing by endosomal cathepsin proteases and promotes cell surface Spike processing (66), consistent with our findings. Furthermore, through mutagenesis, we found T859N and D936H within the B.1.640.1 S2 subunit are independently and additively responsible for high intrinsic fusogenicity of Spike. Both mutations have continuously circulated at a low level (<0.05% of sequences), with T859N most notably being in the Lambda variant and D936H being in a small proportion of omicron subvariants (covSPECTRUM; GISAID). The absence of these mutations in the Delta variant also suggests enhanced fusogenicity can arise through differing mutations. Additionally, B.1.640.1 Spike fusion was increased in the presence of TMPRSS2. Similar levels of inhibition to D614G suggest a preferential cell surface entry route for B.1.640.1, consistent with variants prior to Omicron BA.1 (67).
No mutations are present in the cytoplasmic tail of B.1.640.1, the region containing a suboptimal COPI-binding site which is responsible for surface trafficking of Spike (25). Therefore, surface levels of B.1.640.1 Spike are not impacted compared to other variants, as confirmed by cell surface staining, and do not explain differences in fusion. The S2 subunit also contains the membrane fusion machinery including the hydrophobic FP and the heptad repeat regions 1 and 2 (HR1 and HR2). Mutations can alter this process through different mechanisms. For example, N856K present in BA.1 introduces a charged lysine residue that forms a salt bridge with D568 which reduces shedding of the S1 subunit and fusion (16). In silico analysis reveals that residue 859 resides in close proximity to residue 614 within S1 (Fig. S3B). It would be of interest to study any potential interactions between residue 859 and the S1 subunit and if this mutation impacts S1 shedding. HR1 and HR2 within S2 form a six-helical bundle to bring the membranes together during fusion (68). D936 within HR1 forms a salt bridge with the positively charged R1185 residue of HR2 (69). Thus, the introduction of H936 may also impact the conformational changes Spike undergoes during fusion. Deep mutational scanning may also reveal how these mutations alter the effect of other Spike mutations to understand the evolutionary constraints of fusogenicity on Spike (70).
In silico analyses suggest B.1.640.2 to have greater infectivity than B.1.640.1 (71). We previously showed the E484K mutation, present in B.1.640.2 but not B.1.640.1, significantly reduces fusion (9) and contributes to immune evasion (72). Together with the absence of D936H, this explains the twofold reduction in fusogenicity of B.1.640.2. Further investigation into the evolution of the B.1.640 lineage would be relevant to determine the relationship between B.1.640.1 and 2.
We acknowledge that this study contains limitations. First, the low number of infections caused by B.1.640.1 limits the availability of clinical information on this variant. Further research may involve in vivo models, such as Syrian Hamsters or mice (42, 73), to assess B.1.640.1 pathogenicity. Additionally, we did not explore the role of other mutations outside of the Spike in B.1.640.1 replication and cytopathy which could be studied through reverse genetic approaches. Finally, we were unable to quantify the number of syncytia following infection in the hNEC culture system due to the 3D nature of the tissue. Future work would look to implement a reporter system to give an accurate readout of syncytia in this model.
Overall, the data presented here show a now supplanted SARS-CoV-2 variant B.1.640.1 displaying a highly fusogenic phenotype. Analysis of the humoral response provides insight into how this variant, like many others, was displaced upon the emergence of BA.1. The unusually high fusogenic activity of B.1.640.1, linked to an increased cytopathy, provides insight into the consequences of SARS-CoV-2-induced cell-cell fusion.
ACKNOWLEDGMENTS
The authors thank the patients who contributed to this study, the members of the Virus and Immunity Unit (Institut Pasteur) and other teams for discussion and help, and Nathalie Aulner and the staff at the UtechS Photonic BioImaging (UPBI) core facility (Institut Pasteur).
W.B. was supported by a stipend from the Pasteur - Paris University (PPU) International PhD Program.
Work in the O.S. lab is funded by Institut Pasteur, Urgence COVID-19 Fundraising Campaign of Institut Pasteur, Fondation pour la Recherche Médicale (FRM), ANRS-MIE, the Vaccine Research Institute (ANR-10-LABX-77), Labex IBEID (ANR-10- LABX-62-IBEID), ANR / FRM Flash Covid PROTEO-SARS-CoV-2, ANR Coronamito, HERA European funding, Sanofi, and IDISCOVR. The E.S.-L. laboratory receives funding from the Institut Pasteur, the INCEPTION program (Investissements d’Avenir grant ANR-16-CONV-0005), and NIH PICREID program (Award number U01AI151758). The Opera system was co-funded by Institut Pasteur and the Région ile de France (DIM1Health). The funders of this study had no role in study design, data collection, analysis, and interpretation, or writing of the article.
Conceptualization: W.B., J.B., O.S., T.B., V.M. Methodology: W.B., J.B., O.S., T.B., V.M., D.P., F.G-B., F.P., M.H., I.S. Investigation: W.B., J.B., O.S., T.B., V.M., D.P. Data Collection and Analysis: W.B., J.B., V.M., T.B., D.P., M.H. Cohort design: L.H., T.P. Resources: E.S-L., H.M., M.P., C.P., J-M.P., M.N., C.R., S.F., L.H., T.P. Manuscript Writing and Editing: W.B., T.B., O.S., J.B.
Contributor Information
Olivier Schwartz, Email: olivier.schwartz@pasteur.fr.
Julian Buchrieser, Email: julian.buchrieser@pasteur.fr.
Tom Gallagher, Loyola University Chicago - Health Sciences Campus, Maywood, Illinois, USA.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01351-23.
Additional data on the supplemental movies.
Additional data on the hNEC infections.
Additional data on hNEC infections and mutations.
Video microscopy of A549-ACE2 cells infected with indicated SARS-CoV-2 variants at MOI 0.1. Scale bar = 80 μm.
Video microscopy of HEK293T-GFP1-10 cells transfected with indicated SARS-CoV-2 variant spikes and co-cultured with VeroE6-GFP11 cells over 18 hours. Scale bar = 160 µm.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. El-Shabasy RM, Nayel MA, Taher MM, Abdelmonem R, Shoueir KR, Kenawy ER. 2022. Three waves changes, new variant strains, and vaccination effect against COVID-19 pandemic. Int J Biol Macromol 204:161–168. doi: 10.1016/j.ijbiomac.2022.01.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Planas D, Bruel T, Staropoli I, Guivel-Benhassine F, Porrot F, Maes P, Grzelak L, Prot M, Mougari S, Planchais C, Puech J, Saliba M, Sahraoui R, Fémy F, Morel N, Dufloo J, Sanjuán R, Mouquet H, André E, Hocqueloux L, Simon-Loriere E, Veyer D, Prazuck T, Péré H, Schwartz O. 2023. Resistance of Omicron subvariants BA.2.75.2, BA.4.6, and BQ.1.1 to neutralizing antibodies. Nat Commun 14:824. doi: 10.1038/s41467-023-36561-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruel T, Stéfic K, Nguyen Y, Toniutti D, Staropoli I, Porrot F, Guivel-Benhassine F, Bolland WH, Planas D, Hadjadj J, Handala L, Planchais C, Prot M, Simon-Lorière E, André E, Baele G, Cuypers L, Mouthon L, Mouquet H, Buchrieser J, Sève A, Prazuck T, Maes P, Terrier B, Hocqueloux L, Schwartz O. 2022. Longitudinal analysis of serum neutralization of SARS-CoV-2 Omicron BA.2, BA.4, and BA.5 in patients receiving monoclonal antibodies. Cell Rep Med 3:100850. doi: 10.1016/j.xcrm.2022.100850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D, Scott S, Logan N, Ashraf S, et al. , PITCH Consortium, COVID-19 Genomics UK (COG-UK) Consortium . 2022. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol 7:1161–1179. doi: 10.1038/s41564-022-01241-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, Wang M, Yu J, Bowen AD, Chang JY, Shah JG, Nguyen N, Chen Z, Meyers K, Yin MT, Sobieszczyk ME, Sheng Z, Huang Y, Liu L, Ho DD. 2022. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608:603–608. doi: 10.1038/s41586-022-05053-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zabidi NZ, Liew HL, Farouk IA, Puniyamurti A, Yip AJW, Wijesinghe VN, Low ZY, Tang JW, Chow VTK, Lal SK. 2023. Evolution of SARS-CoV-2 variants: implications on immune escape, vaccination, therapeutic and diagnostic strategies. Viruses 15:944. doi: 10.3390/v15040944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland W-H, Porrot F, Staropoli I, Lemoine F, et al. 2022. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602:671–675. doi: 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 9. Rajah MM, Hubert M, Bishop E, Saunders N, Robinot R, Grzelak L, Planas D, Dufloo J, Gellenoncourt S, Bongers A, Zivaljic M, Planchais C, Guivel-Benhassine F, Porrot F, Mouquet H, Chakrabarti LA, Buchrieser J, Schwartz O. 2021. SARS‐Cov‐2 alpha, beta, and Delta variants display enhanced Spike‐Mediated Syncytia formation. EMBO J 40:e108944. doi: 10.15252/embj.2021108944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira I, Datir R, Collier DA, Albecka A, Singh S, Mavousian A, Lee JH, Bassi J, Silacci-Fegni C, Saliba C, Pinto D, Irie T, Yoshida I, Hamilton WL, Sato K, Bhatt S, Flaxman S, James LC, Corti D, Piccoli L, Barclay WS, Rakshit P, Agrawal A, Gupta RK, et al. , Indian SARS-CoV-2 Genomics Consortium (INSACOG), Genotype to Phenotype Japan (G2P-Japan) Consortium, CITIID-NIHR BioResource COVID-19 Collaboration . 2021. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599:114–119. doi: 10.1038/s41586-021-03944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nyberg T, Ferguson NM, Nash SG, Webster HH, Flaxman S, Andrews N, Hinsley W, Bernal JL, Kall M, Bhatt S, et al. 2022. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 Omicron (B.1.1.529) and Delta (B.1.617.2) variants in England: a cohort study. The Lancet 399:1303–1312. doi: 10.1016/S0140-6736(22)00462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bager P, Wohlfahrt J, Bhatt S, Stegger M, Legarth R, Møller CH, Skov RL, Valentiner-Branth P, Voldstedlund M, Fischer TK, et al. 2022. Risk of hospitalisation associated with infection with SARS-CoV-2 Omicron variant versus Delta variant in Denmark: an observational cohort study. Lancet Infect Dis 22:967–976. doi: 10.1016/S1473-3099(22)00154-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abdullah F, Myers J, Basu D, Tintinger G, Ueckermann V, Mathebula M, Ramlall R, Spoor S, de Villiers T, Van der Walt Z, Cloete J, Soma-Pillay P, Rheeder P, Paruk F, Engelbrecht A, Lalloo V, Myburg M, Kistan J, van Hougenhouck-Tulleken W, Boswell MT, Gray G, Welch R, Blumberg L, Jassat W. 2022. Decreased severity of disease during the first global Omicron variant COVID-19 outbreak in a large hospital in Tshwane, South Africa. Int J Infect Dis 116:38–42. doi: 10.1016/j.ijid.2021.12.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, Kosugi Y, Tsuda M, Orba Y, Sasaki M, Shimizu R, Kawabata R, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Genotype to Phenotype Japan (G2P-Japan) Consortium, Sawa H, Ikeda T, Irie T, Matsuno K, Tanaka S, Fukuhara T, Sato K. 2022. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603:700–705. doi: 10.1038/s41586-022-04462-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hui KPY, Ho JCW, Cheung M-C, Ng K-C, Ching RHH, Lai K-L, Kam TT, Gu H, Sit K-Y, Hsin MKY, Au TWK, Poon LLM, Peiris M, Nicholls JM, Chan MCW. 2022. SARS-Cov-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603:715–720. doi: 10.1038/s41586-022-04479-6 [DOI] [PubMed] [Google Scholar]
- 16. Sun C, Wang H, Yang J, Kong D, Chen Y, Wang H, Sun L, Lu J, Teng M, Xie L. 2023. Mutation N856K in spike reduces fusogenicity and infectivity of Omicron BA.1. Sig Transduct Target Ther 8. doi: 10.1038/s41392-022-01281-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J, COVID-19 Genomics UK Consortium, de Silva TI, Peacock SJ, Barclay WS, de Silva TI, Towers GJ, Robertson DL. 2023. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. doi: 10.1038/s41579-022-00841-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-Cov-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lubinski B, Fernandes MHV, Frazier L, Tang T, Daniel S, Diel DG, Jaimes JA, Whittaker GR. 2022. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (alpha). iScience 25:103589. doi: 10.1016/j.isci.2021.103589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lubinski B, Frazier LE, Phan MVT, Bugembe DL, Cunningham JL, Tang T, Daniel S, Cotten M, Jaimes JA, Whittaker GR. 2022. Spike protein cleavage-activation in the context of the SARS-CoV-2 P681R mutation: an analysis from its first appearance in lineage A.23.1 identified in Uganda. Microbiol Spectr 10:e0151422. doi: 10.1128/spectrum.01514-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O. 2020. Syncytia formation by SARS‐CoV‐2‐infected cells. EMBO J 39:e106267. doi: 10.15252/embj.2020106267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaimes JA, Millet JK, Whittaker GR. 2020. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 23:101212. doi: 10.1016/j.isci.2020.101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peacock TP, Brown JC, Zhou J, Thakur N, Sukhova K, Newman J, Kugathasan R, Yan AWC, Furnon W, De Lorenzo G, Cowton VM, Reuss D, Moshe M, Quantrill JL, Platt OK, Kaforou M, Patel AH, Palmarini M, Bailey D, Barclay WS. 2022. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. Microbiology. doi: 10.1101/2021.12.31.474653 [DOI] [Google Scholar]
- 25. Cattin-Ortolá J, Welch LG, Maslen SL, Papa G, James LC, Munro S. 2021. Sequences in the cytoplasmic tail of SARS-CoV-2 spike facilitate expression at the cell surface and syncytia formation. Nat Commun 12:5333. doi: 10.1038/s41467-021-25589-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rajah MM, Bernier A, Buchrieser J, Schwartz O. 2022. The mechanism and consequences of SARS-Cov-2 spike-mediated fusion and syncytia formation. J Mol Biol 434:167280. doi: 10.1016/j.jmb.2021.167280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura T, Mizuma K, Nasser H, Deguchi S, Padilla-Blanco M, Uriu K, Tolentino JEM, Tsujino S, Suzuki R, Kojima I, Nao N, Shimizu R, Jonathan M, Kosugi Y, Guo Z, Hinay AA, Putri O, Kim Y, Tanaka YL, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Saito A, Ito J, Irie T, Zahradnik J, Ikeda T, Takayama K, Matsuno K, Fukuhara T, Sato K, The Genotype to Phenotype Japan (G2P-Japan) Consortium . 2023. Virological characteristics of the SARS-Cov-2 BA.2.86 variant. Edited bySato KMicrobiology. doi: 10.1101/2023.11.02.565304 [DOI] [PubMed] [Google Scholar]
- 28. Qu P, Xu K, Faraone JN, Goodarzi N, Zheng Y-M, Carlin C, Bednash JS, Horowitz JC, Mallampalli RK, Saif LJ, Oltz EM, Jones D, Gumina RJ, Liu S-L, Davis DM. 2023. Immune evasion, infectivity, and Fusogenicity of SARS-CoV-2 Omicron BA.2.86 and flip variants. bioRxiv. doi: 10.1101/2023.09.11.557206 [DOI] [PMC free article] [PubMed]
- 29. Planas D, Staropoli I, Michel V, Lemoine F, Donati F, Prot M, Porrot F, Guivel-Benhassine F, Jeyarajah B, Brisebarre A, Dehan O, Boland WH, Hubert M, Buchrieser J, Vanhoucke T, Rosenbaum P, Veyer D, Péré H, Lina B, Trouillet-Assant S, Hocqueloux L, Prazuck T, Simon-Loriere E, Schwartz O. 2023. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86 lineages combining increased fitness and antibody evasion. Microbiology. doi: 10.1101/2023.11.20.567873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanders DW, Jumper CC, Ackerman PJ, Bracha D, Donlic A, Kim H, Kenney D, Castello-Serrano I, Suzuki S, Tamura T, Tavares AH, Saeed M, Holehouse AS, Ploss A, Levental I, Douam F, Padera RF, Levy BD, Brangwynne CP. 2021. Sars-CoV-2 requires cholesterol for viral entry and pathological Syncytia formation. Elife 10:e65962. doi: 10.7554/eLife.65962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tirelli G, Tofanelli M, Sacchet E, Bussani R, Shafiei V, Gatto A, Boscolo-Rizzo P, Gardenal N. 2021. Extranodal extension in head and neck squamous cell cancer: is there a role for further stratification? Br J Oral Maxillofac Surg 59:567–572. doi: 10.1016/j.bjoms.2020.09.015 [DOI] [PubMed] [Google Scholar]
- 32. Bussani R, Zentilin L, Correa R, Colliva A, Silvestri F, Zacchigna S, Collesi C, Giacca M. 2023. Persistent SARS-CoV-2 infection in patients seemingly recovered from COVID-19. J Pathol 259:254–263. doi: 10.1002/path.6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clemens DJ, Ye D, Wang L, Kim CSJ, Zhou W, Dotzler SM, Tester DJ, Marty I, Knollmann BC, Ackerman MJ. 2023. Cellular and electrophysiological characterization of Triadin knockout syndrome using induced Pluripotent stem cell-derived cardiomyocytes. Stem Cell Reports 18:1075–1089. doi: 10.1016/j.stemcr.2023.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colson P, Delerce J, Burel E, Dahan J, Jouffret A, Fenollar F, Yahi N, Fantini J, La Scola B, Raoult D. 2022. Emergence in Southern France of a new SARS‐CoV‐2 variant harbouring both N501Y and E484K substitutions in the spike protein. Arch Virol 167:1185–1190. doi: 10.1007/s00705-022-05385-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shu Y, McCauley J. 2017. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buchrieser J, Degrelle SA, Couderc T, Nevers Q, Disson O, Manet C, Donahue DA, Porrot F, Hillion K-H, Perthame E, Arroyo MV, Souquere S, Ruigrok K, Dupressoir A, Heidmann T, Montagutelli X, Fournier T, Lecuit M, Schwartz O. 2019. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science 365:176–180. doi: 10.1126/science.aaw7733 [DOI] [PubMed] [Google Scholar]
- 37. Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C, Buchrieser J, Rajah MM, Bishop E, et al. 2021. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 27:917–924. doi: 10.1038/s41591-021-01318-5 [DOI] [PubMed] [Google Scholar]
- 38.Santé Publique France 2021. Enquêtes Flash: Évaluation de La Circulation Des Variants Du SARS-CoV-2 En France. Santé Publique France. Available from: https://www.santepubliquefrance.fr/etudes-et-enquetes/enquetes-flash-evaluation-de-la-circulation-des-variants-du-sars-cov-2-en-france. Retrieved 12 May 2023.
- 39. Planchais C, Fernández I, Bruel T, de Melo GD, Prot M, Beretta M, Guardado-Calvo P, Dufloo J, Molinos-Albert LM, Backovic M, et al. French COVID Cohort Study Group, CORSER Study Group . 2022. Potent human broadly SARS-Cov-2–neutralizing IgA and IgG antibodies effective against Omicron BA.1 and BA.2. J Exp Med 219:e20220638. doi: 10.1084/jem.20220638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bruel T, Hadjadj J, Maes P, Planas D, Seve A, Staropoli I, Guivel-Benhassine F, Porrot F, Bolland WH, Nguyen Y, Casadevall M, Charre C, Péré H, Veyer D, Prot M, Baidaliuk A, Cuypers L, Planchais C, Mouquet H, Baele G, Mouthon L, Hocqueloux L, Simon-Loriere E, André E, Terrier B, Prazuck T, Schwartz O.. 2022. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med 28:1297–1302. [DOI] [PubMed] [Google Scholar]
- 41. Magazine N, Zhang T, Wu Y, McGee MC, Veggiani G, Huang W. 2022. Mutations and evolution of the SARS‐CoV‐2 spike protein. Viruses 14:640. doi: 10.3390/v14030640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinot R, Hubert M, de Melo GD, Lazarini F, Bruel T, Smith N, Levallois S, Larrous F, Fernandes J, Gellenoncourt S, Rigaud S, Gorgette O, Thouvenot C, Trébeau C, Mallet A, Duménil G, Gobaa S, Etournay R, Lledo P-M, Lecuit M, Bourhy H, Duffy D, Michel V, Schwartz O, Chakrabarti LA. 2021. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun 12:4354. doi: 10.1038/s41467-021-24521-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scheuermann SE, Goff K, Rowe LA, Beddingfield BJ, Maness NJ. 2023. Real time analysis of SARS-CoV-2 induced cytolysis reveals distinct variant-specific replication profiles. Viruses 15:1937. doi: 10.3390/v15091937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morrison CB, Edwards CE, Shaffer KM, Araba KC, Wykoff JA, Williams DR, Asakura T, Dang H, Morton LC, Gilmore RC, O’neal WK, Boucher RC, Baric RS, Ehre C. 2022. Proceedings of the National Academy of sciences. doi: 10.1073/pnas [DOI] [PMC free article] [PubMed]
- 45. Ma H, Zhu Z, Lin H, Wang S, Zhang P, Li Y, Li L, Wang J, Zhao Y, Han J. 2021. Pyroptosis of syncytia formed by fusion of SARS-CoV-2 spike and ACE2-expressing cells. Cell Discov 7:73. doi: 10.1038/s41421-021-00310-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Porter AG, Jänicke RU. 1999. Emerging roles of caspase-3 in apoptosis. Cell Death Differ 6:99–104. doi: 10.1038/sj.cdd.4400476 [DOI] [PubMed] [Google Scholar]
- 47. Jiang M, Qi L, Li L, Li Y. 2020. The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer. Cell Death Discov 6:112. doi: 10.1038/s41420-020-00349-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gellenoncourt S, Saunders N, Robinot R, Auguste L, Rajah MM, Kervevan J, Jeger-Madiot R, Staropoli I, Planchais C, Mouquet H, Buchrieser J, Schwartz O, Chakrabarti LA. 2022. The spike-stabilizing D614G mutation interacts with S1/S2 cleavage site mutations to promote the infectious potential of SARS-CoV-2 variants. J Virol 96:e0130122. doi: 10.1128/jvi.01301-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beaudoin-Bussières G, Chen Y, Ullah I, Prévost J, Tolbert WD, Symmes K, Ding S, Benlarbi M, Gong SY, Tauzin A, Gasser R, Chatterjee D, Vézina D, Goyette G, Richard J, Zhou F, Stamatatos L, McGuire AT, Charest H, Roger M, Pozharski E, Kumar P, Mothes W, Uchil PD, Pazgier M, Finzi A. 2022. A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and Synergizes with a nAb to protect mice from lethal SARS-Cov-2 infection. Cell Rep 38:110368. doi: 10.1016/j.celrep.2022.110368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grant MD, Bentley K, Fielding CA, Hatfield KM, Ings DP, Harnum D, Wang E, Stanton R, Holder KA. 2023. Hybrid immunity elicits potent cross-variant ADCC against SARS-CoV-2 through a combination of anti-S1 and S2 antibodies. BioRxiv. doi: 10.1101/2023.03.09.531709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou J, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo S-YC, Rothlauf PW, Bloyet L-M, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D. 2021. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 184:2332–2347. doi: 10.1016/j.cell.2021.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCarthy KR, Rennick LJ, Nambulli S, Robinson-McCarthy LR, Bain WG, Haidar G, Duprex WP. 2021. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 371:1139–1142. doi: 10.1126/science.abf6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, Adhikari UD, Winkler ML, Mueller AA, Hsu TY-T, Desjardins M, Baden LR, Chan BT, Walker BD, Lichterfeld M, Brigl M, Kwon DS, Kanjilal S, Richardson ET, Jonsson AH, Alter G, Barczak AK, Hanage WP, Yu XG, Gaiha GD, Seaman MS, Cernadas M, Li JZ. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beaudoin-Bussières G, Arduini A, Bourassa C, Medjahed H, Gendron-Lepage G, Richard J, Pan Q, Wang Z, Liang C, Finzi A. 2022. SARS-CoV-2 accessory protein ORF8 decreases antibody-dependent cellular cytotoxicity. Viruses 14:1237. doi: 10.3390/v14061237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Groenheit R, Galanis I, Sondén K, Sperk M, Movert E, Bacchus P, Efimova T, Petersson L, Rapp M, Sahlén V, Söderholm S, Valentin Asin K, Zanetti S, Lind Karlberg M, Bråve A, Klingström J, Blom K. 2023. Rapid emergence of Omicron sublineages expressing spike protein R346t. The Lancet Regional Health - Europe 24:100564. doi: 10.1016/j.lanepe.2022.100564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang Q, Li Z, Ho J, Guo Y, Yeh AY, Mohri H, Liu M, Wang M, Yu J, Shah JG, Chang JY, Herbas F, Yin MT, Sobieszczyk ME, Sheng Z, Liu L, Ho DD. 2022. Resistance of SARS-CoV-2 Omicron subvariant BA.4.6 to antibody neutralisation. Lancet Infect Dis 22:1666–1668. doi: 10.1016/S1473-3099(22)00694-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ai J, Wang X, He X, Zhao X, Zhang Y, Jiang Y, Li M, Cui Y, Chen Y, Qiao R, Li L, Yang L, Li Y, Hu Z, Zhang W, Wang P. 2022. Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell Host Microbe 30:1077–1083. doi: 10.1016/j.chom.2022.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wilhelm A, Widera M, Grikscheit K, Toptan T, Schenk B, Pallas C, Metzler M, Kohmer N, Hoehl S, Marschalek R, Herrmann E, Helfritz FA, Wolf T, Goetsch U, Ciesek S.. 2022. Limited neutralisation of the SARS-CoV-2 Omicron subvariants BA.1 and BA.2 by convalescent and vaccine serum and monoclonal antibodies 10.1016/j. [DOI] [PMC free article] [PubMed]
- 59. Cheng SMS, Mok CKP, Leung YWY, Ng SS, Chan KCK, Ko FW, Chen C, Yiu K, Lam BHS, Lau EHY, Chan KKP, Luk LLH, Li JKC, Tsang LCH, Poon LLM, Hui DSC, Peiris M. 2022. Neutralizing antibodies against the SARS-CoV-2 Omicron variant BA.1 following homologous and heterologous Coronavac or BNT162b2 vaccination. Nat Med 28:486–489. doi: 10.1038/s41591-022-01704-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, De Los Santos M, Leon P, Breban MI, Billig K, Yildirim I, Pearson C, Downing R, Gagnon E, Muyombwe A, Razeq J, Campbell M, Ko AI, Omer SB, Grubaugh ND, Vermund SH, Iwasaki A. 2022. Neutralizing antibodies against the SARS-Cov-2 Delta and Omicron variants following heterologous Coronavac plus BNT162b2 booster vaccination. Nat Med 28:481–485. doi: 10.1038/s41591-022-01705-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jessie B, Dobrovolny HM. 2021. The role of syncytia during viral infections. J Theor Biol 525:110749. doi: 10.1016/j.jtbi.2021.110749 [DOI] [PubMed] [Google Scholar]
- 62. Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, Deng Y, Mao L, Su C, Qiang G, Jiang T, Zhao J, Wu G, Song J, Tan W. 2020. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat Commun 11. doi: 10.1038/s41467-020-17796-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kanai Y, Kawagishi T, Sakai Y, Nouda R, Shimojima M, Saijo M, Matsuura Y, Kobayashi T. 2019. Cell-cell fusion induced by reovirus FAST proteins enhances replication and pathogenicity of non-enveloped dsRNA viruses. PLoS Pathog 15:e1007675. doi: 10.1371/journal.ppat.1007675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zeng C, Evans JP, King T, Zheng Y-M, Oltz EM, Whelan SPJ, Saif LJ, Peeples ME, Liu S-L. 2022. SARS-CoV-2 spreads through cell-to-cell transmission. Proc Natl Acad Sci U S A 119:e2111400119. doi: 10.1073/pnas.2111400119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chaudhary S, Yadav RP, Kumar S, Yadav SC. 2023. Ultrastructural study confirms the formation of single and heterotypic syncytial cells in bronchoalveolar fluids of COVID-19 patients. Virol J 20:97. doi: 10.1186/s12985-023-02062-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lista MJ, Winstone H, Wilson HD, Dyer A, Pickering S, Galao RP, De Lorenzo G, Cowton VM, Furnon W, Suarez N, Orton R, Palmarini M, Patel AH, Snell L, Nebbia G, Swanson C, Neil SJD. 2022. The P681H Mutation in the spike glycoprotein of the alpha variant of SARS-CoV-2 escapes IFITM restriction and is necessary for type I interferon resistance. J Virol 96:e0125022. doi: 10.1128/jvi.01250-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Meng B, Abdullahi A, Ferreira I, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, et al. , CITIID-NIHR BioResource COVID-19 Collaboration, Genotype to Phenotype Japan (G2P-Japan) Consortium, Ecuador-COVID19 Consortium . 2022. Altered Tmprss2 usage by SARS-Cov-2 Omicron impacts infectivity and Fusogenicity. Edited by Motozono C., Nasser H., Shimizu R., Kitazato K., Hasebe H., Irie T., Nakagawa S., Wu J., Takahashi M., Fukuhara T., Shimizu K., Tsushima K., Kubo H., Kazuma Y., Nomura R., Horisawa Y., Nagata K., Kawai Y., Yanagida Y., Tashiro Y., Tokunaga K., Ozono S., Kawabata R., and Morizako N.. Nature 603:706–714. doi: 10.1038/s41586-022-04474-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xia S, Zhu Y, Liu M, Lan Q, Xu W, Wu Y, Ying T, Liu S, Shi Z, Jiang S, Lu L. 2020. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol 17:765–767. doi: 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Oliva R, Shaikh AR, Petta A, Vangone A, Cavallo L. 2021. D936Y and other mutations in the fusion core of the Sars-CoV-2 spike protein heptad repeat 1: frequency, geographical distribution, and structural effect. Molecules 26:2622. doi: 10.3390/molecules26092622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Starr TN, Greaney AJ, Hannon WW, Loes AN, Hauser K, Dillen JR, Ferri E, Farrell AG, Dadonaite B, McCallum M, Matreyek KA, Corti D, Veesler D, Snell G, Bloom JD. 2022. Shifting mutational constraints in the SARS-CoV-2 receptor-binding domain during viral evolution. Microbiology. doi: 10.1101/2022.02.24.481899 [DOI] [PMC free article] [PubMed]
- 71. Pascarella S, Bianchi M, Giovanetti M, Benvenuto D, Borsetti A, Cauda R, Cassone A, Ciccozzi M. 2022. The biological properties of the SARS-CoV-2 cameroon variant spike: an intermediate between the alpha and Delta variants. Pathogens 11:814. doi: 10.3390/pathogens11070814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jangra S, Ye C, Rathnasinghe R, Stadlbauer D, Krammer F, Simon V, Martinez-Sobrido L, García-Sastre A, Schotsaert M, Alshammary H, et al. 2021. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe 2:e283–e284. doi: 10.1016/S2666-5247(21)00068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, et al. 2020. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. PNAS. doi: 10.1073/pnas.2009799117/-/DCSupplemental [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional data on the supplemental movies.
Additional data on the hNEC infections.
Additional data on hNEC infections and mutations.
Video microscopy of A549-ACE2 cells infected with indicated SARS-CoV-2 variants at MOI 0.1. Scale bar = 80 μm.
Video microscopy of HEK293T-GFP1-10 cells transfected with indicated SARS-CoV-2 variant spikes and co-cultured with VeroE6-GFP11 cells over 18 hours. Scale bar = 160 µm.