ABSTRACT
The global evolution of SARS-CoV-2 depends in part upon the evolutionary dynamics within individual hosts with varying immune histories. To characterize the within-host evolution of acute SARS-CoV-2 infection, we sequenced saliva and nasal samples collected daily from vaccinated and unvaccinated individuals early during infection. We show that longitudinal sampling facilitates high-confidence genetic variant detection and reveals evolutionary dynamics missed by less-frequent sampling strategies. Within-host dynamics in both unvaccinated and vaccinated individuals appeared largely stochastic; however, in rare cases, minor genetic variants emerged to frequencies sufficient for forward transmission. Finally, we detected significant genetic compartmentalization of viral variants between saliva and nasal swab sample sites in many individuals. Altogether, these data provide a high-resolution profile of within-host SARS-CoV-2 evolutionary dynamics.
IMPORTANCE
We detail the within-host evolutionary dynamics of SARS-CoV-2 during acute infection in 31 individuals using daily longitudinal sampling. We characterized patterns of mutational accumulation for unvaccinated and vaccinated individuals, and observed that temporal variant dynamics in both groups were largely stochastic. Comparison of paired nasal and saliva samples also revealed significant genetic compartmentalization between tissue environments in multiple individuals. Our results demonstrate how selection, genetic drift, and spatial compartmentalization all play important roles in shaping the within-host evolution of SARS-CoV-2 populations during acute infection.
KEYWORDS: SARS-CoV-2, within-host evolution, viruses, genetic compartmentalization
INTRODUCTION
The large-scale sequencing and phylogenetic analyses of clinical samples during the SARS-CoV-2 pandemic captured the global evolutionary dynamics of the virus with unprecedented speed and resolution. However, our understanding of viral evolutionary dynamics within individual infected hosts remains limited. Most studies of SARS-CoV-2 within-host evolution have focused on chronic infections of immunocompromised individuals, as these patients are more amenable to repeated, longitudinal sampling. It has been hypothesized that chronic infections promote the emergence of novel viral variants by providing a combination of prolonged time for replication and relatively weak immune selection that promotes the emergence of variants with increased fitness to high frequency within the host (1–3). Persistent replication within immunocompromised individuals treated with convalescent sera or therapeutic monoclonal antibodies has also been identified as a potential source of antigenically novel variants (4–6).
Previous studies of SARS-CoV-2 within-host evolutionary dynamics during acute infection of immunocompetent hosts detected low within-host diversity in SARS-CoV-2 populations, with most specimens containing 15 or fewer intra-host single-nucleotide variants (iSNVs) (7–10). Studies of household transmission reaffirm that within-host diversity is low and that iSNVs are rarely transmitted between members of a household (7, 8, 10). Altogether, these data suggest that acute infections typically exhibit low overall levels of within-host genetic diversity and that the selection-driven emergence of iSNVs to high frequency during acute infection is likely rare. However, our understanding of within-host evolutionary dynamics has been hampered by the absence of high-resolution time course data within individuals.
The extent to which pre-existing immunity, elicited either through vaccination and/or prior infection, influences the within-host evolution of SARS-CoV-2 is poorly understood. As vaccine-breakthrough infections have become common, it remains unclear whether conditions of partial immunity may create an evolutionary sandbox where moderate immune selection in the absence of rapid clearance can drive the emergence of immune-escape variants (11, 12). Thus, it is important to characterize the extent of immune selection and potential for escape variant emergence during infections of immune-competent individuals at differing stages of vaccination.
To characterize viral evolutionary dynamics during acute SARS-CoV-2 infection, we sequenced longitudinal nasal swab and saliva samples collected from 31 students, faculty, and staff at the University of Illinois at Urbana-Champaign enrolled during the early stages of infection through an on-campus screening program in late 2020 and early 2021 (13). This cohort included 20 unvaccinated individuals with no known prior infection and 11 individuals with some degree of presumed pre-existing immunity to SARS-CoV-2 resulting from vaccination. By taking repeated measures of iSNV frequencies from two sample sites (mid-turbinate nasal swab and saliva) within individuals, we were able to generate high-resolution profiles of iSNV dynamics between tissue compartments and across time. Our results demonstrate that selection, genetic drift, and spatial compartmentalization all play important roles in shaping the within-host evolution of SARS-CoV-2 populations.
RESULTS
Sample collection
During the 2020–2021 school year, all students, faculty, and staff on the University of Illinois at Urbana-Champaign campus were required to undergo saliva-based PCR testing for SARS-CoV-2 at least twice a week (13). We enrolled individuals who were either (i) within 24 hours of their first positive test result or (ii) within 5 days of exposure to someone else who tested positive. Daily saliva samples and nasal swabs were collected from each enrolled participant for up to 14 days. Sample collection for individuals included in this study spanned from December 2020 to April 2021, a period which captures the early spread of the alpha and gamma variants in the United States. Viral lineages for each sample are reported at github.com/BROOKELAB/SARS-CoV-2-within-host-evolution (14).
The cohort of enrolled individuals was predominantly young, with a median age of 22 and an age range of 18–59. The cohort consisted of 17 male and 14 female participants, and the majority of participants were non-Hispanic white. Details on the dynamics of viral shedding in this cohort have been published previously (15–17). Each participant self-reported vaccination and prior infection status.
Optimization and validation of saliva sample sequencing protocol
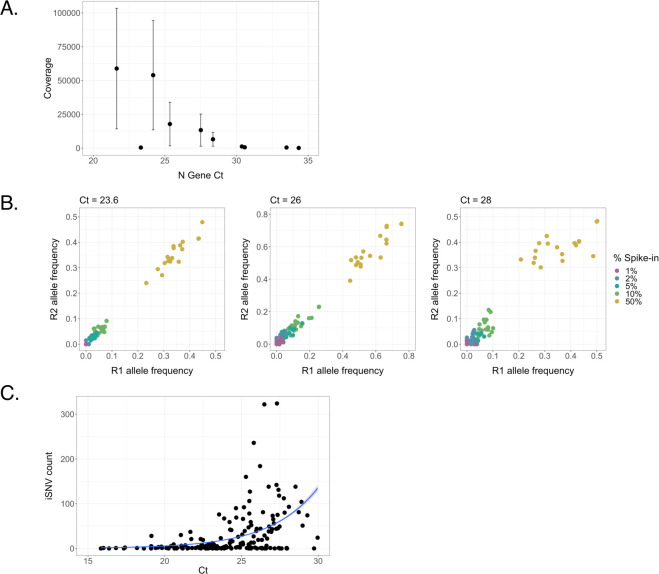
The saliva-based reverse transcription quantitative PCR (RTqPCR) assay used in this study involves a 30-minute treatment at 95°C which partially degrades the viral RNA present in the sample and could potentially compromise sequencing quality (13). To address this concern and determine whether saliva cycle threshold (Ct) values are predictive of sequencing data quality, we examined sequencing depth across samples with a range of Ct values. Over a set of 10 samples that spanned a Ct range of 21.6 to 34.3, we observed a clear negative correlation between N gene Ct value and coverage depth (Pearson correlation, r = −0.702, P = 0.02359) (Fig. 1A). For Ct values below 28, we generally obtained average per-nucleotide read depths of over >10,000 reads, indicating that high-quality sequence data can be obtained from heat-treated samples.
Fig 1.
Relationship between saliva sample Ct values and sequence quality. (A) Linear regression between Ct values of nucleocapsid (N) gene and mean sequence coverage depth. Error bars represent standard deviation. (B) Frequencies of characteristic B.1.1.7 SNPs at Ct values of 23.6, 26, and 28. B.1.1.7 RNA was spiked into B.1.2 RNA at final percentages ranging from 1% to 50% and divided between two replicates (R1 and R2). (C) Relationship between Ct of the SARS-CoV-2 nucleocapsid (N) gene and total iSNV count of the associated sample (Poisson regression, regression coefficient = 0.321, P < 0.001).
We next evaluated the relationship between sample Ct and the reliability of iSNV detection in saliva samples. We generated control samples in which RNA isolated from a B.1.1.7 (alpha) lineage sample was spiked into a B.1.2 lineage sample at defined frequencies of 50%, 10%, 5%, 2%, and 1%. We normalized both B.1.1.7 and B.1.2 samples to Ct values of 23.6, 26, or 28 based on Ct values collected prior to mixing. Spike-ins were then divided into replicate samples and deep sequenced. We compared the measured frequencies of the 17 characteristic B.1.1.7 single nucleotide polymorphisms (SNPs) and indels between technical replicates (Fig. 1B) and found that overall correlation between technical replicates across groups was stronger in samples with a Ct of 23.6 (Pearson correlation, r = 0.996, P < 0.001) or 26 (Pearson correlation, r = 0.992, P < 0.001) than in samples with a Ct of 28; however, the correlation between the Ct = 28 samples was still high (Pearson correlation, r = 0.967, P < 0.001). Across the entire cohort, the number of iSNVs per sample was correlated with the Ct value of the sample (Poisson regression, reg. coef. = 0.321, P < 0.001), further demonstrating that high Ct values can contribute to noise in iSNV detection (Fig. 1C). Based on these results, we used a Ct cutoff or a cycle number (CN) cutoff of <28 in downstream analyses.
To determine a suitable frequency threshold for iSNV detection, we compared frequencies of non-B.1.1.7 iSNVs between technical replicates, using a pooled data set consisting of all spike-in dilutions and all Ct values (Fig. S1). We found that the number of iSNVs present in one replicate but absent in the other (suggestive of base-calling error) declined sharply at an iSNV-calling threshold of 0.03. Therefore, we selected a frequency threshold of 0.03 for initial variant calling.
Identification of “shared” iSNVs
We selected 20 unvaccinated participants who each had multiple saliva samples with Ct <28 spanning several days for further study. We also selected 11 study participants who were vaccinated (fully or partially; definitions in Materials and Methods) and had at least one saliva sample with Ct <30. We chose a higher Ct threshold for vaccinated participants because Ct values were overall higher in this group (unvaccinated mean = 23.8, vaccinated mean = 25.1, Welch two-sample t-test, P = 0.0111) (16, 17). One limitation of this study is that we were unable to empirically measure immune responses in these participants.
We next focused our analysis on iSNVs that appeared within the same individual across multiple samples (“shared iSNVs”). As we expect most sequencing artifacts to be randomly distributed, false variant calls are unlikely to arise in the same genomic site across multiple days. This approach is analogous to the sequencing of paired technical replicates which is commonly used to eliminate false-positive iSNV calls for single samples when repeated sampling is not possible. In our unvaccinated cohort, we also further restricted the pool of shared iSNVs to include only the iSNVs that were present in multiple samples with Ct <26. Because of the small size and higher average Ct values of our vaccinated cohort, we did not impose the same Ct <26 threshold on vaccinee samples during shared variant calling.
To assess whether variation in Ct values among the samples affected the confidence of variant calling for shared iSNVs, we examined the relationship between Ct values and shared iSNV counts in unvaccinated individuals. We observed significant but minor correlations between Ct and shared iSNVs in saliva samples at Ct thresholds of 26 and 24 (Fig. S2A and B). However, due to the minimal association between these variables and our observation that stricter Ct cutoffs did not necessarily reduce the association (the highest significant comparison was observed at a threshold of Ct <23, and a threshold of Ct <28 yielded no correlation), we determined that the link between Ct and shared iSNVs was unlikely to greatly impact the integrity of the unvaccinated saliva sample data set. We observed a minor negative correlation between shared iSNVs and Ct in our vaccinee data set (reg. coef. = −0.0608, P = 0.00919) (Fig. S2C) and no correlation between shared iSNVs and CN values in our nasal sample data set (Fig. S2D).
Because samples were sequenced across multiple runs, we also ran controls to affirm that there was no significant variation in shared iSNV calling between sequencing runs. We re-sequenced samples collected from a single participant (444332) across multiple days (allowing us to identify shared iSNVs for both sequencing runs) and observed that shared iSNV frequencies were strongly correlated between runs (Pearson correlation, r = 0.975, P = 0.0246) (Table 1).
TABLE 1.
Between-run shared iSNV frequencies within a single individual
| SNP | Day | RUN1 | RUN2 |
|---|---|---|---|
| C20178T | 1 | 0.077232 | 0.056249 |
| T9481C | 3 | 0.052526 | 0.054462 |
| C20178T | 3 | 0.157866 | 0.129242 |
| T9481C | 5 | 0.161942 | 0.150735 |
To further account for the role of viral genome load in influencing sequencing fidelity, we re-sequenced samples from various participants, with a range of Ct values from 21.34 to 27.94. Since these samples represented single time points from different participants, shared iSNVs could not be detected from the re-sequenced data alone—therefore, we limited the data set to mutations that had been called as shared iSNVs in the original run. We found that shared iSNV frequencies were highly correlated between runs (Pearson correlation, r = 0.999, P < 0.001) (Table 2).
TABLE 2.
Between-run shared iSNV frequencies across a range of Ct values
| Participant | Day | Ct | SNP | RUN1 | RUN2 |
|---|---|---|---|---|---|
| 432870 | 1 | 21.34 | A9085G | 0.998796 | 0.999204 |
| 432870 | 1 | 21.34 | C11956T | 0.999562 | 0.999367 |
| 432870 | 1 | 21.34 | C12623T | 0.0147206 | 0.0109745 |
| 432870 | 1 | 21.34 | C14805T | 0.999796 | 0.999094 |
| 432870 | 1 | 21.34 | A18424G | 0.999593 | 0.999763 |
| 432870 | 1 | 21.34 | C21304T | 0.999543 | 0.999251 |
| 432870 | 1 | 21.34 | G24933T | 0.10782 | 0.0896666 |
| 432870 | 1 | 21.34 | C28869T | 0.997505 | 0.9992 |
| 444332 | 2 | 23.93 | T4183C | 0.999438 | 0.9989 |
| 444332 | 2 | 23.93 | G25907T | 1 | 0.999919 |
| 449650 | 1 | 24.9 | C11572T | 0.9985 | 1 |
| 449650 | 1 | 24.9 | G25500A | 0.999697 | 0.999744 |
| 435786 | 3 | 25.8 | C5416T | 0.289822 | 0.331139 |
| 435805 | 9 | 27.94 | A23583T | 1 | 1 |
To account for inconsistencies in iSNV frequency reporting originating from variability in template sampling during sequencing, we calculated standard error for iSNV frequency over a range of genome loads and frequencies of detection. We used Ct (saliva samples) or CN (nasal samples) values to estimate the viral genome load for each sample, as described in Ke et al. (16), and then calculated expected error for iSNVs from frequencies of 0.0–1.0, using a range of Ct or CN values spanning 22–30 (Fig. S3). We found that the expected error estimations were low in both saliva and nasal samples (although about one order of magnitude higher in nasal samples). For instance, at a Ct threshold of 28, the expected error in frequency estimation is ≤0.00618 [a 95% confidence interval (CI) of ±0.0121], and at a CN threshold of 28, the expected error is ≤0.0334 (a 95% confidence interval of ±0.0655).
Analysis of within-host diversity
We next examined the diversity within and between individual saliva samples, focusing on iSNVs and short insertions/deletions (indels) present at frequencies between 3% and 97%, with coverage depths of >1,000 reads. To further minimize the potential for false-positive variant calling, we filtered our data set to remove common sequencing artifacts that have previously been described (18), as well as iSNVs that appeared across multiple different individuals within a cohort but were not associated with circulating viral variants present in our cohort.
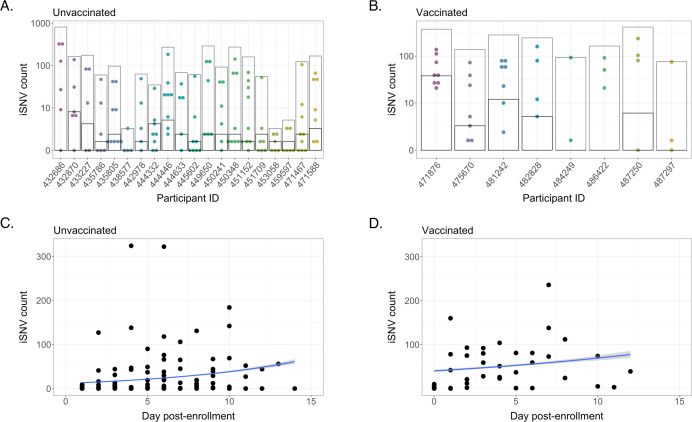
The numbers of iSNVs that fit these criteria varied substantially between samples, generally spanning several orders of magnitude at different points during infection (Fig. 2A and B). However, when limiting our analyses to shared iSNVs only, the numbers of shared iSNVs were similar between participants—averaging 2.35 shared iSNVs per individual, which aligns with previous assessments that within-host diversity is low during acute infection (7–9, 19). Shared iSNV counts were lower in unvaccinated participants than within the vaccinated cohort, with average variant counts of 1.33 and 5.95, respectively (fixed-effect Poisson model, P < 0.001) (Fig. 2A and B). However, the higher observed iSNV counts among vaccinees may be attributable to the less stringent Ct and sample coverage filtering applied to the data set as well as the small size of the vaccinated cohort.
Fig 2.
Intra-host single nucleotide variant (iSNV) diversity compared between samples and individuals. (A) Total iSNV counts for each sample from each unvaccinated participant. Light gray boxes indicate total discrete iSNV count for all samples, and horizontal black lines indicate number of shared iSNVs for each participant. (B) iSNV counts for vaccinated participants. (C) iSNV counts for individual samples from unvaccinated participants as a function of number of days post enrollment (Poisson regression, reg. coef. = 0.117, P < 0.001). (D) iSNV counts for individual samples from vaccinated participants as a function of number of days post enrollment (Poisson regression, reg. coef. = 0.0546, P < 0.001).
We then examined the accumulation of iSNVs over the course of time (we did not restrict the analysis to only shared iSNVs, to avoid violating assumptions of independence between data points). We found only minor correlations between iSNV counts and date of sampling in unvaccinated individuals (reg. coef. = 0.117, P < 0.001) and vaccinated individuals (reg. coef. = 0.0546, P < 0.001) (Fig. 2C and D), and this effect may be due in part to increasing Ct values as infections wane. These data indicate similar overall levels of within-host diversity and mutational accumulation over time in unvaccinated and vaccinated individuals.
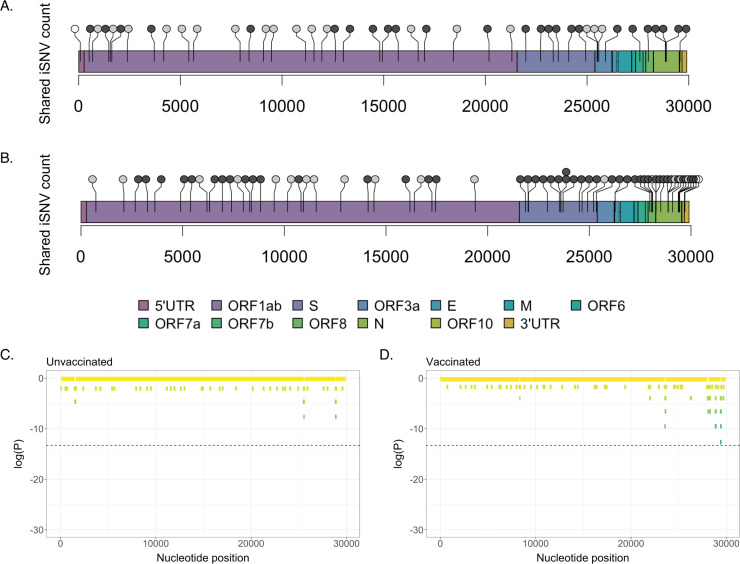
To test whether mutations accumulated at different sites in unvaccinated versus vaccinated individuals, we then plotted the genomic positions of shared iSNVs for each cohort (Fig. 3A and B).
Fig 3.
Locations of shared iSNVs across the SARS-CoV-2 genome. Genome locations of shared iSNVs detected in unvaccinated (A) and vaccinated (B) participants. Number of dots at a locus indicates number of participants in which the shared iSNV was detected. Light gray dots indicate synonymous mutations, dark gray dots indicate nonsynonymous mutations, and white dots indicate UTR mutations. (C) Log(P-values) for shared iSNV counts within 100-nt genomic windows, based on a Poisson distribution derived from the average shared iSNV count for all genomic windows. Plot shows shared iSNV counts in unvaccinated individuals. Dashed line marks significance threshold of 1.68e-06. (D) Log(P-values) for shared iSNV counts within 100-nt genomic windows, in vaccinated individuals.
For our vaccinated cohort, since three participants only had a single sample timepoint and three others had no shared iSNVs, we excluded six individuals from this analysis. To test whether shared iSNVs were unevenly distributed along the viral genome, we used a sliding genomic window of 100 nucleotides to determine the average number of shared iSNVs per 100 nt. We then searched for windows where iSNVs were significantly enriched, relative to what would be expected given a Poisson distribution. There were no significant hotspots of mutation in either the unvaccinated or the vaccinated data set (Fig. 3C and D). We did note that shared iSNVs in the nucleocapsid (N) gene appeared enriched in vaccinated participants compared to unvaccinated individuals—they made up 20.3% of shared variants in vaccinees but only 6.82% of shared variants in unvaccinated participants (Fisher’s exact test, P = 0.0592). However, this enrichment was not statistically significant and was largely driven by samples from one vaccinated individual (471876), who had 10 shared iSNVs in the N gene. While there appear to be certain differences in the genome-wide pattern of mutation accumulation of between vaccinated versus unvaccinated individuals, these differences may be driven by random variation rather than the effects of distinct evolutionary pressures.
Compartmentalization between tissue environments
Previous studies revealed that SARS-CoV-2 replication dynamics can be highly discordant between saliva and nasal swab samples, suggesting strong compartmentalization of virus between different anatomical sites (16, 17, 20, 21). To directly evaluate the extent of compartmentalization between nasal and saliva-associated tissue sites, we compared iSNV frequencies between paired saliva and nasal swab samples collected over the course of infection in 12 individuals from which we were able to generate high-quality sequence data for both saliva and nasal samples.
As in our analysis of saliva samples, we set a frequency threshold of 0.03 and a per-nucleotide depth threshold of 1,000 reads for variant calling in nasal swab samples. Samples with mean genomic coverage depths lower than 1,000 reads per nucleotide were also excluded from the data set, and we once again restricted our analysis to iSNVs present across multiple low-Ct or low-CN samples. To focus on sub-fixation genetic diversity, we only called variants that appeared at frequencies below 0.97 in at least two samples (across all samples from both data sets). This approach was adopted to include variants that were fixed in one compartment but at sub-fixation frequencies in the other.
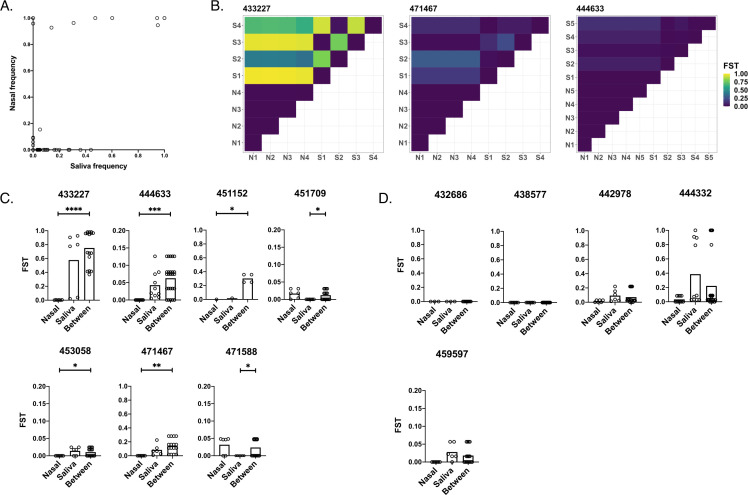
To evaluate genetic differences between viral populations sampled at the two tissue sites, we first simply compared the frequencies of shared iSNVs in saliva versus nasal swab samples (Fig. 4A). In the absence of compartmentalization, we would expect the frequencies of these iSNVs to be highly correlated between sample sites. We observed a correlation of r = 0.615 (P < 0.001) between iSNV frequencies in saliva and nasal samples and observed that many iSNVs were present in one compartment but absent in the other, especially in the case of sub-consensus iSNVs. We also detected fewer shared iSNVs overall in nasal samples (mean = 0.317) than in saliva samples (mean = 0.917) (fixed-effect Poisson model, P < 0.001).
Fig 4.
Quantification of genetic compartmentalization of virus between sample sites. (A) Comparison of iSNV frequencies between matched samples in saliva and nasal environments (Pearson correlation, r = 0.615, P < 0.001). (B) Representative heatmaps exemplifying compartmentalization. Maps show FST values between pairs of samples from nasal (“N”) and/or saliva (“S”) environments (numbered by order of sampling). (C) Participants exhibiting compartmentalization (one set of within-environment FST values is lower than between-environment FST values). (D) Participants exhibiting no significant compartmentalization (neither set of within-environment FST values is lower than between-environment FST values). Scales range from 0 to 1 or 0 to 0.2 depending on the spread of the data. Asterisks indicate levels of significance (*P < .05, **P < .01, ***P < .001, ****P < .0001). P-values are derived from Monte Carlo permutation tests.
To quantify the extent of compartmentalization more precisely, we calculated pairwise fixation indices (FST) within and between environments (Fig. 4B through D). The fixation index measures the ratio of allele frequency variation between sub-populations versus the variation in the total population. FST values range from 0 to 1, and values closer to one indicate higher levels of variation between populations. To account for variation in iSNV calling between sequencing runs, we calculated a threshold FST value of 0.00128 using our between-run sequencing controls (Tables 1 and 2). Any calculated FST value below this threshold was set to 0 to control for the possibility that the observed variation was due to variation between sequencing runs instead of true variation between samples.
We found that the between-environment FST values were significantly higher than the within-nasal FST values in 5 out of 12 individuals (and overall), reflecting a significant degree of genetic compartmentalization. In 2 out of 12 individuals (but not overall), the between-environment FST values were significantly higher than the within-saliva FST values (Fig. 4C), again indicative of genetic compartmentalization between tissue compartments. For some of these participants, there existed a relatively minor disparity in within-environment versus between-environment FST values (see participants 444633, 451709, 453058, and 471588), while other individuals exhibited more extreme levels of variation. We also observed that compartmentalization was driven in part by lowered genetic diversity in the nasal environment, with 5 out of 12 participants exhibiting significantly higher FST values in saliva samples than in nasal samples, and only two participants exhibiting significantly higher nasal FSTs than saliva FSTs. Overall, FST values within paired nasal samples were significantly lower than FST values in saliva samples (nasal mean = 0.0115, saliva mean = 0.144, P = 0.0022). This pattern aligns with our observation of generally low mutational diversity in nasal samples.
Several study participants showed no difference in between-environment versus within-environment variation, consistent with the absence of significant compartmentalization and suggesting that tissue compartmentalization is not a uniform feature of all SARS-CoV-2 infections (Fig. 4D). However, two out of the five participants who fell in this final category had no detectable iSNVs in any saliva or nasal samples—thus, these results can be seen as a failure to detect evidence of compartmentalization rather than clear evidence for the absence of compartmentalization. Finally, we observed no instances where within-environment FST values were significantly higher than between-environment FST values (Fig. 4). Our data suggest that a significant degree of genetic compartmentalization exists between tissue environments in some (7/12), but not all, participants examined.
Within-host evolutionary dynamics
Our dense longitudinal sampling allowed us to examine the evolutionary dynamics of SARS-CoV-2 populations over the course of acute infection. To look for signs of potential positive selection acting on specific sites in the viral genome, we examined changes in the frequencies of shared iSNVs over time. We plotted all detected instances of these shared iSNVs, even if they fell outside of the frequency range of 3% to 97% or fell below our chosen depth threshold of 1,000 reads for certain timepoints.
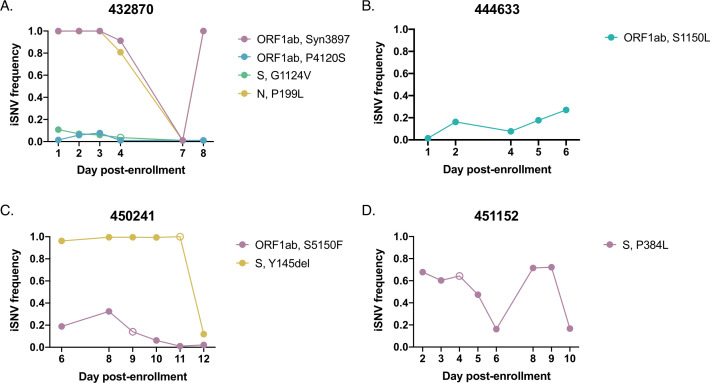
Overall, the longitudinal dynamics of many iSNVs in both unvaccinated and vaccinated individuals appeared highly stochastic, consistent with a dominant role for genetic drift rather than strong selection (Fig. 5 and 6; Fig. S4 to S6). Many iSNVs detected at high frequency in one or more samples fell below the limit of detection (LOD) at other timepoints during the same infection. In one individual (432870), two iSNVs that were initially detected at fixation fell to intermediate frequencies on day 4 of sampling and to undetectable frequencies on day 7, before eventually resurging to high frequencies on day 8 (Fig. 5A). Two sub-consensus mutations (ORF1ab:P4120S and S:G1124V) also fell below the limit of detection on day 7 and did not reemerge. It is unlikely that the observed drops in frequency are due to low sequencing coverage, as all samples considered had a mean genomic sequencing depth of >1,000 reads per nucleotide. An examination of the sites that appeared to revert to wild-type on day 7 revealed that the depth of coverage was greater than 1,000 reads at each locus, with the exception of nucleotide site 24933 (codon site S:1124), which had a depth of 683 reads. We also do not believe that this effect can be explained by template resampling—given the Ct of the sample in question (26.5), the expected error in frequency estimation is less than 0.004 (a 95% CI of ±0.0121) (Fig. S3A).
Fig 5.
iSNV dynamics over time in saliva from unvaccinated individuals. Frequency tracking of selected iSNVs from unvaccinated participants (A) 432870, (B) 444633, (C) 450241, and (D) 451152. Unfilled points mark iSNVs with read depths below the threshold of 1,000 reads.
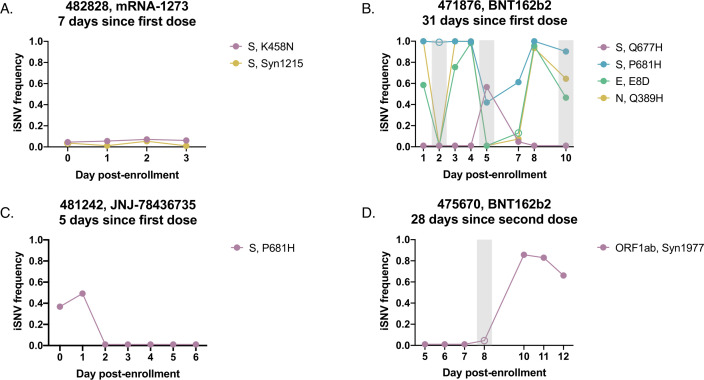
Fig 6.
iSNV dynamics over time in saliva from vaccinated individuals. Frequency tracking of selected iSNVs from vaccinated participants (A) 482828 (newly vaccinated), (B) 471876 (partially vaccinated), (C) 481242 (newly vaccinated), and (D) 475670 (fully vaccinated). Unfilled points mark iSNVs with read depths below the threshold of 1,000 reads. Gray boxes mark samples with mean per-nucleotide coverages below 1,000 reads. Panel headings indicate vaccine received and time between enrollment and last vaccine dose.
To confirm that these dramatic changes in frequency could not be explained by cross-contamination or inadvertent swapping of samples during preparation, we calculated correlations between the iSNV profile of the sample in question (participant 432870, day 7) and the iSNV profiles of each other saliva sample (including samples in the vaccinee data set) (Fig. S7A). We then set a sample correlation threshold equal to the minimum correlation observed between identical samples across sequencing replicates (r = 0.660). With this threshold in place, we found that the only significant correlation resulted from a comparison of the sample against itself—indicating that the sample does not bear a significant resemblance to any other sample in the data set and that cross-contamination or a sample mix-up is unlikely explanation for the observed dynamics.
We then performed the same contamination analysis for three other samples that displayed similar jumps in iSNV frequencies (Fig. S4 and S7B through D). One of these samples (participant 444332, day 7) was only significantly correlated with itself (Fig. S7B). The other samples (participant 433227, day 5, and participant 433227, day 7) were correlated with themselves and with other samples from the same participant but also exhibited correlations with one or more samples from other individuals (Fig. S7C and D). However, the observed correlations between the samples of interest and samples from different participants were weaker than the correlations between the samples of interest and other samples originating from the same individual.
Based on these findings, we concluded that iSNVs observed across the course of infection sometimes exhibit extreme shifts in frequency that cannot be explained by low sample quality, template resampling, or contamination. Interestingly, we observed multiple instances of variants rising from initially undetectable frequencies to high frequencies (over 0.5) before falling back below the limit of detection (as with S:A846G in participant 433227 and ORF1ab:Syn1717 in participant 435786) (Fig. S4). The emergence of a novel variant to a frequency sufficient for forward transmission (even for only a single day of sampling) suggests that acute infections may, in rare cases, support the emergence of new viral variants at the host population level.
While most recurrent iSNVs did not appear to be under strong selection in unvaccinated individuals, we observed certain variants that exhibited consistent patterns of emergence or decline over the course of acute infection that could be indicative of selection. An ORF1ab:S1150L substitution exhibited dynamics suggestive of positive selection, gradually rising from a frequency of 0.01 to 0.270 over the course of infection (Fig. 5B). However, this substitution is observed only sporadically across the global SARS-CoV-2 tree, suggesting the absence of positive selection on the allele at the between-host scale (Fig. S8).
We also observed an example of dynamics consistent with negative selection, in which an iSNV (ORF1ab:S5150F) initially present at a relatively high frequency (>0.3) gradually declined until falling below the LOD (Fig. 5C). The same individual also saw an alpha-associated deletion in spike drop in frequency on the final day of sampling, but this drop may be merely representative of the same iSNV stochasticity observed in other infections (Fig. 5A; Fig. S4). We observed several other spike mutations in our unvaccinated cohort (Fig. 5D; Fig. S4); however, none of them exhibited dynamics suggestive of directional selection.
Overall, these data suggest that in a subset of acute infections, selection may drive the emergence of specific iSNVs, but not generally to high enough frequencies to reliably transmit given the narrow transmission bottlenecks observed across multiple studies (7, 8, 22).
We did not observe any sweeps of antigenically significant spike substitutions in our small cohort of vaccinated participants, suggesting that spike-based vaccination does not impose strong immune selection on the viral populations sampled over the course of acute infection (Fig. 6; Fig. S5). The only recurrent, antigenically significant spike iSNV that we observed within our vaccinated cohort resulted in a K458N substitution in the receptor binding domain, a residue that has been previously associated with monoclonal antibody escape (Fig. 6A) (23, 24). This iSNV remained steady at a low frequency between 3% and 10% over the course of infection however, suggesting the absence of a strong selective advantage and low probability of forward transmission.
Outside of the established antigenic sites, we observed interesting dynamics near the S1/S2 cleavage site in vaccinated individuals. In participant 471876, S:P681H was at or near fixation over the first 4 days of sampling, dropped in frequency on days 5 through 7, and then returned to near fixation at day 8 (Fig. 6B). On the 2 days where S:P681H dropped below 90%, a nearby spike substitution, Q677H, emerged to high frequency before dropping back below the LOD on days 8 and 9. The co-occurrence of the dip in S:P681H frequency with the emergence of S:Q677H and subsequent reversal are consistent with competition between these two substitutions. Both substitutions have proliferated at the global scale, suggesting that in some cases, within-host and global dynamics may be aligned (25–28). Critically, S:Q677H peaked at a time (day 5 post enrollment) when this participant was still viral culture positive [see Fig. 1 in reference (17)], indicating the potential for this de novo variant to be successfully transmitted.
Interestingly, S:P681H was also observed at intermediate frequencies in participant 481242 on days 1 and 2 post enrollment but dropped below the LOD by day 3 and remained undetectable in later timepoints, suggesting a sweep by an S:P681 revertant (Fig. 6C). The within-host transience of a mutation associated with increased fitness at the global scale highlights how within-host evolutionary trends can diverge from global trends.
Overall, we did not detect obvious signs of antibody-mediated immune selection within vaccinated individuals and found that iSNV frequencies often appeared to vacillate stochastically, similarly to in unvaccinated individuals. In participant 471876, several iSNVs (including E:E8D and N:Q389H) fluctuated between fixation and frequencies below the LOD over the course of infection (Fig. 6B). Additionally, in participant 475670, we observed a synonymous mutation (ORF1ab:Syn1977) rapidly rise to high frequencies (Fig. 6D). However, these fluctuations may result from the more relaxed quality filtering applied to samples in the vaccinee data set.
Despite the highly variable frequencies of iSNVs detected across the course of infection in vaccinated individuals, it is also difficult to ascribe all observed dynamics entirely to genetic drift. Our observations of wild-type reversion and competition between iSNVs at the S1/S2 cleavage site (Fig. 6B and C) suggest that selection may drive the within-host fluctuations of iSNVs at non-antigenic sites during acute infection in some individuals.
Finally, in keeping with our compartmentalization analysis, we found that frequencies of shared iSNVs in nasal swab samples over the course of infection often varied from the dynamics observed in saliva samples (Fig. 5; Fig. S6). These variants from paired nasal and saliva samples differed both in frequency and in presence/absence. Following the trend observed in saliva samples, iSNV dynamics in nasal swab samples appeared largely stochastic.
DISCUSSION
By analyzing longitudinal samples collected daily over the course of acute infection, we captured a high-resolution temporal profile of SARS-CoV-2 within-host dynamics in humans. In general, we observed little evidence of strong selection acting on within-host viral populations in our cohort, consistent with previous reports (7–9). This was true even within our group of vaccinated individuals, mirroring a previous study on influenza virus that failed to detect the emergence of antigenic variants during infection of immune individuals (29). However, a key limitation of this study is that we were not able to confirm immune histories of our participants using serology. Thus, our only indicator for the presence/absence of a SARS-CoV-2-specific antibody response in our cohort is self-reporting of vaccination or prior infection detected through frequent on-campus PCR testing.
While signs of strong positive selection were rare in this cohort, we did identify two nonsynonymous substitutions (S:Q677H and S:A846G) that emerged from below the limit of detection to high frequency over the course of infection. Importantly, S:Q677H emerged to 56.5% frequency on a day when the associated study participant had detectable infectious virus in a nasal swab (17), suggesting the potential for this iSNV to be transmitted forward. Substitutions at S:Q677 (including Q677H) have independently emerged in multiple viral sub-lineages around the world, supporting that mutations at this site can be advantageous. We also observed signs of competition between S:Q677H and S:P681H within the same individual, with S:Q677H briefly emerging to a high frequency on a day at which the initially near-fixation S:P681H dipped in frequency. However, the observed reversion to an S:P681H-only genotype after day 7 suggests that the selective advantage conferred by S:P681H is greater than that of S:Q677H. This fitness advantage is supported globally by the more widespread proliferation of S:P681H-containing lineages in comparison to S:Q677H (30). Our data demonstrate how, in rare cases, de novo variants can emerge within individuals to frequencies sufficient for transmission during acute infection.
In addition to the limited number of iSNVs that emerged to high frequency within single individuals, we observed several iSNVs that arose above background for multiple days of infection. Mapping the genomic locations of these mutations revealed a high density of N gene substitutions (although this effect was not statistically significant, and was largely driven by one individual). The detected substitutions include N:R203K and N:G204R, which have been shown to increase the relative fitness of the virus, potentially through increased phosphorylation of the nucleocapsid (31). These substitutions have also been associated with the transcription of an alternate sub-genomic mRNA with anti-interferon activity (32). While spike protein substitutions are clearly the primary drivers of SARS-CoV-2 adaptation to humans, our results are also consistent with previous data suggesting an important role for the N gene during human adaptation.
Variant dynamics in multiple participants exhibited extreme fluctuations where iSNVs at or near fixation abruptly fell below the limit of detection, only to return to high frequencies days later. Given the abruptness of these fluctuations, it is doubtful that they were selection driven. However, they could potentially be explained if there is a significant degree of spatial structuring of within-host viral genetic diversity, as has recently been described for influenza virus (33). Spatial structuring could promote more extreme, drift-driven fluctuations in sampled iSNV frequencies due to bottleneck effects (33–35) and could lead to incomplete sampling of total within-host genetic diversity. Our tracking of variant dynamics also reaffirms previously described variability in iSNV frequency. Previous work on SARS-CoV-2 and influenza virus has revealed extreme shifts in variant frequencies between pairs of samples collected at different time points (or even twice within the same day) (9, 36). Our results build upon these findings with a high-resolution portrait of daily variant dynamics and suggest that large, seemingly stochastic jumps in iSNV frequencies may be a feature of many acute viral infections.
We cannot formally rule out the possibility that these fluctuations are artifactual, potentially arising from poor-quality sampling of the viral population. We think that this explanation is unlikely, however, due to the Ct value and mean coverage thresholds we used for including samples in our analyses. Regardless, either explanation further emphasizes the advantages of longitudinal sampling, as single-timepoint snapshots of viral populations can present misleading views of the within-host landscape.
Supporting the possibility that stochastic within-host SNV dynamics may partially result from spatial structuring, we observed significant compartmentalization between the oral and nasal environments over the course of SARS-CoV-2 infection in a subset of individuals. iSNVs varied in frequency between the two environments, a finding that builds on previous observations that peaks in viral shedding are often offset by several days between saliva and nasal environments (16) and that shedding is sometimes limited to the saliva compartment in immune individuals (17). These findings also align with previous studies that have described compartmentalization of influenza populations (due to within-host bottlenecks) (33) and intra-host adaptation to different tissue sites by polioviruses (37). Our results indicate that the mutational profile of SARS-CoV-2 can differ between tissue environments and reaffirm that sampling of a single tissue site may not provide a complete view of viral population diversity within a host.
A clear advantage of repeated longitudinal sampling is that it allows for higher confidence variant calling compared with single-timepoint sampling. Across individual samples, we measured iSNV counts ranging from zero to several hundred and found that these values could shift rapidly within an individual over short periods of time. However, the number of variants shared across multiple days was remained relatively low in both unvaccinated and vaccinated individuals, with both groups exhibiting shared iSNV counts that align with previous assessments of within-host diversity (7, 8, 10, 19).
Altogether, our results suggest that viral evolution is largely driven by genetic drift during acute infections but that, on rare occasions, selection can drive the emergence of iSNVs capable of forward transmission. Furthermore, our detection of iSNVs that have been successful (or not) at the global scale indicates areas of alignment and discordance between within-host and between-host selective pressures and thus help shed light on the forces that shape global patterns of SARS-CoV-2 evolution.
MATERIALS AND METHODS
Sample collection
To monitor on-campus COVID cases, students and employees at the University of Illinois at Urbana-Champaign were required to submit biweekly saliva samples for reverse transcription quantitative PCR (RTqPCR) testing. Individuals who tested positive were given the option to enroll in a longitudinal sample collection study within 1 day of receiving a positive result. Additionally, individuals who had been in close contact with a positive case were eligible to enroll in the same study within 5 days of their exposure. Enrolled participants then provided saliva samples and mid-turbinate nasal swabs for 14 days after the date of their first positive test [this collection protocol is described in detail in reference (16)]. Within 12 hours of collection, RTqPCR was performed on heat-inactivated saliva samples to assess viral load, as described in Ranoa et al. (38). Nasal swab samples were stored in viral transport media at −80°C and shipped to Johns Hopkins University for RNA extraction.
Participants were designated as fully vaccinated if they had been infected at least 14 days after receiving a single-dose vaccine (JNJ-78436735) or a second dose of a two-dose vaccine (BNT162b2 or mRNA-1273). If at least 14 days had passed since receiving the first dose of a two-dose vaccine, participants were designated as partially vaccinated, and if less than 14 days had passed since receiving a dose of any vaccine, participants were designated as newly vaccinated. Study enrollment was concluded prior to the approval of vaccine boosters.
Participant selection
After RTqPCR analysis, unvaccinated participants with fewer than three saliva samples under the Ct cutoff value of 28 were filtered out of the data set. The remaining participants were sorted and ranked based on their number of quality samples and the range of dates covered by these samples (using a metric that multiplied the two values). The top 20 participants were selected for further analysis and further filtered to remove samples with mean genomic coverage depths under 1,000 reads per nucleotide. All vaccinated participants with saliva samples under a Ct value of 30 were retained, which resulted in a study group of 11 individuals. Due to the small number of samples in the vaccinated cohort, an overall mean genomic coverage cutoff was not applied; however, further analysis was limited to individual iSNVs present above 1,000 reads. Nasal samples from 12 unvaccinated individuals were chosen to evaluate environmental differences between the oral and nasal cavities. Only nasal samples with mean genomic coverage values over 1,000 reads per nucleotide were included in the analysis.
RNA extraction (saliva samples)
A volume of 140 μL from each heat-inactivated saliva sample was processed using the QIAamp viral RNA mini kit. Samples were stored at −80°C until use.
RNA extraction (nasal samples)
Mid-turbinate nasal swab specimen aliquots were maintained at −80°C prior to use. RNA was extracted from 300 µL of clinical specimen using the Chemagic 360 system (Perkin Elmer) according to the manufacturer’s specifications. RNA was eluted with 60 µL elution buffer and stored at −80°C until use.
Sequencing
Viral cDNA was generated from 100 ng of the extracted RNA aliquots, and sequencing libraries were prepared from the cDNA using the Swift SNAP Amplicon SARS-CoV-2 kit. Deep sequencing was then performed on an Illumina NovaSeq. Raw sequences were processed using the nf-core/viralrecon workflow (v1.1.0, v2.2) (39), in order to align sequences to the Wuhan-Hu-1 reference genome and extract frequencies and annotations for variants at frequencies higher than 0.01. Lineages were assigned using Pangolin (v1.2.34) (40).
Analysis of variant dynamics
Variants were extracted from sequences aligned to Wuhan-Hu-1 using iVar (v1.3, v1.3.1) (41), and variant codon effects were annotated using SnpEff (v5.0) (42) and SnpSift (v4.3t) (43). iSNVs were called using a frequency threshold of 0.03–0.97 and a per-nucleotide depth threshold of 1,000 reads. Commonly reported sequencing artifacts (18) were filtered from each sample, along with non-variant-associated SNPs that recurred across multiple participants within either cohort. To identify high-confidence, “shared” iSNVs, variants were extracted if they met the following conditions: present above a frequency of 0.03 with a per nucleotide depth of at least 1,000 reads in at least two samples with Ct <26 (or CN <26), and present below a frequency of 0.97 in at least two samples. For each participant, variants present across two or more days of infection were extracted, and their frequencies were tracked. Although the frequency, depth, and Ct cutoffs described above were used to identify these shared variants, frequency tracking was performed on a data set curated without thresholds, to avoid cases in which variants crossing either threshold may erroneously appear to fall out of the data set. Therefore, all instances of these shared variants were then called from all samples. Variants with per-nucleotide coverage values below the cutoff were specially marked and plotted to indicate their low depth values. SnpEff annotations were used to characterize shared variants as synonymous, nonsynonymous, or untranslated and to assign them to the appropriate region of the SARS-CoV-2 genome.
Estimation of error
Standard error for iSNV frequency detection was calculated using , where p = detected iSNV frequency and V = viral genome load. Genome copies per milliliter was estimated from Ct using log10V = 14.24 − 0.28 * Ct (for saliva samples) and from CN using log10V = 11.35 − 0.25 * CN (for nasal samples) (16). Genome load was then estimated by approximating that 10 µL of extracted RNA went into each sequencing reaction.
Analysis of variant distribution
Genome positions of variants were visualized using trackViewer (v1.28.1) (44) in R. To identify underdispersion in genomic iSNV distribution, a sliding genomic window was used to determine the mean iSNV count per 100 nucleotides. Genomic windows with significantly higher-than-average iSNV counts were identified using a Poisson distribution, and a threshold for significance was calculated by dividing an initial threshold of 0.05 by the number of genomic sites (0.05/29,803 = 0.00000168).
Analysis of genetic compartmentalization between sample sites
To identify high-confidence iSNVs, variants in both data sets were extracted using similar criteria as described above in Analysis of variant dynamics. However, in this analysis, high-frequency variants were called if they fell below a frequency of 0.97 at least twice in a pooled data set of saliva and nasal samples from the same individual (allowing for detection of iSNVs that were fixed in one compartment but not in the other). All instances of high-confidence variants in a given participant were then called from all samples in either environment. We also filtered out commonly reported artifacts (18) and the artifacts previously identified in saliva samples from each data set. For each participant, pairwise FST values were calculated for all possible pairs of samples, including pairs of saliva samples, pairs of nasal samples, and pairs of one saliva sample and one nasal sample. An FST threshold of detection was calculated from paired sequencing replicates (see Tables 1 and 2), using a version of Table 2 that was filtered to only include variants at frequencies of 0.03–0.97 (so as to apply a slightly stricter threshold—0.00128 as opposed to 0.00125). Comparisons of iSNVs where one or both iSNV instances were detected at low read depths (<1,000 reads) were not used to calculate FSTs. Monte Carlo permutation tests were used to perform significance testing between groups of FST values derived from different sample pairings (i.e., nasal-nasal, saliva-saliva, or nasal-saliva). Briefly, FSTs were randomly shuffled across groups, with 10,000 randomizations performed for each individual participant. The between-group differences in FST means were calculated for the 10,000 shuffled data sets as well as for the true data set, and P-values were derived from the fraction of instances in which the difference calculated using a simulated data set was greater than or equal to the difference calculated using the true data set.
Phylogenetic analysis
The metadata file for all sequences present in the GISAID EpiCov database (10.2807/1560-7917.ES.2017.22.13.30494) was downloaded on 10 June 2022. This metadata file was filtered to include only entries from human hosts, only complete and high coverage entries, and only those with complete sampling dates. The filtered metadata entries were downsampled to at most 100 per month. Downsampling was conducted in Python v3.9.4 (45) using Pandas v1.1.4 (10.5281/zenodo.3509134) and Numpy v1.19.4 (10.1038/s41586-020-2649-2). The selected sequences were downloaded from GISAID EpiCov and aligned to Wuhan/WIV04 (EPI_ISL_402124) using MAFFT v7.464 (10.1093/molbev/mst010), removing any insertions to the reference. IQtree v2.1.3 (10.1093/molbev/msaa015) was used to infer a phylogenetic tree of the aligned sequences using a GTR + G4 substitution model, saving with Wuhan/WIV04 as the outgroup. TreeTime v0.8.0 (10.1093/ve/vex042) was used to filter sequences to include only those falling within four interquartile ranges of the best fit molecular clock, rooting at Wuhan/WIV04. Wuhan/WIV04 was forced to be included in the filtered tree, and no other tips were identified as failing this filter.
For the amino acid of interest (ORF1ab S1150), we first identified the corresponding nucleotide position and then identified the nucleotide identity at each of those sites for each sequence in the alignment. These nucleotide identities were used to infer the amino acid for each sequence. Note that this method does not account for the presence of frame shift mutations; however, we expect these to be sufficiently rare as to not bias our results.
We then plotted the downsampled phylogenetic tree, labeling any tips with amino acids that did not match the reference. Any tips in which any of the nucleotides in the codon of interest were deleted or ambiguously genotyped were ignored. Visualization was done in Python using Matplotlib v3.5.1 (10.1109/MCSE.2007.55) and Baltic v0.1.5 (https://github.com/evogytis/baltic).
Substitution frequency analysis
The GISAID EpiCoV “MSA full” alignment was downloaded on 2 June 2022. All sequences in this file have been aligned to Wuhan/WIV04 using MAFFT, retaining any insertions relative to the reference. Full details on how this file were generated are available from GISAID.
For each of the four amino acid positions of interest, we first identified the corresponding nucleotide positions in the gapped alignment and identified the nucleotides at each of these for each position. These nucleotides were used to infer the amino acid for each sequence in the full alignment at each position. Similar to above, this method does not account for frameshift mutations.
For each amino acid position, we identified the percentage of sequences harboring non-reference amino acids per month, ignoring any sequences in each one of the nucleotides was deleted or ambiguously genotyped. All amino acids identified with a maximum monthly frequency ≤0.01% were grouped into an “Other” category. This analysis was conducted in Python using Pandas and visualized with Matplotlib.
ACKNOWLEDGMENTS
We are grateful to Alvaro Hernandez, Chris Wright, and the DNA services team at the Roy J. Carver Biotechnology Center for their assistance in next-generation sequencing.
We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which Fig. S6 is based (Table S1).
This work has been generously supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health award 3U54HL143541-02S2) through the RADx-Tech program to D.D.M., L.L.G., and C.B.B. as well as additional funds generously provided by the Carl R. Woese Institute for Genomic Biology and the Department of Microbiology at the University of Illinois at Urbana-Champaign.
Contributor Information
Christopher B. Brooke, Email: cbrooke@illinois.edu.
Shan-Lu Liu, The Ohio State University, Columbus, Ohio, USA.
DATA AVAILABILITY
All raw sequence data are available in SRA BioProject PRJNA858053. Other associated supplemental data and in-house code for data analysis are posted at github.com/BROOKELAB/SARS-CoV-2-within-host-evolution (14).
ETHICS APPROVAL
This study was approved by the Western Institutional Review Board, and all participants provided informed consent.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01618-23.
Figures S1 to S8.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Baang JH, Smith C, Mirabelli C, Valesano AL, Manthei DM, Bachman MA, Wobus CE, Adams M, Washer L, Martin ET, Lauring AS. 2021. Prolonged severe acute respiratory syndrome Coronavirus 2 replication in an immunocompromised patient. J Infect Dis 223:23–27. doi: 10.1093/infdis/jiaa666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corey L, Beyrer C, Cohen MS, Michael NL, Bedford T, Rolland M. 2021. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med 385:562–566. doi: 10.1056/NEJMsb2104756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Avanzato VA, Matson MJ, Seifert SN, Pryce R, Williamson BN, Anzick SL, Barbian K, Judson SD, Fischer ER, Martens C, Bowden TA, de Wit E, Riedo FX, Munster VJ. 2020. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 183:1901–1912. doi: 10.1016/j.cell.2020.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, et al. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592:277–282. doi: 10.1038/s41586-021-03291-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, Solomon IH, Kuo H-H, Boucau J, Bowman K, et al. 2020. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 383:2291–2293. doi: 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Truong TT, Ryutov A, Pandey U, Yee R, Goldberg L, Bhojwani D, Aguayo-Hiraldo P, Pinsky BA, Pekosz A, Shen L, et al. 2021. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine 67:103355. doi: 10.1016/j.ebiom.2021.103355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Valesano AL, Rumfelt KE, Dimcheff DE, Blair CN, Fitzsimmons WJ, Petrie JG, Martin ET, Lauring AS. 2021. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog 17:e1009499. doi: 10.1371/journal.ppat.1009499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun KM, Moreno GK, Wagner C, Accola MA, Rehrauer WM, Baker DA, Koelle K, O’Connor DH, Bedford T, Friedrich TC, Moncla LH. 2021. Acute SARS-CoV-2 infections harbor limited within-host diversity and transmit via tight transmission bottlenecks. PLoS Pathog 17:e1009849. doi: 10.1371/journal.ppat.1009849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tonkin-Hill G, Martincorena I, Amato R, Lawson ARJ, Gerstung M, Johnston I, Jackson DK, Park N, Lensing SV, Quail MA, et al. 2021. Patterns of within-host genetic diversity in SARS-CoV-2. Elife 10:e66857. doi: 10.7554/eLife.66857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lythgoe KA, Hall M, Ferretti L, de Cesare M, MacIntyre-Cockett G, Trebes A, Andersson M, Otecko N, Wise EL, Moore N, et al. 2021. SARS-COv-2 within-host diversity and transmission. Science 372:eabg0821. doi: 10.1126/science.abg0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saad-Roy CM, Morris SE, Metcalf CJE, Mina MJ, Baker RE, Farrar J, Holmes EC, Pybus OG, Graham AL, Levin SA, Grenfell BT, Wagner CE. 2021. Epidemiological and evolutionary considerations of SARS-CoV-2 vaccine dosing regimes. Science 372:363–370. doi: 10.1126/science.abg8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cobey S, Larremore DB, Grad YH, Lipsitch M. 2021. Concerns about SARS-CoV-2 evolution should not hold back efforts to expand vaccination. Nat Rev Immunol 21:330–335. doi: 10.1038/s41577-021-00544-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ranoa DRE, Holland RL, Alnaji FG, Green KJ, Wang L, Fredrickson RL, Wang T, Wong GN, Uelmen J, Maslov S, et al. 2022. Mitigation of SARS-CoV-2 transmission at a large public University. Nat Commun 13:3207. doi: 10.1038/s41467-022-30833-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mireille Farjo, m-a-martin & Chris Brooke . 2023. BROOKELAB/SARS-CoV-2-within-host-evolution: v1.0_ BROOKELAB/SARS-CoV-2-within-host-evolution [Google Scholar]
- 15. Smith RL, Gibson LL, Martinez PP, Ke R, Mirza A, Conte M, Gallagher N, Conte A, Wang L, Fredrickson R, et al. 2021. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. J Infect Dis 224:976–982. doi: 10.1093/infdis/jiab337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ke R, Martinez PP, Smith RL, Gibson LL, Mirza A, Conte M, Gallagher N, Luo CH, Jarrett J, Zhou R, et al. 2022. Daily longitudinal sampling of SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. Nat Microbiol 7:640–652. doi: 10.1038/s41564-022-01105-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ke R, Martinez PP, Smith RL, Gibson LL, Achenbach CJ, McFall S, Qi C, Jacob J, Dembele E, Bundy C, et al. 2022. Longitudinal analysis of SARS-CoV-2 vaccine breakthrough infections reveals limited infectious virus shedding and restricted tissue distribution. Open Forum Infect Dis 9:fac192. doi: 10.1093/ofid/ofac192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Issues with SARS-CoV-2 sequencing data - SARS-CoV-2 Coronavirus / nCoV-2019 genomic epidemiology. 2020. Virological. https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473.
- 19. Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, Zhou Z, Yang J, Zhong J, Yang D, Guo L, Zhang G, Li H, Xu Y, Chen M, Gao Z, Wang J, Ren L, Li M. 2020. Genomic diversity of severe acute respiratory syndrome–Coronavirus 2 in patients with Coronavirus disease 2019. Clin Infect Dis 71:713–720. doi: 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Savela ES, Viloria Winnett A, Romano AE, Porter MK, Shelby N, Akana R, Ji J, Cooper MM, Schlenker NW, Reyes JA, Carter AM, Barlow JT, Tognazzini C, Feaster M, Goh Y-Y, Ismagilov RF. 2022. Quantitative SARS-CoV-2 viral-load curves in paired saliva samples and nasal swabs inform appropriate respiratory sampling site and analytical test sensitivity required for earliest viral detection. J Clin Microbiol 60:e0178521. doi: 10.1128/JCM.01785-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Viloria Winnett A, Akana R, Shelby N, Davich H, Caldera S, Yamada T, Reyna JRB, Romano AE, Carter AM, Kim MK, Thomson M, Tognazzini C, Feaster M, Goh Y-Y, Chew YC, Ismagilov RF. 2023. Daily SARS-CoV-2 nasal antigen tests miss infected and presumably infectious people due to viral load differences among specimen types. Microbiol Spectr 11:e0129523. doi: 10.1128/spectrum.01295-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin MA, Koelle K. 2021. Comment on “Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2” Sci Transl Med 13:eabh1803. doi: 10.1126/scitranslmed.abh1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Z, VanBlargan LA, Bloyet L-M, Rothlauf PW, Chen RE, Stumpf S, Zhao H, Errico JM, Theel ES, Liebeskind MJ, Alford B, Buchser WJ, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ. 2021. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 29:477–488. doi: 10.1016/j.chom.2021.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hodcroft EB, Domman DB, Snyder DJ, Oguntuyo KY, Van Diest M, Densmore KH, Schwalm KC, Femling J, Carroll JL, Scott RS, Whyte MM, Edwards MW, Hull NC, Kevil CG, Vanchiere JA, Lee B, Dinwiddie DL, Cooper VS, Kamil JP. 2021. Emergence in late 2020 of multiple lineages of SARS-CoV-2 spike protein variants affecting amino acid position 677. medRxiv:2021.02.12.21251658. doi: 10.1101/2021.02.12.21251658 [DOI] [Google Scholar]
- 26. Colson P, Delerce J, Burel E, Beye M, Fournier P-E, Levasseur A, Lagier J-C, Raoult D. 2022. Occurrence of a substitution or deletion of SARS-CoV-2 spike amino acid 677 in various lineages in Marseille, France. Virus Genes 58:53–58. doi: 10.1007/s11262-021-01877-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ghosh AK, Kaiser M, Molla MMA, Nafisa T, Yeasmin M, Ratul RH, Sharif MM, Akram A, Hosen N, Mamunur R, Amin MR, Islam A, Hoque ME, Landt O, Lytton SD. 2021. Molecular and serological characterization of the SARS-CoV-2 Delta variant in Bangladesh in 2021. Viruses 13:2310. doi: 10.3390/v13112310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saxena SK, Kumar S, Ansari S, Paweska JT, Maurya VK, Tripathi AK, Abdel-Moneim AS. 2022. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol 94:1738–1744. doi: 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 29. Debbink K, McCrone JT, Petrie JG, Truscon R, Johnson E, Mantlo EK, Monto AS, Lauring AS. 2017. Vaccination has minimal impact on the intrahost diversity of H3N2 influenza viruses. PLoS Pathog 13:e1006194. doi: 10.1371/journal.ppat.1006194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson BA, Zhou Y, Lokugamage KG, Vu MN, Bopp N, Crocquet-Valdes PA, Kalveram B, Schindewolf C, Liu Y, Scharton D, Plante JA, Xie X, Aguilar P, Weaver SC, Shi P-Y, Walker DH, Routh AL, Plante KS, Menachery VD. 2022. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog 18:e1010627. doi: 10.1371/journal.ppat.1010627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mears HV, Young GR, Sanderson T, Harvey R, Crawford M, Snell DM, Fowler AS, Hussain S, Nicod J, Peacock TP, Emmott E, Finsterbusch K, Luptak J, Wall E, Williams B, Gandhi S, Swanton C, Bauer DL. 2022. Emergence of new subgenomic mRNAs in SARS-CoV-2. Microbiology. doi: 10.1101/2022.04.20.488895 [DOI] [Google Scholar]
- 33. Amato KA, Haddock LA, Braun KM, Meliopoulos V, Livingston B, Honce R, Schaack GA, Boehm E, Higgins CA, Barry GL, Koelle K, Schultz-Cherry S, Friedrich TC, Mehle A. 2022. Influenza A virus undergoes compartmentalized replication in vivo dominated by stochastic bottlenecks. Nat Commun 13:3416. doi: 10.1038/s41467-022-31147-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Orton RJ, Wright CF, King DP, Haydon DT. 2020. Estimating viral bottleneck sizes for FMDV transmission within and between hosts and implications for the rate of viral evolution. Interface Focus 10:20190066. doi: 10.1098/rsfs.2019.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pfeiffer JK, Kirkegaard K. 2006. Bottleneck-mediated quasispecies restriction during spread of an RNA virus from inoculation site to brain. Proc Natl Acad Sci U S A 103:5520–5525. doi: 10.1073/pnas.0600834103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS. 2018. Stochastic processes constrain the within and between host evolution of influenza virus. Elife 7:e35962. doi: 10.7554/eLife.35962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao Y, Dolan PT, Goldstein EF, Li M, Farkov M, Brodsky L, Andino R. 2017. Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses. Nat Commun 8:375. doi: 10.1038/s41467-017-00354-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ranoa DRE, Holland RL, Alnaji FG, Green KJ, Wang L, Brooke CB, Burke MD, Fan TM, Hergenrother PJ. 2020. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. Microbiology. doi: 10.1101/2020.06.18.159434 [DOI] [Google Scholar]
- 39. Patel H. 2023. nf-core/viralrecon: nf-core/viralrecon v2.6.0 - Rhodium Raccoon [Google Scholar]
- 40. O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, Yeats C, du Plessis L, Maloney D, Medd N, Attwood SW, Aanensen DM, Holmes EC, Pybus OG, Rambaut A. 2021. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 7:veab064. doi: 10.1093/ve/veab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ, Tan AL, Paul LM, Brackney DE, Grewal S, Gurfield N, Van Rompay KKA, Isern S, Michael SF, Coffey LL, Loman NJ, Andersen KG. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol 20:8. doi: 10.1186/s13059-018-1618-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6:80–92. doi: 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. 2012. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet 3:35. doi: 10.3389/fgene.2012.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ou J, Zhu LJ. 2019. trackViewer: a bioconductor package for interactive and integrative visualization of multi-omics data. Nat Methods 16:453–454. doi: 10.1038/s41592-019-0430-y [DOI] [PubMed] [Google Scholar]
- 45. van Rossum G, Drake FL. 2009. Python 3 reference manual
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S8.
Data Availability Statement
All raw sequence data are available in SRA BioProject PRJNA858053. Other associated supplemental data and in-house code for data analysis are posted at github.com/BROOKELAB/SARS-CoV-2-within-host-evolution (14).