Abstract
Sepsis, a dysregulated immune response to infection, has been the leading cause of morbidity and mortality in critically ill patients. Multiple studies have demonstrated improved survival outcomes when early treatment is initiated for septic patients. In our previous work, we developed a real-time machine learning algorithm capable of predicting onset of sepsis four to six hours prior to clinical recognition. In this work, we develop AIDEx, an open-source platform that consumes data as FHIR resources. It is capable of consuming live patient data, securely transporting it into a cloud environment, and monitoring patients in real-time. We build AIDEx as an EHR vendor-agnostic open-source platform that can be easily deployed in clinical environments. Finally, the computation of the sepsis risk scores uses a common design pattern that is seen in streaming clinical informatics and predictive analytics applications. AIDEx provides a comprehensive case study in the design and development of a production-ready ML platform that integrates with Healthcare IT systems.
I. INTRODUCTION
A. Sepsis – A Health Crisis
Sepsis is a syndromic, life-threatening condition that occurs when the body exerts an exaggerated response to infection [1]. Sepsis when left untreated progresses to deadly severe sepsis or septic shock that the host triggers injuring internal organs. Nearly 6% of the inpatient hospital population in the United States will carry a diagnosis of sepsis during their stay. 35% of all hospital deaths, in the US, are attributed to sepsis, and it accounts for $23.7 billion in annual costs[2]. Numerous trials have demonstrated dramatic improvements in survival outcomes for sepsis by early recognition of the condition and rapid treatment [3-6]. While there are effective protocols for treating sepsis once it has been diagnosed, there are several challenges in reliably identifying septic patients early in their course owing to the significant variability in the disease’s presentation. The Sepsis-3 guidelines[1] have narrowed the constellation of signs and symptoms of sepsis into a clinical criterion that can be reliably used by clinicians and researchers to identify this life-threatening condition retrospectively. However, this criterion alone cannot help identify a patient, who is experiencing the effects of sepsis early in the disease’s course.
B. Sepsis Prediction using Machine Learning
In recent years, the increased adoption of electronic medical records (EMR) has spurned the development of machine learning based surveillance tools for detection [7-10] and prediction [9-12] of patients with sepsis or septic shock. However, there has been slow progress in real-time implementation of a high-dimensional machine learning model in an Intensive Care Unit (ICU) environment.
Recently, Nemati et al have developed the Artificial Intelligence Sepsis Expert (AISE), a modified Weibull-Cox model that uses data commonly available in the EMR to predict the onset of sepsis four to six hours in advance with an area under the ROC curve (AUC) of 0.85 [11]. The AISE development cohorts contained over 30,000 patients from multiple hospitals in the Emory Healthcare system and was validated using a cohort of 50,000 patients from the MIMIC-III database [13]. In this work, we present a platform, that is used to deploy the AISE algorithm in a real-world setting, using live clinical data.
Our platform fetches patients records from a real-time EMR database and displays hourly sepsis risk score for each patient. The platform, called Artificial Intelligence Decompensation Expert (AIDEx), is scalable, resilient, open-source and developed using the emerging Fast Healthcare Interoperability Resources (FHIR) standard.
The overarching emphasis of our architecture is to examine integration with healthcare IT systems, significant attention has been given to elements such as software quality control, and tracking feature drifts. The user interface has been designed to minimize false-alarms [12] as well as assist in clinical interpretability and workflow integration requirements necessary for a successful clinical decision support (CDS) system [14-15]. AIDEx sheds some light on the processes and development needed to interface with healthcare IT systems and build deployable applications. Thus, AIDEx1 also provides a good case-study in taking ML based predictive analytics algorithms, for healthcare applications, to production.
II. Platform Architecture
ML algorithms are usually developed within controlled environments where researchers can control data, wrangle it into an appropriate format, and once the model has been developed, evaluate, and validate it suitably. The deployment of a trained model, into a real-world setting is a non-trivial activity that can include data-wrangling pipelines that can operate without human intervention; on-demand deployment and scaling of the algorithm; monitoring of the underlying infrastructure; tracking and tuning for performance and latency; user interfaces; and quality control.
In most real-world ML systems, the actual ML algorithm or model, is a lot smaller than the infrastructure needed to support it [16].
ML algorithms and systems therefore incur, what is known as the technical debt of ML. This hidden technical debt, results in a highly incomplete view of the field and, by overly simplifying the process, contributes to the hype-cycles. One of the key contributions of AIDEx, is its ability to work with a real-time stream of live clinical data. It required the design and implementation of a real-world system that is scalable, elastic, and fault-tolerant.
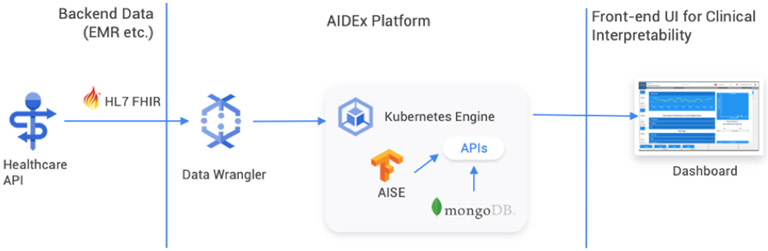
AIDEx adopts a modular architecture where each of its core functionality is captured as a microservice. It builds microservices for preprocessing data, executing the prediction algorithm, storing the prediction outcomes, and visualizing outcomes. Figure 1 illustrates the flow of data in AIDEx.
Fig. 1.
An overview of the AIDEx platform that illustrates how healthcare data is pulled from the FHIR data store and then the results are streamed into a MongoDB based results services. Clients can use the web-based dashboard to review results
It has been deployed on a Cloud Platform, and uses a few managed services and functionalities, including security features such as firewalls and virtual private clouds (VPCs). It is worth noting that while this paper focuses on deployment of AIDEx on the Cloud, AIDEx is also a platform agnostic container-based system that can be deployed on the cloud as well as on-premises.
A. AIDEx Microservices
AIDEx consumes patient data as a series of FHIR resources, computes the risk of developing sepsis in the next four to six hours (sepsis scores), and presents them in an interpretable manner via a web-based dashboard. The environment is secured via a VPC and utilities have been deployed to push data from the institution to the cloud. The use of containerized microservices removes the need to install distinct applications and their associated dependencies on a host machine at various deployment sites. It also allows us to leverage the inherent scalability and fault tolerance. The AIDEx pipeline is unique in its health system agnostic design and its use of a state-of-the-art machine learning algorithm capable of accurately identifying patients with sepsis early in their clinical course. Though this tool provides population-level surveillance of a large cohort of ICU patients, its real strength lies in its ability to provide clinicians with individual patient vital sign trends and the most relevant features contributing to their risk score. Table 1 presents a summary of the services comprising the pipeline and are described in greater detail in the following sections.
TABLE I.
Microservices that make up the AIDEx Platform.
| Name | Service Objective |
|---|---|
| Data Wrangler | Retrieves live data streams as FHIR Resources; prepares data;orchestrates predictive algorithm; saves data |
| Results Store | A time-series data store that stores the data for each patient at each time-point |
| Sepsis Predictor | Algorithm running inference on data for each patient and forecasts the onset of sepsis |
| Clinical Dashboard | Presents the outcomes in an interpretable user-friendly interface |
1). Data Wrangler - Clinical Data Harmonization:
Preprocessing the data is a crucial first step in the machine learning applications. Data arriving from an active EMR is not always ready for use by a machine learning algorithm and requires a series of pre-processing steps. AIDEx consists of a Data Wrangler service that pulls real-time patient data from the host EMR’s FHIR database to the sepsis predictor service, and finally to the results store. Figure 1 illustrates the data flow in the AIDEx platform. The Data Wrangler service starts its execution by querying a live EMR FHIR database capturing the patient features necessary for sepsis prediction. These features include laboratory results, vital signs, and demographic information for all active patients over the last hour. Errors in data entry can result in values that are not physiologically plausible. The Data Wrangler minimizes the impact of erroneous data by limiting all extreme values to a maximum and minimum value based upon the 95% confidence interval for each feature obtained from a pre-collected patient cohort. Following this preprocessing step, the Data Wrangler service then makes a stateless API call to the Sepsis Predictor service. An active patient’s standardized data is then transmitted from the Data Wrangler service to the Sepsis Predictor service.
2). Sepsis Predictor:
The core of the AIDEx framework consists of the prediction algorithm, that predicts the onset of sepsis early on. The Sepsis Predictor service runs the AISE algorithm[11]. When deployed, the algorithm can alert clinicians four to six hours before a patient meets the Sepsis-3 criterion. The output of the Sepsis Predictor Service is a sepsis risk score and the top three factors contributing to the sepsis risk score (see Fig. 2). The data returned by the Sepsis Predictor service along with all patient features are combined into JSON documents to be stored in the Results Store Data Warehouse. The Data Wrangler service’s final function is to provide a standardized interface for reading and writing data as JSON documents to the Results Store. Each JSON document contains timestamp and corresponding patient features, sepsis risk score, change in risk score over the last four hours, demographic information, and the three factors contributing most to the sepsis risk score.
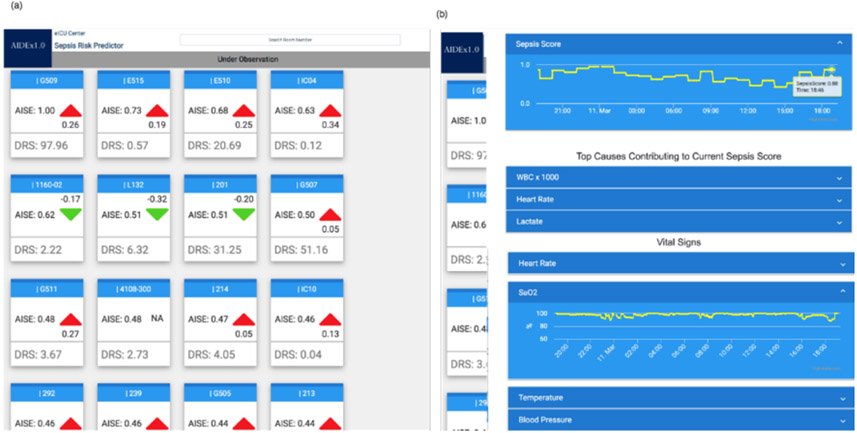
Fig. 2.
A population view of all ICU patients. Each patient is represented by a single card that displays: a sepsis risk score (AISE), discharge readiness score (DRS), the increase in patient’s sepsis score (delta). Patients are listed descending based on their AISE score (on left) The detailed view for a single patient is displayed. Visible is the 12-hour trajectory of the patient’s sepsis risk score, and their vital signs.
3). Results Store:
A data store is necessary to store the outcomes of the Sepsis Predictor. We developed a Results Store service as a time-series data store to store data for the patients at each time-point and offer a standard access interface to the stored data. The AIDEx pipeline is designed to be scalable and capable of managing data streams from a large patient population. The patient data streams and the computed sepsis scores are transformed into a timeseries JSON document. These documents are stored in MongoDB - a well-known NoSQL document store that is highly scalable and has been used in a variety of clinical and research applications. The database is accessed via a REST API that is built using an OSGI based declarative middleware called Bindaas [17-18]. Bindaas is an extensible big data middleware that lets the users create interoperable RESTful interfaces to various data sources. Other services in the AIDEx pipeline, including the user-interface access Results Store via this API.
4). User Interface:
Graphical representations in a user interface supports the clinicians to interpret the outcomes of the AIDEx services. The clinical dashboard retrieves JSON documents generated by the Sepsis Predictor service and displays data in a graphical user interface (UI) for interpretation by clinical team members. As seen in Fig. 2, the UI includes a command center that gives a high-level overview of the ICU population and detailed view that presents detailed information including sepsis scores, clinical interpretations and vital signs. The default view seen in Fig. 2a demonstrates a population-level view of ICU patients. Each patient is represented by a single card, and the front of each card contains the patient’s room number at the top, a sepsis score, a discharge readiness score, and finally a directional arrow with magnitude representing the acceleration (i.e. delta) of a patient’s sepsis risk score over the last four hours.
The patient list is ranked according to the sepsis risk score with the most acute patients at the top of the list and a second UI see in Figure 2b is revealed. This patient centric view reveals the top three factors contributing to the risk score in addition to the vital sign trends for the patient over the last 24 hours. As previously described the Data Warehouse stores patient features in addition to the AISE Algorithm outputs in JSON files inside the Data Warehouse. This approach to data storage makes it simple for the UI to obtain patient data from the Data Warehouse for display in the user interface.
B. Security
We enforce security measures in development and deployment, to ensure the code satisfies the test requirements while the deployed AIDEx platform can be accessed only by the intended users. To prevent unauthorized access to sensitive data and APIs, we must ensure proper access policies and authentication mechanisms are in place. In AIDEx we configured secured access to the services to ensure proper authentication and authorization. We further configured firewall policies at the cloud instances and on-premise servers to ensure only the specified IP addresses can access the services, and only through the explicitly specified ports. In addition to securing against unauthorized accesses, we note that such protected network also minimizes the potential for denial of service attacks, by ignoring the service invocations from the unknown sources.
C. Testing Reliability and Quality Control for Model and Features
Deploying a machine learning model in a real-time environment poses challenges not common in offline experiments [19]. Assessing production readiness level, monitoring and testing the system automatically are key consideration for a real-world ML software system. Real-time ML platforms are greatly dependent on the nature of data, more precisely the features. We have developed a series of automatic tests, specific to real-time ML platform, that run alongside traditional software engineering regression tests. In addition to the unit, integration, and system level tests to evaluate the functionality of the pipeline, AIDEx includes complement set of tests to assess, monitor and track the features and the data[19]. The underlying data distribution may change over time. ML platforms rely on the hidden representation of the feature, so changes in underlying data distribution will affect the model performance. This is a well-known concept of feature drift wherein a model built on stale data becomes inconsistent with newer data [19]. Feature drift affects Model reliability, which is crucial in the clinical environment so monitoring the data distribution and considering the feature drift is the important key in the real-time production ML system.
In AIDEx we have developed a set of automated tests to evaluate the difference in distribution of the features. We performed Kolmogorov–Smirnov test (KS test) to quantify the distance between the distribution of features from source and target data set. Target features are sampled from our ICU population weekly. We store the p-value and the power of each test for our monitoring purposes and for updating the sepsis predictor algorithm in cases of feature drift. Further, we assess the difference between the predicted sepsis score for our ICU population with a 95% confidence interval every week.
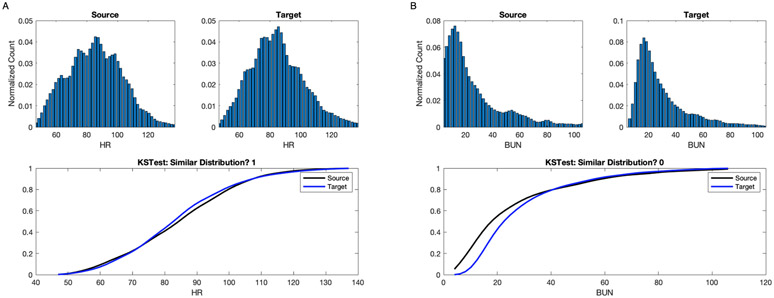
Figure 3 illustrates two example of assessing the difference between the distribution of Heart rate and Blood urea nitrogen (BUN) real-values from source and target ICU patient population. Fig.3 A illustrates the assessing of the difference of the Heart rate distribution between source and target ICU patient population. The upper panels represents the Heart rate distribution from source and target population. The lower panel shows the cumulative distribution function (CDF) of the Heart rate. The KS test reveals that for Heart rate the two distribution are similar with the confidence level of 0.99.Fig.3 B illustrates the assessing of the difference of the BUN real-values distribution between source and target ICU patient population. The upper panels represents the BUN distribution from source and target population. The lower panel shows the cumulative distribution function (CDF) of the BUN. The KS test reveals that for the BUN the two distribution are dissimilar with the same confidence level.
Fig. 3.
An illustrative example of assessing the feature distribution over source and target ICU patient population. A: the upper panels show the histogram of the Heart rate, recorded every 5 minutes, from source and target population. The lower panel shows the CDF for the same population cohorts. The KS test reveals that the Heart rate real-values distribution between the source and target ICU population are similar. B: the upper panel represent the histogram of the BUN from source and target population. The lower shows the CDF for the same population cohorts. The KS test reveals that the BUN real-values distribution between the source and target ICU population are dissimilar.
III. Conclusion
Existing literature on the application of machine learning and deep learning techniques to healthcare applications are narrowly focused on novel algorithms, while in practice it takes coordinated efforts of many teams, including machine learning experts, software engineers, implementation scientists, and hospital IT teams, to bring such systems to the bedside. This is no trivial amount of work and every design and implementation choice makes a difference. AIDEx captures many of the critical elements necessary to take a well-tested and validated machine learning algorithm to production. It adopts a robust testing and quality control methodology that spans the software and the data. This has allowed us to tackle the major healthcare problem of sepsis. Early detection and treatment of sepsis is categorically one of the most important interventions that can be taken in a modern ICU. In this work, we have developed the AIDEx platform,and make it open-source available as a comprehensive way to detect, triage, and inform clinicians of a patient’s risk for developing sepsis.
We are currently undertaking extensive external validations of the algorithm as well as integrations of AISE and gathering data for regulatory approval. AIDEx has allowed us to begin planning a multi-center clinical trial to examine the interventional use and utility of our sepsis prediction algorithms.
Footnotes
The code for AIDEx platform is available online at https://github.com/aise-on-fhir
References
- [1].Singer M et al. , “The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3),” JAMA, vol. 315, no. 8, pp. 801–10, Feb 23 2016, doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rhee C et al. ,“Incidence and trends of sepsis in US hospitals using clinical vs claims data,2009-2014,”Jama, vol. 318, no. 131249, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ferrer R et al. ,“Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program,” Critical care medicine, vol.42,no.8,pp.1749–1755,2014. [DOI] [PubMed] [Google Scholar]
- [4].Rhodes A et al. , “The surviving sepsis campaign bundles and outcome: results from the international multicentre prevalence study on sepsis (the IMPreSS study),” Intensive care medicine, vol. 41, no. 9, pp. 1620–1628, 2015. [DOI] [PubMed] [Google Scholar]
- [5].Sterling SA, Miller WR, Pryor J, Puskarich MA, and Jones AE, “The impact of timing of antibiotics on outcomes in severe sepsis and septic shock: a systematic review and meta-analysis,” Critical care medicine, vol. 43, no. 9, p. 1907, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Venkatesh AK, Avula U, Bartimus H, Reif J, Schmidt MJ, and Powell ES, “Time to antibiotics for septic shock: evaluating a proposed performance measure,” The American journal of emergency medicine, vol. 31, no. 4, pp. 680–683, 2013. [DOI] [PubMed] [Google Scholar]
- [7].Shashikumar SP, Li Q, Clifford GD, and Nemati S, “Multiscale network representation of physiological time series for early prediction of sepsis,” Physiological measurement, vol. 38, no. 12, p. 2235, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Horng S, Sontag DA, Halpern Y, Jernite Y, Shapiro NI, and Nathanson LA, “Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning,” PloS one, vol. 12, no. 4, p. e0174708, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Desautels T et al. , “Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach,” JMIR medical informatics, vol. 4, no. 3, p. e28, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Brown SM, Jones J, Kuttler KG, Keddington RK, Allen TL, and Haug P, “Prospective evaluation of an automated method to identify patients with severe sepsis or septic shock in the emergency department,” BMC emergency medicine, vol. 16, no. 1, p. 31, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, and Buchman TG, “An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU,” Critical care medicine, vol. 46, no. 4, pp. 547–553, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Henry KE, Hager DN, Pronovost PJ, and Saria S, “A targeted real-time early warning score (TREWScore) for septic shock,” Science translational medicine, vol. 7, no. 299, pp. 299ra122–299ra122, 2015. [DOI] [PubMed] [Google Scholar]
- [13].Johnson AEW, Pollard TJ, Shen L, Lehman L, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, and Mark RG,“MIMIC-III, a freely accessible critical care database”,Scientific Data (2016). DOI: 10.1038/sdata.2016.35. Available at: http://www.nature.com/articles/sdata201635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Postelnicu R, Pastores SM, Chong DH, and Evans L, “Sepsis early warning scoring systems: The ideal tool remains elusive!,” ed, 2018. [DOI] [PubMed] [Google Scholar]
- [15].Norrie J, “The challenge of implementing AI models in the ICU,” The Lancet Respiratory Medicine, vol. 6, no. 12, pp. 886–888, 2018.https://www.overleaf.com/project/5e337a2ed5de4e0001f98d29 [DOI] [PubMed] [Google Scholar]
- [16].Sculley D et al. , “Hidden technical debt in machine learning systems,” in Advances in neural information processing systems, 2015, pp. 2503–2511. [Google Scholar]
- [17].Sharma A, Kazerouni A, Saghar N, Commean P, Tarbox L, and Prior F, “Framework for Data Management and Visualization of The National Lung Screening Trial Pathology Images,” Pathology Informatics Summit, pp. 13–16, 2014. [Google Scholar]
- [18].Saltz J et al. , “A containerized software system for generation, management, and exploration of features from whole slide tissue images,” Cancer research, vol. 77, no. 21, pp. e79–e82, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Breck E, Cai S, Nielsen E, Salib M, and Sculley D, “The ml test score: A rubric for ml production readiness and technical debt reduction,” in 2017 IEEE International Conference on Big Data (Big Data), 2017: IEEE, pp. 1123–1132. [Google Scholar]