ABSTRACT
Wastewater-based surveillance (WBS) is a noninvasive, epidemiological strategy for assessing the spread of COVID-19 in communities. This strategy was based upon wastewater RNA measurements of the viral target, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). The utility of WBS for assessing the spread of COVID-19 has motivated research to measure targets beyond SARS-CoV-2, including pathogens containing DNA. The objective of this study was to establish the necessary steps for isolating DNA from wastewater by modifying a long-standing RNA-specific extraction workflow optimized for SARS-CoV-2 detection. Modifications were made to the sample concentration process and included an evaluation of bead bashing prior to the extraction of either DNA or RNA. Results showed that bead bashing reduced detection of RNA from wastewater but improved recovery of DNA as assessed by quantitative polymerase chain reaction (qPCR). Bead bashing is therefore not recommended for the quantification of RNA viruses using qPCR. Whereas for Mycobacterium bacterial DNA isolation, bead bashing was necessary for improving qPCR quantification. Overall, we recommend 2 separate workflows, one for RNA viruses that does not include bead bashing and one for other microbes that use bead bashing for DNA isolation. The experimentation done here shows that current-standing WBS program methodologies optimized for SARS-CoV-2 need to be modified and reoptimized to allow for alternative pathogens to be readily detected and monitored, expanding its utility as a tool for public health assessment.
INTRODUCTION
Since the start of the COVID-19 outbreak in December of 2019, there was a quick response for developing human health surveillance and wastewater-based surveillance (WBS) processes.1, 2, 3, 4, 5, 6, 7, 8, 9 Collected wastewater has a unique capability of assimilating human health markers of the specific community contributing to a sampling point.3, 10 For severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), WBS has been pivotal to addressing its impact on communities worldwide, with its correlative power for clinical cases and hospitalizations4, 6, 7, 9, 11, 12, 13, 14, 15, 16 as well as for realizing SARS-CoV-2’s evolution with emerging variants.16, 17
One consistent message that has echoed from COVID-19 outbreak research has been the importance of method selection for epidemiological sampling strategies. Upstream methodologies for processing collected wastewater are known to impact results from targeted molecular approaches and sequencing. Studies have described the diverse use of molecular biology techniques for assessing water, wastewater, and reclaimed water18, 19, 20, 21, 22, 23, 24 for monitoring pathogens, identifying antibiotic resistance, and utilizing targeted approaches for surveilling specific infectious diseases. A 2021 study of pathogen concentrations in raw sewage compared the reproducibility and sensitivity of 36 sample processing and workflow approaches for optimizing “genetic signal” to quantify SARS-CoV-2 in raw wastewater.25 A few others, such as Lemarchand et al. 200526 and Walden et al. 2017,27 for example, evaluated methodological approaches by addressing upstream concentration processes of wastewater and specific commercial kits, respectively, for the method’s ultimate impact on downstream processing/quantification. In other environmental samples, such as air samples, Luhung et al. 201528 describes key concepts for improving the concentration of DNA downstream and impacts that upstream sample parameters play on the overall recovery of DNA from samples.
Due to the many considerations of quickly adapting COVID-19 WBS, many labs have established unique approaches for sample processing, sometimes incorporating bead bashing (BB) and sometimes not. No studies, to our knowledge, have compared the impacts of BB on RNA and DNA concentrations isolated from wastewater alongside quantitative polymerase chain reaction (qPCR) quantification for viral and bacterial targets, although one study29 investigated the role upstream sample processing (i.e., BB) had on sequencing results. Most methodological studies have compared specific concentration processing methods to one another.2, 14, 25, 30, 31 However, it is rarely discussed how the primary concentration can impact nucleic acid products following extraction and the necessity of certain procedures like BB on downstream molecular investigation.
This study branches from a WBS program at the University of Miami (UM), which began collecting samples during September of 2020.5 Recent experimentation has focused on the expansion of efforts to incorporate quantification of DNA from alternative targets extracted from wastewater.27, 32, 33 The optimization of methods towards DNA-specific extraction processes began with questioning the necessity of BB. We hypothesized that BB was not necessary for the recovery of viral RNA, as detergent lysis typically dissolves viral capsids exposing nucleic acids but would be necessary for the recovery of bacterial DNA because of the differing morphology. This hypothesis stemmed from the concept in which some studies use mechanical lysis techniques for extracting bacterial DNA from environmental and wastewater samples.28, 33 In this study, we evaluated whether BB was necessary for processing viral RNA and determined the benefits of BB for recovering bacterial DNA with targeted qPCR approaches.
Separate dedicated workflows for RNA and DNA, rather than focusing on total nucleic acid extraction, has the advantage of improving the overall quality of RNA and DNA recovered. Our objectives were to establish the effect of BB on the quantity of RNA and DNA downstream that could be utilized for molecular applications and compare upstream concentration methods prior to nucleic acid isolation to ascertain differences from qPCR signals.
METHODS
Wastewater collection and pretreatment
Raw wastewater was collected from either the University’s Hospital, UHealth Tower (sample identifier: 06), or from the downstream wastewater treatment plant located on Virginia Key, Florida, draining the UM campus and central Miami-Dade County (site D). A total 1 L volume of either grab (sample identifier: Dg) or composite (sample identifier: Dc) wastewater (depending upon availability of a composite provided by the wastewater treatment plant) were collected. Samples were collected between October and November 2022 (October 18 [06 and Dc], November 1 [Dg], 2 [Dg], and 18 [Dc]).
Filtration processing was adjusted to determine if the filter selection/concentration method (including and excluding pretreatment) also played a role in the downstream results of practiced methods. This concept was built up from Luhung et al. 201528 in that 2 upstream concentration processes were compared here to identify changes in the concentration of nucleic acids downstream. In the current study, wastewater samples (1 L) were split to assess 2 primary concentration workflows, ultimately either incorporating or foregoing BB of wastewater concentrates, and followed-up with separate RNA and DNA extraction processes to experimentally compare methods (Figure 1). Specifically, these samples were split into 2 equivalent aliquots (500 mL) and pretreated separately for concentration by electronegative filtration (ENF) or vacuum filtration (VF). Pretreatment for both filtration methods involved the addition of viral and bacterial recovery controls, a human coronavirus-OC43 (OC43, [1×106 genomic copies/L (gc/L)] and Mycobacterium smegmatis [2×106 gc/L], respectively.5, 25, 34, 35 The concentrations per spike were chosen to assure detectable levels of the target in downstream qPCR quantification processes. OC43 is a positive-sense, single-stranded RNA coronavirus similar to SARS-CoV-2, which has been justified for use with WBS studies as a measure for percent recovery.32 Prior to use, an aliquot of OC43 was heat inactivated at 56°C for 15 min and briefly vortexed and centrifuged to ensure a homogenized spike in each wastewater sample. M. smegmatis has been growing in use as a nonpathogenic Mycobacterial model for determining host–pathogen interactions of Mycobacterium tuberculosis.36, 37 It has a unique morphology, with a thick lipid cell wall, and capability of dormancy, which can mimic other hard-to-lyse microbes and fungal pathogens such as Candida species.35, 36, 37 Moreover, M. smegmatis’ low replication rate, compared to bacteria like Escherichia coli, minimized the possibility of drastic increase (due to cell division) by the time the wastewater concentrates were created and subjected to molecular isolation/analysis. Similar to the OC43 spike, the M. smegmatis aliquot used per wastewater sample was also vortexed and briefly centrifuged to ensure a homogenized spike in per wastewater sample. The spiked wastewater aliquots (500 mL) were shaken for homogenization prior to pretreatment or filtration by either method.
Figure 1. Laboratory workflow of sample preparation methodology. Experimentation included the splitting of raw wastewater with subsequent pretreatment specific to ENF or VF. BB and the exclusion of BB were performed for each condition evaluated followed by separate RNA and DNA extractions of either bashed or un-bashed concentrates. Image created with BioRender.com.
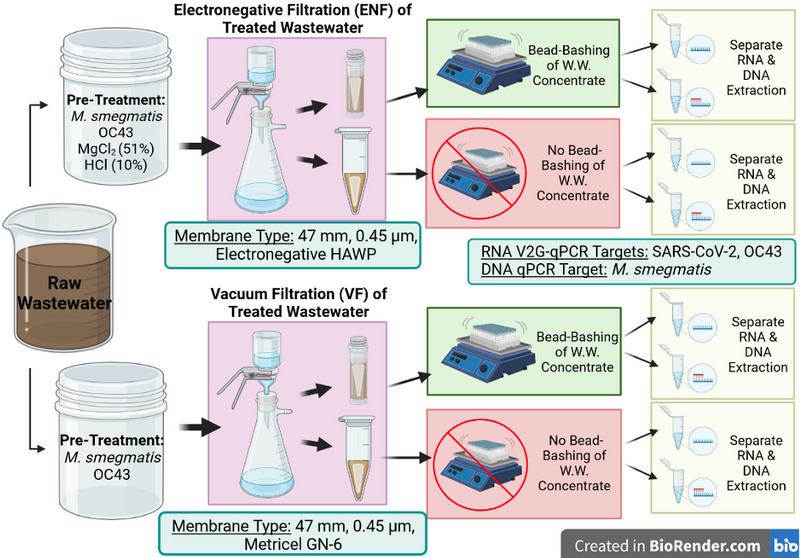
Primary concentration approaches: ENF versus VF
The first method of concentration, used routinely to isolate SARS-CoV-2 and viral RNA downstream, was ENF. This workflow required additional pretreatment, besides spiked-in controls, of the samples to convert the viruses to a positive charge by adding 51% MgCl2 to a concentration of 50 mM and adjusting the pH to a range of 3.5 to 4.5 using 10% HCl. After pH adjustment, samples were filtered through negatively charged mixed cellulose ester membranes (47 mm, 0.45 µm, EMD Millipore #HAWP04700), collecting viruses by size exclusion (those attached to particles larger than the pore size) and by charge (attracting positively charged viral particles to the negatively charged filter membrane).11, 31, 34, 38, 39
The second concentration method, modified for bacterial and fungal pathogens, employed VF with the omission of the MgCl2 and acid adjustment. Furthermore, it substituted the HAWP filter membrane for a GN-6 Metricel filter membrane (47 mm, 0.45 µm, Pall Corp. #66278).39, 40, 41 The pretreatment with salt and acid was removed from the VF methodology. Both filtration methods were performed per wastewater sample for each day of collection. For consistency, 40 mL of wastewater was used for all filtrations, and 2 membranes were prepared per concentration method per day of collection.
After each concentration step, 1 of the 2 filter membranes was split in half, with both halves intended for BB, separately. The second, whole filter was used for the non-BB (nBB) condition as a comparative control. The reason only half filters were used for BB was because of the physical size constraints of the bashing tubes. Qualitative experiments were performed prior to sample collection for this study to determine the “optimum” size of the membrane for BB. See supplemental information for more detail. The BB filters were split by placing the filter membrane from the filter funnel onto a sterile petri dish and then cutting the filter in half with a sterile scalpel. Once cut, the half filters were folded in on themselves 3 times and placed in 1 mL 1X DNA/RNA Shield in a Zymo ZR BashingBead Lysis Tubes (0.1 and 0.5 mm) (cat# S6012-50). The second, whole filter, set to forego BB, was immediately folded in on itself 4 times and placed within a microcentrifuge tube in 2 mL 1X DNA/RNA Shield, keeping the ratio between the filter size and amount of DNA/RNA Shield constant between the RNA and DNA protocols. Generated wastewater concentrates (filters in DNA/RNA Shield) were kept at 4oC until undergoing separate DNA and RNA extractions and subsequent qPCR.
Nucleic acid extraction procedures: RNA and DNA
Modifications to the long-established RNA extraction process at UM5, 13, 31, 34 were performed for this study, with the purpose of expanding workflows to also extract bacterial DNA. The commercial kit used for DNA extraction was the ZymoBIOMICS DNA Miniprep Kit (Zymo Research cat# D4300) and for RNA extraction was the Zymo Research Quick-RNA Viral kit (Zymo Research cat# R1034/1035). For the half-filter concentrates where BB was included prior to extraction, 400 µL of the bashed wastewater concentrate was transferred directly into the first column of the ZymoBIOMICS Miniprep Kit. The DNA Miniprep Kit was used according to the manufacturer’s recommendations with an OMNI Bead-Ruptor12 instrument, except for the increase in the initial centrifugation step (10 000 x g) following BB, wherein the centrifugation speed was increased by 2000 x g to improve the pelleting of ruptured membrane particles.39, 41 The RNA protocol utilized here, corresponding to the Zymo Quick-RNA Viral Kit, starts with BB 400 µL to mimic the DNA Miniprep Kit followed by the recommended 1:2 ratio of starting sample to binding buffer, wherein 800 µL of β-mercaotpethanol binding buffer was utilized in the first column. Due to the increased starting volume (1200 µL), similarly to the DNA Miniprep Kit, 2 centrifugation steps were conducted to fully bind material to the IC spin column at 13 000 rpm for 2 minutes each. The 2 wash steps followed the in-house approach performed by UM,5, 13, 31, 34 wherein 660 µL of wash buffer was added and centrifuged at 13 000 rpm for 30 seconds per wash. Similarly, 660 µL of 100% ethanol was added as the final wash step of the RNA and spun at 13 000 rpm for 1 minute prior to elution with 100 µL of nuclease-free water. Like the DNA Miniprep Kit, the final cleanup Zymo-Spin III-HRC filters (Zymo Research cat# C1058-50) were used on the 100 µL RNA eluate to remove qPCR inhibitors according to the manufacturer’s instructions. For clarity, Zymo-Spin III-HRC filters can be utilized for both RNA and DNA to remove inhibitors. nBB whole-filter concentrates utilized brief vortexing (< 5 seconds) and repeat pipetting to flush suspended solids and homogenize the sample, and 400 µL was immediately transferred to the first column and then followed by the separate nucleic acid extraction procedures described above.
The use of the ZymoBIOMICS DNA Miniprep Kit for DNA extraction included the use of BB as step 1 of the protocol. All other parameters were kept consistent to facilitate comparisons. Thus, the RNA extraction process was adjusted by raising the starting volume of the wastewater concentrate to be the same as the DNA kit (400 µL), increasing the volume of nuclease-free water used to match the total eluate volume between RNA and DNA samples (100 µL), and including the III-HRC Prep Columns, used normally within the DNA Miniprep Kit, to rid the RNA eluate of qPCR inhibitors. Whole-membrane controls, using the long-standing RNA extraction approach of homogenization with repeat pipetting prior to extraction,31, 34 were performed to maintain a set of samples as comparators to the membranes that were bashed.
Nucleic acid concentration analysis
To illustrate the impact BB had on nucleic acid concentration, following extraction, quality assessment tests were performed using the Synergy BioTek Plate Reader instrument to quantify RNA and double-stranded DNA (dsDNA) concentrations in ng/µL. Two nuclease-free water blanks (2 µL each) per 16-well, micro-well plate in addition to 2 µL of sample were assessed per analysis.
Volcano 2nd Generation qPCR and qPCR: RNA and DNA target quantification
The qPCR assay performed for RNA targets, SARS-CoV-2 and OC43, was Volcano 2nd Generation qPCR (V2G-qPCR) described in earlier publications.5, 11, 13, 31, 34 The V2G polymerase utilized in the assay reads both RNA and DNA templates, eliminating the need for prior complementary DNA synthesis before PCR amplification, allowing the assay to be an equivalent reaction to RT-qPCR but reducing overall run time by half and removing additional synthesis steps at the bench. Primers (500 nM) and fluorescent reporter probes (250 nM) for the N3 gene of SARS-CoV-2 and OC43 targets are listed in Table 1 and were utilized in the master mix in addition to V2G buffer (1.1X), deoxyribonucleoside triphosphates (200 nM), V2G polymerase (2 units), anti-Taq antibody (1 unit), nuclease-free water, and ROX reference dye (1X). qPCR cycling for SARS-CoV-2 and OC43 was 95°C for 1 minute followed by 45 cycles of 95°C for 10 seconds, 60°C for 20 seconds, and 72°C for 15 seconds on a BioRad CFX Connect instrument with efficiencies measuring 95% or higher. The V2G polymerase was purchased from myPOLS Biotec (Konstanz, Germany), anti-Taq antibody from Thermo Fisher Scientific (Waltham, Massachusetts), deoxynucleotides from New England Biolabs (Ipswich, Massachusetts), primers and ROX from Sigma-Aldrich, Inc. (St. Louis, Missouri), and reporter probes from Integrated DNA Technologies (Coralville, Iowa).
Table 1. Primer and probe sequence information for molecular targets SARS-CoV-2, OC43, and M. smegmatis quantified by qPCR.
|
Molecular target |
Primer/probe |
Sequence of primer/probe 5’ —> 3’ |
|---|---|---|
|
SARS-CoV-2 (N3) |
CV3b/f CV3c/r CV3.prb |
TGCTAACAAAGACGGCATCA GTAGCACGATTGCAGCATTG 56-FAM/ACATTGGCA/ZEN/CCCGCAATCCTGCT/3IABkFQ |
|
OC43 (NC) |
OC43/f OC43/r OC43.prb |
CAACCAGGCTGATGTCAATAC AAACCTAGTCGGAATAGCCTCA 5HEX/ACATTGTCG/ZEN/ATCGGGACCCAAGT/3IABkFQ |
|
M. smegmatis (katG) |
qMsmKat/f qMsmKat/r MsmKat.prb |
CCGCTCGAAGAGGTCG GTCCAGGTGACCTCGAGAC 5HEX/TCCTTGCCG\/ZEN/ACGCCGGTG/3IABkFQ |
The qPCR assay performed for the bacterial target M. smegmatis was designed to specifically amplify a segment of the catalase-peroxidase gene (katG). The master mix combined 1X TaqMan Fast Universal PCR Master Mix (2X) (Thermo Fisher Scientific cat# 4352042), 400 nM forward and reverse primers (20 µM), and 250 nM fluorescent reporter probes (100 µM) (Table 1) with nuclease-free water.40 qPCR cycling for M. smegmatis was 95°C for 2 minutes followed by 40 cycles of 95°C for 10 seconds and 61°C for 25 seconds on a BioRad CFX Connect instrument with efficiencies measuring 95% or higher.
Master mixes for both assays were set up utilizing appropriate practices for avoiding contamination and loaded into qPCR plates secured in ice blocks (frozen at -20°C). For all qPCR reactions, 4 of 100 µL either RNA or DNA were amplified in 40 µL reactions per well run in replicate. Seven no-template controls of nuclease-free water combined with master mix were added to each qPCR plate to check for cross-contamination. Standards specific to each molecular target were prepared as 10-fold dilutions in ranges of 101 to 105 for developing the standard curve for quantification.
Data analysis
Shapiro–Wilk normality assessments were first performed on qPCR data, per molecular target and experimental condition (ENF versus VF and BB versus nBB), with most data (10 of 12 categories) resulting in normal distributions. Due to this, paired t-tests and Pearson correlations were utilized to assess the difference between the mean values per experimental and control groups. The statistical software package SPSS v28.0.0.0 was utilized for all assessments, wherein α = 0.05 (95% confidence) with P values less than 0.05 were considered significant unless otherwise described. Results from filter halves (half A and half B) were averaged per sampling site per day, and qPCR replicates were averaged and are reported as quantity per 4 µL input, wherein no back calculation was made to copies per liter of sewage. Initial wastewater processing volumes, ratio of filter size to amount of DNA/RNA shield utilized, and quantity of qPCR input remained consistent so that raw qPCR data could be assessed directly with no need for conversion. Figures were created using the free version of the online software BioRender and GraphPad Prism.
RESULTS AND DISCUSSION
Nucleic acid concentrations of BB and nBB wastewater concentrates
Results of the measured concentrations of dsDNA and RNA (Table 2) showed that there was a roughly 5 to 10X increase in the amount quantified between nBB and BB samples. This increase is likely attributable to the increase in DNA and RNA released due to the physical rupturing of microbial cell membranes. DNA present under the nBB condition may have been due to dsDNA viruses and/or free dsDNA from ruptured cells. We believe that a large portion of the quantified increase in RNA was ribosomal, or other: RNA rather than viral RNA. Due to the increase in quantity, we do not recommend the use of BB for viral RNA extraction, as this additional RNA signal may interfere with qPCR results of viral targets.
Table 2. Concentration (ng/µL) of resulting RNA or dsDNA following extraction for each experimental condition. ENF using HAWP membranes compared against VF using GN-6 Metricel membranes shown with or without the use of BB prior to nucleic acid extraction.
|
Experimental condition per WW sample |
ENF (BB) |
ENF (nBB) |
VF (BB) |
VF (nBB) |
||||
|---|---|---|---|---|---|---|---|---|
|
Nucleic acid type per WW sample |
dsDNA |
RNA |
dsDNA |
RNA |
dsDNA |
RNA |
dsDNA |
RNA |
|
06-221018 |
22.8 |
60.2 |
5.6 |
18.0 |
18.2 |
71.0 |
5.5 |
16.8 |
|
Dc-221018 |
35.2 |
125.0 |
3.9 |
20.6 |
37.4 |
142.0 |
5.7 |
22.5 |
|
Dg-221101 |
45.4 |
161.0 |
5.8 |
30.3 |
40.5 |
198.0 |
10.8 |
32.3 |
|
Dg-221102 |
72.2 |
191.0 |
6.2 |
44.7 |
62.8 |
175.0 |
5.6 |
19.5 |
|
Dc-221108 |
44.6 |
197.0 |
7.7 |
26.0 |
40.0 |
195.0 |
5.6 |
24.4 |
|
WW average per condition |
44.0 |
147.0 |
5.8 |
27.9 |
39.8 |
156.0 |
6.6 |
22.7 |
Although qPCR requires low concentrations of nucleic acids (5 to 15 copies of genetic material), sequencing technologies generally require larger amounts of starting nucleic acid template.30, 42 Results from this study show that the concentrations of RNA were about a factor of 4 to 5 greater than for dsDNA. The amount of nucleic acid traditionally needed for approaches like Sanger sequencing has been roughly 1 microgram (i.e., 1000 ng) as described by Dzunkova et al. 2014.43 Current and newer sequencing technologies based upon Illumina (1 ng) and IDT (0.01 ng) do not require these larger amounts within their recommended protocols. However, wastewater is highly complex and abundant with diverse microbial communities; to evaluate highly diverse samples from 0.1 to 1 µg is recommended. To describe in relation to the results generated here, at a nucleic acid concentration of 40 ng/µL, the upper bound of 1 µg can be achieved with only 25 µL of DNA or RNA eluate. The 40 ng/µL level is reached for RNA for both BB and nBB conditions. However, for dsDNA, this level is reached only for the BB condition and not for the nBB condition. These results are in agreement with Luhung et al. 201528 in that one of the recommended strategies put forth within this previous study was the inclusion of heat with sonication to increase the concentration of dsDNA downstream. If nucleic acid concentrations are to be increased, BB is therefore recommended here for DNA isolation from wastewater suspended solids, similarly to its standard use with environmental surveillance for bacteria.
For the BB condition, there was no significant difference between dsDNA or RNA quantified at 95% confidence from either ENF or VF filtration processes (dsDNA: P = 0.083, RNA: P = 0.349). Overall results show that both concentration processes provide sufficient, equivalent nucleic acid concentrations following extraction using BB.
For the nBB condition, there was also no significant difference between the ENF and VF method’s impact on mean nucleic acid concentration (dsDNA: P = 0.550, RNA: P = 0.452). From these results, for the nBB condition, we observed that the mean nucleic acid concentration was not different between the concentration methods.
qPCR analysis of ambient SARS-CoV-2 from wastewater
Results show that for the viral target SARS-CoV-2, the highest recoveries were for VF concentrates that were nBB followed by the ENF concentrates that were also nBB (Figure 2, panel A). The nBB condition showed the highest detections of SARS-CoV-2 by qPCR. Differences between BB and nBB were statistically different at 95% confidence for the SARS-CoV-2 target via the ENF method (P = 0.030) and at 90% confidence for the VF method (P = 0.077). The slope between BB and nBB samples for the ENF process (m = 0.507) suggests that BB decreases the sensitivity of the analysis by 50%. Similarly, for the VF process, the slope between BB and nBB samples (m = 0.252) suggests that BB decreases the sensitivity of analysis by 75%. To further emphasize the negative impact of BB on SARS-CoV-2 recovery, in one sample, 06-221018, collected from the UHealth Tower hospital, SARS-CoV-2 measured below detection limits with the use of BB. However, this same sample was detectable when BB was omitted. This observation provides evidence that using BB for a target such as SARS-CoV-2 decreases the efficiency of downstream qPCR. This decrease could be due to multiple factors; physical rupturing may degrade the viral RNA or increase the quantity of qPCR inhibitors, and vast differences of nucleic acid concentration could have interfered with the quantified signal by V2G-qPCR. Future research is needed to determine the utility of BB for viral RNA extraction and downstream quantification due to these limitations.
Figure 2. Quantitation of molecular targets by qPCR from BB and nBB wastewater concentrates. (A) SARS-CoV-2, (B) OC43, and (C) Mycobacterium smegmatis. HAWP membranes illustrated alongside GN-6 membranes quantified from 4 µL nucleic acid input.

qPCR analysis of spiked-in targets from wastewater
For OC43, the highest recoveries corresponded to nBB VF concentrates; BB resulted in a decrease in recoveries at 90% confidence compared to nBB concentrates (P = 0.075) (Figure 2, panel B). Compared to SARS-CoV-2, larger quantities of the spiked-in OC43 viral target were detected by qPCR following analysis with all method conditions. Results of the paired t-tests showed significant differences at 90% confidence for the VF method (P = 0.075) between BB and nBB sample concentrates. Yet, with the ENF method, an opposite trend was observed; recoveries with BB were higher than for nBB at 95% confidence (P = 0.028). The slope between BB and nBB conditions for VF was m = 0.742, suggesting that BB sample concentrates for measuring the OC43 target downstream reduces the sensitivity by 25%. For the OC43 virus, BB improved the quantity measured with qPCR only when the ENF concentration method was utilized. This suggests that heating by BB does not lead to RNA degradation, although this claim should be evaluated by future research.
These results address one objective of this study, showing that the concentration method impacts the downstream molecular quantification for this target, as consistent with current literature.26, 27, 28 These results may have been related to the pretreatment process incorporating 10% HCl for pH adjustment with the ENF method. These results collectively indicate that the chemical lysis of DNA/RNA shield alone (nBB), which releases viral nucleic acid via disassociation of the viral capsid, evaluated against the coupled mechanical lysis of the DNA/RNA shield wastewater concentrate (with BB) does not necessarily improve the overall recovery of OC43, especially if pretreatment includes acidification.
M. smegmatis had the most drastic difference between BB and nBB in that all samples had at least double the average quantity following BB (Figure 2, panel C). Statistical differences were observed between BB and nBB for both ENF (P = 0.003) and VF (P = 0.009). These significant differences emphasize the necessity of BB for bacterial DNA–containing microbes, especially for those characterized by tough lipid cell walls. Results confirm that the M. smegmatis’s microbial physiology requires mechanical lysis in addition to the chemical lysis, which DNA/RNA shield provides for an adequate result with qPCR. This agrees with the current literature for extracting DNA from bacterial targets from wastewater, sludges, and environmental samples.26, 28, 32 These results support that this Mycobacterial target is more difficult than viral particles to lyse, requiring an additional step prior to extraction for releasing DNA. As the ENF concentration method, incorporating a pH reduction with 10% HCl, had a larger difference between BB and nBB conditions, there is evidence that the incorporation of acid further assisted in the degradation of the cellular membrane of M. smegmatis for nucleic acid release, similarly to what was seen for OC43.
CONCLUSIONS
Emphasized from the COVID-19 pandemic was the narrative of important method selection for the accurate reporting of pathogens. In considering the future applications of WBS, optimized workflows stemming from current programs should be adapted to measure additional targets and expand the role of WBS in protecting public health. This study showed that for the ambient SARS-CoV-2 viral target, BB reduced the effectiveness of RNA quantification by 50 to 75%, and overall, the VF concentration method provided better recoveries as reported by qPCR coupled with nBB. Overall, separate dedicated workflows for RNA and DNA, rather than focusing on total nucleic acid extraction, had the advantage of improving the quality of RNA and DNA recovered. The above results of quantified SARS-CoV-2 between concentrates using or not using BB address an objective of this study in which we do not recommend the use of BB for measuring SARS-CoV-2, as measurable nucleic acid may become undetectable following its use—especially if abundance in a community is low.
Similarly, for the spiked-in OC43 control, differences were noted between the BB and nBB methods prior to extraction. For the optimum concentration method for OC43 (VF), nBB provided the highest recoveries. Again, for viral targets, BB resulted in lower recoveries, potentially because of the physical lysis process that may have degraded the viral RNA and/or released non-viral RNA from the samples, which may have interfered with subsequent qPCR processes. Overall, for both viral targets considered, SARS-CoV-2 and OC43, the optimum method resulting in the highest recoveries was VF with nBB. For both target viruses, the inclusion of BB would decrease recoveries from 25 to 75%.
Another important aspect of this study were the modifications made to the current-standing SARS-CoV-2 program for isolating alternative targets, specifically those containing DNA from microbes with tougher cell walls such as M. smegmatis. BB has been shown here as a necessity when isolating DNA from these microbe types. Also, the amount of DNA was larger when BB was included for both ENF and VF with the M. smegmatis target. Thus, BB can be used to increase DNA concentrations to assure that a sufficient template is available if samples are to be subjected to sequencing.
Limitations of this study include the low quantities of SARS-CoV-2 measured from wastewater. Moreover, the results of the OC43 show a general decreasing trend over time (Figure 2, panel B). The storage conditions as well as heat inactivation and use of OC43 remained consistent throughout the study period, providing that die off of this control could have been occurring. For a longer study period, fresh viral cultures of OC43 are recommended to ensure consistent signal over time; as this coronavirus is typically utilized as a recovery control within WBS research, ensuring minimal degradation of the aliquots utilized for spikes should be addressed by future research. Additional considerations for future work should address the microbiome of samples collected with different sequencing applications to ensure the RNA and DNA isolated from wastewater are representative of the diverse array of organisms found within wastewater. Finally, this study was limited by the small sample size, testing 5 raw sewage samples, in replicate. Future work should focus on replicating these experiments with larger sample sizes or additional parameters for comparison.
This investigation applied proof of principle for current SARS-CoV-2–focused WBS protocols for expansion to alternative pathogens found within wastewater. To our knowledge, this is the first study that addresses the differences between measured viral and bacterial quantities from wastewater following 2 differing primary concentration approaches with and without the incorporation of BB. Approaches must be adapted to the specific target of focus, and expansion of WBS to additional targets will require considerable experimentation among different classes of microbes ranging from RNA to DNA viruses (enveloped and non-enveloped), to bacteria with different types of cell membranes, to fungi and protozoa. For example, the use of this sample processing workflow for DNA isolation has allowed for accurate detection of Mpox and Candida auris pathogens from wastewater in 2 recent publications.39, 41 We envision a menu of methods based upon microbe morphological properties that would be available to guide WBS researchers for preprocessing wastewater samples subjected to molecular methods.
ACKNOWLEDGMENTS
The research reported in this publication was supported by the National Institute on Drug Abuse of the National Institutes of Health under Award Number U01DA053941. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPLEMENTARY MATERIAL
References
- 1.Ahmed W, Bertsch PM, Bibby K, et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ Res. 2020;191:110092. doi: 10.1016/j.envres.2020.110092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed W, Bivins A, Bertsch PM, et al. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimisation and quality control are crucial for generating reliable public health information. Curr Opin Environ Sci Health. Published online September 30, 2020. doi: 10.1016/j.coesh.2020.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bivins A, North D, Ahmad A, et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ Sci Technol. 2020;54(13):7754-7757. doi: 10.1021/acs.est.0c02388 [DOI] [PubMed] [Google Scholar]
- 4.Schmitz BW, Innes GK, Prasek SM, et al. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci Total Environ. 2021;801:149794. doi: 10.1016/j.scitotenv.2021.149794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharkey ME, Kumar N, Mantero AMA, et al. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci Total Environ. 2021;798:149177. doi: 10.1016/j.scitotenv.2021.149177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherchan SP, Shahin S, Ward LM, et al. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci Total Environ. 2020;743:140621. doi: 10.1016/j.scitotenv.2020.140621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Z, Reinke R, Hoxsey M, et al. Detection of SARS-CoV-2 in wastewater: community variability, temporal dynamics, and genotype diversity. ACS ES&T Water. 2021;1(8):1816-1825. doi: 10.1021/acsestwater.1c00119 [DOI] [Google Scholar]
- 8.Yang S, Dong Q, Li S, et al. Persistence of SARS-CoV-2 RNA in wastewater after the end of the COVID-19 epidemics. J Hazard Mater. 2022;429:128358. doi: 10.1016/j.jhazmat.2022.128358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zulli A, Pan A, Bart SM, et al. Predicting daily COVID-19 case rates from SARS-CoV-2 RNA concentrations across a diversity of wastewater catchments. FEMS Microbes. 2021;2:xtab022. doi: 10.1093/femsmc/xtab022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertels X, Demeyer P, Van den Bogaert S, et al. Factors influencing SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage: a systematic review. Sci Total Environ. 2022;820:153290. doi: 10.1016/j.scitotenv.2022.153290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solo-Gabriele HM, Kumar S, Abelson S, et al. Predicting COVID-19 cases using SARS-CoV-2 RNA in air, surface swab and wastewater samples. Sci Total Environ. 2023;857(Pt 1):159188. doi: 10.1016/j.scitotenv.2022.159188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu F, Xiao A, Zhang J, et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentations of new COVID-19 cases. Sci Total Environ. 2022;805:150121. doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan Q, Babler KM, Sharkey ME, et al. Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human waste. ACS ES T Water. 2022;2(11):1992-2003. doi: 10.1021/acsestwater.2c00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philo SE, Keim EK, Swanstrom R, et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci Total Environ. 2021;760:144215. doi: 10.1016/j.scitotenv.2020.144215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peccia J, Zulli A, Brackney DE, et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat Biotechnol. 2020;38(10):1164-1167. doi: 10.1038/s41587-020-0684-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan Q, Solo-Gabriele H, Sharkey M, et al. Correlative analysis of wastewater trends with clinical cases and hospitalizations through five dominant variant waves of COVID-19. ACS ES T Water. 2023;3(9):2849-2862. doi: 10.1021/acsestwater.3c00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vo V, Tillett RL, Chang CL,. Gerrity D, Betancourt WQ, Oh EC. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci Total Environ. 2022;805:149930. doi: 10.1016/j.scitotenv.2021.149930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilbride KA, Lee D-Y, Beaudette LA. Molecular techniques in wastewater: understanding microbial communities, detecting pathogens, and real-time process control. J Microbiol Methods. 2006;66(1):1-20. doi: 10.1016/j.mimet.2006.02.016 [DOI] [PubMed] [Google Scholar]
- 19.Jiang SC, Bischel HN, Goel R, et al. integrating virus monitoring strategies for safe non-potable water reuse. Water (Basel). 2022;14(8):1187. doi: 10.3390/w14081187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemarchand K, Masson L, Brousseau R. Molecular biology and DNA microarray technology for microbial quality monitoring of water. Crit Rev Microbiol. 2004;30(3):145-172. doi: 10.1080/10408410490435142 [DOI] [PubMed] [Google Scholar]
- 21.Lu Y, Redlinger TE, Avitia R, Galindo A, Goodman K. Isolation and genotyping of Helicobacter pylori from untreated municipal wastewater. Appl Environ Microbiol. 2002;68(3):1436-1439. doi: 10.1128/AEM.68.3.1436-1439.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanz JL, Kochling T. Molecular biology techniques used in wastewater treatment: an overview. Process Biochem. 2007;42(2):119-133. doi: 10.1016/j.procbio.2006.10.003 [DOI] [Google Scholar]
- 23.Schwartz T, Kohnen W, Jansen B, Obst U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol Ecol. 2003;43(3):325-335. doi: 10.1111/j.1574-6941.2003.tb01073.x [DOI] [PubMed] [Google Scholar]
- 24.Urrea-Valencia S, de Almeida Melo AL, Potma Goncalves DR, Galvao CW, Etto RM. Molecular techniques to study microbial wastewater communities. Braz Arch Biol Technol. 2021;64:e21200193. doi: 10.1590/1678-4324-2021200193 [DOI] [Google Scholar]
- 25.Pecson BM, Darby E, Haas CN, et al. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ Sci (Camb). 2021;7:504-520. doi: 10.1039/d0ew00946f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemarchand K, Berthiaume F, Maynard C, et al. Optimization of microbial DNA extraction and purification from raw wastewater samples for downstream pathogen detection by microarrays. J Microbiol Methods. 2005;63(2):115-126. doi: 10.1016/j.mimet.2005.02.021 [DOI] [PubMed] [Google Scholar]
- 27.Walden C, Carbonero F, Zhang W. Assessing impacts of DNA extraction methods on next generation sequencing of water and wastewater samples. J Microbiol Methods. 2017;141:10-16. doi: 10.1016/j.mimet.2017.07.007 [DOI] [PubMed] [Google Scholar]
- 28.Luhung I, Wu Y, Ng CK, Miller D, Cao B, Chang VW. Protocol Improvements for low concentration DNA-based bioaerosol sampling and analysis. PLoS One. 2015;10(11):e0141158. doi: 10.1371/journal.pone.0141158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng S, Owens SM, Shrestha A, Poretsky R, Hartmann EM, Wells G. Intensity of sample processing methods impacts wastewater SARS-CoV-2 whole genome amplicon sequencing outcomes. Sci Total Environ. 2023;876:162572. doi: 10.1016/j.scitotenv.2023.162572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed W, Simpson SL, Bertsch PM, et al. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Sci Total Environ. 2022;805:149877. doi: 10.1016/j.scitotenv.2021.149877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babler KM, Amirali A, Sharkey ME, et al. Comparison of electronegative filtration to magnetic bead-based concentration and V2G-qPCR to RT-qPCR for quantifying viral SARS-CoV-2 RNA from wastewater. ACS ES&T Water. 2022;2(11):2004-2013. doi: 10.1021/acsestwater.2c00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourrain M, Achouak W, Urbain V, Heulin T. DNA extraction from activated sludges. Curr Microbiol. 1999;38(6):315-319. doi: 10.1007/pl00006809 [DOI] [PubMed] [Google Scholar]
- 33.Li AD, Metch JW, Wang Y, et al. Effects of sample preservation and DNA extraction on enumeration of antibiotic resistance genes in wastewater. FEMS Microbiol Ecol. 2018;94(2). doi: 10.1093/femsec/fix189 [DOI] [PubMed] [Google Scholar]
- 34.Babler KM, Sharkey ME, Abelson S, et al. Degradation rates influence the ability of composite samples to represent 24-hourly means of SARS-CoV-2 and other microbiological target measures in wastewater. Sci Total Environ. 2023;867:161423. doi: 10.1016/j.scitotenv.2023.161423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada H, Yamaguchi M, Igarashi Y, et al. Mycolicibacterium smegmatis, Basonym Mycobacterium smegmatis, expresses morphological phenotypes much more similar to Escherichia coli than Mycobacterium tuberculosis in quantitative structome analysis and CryoTEM examination. Front Microbiol. 2018;9:1992. doi: 10.3389/fmicb.2018.01992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anuchin AM, Mulyukin AL, Suzina NE, Duda VI, El-Registan GI, Kaprelyants AS.. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiology (Reading). 2009;155(Pt 4):1071-1079. doi: 10.1099/mic.0.023028-0 [DOI] [PubMed] [Google Scholar]
- 37.Fujiwara N, Ohara N, Ogawa M, et al. Glycopeptidolipid of Mycobacterium smegmatis J15cs affects morphology and survival in host cells. PLoS One. 2015;10(5):e0126813. doi: 10.1371/journal.pone.0126813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmed W, Bivins A, Korajkic A, Metcalfe S, Smith WJM, Simpson SL. Comparative analysis of adsorption-extraction (AE) and nanotrap(R) magnetic virus particles (NMVP) workflows for the recovery of endogenous enveloped and non-enveloped viruses in wastewater. Sci Total Environ. 2023;859(Pt 1):160072. doi: 10.1016/j.scitotenv.2022.160072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharkey ME, Babler KM, Shukla BS, et al. Monkeypox viral nucleic acids detected using both DNA and RNA extraction workflows. Sci Total Environ. 2023;890:164289. doi: 10.1016/j.scitotenv.2023.164289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharkey ME, Babler KM, Amirali A, et al. First detection of the Monkeypox virus using wastewater-based surveillance in Miami-Dade County. Research Square. Preprint posted online September 1, 2022. doi: 10.21203/rs.3.rs-2010415/v1 [DOI] [Google Scholar]
- 41.Babler K, Sharkey M, Arenas S, et al. Detection of the clinically persistent, pathogenic yeast spp. Candida auris from hospital and municipal wastewater in Miami-Dade County, Florida. Sci Total Environ. 2023;898:165459. doi: 10.1016/j.scitotenv.2023.165459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahmed W, Bivins A, Bertsch PM, et al. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ Res. 2021;193:110531. doi: 10.1016/j.envres.2020.110531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dzunkova M, Garcia-Garcera M, Martinez-Priego L, D'Auria G, Calafell F, Moya A. Direct squencing from the minimal number of DNA molecules needed to fill a 454 picotiterplate. PLoS One. 2014;9(6):e97379. doi: 10.1371/journal.pone.0097379 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


