Abstract
Proteins present in the outer membrane of chlamydiae that are involved in mucosal epithelial cell infection must clearly be identified and characterized if we are to understand and modify the pathogenic mechanisms utilized by these organisms. We have identified and isolated a family of four genes encoding putative outer membrane proteins (POMPs), a group of proteins of approximately 90 kDa present in the outer membrane of the subtype of Chlamydia psittaci that causes ovine enzootic abortion (strain S26/3). These proteins, although minor components, are major immunogens, as shown by the immunoblotting of chlamydial outer membrane complexes with postabortion sheep sera, and are therefore potential diagnostic and/or protective antigen candidates. Immunoblotting of the expressed amino- and carboxy-terminal halves of one of the POMPs with postabortion sheep sera showed that the major humoral immune response appeared to be directed solely against the amino-terminal half. This result, in combination with the positive immunofluorescence staining of S26/3-infected cells using POMP-specific (specific to the amino-terminal half of the proteins) monoclonal antibodies, suggests the probable surface localization of the POMPs and, more specifically, the surface exposure of the amino-terminal half of these proteins. The four pomp genes are highly homologous, sharing 82 to 100% similarity with each other (two of the genes are identical). Genes with strong and weak homologies were also detected in C. psittaci avian and feline pneumonitis strains, respectively. No pomp homologs were found in strains of C. trachomatis and C. pneumoniae, but this does not preclude their existence. The absence of homology with various subtypes of C. pecorum, which complicate the diagnosis of the ovine abortion subtype, indicates the possible suitability of the these 90-kDa proteins as serodiagnostic antigens.
The elementary body (EB) outer cell envelope of members of the genus Chlamydia resembles that of gram-negative bacteria in that it contains lipopolysaccharide but is atypical in lacking a peptidoglycan layer (9). The structural stability of EBs is attributed to extensive intermolecular disulfide bond cross-linking of the 40-kDa major outer membrane protein (MOMP), one or two large (60-kDa), cysteine-rich proteins (EnvB or Omp2), and a small (12-kDa), cysteine-rich lipoprotein (EnvA or Omp3) forming a supramacromolecular lattice (reviewed in reference 25). While these are the major protein components of the outer membrane, other minor and so far uncharacterized proteins have been identified which may have major functional roles (25, 26).
Since the EB outer surface has such an important role in the infection process, it is not surprising that its outer membrane has been the focus of much research, particularly with regard to serodiagnosis and vaccine design. Currently, our research is focused on trying to improve serodiagnosis (2) and developing a recombinant vaccine (15) against the subtype of Chlamydia psittaci which causes ovine enzootic abortion (OEA). This infection is the most common infectious cause of abortion in sheep in the United Kingdom (UK) (1) and can also cause abortion in women (13). We have been particularly interested in protein antigens located in the outer membrane of OEA C. psittaci which may be involved in protective immunity. The only protein consistently shown to be exposed on the surface of the EB is the MOMP (3, 4, 6, 29). It is the major component of the chlamydial outer membrane complex (COMC), which we have previously shown protects pregnant ewes against experimentally induced OEA (30).
In addition to the MOMP, the C. psittaci COMC contains a group of minor proteins with sizes of approximately 90 kDa which have been shown to react very strongly in immunoblotting experiments (5, 12, 19, 27) and to be important serodiagnostic antigen candidates (27). Recently, we have reported the isolation of two recombinant λgt11 clones (named 31 and 99) which contain fragments of the genes coding for these 90-kDa proteins, as well as the finding that these proteins belong to a multigene family (18). This report describes the isolation and characterization of four genes which belong to this family and provides evidence of the location and possible orientation of their protein products in the outer membrane of C. psittaci.
MATERIALS AND METHODS
Chlamydia.
OEA C. psittaci S26/3 was grown in L929 cells, and C. pecorum strains were cultured in embryonated chicken eggs (20). EBs were purified from infected cells or chicken embryo yolk sacs by density gradient centrifugation, and genomic DNA was extracted as described previously (20). In addition to the C. psittaci OEA strain, genomic DNAs from the following Chlamydia strains were also studied: (i) C. psittaci avian (cockatiel isolate 725; described in reference 20) and feline pneumonitis (FePn; described in reference 21) isolates; (ii) C. pecorum subtypes P787 (causing ovine arthritis), W73 (causing ovine enteritis), PV3056/3 (causing bovine metritis), and JP1751 (ovine normal feces); (iii) C. trachomatis (VW1 serovar D; DNA obtained from J. Treharne, Institute of Ophthalmology, London, UK); and (iv) C. pneumoniae (IOL-207; DNA obtained from J. Treharne).
For immunofluorescence microscopy, McCoy cells were grown on sterile glass coverslips in trac bottles (Bibby Sterilin Ltd., Stone, UK) and infected with OEA C. psittaci (inoculum titer of 5 × 108 inclusion-forming units/ml) as previously described (20).
Hybridization screening.
A partially MboI-digested S26/3 genomic DNA library constructed in lambda replacement vector EMBL-3 (Stratagene Ltd., Cambridge, UK) from S26/3 DNA (14) was screened by standard techniques using digoxigenin (DIG)-labelled p90f31 (216-bp fragment) and p91Bf99 (450-bp EcoRI fragment) probes (clone 31 and 99 DNA inserts, respectively; described in reference 18). Recombinant phage were plated by using the restrictive host strain Escherichia coli Q359. Recombinant lambda DNA was isolated from hybridization-positive clones by plate lysates using the permissive strain E. coli LE392 and purified from lambda phage particles by polyethylene glycol 6000 precipitation in accordance with standard procedures.
Sequencing and gene analysis.
The recombinant lambda DNA clones were Southern blotted by using p90f31 and p91Bf99 DIG-labelled probes, and probe-positive restriction enzyme fragments were cloned into pBluescript (Stratagene) as described previously (18). PCR amplification of recombinant lambda DNA was performed by using the Expand Long Template PCR System (Boehringer Mannheim, Lewes, UK) and primers which flanked the cloning site of λEMBL-3 (5′-GCGCAACTCGTGAAAGGTAGGCGGA-3′ and 5′-TGAACACTCGTCCGAGAATAACGAGTGGAT-3′) and those based on sequence information obtained from the cloned restriction enzyme fragments. PCR products were subcloned into vector pGEM-T (Promega, Southampton, UK) as previously described (18). Recombinant plasmid DNA was purified by using a QIAprep Spin Plasmid Miniprep Kit (QIAGEN Ltd., Crawley, UK) for manual sequencing by the dideoxy-chain termination method using the Sequenase Version 2.0 DNA Sequencing Kit (Amersham International plc, Little Chalfont, UK) and using a QIAGEN Plasmid Kit (QIAGEN Ltd.) for automated Taq FS cycle sequencing (at The Advanced Biotechnology Centre, The Charing Cross and Westminster Medical School, London, UK). All internal primers were synthesized by Oswel DNA Service (Medical and Biological Sciences Building, University of Southampton, Southampton, UK). Recombinant plasmid DNA was sequenced manually at least once in each direction. All sequencing problems, primarily due to secondary structure, were resolved by automated cycle sequencing.
DNA and deduced amino acid sequences were analyzed by using the Wisconsin GCG software package on Seqnet (Daresbury Laboratory) and DNASTAR software (DNASTAR Inc.). The protein sequences were also analyzed by using the program PSORT (on the World Wide Web at URL http://psort.nibb.ac.jp) (23), which predicts the presence of signal sequences by the methods of McGeoch (22) and von Heijne (31) and detects potential transmembrane domains by the method of Klein et al. (17). DNA and amino acid sequences were compared with those in the GenBank/EMBL and SwissProt databases by using the GCG FASTA program and the Genome Sequence Database (GSDB) (on the World Wide Web at URL http://www.tigr.org) (8, 10).
PCR analysis.
C. psittaci S26/3 genomic DNA fragments containing the four pomp genes were amplified by PCR by using the Expand Long Template PCR System and mutually exclusive pomp flanking region primers: primer pairs S5766 (5′-GTAAGCAAGAACGCCGCAAACG-3′) plus P0787 (5′-CTAGTGGAGCCGTTGAATTCAGTTGC-3′) and S5765 (5′-TGAGCCAAATCCTTCTACAGCAAT-3′) plus S2429 (5′-AGTCTTAGCGTCACAGGCCGAG-3′) for regions A and B, respectively (see Fig. 2). PCR products were analyzed by restriction endonuclease digestion (EcoRI, PstI, and XbaI; single and double digests).
FIG. 2.
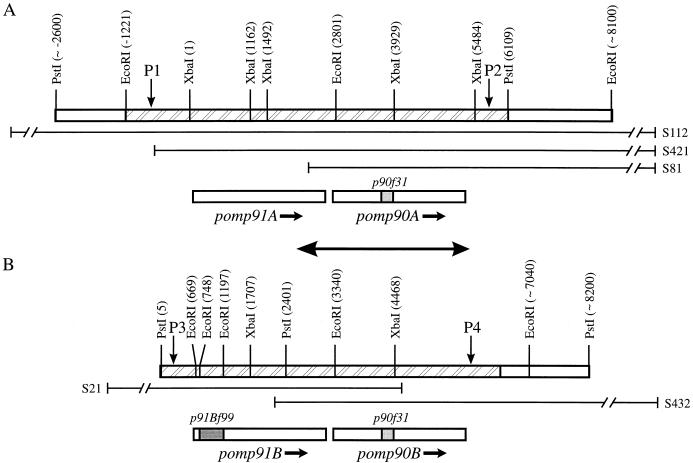
Restriction enzyme maps for the family of 90-kDa pomp genes. The positions of the restriction enzyme sites (expressed in base pairs), deduced from the sequencing (hatched areas) and Southern blotting results, are in parentheses. The locations of the p90f31 and p91Bf99 sequences within the pomp genes are denoted by the shaded regions. A and B represent two separate regions of the OEA C. psittaci genome. The sequences encoded by λEMBL-3 clones S81, S112, and S421 for region A (A) and by clones S21 and S432 for region B (B) are as illustrated. The sequences incorporating pomp91A and pomp90A (positions 1 to 6110 in A) and pomp91B and pomp90B (positions 1 to 6234 in B) have been submitted to the GenBank/EMBL databases and assigned accession no. U65942 and U65943, respectively. The double-headed arrow depicts the region of complete identity in A (positions 2111 to 5312) and B (positions 2650 to 5851). The pomp flanking region forward primers S5766 (P1) and S5765 (P3) and reverse primers P0787 (P2) and S2429 (P4) bind at the indicated positions.
Immunoblotting.
C. psittaci S26/3 EBs were purified from infected L929 cells by density gradient centrifugation as described previously (20). COMCs prepared from EBs as the Sarkosyl-dithiothreitol (DTT)-insoluble fraction (30) were solubilized in n-octyl-α-d-glucopyranoside–DTT (19) and subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on 7.5% gels after boiling in sample buffer containing 2% SDS and 5% 2-mercaptoethanol. Protein was transferred to a nitrocellulose membrane and incubated with P90F31 and P91BF99 (protein products of gene fragments p90f31 and p91Bf99, respectively) affinity-purified antibodies or a pool of postabortion sera from 10 ewes experimentally infected with OEA C. psittaci, and antibody binding was detected as described previously (18).
Expression analysis.
The PCR-generated 5′ and 3′ halves of pomp90A were cloned in frame into expression vector pET-22b(+) (Novagen, R&D Systems Europe Ltd., Abingdon, UK) via engineered NdeI/HindIII and NdeI/EcoRI restriction enzyme sites, respectively, in the primers by using procedures described previously (18). The 1.16-kb 5′ and 1.26-kb 3′ fragments were generated by using primer pairs P0786 (5′-CCAAAGCATGAACATATGAAACATCCAGTC-3′) plus V8704 (5′-AAGAAAGCTTCTGTAATGTGGTCCCTAG-3′) and T3252 (5′-AACGCTGTCCATATGGTCGATGCTGATGGC-3′) plus V8703 (5′-GAGGTGGATGAGAATTCAACTGGATCTTCG-3′), respectively. Initial cloning was performed in recA mutant strain JM109, which lacks the gene for T7 RNA polymerase, to allow examination of the construct sequences. The pET constructs were subsequently transformed into expression host BL21(DE3), which contains the T7 RNA polymerase gene, and induced at 37°C for 3 h with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. Expressed products were purified from inclusion bodies by a differential centrifugation procedure as detailed in Novagen’s pET System Manual (24) and analyzed by Western blotting as described in the paragraph on immunoblotting but also by using 12 anti-90-kDa protein monoclonal antibodies raised against whole EBs from C. psittaci abortion strains (three monoclonal antibodies) and a parakeet strain (nine monoclonal antibodies). All of these monoclonal antibodies were obtained from E. Vretou, Institut Pasteur Hellenique, Athens, Greece (the original source of the avian monoclonal antibodies was W. Schuy, Behringwerke, Marburg, Germany), and have been described previously (32). In particular, the avian strain monoclonal antibodies are known to cross-react with the ovine abortion strain 90-kDa antigen triplet. A summary of the various antibodies used in this study is shown in Table 1.
TABLE 1.
Antibody reagents used in this studya
| Antibody | Antibody type | Antigen source | Antigen recognition | Source or reference |
|---|---|---|---|---|
| 181 | Ascites | C. psittaci abortion strain EBs | 90-kDa triplet; 49-kDa amino-terminal fragment of POMP90A | 32 |
| 191 | ||||
| 192 | ||||
| 89-79/0040 | Tissue culture supernatant | C. psittaci avian (parakeet) strain EBs | 90-kDa triplet; P91BF99 | W. Schuy, Behringwerke, Marburg, Germany; 32 |
| 89-84/019 | ||||
| 89-76/073 | 90-kDa triplet; P90F31 | |||
| 89-76/049 | ||||
| 89-76/030 | ||||
| 89-73/053 | 90-kDa triplet; 49-kDa amino-terminal fragment of POMP90A | |||
| 89-73/04 | ||||
| 89-73/0200 | ||||
| 89-73/0248 | ||||
| Anti-P90F31 | Affinity-purified monospecific antiserum | P90F31 | 90-kDa triplet; P90F31 | 18 |
| Anti-P91BF99 | P91BF99 | 90-kDa triplet; P91BF99 |
Antibodies 181, 191, and 192 are serotype 1 specific (32). Either mouse monoclonal antibodies or antibodies affinity purified by using pooled postabortion sera from ewes experimentally infected with OEA C. psittaci were used. The antigen source for antibody production or purification was either formalin-inactivated whole EBs or SDS-PAGE-denatured antigen.
Immunizations.
Two 500-μg doses of the purified carboxyl half of POMP90A in adjuvant Montanide ISA 773 (SEPPIC, Paris, France) were administered subcutaneously 2 weeks apart to four hoggs (1- to 2-year-old sheep). All animals were bled prevaccination and 3 weeks after the second vaccination. Sera were analyzed by Western blotting of whole EB preparations as described previously (18).
Immunofluorescence.
OEA strain-infected McCoy cells, grown on coverslips for various times (18 to 72 h) postinfection, were briefly washed in phosphate-buffered saline (PBS) and fixed with acetone for 10 min. After air drying for 15 min, coverslips were stored at −70°C. Upon removal from the freezer, coverslips were washed in PBS and incubated with 2% bovine serum albumin in PBS for 30 min and then incubated for 1 h at 37°C in primary antibody diluted in 2% bovine serum albumin–PBS. Coverslips were washed three times for 5 min each time with PBS and then incubated for 1 h at 37°C in fluorescein-conjugated sheep anti-mouse secondary antibody (Scottish Antibody Production Unit, Law Hospital, Carluke, Scotland, United Kingdom) diluted 1/80 in PBS. After a further three washes, coverslips were inverted onto a drop of Citifluor (glycerol-PBS mixture) (Citifluor, Canterbury, UK) and examined under an Olympus BX50 microscope using a 40× objective.
Southern blot analysis.
Genomic DNA (500-ng aliquots) prepared from C. psittaci cockatiel and FePn isolates, as well as C. trachomatis (VW1 serovar D), C. pneumoniae (IOL-207), and various C. pecorum (P787, W73, PV3056/3, and JP1751) subtypes were Southern blotted by using DIG-labelled p91Bf99 (450-bp EcoRI fragment) and pomp90A (1.1-kb EcoRI/XbaI and 1.6-kb XbaI fragments) probes (probe concentration of 25 ng/ml) as previously described (18). The hybridization buffer consisted of 25% deionized formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.02% SDS, and 2% blocking reagent (Boehringer Mannheim). Hybridization was performed at 37°C for 17 h. The most stringent wash was in 2× SSC–0.1% SDS at 37°C. Detection of probe binding was with the chemiluminescent substrate CSPD (Boehringer Mannheim).
Nucleotide sequence accession numbers.
The DNA sequences reported here were submitted to the GenBank/EMBL databases and assigned accession no. U65942 and U65943.
RESULTS
Cloning and sequence analysis of the genes for the 90-kDa proteins.
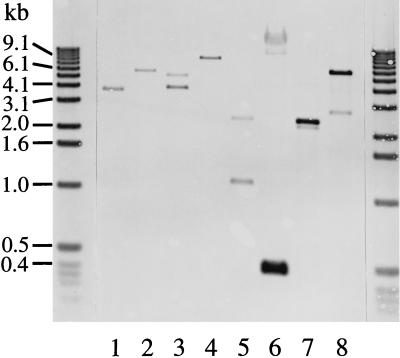
Five positive clones were isolated from the λEMBL-3 library, and restriction endonuclease-digested recombinant lambda DNA was analyzed by Southern blotting by using p90f31 (data not shown) and p91Bf99 (Fig. 1) probes. All of the fragments previously observed by Southern blotting of digests of S26/3 genomic DNA with these probes (18) were identified. Initially, the 0.45-, 3.7-, 4.0-, and 5.3-kb EcoRI fragments were subcloned and sequenced. Further sequence analysis was performed on PCR-derived DNA fragments, although ultimately all of the sequence information presented here was obtained from λEMBL-3 clone-derived restriction enzyme fragments. The combination of information obtained from the Southern blotting, mapping, and sequencing of these EMBL-3 clones enabled the maps shown in Fig. 2 to be constructed. The maps were verified further by the restriction enzyme analysis of PCR-generated 6.5- and 5.6-kb DNA fragments of the two OEA strain genomic DNA regions (A and B in Fig. 2) containing the pomp91A and pomp90A and the pomp91B and pomp90B genes, respectively (results not shown).
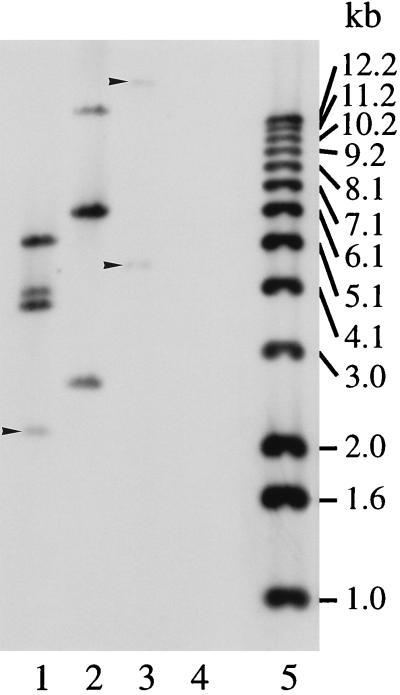
FIG. 1.
Southern blot analysis of ovine abortion strain S26/3 λEMBL-3 clone DNA. Aliquots (50 ng) of recombinant lambda DNA prepared from clones S432 (lanes 1 and 2), S112 (lanes 3 to 5), and S21 (lanes 6 to 8) were digested with EcoRI (lanes 1, 3, and 6), PstI (lanes 2, 4, and 7), and XbaI (lanes 5 and 8) and Southern blotted by using a DIG-labelled p91Bf99 probe as described in Materials and Methods. DNA size markers are indicated on the left.
Sequence analysis identified four possible open reading frames (ORFs) designated as follows: pomp for putative outer membrane protein, 90 or 91 for the predicted molecular mass (in kilodaltons) of the translated product, and A or B to distinguish the two different locations of these genes in the chlamydial genome (Fig. 2). Genes pomp90A and pomp90B are identical, each having an ORF of 2,520 bp encoding a protein (POMP90A and POMP90B, respectively) with a predicted molecular mass of 89,836 Da and pI 5.27. Gene pomp91A has an ORF of 2,544 bp encoding a protein (POMP91A) with a predicted mass of 90,707 Da and pI 5.45, and pomp91B has an ORF of 2,541 bp encoding a protein (POMP91B) with a predicted mass of 90,847 Da and pI 6.08. These predicted molecular masses and pI values correlate well with those determined by two-dimensional electrophoretic analysis (33). Overall, the pomp91A and pomp91B sequences are 85% similar and both are 82% similar to that of pomp90A or pomp90B. Consensus promoter sequences, ribosomal binding sites, and transcription termination sequences could be identified for each gene (see GenBank database entries U65942 and U65943). In addition, polyguanine tracts, consisting of 13 to 15 dGTP groups, could be found located centrally in each of the four genes (Fig. 3). Analysis of the sequences also confirmed our previous suggestion that p90f31 and p91Bf99 were derived from different genes (18), with p90f31 originating from either pomp90A or pomp90B and p91Bf99 originating from pomp91B (Fig. 2). Searches in the GenBank/EMBL, GSDB, SwissProt databases failed to find any related DNA or protein sequences.
FIG. 3.
The polyguanine tract regions of the pomp genes. These sequences can be found under GenBank/EMBL accession no. U65942 for pomp91A (positions 1283 to 1324) and pomp90A (positions 3942 to 3980) and U65943 for pomp91B (positions 1828 to 1866) and pomp90B (positions 4481 to 4519). Dots represent gaps introduced into the sequences to maximize alignment.
Figure 4 shows a multiple sequence alignment of the four proteins and highlights the similarity of the sequences. POMP91A and POMP91B were found to be 89% similar to each other and 86% and 87% similar, respectively, to POMP90A and POMP90B. Analysis of the N-terminal protein sequences by the PSORT protein localization prediction program showed that all of the POMPs have predicted signal sequences and, with the exception of POMP91A, signal peptidase I cleavage sites (Fig. 4). Furthermore, no hydrophobic transmembrane regions were identified and the overall prediction, again with the exception of POMP91A, was that the proteins are located in the outer membrane. This correlated with secondary-structure analyses (11) predicting that they consist almost entirely of β-sheet, a normal conformation for bacterial outer membrane proteins where the membrane-spanning regions are formed by the β strands. In addition, the last residue of all four proteins is phenylalanine, a feature found in the majority of bacterial outer membrane proteins (28).
FIG. 4.
Multiple sequence alignment of the predicted amino acid sequences of the pomp genes. Sequences were aligned by using the PILEUP program of the Wisconsin GCG package. Dots represent gaps introduced into the sequences to maximize sequence similarity. The blackened areas indicate amino acid identity, whereas the shaded residues indicate different levels of similarity (the darker shading represents a greater degree of similarity). P90F31 and P91BF99 are represented by residues 311 to 382 in POMP90A and POMP90B and residues 44 to 195 in POMP91B, respectively. Signal peptidase I cleavage sites were predicted for POMP90A, POMP90B, and POMP91B between positions 16 and 17 (arrowhead).
Immunoblotting analysis of COMCs.
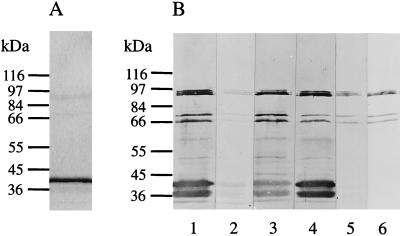
When COMCs were immunoblotted by using a pool of postabortion sera from experimentally infected ewes, the major reaction was with the MOMP, which comprises about 90% of the protein detected by Coomassie blue staining and typically occurs in these preparations as a double band of about 38 and 36 kDa. However, a triplet of bands with molecular masses in the region of 90 to 95 kDa was also clearly recognized, together with proteins of 70 and 66 kDa (Fig. 5B, lane 1). Two smaller bands have also been observed, in addition to the 90-kDa protein triplet, in whole-EB preparations (18), although these bands were larger than those reported here in COMCs. The reduction in size of these smaller proteins possibly resulted from proteolytic cleavage occurring during the preparation of the COMCs, an effect which could also account for the double MOMP band.
FIG. 5.
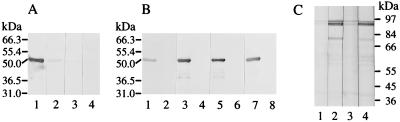
Immunoblot analysis of EB outer membrane preparations. COMCs prepared as detailed in Materials and Methods were subjected to SDS-PAGE on 7.5% gels under reducing conditions and Coomassie stained (A) or immunoblotted (B) with the pool of postabortion sera used to screen the λgt11 library from which p90f31 and p91Bf99 were isolated (lane 1); a pool of negative control sera from a flock free of chlamydial infection (lane 2); sera from two individual ewes, 3205 (lane 3) and 3210 (lane 4); and P91BF99 (lane 5) and P90F31 (lane 6) affinity-purified antibodies. Molecular masses are indicated.
Coomassie staining indicated that the proteins were present in low amounts in COMCs (Fig. 5A) and thus appear to be highly immunogenic and immunodominant: they were recognized by a large proportion of the postabortion sera obtained from individual ewes (for example, Fig. 5B, lanes 3 and 4). The bands were also recognized by the P90F31 and P91BF99 affinity-purified antibodies (Fig. 5B, lanes 5 and 6), confirming that these proteins are the products of the genes from which p90f31 and p91Bf99 originated.
Expression of the amino- and carboxy-terminal halves of POMP90A.
Immunoblotting of the expressed and purified products of both the 5′ and 3′ fragments of pomp90A showed that all 12 abortion and avian strain monoclonal antibodies and the pool of postabortion sera only detected the amino-terminal half (Fig. 6A and B). There was no reaction with the carboxy-terminal half, even when further postabortion sera were tested. In contrast, sera from sheep that were vaccinated with the purified carboxy-terminal product showed clear recognition of the 90-kDa protein triplet in immunoblotting of a whole-EB preparation (Fig. 6C).
FIG. 6.
Immunoblot analysis of the expressed amino- and carboxy-terminal halves of POMP90A. (A and B) Purified amino (A, lanes 1 and 2; B, lanes 1, 3, 5, and 7) and carboxyl (A, lanes 3 and 4; B, lanes 2, 4, 6, and 8) fragments of POMP90A (0.3- and 0.2-μg aliquots in A and B, respectively) were subjected to SDS-PAGE on 10% gels under reducing conditions and immunoblotted with the pool of postabortion sera (A, lanes 1 and 3); the pool of negative control sera (A, lanes 2 and 4); and anti-90-kDa monoclonal antibodies (B) 89-76/049 (lanes 1 and 2), 89-84/019 (lanes 3 and 4), 181 (lanes 5 and 6), and 89-73/053 (lanes 7 and 8). (C) Whole-EB preparations were subjected to SDS-PAGE on 7.5% gels under reducing conditions and immunoblotted with sera from ewes immunized with the carboxy-terminal half of POMP90A. Sera from ewes 2631 (lanes 1 and 2) and 2637 (lanes 3 and 4) collected preimmunization (lanes 1 and 3) and postimmunization (lanes 2 and 4) were used. Molecular masses are indicated.
As controls, the P90F31 and P91BF99 affinity-purified antibodies only recognized the amino-terminal product (data not shown), and a pool of negative control sera from a flock free of chlamydial infection (Fig. 6A, lanes 2 and 4) and anti-MOMP monoclonal antibody 4/11 (19) (data not shown) recognized neither product. Further analysis of the expressed products of p90f31 and p91Bf99 with the avian monoclonal antibodies showed that three (including 89-76/049 in Fig. 6B, lane 1) recognized P90F31, while two (including 89-84/019 in Fig. 6B, lane 3) recognized P91BF99 (data not shown). The remaining five avian strain (including 89-73/053 in Fig. 6B, lane 7) and three abortion strain (including 181 in Fig. 6B, lane 5) monoclonal antibodies must recognize the regions outside the P90F31 and equivalent P91BF99 regions of the amino-terminal half of POMP90A. Although the size of the carboxy-terminal product, estimated by SDS-PAGE analysis, correlates well with its theoretical molecular mass of 48 kDa, the amino-terminal product ran anomalously, giving an estimated mass of 49 kDa, which is larger than its theoretical size of 41 kDa.
Immunofluorescence.
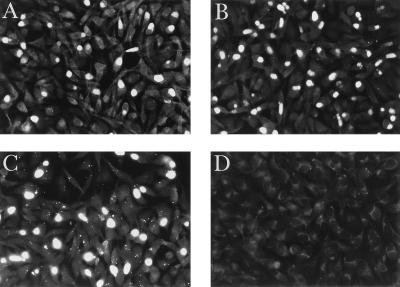
Immunofluorescence microscopy of S26/3-infected McCoy cells at different times throughout the chlamydial growth cycle (every 6 h from 18 to 54 h plus 72 h postinfection) revealed weak immunofluorescent staining of very small inclusions at 18 h postinfection when anti-MOMP monoclonal antibody 4/11 was used. This staining increased in intensity by 24 h, when inclusions were clearly recognizable within the cytoplasm of the McCoy cells, and by 30 h, inclusions were highly fluorescent. The inclusions remained highly fluorescent at all subsequent times up to 72 h postinfection. In contrast, with anti-POMP monoclonal antibodies 181 and 0040 (Table 1), weak immunofluorescent staining was not observed until 30 to 36 h postinfection and it was not until 42 h that inclusions were highly fluorescent. This staining intensity was maintained at all subsequent times up to 72 h postinfection. Figure 7 shows the immunofluorescent staining of inclusions at 48 h postinfection with both anti-POMP (A and B) and anti-MOMP (C) monoclonal antibodies (approximately one cell in three was infected). Nonspecific staining was not observed with any of these antibodies on uninoculated McCoy cells, and no staining was observed on infected cells with a nonspecific monoclonal antibody (Fig. 7D).
FIG. 7.
Immunofluorescence microscopy of C. psittaci S26/3-infected McCoy cells. C. psittaci S26/3-infected McCoy cells at 48 h postinfection were probed with putative outer membrane protein-specific monoclonal antibodies 181 (A) and 0040 (B) and MOMP-specific monoclonal antibody 4/11 (C) as detailed in Materials and Methods. A nonspecific (antileukotoxin) monoclonal antibody (D) was used as a negative control (leukotoxin is a cytolytic pore-forming protein from Pasteurella haemolytica serotype A1).
Homologous genes in other chlamydial species.
Analysis of genomic DNAs from C. trachomatis (VW1 serovar D), C. pneumoniae (IOL-207), and four subtypes of C. pecorum (P787, W73, PV3056/3, and JP1751) by Southern blotting with p91Bf99 and pomp90A probes under low-stringency hybridization and washing conditions failed to reveal any related sequences (Fig. 8, lane 4 for P787). With the FePn strain of C. psittaci, EcoRI fragments of 4.4 and ∼20 kb hybridized weakly under low-stringency conditions with the pomp90A (Fig. 8, lane 3), but not the p91Bf99 (data not shown), probe. However, with the avian strain of C. psittaci, EcoRI fragments of 2.5, 6.2, and ∼14 kb showed strong homology with both the pomp90A (Fig. 8, lane 2) and p91Bf99 (data not shown) probes, even under high-stringency conditions. No extra bands were identified by exposing blots to X-ray film for longer periods.
FIG. 8.
Southern blot analysis of other Chlamydia subtypes and species. EcoRI-digested genomic DNAs from C. psittaci OEA (lane 1), avian (lane 2), and FePn (lane 3) strains and C. pecorum ovine arthritis strain P787 (lane 4) were Southern blotted and probed with DIG-labelled 1.1- and 1.6-kb fragments of pomp90A as described in Materials and Methods. Weak hybridization signals are indicated by arrowheads. DNA size markers (lane 5) are indicated.
DISCUSSION
The chlamydial outer membrane surface must play an important role in mucosal epithelial cell infection, being involved in the attachment of EBs to the host cell membrane, their subsequent uptake, and possibly the prevention of lysosome fusion with the inclusion membrane. The abundance and immunogenicity of the MOMP have caused most research to be focused on this protein. However, antibodies against other chlamydial proteins are produced during the course of a natural or experimental infection, particularly a group of three proteins with molecular masses of about 90 kDa (12, 18, 27). Additionally, a monoclonal antibody specifically recognizing an 89-kDa protein present in COMCs partially reduced infectivity in vitro (5). This 89-kDa protein band was reported later to consist of a triplet of proteins of 80, 85, and 90 kDa (27), which are the same as the 90-kDa protein triplet described in the present study from comparison of reactivities of antigen-specific monoclonal antibodies, affinity-purified antibodies, and a series of anti-90-kDa chlamydial parakeet strain monoclonal antibodies (32). These 90-kDa proteins are minor but highly immunogenic components of the COMC (Fig. 5) that we have previously shown to protect sheep from experimental chlamydial infection (30). They have also been suggested to be important for the diagnosis of OEA strain infection (27). For these reasons, it was considered extremely important to characterize these proteins at the molecular level since they may play an important role in both the serodiagnosis of, and protection from, OEA C. psittaci.
Previously, we have isolated two λgt11 clones, containing p90f31 and p91Bf99, which were shown to be part of a gene or genes coding for a group of three 90-kDa proteins (18). These bands could have resulted from separate related genes or from the posttranslational modification of one or more genes. Southern blot analysis of genomic DNA suggested several genes (18), a finding that was confirmed in this study, in which four genes have been identified and isolated. The genes were located as two pairs in different regions of the chlamydial genome (Fig. 2). Since the OEA C. psittaci genome has not been mapped, the genomic distance separating these two regions has not been determined, although it could be estimated by pulsed-field gel electrophoresis. The 3,202 bp of complete identity that exists between the two regions (Fig. 2) encompasses the pomp90A and pomp90B genes, as well as the 42 bp downstream and 640 bp upstream (includes the 130-bp intergenic region) DNA sequences (Fig. 2). This may indicate the point at which divergence in the sequence has occurred during evolution, resulting in the duplication of the genes. This duplication was shown to be genuine and not due to cloning artifacts caused by rearrangements occurring during the construction of the λEMBL-3 library, by comparison of Southern hybridization restriction enzyme banding patterns of the lambda clones (Fig. 1) with those of genomic DNA (18), as well as by restriction endonuclease analysis of PCR-generated genomic DNA fragments containing the pomp gene sequences.
Sequence analysis predicts that the protein products of these pomp genes are located in the outer membrane. But of more importance are the immunofluorescence results obtained by using POMP-specific monoclonal antibodies (Fig. 7) and the immunoblotting results obtained with the Sarkosyl-DTT-insoluble COMC fraction (Fig. 5), since they provide experimental evidence of the localization of the 90-kDa proteins in the outer membrane. These results also imply a direct correlation between the surface accessibility of the proteins and in vivo immune recognition. Further evidence supporting the outer membrane location of these proteins came recently from Everett and Hatch, who investigated the probable cellular location of the cysteine-rich proteins (7). They clearly demonstrated the presence of the characteristic triplet of 90-kDa proteins in COMCs of C. psittaci 6BC (Fig. 1 in reference 7), as well as the surface labelling, albeit weak labelling, of these proteins using the hydrophobic affinity probe 3-(trifluoromethyl)-3-(m-[125I]iodophenyl)-diazirine (Fig. 4 in reference 7), although they did not discuss or refer to these results in their report.
Since the number of chlamydiae increases dramatically during infection, the inability to detect fluorescent staining at times earlier than 30 to 36 h postinfection with the anti-POMP monoclonal antibodies may be a consequence of the proteins being minor components of COMCs (i.e., due to detection levels). An alternative explanation could be that the proteins are products of late genes. Although the latter part of the cycle is asynchronous, with cells containing both dividing reticulate bodies and mature EBs, at 48 h chlamydiae are predominantly in the form of EBs, as judged by transmission electron microscopy. This implies that the fluorescence observed at this time (Fig. 7A and B), and at subsequent times postinfection, is due to staining of EBs and, possibly, reticulate bodies just prior to their reorganization into EBs. This requires further extensive investigation by electron microscopy at times early and late in the chlamydial cycle.
The cysteine content of the proteins ranges from 1.6 to 2% (POMP90A/B, POMP91A, and POMP91B contain 13, 15, and 17 cysteine residues, respectively), which is similar to the 1.9% of the MOMP and may indicate that the proteins are also involved in the disulfide-bonded cross-linking which is responsible for the structural rigidity of EBs, as was also suggested by Vretou et al. (33). Furthermore, the immunoblotting results obtained with the amino- and carboxy-terminal halves of POMP90A (Fig. 6), which show that the major immunological response was to the amino-terminal half of the protein, could indicate that the proteins form two domains with the carboxy-terminal domain inserted in the outer membrane and the amino-terminal domain exposed on the surface. Indeed, the short polyglycine region located centrally in each protein, coded for by the polyguanine tract (Fig. 3), may reflect a flexible linking region between two such protein domains. The lack of immune recognition of the carboxy-terminal fragment was investigated further by attempting to raise antibodies to the recombinant protein in sheep. Identification of the 90-kDa protein triplet by immunoblot analysis with sera from these immunized animals shows that the carboxyl-terminal fragment is, indeed, immunogenic, and so the lack of recognition with postabortion sheep sera is possibly due to the internal localization of this part of the protein. A further explanation could be that part of the carboxy-terminal end of the protein is surface exposed but inaccessible to antibody. The surface exposure of the amino-terminal fragment, suggested by immunofluorescence microscopy studies using monoclonal antibodies which recognize this part of the protein (Fig. 7), and the possible internal localization of the carboxy-terminal fragment require further investigation by immunoelectron microscopy using the anti-90-kDa monoclonal antibodies and monospecific antisera prepared against the carboxy-terminal protein fragment, respectively.
Since this group of proteins may play an important role in protective immunity against and/or the serodiagnosis of chlamydial infection, we searched for homologous genes in other chlamydial species (isolates of C. pecorum, C. trachomatis, and C. pneumoniae) but failed to detect any related gene sequences (Fig. 8). However, this does not necessarily imply that such genes are absent but that the homologies are too low for identification. The possible absence, or low homologies, of these genes in the C. pecorum subtypes should make the 90-kDa proteins suitable as serodiagnostic antigens, enabling ewes infected with the OEA agent to be identified without the problem of false-positive values due to widespread C. pecorum infection. The antigens are currently undergoing assessment for serodiagnostic potential, the preliminary results of which look promising (16). The role of the proteins in protection remains an open question, although they are highly immunogenic components of the subcellular vaccine that we have demonstrated to be protective (30).
This is the first report describing the isolation from Chlamydia species of duplicated genes coding for a cluster of immunogenic envelope proteins which may participate in key steps of the infection process. As research on these proteins is in its infancy and there are no homologous genes or proteins in the various databases, no function can be assigned. This is compounded by the constraints we have on the growth and genetic manipulation of the organism, which is perhaps the reason that the functions of other chlamydial proteins, including the MOMP and Omp2, have yet to be definitively demonstrated. Regardless of this, the proteins are important serodiagnostic and vaccine antigen candidates that we believe will eventually have a broad impact on most chlamydial species.
ACKNOWLEDGMENTS
We thank J. Machell for providing EBs for Southern blot analysis, S. Wyllie for preparing the outer membrane EB fraction, I. E. Anderson for assistance in screening the λEMBL-3 library, M. Livingstone for assistance with the sheep vaccinations, and B. J. Easter for photography. We are grateful to D. Buxton for helpful discussions. We also thank E. Vretou, Institut Pasteur Hellenique, Athens, Greece, for supplying the anti-POMP monoclonal antibodies; W. Donachie for the antileukotoxin antibody; and J. Treharne, Institute of Ophthalmology, London, UK, for the C. trachomatis and C. pneumoniae isolates.
This work was supported by grants from the European Commission, Division of Agricultural Research, project CT93-0957 (AIR 3), and the Scottish Office Agriculture, Environment and Fisheries Department.
ADDENDUM
Since this report was submitted for publication, nine genes with protein sequences homologous to those of the POMPs have been identified in C. trachomatis serovar D (D/UW-3/Cx) trachoma biovar by the Chlamydia Genome Project (on the World Wide Web at URL http://chlamydia-www.berkeley.edu :4231/index.html). The genes have been named pmpA to pmpI.
REFERENCES
- 1.Aitken I D. Enzootic (chlamydial) abortion. In: Martin W B, Aitken I D, editors. Diseases of sheep. Oxford, England: Blackwell Scientific Publications; 1991. pp. 43–48. [Google Scholar]
- 2.Anderson I E, Herring A J, Jones G E, Low J C, Greig A. Development and evaluation of an indirect ELISA to detect antibodies to abortion strains of Chlamydia psittaci in sheep sera. Vet Microbiol. 1995;43:1–12. doi: 10.1016/0378-1135(94)00083-9. [DOI] [PubMed] [Google Scholar]
- 3.Baehr W, Zhang Y-X, Joseph T, Su H, Nano F E, Everett K D E, Caldwell H D. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc Natl Acad Sci USA. 1988;85:4000–4004. doi: 10.1073/pnas.85.11.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cevenini R, Donati M, Brocchi E, De Simone F, La Placa M. Partial characterisation of an 89-kDa highly immunoreactive protein from Chlamydia psittaci A/22 causing ovine abortion. FEMS Microbiol Lett. 1991;81:111–116. doi: 10.1016/0378-1097(91)90481-o. [DOI] [PubMed] [Google Scholar]
- 6.Collett B A, Newhall V W J, Jersild R A, Jr, Jones R B. Detection of surface-exposed epitopes on Chlamydia trachomatis by immune electron microscopy. J Gen Microbiol. 1989;135:85–94. doi: 10.1099/00221287-135-1-85. [DOI] [PubMed] [Google Scholar]
- 7.Everett K D E, Hatch T P. Architecture of the cell envelope of Chlamydia psittaci 6BC. J Bacteriol. 1995;177:877–882. doi: 10.1128/jb.177.4.877-882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 9.Fox A, Rogers J C, Gilbart J, Morgan S, Davis C H, Knight S, Wyrick P B. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect Immun. 1990;58:835–837. doi: 10.1128/iai.58.3.835-837.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrmann J, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of a simple method for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths P C, Philips H L, Dawson M, Clarkson M J. Antigenic and morphological differentiation of placental and intestinal isolates of Chlamydia psittaci of ovine origin. Vet Microbiol. 1992;30:165–177. doi: 10.1016/0378-1135(92)90111-6. [DOI] [PubMed] [Google Scholar]
- 13.Herring A J, Anderson I E, McClenaghan M, Inglis N F, Williams H, Matheson B A, West C P, Rodger M, Brettle R P. Restriction endonuclease analysis of DNA from two isolates of Chlamydia psittaci obtained from human abortions. Br Med J. 1987;295:1239. doi: 10.1136/bmj.295.6608.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herring A J, Tan T W, Baxter S, Inglis N F, Dunbar S. Sequence analysis of the major outer membrane protein gene of an ovine abortion strain of Chlamydia psittaci. FEMS Microbiol Lett. 1989;65:153–158. doi: 10.1016/0378-1097(89)90383-2. [DOI] [PubMed] [Google Scholar]
- 15.Herring A J, McCafferty M C, Jones G E, Dunbar S, Andersen A A. Vaccination against chlamydial abortion in sheep: problems and progress with a recombinant vaccine. In: Orfila J, Byrne G I, Chernesky M A, Grayston J T, Jones R B, Ridgway G L, Saikku P, Schachter J, Stamm W E, Stephens R S, editors. Chlamydial infections. Bologna, Italy: Societa Editrice Esculapio; 1994. pp. 118–121. [Google Scholar]
- 16.Jones, G. E., J. Machell, E. Vretou, and D. Longbottom. Unpublished data.
- 17.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 18.Longbottom D, Russell M, Jones G E, Lainson F A, Herring A J. Identification of a multigene family coding for the 90 kDa proteins of the ovine abortion subtype of Chlamydia psittaci. FEMS Microbiol Lett. 1996;142:277–281. doi: 10.1111/j.1574-6968.1996.tb08443.x. [DOI] [PubMed] [Google Scholar]
- 19.McCafferty M C, Herring A J, Andersen A A, Jones G E. Electrophoretic analysis of the major outer membrane protein of Chlamydia psittaci reveals multimers which are recognized by protective monoclonal antibodies. Infect Immun. 1995;63:2387–2389. doi: 10.1128/iai.63.6.2387-2389.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClenaghan M, Herring A J, Aitken I D. Comparison of Chlamydia psittaci isolates by DNA restriction endonuclease analysis. Infect Immun. 1984;45:384–389. doi: 10.1128/iai.45.2.384-389.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClenaghan M, Inglis N F, Herring A J. Comparison of isolates of Chlamydia psittaci of ovine, avian and feline origin by analysis of polypeptide profiles from purified elementary bodies. Vet Microbiol. 1991;26:269–278. doi: 10.1016/0378-1135(91)90020-g. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D J. On the predictive recognition of signal peptide sequences. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 23.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 24.Novagen Inc. pET system manual. 6th ed. Madison, Wis: Novagen Inc.; 1995. [Google Scholar]
- 25.Raulston J E. Chlamydial envelope components and pathogen-host cell interactions. Mol Microbiol. 1995;15:607–616. doi: 10.1111/j.1365-2958.1995.tb02370.x. [DOI] [PubMed] [Google Scholar]
- 26.Salari S H, Ward M E. Polypeptide composition of Chlamydia trachomatis. J Gen Microbiol. 1981;123:197–207. doi: 10.1099/00221287-123-2-197. [DOI] [PubMed] [Google Scholar]
- 27.Souriau A, Salinas J, De Sa C, Layachi K, Rodolakis A. Identification of subspecies- and serotype 1-specific epitopes on the 80- to 90-kilodalton protein region of Chlamydia psittaci that may be useful for diagnosis of chlamydial induced abortion. Am J Vet Res. 1994;55:510–514. [PubMed] [Google Scholar]
- 28.Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 29.Su H, Zhang Y-X, Barrera O, Watkins N G, Caldwell H D. Differential effect of trypsin on infectivity of Chlamydia trachomatis: loss of infectivity requires cleavage of major outer membrane protein variable domains II and IV. Infect Immun. 1988;56:2094–2100. doi: 10.1128/iai.56.8.2094-2100.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan T-W, Herring A J, Anderson I E, Jones G E. Protection of sheep against Chlamydia psittaci infection with a subcellular vaccine containing the major outer membrane protein. Infect Immun. 1990;58:3101–3108. doi: 10.1128/iai.58.9.3101-3108.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vretou E, Loutrari H, Mariani L, Costelidou K, Eliades P, Conidou G, Karamanou S, Mangana O, Siarkou V, Papadopoulos O. Diversity among abortion strains of Chlamydia psittaci demonstrated by inclusion morphology, polypeptide profiles and monoclonal antibodies. Vet Microbiol. 1996;51:275–289. doi: 10.1016/0378-1135(96)00048-x. [DOI] [PubMed] [Google Scholar]
- 33.Vretou E, Yannikopoulou P, Psarrou E, Papavassiliou P, Pallini V, Bini L. 2-D electrophoretic and antigenic characterisation of the protein family at 90 kDa in the envelope of C. psittaci serotype 1. In: Stary A, editor. Proceedings to the Third Meeting of the European Society for Chlamydia Research. Bologna, Italy: Societa Editrice Esculapio; 1996. p. 52. [Google Scholar]