Summary
The immune response is a fundamental process in the treatment of solid tumors. Here, we present a protocol for implanting diffuse midline glioma cells in the brain of immune-competent mice and characterizing the different immune populations in the tumor microenvironment in a flow cytometry panel. We describe steps for processing of brain tumors, isolating the immune cells, and subsequent staining with antibodies for flow cytometry. We then detail procedures for implementing the gating strategy.
For complete details on the use and execution of this protocol, please refer to Ausejo-Mauleon et al.1
Subject areas: Flow Cytometry, Cancer, Immunology
Graphical abstract
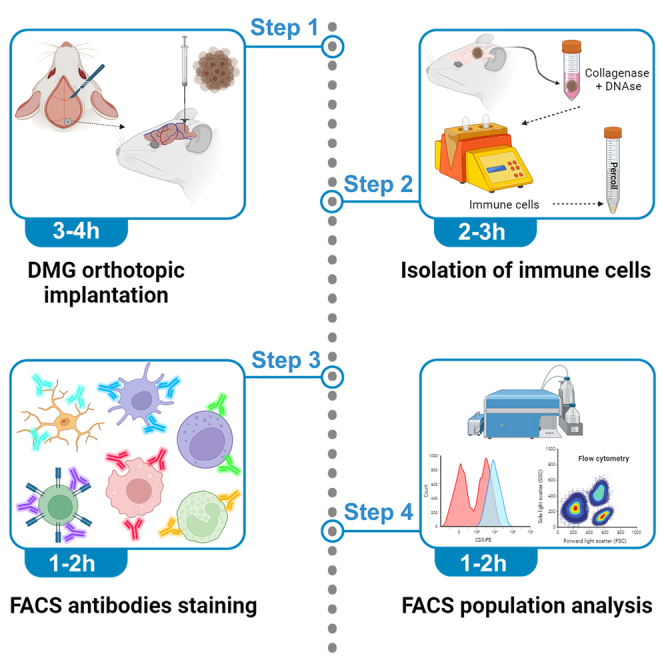
Highlights
-
•
Development of an orthotopic pontine diffuse midline glioma model
-
•
Implantable guide-screw system for injection of tumor cells and drugs into the brain
-
•
Steps described for isolating immune cells from orthotopic DMG mouse model
-
•
Characterization of the brain immune populations with a single flow cytometry panel
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The immune response is a fundamental process in the treatment of solid tumors. Here, we present a protocol for implanting diffuse midline glioma cells in the brain of immune-competent mice and characterizing the different immune populations in the tumor microenvironment in a flow cytometry panel. We describe steps for processing of brain tumors, isolating the immune cells, and subsequent staining with antibodies for flow cytometry. We then detail procedures for implementing the gating strategy.
Before you begin
The protocol below describes the steps to develop a pontine DMG orthotopic mouse model using an implantable guide-screw system previously developed in our lab.2 In addition, we provide information on how to isolate and analyze the tumor microenvironment immune population as in Ausejo-Mauleon et al.1 and supported in other publications.3,4
Institutional permissions
Ethical approval for the animal studies was granted by the Animal Ethical Committee of the University of Navarra (CEEA; Comité Etico de Experimentación Animal) under license number CEEA/004–21. All animal studies were performed at the animal facilities of the Center for Applied Medical Research in accordance with institutional, regional, and national laws and ethical guidelines for experimental animal care. Mice were housed in an individually ventilated cage (IVC) system under a 12-h light/ dark schedule at 22°C, in the presence of 1–4 cage mates (2–5 mice per cage). We used standard IVC cages with a regular wire lid. We used standard cage enrichment, including paper or plastic tunnels. Mice adapted quickly (< 3 days) to the surgery and were perfectly able to navigate through their cage enrichment materials.
Confirmation that the operation is being performed at the proper coordinates
Timing: 1–2 h
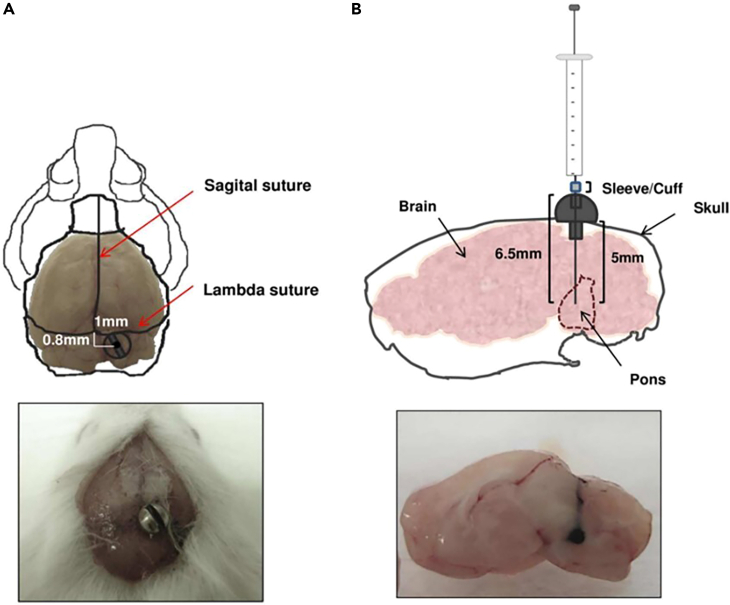
The development of the pontine DMG (DIPG) model was previously described by Marigil et al.2 Due to the complexity of the surgery; it is necessary to confirm that it is fine-tuned the first time that is performed in a laboratory. We highly recommend proper training before performing surgery on mice. This can be achieved by injecting Indian ink and immediately sacrificing the animal to ensure proper anatomical placement (Figure 1).
Figure 1.
Development of pontine DMG model
(A) Up panel, Bolt coordinates in relation to lambda and sagittal sutures. Down panel, photo of mouse skull with bolt positioned in DIPG coordinates.
(B) Up panel, schematic drawing of screw guide components used in this administration method. Down panel, a macroscopic photograph of ink solution injected at 6.5 mm of depth with a Hamilton syringe through the bolt. Adapted from Marigil et al.2
Depilation of mice two days before the surgery
Timing: 1 h
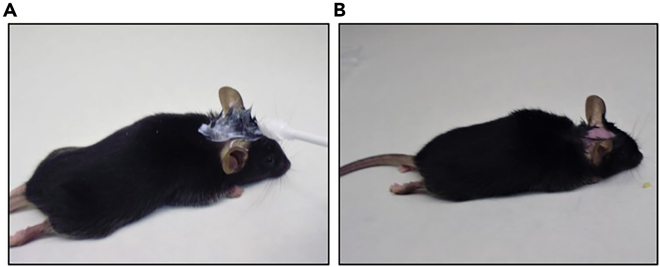
This section describes how to depilate the mice a few days before surgery to avoid infections and to facilitate the work during the implantation of the guide screw system (Figure 2).
-
1.
Anesthetize the mouse with ketamine (Ketamidor)/xylazine (Rompun) solution (See “materials and equipment”) at a dosage of 10 μL of solution (0.375 mg)/5 g body weight.
-
2.
Add depilatory cream on the head of the mouse and rub using a cotton swab.
-
3.
Wait for 5 min for it to take effect.
-
4.
Remove the cream and the hair with a cotton-tipped applicator, applying plenty of water or saline solution to avoid burns.
-
5.
Place the mouse on a heating pad.
-
6.
Wait for the mouse to recover completely before putting it back in its cage.
Note: You can speed up waking the mice earlier by using a selective alpha-2 adrenergic receptor antagonist (antisedan).
Figure 2.
Prepare the animals for the surgery
(A) Add depilatory cream on the head with a swab.
(B) Remove the cream applying water with a pad.
Preparation of buffers and operative instruments for surgery
Timing: 30 min
-
7.
See “materials and equipment” for preparation of needed materials for surgery.
-
8.
Disinfect scissors, tweezers, and operative instruments for surgery with 70% ethanol.
-
9.
Prepare for the guide-screw system implantation surgery by disinfecting all the instruments with 70% ethanol.
Preparation of buffers and operative instruments for immune cell isolation
Timing: 30 min
-
10.
See “materials and equipment” for preparation of needed materials.
-
11.
Keep the FACS buffer on ice during all protocol processes.
-
12.
Disinfect scissors and tweezers with 70% ethanol.
-
13.
Place a 24-plate with sterile DPBS on ice for a minimum of 10 min to leave the brains after taking them from the mice.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AF750 anti-mouse CD45 (30-F11), dilution (1:200) | BioLegend | Cat#103128; RRID: AB_493715 |
| APC anti-mouse F4/80 (BM8), dilution (1:200) | BioLegend | Cat#123116; RRID: AB_893481 |
| BUV395 anti-mouse CD11b (M1/70), dilution (1:200) | BD Biosciences | Cat#563553; RRID: AB_2738276 |
| BUV496 anti-mouse CD4 (GK1.5), dilution (1:200) | BioLegend | Cat#612952; RRID: AB_2813886 |
| BV421 anti-mouse CD19 (6D5), dilution (1:400) | BioLegend | Cat#115538; RRID: AB_11203527 |
| BV510 anti-mouse CD8a (53-6.7), dilution (1:100) | BioLegend | Cat#107520 |
| BV605 anti-mouse NK1.1 (PK136), dilution (1:200) | BioLegend | Cat#141721; RRID: AB_2562273 |
| BV650 anti-mouse IA/IE (M5/114.15.2), dilution (1:300) | BioLegend | Cat#108739; RRID: AB_2565975 |
| BV785 anti-mouse TCRb (H57-597), dilution (1:100) | BioLegend | Cat#107641; RRID: AB_2810347 |
| FITC anti-mouse Ly6C (1A8), dilution (1:100) | BioLegend | Cat#151211; RRID: AB_1186135 |
| PECy7 anti-mouse CD11c (N418), dilution (1:200) | BioLegend | Cat#119703; RRID: AB_493568 |
| PE-eFluor 610 anti-mouse FOXP3 (FJK-16S), dilution (1:200) | Thermo Fisher Scientific | Cat#124227; RRID: AB_2574624 |
| PerCP/Cy5.5 anti-mouse Ly6G (PK136), dilution (1:200) | BioLegend | Cat#505821; RRID: AB_1877271 |
| PromoFluor-840 maleimide, dilution (1:10,000) | AAT Bioquest | Cat#1402 |
| Chemicals, peptides, and recombinant proteins | ||
| Cell culture water | Sigma-Aldrich | Cat#W3500-500ML |
| DPBS (with calcium, magnesium, glucose, pyruvate) | Thermo Fisher Scientific | Cat#14287080 |
| Fetal bovine serum (FBS) | Gibco | Cat#10270106 |
| RPMI medium | Gibco | Cat#61870-010 |
| Penicillin-Streptomycin (10.000 U/mL) | Thermo Fisher Scientific | Cat#15140122 |
| EDTA, 0.5 M sterile solution, pH 8.0 | VWR | Cat#E177-100ML |
| FcR Block | BD Biosciences | Cat#553141 |
| Percoll stock solution | Sigma-Aldrich | Cat#GE17-0891-01 |
| Scalpel | Salunatur | #Cat0210 |
| Ophthalmic “lipolac” | Bausch + Lomb | #Cat764118.6 |
| Ketamidor (ketamine) | Richter Pharma | N/A |
| Rompun (xylazine) | Bayer | N/A |
| Buprenorphine | Acofarma | #Cat961425 |
| Antisedan | Pfizer | #CatPF61 |
| Hystoacryl | B. Braun | #Cat10520010 |
| Critical commercial assays | ||
| Foxp3/transcription factor staining buffer | eBioscience | Cat#00-5523-00 |
| Experimental models: Cell lines | ||
| Murine DIPG: NP53 | Laboratory of Dr. Oren Becher | N/A |
| Murine DIPG: XFM | Laboratory of Dr. Oren Becher | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: NP53 fl/fl (NTV-a p53floxed); age: 4 weeks; gender: male and female (1:1) | Laboratory of Dr. Oren Becher | N/A |
| Mouse: BALB/cJ; strain #:000651; age: 4 weeks; gender: male and female (1:1) | The Jackson Laboratory | Strain #:000651 |
| Software and algorithms | ||
| FlowJo 10.8.1 | BD Biosciences | N/A |
| Prism software 9.3.1 | GraphPad by Domatics | N/A |
| Other | ||
| Bolts (guide-screw system) | Plastics One | #CatC212SG |
| Hamilton syringe | Thermo Fisher Scientific | #Cat87900 |
| PHD 2000 infusion syringe pump multiple syringe holder | Harvard Apparatus | #CatHPHD2000IWMSRR |
| 1 mm diameter drill bit | Plastics One drill HSS | #Cat8J60 |
| Screwdriver | Plastics One | N/A |
| 96-well clear V-bottom sterile microplate | Corning | Cat#P-96-450V-C |
| Cell strainer, pore size 70 mm | Sarstedt | Cat#83.3945.070 |
| 5 mL round bottom polystyrene tubes (FACS tubes) | Corning | Cat#352054 |
| 50 mL Falcon tubes | Sarstedt | Cat#62547004 |
| 15 mL Falcon tubes | Sarstedt | Cat#430791 |
| gentleMACS C tubes | Miltenyi | Cat#130-093-237 |
| gentleMACS Octo dissociator with heaters | Miltenyi | Cat#130-096-427 |
| MACSmix tube rotator | Miltenyi | Cat#130-090-753 |
| CytoFLEX LX | Beckman Coulter | N/A |
Materials and equipment
Here, we report the preparation of materials specific to tumor processing and cytometry analysis. The equipment needed for the execution of this assay is described in the section ‘‘Other’’ in the key resources table.
Anesthesia
| Reagent | Final concentration | Amount |
|---|---|---|
| Ketamidor (100 mg/ml) | 37.5 mg/mL | 450 mL |
| Rompum (20 mg/mL) | 5 mg/mL | 300 mL |
| Saline solution | N/A | 450 mL |
| Total | N/A | 1200 mL |
Prepare the solution freshly every day.
Dissociation medium
| Reagent | Final concentration | Amount |
|---|---|---|
| X GI | N/A | 500 mL |
| Collagenase IV | 1 mg/mL | 500 mg |
| DNase I | 40 μg/mL | 20 mg |
| Total | N/A | 500 mL |
Store at −20°C for 6 months, pre-warm in a water bath at 37°C before use.
FACS Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS | N/A | 487.5 mL |
| Fetal bovine serum | 1% (v/v) | 5 mL |
| Penicillin/streptomycin | 1% (v/v) | 5 mL |
| EDTA 0,5 M | 2.5 mM | 2.5 mL |
| Total | N/A | 500 mL |
Store at 4°C for up to 1 week.
Percoll solution (per sample)
| Reagent | Final concentration | Amount |
|---|---|---|
| Percoll | 30% (v/v) | 3 mL |
| DPBS 10× | 1× (v/v) | 1 mL |
| X GI | N/A | 6 mL |
| Total | N/A | 10 mL |
Prepare the solution freshly every day.
The following amount refers to a total well volume of 100 μL per sample. Multiply the following reagents described in the table by the total number of samples/conditions according to the experimental design.
FACS staining surface antibodies
| Antibody | Clone | Fluorochrome | Dilution factor | Amount/100 μL staining mix |
|---|---|---|---|---|
| Ly6G | PK136 | PrCPC5.5 | 1:200 | 0.5 μL |
| Ly6C | 1A8 | FITC | 1:200 | 0.5 μL |
| CD11c | N418 | PE-Cy7 | 1:100 | 1 μL |
| CD45 | 30-F11 | AF700 | 1:200 | 0.5 μL |
| F4/80 | BM8 | APC | 1:200 | 0.5 μL |
| CD19 | 6D5 | BV421 | 1:400 | 0.25 μL |
| CD8 | 53–6.7 | BV510 | 1:100 | 1 μL |
| NK1.1 | PK136 | BV605 | 1:100 | 1 μL |
| IA/IE | M5/114.15.2 | BV650 | 1:400 | 0.25 μL |
| TCRβ | H57-597 | BV785 | 1:200 | 0.5 μL |
| CD11b | M1/70 | BUV395 | 1:100 | 1 μL |
| CD4 | GK1.5 | BUV496 | 1:200 | 0.5 μL |
| FcR Block | 2.4G2 | N/A | 1:100 | 1 μL |
| FACS Buffer | N/A | N/A | N/A | 92 μL |
| Total | N/A | N/A | N/A | 100 μL |
Prepare the solution immediately before use, protect it from light, and keep it on ice.
FACS staining intracellular antibody
| Antibody | Clone | Fluorochrome | Dilution factor | Amount/100 μL staining mix |
|---|---|---|---|---|
| FOXP3 | FJK-16S | PE-eFluor 610 | 1:100 | 1 μL |
| PermWash 1× | N/A | N/A | N/A | 99 μL |
| Total | N/A | N/A | N/A | 100 μL |
Prepare the solution immediately before use, protect it from light, and keep it on ice.
Equipment and reagent alternatives
-
•
GentleMACS Dissociator (Miltenyi Biotec, Cat#130-093-235) can be replaced simply by a set of 3 glass Pasteur Pipettes. The Pipette’s diameter must be adjusted manually getting smaller and smaller using a flame (diameters: 1.2 mm, 0.8 mm, and 0.4 mm).
-
•
As mentioned above, we recommend using the listed antibodies. However, different antibodies of choice can replace the ones we mention. It is of vital importance to perform optimization of each antibody for each panel.
-
•
The CytoFLEX LX was used to perform the acquisition of samples. The CytoFLEX LX expands research possibilities with up to six lasers and 21 color parameters. It is important to NOTE that the settings are specific for the brand and model of the flow cytometer and optimization of the procedure always needs to be performed.
Step-by-step method details
Mouse preparation
Timing: 15–20 min
This section will explain how to anesthetize and prepare the mouse before initiating the surgery.
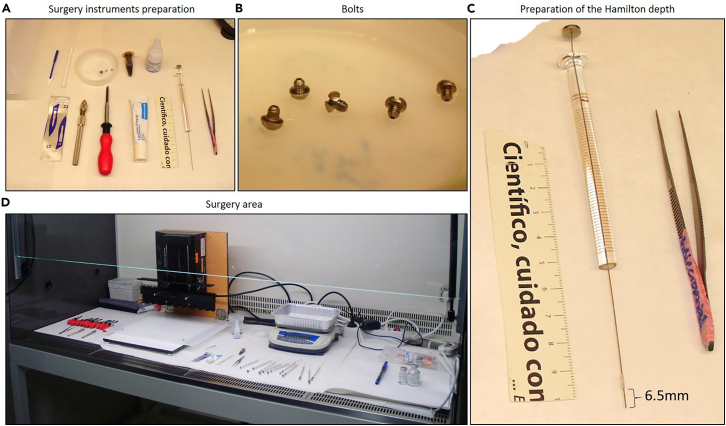
Note: Before starting, prepare all sterilized surgical tools and the other materials required during the surgery and place them next to the pump apparatus. The same surgical tools can be used for multiple surgeries, but make sure to extensively clean them with 70% ethanol and sterilize them in a glass bead sterilizer before each surgery (Figure 3).
-
1.
Prepare the Hamilton syringes (10 μL Gastight Syringe Model 1701, 26s gauge; Cat#80030) with a plastic lock to the corresponding height (6.5 mm Depth), Betadine, scalpel blades, anesthesia, ophthalmic gel, surgery glue, and buprenorphine.
-
2.
Anesthetize the mouse with ketamine (Ketamidor)/xylazine (Rompun 2% Bayer) (80 mg/kg//10 mg/kg) solution at a dosage of 0.15 mg/10 g body weight.
CRITICAL: To ensure the mouse is fully anesthetized, pinch the toes with surgical tweezers. If fully asleep, the mouse will not react. Heat loss is rapid in anesthetized mice. Providing a safe supplemental thermal pad heat source (Cat#5102 Daga S.L.) and preventing contact with cold surfaces is critical for maintaining average body temperature during anesthesia.
-
3.
Place the mouse on a heating pad and apply betadine to the surgical site to sterilize.
-
4.
Apply ophthalmic gel to prevent the mouse’s eyes from drying out.
Figure 3.
Preparation for the guide-screw system implantation surgery
(A) Prepare the surgery preparation area by placing the necessary items.
(B) Detailed image of the bolts used for the guide-screw system.
(C) Detailed image of the plastic stopper used to set the depth of the Hamilton.
(D) Prepare the surgery area in an orderly manner and with all instruments adequately disinfected and sterilized.
Implantation of the guide-screw system and injection of DMG tumor cells
Timing: 3–4 h
This section describes the implantation of DMG tumor cells in the brain of immunocompetent adult mice using a guide-screw system previously described by our lab2 (Figure 4).
-
5.
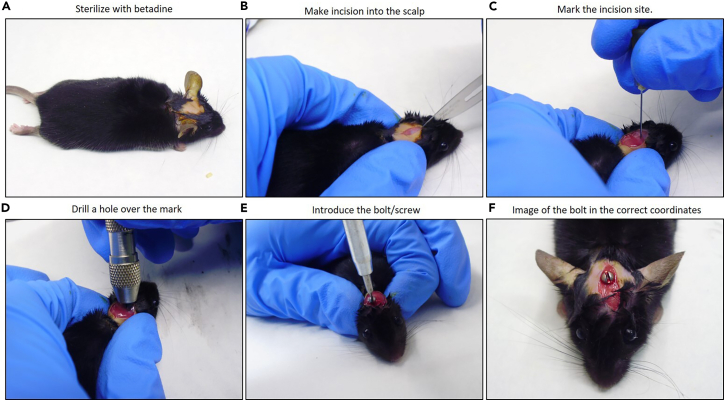
Prepare the head skin with betadine (Figure 4A). Use the scalpel to make an incision into the skin. Cut longitudinally between the eyes as far as the back of the skull (Figure 4B).
-
6.
Localize our entry point 1.0 mm right to lambda and just posterior (0.8 mm) to lambdoid suture.
-
7.
Make a small mark according to the coordinates mentioned above with a small 15-gauge needle (Figure 4C).
Note: This mark can also be done using a fine marker pen simply to have a reference of where to make the hole.
-
8.
Drill a hole (1.60 mm diameter) over the mark with a hand-held twist drill (Drill HSS, #8J60 Plastics One) to the skull until it reaches the dura mater (Figure 4D).
Note: At this point, some bleeding from epidural space venous plexus could appear in some animals although constant pressure applied over the entry point with swabs should stop the bleeding.
-
9.
Introduce the screw/bolt (Head Diameter: 2.5 mm; Shaft Diameter: 1.57 mm; Cat# C212SG, Bilaney) with its specific screwdriver by applying slight pressure throughout the previous twist hole (Figure 4E).
-
10.
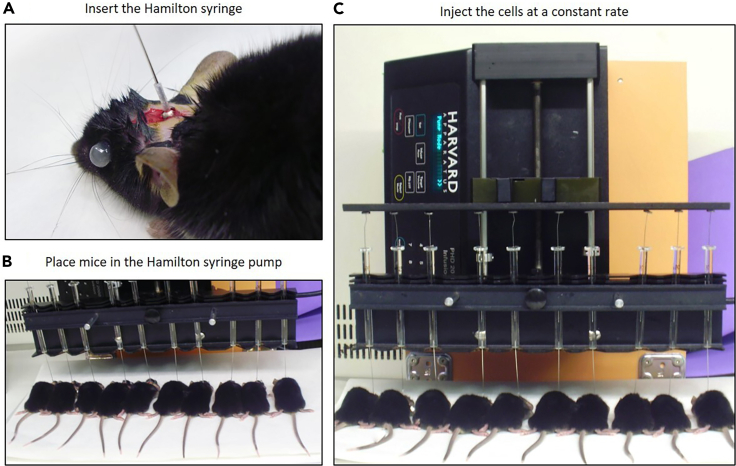
Slowly insert the Hamilton syringe into the hole applying light pressure until the syringe sleeve reaches the surface of the screw and the desired depth (6.5 mm) is reached (Figure 5A).
-
11.
Place mice (up to 10) in the pump for cell injection (PHD 2000 Infusion syringe pump, Harvard apparatus) (Figure 5B).
-
12.
Inject the cells at a constant rate of 0.25 μL/min (Figure 5C).
Note: The number of cells injected depends on the model being used. In our case, we injected 10,000 NP53 or 1,000 XFM.
CRITICAL: We recommend injecting the cells in a volume of 2 or 3 μL, depending on the number of cells, as the solution should always be liquid. It is important to put the number of cells we want to inject in a maximum volume of 3 μL if we inject in the brainstem zone or 5 μL in the cortex zone.
-
13.
Wait for at least 5 min without removing the Hamilton syringe after finishing the injection with the pump to avoid backflow of cells through the hole left by the syringe.
CRITICAL: Failure to wait may result in the cells being distributed to other unwanted parts of the brain.
-
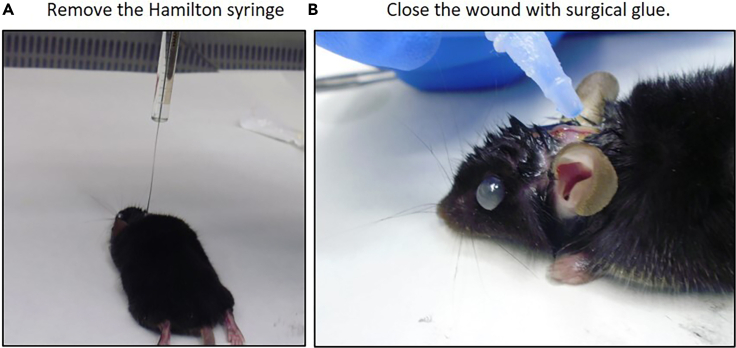
14.
Remove the Hamilton syringe (Figure 6A).
-
15.
Close the wound with special surgical glue (Figure 6B).
Note: Suture thread can also be used in this case, but it can be very tedious to do so with a large number of animals.
-
16.
Weigh the mouse to have a baseline measurement for subsequent weight monitoring.
-
17.
Place the mouse on a heating pad and administer buprenorphine (0.3 mg/mL) as analgesia (dose: 1.2 μg/mice).
-
18.
Wait for the mouse to recover completely before putting it back in its cage.
Note: We consider mice to be recovered when they can walk upright on all four paws.
Note: You can speed up the process so that they wake up earlier by using a selective alpha-2 adrenergic receptor antagonist (anti sedan).
CRITICAL: Since animals were exposed to a highly invasive surgery they needed a minimum of two days of analgesics (every 8 hours). In addition, we administered recovery gels and gathered their food on the cage floor to facilitate food ingestion and accelerate mice recovery.
Note: A few days after implanting the cells, treatment can also be given orthotopically using the same system. This will allow us to study the populations depending on the treatment.
Figure 4.
Implantation of the guide-screw system
(A) Prepare the surgery by sterilizing the head of the mouse with betadine.
(B) Prepare the mouse for the implantation surgery by making an incision into the shaved and clean scalp using a fresh scalpel blade.
(C) Make a small mark according to the coordinates mentioned (1.0 mm right to lambda and 0.8 mm posterior) with a small 15-gauge needle.
(D) Drill a hole over the mark with a hand-held twist drill the skull.
(E) Introduce the bolt/screw with its specific screwdriver by applying slight pressure throughout the previous twist hole.
(F) Representative image of how it should fit implanted in the skull.
Figure 5.
Tumor cells orthotopic injection
(A) Insert the Hamilton syringe into the hole applying light pressure until the syringe sleeve reaches the surface of the screw and the desired depth (6.5 mm) is reached.
(B) Place mice (up to 10) in the Hamilton syringe pump for cell injection.
(C) Representative image of ten mice being injected in the pump at the same time.
Figure 6.
Completion of surgery
(A) Carefully remove the Hamilton syringe.
(B) Close the wound using special surgical glue.
Isolation of immune cells from brain tumor adult mice
Timing: 3–4 h
Animals implanted with orthotopic tumors can be used for different survival or mechanistic studies. In this protocol, we describe their use for the characterization of the tumor immune microenvironment in a single cytometry panel. Therefore, we recommend that everyone perform this protocol at the time they consider appropriate according to the kinetics of their cells. In our case, we measured the immune response 3 (early response), 7 (full response), and 14 (late response) days after our treatment.
This section describes the isolation of mouse brain immune cells, which will be used to characterize the tumor microenvironment.
-
19.
Sacrifice animals according to your institutional permissions.
-
20.
Remove the brain, collect tumors, and place them on cold DPBS until processing.
Note: To collect the tumors we cut with a scalpel trying to take only the tumor part and trying to take the least amount of healthy tissue surrounding the tumor.
-
21.
Add the tumor into a gentleMACS C Tubes (Miltenyi, C130-093-237) with 2.5 mL of the digestion mix. Chop the tumor by mechanics dissociation using sterile scissors.
Note: We use our digestion mix compound by collagenase IV/DNase I (17018-029 Gibco/11284932001 Roche). You can also use the Miltenyi’s adult brain dissociation kit (#Cat130-107-677)
-
22.
Process the sample using the gentleMACS Dissociator (#Cat130-093-235) m_brain protocol.
-
23.
Incubate tumors for 20 min at 37°C in rotation.
Note: We use MACSmix Tube Rotator (#Cat130-090-753).
-
24.
Spin the tube in a centrifuge for 5–10 s so that all cells fall to the bottom after rotation.
-
25.
Transfer the suspension into a 50 mL falcon tube by filtering through a 70 μm cell strainer using 5 mL of cold DPBS.
Note: To break up large pieces of tissue that may remain in the filter, we use the arm of a sterile syringe to dissociate it and get single-cells.
-
26.
Centrifugate at 1800 × g for 5 min at 4°C.
-
27.
Remove the supernatant and resuspend the pellet in 10 mL of 30% Percoll solution.
Note: We prepare the Percoll solution with X GI medium and 10× DPBS.
-
28.
Centrifuge at 500 g for 15 min at 22°C (room temperature).
-
29.
Aspirate the supernatant and wash the pellet once in 5 mL of DPBS.
Note: By aspirating the supernatant, tumor cells are also discarded. We use an aspiration pump to improve sample cleanliness.
-
30.
Centrifuge at 1.800 × g for 5 min at 4°C.
-
31.
Remove the supernatant and resuspend the pellet in 200 μL of cold DPBS or FACS buffer.
Note: We resuspend the pellet in cold DPBS because our dead marker needs to be used in DPBS. If your dead marker can be used in the FACS buffer, you should use the FACS buffer in this step.
FACS staining protocol
Timing: 2–3 h
The following describes the staining of the immune cells that have been isolated previously. It should be added that we recover enough cells from each tumor to perform two different cytometry panels. In this protocol, we continue only with the part in which we characterize the immune populations employing general population markers.
-
32.
Place the suspension with DPBS in a 96 well plate to start staining.
Note: You can also use tubes to perform the staining. We use 96 well plates because we consider it more convenient when you have many samples to stain.
-
33.
Centrifugate the plate at 2000 × g for 3 min.
-
34.
Discard the supernatant and resuspend the cells in 100 μL of PromoFluor-840 in DPBS.
-
35.
Incubate in darkness for 10 min at 4°C.
-
36.
Wash with 100 μL of cold DPBS and centrifugate at 2.000 × g for 3 min.
-
37.
Discard the supernatant and resuspend the pellet with 100 μL of the surface antibodies mix (including Fc block) diluted in FACS buffer.
-
38.
Incubate in darkness for 15 min at 4°C.
-
39.
Add 150 μL of FACS buffer to wash.
-
40.
Centrifugate at 2000 × g for 3 min. Discard the supernatant and repeat the wash.
For intracellular staining of FOXP3: It continues…
-
41.
Fix samples using 100 μL of Fix buffer (1×) from BD Bioscience (Foxp3 / Transcription Factor Staining Buffer, Cat#560409) for 30 min at 22°C (room temperature).
-
42.
Add 100 μL of PermWash buffer (1× in water) and centrifuge 2.000 × g for 3 min.
-
43.
Discard supernatant and incubate samples with 100 μL of FOXP3 antibody diluted in PermWash buffer 1×.
Note: In this step, you can use any intracellular antibody. The pellet may turn transparent upon fixation.
-
44.
Incubate for 30 min at 22°C (room temperature).
-
45.
Wash samples by adding 100 μL of PermWash and centrifugate at 2.000 × g for 3 min.
-
46.
Discard the supernatant and repeat the wash.
-
47.
Remove the supernatant and resuspend samples in 150 μL of FACS buffer to acquire them in the cytometer. Transfer the sample to a FACS tube and proceed immediately to analyze the sample by Flow cytometry.
Note: The volume in which to resuspend depends on the pellet size that exists in all samples.
Note: As the samples have been fixed during processing, they can be passed through the cytometer the next day. However, we do not recommend waiting more than a day since cytometry antibodies (especially tandem ones) can degrade.
FACS analysis
Timing: 1-2 h
-
48.
First, check that the time gate is constant and that there are no clogging events or issues during acquisition.
-
49.
Draw a gate in order to select the acquired cells based on their cytoplasmic granularity (Side Scatter, SSC-A) and size (Forward, FSC-A) trying to avoid the debris and dead cells.
-
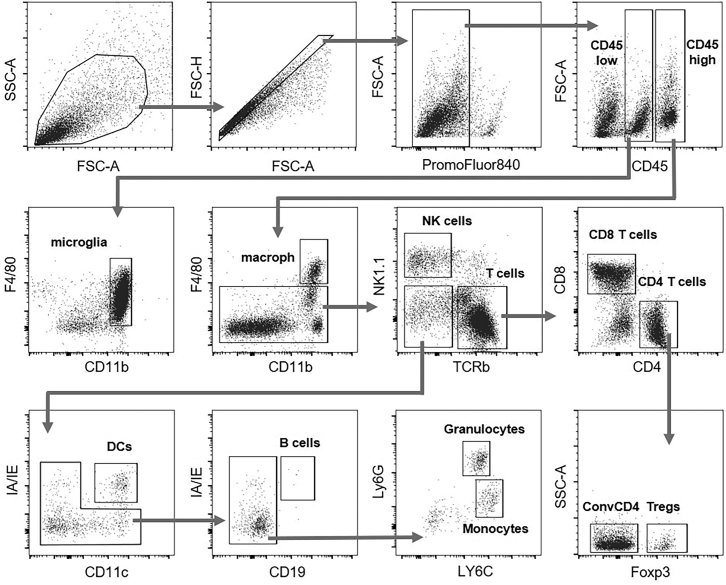
50.
Continue drawing the following gates as shown in Figure 7.
-
51.
Quantify the frequencies and number of cells from each population: Once the gating strategy is completed (Figure 7), obtain the percentage (frequency) of each immune cell subtype from the total of CD45 positive cells (CD45+), which corresponds to the total immune cells infiltrating the tumor.
Figure 7.
Flow gating strategy used for brain tumor microenvironment immune population characterization
Expected outcomes
Following this protocol, we expect to obtain a clear differentiation of all immune cell types present in the brain with specific markers for each one.
The expected results of this protocol are shown in Figure 8. The percentage of each cell type will depend on the model used and the time point chosen for the tumor microenvironment analysis. In addition, these numbers may differ depending on the treatment used in each case. Therefore, different quantification methods can be used based on flow cytometry (count, percentage).
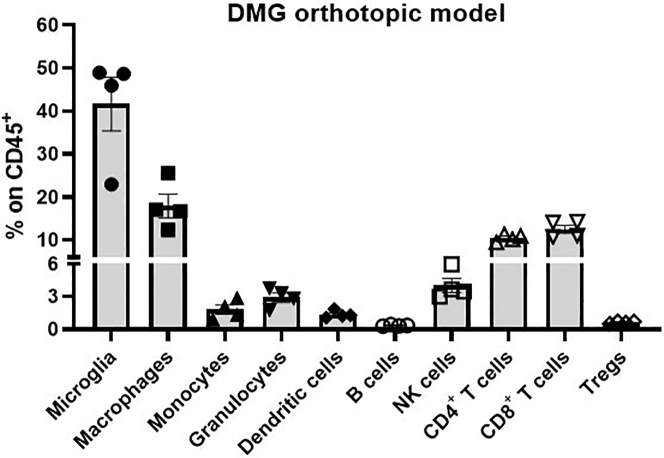
Figure 8.
Example of immune tumor microenvironment composition analysis for the NP53 DMG model
Presents the results obtained using the antibodies panel and gating strategies described in the protocol.
For our NP53 DMG model, the percentage of the different immune populations in the tumor microenvironment is as follows (Figure 8): Microglia (41.66%), macrophages (17.9%), monocytes (1.36%), granulocytes (2.92%), dendritic cells (1.38%), B cells (0.34%), NK cells (4.03%), CD4+ T cells (10.51%), CD8+ T cells (12.54%), Tregs (0.52%).
Quantification and statistical analysis
Flow cytometry data analysis of translation rate in adult microglia can be carried out with FlowJo software (v10, BD Biosciences). A general guideline for data analysis is described below:
-
1.
Run FlowJo (or alternative flow cytometry analysis software) on a computer.
-
2.
Import .fcs files exported from the flow cytometer in FlowJo.
-
3.
Apply for compensation if needed.
-
4.
Gate cells according to the gating strategy described in Figure 7.
Limitations
It is important to know that there is a limitation in the volume that you can inject into each part of the brain. Therefore, it may be that in models in which you need many cells to implant the tumor, the maximum injection volume is a very large limitation.
The methodology to quantify the immune cells infiltrating the tumors may be different according to the characteristics of the cytometer used. It is important to add that this type of characterization is done on a CytoFLEX with 6 lasers and 21 different colors. Considering that 14 different colors are used in the panel, a cytometer without violet, ultraviolet, and infrared lasers could not be used. Therefore, our protocol is not suitable for 4 or fewer laser cytometers.
Troubleshooting
Problem 1
Some animals may need to be euthanized some days after anesthesia due to their aggressiveness (related to step 18 of the “implantation of the guide-screw system and injection of DMG tumor cells” protocol).
Potential solution
-
•
A key point is the experience and skills of the person doing the surgery. After some time practicing, you should not have to sacrifice animals post-surgery. However, during the first surgeries due to the lack of expertise of the handler, it is not strange that some animals need to be euthanized.
-
•
It is important to be very careful throughout the entire operation process, but also during post-surgery care. Additional, supportive care should be provided to the mice if needed.
-
•
It is necessary to take this into account during the design of the experiments and include a sufficient number of animals so that, if this happens, we continue to maintain an adequate number that give us enough statistical power for the characterization of the microenvironment.
Problem 2
Timing of the sampling. The schedule used for the experiment is important not only for the treatment procedures but also for the analysis of the samples (related to the entire protocol).
Potential solution
-
•
For this, it would be best to perform the study at different times. In this way, we can better characterize the immune response of our model, or treatment, and then focus on the section that matters most to us.
Problem 3
Difficult to cut only the tumor without taking healthy tissue in the brain (related to step 2 of the “isolation of immune cells” protocol).
Potential solution
-
•
Sometimes, depending on where the tumor is located, it is not clearly visible. In our case, the wound from the operation with which we injected the cells is visible. Therefore, in case it is not clearly visible, we cut the tissue around this signal.
-
•
In other types of tumors, GFP+ tumor cells can be used and then looked at with a fluorescence microscope to collect the entire tumor area.
Problem 4
Excessive red blood cells after Percoll step (related to step 12 of the “isolation of immune cells” protocol).
Potential solution
-
•
If after making the Percoll step the pellet is very red due to a large number of red blood cells, we should treat it with ACK buffer lysis. It depends on the pellet size, but the standard treatment would be 500 μL for 3 min. After this process, it is important to wash the pellet very well with cold DPBS.
Problem 5
Low number of infiltrated cells in the tumor (related to step 14 of the “isolation of immune cells” protocol).
Potential solution
-
•
In case we have a very low number of tumor-infiltrating immune cells, we can use all the cells in a brain for a single panel.
-
•
In case there are too few for a single panel, we can perform pulls with mice of the same model or treatment group in order to have a sufficient number.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marta Alonso (mmalonso@unav.es).
Technical contact
Please direct technical questions regarding this protocol to the technical contact, Iker Ausejo-Mauleon (iausejo@alumni.unav.es).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
The performed work was supported through Plan de colaboración Internacional (PCI2021-122084-2B) Spanish Ministry of Science and Innovation and Blanca Morell Foundation (S.L.); ChadTough Defeat DIPG (M.M.A.), AECC General Projects (PRYGN21937; M.M.A.), and Instituto de Salud Carlos III y Fondos Feder (PI19/01896 M.M.A. “A way to make Europe”); Fundación El sueño de Vicky; Asociación Pablo Ugarte-FuerzaJulen, Fundación ADEY, Fundación ACS, (M.M.A.), and Fundación Hay que tomarse la vida con tumor; Fundación + Investigación + Vida (La Guareña); and Red Española de Terapias Avanzadas TERAV ISCIII (RD21/0001/0022; Financiado por la Unión Europea-Next GenerationEU. Plan de Recuperación Transformación y Resiliencia). This project also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Programme (817884 ViroPedTher to M.M.A.).
Author contributions
I.A.-M. designed the experiments, performed the experiments, designed the cytometry panels, optimized protocols for cell isolation, and wrote the first draft. S.L. established the initial protocols, designed the cytometry panels, supervised experimental work, and amended the text. M.M.A. designed the experiments, supervised work, secured funding, and amended the text.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sara Labiano, Email: slalminana@unav.es.
Marta M. Alonso, Email: mmalonso@unav.es.
References
- 1.Ausejo-Mauleon I., Labiano S., de la Nava D., Laspidea V., Zalacain M., Marrodán L., García-Moure M., González-Huarriz M., Hervás-Corpión I., Dhandapani L., et al. TIM-3 blockade in diffuse intrinsic pontine glioma models promotes tumor regression and antitumor immune memory. Cancer Cell. 2023;41:1911–1926.e8. doi: 10.1016/j.ccell.2023.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marigil M., García-Moure M., Domínguez P.D., Idoate M.A., Xipell E., Patiño-García A., Tejada-Solís S., García-Moure M., Junier M.P., Chneiweiss H., et al. Development of a DIPG Orthotopic Model in Mice Using an Implantable Guide-Screw System. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laspidea V., Puigdelloses M., Labiano S., Marrodán L., Garcia-Moure M., Zalacain M., Gonzalez-Huarriz M., Martínez-Vélez N., Ausejo-Mauleon I., de la Nava D., et al. Exploiting 4-1BB immune checkpoint to enhance the efficacy of oncolytic virotherapy for diffuse intrinsic pontine gliomas. JCI Insight. 2022;7 doi: 10.1172/jci.insight.154812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Moure M., Gonzalez-Huarriz M., Labiano S., Guruceaga E., Bandres E., Zalacain M., Marrodan L., de Andrea C., Villalba M., Martinez-Velez N., et al. Delta-24-RGD, an oncolytic adenovirus, increases survival and promotes proinflammatory immune landscape remodeling in models of AT/RT and CNS-PNET. Clin. Cancer Res. 2021;27:1807–1820. doi: 10.1158/1078-0432.CCR-20-3313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.