Abstract
An ancient conflict between hosts and pathogens has driven the innate and adaptive arms of immunity. Knowledge about this interplay can not only help us identify biological mechanisms but also reveal pathogen vulnerabilities that can be leveraged therapeutically. The humoral response to SARS-CoV-2 infection has been the focus of intense research, and the role of the innate immune system has received significantly less attention. Here, we review current knowledge of the innate immune response to SARS-CoV-2 infection and the various means SARS-CoV-2 employs to evade innate defense systems. We also consider the role of innate immunity in SARS-CoV-2 vaccines and in the phenomenon of long COVID.
Keywords: SARS-CoV-2, Innate immune response, Interferon, Cytokines, Inflammation, Antiviral targets
Subject terms: Immune evasion, RIG-I-like receptors, Interferons
Introduction
In late 2019, a novel respiratory disease named coronavirus disease 2019 (COVID-19) struck with ferocity, quickly becoming a global pandemic; this disease was later found to be caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1–3]. There are multiple CoVs that infect humans, including common seasonal human coronaviruses (hCoVs), such as Betacoronaviruses HKU1 and OC43 and Alphacoronaviruses NL63 and 229E, and more uncommon CoVs, such as Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV) [4–6]. SARS-CoV-2, consistent with other members of its genus, has a positive-sense, single-stranded RNA genome of ~30,000 nucleotides and produces approximately 30 proteins [7]. Starting from the 5′ terminus, the genes for the replicase and nonstructural proteins (ORF1a and ORF1ab) are present, followed by the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, with various intergenic accessory factors interspaced within this framework [8]. Two large polyproteins, ORF1a and ORF1ab (pp1a and pp1ab), are cleaved by viral proteases (PLpro and 3CLpro) into 16 nonstructural proteins (NSP) that predominantly constitute the RNA-dependent viral replicase, including four proteins that form the virion (S, M, N, E), and seven accessory proteins that play a pivotal role in manipulating cell biology and modulating viral pathogenesis (ORF3a, ORF6, ORF7a/b, ORF8, ORF9, and ORF10) [3, 7, 9]. COVID-19 manifests in a wide range of disease states, from mild nonspecific symptomology and even asymptomatic disease to moderate and severe illness requiring hospitalization [10–13]. Cytokine release syndrome (CRS) is a systemic condition seen in some severe COVID-19 cases that results from an overwhelming release of cytokines, which causes severe inflammation and induces acute respiratory distress syndrome (ARDS) and secondary hemophagocytic lymphohistiocytosis (sHLH) [14, 15]. To mitigate CRS, the innate immune system maintains endogenous feedback networks using cytokines such as IL-4, IL-10, IL-11, and IL-13 to drive anti-inflammatory phenotypes [16].
Innate immunity is an ancient system conserved from fish to mammals that provides the time necessary for the adaptive immune response to commence [17]. Various factors play a role in determining the severity of infection, and our innate immune response is central. Broadly, the role of the innate immune response to viruses is (1) to limit viral entry into a cell, block the translation of viral elements, the replication of the viral genome, and prevent egress of new infectious virions; (2) to identify and purge infected cells; and (3) to accelerate the development of a targeted adaptive immune response [18–20]. Activation of the inflammatory cascade, including the broad interferon response, plays a critical role in the clinical manifestation of SARS-CoV-2 [21, 22]. Various cell surface cytosolic and endosomal pattern recognition receptors (PRRs) in the cell surface and cytosol are activated once signaled by pathogen-associated molecular patterns (PAMPs), thereby triggering an inflammatory cascade and controlled cell death in affected cells [23–25]. As might be anticipated, excessive activation of this highly regulated system can lead to severe systemic inflammation [26, 27].
The release of IFN-I, along with various other inflammatory molecules following PRR activation, initiates antiviral defenses in neighboring cells in an attempt to limit further viral replication and spread [28–30]. While recent reports indicate that SARS-CoV-2 is sensitive to IFNs in vitro [31], even more so than its relative virus, SARS-CoV, the relationship between IFN release and disease presentation remains an area of interest. Innate immune cells, such as macrophages, dendritic cells, neutrophils, monocytes, and natural killer (NK) cells, modulate this response, as they are equipped with an array of PRRs capable of recognizing PAMPs and damage-associated molecular patterns (DAMPs) to initiate inflammatory pathways and foster these immune responses [32]. Although IFN-λ is primarily produced by epithelial cells rather than monocytes, IFN-λ appears to be more protective against infection and disease progression [33, 34].
Calibration of the type I interferon (IFN-I) response is critical to disease outcomes, as excessive or insufficient activation of IFN signaling can be life-threatening [35–37]. Mounting a robust IFN response at the onset of SARS-CoV-2 infection is of critical importance for developing a protective immune response, and suppression of IFN signaling contributes to severe COVID-19 disease states [14, 37]. Extended IFN production exacerbates disease progression by impeding the regeneration of lung epithelial cells [38, 39]. Evidence for the critical nature of the timing and magnitude of innate responses comes from data showing that although IFN-I can block infection in vitro, IFN-β may fail to provide therapeutic benefits if administered late in severe cases [31, 40–42]. Furthermore, the broad unbridled activity of the innate immune response and autoantibodies against IFN-I has been associated with severe COVID-19 [43–45].
Like many viruses, SARS-CoV-2 can evade the innate immune system through multiple strategies, including viral antagonism, avoidance of detection, and inflammatory response modulation (Table 1) [46–49]. Over- or under-activation of the innate immune response is detrimental in efforts to clear the infection; thus, a balanced response is needed as part of a “goldilocks” just-right scenario. In this review, we aim to summarize the innate immune response to SARS-CoV-2 infection with regard to disease modulation and immune system evasion and determine how we might manipulate this response for therapeutic benefit.
Table 1.
Key SARS-CoV-2 proteins involved in counteracting host innate immune responses
| SARS-CoV-2 protein | Mechanism of antagonism | Effect on the host’s innate immune response | References |
|---|---|---|---|
| NSP1 | Inhibits IFN response through the depletion of key signaling factors | Reduces IFN production and signaling | (Kumar et al.) [186] |
| NSP3 | Inhibits IFN-I production through the cleavage of IRF3 | Reduces IFN production and signaling |
(Moustaqil et al.) [262] (Alhammad et al.) [195] (Taha et al.) [192] |
| NSP13, NSP14, and NSP15 | Inhibit nuclear localization of IRF3 | IFN action and signaling antagonism |
(Yuen et al.) [181] (Feng et al.) [263] (Fung et al.) [264] |
| ORF3a | Inhibits fusion of autophagosomes with lysosomes | Manipulates and antagonizes autophagy | (Zhang et al.) [162] |
| ORF3b | Inhibits the nuclear localization of IRF3 | Acts as a potent interferon antagonist | (Konno et al.) [179] |
| ORF3c | Interacts with PGAM5 to induce caspase-3 cleavage of MAVS | Antagonizes IFN-β production, alters mitochondrial metabolism, blocks, autophagy |
(Mozzi et al.) [163] (Stewart et al.) [174] |
| ORF6 | Interacts with Nup98-Rae1 to blocks STAT1 and STAT2 nuclear translocation | Acts as a potent interferon antagonist | (Miorin et al.) [182] |
| ORF7a | Blocks SERINC5 incorporation into the virion and interferes with autophagosome acidification | Antagonizes autophagy and antiviral action |
(Timilsina et al.) [86] (Hayn et al.) [48] |
| ORF8 | Decreases nuclear translocation of IRF3 and mimics IL-17A | Antagonizes IFN production and downregulates MHC-I |
(Rashid et.) [183] (Wu et al.) [209] |
| ORF9b | Interrupts the K63-linked ubiquitination of the interferon signaling modulator NEMO | Antagonize RIG-I-MAVS antiviral IFN-I response | (Wu et al.) [172] |
| ORF10 | Triggers NIX-dependent (Nip3-like protein X) mitophagy leading to the degradation of MAVS | Degrades MAVs through mitophagy | (Li et al.) [173] |
Pattern recognition receptors (PRRs) of SARS-CoV-2
Consistent with most viral infections, one of the first steps initiated by the host’s innate immune response at the start of SARS-CoV-2 infection is the production and release of type I and type III interferons (IFN-I and IFN-III, respectively) [35, 50]. As SARS-CoV-2 infects a cell and viral replication commences, viral RNA is detected through a series of cellular PRRs, such as RIG-I-like receptors (RLRs), nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), and Toll-like receptors (TLRs), which are highly evolved cellular surveillance systems that specialize in detecting PAMPs associated with viruses or other pathogens [25, 51, 52].
Cytoplasmic RNA sensors, including TLR3 and RLRs such as MDA5 and LGP2, play a critical role in innate immunity by recognizing viral RNA [53]. In particular, RIG-I detects short dsRNA or 5′-pppRNA, and MDA5 detects long dsRNA. LGP2 also detects viral RNA and is known to be a positive regulator of MDA5- and RIG-I-mediated antiviral responses [54, 55]. Another pathway called 2′,5′-oligodenylate synthetase (OAS1)-ribonuclease L (RNase L) senses non-self dsRNA, cleaves ssRNA, and induces cell death [56]. In SARS-CoV-2 infections, OAS1 efficacy relies on the post-translational modification of prenylation and is significantly linked with severe COVID-19 prevention [57]. Prenylation is the covalent addition of a lipid near the C-terminus of a protein, this addition allows the protein to anchor to the cell membrane. Notably, this defense mechanism is absent in horseshoe bats, a possible reservoir for CoVs, indicating that horseshoe bats may have evolved to be optimal reservoirs of SARS-like CoVs [57]. Additionally, inborn errors of OAS-RNase L have been found to be associated with multisystem inflammatory syndrome in children (MIS-C), which involves the release of an excessive amount of inflammatory cytokines upon SARS-CoV-2 infection [58, 59]; this downstream effect highlights the importance of this pathway in IFN stimulation and disease outcome.
Interferon inducible antiviral restriction factors
Interferon-induced transmembrane proteins (IFITMs) are proteins embedded in the lipid membrane of cells with the primary function of inhibiting fusion between the viral envelope and the host cell membrane [60, 61]. These IFITMs likely prevent fusion by changing membrane curvature and decreasing membrane fluidity; IFITMs have been shown to inhibit the entry of several viruses, including the Ebola virus, the influenza virus, and HIV-1 [62–64]. Three main IFITMs exhibit antiviral properties; IFITM1 is found on the plasma membrane, IFITM2 is found on late endosomes, and IFITM3 is found on early endosomes [65–67]. Interestingly, there have been conflicting findings on the role of IFITMs and SARS-CoV-2 infection. While IFITMs have widely been demonstrated to inhibit infection, in hCoV, OC43, IFITM2, and IFITM3 have been shown to increase viral entry into cells [68, 69]. Alternatively, in SARS-CoV-2 infections, early in vitro studies demonstrated that IFITM2 and IFITM3 but not IFITM1 sufficiently restricted viral entry into the cell [70, 71]. Additional studies showed that IFITM2 restricts viral entry more than IFITM3, likely because of the route of viral entry [72]. SARS-CoV-2 enters cells through membrane fusion and/or endocytosis mediated by the spike protein. The spike protein contains a receptor-binding domain (RBD) that interacts with the cellular receptor angiotensin-converting enzyme 2 (ACE2) and a polybasic cleavage site (PBCS) S1/S2 that is proteolytically cleaved by transmembrane serine protease 2 (TMPRSS2) and cellular cathepsin L [73, 74]. Depending on the status of spike cleavage and the relative abundance of TMPRSS2 on the plasma membrane, the virion enters through TMPRSS2-mediated membrane fusion or late endosomal entry via secondary cleavage [74–77]. Importantly, this cleavage is impacted by allostery; for example, the allostery between the NTD and the PBCS [73, 78, 79]. TMPRSS2 is found on lung and primary human airway epithelial cells and enables endosome-independent viral entry that avoids the antiviral actions of IFITM2 and IFITM3 [72, 80, 81]. This suggests that different SARS-CoV-2 variants have different sensitivities to the TMPRSS2-mediated entry pathway; thus, they may have different susceptibility to antiviral IFITMs [73, 82, 83]. The Delta variant appeared to have evolved toward plasma membrane fusion, and somewhat unexpectedly, the Omicron variant has evolved to primarily use endosomal entry; this may be the result of immune evasion and changes in spike cleavage efficiency [73, 84]. The Omicron variant had decreased cleavage efficiency in certain cell types and increased susceptibility to IFITM2 and IFITM3 [85], possibly contributing to the milder disease states seen with Omicron infections.
In addition to IFITMs that exert their antiviral function at membranous structures, SERINC5 is a cellular multipass transmembrane protein that is involved in lipid transport and biosynthesis and is most well known for its inhibition of human immunodeficiency virus (HIV) infection and for being a target of the HIV antagonist protein Nef [86–89]. SERINC5 is incorporated into budding virions and prevents viral entry by blocking virus‒cell fusion; SARS-CoV-2 ORF7a has been demonstrated to block the incorporation of SERINC5 in budding SARS-CoV-2 virions, thus antagonizing the antiviral action of SERINC5 [86, 87].
cGAS-STING Axis
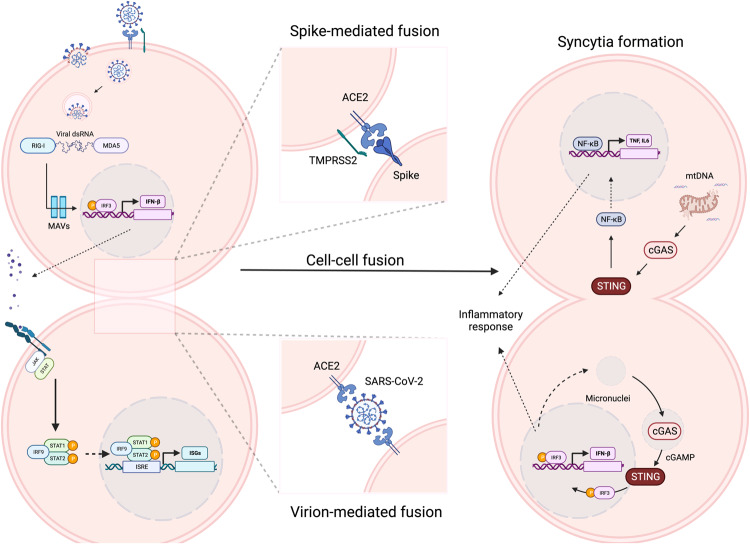
Cytoplasmic DNA sensors such as IFI16, AIM2, and cGAS play a critical role in identifying viral signatures and sensing cellular mitochondrial DNA (mtDNA) during RNA virus infection [36, 90, 91]. The Cyclic-GMP-AMP synthase STimulator of INterferon Genes (cGAS-STING) system [92] is broadly manipulated during SARS-CoV-2 infection; this system is involved in syncytial pneumocyte formation, cell-to-cell fusion, and reduced cytokine signaling [93–96]. Papain-like protease (PLpro), one of the virally encoded proteases whose activity may correlate with pathogenicity [97], also activates the antiviral IFN response by deubiquitinating K63-linked polyubiquitin chains of STING, therefore disrupting the cGAS-STING axis for the induction of IFNs and interferon-stimulated genes (ISGs) [98]. Cell‒cell fusion, where plasma membranes fuse to form multinuclei cells, is a well-documented phenomenon mediated by the SARS-CoV-2 spike protein and its receptor ACE2, a process triggered by spike cleavage to expose the fusion peptide. Syncytium formation has been observed in postmortem tissues associated with severe COVID-19 [99, 100] and extensively demonstrated in vitro [64, 73, 79, 101]. Transcriptomic analysis of spike-mediated fused cells showed that the IFN response was one of the most upregulated processes [94]. Further analysis showed that IFN is induced by cGAS, which was colocalized with large DNA aggregates due to nuclear membrane rupture, leading to the activation of IRF3. This IFN induction is dependent on spike cleavage and is increased in cells that overexpress host proteases, such as TMPRSS2, that are drug targets; furthermore, mutating the cleavage site completely abrogates IFN stimulation in fused cells [94]. Interestingly, damaged DNA released from the ruptured nucleus is not the only source for cGAS sensing, as SARS-CoV-2 infection also causes direct mtDNA damage [102]. However, as cGAS signaling requires cGAS-G3BP1 coassembly on dsDNA to form stress granules, the viral nucleocapsid restricts cGAS signaling by competitively binding G3BP1 to divert dsDNA into alternative liquid‒liquid phase-separation condensates (Fig. 1) [102].
Fig. 1.
SARS-CoV-2 mediated cell-cell fusion and impact on cGAS-STING and IFN signaling
RNA editing-dependent antiviral innate responses
RNA viruses tend to have high mutation rates due to the high mutation rate of viral RNA-dependent RNA polymerase (RdRp), which limits their genome length as extensive mutations can lead to error catastrophe [103]. CoVs manage the largest genomes (~30 kb) among RNA viruses by encoding 3′-5′ exoribonucleases with proofreading activity that lower mutation rates up to 100-fold compared to that of other RNA viruses, such as influenza, HCV, and HIV [104–107]. RNA editing is a crucial innate immune response to endogenous and exogenous RNA sequences in both health and disease states; these modifications had an unintended impact on SARS-CoV-2 genome diversity [108–116]. In particular, one mRNA editing enzyme, apolipoprotein B (ApoB), which is part of the catalytic polypeptide-like (APOBEC) family that is further explored below, has been hypothesized to be increased in SARS-CoV-2-induced senescence alveolar type II (ATII) cells, which are a fertile source for generating hypermutated progeny [117, 118]. As new strains emerge, alterations in tissue tropism can redirect viral evolution. This is evident from the Omicron variant; the transition to upper respiratory tract replication occurred with a significant reduction of G > T in the mutational spectra compared to previous variants [119], which replicate both in the upper and lower respiratory tract.
In SARS-CoV-2 infection, cytoplasmic viral dsRNA from the transcription-replication complex is recognized as pathogenic non-self RNA by host antiviral proteins and sensors [57]. As the amount of adenosine deaminase that acts on RNA type I (ADAR1) and APOBEC3 is increased in the interferon innate immune antiviral response, modulation by the viral genome can suppress excessive immune reactions initiated by intracellular dsRNA sensors such as MDA5, OAS-RNase L, and protein kinase R (PKR) [120–122]. In addition, the recent discovery that dsRNA can trigger pyroptosis via NLRP1 sensing further suggests that the regulation of dsRNA signaling plays a role in SARS-CoV-2 pathogenesis [123].
APOBEC
The APOBEC family includes restriction factors for a diverse range of viruses, including retroviruses, hepatitis C virus (HCV), herpesviruses, and foot and mouth disease virus [124, 125]. The key members of this family that are capable of single-stranded RNA deamination are APOBEC1, APOBEC3A, and APOBEC3G [126]. Aberration of viral replication is achieved by performing lethal editing via cytidine deaminase activity in a sequence- and structure-dependent context [127]. While the exoribonuclease grants CoVs a level of “immunity” against APOBEC editing-induced mutagenesis, studies of global SARS-CoV-2 consensus sequences deposited in Global Initiative on Sharing All Influenza Data (GISAID) revealed signatures of APOBEC-mediated C-to-U transition. This change is the most abundant SARS-CoV-2 mutation, accounting for up to 46% of nucleotide substitutions [103, 114, 128]. The frequency of this mutation is supported by the asymmetric abundance in C-to-U transitions in specific dinucleotide contexts (TC, AC, or CC) and the high non-synonymous mutation to synonymous mutation ratio, which suggests non-neutral evolution driven by additional mutational mechanisms beyond random changes seen in genetic drift [109, 129–132].
ADAR
Another family of RNA editing enzymes is the ADAR gene family. In contrast to the APOBEC family, ADAR recognizes dsRNA, a common structure found in replication complexes and secondary RNA structures [133]. Within the ADAR family, only ADAR1, and more specifically, ADAR1 isoform p150, is interferon-inducible and shuttles between the nucleus and cytoplasm [134]. ADAR has the important function of regulating cytoplasmic innate immunity by deaminating adenosine to inosines. This destabilizes the dsRNA structure between complementary strands or secondary RNA structures, such as hairpin loops formed by retrotransposons known as Alu repeats [135]. Failure to eliminate endogenous dsRNA leads to a constitutive antiviral response and inflammation, as seen in the autoimmune disorder Aicardi-Goutiéres Syndrome [135, 136].
The interest in ADAR1 sparked after the acquisition of the A23403G mutation that led to the spike D614G amino acid change [137]. While there is evidence of upregulated expression and RNA‒protein interactions involving ADAR1, such as interactions with APOBEC3A, IFN-responsive SARS-CoV-2-infected human cells and RNA-seq of patient bronchoalveolar lavage samples showed A-to-G/T-to-C biases; there has been limited emergence of ADAR1-related mutations in in vitro models and the general population [8, 110, 138–140]. Nevertheless, deep sequencing analysis showed ADAR1 mutation signatures in minor viral populations that were inversely correlated with viral load, mortality, and incidence, suggesting that ADAR1 mutations may be significant [141].
ZAP
Last, zinc-finger antiviral protein (ZAP, also known as poly(ADP-ribose) polymerase 13 (PARP13)) binds to CG dinucleotides and recruits the cofactors KHNYN and TRIM25 to degrade viral RNA [142–146]. The antiviral activity of ZAP can be potentiated by a cellular polynucleotide poly(ADP-ribose) [147]. ZAP expression is upregulated in the SARS-CoV-2 innate immune antiviral response [148], and thus, the SARS-CoV-2 virus has shown reduced CG content since its emergence; this change occurred as an adaptation to circulation in human hosts [149].
Programmed cell death
Autophagy is an evolutionarily conserved cellular process that can flip between beneficial and harmful actions amidst an active viral infection [150, 151]. Upon SARS-CoV-2 infection, abundant cellular machinery and organelles serve as sanctuaries for viral replication, thereby enabling extended replication and continuous infection. Activation of well-timed and appropriate autophagy mounts a counterattack on virus-producing compartments by initiating cell death and degrading the viral particles within infected cell [152–154]. However, there is a molecular arms race between viruses and humans, as some viruses, such as poliovirus, HIV, HCV, and SARS-CoV-2, can manipulate autophagy for their benefit [153, 155, 156]. Autophagy requires a delicate balance, and perturbations, such as those induced by viral antagonistic proteins, can tilt the scale in favor of viral success. However, inappropriate or excessive autophagy can contribute to cellular damage and systemic inflammation, disrupt normal cell function and exacerbate cytokine responses often associated with severe COVID-19 [48, 153].
In SARS-CoV-2, the NSP15, ORF3a, ORF3c, ORF7a, ORF10, E, and M proteins have been reported to manipulate and antagonize autophagy [157–160], with the ORF3a and ORF7a proteins appearing to be dominant [48]. Although these proteins broadly prevent autophagy, they do so in a myriad of mechanisms. ORF3a prevents autophagy by inhibiting the fusion of autophagosomes with lysosomes by sequestering the homotypic fusion and protein sorting (HOPS) component VPS39, which prevents assembly of the SNARE complex [161, 162]. On the other hand, ORF7a was shown to prevent autophagy by reducing the acidity of the lysosome by increasing the pH [48]. This is complemented by ORF3c hyperactivation of oxidative phosphorylation, which induces the overproduction of ROS and compromises lysosomal acidity and autophagy [163].
Pyroptosis, a proinflammatory form of programmed cell death utilized in the innate immune response, was observed in the lung tissues of patients with severe COVID-19 [164]. Using transcriptome analysis, NSP6 was demonstrated to trigger NLRP3-dependent pyroptosis by targeting ATP6AP1, a vacuolar ATPase proton pump component [165]. Interestingly, pyroptosis observed in FcγR-mediated SARS-CoV-2 infection of monocytes and macrophages led to abortive replication and systemic inflammation, which contributed to COVID-19 severity [166, 167]. However, pyroptosis can be blocked through N binding to the Gasdermin D (GSDMD) linker region, which prevents GSDMD cleavage that is needed for the initiation of pyroptosis [168, 169]. This finding indicates that a GSDMD inhibitor could be potential approach to counter excessive inflammation in severe COVID-19.
Viral antagonism of cytokine and IFN signaling
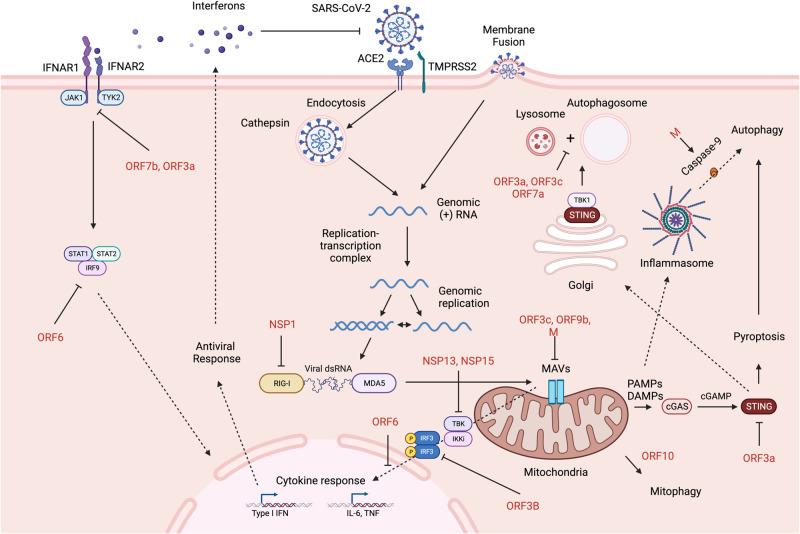
Upon the detection of SARS-CoV-2 viral RNA through one of the cytoplasmic RNA sensors, downstream activation via mitochondrial anti-viral signaling protein (MAVs) induces the phosphorylation of interferon regulatory factor (IRF) and nuclear translocation (Fig. 2). This cascade event can lead to the activation of immune genes, including IFN-I and IFN-III, and an antiviral response. In SARS-CoV-2 infections, a deficiency in MDA5, MAVS, or RIG-I can lead to enhanced viral replication [170]. The SARS-CoV-2 glycoprotein M was demonstrated to impair MAVS aggregation and the further recruitment of TNF receptor associated Factor 3 (TRAF3), TANK binding kinase 1 (TBK1), and IRF3 sufficiently attenuated the innate antiviral response [171]. Through a different mechanism, SARS-CoV-2 ORF9b was also shown to antagonize the RIG-I-MAVS antiviral IFN-I response by preventing the K63-linked ubiquitination of the interferon signaling modulator NEMO [172]. Alternatively, ORF10 was also shown to suppress the antiviral innate immune response by triggering NIX-dependent (Nip3-like protein X) mitophagy, which led to the degradation of MAVS [173]. Finally, ORF3c, a recently discovered 41-aa peptide product of leaky ribosomal scanning (+1 reading frame) entirely nested within the ORF3a sgRNA, localizes and interacts with both MAVS and mitochondrial protein phosphoglycerate mutant family member 5 (PGAM5) to induce caspase-3-mediated cleavage of MAVS [174]. RIG-I may also have noncanonical action against SARS-CoV-2 by competitively inhibiting RdRp via binding to the SARS-CoV-2 3′ UTR without triggering ATPase and thus downstream activity [175].
Fig. 2.
SARS-CoV-2 cell entry and early life cycle, innate immune pathways, and virus-encoded antagonists
Betacoronavirus CoVs, including SARS-CoV and MERS-CoV, use several strategies also used by SARS-CoV-2 to avoid innate immune detection. MERS-CoV, consistent with the IFN antagonism employed by SARS-CoV-2, regulates IRF3 nuclear trafficking with ORF4a, ORF4b, ORF5, and the membrane protein [176]. Similarly, SARS-CoV-2 ORF6 was shown to interfere less efficiently with interferon signaling than SARS-CoV ORF6 [177]. Collectively, despite marked differences in entry mechanisms, transmissibility, and pathogenicity among coronaviruses, these viruses have evolutionarily converged strategies for evading our immune system [89].
IFN responses in SARS-CoV-2 infections appear to remain weak overall, potentially indicating efficient antiviral evasion/antagonism of PRRs [37, 178–180]. The NSP13, NSP14, NSP15, ORF8, ORF3b, and ORF6 proteins act as potent interferon antagonists and contribute to suppression of the primary interferon response [179–181]. NSP14 targets the IFN-I receptor (IFNAR1) for lysosomal degradation [48]. Alternatively, ORF6 inhibits the IFN-I response by blocking STAT, and ORF8 inhibits IFN production by reducing the nuclear translocation of IRF3 [180, 182, 183]. Another study demonstrated that the D61L mutation in ORF6 is responsible for the decrease in IFN-β secretion in vitro [184], which might account for the reduced clinical severity seen in Omicron induced diseases [185]. Likewise, more recently, the SARS-CoV-2 ORF3b protein has also been demonstrated to be a potent interferon antagonist; this protein suppresses IFN-I induction more efficiently than its ORF3b ortholog in SARS-CoV [179].
Some SARS-CoV-2 proteins play multiple roles in the manipulation of the IFN response, such as NSP1, which prevents IFN induction through the blockage of IRF3 phosphorylation and the depletion of the IFN-I signaling components, TYK2, and STAT2 [186]. Experimental evidence also demonstrated that NSP3 antagonizes IFN-I production through the cleavage of ISG15 from IRF3, which is associated with MDA5 signaling [187, 188]. NSP3 contains a pancoronavirus-conserved macrodomain (Macro1) that is essential to pathogenesis [189–192], although slight differences in residues define their specificity [193]. Depending on the ADP-ribosylation target, interferon-responsive poly(ADP-ribose) polymerases can act as positive and negative regulators of the innate antiviral response [194]. SARS-CoV-2 virus bearing Macro1 deletion of a single site in its catalytic domain increased the levels of IFNs and ISGs both in cell lines and mice, indicating that Macro1 is an antagonist of IFNs and explaining the attenuated pathology in mice [192, 195]. Macro1 reverses global PARP9/DTX3L ADP-ribosylation upregulation of IFN-I and -II expression without preventing the induction of ISGs. As PARP9/DTX3L targets host histones to promote IFN signaling and viral proteins for degradation [196], ADP-ribosylation likely plays an undiscovered role in SARS-CoV-2 infection.
NSP6 was shown to bind TBK1, which also suppresses IRF3 phosphorylation [197]. In addition to their primary role in facilitating RdRp activity, NSP7 and NSP8 have also been demonstrated to suppress IFN-I responses, with NSP8 binding to MDA5 CARD to block K63-linked polyubiquitination, a key step involved in the regulation of the innate immune response [198, 199].
Among all proteins, SARS-CoV-2 ORF8 is the most hypervariable in SARS-CoV-2, following the spike protein, and shares the least homology among earlier major human coronaviruses [200]. At the sequence level, ORF8 shares greater homology with bat and pangolin coronaviruses than SARS-CoV (~90% sequence homology with BAT-SL-CoVZC45 vs. ~30% with SARS-CoV) [201, 202]. In addition, SARS-CoV gained a 29-nt deletion, splitting into ORF8a and ORF8b [203]. SARS-CoV-2 ORF8 has de novo functions not seen in SARS-CoV or MERS-CoV. Although paralogs that share the immunoglobulin (Ig)-fold-like structure can be found, such as ORF7b, SARS-CoV-2 ORF8 is unique in its ability to dimerize and has lost its C-terminal transmembrane domain, allowing secretion from infected cells [200, 204, 205].
Interestingly, milder infection was observed in 39 Singaporean patients infected by a virus with a 382-nucleotide deletion (∆382), which truncated ORF7b and eliminated the ORF8 leader transcriptional regulatory sequence (TRS-L) [206]. ORF8 ablation either by truncation or TRS deoptimization has been shown to be under positive selection and has increased transmissibility in certain lineages, including the B.1.1.7/Alpha (Q27*), BA.5 (C27889T), XBC (G27890T), and XBB.1 (G8*) sublineages, and reached a 80% global prevalence [207]. Although there is a de facto loss of ORF8 function, more transmissible variants often have mutations in the spike protein and other regions, possibly explaining the increased pathogenicity of the Alpha variant despite ORF8 truncation [208].
ORF8 is an interferon antagonist that disrupts epigenetic-related posttranslational modifications (PTMs) of histones; this disruption prevents detection by MHC-I, abolishes interferon production and signaling, and mimicks cytokine signaling [209, 210]. Wuhan-1 and VoCs of SARS-CoV-2 ORF8 possess an ARKSAP motif, which has not been previously observed in SARS-CoV but is present in BAT-SL-CoVZC45. The ARKS motif is a PTM site critical to histone H3 at lysine 9 and lysine 27 (H3K9 and H3K27) [210]. Kee and colleagues demonstrated that the histone mimic ORF8 disrupts chromatin accessibility and results in significant differences in gene expression [210].
Dimerized ORF8 also mediates MHC-I degradation through autophagy pathways, thereby providing protection against CTLs [202, 211]. ORF8 also inhibits global protein synthesis by interfering with the ER-Golgi process and induces ER stress by activating the transcription factor (ATF6) and inositol-requiring enzyme 1 (IRE1) pathways [212, 213]. The expression of antiviral interferon-stimulated genes is further downregulated by inhibiting the phosphorylation of IRF3 and thus limiting nuclear translocation [213, 214].
SARS-CoV-2 evolution, variants of concern, and increased antagonism
SARS-CoV-2 VoC has emerged sequentially from the ancestral B.1 lineage throughout the pandemic and often harbors mutations in key viral proteins that modulate immune antagonism and evasion [215, 216]. Although infectivity and antibody evasion are the two primary factors that drive SARS-CoV-2 evolution [217], it has become increasingly clear that IFN resistance may also play a critical role in shaping the trajectory of SARS-CoV-2 [216, 218].
The Alpha variant was shown to suppress innate immune responses in vitro to a greater extent than first-wave isolates. Furthermore, there was an increase in the subgenomic RNA and protein levels of ORF9b, ORF6, and N, which are known to antagonize the innate immune response [47]. Interestingly, the Alpha variant had reduced secretion of IFN-β due to lower amounts of dsRNA intermediates sensed by cells [47, 219]. These innate immune evasion strategies may contribute to the increased transmissibility and enhanced innate immune evasion seen with the alpha variant. While the Alpha variant demonstrated high transmissibility in the human population [220], this level of transmissibility is yet to be fully established in vitro [221], with one study showing a spike-dependent replication advantage in low ACE2-expressing bronchial cell lines compared with the ancestral B.1 [222]. Interestingly, the Alpha variant contains a P681H mutation within PBCS in the spike protein that enables IFITM escape and increases IFN-I resistance [223].
Like IFITM, which is a broad ISG that can restrict viral entry, interferon-inducible restriction factor guanylate-binding proteins (GBPs) have been demonstrated to inhibit furin-mediated processing of viral envelope proteins, including SARS-CoV-2 [85]. Notably, in 2020, the evolution of a D614G substitution on the spike protein of SARS-CoV-2 allowed the mutated virus to escape GBP restriction [137]. Early lineage isolates of Wuhan-Hu-1 and BetaCoV/Australia/VIC01/2020 (VIC) remain susceptible to GBP2 and GBP5, but VoC such as Alpha and Delta escaped GBP2/5 restriction [85]. Interestingly, the Omicron variant remains sensitive to GBP2/5 and endosomal IFITMs [85], likely because of the alternate cell entry pathways associated with different selective pressures and spike mutations [73, 224, 225].
Sensitivity to interferon also varied greatly among patients with VoC. The Omicron variant [226] showed reduced antagonism of the host interferon response compared to the Delta variant [227, 228]; however, Omicron maintained resistance to interferon treatments, in contrast with Delta [229, 230]. Likewise, through integrative computational analyses, the Alpha, Beta, Gamma, and Delta VoC suppressed ISGs, yet the Omicron variant did not [231]. Overall, there is increased interferon resistance among VoC lineages when compared with ancestral isolates, suggesting that innate immune evasion may play a critical role in driving and shaping SARS-CoV-2 evolution [218].
The innate immune response to vaccines
The goal of vaccination is to sufficiently prime the immune system against an infectious agent to prevent future disease. An effective vaccination program has proven to be critical in the fight against the COVID-19 pandemic, with the following four main methods leading the charge: mRNA vaccines, viral vector vaccines, inactivated vaccines, and protein subunit vaccines [232–234]. Since the start of the pandemic, primary T and B-cell responses following vaccination have been abundantly characterized, yet the role of the innate immune system in protection following vaccination has received less attention.
Following BNT162b1 vaccination, there is preferential stimulation of classical (CD14bright CD16−) and intermediate (CD14dim/CD16dim) monocytes but a reduction in nonclassical (CD14dim/CD16bright) monocytes compared to their baseline levels [235]. Even 6 months after the first booster, the percentage of nonclassical monocytes was reduced compared to baseline levels. These vaccine-induced changes shown in the monocyte subpopulations also highlight activation of the protective innate immune response by vaccination, as classical monocytes are critical for the initial inflammatory response, whereas nonclassical monocytes have an anti-inflammatory role and respond poorly to TLR stimulation [235, 236].
Interestingly, BNT162b1 vaccine-induced antibodies have been shown to activate CD107a by NK cells at a greater rate than antibodies generated by natural infection [237]. Despite the diminished CD16 expression, the increased NK cell activity may be explained by the differences in the activation of stimulatory and inhibitory receptors on NK cells following BNT162b1 vaccination. Stimulatory 2DS2-expressing NK cells were significantly augmented after 3 doses of vaccine compared to the baseline levels, while the opposite effect was observed in terms of inhibitory KIR receptor expression. The amount of ILT-2-expressing NK cells, in particular, was significantly reduced [235].
Not only have mRNA-based vaccines been observed to provide enhanced innate immune protection against SARS-CoV-2 infection through the differential activation of innate immune cells, but recent data have also shown an increased innate immune response following dose 2 compared to that following dose 1 [238]. The frequency of intermediate monocytes (CD14+ and CD16+) increased significantly two days after vaccination dose 1 and was substantially higher two days after dose 2. In addition, there were increased levels of pSTAT3 and pSTAT1 in multiple cell types on day one after dose 2, relative to that on day one after dose 1 vaccination. This suggests that BNT162b2 vaccination induced a more substantial innate immune response after dose 2 [238]. These findings are particularly revealing when considering that one of the hallmarks of SARS-CoV-2 infection is impaired IFN-I and III production and responses [239].
Enhancement of innate immune responses was also observed post-BNT162b2 vaccination in individuals previously vaccinated with two doses of the AZD1222 course [240]; Ferreira et al. observed a greater increase in interferon alpha and gamma mRNA signatures and lymphocyte costimulatory signatures by scRNA-seq one month following the mRNA booster dose in comparison to a similar time point post dose 2 of AZD1222 [240]. These innate responses appeared somewhat blunted in elderly individuals [240], and this may underlie reduced spike-specific B and T-cell responses in this vulnerable group [240, 241]. Similarly, reduced germinal center function has been observed in aged mice following vaccination [242], likely due to the microenvironment rather than intrinsic defects [242, 243]. Consistent with the critical role of innate signaling in driving robust vaccine responses, TLR4 can boost germinal center responses to immunization in aged mice by promoting innate immune signaling [244].
Neutralizing antibody is known to predict immune protection [245]. Although showing a lower level of neutralizing antibody in comparison to mRNA vaccines [73, 246–249], inactivated vaccines provided similar protection from this disease at three doses [250]. Unlike mRNA- or vector-based vaccines where the spike protein is the only immunogen, inactivated vaccines produce broader immune responses due to the presence of other structural proteins (M, N, and E). Therefore, it is not too surprising that although the inactivated vaccine elicited a lower magnitude spike-specific T-cell response, it produced a broader multiprotein-specific T-cell response with M, N, and spike-specific T cells, which can more efficiently target highly mutated variants, such as Omicron, compared with the response generated from mRNA vaccines [250]. Moreover, a higher frequency of HLA-DRhi classical and nonclassical monocytes was found to be positively correlated with asymptomatic Omicron breakthrough infection in patients who had three doses of inactivated vaccines [251]. It was suggested that a booster vaccine could train immunity by priming monocytic activation and differentiation rather than suppressed monocytes upon SARS-CoV-2 infection.
Long COVID-19 and the innate immune response
Long COVID-19, also known as the postacute sequelae of COVID-19 (PASC), is a chronic multisystemic condition with a wide range of symptoms that can occur following the resolution of SARS-CoV-2 infection. While substantial progress has been made in characterizing the illness and identifying pathophysiological changes, the etiology of this condition has yet to be revealed, and dysregulation of the innate immune response may play a critical role in the manifestation of associated symptoms.
Early studies reported that some individuals with long COVID had highly activated innate immune cells with elevated expression of IFN-I and IFN-III persisting 8 months post infection [252]. Likewise, other studies have noted elevated levels of cytokines, such as TNF, IP10, IL-1β, and IL-6, in long COVID individuals [253, 254]. These elevated levels of interferons and other proinflammatory cytokines may contribute to the chronic inflammation and immune dysfunction seen in individuals with long COVID.
Various forms of mitochondrial dysfunction were also identified in individuals with long COVID, such as altered fatty acid metabolism [255] and the loss of mitochondrial membrane potential [256]. Abnormal levels of mitochondrial proteins, along with abnormal levels of the S and N proteins, were also found in the central nervous system [257]. Reactivation of latent infections, including human herpesvirus (HHV)-6 and Epstein Bar virus (EBV), has also been identified in individuals with long COVID; this reactivation can induce mitochondrial fragmentation [254, 258–261]. Conceivably, mitochondrial dysfunction and disruption seen in individuals with long COVID-19 may contribute to increased activation of the MAV-IFN signaling pathway, which would inform disease presentation.
Conclusions and perspectives
The host‒virus arms race is complex and involves both the innate and adaptive arms of the immune system. For SARS-CoV-2, the emergence of VoC has allowed us to witness how evolution is used to evade immunity in an unprecedented way. In the future, we could develop pharmacological approaches to exploit or harness our knowledge regarding innate immunity and viral evasion. For example, when a virus encodes a protein such as ORF8 that mimics a human protein, we could develop a specific antagonist of the viral protein. Similarly, pharmacological blockade of ORF3a and ORF7a, which would eliminate their contribution to the persistence of virally infected cells, might tip the balance toward expedited viral clearance. Alternatively, manipulation of PRRs or the cGAS-STING pathway might reduce systemic inflammation and improve COVID-19 outcomes. In particular, future work needs to consider the delicate balance between early protective innate responses and delayed chronic inflammatory responses and how humans and other mammalian hosts, such as bats, find the so-called “goldilocks” zone. Specific bat adaptations, such as mutations in ASC that dampen inflammasome activity and allow the asymptomatic carriage of viruses, may shed light on the drivers of pathological inflammation in humans that are amenable to pharmacological manipulation.
Author contributions
BLS, MTKC, KC, BM, and RKG wrote, revised, and edited the review article.
Competing interests
The authors declare no competing interests.
Contributor Information
Bo Meng, Email: bm432@cam.ac.uk.
Ravindra K. Gupta, Email: rkg20@cam.ac.uk
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu WJ, Liu P, Lei W, Jia Z, He X, Shi W, et al. Surveillance of SARS-CoV-2 at the Huanan Seafood Market. Nature. 2023:1–3. 10.1038/s41586-023-06043-2. [DOI] [PubMed]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–9. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edridge AWD, Kaczorowska J, Hoste A, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med. 2020;26:1691–3. doi: 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 5.Dijkman R, Jebbink MF, Gaunt E, Rossen JW, Templeton KE, Kuijpers TW, et al. The dominance of human coronavirus OC43 and NL63 infections in infants. J Clin Virol. 2012;53:135–9. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson EM, Goodwin EC, Verma A, Arevalo CP, Bolton MJ, Weirick ME, et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–64.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet Lond Engl. 2020;395:565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–21.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433:166725. doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen NT, Chinn J, Nahmias J, Yuen S, Kirby KA, Hohmann S, et al. Outcomes and mortality among adults hospitalized with COVID-19 at US medical centers. JAMA Netw Open. 2021;4:e210417. doi: 10.1001/jamanetworkopen.2021.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tjendra Y, Al Mana AF, Espejo AP, Akgun Y, Millan NC, Gomez-Fernandez C, et al. Predicting disease severity and outcome in COVID-19 patients: a review of multiple biomarkers. Arch Pathol Lab Med. 2020;144:1465–74. doi: 10.5858/arpa.2020-0471-SA. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Beltran WF, Lam EC, Astudillo MG, Yang D, Miller TE, Feldman J, et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184:476–88.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergamaschi L, Mescia F, Turner L, Hanson AL, Kotagiri P, Dunmore BJ, et al. Longitudinal analysis reveals that delayed bystander CD8 + T cell activation and early immune pathology distinguish severe COVID-19 from mild disease. Immunity. 2021;54:1257–75.e8. doi: 10.1016/j.immuni.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore JB, June CH. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–4. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 15.Quan C, Li C, Ma H, Li Y, Zhang H. Immunopathogenesis of coronavirus-induced acute respiratory distress syndrome (ARDS): potential infection-associated hemophagocytic lymphohistiocytosis. Clin Microbiol Rev. 2020;34:e00074–20. doi: 10.1128/CMR.00074-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J-M, An J. Cytokines, inflammation and pain. Int Anesthesiol Clin. 2007;45:27–37. doi: 10.1097/AIA.0b013e318034194e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riera Romo M, Pérez‐Martínez D, Castillo Ferrer C. Innate immunity in vertebrates: an overview. Immunology. 2016;148:125–39. doi: 10.1111/imm.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann H-H, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–38. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotenko SV, Durbin JE. Contribution of type III interferons to antiviral immunity: location, location, location. J Biol Chem. 2017;292:7295–303. doi: 10.1074/jbc.R117.777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy DE, Marié IJ, Durbin JE. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol. 2011;1:476–86. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–62. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Channappanavar R, Perlman S. Age-related susceptibility to coronavirus infections: role of impaired and dysregulated host immunity. J Clin Invest. 2020;130:6204–13. doi: 10.1172/JCI144115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasuga Y, Zhu B, Jang K-J, Yoo J-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp Mol Med. 2021;53:723–36. doi: 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H, Lyu Y, Hou F. SARS-CoV-2 infection and the antiviral innate immune response. J Mol Cell Biol. 2020;12:963–7. doi: 10.1093/jmcb/mjaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol Sin. 2020;35:266–71. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasrija R, Naime M. The deregulated immune reaction and cytokines release storm (CRS) in COVID-19 disease. Int Immunopharmacol. 2021;90:107225. doi: 10.1016/j.intimp.2020.107225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Streicher F, Jouvenet N. Stimulation of innate immunity by host and viral RNAs. Trends Immunol. 2019;40:1134–48. doi: 10.1016/j.it.2019.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Schoggins JW. Interferon-stimulated genes: what do they all do? Annu Rev Virol. 2019;6:567–84. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 30.Crouse J, Kalinke U, Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol. 2015;15:231–42. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 31.Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J Virol. 2020;94. 10.1128/jvi.01410-20. [DOI] [PMC free article] [PubMed]
- 32.Diamond MS, Kanneganti T-D. Innate immunity: the first line of defense against SARS-CoV-2. Nat Immunol. 2022;23:165–76. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santer DM, Li D, Ghosheh Y, Zahoor MA, Prajapati D, Hansen BE, et al. Interferon-λ treatment accelerates SARS-CoV-2 clearance despite age-related delays in the induction of T cell immunity. Nat Commun. 2022;13:6992. doi: 10.1038/s41467-022-34709-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portela Sousa C, Brites C. Immune response in SARS-CoV-2 infection: the role of interferons type I and type III. Braz J Infect Dis. 2020;24:428–33. doi: 10.1016/j.bjid.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–24. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20:397–8. doi: 10.1038/s41577-020-0346-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–12. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Major J, Crotta S, Llorian M, McCabe TM, Gad HH, Priestnall SL, et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–7. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalil AC, Mehta AK, Patterson TF, Erdmann N, Gomez CA, Jain MK, et al. Efficacy of interferon beta-1a plus remdesivir compared with remdesivir alone in hospitalised adults with COVID-19: a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Respir Med. 2021;9:1365–76. doi: 10.1016/S2213-2600(21)00384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Wijst MGP, Vazquez SE, Hartoularos GC, Bastard P, Grant T, Bueno R, et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021;13:eabh2624. doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minkoff JM, tenOever, B. Innate immune evasion strategies of SARS-CoV-2. Nat Rev Microbiol. 2023;21:178–94. doi: 10.1038/s41579-022-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorne LG, Bouhaddou M, Reuschl AK, Zuliani-Alvarez L, Polacco B, Pelin A, et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature. 2022;602:487–95. doi: 10.1038/s41586-021-04352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayn M, Hirschenberger M, Koepke L, Nchioua R, Straub JH, Klute S, et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep. 2021;35:109126. doi: 10.1016/j.celrep.2021.109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong L-YR, Perlman S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses—are we our own worst enemy? Nat Rev Immunol. 2022;22:47–56. doi: 10.1038/s41577-021-00656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JS, Shin E-C. The type I interferon response in COVID-19: implications for treatment. Nat Rev Immunol. 2020;20:585–6. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanneganti T-D. Intracellular innate immune receptors: Life inside the cell. Immunol Rev. 2020;297:5–12. doi: 10.1111/imr.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–96. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–51. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci USA. 2010;107:1512–7. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains. J Biol Chem. 2009;284:17465–74. doi: 10.1074/jbc.M109.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, et al. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7:e00258. doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wickenhagen A, Sugrue E, Lytras S, Kuchi S, Noerenberg M, Turnbull ML, et al. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021;374:eabj3624. doi: 10.1126/science.abj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee D, Le Pen J, Yatim A, Dong B, Aquino Y, Ogishi M, et al. Inborn errors of OAS–RNase L in SARS-CoV-2–related multisystem inflammatory syndrome in children. Science. 2022;379:eabo3627. doi: 10.1126/science.abo3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Q, Matuozzo D, Le Pen J, Lee D, Moens L, Asano T, et al. Recessive inborn errors of type I IFN immunity in children with COVID-19 pneumonia. J Exp Med. 2022;219:e20220131. doi: 10.1084/jem.20220131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diamond MS, Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey CC, Zhong G, Huang I-C, Farzan M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu Rev Virol. 2014;1:261–83. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Foster TL, Pickering S, Neil SJD. Inhibiting the Ins and Outs of HIV replication: cell-intrinsic antiretroviral restrictions at the plasma membrane. Front Immunol. 2018;8:1853. doi: 10.3389/fimmu.2017.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao X, Li J, Winkler CA, An P, Guo J-T. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front Microbiol. 2019;9:3228. doi: 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2021;40:e107405. doi: 10.15252/embj.2020107405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, et al. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe. 2016;20:429–42. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi G, Schwartz O, Compton AA. More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology. 2017;14:53. doi: 10.1186/s12977-017-0377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mudhasani R, Tran JP, Retterer C, Radoshitzky SR, Kota KP, Altamura LA, et al. IFITM-2 and IFITM-3 but Not IFITM-1 restrict Rift Valley fever virus. J Virol. 2013;87:8451–64. doi: 10.1128/JVI.03382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao X, Sehgal M, Hou Z, Cheng J, Shu S, Wu S, et al. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J Virol. 2018;92:e01535–17. doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, et al. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci USA. 2014;111:6756–61. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zang R, Case JB, Yutuc E, Ma X, Shen S, Gomez Castro MF, et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc Natl Acad Sci USA. 2020;117:32105–13. doi: 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi G, Kenney AD, Kudryashova E, Zani A, Zhang L, Lai KK, et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40:e106501. doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winstone H, Lista MJ, Reid AC, Bouton C, Pickering S, Galao RP, et al. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol. 2021;95:e02422–20. doi: 10.1128/JVI.02422-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng B, Abdullahi A, Ferreira I, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–14. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci. 2009;106:5871–6. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park J-E, Li K, Barlan A, Fehr AR, Perlman S, McCray PB JR, et al. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci. 2016;113:12262–7. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ou T, Mou H, Zhang L, Ojha A, Choe H, Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLOS Pathog. 2021;17:e1009212. doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qing E, Li P, Cooper L, Schulz S, Jäck HM, Rong L, et al. Inter-domain communication in SARS-CoV-2 spike proteins controls protease-triggered cell entry. Cell Rep. 2022;39:110786. doi: 10.1016/j.celrep.2022.110786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meng B, Datir R, Choi J, CITIID-NIHR Bioresource COVID- C, Bradley JR, Smith K, et al. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. Cell Rep. 2022;40:111220. doi: 10.1016/j.celrep.2022.111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 81.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA. 2020;117:7001–3. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung C, Kmiec D, Koepke L, Zech F, Jacob T, Sparrer K, et al. Omicron: what makes the latest SARS-CoV-2 variant of concern so concerning? J Virol. 2022;96:e02077–21. doi: 10.1128/jvi.02077-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao H, Lu L, Peng Z, Chen LL, Meng X, Zhang C, et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect. 2022;11:277–83. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira I, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–9. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mesner D, Reuschl AK, Whelan M, Bronzovich T, Haider T, Thorne LG, et al. SARS-CoV-2 evolution influences GBP and IFITM sensitivity. Proc Natl Acad Sci USA. 2023;120:e2212577120. doi: 10.1073/pnas.2212577120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Timilsina U, Umthong S, Ivey EB, Waxman B, Stavrou S. SARS-CoV-2 ORF7a potently inhibits the antiviral effect of the host factor SERINC5. Nat Commun. 2022;13:2935. doi: 10.1038/s41467-022-30609-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meseguer S, Rubio MP, Lainez B, Pérez-Benavente B, Pérez-Moraga R, Romera-Giner S, et al. SARS-CoV-2-encoded small RNAs are able to repress the host expression of SERINC5 to facilitate viral replication. Front Microbiol. 2023;14:1066493. doi: 10.3389/fmicb.2023.1066493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Usami Y, Wu Y, Göttlinger HG. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature. 2015;526:218–23. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perlman S, Peiris M. Coronavirus research: knowledge gaps and research priorities. Nat Rev Microbiol. 2023;21:125–6. doi: 10.1038/s41579-022-00837-3. [DOI] [PubMed] [Google Scholar]
- 90.Zhou Z, Zhang X, Lei X, Xiao X, Jiao T, Ma R, et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal Transduct Target Ther. 2021;6:382. doi: 10.1038/s41392-021-00800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS–STING pathway in health and disease. Nat Rev Genet. 2019;20:657–74. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 92.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma Z, Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe. 2016;19:150–8. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu X, Wei L, Xu F, Zhao F, Huang Y, Fan Z, et al. SARS-CoV-2 spike protein-induced cell fusion activates the cGAS-STING pathway and the interferon response. Sci Signal. 2022;15:eabg8744. doi: 10.1126/scisignal.abg8744. [DOI] [PubMed] [Google Scholar]
- 95.Li H, Zhou F, Zhang L. STING, a critical contributor to SARS-CoV-2 immunopathology. Signal Transduct Target Ther. 2022;7:1–3. doi: 10.1038/s41392-022-00967-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Domizio JD, Gulen MF, Saidoune F, Thacker VV, Yatim A, Sharma K, et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603:145–51. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiong Y, Huang B, Yang Y, Fu X, Fu Z, Xu H, et al. The substrate selectivity of papain-like proteases from human-infecting coronaviruses correlates with innate immune suppression. Sci Signal. 2023;16:eade1985. doi: 10.1126/scisignal.ade1985. [DOI] [PubMed] [Google Scholar]
- 98.Cao D, Duan L, Huang B, Xiong Y, Zhang G, Huang H. The SARS-CoV-2 papain-like protease suppresses type I interferon responses by deubiquitinating STING. Sci Signal. 2023;16:eadd0082. doi: 10.1126/scisignal.add0082. [DOI] [PubMed] [Google Scholar]
- 99.Bussani R, Schneider E, Zentilin L, Collesi C, Ali H, Braga L, et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61:103104. doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Z, Zheng Y, Niu Z, Zhang B, Wang C, Yao X, et al. SARS-CoV-2 spike protein dictates syncytium-mediated lymphocyte elimination. Cell Death Differ. 2021;28:2765–77. doi: 10.1038/s41418-021-00782-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Papa G, Mallery DL, Albecka A, Welch LG, Cattin-Ortolá J, Luptak J, et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLOS Pathog. 2021;17:e1009246. doi: 10.1371/journal.ppat.1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cai S, Zhang C, Zhuang Z, Zhang S, Ma L, Yang S, et al. Phase-separated nucleocapsid protein of SARS-CoV-2 suppresses cGAS-DNA recognition by disrupting cGAS-G3BP1 complex. Signal Transduct Target Ther. 2023;8:170. doi: 10.1038/s41392-023-01420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith EC, Blanc H, Surdel MC, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Amicone M, Borges V, Alves MJ, Isidro J, Zé-Zé L, Duarte S, et al. Mutation rate of SARS-CoV-2 and emergence of mutators during experimental evolution. Evol Med Public Health. 2022;10:142–55. doi: 10.1093/emph/eoac010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. 2021;22:757–73. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Markov PV, Ghafari M, Beer M, Lythgoe K, Simmonds P, Stilianakis NI, et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. 2023;21:361–79. doi: 10.1038/s41579-023-00878-2. [DOI] [PubMed] [Google Scholar]
- 107.Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–76. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 108.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Picardi E, Manzari C, Mastropasqua F, Aiello I, D'Erchia AM, Pesole G. Profiling RNA editing in human tissues: towards the inosinome Atlas. Sci Rep. 2015;5:14941. doi: 10.1038/srep14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Di Giorgio S, Martignano F, Torcia MG, Mattiuz G, Conticello SG. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv. 2020;6:eabb5813. doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim K, Calabrese P, Wang S, Qin C, Rao Y, Feng P, et al. The roles of APOBEC-mediated RNA editing in SARS-CoV-2 mutations, replication and fitness. bioRxiv 2021.12.18.473309. 2022. 10.1101/2021.12.18.473309. [DOI] [PMC free article] [PubMed]
- 112.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Asaoka M, Ishikawa T, Takabe K, Patnaik SK. APOBEC3-mediated RNA editing in breast cancer is associated with heightened immune activity and improved survival. Int J Mol Sci. 2019;20:5621. doi: 10.3390/ijms20225621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Azgari C, Kilinc Z, Turhan B, Circi D, Adebali O. The mutation profile of SARS-CoV-2 is primarily shaped by the host antiviral defense. Viruses. 2021;13:394. doi: 10.3390/v13030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peng X, Luo Y, Li H, Guo X, Chen H, Ji X, et al. RNA editing increases the nucleotide diversity of SARS-CoV-2 in human host cells. PLoS Genet. 2022;18:e1010130. doi: 10.1371/journal.pgen.1010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Graudenzi A, Maspero D, Angaroni F, Piazza R, Ramazzotti D. Mutational signatures and heterogeneous host response revealed via large-scale characterization of SARS-CoV-2 genomic diversity. iScience. 2021;24:102116. doi: 10.1016/j.isci.2021.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Evangelou K, Veroutis D, Paschalaki K, Foukas PG, Lagopati N, Dimitriou M, et al. Pulmonary infection by SARS-CoV-2 induces senescence accompanied by an inflammatory phenotype in severe COVID-19: possible implications for viral mutagenesis. Eur Respir J. 2022;60:2102951. doi: 10.1183/13993003.02951-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karakasiliotis I, Lagopati N, Evangelou K, Gorgoulis VG. Cellular senescence as a source of SARS-CoV-2 quasispecies. FEBS J. 2023;290:1384–92. doi: 10.1111/febs.16230. [DOI] [PubMed] [Google Scholar]
- 119.Ruis C, Peacock TP, Polo LM, Masone D, Alvarez MS, Hinrichs AS, et al. A lung-specific mutational signature enables inference of viral and bacterial respiratory niche. Microb Genom. 2023;9:mgen001018. doi: 10.1099/mgen.0.001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Samuel CE. Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. J Biol Chem. 2019;294:1710–20. doi: 10.1074/jbc.TM118.004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei Y, Silke JR, Aris P, Xia X. Coronavirus genomes carry the signatures of their habitats. PloS One. 2020;15:e0244025. doi: 10.1371/journal.pone.0244025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sharma S, Patnaik SK, Taggart RT, Kannisto ED, Enriquez SM, Gollnick P, et al. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun. 2015;6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bauernfried S, Scherr MJ, Pichlmair A, Duderstadt KE, Hornung V. Human NLRP1 is a sensor for double-stranded RNA. Science. 2021;371:eabd0811. doi: 10.1126/science.abd0811. [DOI] [PubMed] [Google Scholar]
- 124.Simmonds P, Ansari MA. Extensive C- > U transition biases in the genomes of a wide range of mammalian RNA viruses; potential associations with transcriptional mutations, damage- or host-mediated editing of viral RNA. PLoS Pathog. 2021;17:e1009596. doi: 10.1371/journal.ppat.1009596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cheng AZ, Moraes SN, Shaban NM, Fanunza E, Bierle CJ, Southern PJ, et al. APOBECs and herpesviruses. Viruses. 2021;13:390. doi: 10.3390/v13030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nakata Y, Ode H, Kubota M, Kasahara T, Matsuoka K, Sugimoto A, et al. Cellular APOBEC3A deaminase drives mutations in the SARS-CoV-2 genome. Nucleic Acids Res. 2023;51:783–95. doi: 10.1093/nar/gkac1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McDaniel YZ, Wang D, Love RP, Adolph MB, Mohammadzadeh N, Chelico L, et al. Deamination hotspots among APOBEC3 family members are defined by both target site sequence context and ssDNA secondary structure. Nucleic Acids Res. 2020;48:1353–71. doi: 10.1093/nar/gkz1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rice AM, Castillo Morales A, Ho AT, Mordstein C, Mühlhausen S, Watson S, et al. Evidence for strong mutation bias toward, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol Biol Evol. 2021;38:67–83. doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Dorp L, Richard D, Tan C, Shaw LP, Acman M, Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun. 2020;11:5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ratcliff J, Simmonds P. Potential APOBEC-mediated RNA editing of the genomes of SARS-CoV-2 and other coronaviruses and its impact on their longer term evolution. Virology. 2021;556:62–72. doi: 10.1016/j.virol.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simmonds P. Rampant C → U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5:e00408–20. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yi K, Kim SY, Bleazard T, Kim T, Youk J, Ju YS. Mutational spectrum of SARS-CoV-2 during the global pandemic. Exp Mol Med. 2021;53:1229–37. doi: 10.1038/s12276-021-00658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang Q, Li X, Qi R, Billiar T. RNA editing, ADAR1, and the innate immune response. Genes. 2017;8:41. doi: 10.3390/genes8010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–15. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chung H, Calis J, Wu X, Sun T, Yu Y, Sarbanes SL, et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell. 2018;172:811–24.e14. doi: 10.1016/j.cell.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet. 2012;44:1243–8. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–27.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mourier T, Sadykov M, Carr MJ, Gonzalez G, Hall WW, Pain A. Host-directed editing of the SARS-CoV-2 genome. Biochem Biophys Res Commun. 2021;538:35–9. doi: 10.1016/j.bbrc.2020.10.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schmidt N, Lareau CA, Keshishian H, Ganskih S, Schneider C, Hennig T, et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat Microbiol. 2021;6:339–53. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pfaller CK, George CX, Samuel CE. Adenosine deaminases acting on RNA (ADARs) and viral infections. Annu Rev Virol. 2021;8:239–64. doi: 10.1146/annurev-virology-091919-065320. [DOI] [PubMed] [Google Scholar]
- 141.Ringlander J, Fingal J, Kann H, Prakash K, Rydell G, Andersson M, et al. Impact of ADAR-induced editing of minor viral RNA populations on replication and transmission of SARS-CoV-2. Proc Natl Acad Sci USA. 2022;119:e2112663119. doi: 10.1073/pnas.2112663119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–6. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 143.Zhu Y, Gao G. ZAP-mediated mRNA degradation. RNA Biol. 2008;5:65–67. doi: 10.4161/rna.5.2.6044. [DOI] [PubMed] [Google Scholar]
- 144.Zheng X, Wang X, Tu F, Wang Q, Fan Z, Gao G. TRIM25 is required for the antiviral activity of zinc finger antiviral protein. J Virol. 2017;91:e00088–17. doi: 10.1128/JVI.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ficarelli M, Wilson H, Pedro Galão R, Mazzon M, Antzin-Anduetza I, Marsh M, et al. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. eLife. 2019;8:e46767. doi: 10.7554/eLife.46767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li MM, Lau Z, Cheung P, Aguilar EG, Schneider WM, Bozzacco L, et al. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP) PLoS Pathog. 2017;13:e1006145. doi: 10.1371/journal.ppat.1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xue G, Braczyk K, Gonçalves-Carneiro D, Dawidziak DM, Sanchez K, Ong H, et al. Poly(ADP-ribose) potentiates ZAP antiviral activity. PLOS Pathog. 2022;18:e1009202. doi: 10.1371/journal.ppat.1009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kamel W, Noerenberg M, Cerikan B, Chen H, Järvelin AI, Kammoun M, et al. Global analysis of protein-RNA interactions in SARS-CoV-2-infected cells reveals key regulators of infection. Mol Cell. 2021;81:2851–67.e7. doi: 10.1016/j.molcel.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kumar A, Goyal N, Saranathan N, Dhamija S, Saraswat S, Menon MB, et al. The slowing rate of CpG depletion in SARS-CoV-2 genomes is consistent with adaptations to the human host. Mol Biol Evol. 2022;39:msac029. doi: 10.1093/molbev/msac029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Green DR. The end and after: how dying cells impact the living organism. Immunity. 2011;35:441–4. doi: 10.1016/j.immuni.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 151.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–77. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen T, Tu S, Ding L, Jin M, Chen H, Zhou H. The role of autophagy in viral infections. J Biomed Sci. 2023;30:5. doi: 10.1186/s12929-023-00899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Choi Y, Bowman JW, Jung JU. Autophagy during viral infection—a double-edged sword. Nat Rev Microbiol. 2018;16:341–54. doi: 10.1038/s41579-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, Baumgart GJ, et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat Microbiol. 2017;2:1543–57. doi: 10.1038/s41564-017-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature. 2017;541:412–6. doi: 10.1038/nature21032. [DOI] [PubMed] [Google Scholar]
- 156.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–9. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Su J, Shen S, Hu Y, Chen S, Cheng L, Cai Y, et al. SARS-CoV-2 ORF3a inhibits cGAS-STING-mediated autophagy flux and antiviral function. J Med Virol. 2023;95:e28175. doi: 10.1002/jmv.28175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Han L, Zheng Y, Deng J, Nan ML, Xiao Y, Zhuang MW, et al. SARS‐CoV‐2 ORF10 antagonizes STING‐dependent interferon activation and autophagy. J Med Virol. 2022;94:5174–88. doi: 10.1002/jmv.27965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koepke L, Hirschenberger M, Hayn M, Kirchhoff F, Sparrer KM. Manipulation of autophagy by SARS-CoV-2 proteins. Autophagy. 2021;17:2659–61. doi: 10.1080/15548627.2021.1953847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ren Y, Shu T, Wu D, Mu J, Wang C, Huang M, et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol. 2020;17:881–3. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Miao G, Zhao H, Li Y, Ji M, Chen Y, Shi Y, et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56:427–42.e5. doi: 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhang Y, Sun H, Pei R, Mao B, Zhao Z, Li H, et al. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021;7:1–12. doi: 10.1038/s41421-021-00268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mozzi A, Oldani M, Forcella ME, Vantaggiato C, Cappelletti G, Pontremoli C, et al. SARS-CoV-2 ORF3c impairs mitochondrial respiratory metabolism, oxidative stress, and autophagic flux. iScience. 2023;26:107118. doi: 10.1016/j.isci.2023.107118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang J, Wu H, Yao X, Zhang D, Zhou Y, Fu B, et al. Pyroptotic macrophages stimulate the SARS-CoV-2-associated cytokine storm. Cell Mol Immunol. 2021;18:1305–7. doi: 10.1038/s41423-021-00665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sun X, Liu Y, Huang Z, Xu W, Hu W, Yi L, et al. SARS-CoV-2 non-structural protein 6 triggers NLRP3-dependent pyroptosis by targeting ATP6AP1. Cell Death Differ. 2022;29:1240–54. doi: 10.1038/s41418-021-00916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Junqueira C, Crespo Â, Ranjbar S, de Lacerda LB, Lewandrowski M, Ingber J, et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature. 2022;606:576–84. doi: 10.1038/s41586-022-04702-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sefik E, Qu R, Junqueira C, Kaffe E, Mirza H, Zhao J, et al. Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature. 2022;606:585–93. doi: 10.1038/s41586-022-04802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ma J, Zhu F, Zhao M, Shao F, Yu D, Ma J, et al. SARS-CoV-2 nucleocapsid suppresses host pyroptosis by blocking Gasdermin D cleavage. EMBO J. 2021;40:e108249. doi: 10.15252/embj.2021108249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6:1–21. doi: 10.1038/s41392-021-00507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang D-M, Geng T-T, Harrison AG, Wang P-H. Differential roles of RIG-I like receptors in SARS-CoV-2 infection. Mil Med Res. 2021;8:49. doi: 10.1186/s40779-021-00340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fu Y-Z, Wang SY, Zheng ZQ, Yi H, Li WW, Xu ZS, et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cell Mol Immunol. 2021;18:613–20. doi: 10.1038/s41423-020-00571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wu J, Shi Y, Pan X, Wu S, Hou R, Zhang Y, et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li X, Hou P, Ma W, Wang X, Wang H, Yu Z, et al. SARS-CoV-2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cell Mol Immunol. 2022;19:67–78. doi: 10.1038/s41423-021-00807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stewart H, Lu Y, O'Keefe S, Valpadashi A, Cruz-Zaragoza LD, Michel HA, et al. The SARS-CoV-2 protein ORF3c is a mitochondrial modulator of innate immunity. iScience. 2023;26:108080. doi: 10.1016/j.isci.2023.108080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yamada T, Sato S, Sotoyama Y, Orba Y, Sawa H, Yamauchi H, et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat Immunol. 2021;22:820–8. doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 176.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–34. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schroeder S, Pott F, Niemeyer D, Veith T, Richter A, Muth D, et al. Interferon antagonism by SARS-CoV-2: a functional study using reverse genetics. Lancet Microbe. 2021;2:e210–8. doi: 10.1016/S2666-5247(21)00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–45.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li J-Y, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074. doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yuen C-K, Lam JY, Wong WM, Mak LF, Wang X, Chu H, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418–28. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci USA. 2020;117:28344–54. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rashid F, Suleman M, Shah A, Dzakah EE, Wang H, Chen S, et al. Mutations in SARS-CoV-2 ORF8 altered the bonding network with interferon regulatory factor 3 to evade host immune system. Front Microbiol. 2021;12:703145. doi: 10.3389/fmicb.2021.703145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gori Savellini G, Anichini G, Cusi MG. SARS-CoV-2 omicron sub-lineages differentially modulate interferon response in human lung epithelial cells. Virus Res. 2023;332:199134. doi: 10.1016/j.virusres.2023.199134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hyams C, Challen R, Marlow R, Nguyen J, Begier E, Southern J, et al. Severity of Omicron (B.1.1.529) and Delta (B.1.617.2) SARS-CoV-2 infection among hospitalised adults: a prospective cohort study in Bristol, United Kingdom. Lancet Reg Health—Eur. 2023;25:100556. doi: 10.1016/j.lanepe.2022.100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kumar A, Ishida R, Strilets T, Cole J, Lopez-Orozco J, Fayad N, et al. SARS-CoV-2 nonstructural protein 1 inhibits the interferon response by causing depletion of key host signaling factors. J Virol. 2021;95:e0026621. doi: 10.1128/JVI.00266-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liu G, Lee JH, Parker ZM, Acharya D, Chiang JJ, van Gent M, et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol. 2021;6:467–78. doi: 10.1038/s41564-021-00884-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–62. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Alhammad YMO, Kashipathy MM, Roy A, Gagné JP, McDonald P, Gao P, et al. The SARS-CoV-2 conserved macrodomain is a mono-ADP-ribosylhydrolase. J Virol. 2021;95:e01969–20. doi: 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2014;89:1523–36. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fehr AR, Channappanavar R, Jankevicius G, Fett C, Zhao J, Athmer J, et al. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7:e01721–16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Taha TY, Suryawanshi RK, Chen IP, Correy GJ, McCavitt-Malvido M, O'Leary PC, et al. A single inactivating amino acid change in the SARS-CoV-2 NSP3 Mac1 domain attenuates viral replication in vivo. PLOS Pathog. 2023;19:e1011614. doi: 10.1371/journal.ppat.1011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ortega Granda O, Alvarez K, Mate-Perez MJ, Canard B, Ferron F, Rabah N. Macro1 domain residue F156: a hallmark of SARS-CoV-2 de-MARylation specificity. Virology. 2023;587:109845. doi: 10.1016/j.virol.2023.109845. [DOI] [PubMed] [Google Scholar]
- 194.Hoch NC. Host ADP-ribosylation and the SARS-CoV-2 macrodomain. Biochem Soc Trans. 2021;49:1711–21. doi: 10.1042/BST20201212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Alhammad YM, Parthasarathy S, Ghimire R, O'Connor JJ, Kerr CM, Pfannenstiel JJ, et al. SARS-CoV-2 Mac1 is required for IFN antagonism and efficient virus replication in mice. BioRxiv Prepr Serv Biol. 2023.04.06.535927. 2023. 10.1101/2023.04.06.535927. [DOI] [PMC free article] [PubMed]
- 196.Zhang Y, Mao D, Roswit WT, Jin X, Patel AC, Patel DA, et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat Immunol. 2015;16:1215–27. doi: 10.1038/ni.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yang Z, Zhang X, Wang F, Wang P, Kuang E, Li X. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. 2020.08.12.247767. 2020. Preprint at 10.1101/2020.08.12.247767.
- 199.Madiraju C, Novack JP, Reed JC, Matsuzawa S. K63 ubiquitination in immune signaling. Trends Immunol. 2022;43:148–62. doi: 10.1016/j.it.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Neches RY, Kyrpides NC, Ouzounis CA. Atypical divergence of SARS-CoV-2 Orf8 from Orf7a within the coronavirus lineage suggests potential stealthy viral strategies in immune evasion. mBio. 2021;12. 10.1128/mbio.03014-20. [DOI] [PMC free article] [PubMed]
- 201.Hassan SS, Aljabali A, Panda PK, Ghosh S, Attrish D, Choudhury PP, et al. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput Biol Med. 2021;133:104380. doi: 10.1016/j.compbiomed.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhang Y, Chen Y, Li Y, Huang F, Luo B, Yuan Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc Natl Acad Sci USA. 2021;118:e2024202118. doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Muth D, Corman VM, Roth H, Binger T, Dijkman R, Gottula LT, et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci Rep. 2018;8:15177. doi: 10.1038/s41598-018-33487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Arduini A, Laprise F, Liang C. SARS-CoV-2 ORF8: a rapidly evolving immune and viral modulator in COVID-19. Viruses. 2023;15:871. doi: 10.3390/v15040871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vinjamuri S, Li L, Bouvier M. SARS-CoV-2 ORF8: one protein, seemingly one structure, and many functions. Front Immunol. 2022;13:1035559. doi: 10.3389/fimmu.2022.1035559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Young BE, Fong SW, Chan YH, Mak TM, Ang LW, Anderson DE, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–11. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gangavarapu K, Latif AA, Mullen JL, Alkuzweny M, Hufbauer E, Tsueng G, et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat Methods. 2023;20:512–22. doi: 10.1038/s41592-023-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pereira F. SARS-CoV-2 variants combining spike mutations and the absence of ORF8 may be more transmissible and require close monitoring. Biochem Biophys Res Commun. 2021;550:8–14. doi: 10.1016/j.bbrc.2021.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wu X, Xia T, Shin WJ, Yu KM, Jung W, Herrmann A, et al. Viral mimicry of interleukin-17A by SARS-CoV-2 ORF8. mBio. 2022;13:e0040222. doi: 10.1128/mbio.00402-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kee J, Thudium S, Renner DM, Glastad K, Palozola K, Zhang Z, et al. SARS-CoV-2 disrupts host epigenetic regulation via histone mimicry. Nature. 2022;610:381–8. doi: 10.1038/s41586-022-05282-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chaudhari AM, Singh I, Joshi M, Patel A, Joshi C. Defective ORF8 dimerization in SARS-CoV-2 delta variant leads to a better adaptive immune response due to abrogation of ORF8-MHC1 interaction. Mol Divers. 2023;27:45–57. doi: 10.1007/s11030-022-10405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kim I-J, Lee YH, Khalid MM, Chen IP, Zhang Y, Ott M, et al. SARS-CoV-2 protein ORF8 limits expression levels of Spike antigen and facilitates immune evasion of infected host cells. J Biol Chem. 2023;299:104955. doi: 10.1016/j.jbc.2023.104955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rashid F, Dzakah EE, Wang H, Tang S. The ORF8 protein of SARS-CoV-2 induced endoplasmic reticulum stress and mediated immune evasion by antagonizing production of interferon beta. Virus Res. 2021;296:198350. doi: 10.1016/j.virusres.2021.198350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu P, Wang X, Sun Y, Zhao H, Cheng F, Wang J, et al. SARS-CoV-2 ORF8 reshapes the ER through forming mixed disulfides with ER oxidoreductases. Redox Biol. 2022;54:102388. doi: 10.1016/j.redox.2022.102388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Walensky RP, Walke HT, Fauci AS. SARS-CoV-2 variants of concern in the United States—challenges and opportunities. JAMA. 2021;325:1037–8. doi: 10.1001/jama.2021.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Telenti A, Hodcroft EB, Robertson DL. The evolution and biology of SARS-CoV-2 variants. Cold Spring Harb Perspect Med. 2022;12:a041390. doi: 10.1101/cshperspect.a041390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–82. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Guo K, Barrett BS, Morrison JH, Mickens KL, Vladar EK, Hasenkrug KJ, et al. Interferon resistance of emerging SARS-CoV-2 variants. Proc Natl Acad Sci USA. 2022;119:e2203760119. doi: 10.1073/pnas.2203760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nchioua R, Schundner A, Klute S, Koepke L, Hirschenberger M, Noettger S, et al. Reduced replication but increased interferon resistance of SARS-CoV-2 Omicron BA.1. Life Sci Alliance. 2023;6:e202201745. doi: 10.26508/lsa.202201745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hill V, Du Plessis L, Peacock TP, Aggarwal D, Colquhoun R, Carabelli AM, et al. The origins and molecular evolution of SARS-CoV-2 lineage B.1.1.7 in the UK. Virus Evol. 2022;8:veac080. doi: 10.1093/ve/veac080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022;39:110829. doi: 10.1016/j.celrep.2022.110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Niemeyer D, Stenzel S, Veith T, Schroeder S, Friedmann K, Weege F, et al. SARS-CoV-2 variant Alpha has a spike-dependent replication advantage over the ancestral B.1 strain in human cells with low ACE2 expression. PLoS Biol. 2022;20:e3001871. doi: 10.1371/journal.pbio.3001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lista MJ, Winstone H, Wilson HD, Dyer A, Pickering S, Galao RP, et al. The P681H mutation in the spike glycoprotein of the alpha variant of SARS-CoV-2 escapes IFITM restriction and is necessary for type I interferon resistance. J Virol. 2022;96:e0125022. doi: 10.1128/jvi.01250-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022;7:1161–79. doi: 10.1038/s41564-022-01143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Peacock TP, Brown JC, Zhou J, Thakur N, Sukhova K, Newman J, et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. 2021.12.31.474653. 2022. Preprint at 10.1101/2021.12.31.474653.
- 226.Viana R, Moyo S, Amoako DG, Tegally H, Scheepers C, Althaus CL, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–86. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Dhar MS, Marwal R, Vs R, Ponnusamy K, Jolly B, Bhoyar RC, et al. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. Science. 2021;374:995–9. doi: 10.1126/science.abj9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ferreira IATM, Kemp SA, Datir R, Saito A, Meng B, Rakshit P, et al. SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J Infect Dis. 2021;224:989–94. doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Bojkova D, Rothenburger T, Ciesek S, Wass MN, Michaelis M, Cinatl J JR. SARS-CoV-2 Omicron variant virus isolates are highly sensitive to interferon treatment. Cell Discov. 2022;8:42. doi: 10.1038/s41421-022-00408-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bojkova D, Widera M, Ciesek S, Wass MN, Michaelis M, Cinatl J JR. Reduced interferon antagonism but similar drug sensitivity in Omicron variant compared to Delta variant of SARS-CoV-2 isolates. Cell Res. 2022;32:319–21. doi: 10.1038/s41422-022-00619-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Bouhaddou M, Reuschl AK, Polacco BJ, Thorne LG, Ummadi MR, Ye C, et al. SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell. 2023;186:4597–614. doi: 10.1016/j.cell.2023.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–27. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 233.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med. 2020;383:1920–31. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Creech CB, Walker SC, Samuels RJ. SARS-CoV-2 vaccines. JAMA. 2021;325:1318–20. doi: 10.1001/jama.2021.3199. [DOI] [PubMed] [Google Scholar]
- 235.Saresella M, Piancone F, Marventano I, Hernis A, Trabattoni D, Invernizzi M, et al. Innate immune responses to three doses of the BNT162b2 mRNA SARS-CoV-2 vaccine. Front Immunol. 2022;13:947320. doi: 10.3389/fimmu.2022.947320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–86. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hagemann K, Riecken K, Jung JM, Hildebrandt H, Menzel S, Bunders MJ, et al. Natural killer cell-mediated ADCC in SARS-CoV-2-infected individuals and vaccine recipients. Eur J Immunol. 2022;52:1297–307. doi: 10.1002/eji.202149470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Arunachalam PS, Scott M, Hagan T, Li C, Feng Y, Wimmers F, et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–6. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Matsuyama T, Kubli SP, Yoshinaga SK, Pfeffer K, Mak TW. An aberrant STAT pathway is central to COVID-19. Cell Death Differ. 2020;27:3209–25. doi: 10.1038/s41418-020-00633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ferreira IATM, Lee C, Foster WS, Abdullahi A, Dratva LM, Tuong ZK, et al. Atypical B cells and impaired SARS-CoV-2 neutralization following heterologous vaccination in the elderly. Cell Rep. 2023;42:112991. doi: 10.1016/j.celrep.2023.112991. [DOI] [PubMed] [Google Scholar]
- 241.Collier DA, Ferreira I, Kotagiri P, Datir RP, Lim EY, Touizer E, et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. 2021;596:417–22. doi: 10.1038/s41586-021-03739-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lee JL, Innocentin S, Silva-Cayetano A, Guillaume SM, Linterman MA. B cells from aged mice do not have intrinsic defects in affinity maturation. 2023.04.24.538044. 2023. Preprint at 10.1101/2023.04.24.538044. [DOI] [PMC free article] [PubMed]
- 243.Lee JL, Fra-Bido SC, Burton AR, Innocentin S, Hill DL, Linterman MA. B cell-intrinsic changes with age do not impact antibody-secreting cell formation but delay B cell participation in the germinal centre reaction. Aging Cell. 2022;21:e13692. doi: 10.1111/acel.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Denton AE, Dooley J, Cinti I, Silva-Cayetano A, Fra-Bido S, Innocentin S, et al. Targeting TLR4 during vaccination boosts MAdCAM-1+ lymphoid stromal cell activation and promotes the aged germinal center response. Sci Immunol. 2022;7:eabk0018. doi: 10.1126/sciimmunol.abk0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Khoury DS, Cromer D, Reynaldi A, Schlub TE, Wheatley AK, Juno JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. 2021;27:1205–11. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 246.Rosa Duque JS, Wang X, Leung D, Cheng S, Cohen CA, Mu X, et al. Immunogenicity and reactogenicity of SARS-CoV-2 vaccines BNT162b2 and CoronaVac in healthy adolescents. Nat Commun. 2022;13:3700. doi: 10.1038/s41467-022-31485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Peng Q, Zhou R, Wang Y, Zhao M, Liu N, Li S, et al. Waning immune responses against SARS-CoV-2 variants of concern among vaccinees in Hong Kong. eBioMedicine. 2022;77:103904. doi: 10.1016/j.ebiom.2022.103904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lim WW, Mak L, Leung GM, Cowling BJ, Peiris M. Comparative immunogenicity of mRNA and inactivated vaccines against COVID-19. Lancet Microbe. 2021;2:e423. doi: 10.1016/S2666-5247(21)00177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chen Y, Shen H, Huang R, Tong X, Wu C. Serum neutralising activity against SARS-CoV-2 variants elicited by CoronaVac. Lancet Infect Dis. 2021;21:1071–2. doi: 10.1016/S1473-3099(21)00287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.McMenamin ME, Nealon J, Lin Y, Wong JY, Cheung JK, Lau E, et al. Vaccine effectiveness of one, two, and three doses of BNT162b2 and CoronaVac against COVID-19 in Hong Kong: a population-based observational study. Lancet Infect Dis. 2022;22:1435–43. doi: 10.1016/S1473-3099(22)00345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Yu S, Lin Y, Li Y, Chen S, Zhou L, Song H, et al. Systemic immune profiling of Omicron-infected subjects inoculated with different doses of inactivated virus vaccine. Cell. 2023;186:4615–31.e16. doi: 10.1016/j.cell.2023.08.033. [DOI] [PubMed] [Google Scholar]
- 252.Phetsouphanh C, Darley DR, Wilson DB, Howe A, Munier C, Patel SK, et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat Immunol. 2022;23:210–6. doi: 10.1038/s41590-021-01113-x. [DOI] [PubMed] [Google Scholar]
- 253.Schultheiß C, Willscher E, Paschold L, Gottschick C, Klee B, Henkes SS, et al. The IL-1β, IL-6, and TNF cytokine triad is associated with post-acute sequelae of COVID-19. Cell Rep Med. 2022;3:100663. [DOI] [PMC free article] [PubMed]
- 254.Peluso MJ, Lu S, Tang AF, Durstenfeld MS, Ho HE, Goldberg SA, et al. Markers of immune activation and inflammation in individuals with postacute sequelae of severe acute respiratory syndrome coronavirus 2 infection. J Infect Dis. 2021;224:1839–48. doi: 10.1093/infdis/jiab490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Guntur VP, Nemkov T, de Boer E, Mohning MP, Baraghoshi D, Cendali FI, et al. Signatures of mitochondrial dysfunction and impaired fatty acid metabolism in plasma of patients with post-acute sequelae of COVID-19 (PASC) Metabolites. 2022;12:1026. doi: 10.3390/metabo12111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Díaz-Resendiz KJG, Benitez-Trinidad AB, Covantes-Rosales CE, Toledo-Ibarra GA, Ortiz-Lazareno PC, Girón-Pérez DA, et al. Loss of mitochondrial membrane potential (ΔΨm) in leucocytes as post-COVID-19 sequelae. J Leukoc Biol. 2022;112:23–29. doi: 10.1002/JLB.3MA0322-279RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Peluso MJ, Deeks SG, Mustapic M, Kapogiannis D, Henrich TJ, Lu S, et al. SARS-CoV-2 and mitochondrial proteins in neural-derived exosomes of COVID-19. Ann Neurol. 2022;91:772–81. doi: 10.1002/ana.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Schreiner P, Harrer T, Scheibenbogen C, Lamer S, Schlosser A, Naviaux RK, et al. Human herpesvirus-6 reactivation, mitochondrial fragmentation, and the coordination of antiviral and metabolic phenotypes in myalgic encephalomyelitis/chronic fatigue syndrome. ImmunoHorizons. 2020;4:201–15. doi: 10.4049/immunohorizons.2000006. [DOI] [PubMed] [Google Scholar]
- 259.Zubchenko S, Kril I, Nadizhko O, Matsyura O, Chopyak V. Herpesvirus infections and post-COVID-19 manifestations: a pilot observational study. Rheumatol Int. 2022;42:1523–30. doi: 10.1007/s00296-022-05146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Peluso MJ, Deveau TM, Munter SE, Ryder D, Buck A, Beck-Engeser G, et al. Chronic viral coinfections differentially affect the likelihood of developing long COVID. J Clin Invest. 2023;133:e163669. [DOI] [PMC free article] [PubMed]
- 261.Su Y, Yuan D, Chen DG, Ng RH, Wang K, Choi J, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185:881–95.e20. doi: 10.1016/j.cell.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Moustaqil M, Ollivier E, Chiu HP, Van Tol S, Rudolffi-Soto P, Stevens C, et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infect. 2021;10:178–95. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Feng K, Zhang HJ, Min YQ, Zhou M, Deng F, Wang HL, et al. SARS-CoV-2 NSP13 interacts with host IRF3, blocking antiviral immune responses. J Med Virol. 2023;95:e28881. doi: 10.1002/jmv.28881. [DOI] [PubMed] [Google Scholar]
- 264.Fung S-Y, Siu KL, Lin H, Chan CP, Yeung ML, Jin DY. SARS-CoV-2 NSP13 helicase suppresses interferon signaling by perturbing JAK1 phosphorylation of STAT1. Cell Biosci. 2022;12:36. doi: 10.1186/s13578-022-00770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]