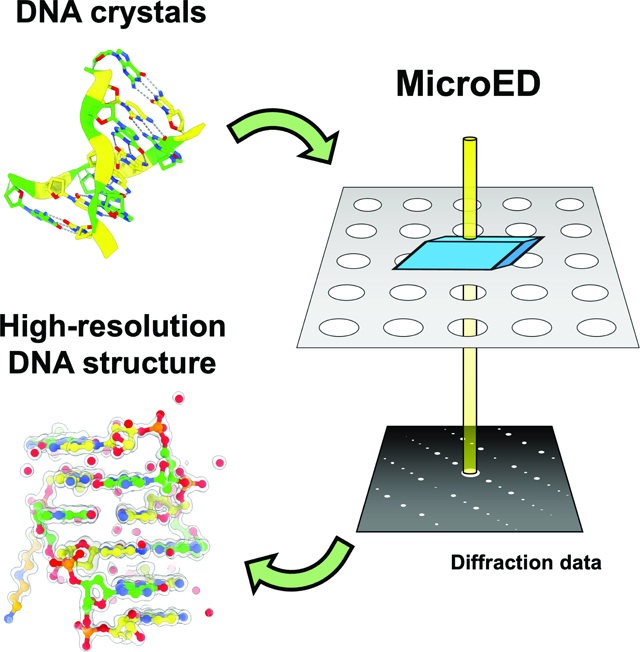
SUMMARY
Microcrystal electron diffraction (MicroED) is a powerful tool for determining high-resolution structures of microcrystals from a diverse array of biomolecular, chemical, and material samples. In this study, we apply MicroED to DNA crystals, which have not been previously analyzed using this technique. We utilized the d(CGCGCG)2 DNA duplex as a model sample and employed cryo-FIB milling to create thin lamella for diffraction data collection. The MicroED data collection and subsequent processing resulted in a 1.10 Å resolution structure of the d(CGCGCG)2 DNA, demonstrating the successful application of cryo-FIB milling and MicroED to the investigation of nucleic acid crystals.
Keywords: Microcrystal electron diffraction (MicroED), DNA, Nucleic Acids, cryo-electron microscopy, cryo-focused ion beam milling
eTOC
Haymaker et al. used microcrystal electron diffraction (MicroED) to determine the structure of a model DNA crystal to 1.1 Å. This study shows the applicability of MicroED to nucleic acid crystallography.
Graphical Abstract
INTRODUCTION
Determining the high-resolution structures of nucleic acids is a crucial step in understanding the structure-function relationships of these essential biomolecules. X-ray crystallography has been the primary method for studying DNA structure, RNA structure, and designed systems from DNA nanotechnology 1–5. However, this method typically requires large, well-ordered crystals for collecting high-resolution data, which can pose a significant challenge for many targets. Newer beamlines and free electron lasers promise to collect X-ray diffraction data from smaller and smaller crystals 6–8, on the order of hundreds of nanometers to a few microns; however, currently access to these sources can be limiting. An alternative approach leverages the strong interaction of electrons with matter and their relatively lower damage to the sample 9. Electron diffraction, which enables the collection of high-resolution diffraction data from much smaller crystals than those needed for conventional X-ray crystallography, is one such approach. The cryo-electron microscopy technique of microcrystal electron diffraction (MicroED) was initially developed for studying protein structures from very small microcrystals 10,11. Since then, MicroED has been used to solve the structures of various soluble proteins, membrane proteins, and peptides 12–14.
The preparation of samples is crucial for obtaining high-resolution MicroED structures. To achieve high-quality diffraction data, it is essential to maintain the integrity of the crystal lattice while reducing the sample thickness, as the ideal thickness of a sample is only approximately 250 nanometers thick 15. While there are several methods available to fragment larger crystals 16 or attempt to grow small crystals, they often involve harsh treatment of the crystals or multiple rounds of sample optimization. Cryo-focused ion beam (cryo-FIB) milling has emerged as an effective technique for preparing thin lamella from biological samples as it minimizes structural damage and preserves sample hydration 17,18. Cryo-FIB milling has been successfully used for various sample types, including protein crystals and other biological materials 19,20. The combination of cryo-FIB milling and MicroED ensures the preservation of native structures while providing high-resolution data for structure determination.
Recent studies have shown that MicroED and cryo-FIB milling are highly effective techniques for determining the structure of other biomolecular systems. For example, Martynowycz et al. (2022) reported the successful determination of the ab initio structure of triclinic lysozyme, a well-characterized model system, using MicroED at 0.87 Å resolution 21. These results demonstrated the potential of MicroED to achieve atomic resolution structures from small protein crystals. Additionally, other studies such as those by Duyvesteyn et al. (2018), Martynowycz et al. (2019), Zhou et al. (2019), Polovinkin et al. (2020), and Li et al. (2018) have utilized cryo-FIB milling to prepare thin lamellae of protein microcrystals, which were then used for high-resolution structure determination by MicroED. Together with the present study, these works highlight the versatility and broad applicability of MicroED and cryo-FIB milling for determining high-resolution structures of a variety of biomolecules.
In this study, we utilized MicroED to obtain high-resolution electron diffraction data from DNA crystals, focusing on the d(CGCGCG)2 DNA duplex as our model target. This Z-DNA hexameric duplex has been extensively studied in crystallography for many years, and has been shown to diffract to very high resolution 22–35. By combining cryo-focused ion beam (cryo-FIB) milling with MicroED, we obtained a 1.10 Å resolution structure from three crystalline lamellae of this DNA duplex, demonstrating the potential of cryo-FIB milling and MicroED for nucleic acid structure determination.
RESULTS
Crystals of the d(CGCGCG)2 duplex were grown using hanging drop crystallization with previously reported conditions 31, yielding large crystals measuring hundreds of microns in approximately one week. Electron diffraction necessitates a sample thickness of less than a micron for beam penetration, and around a few hundred nanometers for optimal data collection 15. Cryo-FIB milling has been established as a powerful technique for thinning crystals that are too thick for MicroED analysis 36–40. FIB milling employs an ion beam to remove material layers without significantly damaging the unmilled sample regions. The thin crystalline lamella produced by milling can generate high-resolution electron diffraction 15,21,41. Consequently, we utilized cryo-FIB milling to thin the DNA crystals for MicroED analysis.
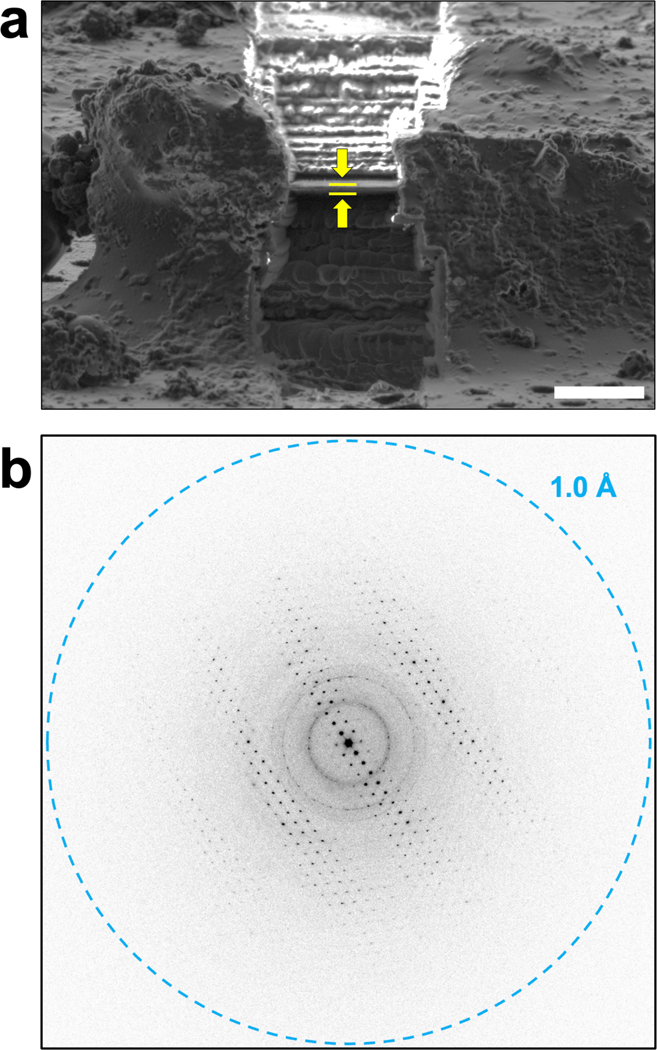
To expedite the process of cryo-FIB milling, the large DNA crystals were initially broken into smaller fragments by pipetting them vigorously within the drops 16. These fragments were promptly placed on holey carbon EM grids and vitrified using conventional MicroED sample preparation techniques 42. Upon examination of the grids in the cryo-FIB/SEM, several crystal fragments were identified on the grid. Out of these fragments, three pieces, with dimensions of approximately 30 × 15 × 10 μm, situated in the center of their respective grid squares, were selected for FIB milling, which was carried out using standard cryo-FIB milling procedures 43. The resulting thin lamella had a thickness of about 250 nm (Fig. 1A).
Figure 1. Cryo-FIB milling and MicroED data collection on d(CGCGCG)2 duplex crystals.
a) Cryo-FIB milling was used on fragmented d(CGCGCG)2 crystals to prepare thin crystalline lamella of approximately 250 nm for MicroED data collection. The location of the crystalline lamella is indicated by the yellow arrows and lines, and the scale bar represents 5 μm. b) The thin DNA crystalline lamella produced by cryo-FIB milling produced high-resolution diffraction data when MicroED was performed, and continuous rotation data sets were collected from these lamellae.
FIB milled crystals were used for energy filtered MicroED data collection with a Falcon4 direct electron detector, which produced high-resolution diffraction patterns for each lamella, indicating that the diffraction power of the DNA crystals was preserved following the cryo-FIB milling process. The datasets from three lamellae were merged, resulting in a 1.10 Å resolution dataset with 98.9% completeness. The space group was P 21 21 21 with unit cell parameters of 18.28 Å, 32.00 Å, and 41.67 Å (Table 1), in good agreement with previous studies on d(CGCGCG)2 crystals found in the PDB 22–35. Molecular replacement was used to phase the data, employing a search model of 3p4j, followed by refinement of the structure using electron scattering factors. The final structure of the d(CGCGCG)2 DNA duplex, including a spermine molecule and 33 water molecules, was determined at 1.10 Å with an overall Rwork/Rfree of 20.98 / 22.57% (Fig. 2, Table 1). Consistent with similar structures, the hexamer duplexes were found to stack along the c-axis, while the length of the a-axis corresponded to the helix’s diameter 31. Notably, the structure exhibited an alternative conformation of the phosphate backbone of the third residue of chain A (Fig. 2d), suggesting some flexibility at this point in an otherwise rigid structure. The MicroED structure of the d(CGCGCG)2 was found to be in good agreement with previous studies on this target, and the resulting potential maps were of high quality.
Table 1.
Data collection and refinement statistics
| Data Collection | |
| Excitation voltage | 300kV |
| Electron Source | Field Emission Gun |
| Wavelength (Å) | 0.019687 |
| Total dose per crystal | ~1 e−/Å2 |
| Frame rate | 2 fps |
| Rotation rate | 0.15 °/s |
| Data Processing | |
| Number of crystals | 3 |
| Space group | P212121 |
| Unit cell dimensions | |
| a, b, c (Å) | 18.28 32.00 41.67 |
| α=β=γ (°) | 90.000 90.000 90.000 |
| Resolution (Å) | 1.10 |
| Total reflections | 95,258 |
| Total Unique Reflections | 10,374 |
| Rmerge (%) | 0.258 (0.794)a |
| CC1/2 | 0.977 (0.810)a |
| Multiplicity | 9.2 (9.3)a |
| Completeness (%) | 98.9 (99.6)a |
| Mean (I/σ(I)) | 5.3 (1.9)a |
| Data Refinement | |
| Rwork/Rfree (%) | 20.98/22.57 |
| RMSD Bonds (Å) | 0.020 |
| RMSD Angles (°) | 1.759 |
Values for highest resolution shell of 1.13 Å – 1.10 Å
Figure 2. MicroED structure of the d(CGCGCG)2 duplex.
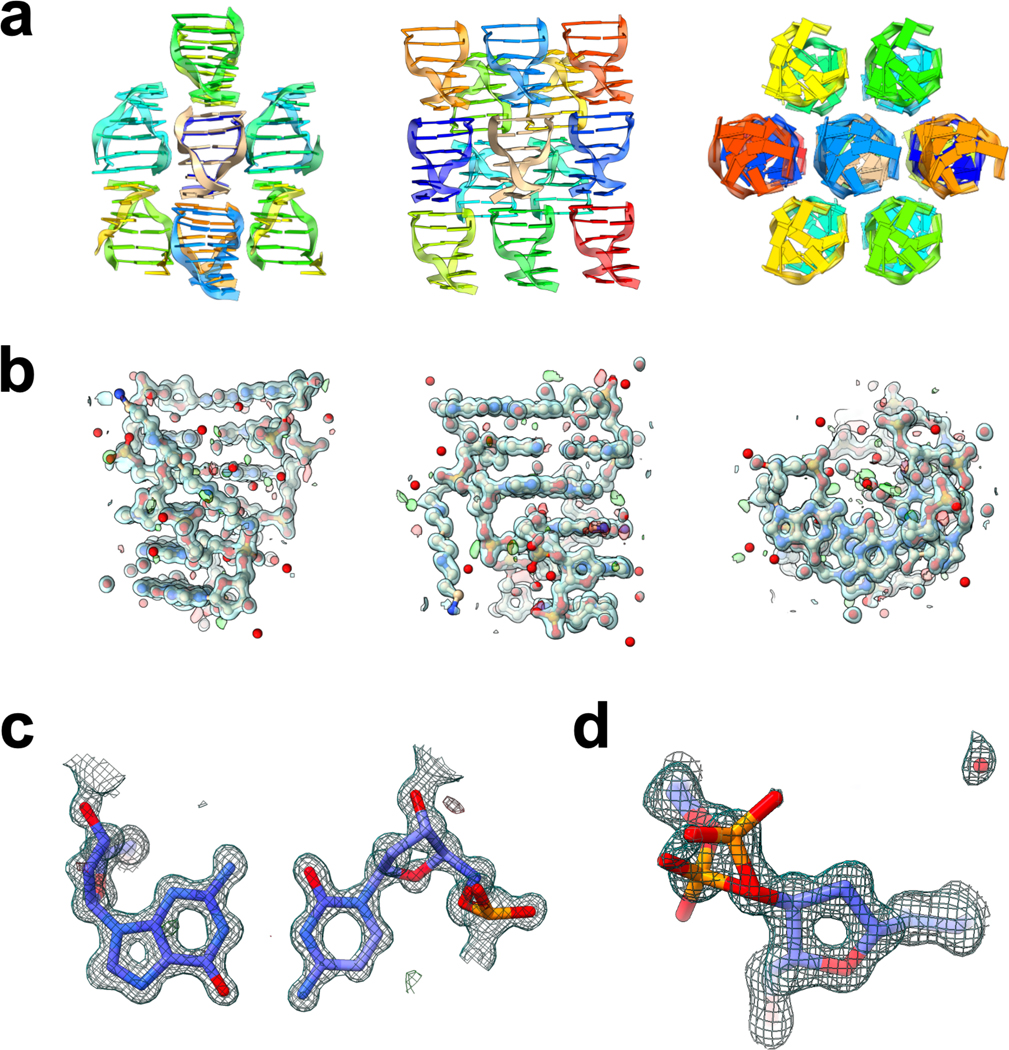
a) The data collected from the cryo-FIB milled crystals yielded a 1.10 Å structure of the Z-DNA hexamer duplex, with packing consistent with previous studies. b) Each asymmetric unit consists of a helix of two dCGCGCG hexamers, and density is seen for each residue of the hexamers, as well as the spermine present in the structure, and several water molecules. c) the quality of the map can be clearly seen when viewing the interacting base pairs between the two hexamers. Here a representative view of residue 4 from chain A with residue 3 from chain B is shown. d) In the structure determined here, an alternative conformation of the phosphate backbone at residue 3 in chain A was modeled, where the occupancy of the two conformations were refined to values of 53% and 47%. Structures shown in (a) and (b) are viewed along the a, b, and c axes (left, middle, right, respectively). 2Fo – Fc maps shown in (b), (c), and (d) are contoured at 1.5σ (blue), and Fo – Fc maps are contoured at ± 3σ (green and red for positive and negative, respectively).
DISCUSSION
The capability of MicroED for studying biomolecular structures from smaller crystals than those used for conventional X-ray crystallography makes the method a valuable tool for structural biology. This study highlights the applicability of MicroED to nucleic acids, which can be prepared using cryo-FIB milling. While the success of this approach in determining the structure of DNA is not surprising, it serves as a compelling demonstration of the quality of structures that can be obtained through electron diffraction data. Moreover, it expands the range of biological molecules that can be studied using MicroED, which has significant implications for future research in this field.
Compared to X-ray diffraction, electron diffraction is highly sensitive to charge effects (Wu and Spence, 2003; Yonekura et al., 2015; Yonekura and Maki-Yonekura, 2016; Yonekura et al., 2018), and nucleic acids, with their highly charged phosphate backbone, are especially affected. Nevertheless, this study demonstrates that, even with neutral electron scattering factors, the potential maps and R-factors of nucleic acids are comparable to those of protein and peptide structures determined by MicroED. However, the potential for improving the quality of nucleic acid structures using charged electron scattering factors is a potential avenue for future research as more nucleic acid structures are determined by MicroED.
The successful determination of the structure of the d(CGCGCG)2 DNA duplex using MicroED in this study suggests that MicroED can be a valuable tool for the structural analysis of nucleic acids. Further investigations on other DNA and RNA samples using MicroED could potentially yield important insights into the structural characteristics of these biomolecules. Additionally, improvements in sample preparation, data acquisition, and data processing techniques may enhance the resolution and quality of nucleic acid structures obtained using MicroED. Overall, this study highlights the potential of MicroED for nucleic acid research and encourages further exploration in this area.
STAR METHODS
RESOURCE AVAILABILITY
LEAD CONTACT
Brent L. Nannenga (Brent.Nannenga@asu.edu)
Further information and requests for resources and reagents may be directed to and will be fulfilled by the Lead Contact, Brent L. Nannenga (Brent.Nannenga@asu.edu).
MATERIALS AVAILABILITY
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
DATA AND CODE AVAILABILITY
This study did not generate new software. Details of deposited coordinates and density are provided in the Key Resources Table. Coordinates and structure factors were deposited in the Protein Data Bank (PDB) under the accession code 8skw. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Sodium chloride | Hampton Research | Cat# HR2–637 |
| (+/−)-2-Methyl-2,4-pentanediol | Hampton Research | Cat# HR2-627 |
| sodium cacodylate, pH 7.0 | Hampton Research | Cat# HR2-939-20 |
| Spermine tetrachloride | Sigma | Cat# 85605 |
| Deposited Data | ||
| d(CGCGCG)2 duplex | This Study | PDB: 8skw |
| Oligonucleotides | ||
| dCGCGCG DNA | Genscript | N/A |
| Software and algorithms | ||
| XDS | 44 | https://xds.mr.mpg.de/ |
| Phaser | 45 | https://www.phenix-online.org/ |
| COOT | 46 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | 47 | https://www.phenix-online.org/ |
| Other | ||
| QUANTIFOIL Holey Carbon R 2/4, 200 Mesh, Copper | Electron Microscopy Sciences | Cat# Q2100CR-4 |
METHOD DETAILS
DNA crystallization and grid preparation
Crystals of the Z-DNA duplex were grown following previously reported procedures 31. dCGCGCG DNA was synthesized by Genscript, and a 1.5 mM solution of DNA in water was annealed at a temperature of 65 °C for 12 minutes. Following annealing, several hanging drop crystallization experiments were set up using identical conditions. 3 μL of annealed DNA was mixed with 3 μL of precipitant solution that consisted of 80 mM NaCl, 10% 2-methyl-2,4-pentanediol (MPD), 12 mM spermine tetrachloride, and 40 mM sodium cacodylate, pH 7.0. Each drop was equilibrated over 1 mL of 35% MPD at room temperature. Crystals began to appear after a few days and grew to full size in 1 to 2 weeks. For MicroED sample preparation, crystals from two drops were combined, and an equal volume reservoir solution was added to the combined drops. Crystals were fragmented by vigorous pipetting as they were monitored using a light microscope until most of the large crystals were broken down into smaller fragments. The solution containing the fragmented crystals (3 μL) was applied to the carbon side of glow discharged holey carbon grid (quantifoil 2/4, 200 mesh). Using a Leica GP2 plunge freezer, the grids were each blotted for 10 to 20 seconds prior to plunge freezing in liquid ethane.
Cryo-FIB milling
Cryo-FIB milling was performed using an Aquilos dual beam FIB/SEM (Thermo Fisher), operating at liquid nitrogen temperatures using standard procedures for microcrystals 43. Briefly, vitrified grids were cryo transferred into the instrument. Whole-grid montaging using the scanning electron beam was conducted to locate crystals near the center of the grid and not near a grid bar. Each target crystal was identified using the electron beam and brought to the eucentric position. Milling was conducted at 18 degrees in steps down to a final thickness of 200 – 300 nm. The initial, rough milling, or trenching, was conducted using a 1 nA beam current to create initial lamellae of 10um thickness. Intermediate milling was conducted by lowering the beam current as the thinned lamellae became thinner, with intermediate currents of 0.3 nA used until 3 μm, 0.1 nA until 1 μm. The 1 μm lamellae were then polished to final thicknesses using a 50 pA beam. The entire milling process for each crystal took approximately 1 hour, with the final polishing step taking about 10 minutes. All milling was conducted at an acceleration voltage of 30 kV, and all SEM images were acquired using a beam current of 6.1 pA and 5 kV acceleration voltage. Grids were stored in liquid nitrogen prior to future experiments.
MicroED data collection
Following cryo-FIB milling, the grid containing the crystalline lamellae was loaded into a Titan Krios equipped with a Falcon4 direct electron detector behind a selectris energy filter operating at an acceleration voltage of 300 kV. The location of the lamella were identified at low magnification, and following alignment and setting the eucentric height, continuous rotation MicroED data were collected from each lamella 11. The data were collected with a rotation rate of 0.15 °/s and a camera frame rate of 0.5 frames per second, which lead to each frame in the data set spanning 0.075°. Each dataset took approximately 10 minutes to collect, was collected at a nominal camera length of 1200 mm, and all images were saved in MRC format and binned by 2.
Data processing and structure determination
MicroED data collected from each of the 3 DNA crystals were indexed, integrated, scaled and merged using XDS 44. Phaser 45 was used for molecular replacement with PDB ID: 3p4j 31 used as a search model. The structure was refined using phenix.refine in the phenix crystallographic suite 47,48 along with manual refinement using coot 49. The scattering factors used in refinement were the default neutral electron scattering factors implemented in phenix.refine. Structures were visualized and figures generated using ChimeraX 50.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data collection and refinement statistics are reported in Table 1.
Highlights.
Microcrystal electron diffraction (MicroED) was applied to determine structure of DNA
A d(CGCGCG)2 DNA duplex was used as a model target and its 1.1 Å structure was determined
Cryo-FIB milling was employed to create thin crystalline lamella for MicroED
This study demonstrates the applicability of MicroED for nucleic acid crystallography
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health P41GM136508 and R01GM124152, and the Department of Defense grant MCDC-2202-002. The Gonen laboratory is supported by funds from the Howard Hughes Medical Institute.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Subirana JA (2003). DNA discoveries through crystallography. Nature 423, 683-683. 10.1038/423683b. [DOI] [PubMed] [Google Scholar]
- 2.Coimbatore Narayanan B, Westbrook J, Ghosh S, Petrov AI, Sweeney B, Zirbel CL, Leontis NB, and Berman HM (2013). The Nucleic Acid Database: new features and capabilities. Nucleic Acids Res 42, D114–D122. 10.1093/nar/gkt980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westhof E. (2015). Twenty years of RNA crystallography. Rna 21, 486–487. 10.1261/rna.049726.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paukstelis PJ, and Seeman NC (2016). 3D DNA Crystals and Nanotechnology. Crystals 6, 97. [Google Scholar]
- 5.Simmons CR., Zhang F, Birktoft JJ, Qi X, Han D, Liu Y, Sha R, Abdallah HO, Hernandez C, Ohayon YP, et al. (2016). Construction and Structure Determination of a Three-Dimensional DNA Crystal. Journal of the American Chemical Society 138, 10047–10054. 10.1021/jacs.6b06508. [DOI] [PubMed] [Google Scholar]
- 6.Crawshaw AD, Beale EV, Warren AJ, Stallwood A, Duller G, Trincao J, and Evans G. (2021). A Sample Preparation Pipeline for Microcrystals at the VMXm Beamline. J Vis Exp. 10.3791/62306. [DOI] [PubMed] [Google Scholar]
- 7.Zatsepin NA, Li C, Colasurd P, and Nannenga BL (2019). The complementarity of serial femtosecond crystallography and MicroED for structure determination from microcrystals. Current Opinion in Structural Biology. 10.1016/j.sbi.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spence JCH (2018). X-ray lasers for structure and dynamics in biology. IUCrJ 5, 236–237. 10.1107/S2052252518005365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson R. (1995). The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys 28, 171–193. [DOI] [PubMed] [Google Scholar]
- 10.Shi D, Nannenga BL, Iadanza MG, and Gonen T. (2013). Three-dimensional electron crystallography of protein microcrystals. Elife 2, e01345. 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nannenga BL, Shi D, Leslie AG, and Gonen T. (2014). High-resolution structure determination by continuous-rotation data collection in MicroED. Nat Methods 11, 927–930. 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nannenga BL, and Gonen T. (2019). The cryo-EM method microcrystal electron diffraction (MicroED). Nat Methods 16, 369–379. 10.1038/s41592-019-0395-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark LJ, Bu G, Nannenga BL, and Gonen T. (2021). MicroED for the study of protein–ligand interactions and the potential for drug discovery. Nature Reviews Chemistry 5, 853–858. 10.1038/s41570-021-00332-y. [DOI] [PubMed] [Google Scholar]
- 14.Clabbers MTB, and Xu H. (2021). Macromolecular crystallography using microcrystal electron diffraction. Acta Crystallogr D Struct Biol 77, 313–324. 10.1107/S2059798320016368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martynowycz MW, Clabbers MTB, Unge J, Hattne J, and Gonen T. (2021). Benchmarking the ideal sample thickness in cryo-EM. Proceedings of the National Academy of Sciences 118, e2108884118. 10.1073/pnas.2108884118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Cruz MJ, Hattne J, Shi D, Seidler P, Rodriguez J, Reyes FE, Sawaya MR, Cascio D, Weiss SC, Kim SK, et al. (2017). Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED. Nat Methods 14, 399–402. 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marko M, Hsieh C, Schalek R, Frank J, and Mannella C. (2007). Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods 4, 215–217. 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 18.Rigort A, Bauerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, and Plitzko JM (2012). Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A 109, 4449–4454. 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duyvesteyn HME, Kotecha A, Ginn HM, Hecksel CW, Beale EV, de Haas F, Evans G, Zhang P, Chiu W, and Stuart DI (2018). Machining protein microcrystals for structure determination by electron diffraction. Proc Natl Acad Sci U S A 115, 9569–9573. 10.1073/pnas.1809978115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam V, and Villa E. (2021). Practical Approaches for Cryo-FIB Milling and Applications for Cellular Cryo-Electron Tomography. In cryoEM: Methods and Protocols T. Gonen, and Nannenga BL, eds. (Springer US; ), pp. 49–82. 10.1007/978-1-0716-0966-8_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martynowycz MW, Clabbers MTB, Hattne J, and Gonen T. (2022). Ab initio phasing macromolecular structures using electron-counted MicroED data. Nat Methods 19, 724–729. 10.1038/s41592-022-01485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang AH-J, Quigley GJ, Kolpak FJ, Crawford JL, Van Boom JH, Van Der Marel G, and Rich A. (1979). Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature 282, 680–686. [DOI] [PubMed] [Google Scholar]
- 23.Gessner R, Frederick CA, Quigley G, Rich A, and Wang A. (1989). The molecular structure of the left-handed Z-DNA double helix at 1.0-Å atomic resolution: Geometry, conformation, and ionic interactions of d (CGCGCG). Journal of Biological chemistry 264, 7921–7935. [DOI] [PubMed] [Google Scholar]
- 24.Ohishi H, Kunisawa S, van der Marel G, van Boom JH, Rich A, Wang AH-J, Tomita K. i., and Hakoshima T. (1991). Interaction between the left-handed Z-DNA and polyamine The crystal structure of the d (CG) 3 and N-(2-aminoethyl)-1, 4-diamino-butane complex. FEBS letters 284, 238–244. [DOI] [PubMed] [Google Scholar]
- 25.Ohishi H, Nakanishi I, Inubushi K, Van Der Marel G, Van Boom JH, Rich A, Wang AH, Hakoshima T, and Tomita K. i. (1996). Interaction between the left-handed Z-DNA and polyamine-2: The crystal structure of the d (CG) 3 and spermidine complex. FEBS letters 391, 153–156. [DOI] [PubMed] [Google Scholar]
- 26.Ohishi H, Terasoma N, Nakanishi I, Van Der Marel G, Van Boom JH, Rich A, Wang AH-J, Hakoshima T, and Tomita K. i. (1996). Interaction between left-handed Z-DNA and polyamine—3 The crystal structure of the d (CG) 3 and thermospermine complex. FEBS letters 398, 291–296. [DOI] [PubMed] [Google Scholar]
- 27.Dauter Z, and Adamiak DA (2001). Anomalous signal of phosphorus used for phasing DNA oligomer: importance of data redundancy. Acta Crystallographica Section D: Biological Crystallography 57, 990–995. [DOI] [PubMed] [Google Scholar]
- 28.Ohishi H, Suzuki K, Ohtsuchi M, Hakoshima T, and Rich A. (2002). The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1, 2-diamine (polyamines) binding to the minor groove of d (CGCGCG) 2, hexamer at room temperature. FEBS letters 523, 29–34. [DOI] [PubMed] [Google Scholar]
- 29.Ohishi H, Tozuka Y, Da-Yang Z, Ishida T, and Nakatani K. (2007). The rare crystallographic structure of d (CGCGCG) 2: the natural spermidine molecule bound to the minor groove of left-handed Z-DNA d (CGCGCG) 2 at 10 C. Biochemical and biophysical research communications 358, 24–28. [DOI] [PubMed] [Google Scholar]
- 30.Ohishi H, Odoko M, Grzeskowiak K, Hiyama Y, Tsukamoto K, Maezaki N, Ishida T, Tanaka T, Okabe N, and Fukuyama K. (2008). Polyamines stabilize left-handed Z-DNA: Using X-ray crystallographic analysis, we have found a new type of polyamine (PA) that stabilizes left-handed Z-DNA. Biochemical and biophysical research communications 366, 275–280. [DOI] [PubMed] [Google Scholar]
- 31.Brzezinski K, Brzuszkiewicz A, Dauter M, Kubicki M, Jaskolski M, and Dauter Z. (2011). High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 ņ. Nucleic Acids Res 39, 6238–6248. 10.1093/nar/gkr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egli M, Williams LD, Gao Q, and Rich A. (1991). Structure of the pure-spermine form of Z-DNA (magnesium free) at 1-. ANG. resolution. Biochemistry 30, 11388–11402. [DOI] [PubMed] [Google Scholar]
- 33.Bancroft D, Williams LD, Rich A, and Egli M. (1994). The low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry 33, 1073–1086. [DOI] [PubMed] [Google Scholar]
- 34.Tereshko V, Wilds CJ, Minasov G, Prakash TP, Maier MA, Howard A, Wawrzak Z, Manoharan M, and Egli M. (2001). Detection of alkali metal ions in DNA crystals using state-of-the-art X-ray diffraction experiments. Nucleic Acids Res 29, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatake T, Tanaka I, Umino H, Arai S, and Niimura N. (2005). The hydration structure of a Z-DNA hexameric duplex determined by a neutron diffraction technique. Acta Crystallographica Section D: Biological Crystallography 61, 1088–1098. [DOI] [PubMed] [Google Scholar]
- 36.Duyvesteyn HME, Kotecha A, Ginn HM, Hecksel CW, Beale EV, de Haas F, Evans G, Zhang P, Chiu W, and Stuart DI (2018). Machining protein microcrystals for structure determination by electron diffraction. Proceedings of the National Academy of Sciences 115, 9569–9573. doi: 10.1073/pnas.1809978115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polovinkin V, Khakurel K, Babiak M, Angelov B, Schneider B, Dohnalek J, Andreasson J, and Hajdu J. (2020). Demonstration of electron diffraction from membrane protein crystals grown in a lipidic mesophase after lamella preparation by focused ion beam milling at cryogenic temperatures. J Appl Crystallogr 53, 1416–1424. 10.1107/S1600576720013096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Zhang S, Zhang J, and Sun F. (2018). In situ protein micro-crystal fabrication by cryo-FIB for electron diffraction. Biophysics reports 4, 339–347. 10.1007/s41048-0180075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martynowycz MW, Zhao W, Hattne J, Jensen GJ, and Gonen T. (2019). Collection of Continuous Rotation MicroED Data from Ion Beam-Milled Crystals of Any Size. Structure 27, 545–548.e542. 10.1016/j.str.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H, Luo Z, and Li X. (2019). Using focus ion beam to prepare crystal lamella for electron diffraction. Journal of Structural Biology 205, 59–64. 10.1016/j.jsb.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Martynowycz MW, Shiriaeva A, Ge X, Hattne J, Nannenga BL, Cherezov V, and Gonen T. (2021). MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP. Proceedings of the National Academy of Sciences 118, e2106041118. 10.1073/pnas.2106041118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bu G, and Nannenga BL (2021). MicroED Sample Preparation and Data Collection For Protein Crystals. Methods Mol Biol 2215, 287–297. 10.1007/978-1-0716-0966-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martynowycz MW, and Gonen T. (2021). Protocol for the use of focused ion-beam milling to prepare crystalline lamellae for microcrystal electron diffraction (MicroED). STAR Protoc 2, 100686. 10.1016/j.xpro.2021.100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new software. Details of deposited coordinates and density are provided in the Key Resources Table. Coordinates and structure factors were deposited in the Protein Data Bank (PDB) under the accession code 8skw. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Sodium chloride | Hampton Research | Cat# HR2–637 |
| (+/−)-2-Methyl-2,4-pentanediol | Hampton Research | Cat# HR2-627 |
| sodium cacodylate, pH 7.0 | Hampton Research | Cat# HR2-939-20 |
| Spermine tetrachloride | Sigma | Cat# 85605 |
| Deposited Data | ||
| d(CGCGCG)2 duplex | This Study | PDB: 8skw |
| Oligonucleotides | ||
| dCGCGCG DNA | Genscript | N/A |
| Software and algorithms | ||
| XDS | 44 | https://xds.mr.mpg.de/ |
| Phaser | 45 | https://www.phenix-online.org/ |
| COOT | 46 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | 47 | https://www.phenix-online.org/ |
| Other | ||
| QUANTIFOIL Holey Carbon R 2/4, 200 Mesh, Copper | Electron Microscopy Sciences | Cat# Q2100CR-4 |