Abstract
Granulocytic Ehrlichia was isolated from canine blood obtained from animals challenged with field-collected Ixodes scapularis and propagated in HL60 cells. PCR primers specific for the 16S ribosomal DNA (rDNA) of the Ehrlichia genogroup comprising E. equi, E. phagocytophila, and the agent of human granulocytic ehrlichiosis (HGE) amplified DNA from extracts of these cells. Sequence analysis of this amplified DNA revealed that it is identical to the 16S rDNA sequence of the HGE agent. A genomic library was constructed with DNA from granulocytic Ehrlichia and screened with pooled sera from tick-challenged, granulocytic Ehrlichia-infected dogs. Several clones were isolated and sequenced. Three complete genes encoding proteins with apparent molecular masses of 100, 130, and 160 kDa were found. The recombinant proteins reacted with convalescent-phase sera from dogs and human patients recovering from HGE. This approach will be useful for identifying candidate diagnostic and vaccine antigens for granulocytic ehrlichiosis and aid in the classification of genogroup members.
Members of the genus Ehrlichia include species which have a tropism for mononuclear phagocytes (E. canis, E. chaffeensis, E. muris, E. sennetsu, and E. risticii) (33) and those which infect granulocytes (E. ewingii [33], E. phagocytophila [10, 33], E. equi [14, 26], and the recently discovered agent of human granulocytic ehrlichiosis [HGE] [3, 7]). Disease caused by granulocytic Ehrlichia (GE) is manifested by fever, lethargy, thrombocytopenia, and death, and many species from diverse geographical locations have shown evidence of natural infection, including horses (25, 27, 29, 38), dogs (24, 34, 35), small mammals (39, 40), and humans (4, 15).
The similar host range and near identity of the 16S rRNA genes of E. phagocytophila, E. equi, and the HGE agent (7) have raised the possibility that these organisms represent a single species (2, 28). In addition, Dumler et al. (9) have shown that they share significant antigenicity by immunofluorescence and immunoblot assays. Objective methods for species classification, e.g., molecular genetic analysis, have not been readily available, primarily because of an inability to culture these ehrlichiae in vitro. However, we (reference 43 and unpublished data) and others (13) have recently demonstrated successful cultivation of GE isolates from dogs and humans, respectively.
In this paper, we describe the use of purified GE obtained from in vitro culture of infected HL60 cells, a promyelocytic human cell line, to generate a genomic DNA library for expression screening with sera obtained from dogs experimentally infected with GE. The screening resulted in the isolation of recombinant clones containing complete genes encoding three putative proteins of GE, GE 160, GE 130, and GE 100 (named for apparent molecular mass in kilodaltons). One of these proteins, the 100-kDa protein, is similar in both glutamic acid content and repeated amino acid structure to an immunodominant 120-kDa E. chaffeensis protein (45). Both the 100- and 130-kDa granulocytic Ehrlichia proteins share some amino acid sequence homology to the 120-kDa E. chaffeensis protein.
MATERIALS AND METHODS
Isolation of GE in cell culture.
The GE agent (strain USG3) was isolated as described previously (43). Briefly, blood from a dog experimentally infected by adult Ixodes scapularis ticks collected from both Westchester County, N.Y., and Montgomery County, Pa., was inoculated into a suspension culture of the human promyelocytic cell line HL60 (ATCC CCL 240). The cells were cultured in RPMI 1640 medium supplemented with 20% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and 0.1 mM nonessential amino acids and maintained at 37°C and 6% CO2 in a humidified chamber. The fetal bovine serum was reduced to 10% once the culture was established. Ehrlichial infection was indicated by lysis of the HL60 cells. Cultures of USG3 (passages 7 to 21) were grown for 5 days prior to GE purification.
Purification of GE.
USG3 cultures at approximately 80% cell lysis (monitored microscopically) were centrifuged at 840 × g for 15 min at 4°C to remove host HL60 cell debris. The supernatant was filtered through a Poretics (Livermore, Calif.) 5-μm-pore-diameter polycarbonate membrane, 47 mm in diameter, followed by a Poretics 3-μm-pore-diameter filter under negative pressure. The USG3 filtrate was centrifuged at 9460 × g in a Sorvall centrifuge for 30 min at 4°C. Following centrifugation, the GE pellet was resuspended in 5 ml of 25 mM Tris (pH 8.0)–10 mM MgCl2–0.9% NaCl. DNase I (Life Technologies, Gaithersburg, Md.) was added to a final concentration of 9 μg per ml, and the solution was incubated for 15 min at 37°C. Following incubation, the DNase was inactivated by the addition of 0.5 ml of 0.5 M EDTA, and the GE was pelleted at 14,000 × g in a Sorvall centrifuge for 30 min at 4°C.
PCR amplification and cloning of GE 16S ribosomal DNA.
Universal eubacterial primers for the 16S rRNA gene (41) were modified to include restriction enzyme recognition sites as follows: forward primer, 5′ CTGCAGGTTTGATCCTGG 3′ (PstI site); reverse primer, 5′ GGATCCTACCTTGTTACGACTT 3′ (BamHI site). These primers (0.5 μM) were added to a 100-μl reaction mixture containing 1× PCR buffer II (Perkin-Elmer Corp.); 1.5 mM MgCl2 (Perkin-Elmer Corp.); 200 μM (each) dATP, dGTP, dCTP, and dTTP; 2.5 U of Amplitaq DNA polymerase; and 20 μl of USG3 DNA. Amplification was performed under the following conditions: 35 cycles at 94°C for 1 min, 53°C for 1 min, and 72°C for 2 min, followed by a 72°C incubation for 10 min. The amplified 1,500-bp fragment was digested with PstI and BamHI and ligated to pUC19 linearized with the same enzymes. The resulting clone, pUCHGE16S, was sequenced as described below.
Construction of a GE genomic library.
Genomic DNA was isolated from purified GE with the QIAamp Genomic DNA kit (Qiagen, Chatsworth, Calif.) for library preparation (Stratagene, La Jolla, Calif.). The DNA was mechanically sheared to a 4- to 10-kb size range and ligated to EcoRI linkers. Linkered fragments were ligated into the EcoRI site of Lambda Zap II, and the library was amplified in Escherichia coli XL1-Blue MRF′ to a titer of 1010 PFU/ml.
Expression screening of the genomic library.
Bacteriophage were plated with XL1-Blue MRF′ and induced to express protein with 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma, St. Louis, Mo.). Proteins were transferred to nitrocellulose filters, and the filters were washed with Tris-buffered saline (TBS; 25 mM Tris HCl [pH 7.5], 0.5 M NaCl). Washed filters were blocked in TBS containing 0.1% polyoxyethylene 20 cetyl ether (Brij 58) and incubated with a 1:50 dilution of pooled sera (depleted of anti-E. coli antibodies) taken from four dogs experimentally infected by exposure to field-collected adult I. scapularis ticks (8). The filters were washed and incubated with rabbit anti-dog horseradish peroxidase-conjugated immunoglobulin (Ig) antibody, rewashed, and developed with 4-chloronaphthol. Positive plaques were isolated, replated, and screened again. Plasmid DNA containing the putative recombinant clones was obtained by plasmid rescue (Stratagene).
DNA sequencing and sequence analysis.
DNA sequencing of recombinant clones was performed by the primer walking method and with an ABI 373A DNA sequencer (ACGT, Northbrook, Ill.; Lark Technologies, Houston, Tex.; and Sequegen, Shrewsbury, Mass.). Sequences were analyzed by the MacVector (Oxford Molecular Group) sequence analysis program, version 6.0. The BLAST algorithm, D version 1.4 (19, 20), was used to search for homologous nucleic acid and protein sequences available on the National Center for Biotechnology Information (NCBI) server.
PCR analysis of USG3 and HL60 DNA.
PCR primer sets were designed based on the sequences of each of the three GE clones and are as follows: S2 (forward, 5′-GCGTCTCCAGAACCAG-3′; reverse, 5′-CCTATATAGCTTACCG-3′), S7 (forward, 5′-GATGTTGCTTCGGGTATGC-3′; reverse, 5′-CAGAGATTACTTCTTTTTGCGG-3′), and S22 (forward, 5′-CACGCCTTCTTCTAC-3′; reverse, 5′-CTCTGTTGCTATAGGGGC-3′). Each 50-μl reaction mixture contained 0.5 μM each primer, 1× PCR Supermix (Life Technologies, Gaithersburg, Md.) and either 100 ng of USG3 DNA, 100 ng of HL60 DNA, or 200 ng of plasmid DNA. PCR amplification was performed under the following conditions: 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. After 30 cycles, a single 10-min extension at 72°C was done. PCR products were analyzed on 4% Nusieve 3:1 agarose gels (FMC Bioproducts, Rockland, Me.).
Western blot analysis.
Individual recombinant plasmid-containing cultures were induced to express protein with 5 mM IPTG. Bacterial cells were pelleted by centrifugation and resuspended in 5× Laemmli buffer (12% glycerol, 0.2 M Tris-HCl [pH 6.8], 5% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol) at 200 μl per optical density unit of culture. Samples were boiled for 5 min, and 10 μl of each was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (for dog sera) or NuPage (Novex, San Diego, Calif.) electrophoresis (for human sera). Proteins were transferred to nitrocellulose filters, the filters were blocked in TBS-Brij 58, and the blots were probed with either the pooled dog sera referenced above or 1:1,000 dilutions of human sera. The sera used in this study were two convalescent-phase serum samples from patients (no. 2 and 3 from New York, kindly provided by M. Aguero-Rosenfeld) and one sample from an individual in Wisconsin who was part of a seroprevalence study (no. 1, kindly provided by J. Bakken). Blots were washed and incubated with biotin-labeled goat anti-dog IgG (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) followed by peroxidase-labeled streptavidin (Kirkegaard & Perry Laboratories, Inc.) or horseradish peroxidase-conjugated antihuman IgG (Bio-Rad, Hercules, Calif.). After several additional washes, the dog serum blots were developed with 4-chloronaphthol (Bio-Rad), and the human serum blots were detected by enhanced chemiluminescence (Pierce, Rockford, Ill.) and viewed by autoradiography.
Nucleotide sequence accession number.
The nucleotide sequences of the GE genes described here have been assigned the following GenBank accession numbers: ank (GE 160), AF020521; rea (GE 130), AF020522; and gra (GE 100), AF020523.
RESULTS
Cloning and sequencing of the USG3 16S rRNA gene.
Sources of GE used to generate a cell culture isolate have included blood from either infected humans (13) or dogs (32, 43). To assess the relatedness of the USG3 isolate to other GE isolates, including the HGE agent, the 16S rRNA gene was amplified by PCR with universal eubacterial primers (41). A 1,500-bp DNA fragment was isolated and cloned into pUC19 (see Materials and Methods), and the insert DNA was sequenced. The USG3 16S rRNA gene sequence was found to be identical to the GenBank sequence of the HGE agent (accession no. U02521). The sequences of other 16S ribosomal DNA PCR fragments generated with USG3 DNA as the template were also identical to the HGE agent sequence (reference 43 and data not shown).
Isolation of recombinant clones identified with GE-positive dog sera.
Using pooled sera from adult I. scapularis-challenged, GE-infected dogs, positive clones were identified and purified as single plaques. pBluescript plasmids were rescued according to the Stratagene protocol, and DNA was purified with Qiagen plasmid purification kits. A number of restriction digests were performed with each clone to assess their relatedness (data not shown). Based on these digests, restriction maps were generated which showed that all of the clones represented just three different GE genes. One clone from each of the three gene groups (clones S2, S7, and S22) was chosen for further analysis.
Analysis of proteins encoded by GE clones.
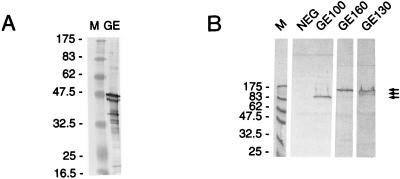
Because recombinant clones were selected based on immunoreactivity, bacterial lysates of each clone were analyzed by SDS-PAGE and Western blotting to identify the immunoreactive proteins. The blots were probed with the same pooled canine sera used to screen the library. Figure 1A shows the reactivity of the antisera against purified USG3. Several immunodominant proteins with sizes of 36, 43, and 45 kDa were observed, and other less-immunoreactive proteins of various molecular masses are shown. Figure 1B shows the Western blot of the library clones, S2, S7, and S22. A single high-molecular-mass protein (indicated by arrows) was detected by the sera for each isolated clone. These proteins were not expressed in the absence of IPTG induction and were not detected with preimmune dog sera (data not shown). The approximate molecular masses of each protein are as follows: 160 (S2), 100 (S7), and 130 (S22) kDa. These proteins are not the major immunodominant antigens of the purified cell culture isolate USG3, two of which migrate at about 43 to 45 kDa (Fig. 1A), but they could be present among the higher-molecular-mass immunoreactive proteins.
FIG. 1.
Expression of GE proteins by Western blotting. (A) Purified USG3 disrupted in SDS (lane GE). (B) Individual recombinant clones containing the genes coding for GE 100, GE 160, GE 130, and a negative control (NEG [no insert]) were grown and incubated with IPTG to induce protein expression as described in Materials and Methods. Samples of each were electrophoresed on SDS-PAGE gels and transferred to nitrocellulose for Western blotting. Blots were probed with convalescent-phase dog sera. Molecular mass markers (M) (in kilodaltons) are shown to the left of each panel.
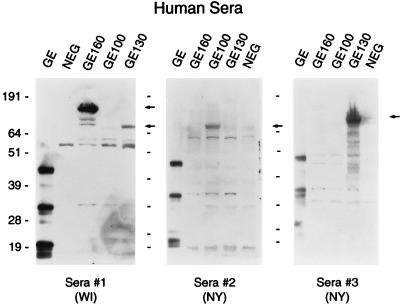
The three GE proteins were also analyzed by Western blotting for their reactivity with human sera. Figure 2 shows three different human serum samples tested against purified USG3 antigen and the three GE recombinant proteins. Each serum sample reacted strongly with one of the three GE proteins: no. 1 with GE 160 and less strongly with GE 130, no. 2 with GE 100, and no. 3 with GE 130. All three serum samples reacted with several proteins in the purified USG3 lanes (GE lanes). A serum sample from a Rhode Island HGE patient recognized two of the three GE proteins, and other HGE patient sera tested also recognized one or two of the proteins (data not shown). None of the samples we have tested to date react strongly with all three recombinant GE proteins. Negative control human sera did not react with the GE proteins on Western blots (data not shown).
FIG. 2.
Expression of GE proteins by Western blotting. Three different human serum samples were used to probe Western blots containing SDS-disrupted USG3 (GE lanes), GE 160, GE 100, and GE 130. A pBluescript library clone containing no insert was used as a negative control (NEG). The origin of the sera is indicated at the bottom of each panel (WI, Wis.; NY, N.Y.). Molecular mass markers (in kilodaltons) are shown to the left of each panel.
It is interesting to note that these human sera do not strongly recognize proteins with sizes of 100, 130, and 160 kDa with USG3 lysates as the antigen source. This may be due to underrepresentation of these proteins in the purified cell culture antigen, either because they are secreted or because they are expressed only after infection of a host species.
DNA sequencing and database analysis of recombinant clones.
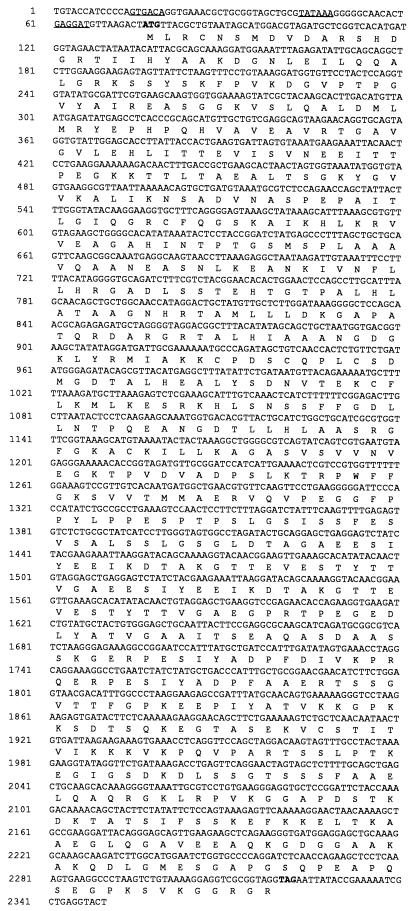
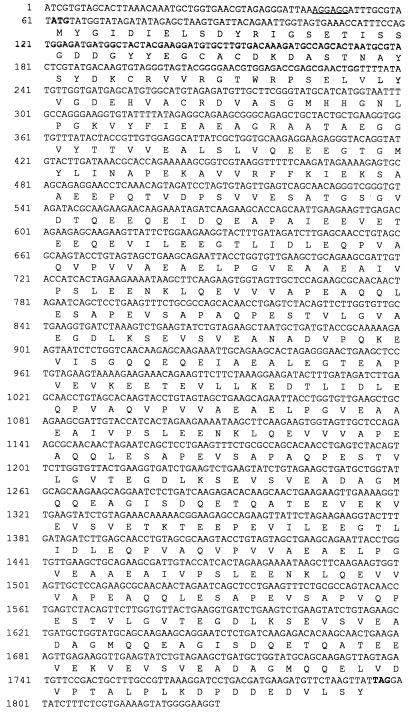
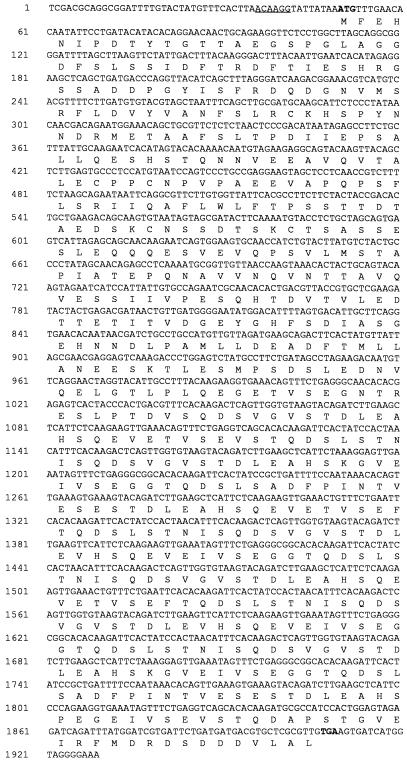
The inserts from each of the recombinant plasmids S2, S7, and S22 were completely sequenced by the primer walking method. Sequence analysis (MacVector 6.0; Oxford Molecular Group) showed that each clone contained a single large open reading frame encoded by the plus strand of the insert and that each one appeared to be a complete gene. The portions of the DNA sequence containing the open reading frame and corresponding amino acid sequence of each of the clones are shown as follows: Fig. 3, S2, GE 160; Fig. 4, S7, GE 100; and Fig. 5, S22, GE 130. A possible ribosome binding site sequence upstream from the initiating ATG codon is underlined in each figure.
FIG. 3.
Sequence of the 160-kDa protein gene. Nucleotide numbers are indicated to the left. The ATG start codon and TAA stop codon are shown in boldface type. The translated amino acid sequence for the open reading frame is displayed underneath the DNA sequence according to the single-letter amino acid code. Upstream regulatory sequences are underlined and represent the −35, −10, and ribosome binding sites.
FIG. 4.
Sequence of the 100-kDa protein gene. Nucleotide numbers are indicated to the left. The ATG start codon and TAA stop codon are shown in boldface type. The translated amino acid sequence for the open reading frame is displayed underneath the DNA sequence according to the single-letter amino acid code. A possible ribosome binding site is underlined.
FIG. 5.
Sequence of the 130-kDa protein gene. Nucleotide numbers are indicated to the left. The ATG start codon and TAA stop codon are shown in boldface type. The translated amino acid sequence for the open reading frame is displayed underneath the DNA sequence according to the single-letter amino acid code. A possible ribosome binding site is underlined.
The numbers of nucleotides sequenced upstream of each reading frame were 1,500 bp for the S2 insert, 500 bp for the S22 insert, and 200 bp for the S7 insert. These sequences were examined for likely ehrlichial promoter sequences based on homology to the E. coli −35 and −10 consensus sequences (16). Of the three clones, S2 had the closest homology to the E. coli promoter sequences, and these are underlined in Fig. 3. A number of possible promoter sequences were found for the S22 and S7 genes, but in the absence of direct experimental evidence, it is difficult to determine which sequence elements are necessary for promoter function. Protein expression in all three clones was IPTG dependent, and thus it is likely that the vector lacZ promoter is used for transcription of the Ehrlichia RNA in E. coli.
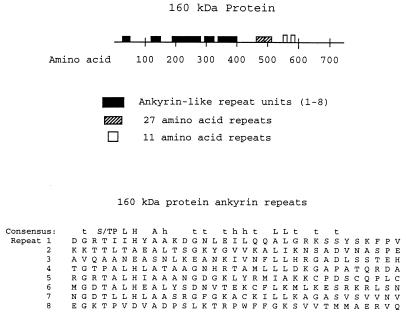
Amino acid sequence analysis of the proteins encoded by the three gene clones showed that all contain regions of repeated amino acids. A schematic version of these repeat structures is shown in Fig. 6 and 7. The 160-kDa protein (Fig. 6) has three groups of repeats. The first set consists of a number of ankyrin-like repeat units of 33 amino acids, the second consists of repeat units of 27 amino acids, and the third consists of repeat units of 11 amino acids. The ankyrin repeats were revealed by a BLAST database search for protein homologies. Ankyrin repeats occur in at least four consecutive copies and are present in yeast, plants, bacteria, and mammals (5, 6). Figure 6 shows a multiple alignment of the 160-kDa protein ankyrin repeats under a consensus sequence derived from the analysis of several hundred similar ankyrin-like motifs (6). The eighth repeat sequence holds to the consensus only through the first half of the repeat unit and may not represent a full ankyrin-like repeat.
FIG. 6.
Schematic diagram of the GE 160-kDa protein. Repeat regions are indicated by the boxes. Sequences of proposed ankyrin repeats, numbered 1 to 8, are aligned according to the consensus sequence at the top (6). h, hydrophobic; t, turn-like or polar; S/T, serine or threonine; capital letters, conserved amino acids.
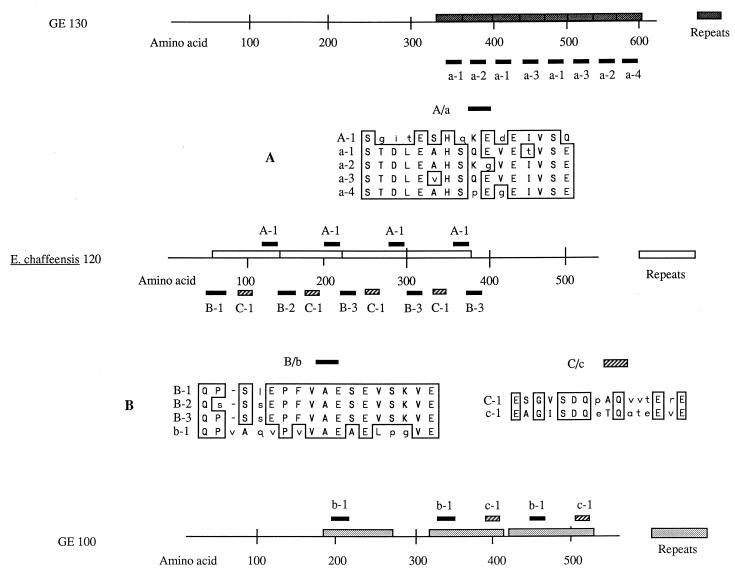
FIG. 7.
Amino acid sequence alignments of selected regions of GE 130-kDa and E. chaffeensis 120-kDa proteins (A) and GE 100-kDa and E. chaffeensis 120-kDa proteins (B). Each protein is shown as a linear amino acid sequence, and amino acids are numbered in hundreds. Boxed regions on the linear sequence represent repeated amino acids. (A) Amino acid alignments of a sequence which occurs four times in the E. chaffeensis protein (45) (top line of alignment, A-1) and eight times in the GE 130-kDa protein (a-1 to a-4). Sequence a-1 is repeated three times, related sequences a-2 and a-3 are each repeated twice, and related sequence a-4 is found once. The positions of these sequences in the proteins are indicated by the small bold lines. (B) Alignments of two different sequence motifs which occur in the E. chaffeensis 120-kDa protein (B-1 to B-3 and C-1) and the GE 100-kDa protein (b-1 and c-1). Bold and cross-hatched boxes indicate the positions of these sequences in the proteins. Identical amino acids are surrounded by boxes, and conserved amino acids are in capital letters.
The 130-kDa protein (Fig. 7) has a repeat unit of 26 to 34 amino acids which occurs eight times in the carboxy-terminal half of the protein. The sequence varies somewhat from repeat to repeat. A database homology search with the NCBI BLAST algorithm revealed that the 130-kDa protein has limited homology to the E. chaffeensis 120-kDa protein (45). An amino acid sequence alignment of a motif common to both proteins is shown in Fig. 7A. This motif is represented by a bold line and occurs four times in an identical fashion in the E. chaffeensis protein (designated A-1) and eight times with four variations in the GE 130-kDa protein (a-1, a-2, a-3, and a-4).
The 100-kDa protein (Fig. 7) has three large repeat units, which differ somewhat in length. A database search revealed that it is similar to the 120-kDa E. chaffeensis protein, which contains four repeats of 80 amino acids each (45). Both proteins contain large amounts of glutamic acid: 18% for the 100-kDa protein and 17% for the 120-kDa protein. When the two protein sequences are aligned, most of the homology occurs in the repeat regions. Figure 7B shows alignments for two homologous groups of amino acid motifs from the two proteins (designated B/b and C/c) found with the BLAST algorithm. These are not the only possible alignments of the two proteins but do provide an example of their similarities. The locations of the homologous sequences are indicated by bold or hatched lines above (GE 100) or below (E. chaffeensis 120) the respective proteins. The B sequence represented by the bold line varies slightly in the E. chaffeensis protein (shown as B-1, B-2, and B-3) and occurs a total of five times. The GE 100 protein equivalent, b-1, is invariant and occurs three times. The sequence represented by the hatched line occurs four times in E. chaffeensis 120 (C-1) and two times in GE 100 (c-1).
All of the GE proteins migrated at higher apparent molecular masses under reducing conditions than calculations would predict. The GE 160 protein consists of 748 amino acids with a calculated molecular mass of 78.8 kDa. The GE 100 protein has 578 amino acids, and its calculated molecular mass is 61.4 kDa. The GE 130 protein contains 619 amino acids and has a calculated molecular mass of 66.1 kDa.
Verification that clones are GE derived with PCR analysis.
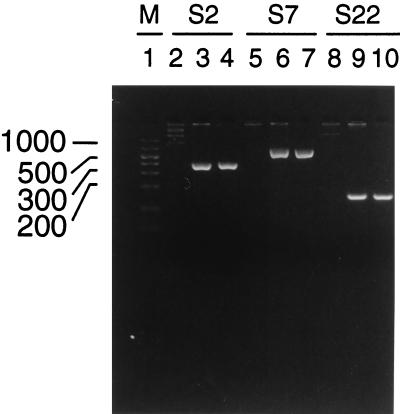
Based on the DNA sequences of each clone, PCR primers were designed to amplify specific regions of each open reading frame (see Materials and Methods). Primer pairs specific for S22, S2, and S7 were used in separate PCRs to amplify three different templates: GE DNA (from USG3), HL60 DNA, or the purified plasmid DNA of each clone. Figure 8 shows the results obtained for primers of S2 (lanes 2 to 4), S7 (lanes 5 to 7), and S22 (lanes 8 to 10). All three clones were specific to GE and were not present in HL60 DNA (lanes 2, 5, and 8). In each case, the size of the PCR product determined with genomic DNA as the template was the same as that generated by the purified plasmid DNA.
FIG. 8.
PCR analysis of GE genes. PCRs were performed as described in Materials and Methods, and the products were analyzed with 4% Nusieve gels. S2 primers were used to amplify a 395-bp region of S2 DNA with HL60 DNA (lane 2), S2 plasmid DNA (lane 3), and USG3 DNA (lane 4) used as templates. S7 primers were used to amplify a 643-bp region of S7 DNA with HL60 DNA (lane 5), S7 plasmid DNA (lane 6), and USG3 DNA (lane 7) used as templates. S22 primers were used to amplify a 159-bp region of S22 DNA with HL60 DNA (lane 8), S22 plasmid DNA (lane 9), and USG3 DNA (lane 10) used as templates. DNA molecular size markers (M) (50 to 1,000 bp [FMC, Rockland, Maine]) are present in lane 1.
DISCUSSION
Ehrlichiae which have a tropism for peripheral blood granulocytes and are closely related based on the sequence of the 16S rRNA gene have been classified as either E. phagocytophila, E. equi, or the agent of HGE, depending on the species and geographical location of the infected host. E. phagocytophila is responsible for a tick-borne fever of cattle, sheep, and goats and occurs primarily in Europe (10, 33), and E. equi is found in horses in North and South America (14, 26). The agent of HGE has been identified as the organism responsible for disease in patients in the upper midwestern and northeastern United States (3, 7). However, classification of GE has been difficult because of a lack of genetic information.
The GE isolate we describe here is indistinguishable from the HGE agent based on the sequence of the 16S rRNA gene. We also describe three previously unidentified proteins of GE, the 160-, 130-, and 100-kDa proteins. Aside from the 16S rRNA gene and the relatively conserved groEL homolog (22), the genes coding for these proteins are the first sequenced genes reported for this organism. Using PCR primers based on the sequences of the three genes, we have confirmed that the genes are specific for GE and that they are not derived from the HL60 cell line used to culture the organism. Preliminary results also show that these genes are present in other members of the GE genogroup, including E. phagocytophila (31a).
The three GE proteins described in this study exhibit several interesting features. All three of the proteins contain repeated regions of amino acids. In the 100-kDa protein, the repeats comprise the majority of the protein, with the exception of the amino terminus. The direct repeats in both the 160- and 130-kDa proteins occur in the carboxyl half of the molecules. Various biological functions have been demonstrated or implicated for repeat regions found in other proteins. These include antigenic variation (17, 31) and ligand interaction (11, 42), both of which are thought to occur in several species of pathogenic bacteria (42). In several proteins of the malaria parasite Plasmodium falciparum, repeated regions are often the major targets for the host antibody response (21). Proteins with several repeated domains are also often found associated with the cell surface (12, 42). In addition to the direct repeats, the 160-kDa protein contains seven to eight ankyrin-like repeats. These domains have been found in over 90 different proteins from nearly all phyla, including both E. coli and humans (6). They are present in functionally diverse proteins, such as enzymes, toxins, and transcription factors, and are thought to play a role in protein-protein interactions (5). Human erythrocyte ankyrin contains 22 repeats, some of which may be involved in binding tubulin, spectrin, or vimentin. The repeats could thus enable the protein to act like a bridge between cytoskeleton and membrane components (23). The myriad functions ascribed to proteins containing repeated regions may imply that the GE proteins described here will be found to have roles in host cell binding, antigenic variation, or some other cell surface function.
The presence of repeated regions in the 100-, 130-, and 160-kDa proteins may in part explain the differences between the expected molecular masses and the observed molecular masses on SDS-PAGE, which are much higher. This phenomenon has been reported for other proteins containing repeated regions, including rickettsial proteins (1, 36), malaria antigens (21), the fibronectin-binding protein from Staphylococcus aureus (18, 37), and the E. chaffeensis 120-kDa protein (45). The difference between calculated and observed molecular mass varies from 10 to 30% for the malaria antigens to almost 100% for the S. aureus and E. chaffeensis proteins. Investigators have speculated that high percentages of certain amino acids in these proteins or posttranslational modifications may contribute to the aberrant migration in an SDS-polyacrylamide gel system (18, 37, 45).
A search of the major protein and nucleotide databases with the NCBI BLAST algorithm did not reveal any significant similarities of the GE proteins to any known proteins, other than the ankyrin repeats of the 160-kDa protein and the homology between the 100-kDa protein or 130-kDa protein and the 120-kDa protein of E. chaffeensis. The function of the E. chaffeensis 120-kDa protein is not known, but the hydrophilic nature of its repeat domain suggests that it may be located on the surface of E. chaffeensis (45). The GE 100-kDa protein is also hydrophilic in the repeat region (data not shown) and may also be surface associated. Although both the 100- and 130-kDa proteins have some homology to the E. chaffeensis 120-kDa protein, they are not homologous to one another. It is possible that whatever the function(s) of the 120-kDa protein is for E. chaffeensis, similar functions may be carried out in GE through the proteins GE 100 and GE 130.
It is unclear whether the three GE proteins described in this report may be useful as potential diagnostic reagents or vaccine candidates. Although they are not well represented in the purified-culture-grown organism, either because they are secreted into the culture medium or their expression is tied more closely to natural infection, they are clearly important immunologically. All three react with convalescent-phase sera from dogs, sheep (data not shown), and humans diagnosed with HGE. The E. chaffeensis 120-kDa protein, the counterpart of the 100-kDa GE protein, has shown sensitivity and specificity as a diagnostic reagent with a panel of E. chaffeensis patient sera (44). However, Western blots with convalescent-phase sera from HGE patients do not show a consistent response to the GE 100-kDa protein. It is not known whether there is any immunologic cross-reactivity between the GE 100-kDa protein and the E. chaffeensis 120-kDa protein or whether the two could be used to distinguish monocytic from granulocytic ehrlichiosis. Some human antisera have been reported to react with both E. chaffeensis and HGE antigens, but it is not known whether this represents cross-reactive epitopes or separate exposures to the two ehrlichial agents (30, 32).
In summary, the expression library approach for the isolation and characterization of GE proteins has resulted in the discovery of three previously unidentified genes which express high-molecular-weight, immunoreactive proteins. Further study of these proteins will aid in the classification of the members of the GE genogroup as corresponding genes are sequenced and compared and will be important in elucidating the role of these proteins in immunity and pathogenesis.
ACKNOWLEDGMENTS
We thank Durland Fish (Yale University) for the experimental challenge work with dogs which led to the isolation of GE in cell culture and Johan Bakken and Maria Aguero-Rosenfeld for the kind gift of human sera.
REFERENCES
- 1.Allred D R, McGuire T C, Palmer G H, Leib S R, Harkins T M, McElwain T F, Barbet A F. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc Natl Acad Sci USA. 1990;87:3220–3224. doi: 10.1073/pnas.87.8.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson B E, Dawson J E, Jones D C, Wilson K H. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J Clin Microbiol. 1991;29:2838–2842. doi: 10.1128/jcm.29.12.2838-2842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakken J S, Dumler J S, Chen S-M, Eckman M R, Van Etta L L, Walker D H. Human granulocytic ehrlichiosis in the upper Midwest. A new species emerging? JAMA. 1994;272:212–218. [PubMed] [Google Scholar]
- 4.Bakken J S, Krueth J, Wison-Nordskog C, Tilden R L, Asanovich K, Dumler J S. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. JAMA. 1996;275:199–205. [PubMed] [Google Scholar]
- 5.Blank V, Kourilsky P, Israel A. NF-kB and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci. 1992;17:135–140. doi: 10.1016/0968-0004(92)90321-y. [DOI] [PubMed] [Google Scholar]
- 6.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins Struct Funct Genet. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 7.Chen S-M, Dumler J S, Bakken J S, Walker D H. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–595. doi: 10.1128/jcm.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coughlin R T, Fish D, Mather T N, Ma J, Pavia C, Bulger P. Protection of dogs from Lyme disease with a vaccine containing outer surface protein (Osp) A, Osp B, and the saponin adjuvant QS-21. J Infect Dis. 1995;171:1049–1052. doi: 10.1093/infdis/171.4.1049. [DOI] [PubMed] [Google Scholar]
- 9.Dumler J S, Asanovich K M, Bakken J S, Richter P, Kimsey R, Madigan J E. Serologic cross-reactions among Ehrlichia equi, Ehrlichia phagocytophila, and human granulocytic ehrlichia. J Clin Microbiol. 1995;33:1098–1103. doi: 10.1128/jcm.33.5.1098-1103.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foggie A. Studies on the infectious agent of tick-borne fever in sheep. J Pathol Bacteriol. 1951;63:1–15. doi: 10.1002/path.1700630103. [DOI] [PubMed] [Google Scholar]
- 11.Gaillard J-L, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from Gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore R D. Comparison of the rompA gene repeat regions of Rickettsiae reveals species-specific arrangements of individual repeating units. Gene. 1993;125:97–102. doi: 10.1016/0378-1119(93)90752-o. [DOI] [PubMed] [Google Scholar]
- 13.Goodman J L, Nelson C, Vitale B, Madigan J E, Dumler J S, Kurtti T J, Munderloh U G. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med. 1996;334:209–215. doi: 10.1056/NEJM199601253340401. [DOI] [PubMed] [Google Scholar]
- 14.Gribble D H. Equine ehrlichiosis. J Am Vet Med Assoc. 1969;155:462–469. [PubMed] [Google Scholar]
- 15.Hardalo C J, Quagliarello V, Dumler J S. Human granulocytic ehrlichiosis in Connecticut: report of a fatal case. Clin Infect Dis. 1995;21:910–914. doi: 10.1093/clinids/21.4.910. [DOI] [PubMed] [Google Scholar]
- 16.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoft D F, Kim K S, Otsu K, Moser D R, Yost W J, Blumin J H, Donelson J E, Kirchhoff L V. Trypanosoma cruzi expresses diverse repetitive protein antigens. Infect Immun. 1989;57:1959–1967. doi: 10.1128/iai.57.7.1959-1967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollingshead S K, Fischetti V A, Scott J R. Complete nucleotide sequence of type 6M protein of the group A Streptococcus. J Biol Chem. 1986;261:1677–1686. [PubMed] [Google Scholar]
- 19.Karlin S, Altschul S F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87:2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlin S, Altschul S F. Applications and statistics for multiple high scoring segments in molecular sequences. Proc Natl Acad Sci USA. 1993;90:5873–5877. doi: 10.1073/pnas.90.12.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kemp D J, Coppel R L, Anders R F. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 22.Kolbert C P, Bruinsma E S, Abdulkarim A S, Hofmeister E K, Tompkins R B, Telford III S R, Mitchell P D, Adams-Stich J, Persing D H. Characterization of an immunoreactive protein from the agent of human granulocytic ehrlichiosis. J Clin Microbiol. 1997;35:1172–1178. doi: 10.1128/jcm.35.5.1172-1178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lux S E, John K M, Bennett V. Analysis of cDNA for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature (London) 1990;344:36–42. doi: 10.1038/344036a0. [DOI] [PubMed] [Google Scholar]
- 24.Madewell B R, Gribble D H. Infection in two dogs with an agent resembling Ehrlichia equi. J Am Vet Med Assoc. 1982;180:512–514. [PubMed] [Google Scholar]
- 25.Madigan J E, Gribble D H. Equine ehrlichiosis in northern California: 49 cases (1968–1981) J Am Vet Med Assoc. 1987;190:445–448. [PubMed] [Google Scholar]
- 26.Madigan J E, Hietala S, Chalmers S, DeRock E. Seroepidemiologic survey of antibodies to Ehrlichia equi in horses of northern California. J Am Vet Med Assoc. 1990;196:1962–1964. [PubMed] [Google Scholar]
- 27.Madigan J E. Equine ehrlichiosis. In: Woldehiwet Z, Ristic M, editors. Rickettsial and chlamydial diseases of domestic animals. Oxford, United Kingdom: Pergamon Press; 1993. pp. 209–214. [Google Scholar]
- 28.Madigan J E, Richter P J, Kimsey R B, Barlough J E, Bakken J S, Dumler J S. Transmission and passage in horses of the agent of human granulocytic ehrlichiosis. J Infect Dis. 1995;172:1141–1144. doi: 10.1093/infdis/172.4.1141. [DOI] [PubMed] [Google Scholar]
- 29.Madigan J E, Barlough J E, Dumler J S, Schankman N S, DeRock E. Equine granulocytic ehrlichiosis in Connecticut caused by an agent resembling the human granulocytotropic ehrlichia. J Clin Microbiol. 1996;34:434–435. doi: 10.1128/jcm.34.2.434-435.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magnarelli L A, Dumler J S, Anderson J F, Johnson R C, Fikrig E. Coexistence of antibodies to tick-borne pathogens of babesiosis, ehrlichiosis, and Lyme borreliosis in human sera. J Clin Microbiol. 1995;33:3054–3057. doi: 10.1128/jcm.33.11.3054-3057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto M, Tange Y, Okada T, Inoue Y, Horiuchi T, Kobayashi Y, Fujita S. Deletion in the 190 kDa antigen repeat region of Rickettsia rickettsii. Microb Pathog. 1996;20:57–62. doi: 10.1006/mpat.1996.0005. [DOI] [PubMed] [Google Scholar]
- 31a.Murphy, C. I., et al. Unpublished observations.
- 32.Nicholson W L, Comer J A, Sumner J W, Gingrich-Baker C, Coughlin R T, Magnarelli L A, Olson J G, Childs J E. An indirect immunofluorescence assay using a cell culture-derived antigen for detection of antibodies to the agent of human granulocytic ehrlichiosis. J Clin Microbiol. 1997;35:1510–1516. doi: 10.1128/jcm.35.6.1510-1516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rikihisa Y. The tribe Ehrlichiae and ehrlichial diseases. Clin Microbiol Rev. 1991;4:286–308. doi: 10.1128/cmr.4.3.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rikihisa Y, Ewing S A, Fox J C, Siregar A G, Pasaribu F H, Malole M B. Analyses of Ehrlichia canis and a canine granulocytic Ehrlichia infection. J Clin Microbiol. 1992;30:143–148. doi: 10.1128/jcm.30.1.143-148.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodgers S J, Morton R J, Baldwin C A. A serological survey of Ehrlichia canis, Ehrlichia equi, Rickettsia rickettsii, and Borrelia burgdorferi in dogs in Oklahoma. J Vet Diagn Invest. 1989;1:154–159. doi: 10.1177/104063878900100212. [DOI] [PubMed] [Google Scholar]
- 36.Schuenke K W, Walker D H. Cloning, sequencing, and expression of the gene coding for an antigenic 120-kilodalton protein of Rickettsia conorii. Infect Immun. 1994;62:904–909. doi: 10.1128/iai.62.3.904-909.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Signas C, Raucci G, Jonsson K, Lindgren P, Anantharamaiah G M, Hook M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stannard A A, Gribble D H, Smith R S. Equine ehrlichiosis: a disease with similarities to tick-borne fever and bovine petechial fever. Vet Rec. 1969;84:149–150. doi: 10.1136/vr.84.6.149. [DOI] [PubMed] [Google Scholar]
- 39.Telford S R, III, Dawson J E, Katavolos P, Warner C K, Kolbert C P, Persing D H. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci USA. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walls J J, Greig B, Neitzel D F, Dumler J S. Natural infection of small mammal species in Minnesota with the agent of human granulocytic ehrlichiosis. J Clin Microbiol. 1997;35:853–855. doi: 10.1128/jcm.35.4.853-855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson K H, Blitchington R B, Greene R C. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–1946. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wren B W. A family of clostridial and streptococcal ligand-binding proteins with conserved C-terminal repeat sequences. Mol Microbiol. 1991;5:797–803. doi: 10.1111/j.1365-2958.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- 43.Yeh M-T, Mather T N, Coughlin R T, Gingrich-Baker C, Sumner J W, Massung R F. Serologic and molecular detection of granulocytic ehrlichiosis in Rhode Island. J Clin Microbiol. 1997;35:944–947. doi: 10.1128/jcm.35.4.944-947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X-J, Crocquet-Valdes P, Cullman L C, Walker D H. The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J Clin Microbiol. 1996;34:2853–2855. doi: 10.1128/jcm.34.11.2853-2855.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X-J, Crocquet-Valdes P, Walker D H. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene. 1997;184:149–154. doi: 10.1016/s0378-1119(96)00586-0. [DOI] [PubMed] [Google Scholar]