Major depressive disorder (MDD) and type 2 diabetes (T2D) are two common complex multifactorial disorders that share several genetic and environmental risk factors such as hypercortisolism and related genes' risk variants within the stress response and the neuroendocrine hypothalamic-pituitary axis. Under stress, the pituitary gland releases prolactin (PRL), whose effects are pleiotropic and include mood control and insulin secretion from the beta cells.1 Variations in the prolactin receptor (PRLR) gene are associated in rodent models with stress level, depression-like behavior,2 and hepatic insulin sensitivity3 and in humans with maternal glucose homeostasis and gestational diabetes.4 Furthermore, prolonged breastfeeding has been associated with reduced incidence of T2D, potentially related to PRL action. To our knowledge, no studies are reporting PRLR as a risk gene for MDD or T2D. Therefore, we aimed to investigate if the PRLR gene encoding for the PRLR plays a role in the familial comorbidity of MDD and T2D.
In 212 Italian families phenotyped for T2D and MDD, we investigated 41 microarray-based single nucleotide polymorphisms (SNPs) in the PRLR gene. We tested the variants for linkage/linkage-disequilibrium (LD, i.e., association) to/with T2D and MDD using recessive and dominant models with incomplete penetrance. To test whether the risk variants were linked/in LD under complete penetrance a s well, we ran a secondary parametric analysis. We performed genotyping and Mendelian error exclusion using PLINK. A P-value < 0.05 was used as the cut-off for the level of statistical significance. The study was approved by the Bios Ethical Committee.
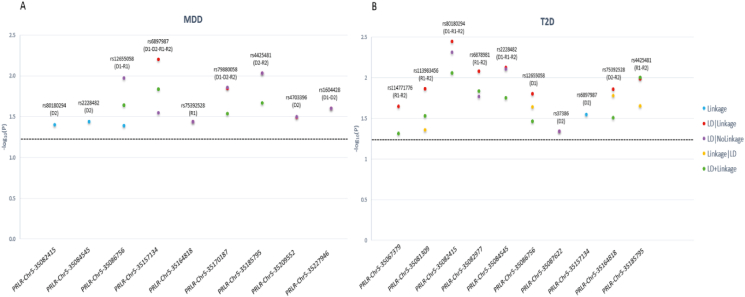
We found a total of 13 risk variants significantly linked/in LD to/with T2D and/or MDD (P < 0.05) (Table S1). Three risk variants (rs79880058, rs4703396, and rs1604428) were significantly linked/in LD to/with MDD (Fig. S2); four risk variants (rs114771776, rs113983456, rs6878981, and rs37386) were significantly linked/in LD to/with T2D (Fig. 1); and six risk variants (rs80180294, rs2228482, rs12655058, rs6897987, rs75392528, and rs4425481) were significantly linked/in LD to/with both MDD and T2D, three of which have concordant risk alleles. The T2D-risk rs114771776 and rs113983456 were within a specific LD block, and the T2D-risk rs6878981 and MDD-T2D-risk rs2228482 were in another specific LD block. Significant variants within a specific LD block replicate each other and perpetuate the pleiotropic effect of the PRLR gene in mediating MDD-T2D co-morbidity. All these risk variants are novel and were not previously reported in MDD, T2D, or another related phenotype. In silico analysis predicted that the comorbid MDD-T2D rs6897987 risk SNP has a high regulatory potential (RegulomeDB score 1 [0–1]) and is located at a peak of chromatin immunoprecipitation (Chip-seq) site, suggesting that it is a binding site for transcription factors without, however, a known transcription factor predicted to bind to it. This is the first study to link the PRLR gene to the risk of morbidity and comorbidity of MDD and T2D. The PRLR gene may play a mental-metabolic role, possibly through activation of the Janus Kinase/Signal Transducers and Activators of Transcription pathway which is involved in stress response and metabolic homeostasis.5 Functional studies are needed to confirm these results.
Figure 1.
MDD and T2D PRLR risk SNPs linkage and LD analysis results. For each significant major depressive disorder (A) and type 2 diabetes (B)-risk SNP in the PRLR gene, we present the −log10(P) as a function of each significant test statistic (Linkage, LD|Linkage, LD|NoLinkage, Linkage|LD, and LD + Linkage) across the significant inheritance model(s): D1: dominant, complete penetrance; D2: dominant, incomplete penetrance; R1: recessive, complete penetrance; R2: recessive, incomplete penetrance.
Ethics declaration
Families were recruited following the Helsinki Declaration guidelines. The Bios Ethical Committee approved this study. Individuals provided written informed consent before participation.
Author contributions
C.G. conceived and supervised the project, including statistical analysis and manuscript drafting. M.A. helped with the bioinformatic analysis, literature search, and manuscript drafting. R.W. and T.T.P. critically helped in data interpretation and critical revision of the manuscript. All authors approved the final manuscript.
Conflict of interests
The authors have declared that they have no conflict of interests.
Funding
This publication was supported in part by the funds received under Nebraska Laws 2021, LB 380, Section 109 awarded to C.G. (PI), Creighton University School of Medicine, through the Nebraska Department of Health & Human Services (DHHS). Its contents represent the views of the authors and do not necessarily represent the official views of the State of Nebraska or DHHS.
Data availability
The study data are available upon reasonable request, and due to lacking specific patients' consent and privacy restrictions, they are not publicly available.
Acknowledgements
We thank the families who participated in the study, and we thank Bios Biotech Multi-Diagnostic Health Center, Rome, Italy, for data access and financial, medical, and laboratory staff support.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2023.06.018.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Brelje T.C., Scharp D.W., Lacy P.E., et al. Effect of homologous placental lactogens, prolactins, and growth hormones on islet B-cell division and insulin secretion in rat, mouse, and human islets: implication for placental lactogen regulation of islet function during pregnancy. Endocrinology. 1993;132(2):879–887. doi: 10.1210/endo.132.2.8425500. [DOI] [PubMed] [Google Scholar]
- 2.de Jong T.V., Kim P., Guryev V., et al. Whole genome sequencing of nearly isogenic WMI and WLI inbred rats identifies genes potentially involved in depression and stress reactivity. Sci Rep. 2021;11(1):14774. doi: 10.1038/s41598-021-92993-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu J., Xiao F., Zhang Q., et al. PRLR regulates hepatic insulin sensitivity in mice via STAT5. Diabetes. 2013;62(9):3103–3113. doi: 10.2337/db13-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le T.N., Elsea S.H., Romero R., Chaiworapongsa T., Francis G.L. Prolactin receptor gene polymorphisms are associated with gestational diabetes. Genet Test Mol Biomarkers. 2013;17(7):567–571. doi: 10.1089/gtmb.2013.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian R.H., Bai Y., Li J.Y., Guo K.M. Reducing PRLR expression and JAK2 activity results in an increase in BDNF expression and inhibits the apoptosis of CA3 hippocampal neurons in a chronic mild stress model of depression. Brain Res. 2019;1725:146472. doi: 10.1016/j.brainres.2019.146472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The study data are available upon reasonable request, and due to lacking specific patients' consent and privacy restrictions, they are not publicly available.