Abstract
The environment is a natural reservoir of Clostridioides difficile, and here, we aimed to isolate the pathogen from seven locations in northern Iraq. Four of the sites yielded thirty-one isolates (ten from soils, twenty-one from sediments), which together represent ribotypes (RTs) 001 (five), 010 (five), 011 (two), 035 (two), 091 (eight), and 604 (nine). Twenty-five of the isolates (∼81%) are non-toxigenic, while six (∼19%) encode the toxin A and B genes. The genomes of eleven selected isolates represent six sequence types (STs): ST-3 (two), ST-15 (one), ST-107 (five), ST-137 (one), ST-177 (one), and ST-181 (one). Five novel RT/ST associations: RT011/ST-137, RT035/ST-107, RT091/ST-107, RT604/ST-177, and RT604/ST-181 were identified, and the first three are linked to RTs previously uncharacterized by multilocus sequence typing (MLST). Nine of the genomes belong to Clade 1, and two are closely related to the cryptic C-I clade. Diverse multiple prophages and CRISPR-Cas systems (class 1 subtype I-B1 and class 2 type V CRISPR-Cas systems) with spacers identical to other C. difficile phages and plasmids were detected in the genomes. Our data show the broader diversity that exists within environmental C. difficile strains from a much less studied location and their potential role in the evolution and emergence of new strains.
Keywords: Clostridioides difficile, multilocus sequence typing, whole-genome sequencing, phylogenetic analysis, CRISPR-Cas system, prophage
Lactobacillus casei condition medium downregulates miR-21 relative expression in HT-29 colorectal cancer cell line, which can potentially be related to induction of apoptosis in cancerous cells rather than in healthy cells.
Introduction
The nosocomial Clostridioides difficile infection (CDI) is characterised by antibiotic induced diarrhoea and pseudomembranous colitis (Czepiel et al. 2019). The identification of clinical C. difficile ribotypes (RTs) in environmental settings indicates putative connections with humans and animals and could contribute to the emergence of new strains in hospital and thus pose a significant health risk (Janezic et al. 2016, Czepiel et al. 2019, Williamson et al. 2022). Environmental C. difficile strains that are genetically related to those isolated from human in clinical settings suggest that the same strains can inhabit multiple niches and the environment is a reservoir of CDI (Knight et al. 2017, Rodriguez Diaz et al. 2018, Janezic et al. 2020, Lim et al. 2020, Williamson et al. 2022). Core genome single nucleotide variant (SNV) analysis revealed that 42% of human strains showed clonal relationship (separated by ≤2 SNVs in their core genome) with one or more strains from environmental samples (Knight et al. 2017, Janezic et al. 2020). This strongly supports a persistent community reservoir with long-range dissemination. Since the sources/reservoirs outside the hospital setting play a significant role in the transmission of CDI, continuing molecular and genomic surveillance of strains from these sources is vital to find opportunities to reduce the overall CDI burden (Knight et al. 2017, Lim et al. 2020).
Clostridioides difficile diversity is mainly characterized using PCR ribotyping, which distinguishes the strains based on the size and copy number of the 16S-23S rRNA intergenic spacer region (Indra et al. 2008, Chatterjee and Raval 2019). Polymerase chain reaction (PCR) ribotyping is clearly useful for outbreak investigations (Seth-Smith et al. 2021) and has relatively equal discriminatory power to multilocus sequence typing (MLST), which identifies C. difficile strains based on the combinations of seven unique housekeeping genes that allow designation of allele profiles or sequence types (ST) to represent a genotype (Griffiths et al. 2010, Knight et al. 2017, Janezic and Rupnik 2019). Whole genome sequencing (WGS), however, permits single nucleotide-level strain resolution over all genomic space, thus, it is essential for long-term epidemiological, evolution, and population dynamics studies (Dominguez et al. 2016, Dingle et al. 2017, Muñoz et al. 2017, Uelze et al. 2020). WGS is currently accessible due to the low sequencing cost and availability of publicly available genome data, which provide valuable resources for more in-depth genome comparisons than ribotyping.
Clostridioides difficile surveillance is more effective in western countries but very few epidemiological studies are reported in northern Iraq, which leaves a significant geographic lack of awareness of this bacterium in this part of the world. We have reported the genomes of three novel Clostridium sp. strains isolated from the environment in northern Iraq (Rashid et al. 2016), but, to date, no other environmental C. difficile genomes from this region have been reported. This highlights the paucity of knowledge that exists on strains such as RTs 001, 010, 011, and 035 that are circulating in the environment in this part of the globe and their potential role in clinical settings (Hargreaves et al. 2013, Hargreaves et al. 2016, Janezic et al. 2016). To further knowledge in this area and strengthen the existence of clinically relevant C. difficile strains in the natural environments, here, we isolated and genetically characterized environmental isolates from northern Iraq. We included strains from our previous studies to conduct whole-genome analyses to ascertain their RT/strain type relationships. Furthermore, we analysed the diverse CRISPR-Cas systems found within the strains and compared these features to strains from other regions to better ascertain possible genetic interactions that occurred through horizontal gene transfer via prophage elements and their role in C. difficile evolution.
Materials and methods
Sampling sites
To isolate C. difficile from northern Iraq, soil (seven) and sediment (five) samples were collected from seven sites: Hamamok, Dokan, Jalee, Chnarok, Taq Taq rivers, and Safeen and Haibat Sultan mountains between 2012 and 2013 (Supplementary Table S1). Samples were collected into screwed-capped, sterile falcon tubes, immediately stored at 4°C, and processed within 2 weeks of collection.
Recovery of C. difficile isolates from environmental samples
Clostridium difficile was isolated using previously described enrichment procedures (Hargreaves et al. 2013). Briefly, ∼1 g of soil/sediment was mixed with 10 mL of fastidious anaerobic broth supplemented with 250 µg mL−l cycloserine and 8 µg mL−1 cefoxitin (Bioconnections, Leeds, UK) to select for C. difficile. Also, 0.1% sodium taurocholate (Sigma–Aldrich, Dorset, UK) was added to the enrichment to enhance spore germination (Foster and Riley 2012). The cultures were incubated for 10 days in a MiniMACS anaerobic chamber (Don Whitley Scientific, West Yorkshire, UK; 10% H2, 5% CO2, and 85% N2) at 37°C, then centrifuged for 10 min at 5000 × g. To further select for C. difficile spores and reduce other bacterial contaminants, the pellet was treated with an equal volume of industrial methylated spirit and incubated for 30 min at room temperature. A loopful of the mixture was spread on Brazier’s cycloserine, cefoxitin, and egg yolk (CCEY) selective agar plates and incubated anaerobically for 48 hours. Clostridioides difficile colonies were purified through three further rounds of sub-culturing on Brain Heart Infusion (BHI) agar (Oxoid, Ltd., UK) supplemented with 7% defibrinated horse blood (TCS Biosciences, Ltd., UK). The presumptive colonies were identified by the characteristic horse manure smell, colony morphology, and yellow-green fluorescence under the long-wave ultraviolet light (Delmée 2001). The isolates were confirmed by PCR targeting the C. difficile 16S rRNA gene, as described by Rinttilä et al. (2004). Bacterial isolates were stored in Protect bacterial preservers (Technical Service Consultants, Ltd., Heywood, UK) at −80°C.
Ribotyping and toxin gene characterization C. difficile isolates
To determine if multiple RTs were found in each site or sample, ten randomly selected bacterial isolates from each sample were subjected to conventional and capillary PCR ribotyping targeting the intergenic spacer 16S-23S rRNA genes using primers GTGCGGCTGGATCACCTCCT-3′ and 5′-CCCTGCACCCTTAATAACTTGACC-3′ (Indra et al. 2008). DNA was extracted from broth cultures that were grown for 18–24 hours anaerobically using 5% Chelex® (BioRad Laboratories, California, USA). The PCR ribotyping conditions were denaturation at 95°C for 120 s, followed by 30 cycles of denaturation at 92°C for 60 s, annealing at 55°C for 60 s, elongation at 72°C for 90 s, and a final extension at 72°C for 5 min. PCR products alongside a 100-bp DNA ladder (Fermentas, York, UK) were resolved at 3%. Response regular agarose gel (Geneflow, Staffordshire, UK) prepared in 1 × Tris-acetate—EDTA buffer and stained with GelRed (Biotium, Hayward, California, USA; Nale et al. 2012). Images were visualized using the SynGene application in a UV transilluminator. Fragments from capillary ribotyping were analysed using Peak Scanner software v1.0 (Applied Biosystems, UK). The similarity of the strains was assessed using a MultiVariate Statistical Package (MVSP, Kovach Computing Services, Anglesey, UK) based on the presence of amplicons of a particular size. Sorensen’s distance was calculated between each combination of isolates and clustered (Supplementary Fig. S1 and B; Shan et al. 2012, Nale et al. 2016). Eleven strains from the sites with six distinct patterns of amplicons were submitted to Leeds Reference Laboratory, UK for further confirmation of RT designation and toxin genes presence. All the isolates were further screened for the presence of the toxin genes using multiplex PCR with primer pairs NK2 and NK3, which amplified the partial sequences of tcdA, NK9, and NK11 targeting the essential repeat region within the tcdA (Kato et al. 1998) and NK-104 and NK-105 for the toxin B gene (Barroso et al. 1990). Amplification conditions for the multiplex PCR for the NK primers were initial denaturation at 95°C for 5 min, followed by 32 cycles of denaturation at 95°C for 20 s, annealing at 62°C for 120 s, elongation at 72°C for 2 min, and a final extension step at 72°C for 5 min. Binary toxin genes cdtA and cdtB presence were determined using primers and procedures previously described (Barroso et al. 1990, Stubbs et al. 2000). PCR reaction conditions were initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 60 s, elongation at 72°C for 2 min, and a final extension stage for 5 min at 72°C. PCR amplicons were resolved in a 1% molecular-grade agarose gel (Bioline, UK) in 1xTAE with GelRed and visualized as described above.
Whole-genome sequencing
To further assess the genome diversity within the isolates, a total of eleven isolates comprising of three isolates of RT091, two isolates each of RT001, RT035, and RT604, and one isolate each of RT010 and RT011 were sequenced using the Illumina MiSeq (2 × 250 bp paired end) platform following NexteraXT library preparation. The genomic DNA was prepared from broth cultures that were grown for 18–24 hours anaerobically in BHI broth (Oxoid, Hampshire, UK) using a QIAGEN Genomic Kit according to manufacturer’s instructions. Approximately 1 ng of DNA was used in the Nextera XT DNA sample preparation (Illumina, San Diego, California, USA), following the manufacturer’s instructions. Libraries were sequenced using a MiSeq V2 reagent kit (2 × 250 bp). Genomes were assembled using SPAdes 2.0 with the following parameters: ‘-k 21, 33,55,77,99127 –careful’. All genomes were submitted to the European Bioinformatic Institute (EBI) and Enterobase under the project accession PRJEB8702. The genome can be accessed online at: https://www.ebi.ac.uk/ena/browser/view/PRJEB8702. Contigs were ordered against the reference strain C. difficile CD630 (NC_009089) using MAUVE v2.3.1 (Darling et al. 2004). Genomes were annotated using PROKKA v1.14.5 with the following settings: ‘– compliant – genus Clostridium use genus’ (Seemann 2014). Clostridioides difficile isolates were sequence-typed as previously described by Griffiths et al. (2010), utilizing seven regions within conserved the housekeeping genes (adk, atpA, dxr, glyA, recA, sodA, and tpi). Alleles from the assembled genomes were extracted and queried against the curated C. difficile database (https://doi.org/10.12688/wellcomeopenres.14826.1; Jolley et al. 2018). To ascertain the phylogenetic relationships between the new isolates and strains from different global locations and relevant additional C. difficile strains, we chose the genomes of 78 C. difficile strains that are publicly available on Enterobase (http://enterobase.warwick.ac.uk) and NCBI based on their strain types, diverse sources, and geographic locations (Supplementary Table S2). A maximum likelihood tree was constructed using PhyML (Guindon et al. 2010) as described previously (Didelot and Wilson 2015). Recombination was accounted for using ClonalFrameML (Didelot and Wilson 2015). The tree was visualized in iTOL software v6.4.3 (Letunic and Bork 2019).
Prophage carriage prediction
Predictions of prophage encoded in the genomes of the 11 sequenced strains were determined using PHASTER (PHAge Search Tool Enhanced Release; using default parameters; Zhou et al. 2011, Arndt et al. 2019). Prophages were detected by querying of contigs against viral and prophage databases in Genbank. PHASTER hits were automatically classified into intact (score > 90), questionable (score 70–90), and incomplete (score < 70) prophages based on their sizes, similarity to known phages, and the presence of phage-like and phage cornerstone genes (e.g. ‘capsid’, ‘head’, ‘plate’, ‘tail’, ‘coat’, ‘portal’, and ‘holin’; Greenrod et al. 2022).
CRISPR arrays prediction
To establish the diversity of the CRISPR-Cas system within the genomes of the isolates, array prediction was conducted using PILERC-CR 1.06 with default settings (Edgar 2007, Ekseth et al. 2013). Direct repeat (DR) sequences were aligned in the Clustal Omega (Sievers et al. 2011) to establish consensus sequences and viewed with Jalview v2 (Waterhouse et al. 2009). The webserver PADLOC was used to determine the CRISPR-Cas system types within the genomes of the isolates based on profile Hidden Markov Models (Payne et al. 2021). Identified spacers were searched against Genbank and NCBI nucleotide BLAST and RefSeq-Plasmid databases to identify a possible extrachromosomal origin using the CRISPRTarget tool (Biswas et al. 2013). The default values used by NCBI BLASTn for short sequences, <30 bases (defaults for long sequences are in brackets) are: gap open −5(−5), gap extend −2(−2), match + 1(+1), mismatch −10(−10), minimum score 30 (Biswas et al. 2013).
Results
Clostridioides difficile was isolated from four of the seven sampling sites
Of the seven sites sampled, only four (Dokan, Jalee, Hamamok, and Chnarok) yielded C. difficile, of which 31 isolates were recovered from these samples (Table 1, Supplementary Table S1, Fig. S1). We did not isolate C. difficile from Taq Taq river (one soil and one sediment samples), Safien mountain (two soil samples), and Haibat sultan mountain (one soil sample) despite sampling Safien mountain twice in the summer and winter of 2012 and 2013, respectively.
Table 1.
RT designation and toxin gene carriage of isolates examined in this study.
| RT | Isolates | Site | Sample | Genotype | ST | Clade |
|---|---|---|---|---|---|---|
| 604 | K3 | Hamamok | Soil | A¯B¯CDT¯ | ND | ND |
| K1 | Hamamok | Soil | A¯B¯CDT¯ | ND | ND | |
| K6 | Hamamok | Soil | A¯B¯CDT¯ | ND | ND | |
| K7 | Hamamok | Soil | A¯B¯CDT¯ | ND | ND | |
| K9 | Hamamok | Soil | A¯B¯CDT¯ | ND | ND | |
| K10 | Hamamok | Soil | A¯B¯CDT¯ | ND | ND | |
| 7F | Hamamok | Soil | A¯B¯CDT¯ | ND | ND | |
| CD105KSO7 | Hamamok | Soil | A¯B¯CDT¯ | 177 | C-I | |
| CD105KSO8 | Hamamok | Soil | A¯B¯CDT¯ | 181 | ||
| 091 | DF6 | Dokan | Sediment | A¯B¯CDT¯ | ND | ND |
| DF7 | Dokan | Sediment | A¯B¯CDT¯ | ND | ND | |
| DF10 | Dokan | Sediment | A¯B¯CDT¯ | ND | ND | |
| DF11 | Dokan | Sediment | A¯B¯CDT¯ | ND | ND | |
| DF4 | Dokan | Sediment | A¯B¯CDT¯ | ND | ND | |
| CD105KSE1 | Dokan | Sediment | A¯B¯CDT¯ | 107 | 1 | |
| CD105KSE2 | Dokan | Sediment | A¯B¯CDT¯ | 107 | ||
| CD105KSO10 | Chnarok | Soil | A¯B¯CDT¯ | 107 | ||
| 001 | F1 | Dokan | Sediment | A + B + CDT¯ | ND | ND |
| F5 | Dokan | Sediment | A + B + CDT¯ | ND | ND | |
| F7 | Dokan | Sediment | A + B + CDT¯ | ND | ND | |
| CD105KSE3 | Dokan | Sediment | A + B + CDT¯ | 3 | 1 | |
| CD105KSE4 | Dokan | Sediment | A + B + CDT¯ | 3 | ||
| 010 | CD105KSE9 | Hamamok | Sediment | A¯B¯CDT¯ | 15 | 1 |
| 7M | Hamamok | Sediment | A¯B¯CDT¯ | ND | ND | |
| 2M | Hamamok | Sediment | A¯B¯CDT¯ | ND | ND | |
| 12M | Hamamok | Sediment | A¯B¯CDT¯ | ND | ND | |
| 6M | Hamamok | Sediment | A¯B¯CDT¯ | ND | ND | |
| 011 | F9 | Dokan | Sediment | A + B + CDT¯ | ND | ND |
| CD105KSE6 | Jalee | Sediment | A¯B¯ CDT¯ | 136 | 1 | |
| 035 | CD105KSE5 | Jalee | Sediment | A¯B¯CDT¯ | 107 | 1 |
| CD105KSE11 | Jalee | Sediment | A¯B¯CDT¯ | 107 |
The RT designation was ascertained using capillary ribotyping targeting the 16S-23S rRNA intergenic spacer. Toxin profiles were determined using PCR to amplify the partial and essential repeat regions of toxin A genes, toxin B, and binary toxin genes.
Diversity of the isolates based on ribotypes and toxin genes carriage
Six RTs: RT001 (five isolates), RT010 (five isolates), RT011 (two isolates), RT035 (two isolates), RT091 (eight isolates), and RT604 (nine isolates) were identified. Although the study only examined a small number of isolates, diverse RTs both the sites and within specific samples were observed (Table 1, Supplementary Table S1, Fig. S1). Characterizing the isolates based on the presence or absence of C. difficile toxin genes showed that of the 31 isolates, 25 (81%) were negative for both tcdA and tcdB (A¯B¯) genes, while the remaining six (19%) isolates encode the toxigenic (A+ B+) genes. The RT011 isolates from Dokan (F9) and Jalee (CD105KSE6) had contrasting toxin profiles with the latter being toxin negative, while the F9 strain encodes both toxin genes tcdA and tcdB (Table 1). Furthermore, all the strains were binary toxin-negative (CDT¯; Table 1). PCR amplicons were sequenced and shown to match the genome data.
Diverse MLST profiles exist among the strains
To gain a detailed understanding of the genome characteristics of the representative RT isolates, a total of eleven isolates representing RT091 (three isolates), RT001, RT035, RT604 (two isolates each), and one isolate each from RT010 and RT011 were sequenced. The assembled genomes ranged from 49 to 458 contigs and 2594–2823 open reading frames per genome (as of February 2023; Supplementary Table S3), and their completeness and contamination level are shown in Supplementary Table S4.
Six distinct ST (ST-3, ST-15, ST-107, ST-137, ST-177, and ST-181) and seven RT/ST associations were identified from the isolates (Table 2). Five of the RT/ST associations (RT011/ST-137, RT035/ST-107, RT091/ST-107, RT604/ST-177, and RT604/ST-181) are novel, and the latter three are linked to RTs previously uncharacterized by MLST (Griffiths et al. 2010, Stabler et al. 2012, Dingle et al. 2014, Gawlik et al. 2015). The other two of the seven associations, RT001/ST-3 and RT010/ST-15, identified here were previously described among UK isolates (Stabler et al. 2012, Janezic and Rupnik 2015). Multiple STs, such as ST-177 and ST-181 were uniquely associated with RT604 (Table 2). In contrast, two RTs, RT091 and RT035, were associated with a single ST, ST-107, as shown in Table 2.
Table 2.
Characteristic of allelic profiles (STs) of the 11 strains of C. difficile isolated in this study, and ST/RT associations.
| MLST alleles | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolates | RT | ST | Clade | adk | atpA | dxr | glyA | recA | sodA | tpi |
| CD105KSE1 | 091 | 107 | 1 | 4 | 1 | 6 | 1 | 3 | 1 | 1 |
| CD105KSE2 | 091 | 107 | 1 | 4 | 1 | 6 | 1 | 3 | 1 | 1 |
| CD105KSE3 | 001 | 3 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| CD105KSE4 | 001 | 3 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
| CD105KSE5 | 035 | 107 | 1 | 4 | 1 | 6 | 1 | 3 | 1 | 1 |
| CD105KSE6 | 011 | 137 | 1 | 1 | 1 | 2 | 3 | 1 | 3 | 1 |
| CD105KS07 | 604 | 177 | C-I | 13 | 18 | 22 | 33 | 18 | 31 | 28 |
| CD105KS08 | 604 | 181 | C-I | 13 | 18 | 22 | 31 | 18 | 31 | 26 |
| CD105KSE9 | 010 | 15 | 1 | 1 | 1 | 6 | 1 | 8 | 5 | 1 |
| CD105KSO10 | 091 | 107 | 1 | 1 | 4 | 1 | 6 | 1 | 3 | 1 |
| CD105KSE11 | 035 | 107 | 1 | 1 | 4 | 1 | 6 | 1 | 3 | 1 |
Five novel RT/ST associations: 011/137, 035/107, 091/107, 604/ST177, and 604/181 are reported in this study.
Phylogenetic relationships based on core genome
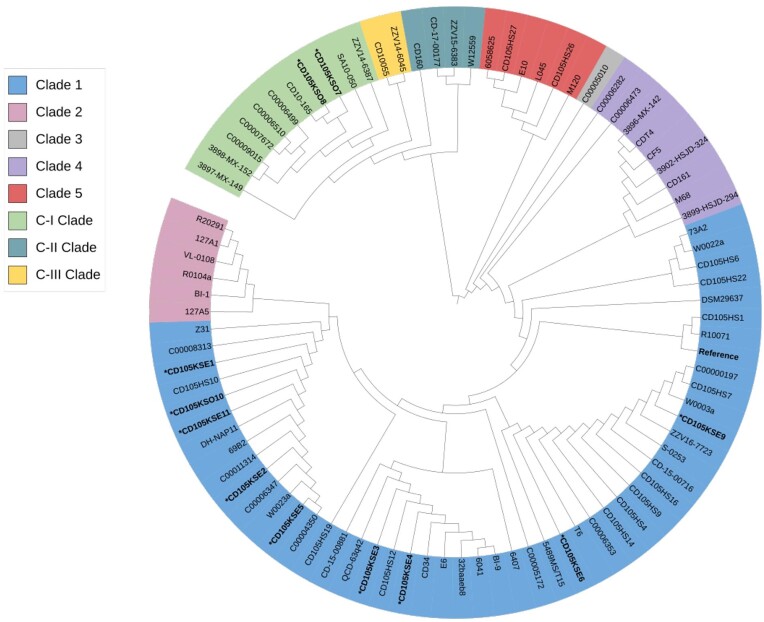
We explored the phylogenetic relationships and diversity of our eleven sequenced strains in the context of other 78 publicly available C. difficile genomes from diverse geographical regions comprising of 28 RTs and 29 strain types of the known eight clades (1, 2, 3, 4, 5, C-I, C-II, and C-III; Supplementary Table S2). Phylogenetic analysis based on whole-genome alignment revealed eight discrete (Clades 1, 2, 3, 4, and 5) and the three previously observed deeply branching clades (Clades C-I, C-II, and C-III; Squire et al. 2015, Ramírez-Vargas et al. 2018, Knight et al. 2021; Fig. 1).
Figure 1.
Phylogenetic tree showing five clades (1, 2, 3, 4, and 5) and three cryptic clades (C-I, C-II, and C-III) of C. difficile isolates examined based on core genome comparison. Maximum likelihood tree was constructed based on the core genes of 11 strains examined in this study (*) and 78 other reference C. difficile using PhyML as described previously. Single nucleotide polymorphisms (SNPs) in core genes were utilized for the phylogeny, and recombination was accounted for with clonalframeML. Tree visualized in an iTOL software v6.4.3.
Consistent with other findings, Clade 1 is the most diverse comprising of seventeen RTs, sixteen STs, and includes toxigenic and non-toxigenic isolates (Supplementary Table S2; Janezic and Rupnik 2015, Janezic et al. 2016). Nine of the isolates characterized in this study (CD105KSE1, CD105KSE2, CD105KSE3, CD105KSE4, CD105KSE5, CD105KSE6, CD105KSE9, CD105KSO10, and CD105KSE11) belonged to Clade 1. Clostridioides difficile strains CD105KSE3, CD105KSE4, CD105KSE5, and CD105KSE11 closely cluster with the other strains of the same RT. However, CD105KSE6 (RT035) clusters distantly from strains of the similar RT.
The Clade C-I, reclassified as a novel independent Clostridioides genomospecies with C-II and C-II clades comprised of RT206, RT289, RT290, RT127, and RT604 isolates, in addition to six other isolates. The RT604 isolates from this study (CD105KSO7 and CD105KSO8) are new additions to the C-I Clade and the only strains from environmental source with the rest being of clinical origin within this clade (Fig. 1).
Multiple prophage carriage detected in environmental strains of C. difficile
We explored the genomes of the isolates and multiple intact and partial prophages were detected within the genomes of the strains (Supplementary Table S5). The size of the intact prophages ranged from 20.6 to 137.9 kb, while the incomplete prophages ranged from 6.8 to 62.1 kb (SupplementaryTable S5). Two intact prophages were identified in CD105KSE1, CD105KSE2, and CD105KSO10, while three intact prophages were found in CD105KSE3, CD105KSE4, CD105KSE5, CD105KSO7, and CD105KSE11. Strains CD105KSE9, CD105KSO8, and CD105KSSE6 had five, four, and one intact prophages predicted in their genomes, respectively. Further analysis of all the predicted regions of the intact prophages in the isolates using BLAST showed similarity to other C. difficile phages (Supplementary Table S5).
CRISPR-Cas system diversity in environmental C. difficile strains
The genomes of the eleven strains were also screened for the presence of CRISPR-Cas systems and found to encode multiple CRISPR arrays, ranging from three to twelve per genome (Fig. 2), with a variable number of DR (average length ∼29 bp) separated by variable spacer contents, ranging from 45 to 112 per strain. A total of 97 DRs were extracted from the CRISPR arrays of the 11 strains, and 22 different DR consensus sequences were identified. Of these, six consensus DRs were unique (Supplementary Fig. S2). SNPs and identical DR sequences are also observed within the arrays of multiple strains (Supplementary Fig. S2). The observed variation is possibly expected assuming how widespread the system is (Rath et al. 2015).
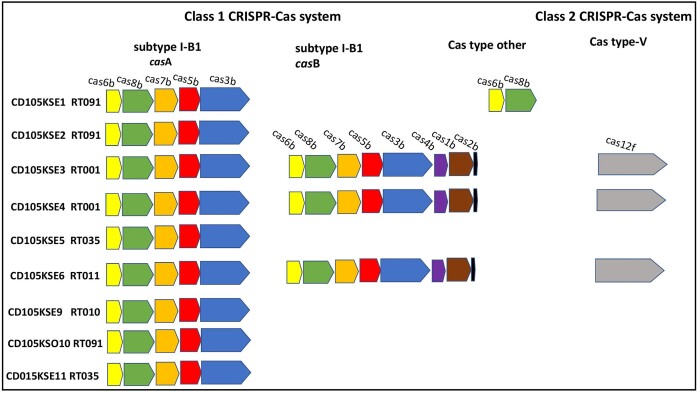
Figure 2.
Schematic diagram showing the CRISPR-Cas systems carried by environmental C. difficile isolates examined in this study. Typical operon organization is shown for each CRISPR-Cas system. Class 1 subtype I-B1 Cas system identified in 81.8% of the genomes code for the two mainly conserved clusters of cas genes that identified in this type. Cluster casA that encodes partial cas gene set (casb6, casb8, casb7, casb5, and casb3), and cluster casB codes for a complete set of subtype I-B1 cas genes (casb6, casb8, casb7, casb5, and casb3) and (casb1, casb2, and casb4). Class 2 V-type Cas system founded in 27.3% of the genomes encode for a single large effector protein (cas12f). Cas type other founded in only 9% of the genomes has only two genes homologous to cas6b and cas8b. Homologous cas genes are shown with coloured arrows. Colour coding is the same for homologous cas genes.
CRISPR-Cas systems were defined within the genomes of C. difficile strains, two known classes of CRISPR-Cas systems (class 1 subtype I-B1 and class 2 type V CRISPR-Cas systems), and a CRISPR-Cas type that has two genes homologous to cas6b and cas8b only were identified (Fig. 2; Payne et al. 2021). Class 1 subtype I-B1 CRISPR-Cas system is described by multi-subunit protein effectors and was previously observed in all queried genomes of C. difficile strains (Hargreaves et al. 2014, Boudry et al. 2015, Andersen et al. 2016, Maikova et al. 2018, Maikova et al. 2019). Class 2 type V CRISPR-Cas systems possess single, large protein effectors (Makarova et al. 2015), observed in several bacterial genomes (Schunder et al. 2013, Vestergaard et al. 2014), and also known as genome editing system that comprises of crRNA and Cas12a protein (Liu et al. 2020). No cas genes were identified in the genome of RT604 strains CD105KSO7 and CD105KSO8. This lack of cas genes could be due to the deletion through horizontal gene transfer resulting in several independent deletions of the complete set of cas genes as shown in enterococcal strains (Palmer and Gilmore 2010).
In the subtype I-B1 CRISPR-Cas system, the two mainly conserved clusters of cas genes were identified, consistent with an earlier report (Andersen et al. 2016). The first cas gene cluster, termed casA, encodes a partial cas gene set (casb6, casb8, casb7, casb5, and casb3) lacking casb1, casb2, and casb4 and was identified in 81.8% of the genomes. The second cas gene cluster is casB, which encodes a complete set of subtype I-B1 cas genes (casb6, casb8, casb7, casb5, and casb3) as well as (casb4, casb1, and casb2) identified in 27.3% of sequenced strains (Andersen et al. 2016, Maikova et al. 2018; Fig. 2). Cas operons incidence was found to be associated with the RT profiles; for example, strains of RT091, RT035 and RT010 have similar cas gene clusters, casA, that encode a partial cas gene set (Fig. 2). Class 2 V-type Cas system with a single large effector protein with 1380 amino acid lengths (Cas12f) has been found in 27.3% of the studied genomes (Pyzocha and Chen 2018, Xiao et al. 2020). Diversity was observed within the strains based on the multiple CRISPR-Cas types; 36.4% of the strains encode two different types of CRISPR-Cas systems within a single genome. For example, both strains of RT001 and strain of RT010 have class 1 subtype I-B1 and class 2 type V CRISPR-Cas systems. Interestingly, the RT091 strain, CD105KSE1, encodes subtype-I-B1 with a casA gene cluster and another CRISPR-Cas type with an unknown Cas-type that has only two cas genes encoded for casb6 and casb8 (Fig. 2).
CRISPR spacers homology among the C. difficile strains
To determine if the CRISPR-Cas systems of the 11 characterized strains could target known phages, the spacers of the arrays within the genomes of the strains were searched against Genbank and BLAST nucleotide databases and RefSeq-Plasmid databases using the CRISPRTarget tool (Biswas et al. 2013). In total, 1054 spacers were identified from the genome of our strains, of which 185 were identical to other published C. difficile phages and plasmid sequences from a diverse range of geographical locations, and 869 spacers were novel (Supplementary Table S6). From the 185 identical spacers, 118 spacers were identical to other published C. difficile phages, 67 spacers identical to plasmid sequences (as of March 2023; Fig. 3, Supplementary Table S6). Strains of RT001, RT091, and RT604 share a similar spacer sequence identity consistent with their evolutionary relationships (Boudry et al. 2015). Both RT604 isolates have the lowest number of spacers (45 spacers), and both strains have three CRISPR arrays with the same number of spacers in each array. However, only two spacers from the arrays of both strains have sequences similar to other published C. difficile phages. This suggests that the majority of the spacers might be derived from unknown phages that have yet to be isolated or characterized. Spacer numbers 13 and 39 in CD105KSO7 have sequences similar to spacer numbers 40 and 22 of strain CD105KSO8, respectively. We have observed conserved numbers of arrays and spacers among the three strains of RT091, but strain CD105KSE2 has only one extra array with four spacers (Fig. 3, Supplementary Table S6). Whilst RT035 strains shared some common spacers, CD105KSE5 lacked the spacer for phiCD146, suggesting that dynamic changes in the CRISPR array content had occurred, possibly through interactions with foreign DNA elements (Hargreaves et al. 2014). It was observed that more than one CRISPR spacer within a strain from all RTs targeted the same phage. For example, two spacers (31, 73) from different CRISPR arrays from strain CD105KSE1 targeted phiCD27, signifying constant interactions between this strain and the corresponding phage (Boudry et al. 2015). Some of the strains carry multiple spacers for the same phage, such as CD105KSE3, which has spacer 12 and spacer 79 showing identical matches to phiCD146. Spacers for phiCDHM19 were only observed in two strains, CD105KSE5 and CD105KSE11, suggesting a less widespread predicted immunity of this strain to this phage (Mayer et al. 2008, Horgan et al. 2010, Meessen-Pinard et al. 2012, Sekulovic et al. 2014; Fig. 3). The shared spacers could also imply probable hot spots of phage genome evolution loci in which bacterial strains are more exposed to these phages, which have counter-evolved through infections.
Figure 3.
CRISPR arrays and the corresponding identical spacers encoded in C. difficile isolates examined in this study. Identical sequences between spacers and phage sequences are indicated by matching colours; each colour represents the similarity with a particular C. difficile phage (see legend). Coloured spacers with numbers correspond to multiple of protospacers that match C. difficile phages and plasmids sequences as shown in Supplementary Table S6. ( refers to the spacers that match to plasmid and C. difficile phage sequences, the numbers refer to the number of matched plasmid and C. difficile phage sequences,
refers to the spacers that match to plasmid and C. difficile phage sequences, the numbers refer to the number of matched plasmid and C. difficile phage sequences,  refers to the spacers that match to C. difficile phage sequences, the numbers refer to the number of matched phages, and
refers to the spacers that match to C. difficile phage sequences, the numbers refer to the number of matched phages, and  refers to spacers that target multiple plasmid sequences, the numbers refer to the number of matched plasmids). White boxes represent spacers that have no homology to any C. difficile phages.
refers to spacers that target multiple plasmid sequences, the numbers refer to the number of matched plasmids). White boxes represent spacers that have no homology to any C. difficile phages.
Discussion
The paucity of information on environmental C. difficile strains in the Middle East compared to strains from western countries reflects the lack of study in this area. Typically, this pathogen has been considered to be a problem in the western world, and thus, it has not been a priority in other places. However, new geographical areas such as Slovenia and Thailand are beginning to explore this pathogen from different environmental sources to understand the diversity that exists amongst the strains (Janezic et al. 2016, Putsathit et al. 2017, Imwattana et al. 2020, Tkalec et al. 2020).
The work conducted in Slovenia reported C. difficile prevalence from puddle water (14.4%, n = 104), soil (36.7%, n = 79), meat (3.6%, n = 336), and raw vegetables (6.1%, n = 98). Majority of the isolates identified represent RTs (such as RT002, RT005, RT010, RT014/020, RT015, and RT023), which may suggest their association with humans and animals (Janezic et al. 2016, Tkalec et al. 2020). The studies from Thailand focussed on clinical settings and reported prevalence of 9.2% and 15.6% (n = 422) of toxigenic and nontoxigenic isolates, respectively, from the samples (Imwattana et al. 2020, Tkalec et al. 2020).
The very limited studies on this pathogen in the Middle East have focused on characterizing isolates from clinical/hospitals sources (Khalil et al. 2019, Shoaei et al. 2019, Al-Tawfiq et al. 2020, Azimirad et al. 2020, Baghani et al. 2020, Williamson et al. 2022), raw meat (Esfandiari et al. 2014, Bakri 2018, Ersoz Seyma and Cosansu 2018), food products (Rahimi et al. 2015, Bakri 2016), and supermarket environments (Sadeghifard et al. 2010, Shoaei et al. 2019). In the Middle East region, the reported prevalence rates of CDI are 23.8% in Jordan, 8%–10% in Kuwait, and 5.15% in Saudi Arabia (Alzouby et al. 2020). However, there are no surveillance strategies to show the occurrence of CDI in northern Iraq. The lack of information on the strains that are found in the region’s natural environment, or potentially transmitted by human and animal activities may greatly affect the control of this infection in this region and the world at large. Also, environmental C. difficile strains have been reported to encode several genetic elements that could contribute to the emergence of novel clinical strains in hospitals, as previously reported (Hargreaves et al. 2015).
We previously reported the genome characteristic of three novel species of Clostridia from the natural habitats of this region, in which all three isolates encode multiple prophage elements and the CRISPR Cas-system was found in two of the isolates (Rashid et al. 2016). Here, we went further to isolate and characterize C. difficile isolates from river sediments and soils in northern Iraq for further work in this area.
In the current study, we isolated C. difficile from the sediment and soil samples of four of the seven examined sites in northern Iraq. This indicates that these sources are important habitats from which to study C. difficile presence and diversity, which concurs with previous work conducted in our laboratory and elsewhere (al Saif and Brazier 1996, Hargreaves et al. 2013, Janezic et al. 2016, Rodriguez et al. 2019, Williamson et al. 2022). Again, consistent with previous work, here, the highest number of isolates that yielded C. difficile (21/31 isolates, ∼68%) were from sediment samples. This may be attributed to the dormant spores, which protect the bacteria and therefore may contribute to the transmission and persistence of C. difficile in the marine ecosystem (Zidaric et al. 2010, Hargreaves et al. 2013, Xu et al. 2014).
Despite the small sample size, we had good recovery rates of C. difficile, which we isolated from four of the seven examined sites (∼60%), suggesting it is abundant in the types of areas. This rate of C. difficile isolation is comparable to a previous study, in which were 54% and 60% recovery were observed in two consecutive years (Hargreaves et al. 2013). However, one other study was unable to detect C. difficile from sediment samples (Pasquale et al. 2011), and others detected only 24.0% from environmental samples (Janezic et al. 2016). The bacterial recovery observed in our study may also be attributed to enrichment procedures carried out on the samples before isolation, which greatly enhanced the isolation of the bacterium (Hargreaves et al. 2013).
Both environmentally associated RTs (RT010, RT035, RT091, and RT604) isolates and those associated with an important clinical RT (RT001, RT011) were detected in our sample sites, which concurs with previous studies (Hargreaves et al. 2013, Hargreaves et al. 2016, Janezic et al. 2016). Whilst RT001, RT010, and RT035 have previously been isolated from the environments of Europe, all studies conducted in the Middle East have associated these RTs with clinical samples (Al-Tawfiq and Abed 2010, Hargreaves et al. 2013, Al-Thani et al. 2014, Azimirad et al. 2020, Baghani et al. 2020). To our knowledge, this is the first time that these RTs have been found to be associated with the environmental sources in these parts of the country. This may be linked to human activities such as recreational activities in all the sample areas and agricultural runoff found in Dokan. In contrast, none of the environmental RT strains reported by other researchers from the Middle East were isolated in this study. This may be attributed to the fact that some of the previously reported strains were not ribotyped, hence their identities are unknown (Jamal et al. 2002, Rotimi et al. 2003, Rahimi et al. 2015). In addition, pathogenic toxin strains that are associated with community-acquired infections were isolated from retail surfaces, which enhanced the need to understand their medical impact and to enact any necessary preventative measures (Alqumber 2014). We found uncommon RTs, RT604 and RT091 from the West to be prevalent in the examined areas at the time of our sampling. In contrast, RT010, which is common in both Europe and the United Kingdom, was found to be rare in the region examined (Rotimi et al. 2003, Al-Thani et al. 2014, Baghani et al. 2020). This suggests that certain C. difficile strains are more prevalent in certain regions of the world than others are.
The isolation of both toxigenic and nontoxigenic isolates in this study is consistent with previous studies (Rotimi et al. 2003, Hargreaves et al. 2013, Janezic et al. 2016). The diverse toxin gene profiles observed within a RT show that the pathogenicity locus is variable and may not be a feature of clonality, and could be readily lost (Dingle et al. 2014). We did not isolate any binary toxin-positive environmental isolates. However, RT078 and RT027 isolates, which encode for a binary toxin, have previously been identified in environmental samples from England and Saudi Arabia and may be attributed to human or animal activities (Hargreaves et al. 2013, Bakri 2016).
The isolation of six RTs that are associated with six MLST profiles also concurs with previous studies that showed that although MLST are normally associated with a specific RT, but may not always predict the strain types and vice versa (Griffiths et al. 2010, Wang et al. 2018, Zhao et al. 2021). Multiple RTs RT091 and RT0235 are associated with ST-107, and multiple STs ST-181 and ST-177 are related to RT604. The associations of RTs with multiple STs or vice versa have previously been reported and may suggest the constant divergent nature of C. difficile genomes (Dingle et al. 2011, Stabler et al. 2012, Janezic and Rupnik 2015, Janezic et al. 2016, Knight et al. 2017). Phylogenetic analysis of these strains isolated in this study identified a lineage (Clade C-I) that is highly divergent from the other five established clades. In line with earlier studies, Clade 1 is diverse in term of RTs and STs, and comprised of both toxigenic and nontoxigenic strains (Stabler et al. 2012, Janezic and Rupnik 2015, Janezic et al. 2016, Ramírez-Vargas et al. 2018).
The occurrence of multiple and diverse prophage carriage within C. difficile is high and has been previously isolated from environmental strains (Shan et al. 2012, Hargreaves et al. 2013, Hargreaves et al. 2015, Mullany et al. 2015). Here, we detected up to six intact prophages in a single C. difficile genome, and this complex network of prophages within environmental strains could contribute to the evolution of new pathogenic strains. Further work is required to ascertain if all the six prophages are inducible, as observed in previous work (Fortier and Sekulovic 2013, Hargreaves et al. 2015).
Evidence of the interplay between hosts and phages can be seen from the CRISPR arrays detected here. The CRISPR-Cas system is a form of adaptive immunity that bacteria use to resist phage infection (Hargreaves et al. 2014, Maikova et al. 2018). Our results show that this system is diverse within our strains (Soutourina et al. 2013, Hargreaves et al. 2014, Hargreaves et al. 2016). Here, we showed for the first time the prevalence of the class 2 type V CRISPR-Cas system in C. difficile strains. To date, class 1 subtype I-B is the native CRISPR-Cas system in C. difficile (Boudry et al. 2015, Maikova et al. 2018, Maikova et al. 2019). Both strains of RT001 possess a class 2 type V CRISPR-Cas system present within the mobile genetic region in both strains, this perhaps had been acquired via horizontal gene transfer as studies proposed that class 2 effectors originated from nuclease encoded by different mobile genetic elements (MGE; Koonin and Makarova 2019). We have also reported the presence of two or more CRISPR-Cas types within the genome of a single strain; 27.3% of the sequenced strains carry class 2 type V CRISPR-Cas systems beside the native subtype I-B CRISPR-Cas systems (Fig. 2). Multiple CRISPR-Cas systems have been found in some organisms that occur naturally (Carte et al. 2014). Consistent with the earlier reports, both cas gene sets (casA and casB) of the I-B subtype were found within the sequenced strain (Boudry et al. 2015, Maikova et al. 2018). The occurrence of cas operons found to be associated with the RT profiles. The variation of the CRISPR-Cas system types and the contents within RT strains could affect their susceptibility to infection by phages (Hargreaves et al. 2014). The spacer contents of CRISPR arrays are identical to known phage sequences and are particularly insightful since it was previously shown that 100% identity between spacer and proto-spacer sequences is required to provide immunity (Boudry et al. 2015, Maikova et al. 2018, Deem 2020). Although small numbers of mismatches could confer a degree of immunity during infection through target cleavage (Michael et al. 2022). Our data are in line with earlier studies and supports the potential role of phages to drive the evolution of epidemic strains (Hargreaves and Clokie 2014).
Conclusions
To conclude, C. difficile strains were found to be present in the natural environment of northern Iraq and were readily isolated from 57% samples obtained. Genome analysis showed that these strains are diverse and distinct from those found elsewhere, and as is seen in all C. difficile genomes, these strains had multiple prophage carriages with diverse CRISPR-Cas system types that have arrays containing diverse spacers. We have showed for the first time instance of the class 2 type V CRISPR-Cas system in C. difficile strains that has been described in other bacterial genomes. Although this was a small-scale study, the observations of RT and genome diversity in this region would provide an overall understanding of the diversity of this organism. Studies in new geographies will further reveal insights into how this pathogen can evolve and increase our understanding on the relationship between strains observed in patients and those found in the environment.
Supplementary Material
Contributor Information
Srwa J Rashid, Medical Laboratory Technology Department, Koya Technical Institute, Erbil Polytechnic University, Erbil, Kurdistan, Iraq.
Janet Y Nale, Centre for Epidemiology and Planetary Health, Scotland’s Rural College, Inverness IV2 5NA, UK.
Andrew D Millard, Department of Genetics and Genome Biology, College of Life Sciences, University of Leicester, University Road, Leicester LE1 7RH, UK.
Martha R J Clokie, Department of Genetics and Genome Biology, College of Life Sciences, University of Leicester, University Road, Leicester LE1 7RH, UK.
Funding
This project was supported by Human Capacity Development Program (HCDP) sponsored by Kurdistan Regional Government-Iraq. M. R.J.C.’s work was supported by an MRC New Investigator Research Grant [G0700855]. The funders had no role in study design, data collection, and analysis or preparation of the manuscript.
Conflicts of interest
No conflict of interest exists.
References
- Al Saif N, Brazier JS. The distribution of Clostridium difficile in the environment of South Wales. J Med Microbiol. 1996;45:133–7. [DOI] [PubMed] [Google Scholar]
- Al-Tawfiq JA, Abed MS. Clostridium difficile-associated disease among patients in Dhahran, Saudi Arabia. Travel Med Infect Dis. 2010;8:373–6. [DOI] [PubMed] [Google Scholar]
- Al-Tawfiq JA, Rabaan AA, Bazzi AMet al. Clostridioides (Clostridium) difficile-associated disease: epidemiology among patients in a general hospital in Saudi Arabia. Am J Infect Control. 2020;48:1152–7. [DOI] [PubMed] [Google Scholar]
- Al-Thani AA, Hamdi WS, Al-Ansari NAet al. Polymerase chain reaction ribotyping of Clostridium difficile isolates in Qatar: a hospital-based study. BMC Infect Dis. 2014;14:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqumber MA. Clostridium difficile in retail baskets, trolleys, conveyor belts, and plastic bags in Saudi Arabia. Saudi Med J. 2014;35:1274–7. [PMC free article] [PubMed] [Google Scholar]
- Alzouby S, Baig K, Alrabiah Fet al. Clostridioides difficile infection: incidence and risk factors in a tertiary care facility in Riyadh, Saudi Arabia. J Infect Public Health. 2020;13:1012–7. [DOI] [PubMed] [Google Scholar]
- Andersen JM, Shoup M, Robinson Cet al. CRISPR diversity and microevolution in Clostridium difficile. Genome Biol Evol. 2016;8:2841–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D, Marcu A, Liang Yet al. PHAST, PHASTER and PHASTEST: tools for finding prophage in bacterial genomes. Brief Bioinf. 2019;20:1560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimirad M, Krutova M, Yadegar Aet al. Clostridioides difficile ribotypes 001 and 126 were predominant in Tehran healthcare settings from 2004 to 2018: a 14-year-long cross-sectional study. Emerg Microbes Infect. 2020;9:1432–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghani A, Mesdaghinia A, Kuijper EJet al. High prevalence of Clostridiodes diffiicle PCR ribotypes 001 and 126 in Iran. Sci Rep. 2020;10:4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakri M. Prevalence of Clostridium difficile in raw cow, sheep, and goat meat in Jazan, Saudi Arabia. Saudi J Biol Sci. 2018;25:783–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakri MM. Investigating the presence of Clostridium difficile in vegetables in Jazan markets, Saudi Arabia. Sky J Microbiol Res. 2016;4:060–4. [Google Scholar]
- Barroso LA, Wang SZ, Phelps CJet al. Nucleotide sequence of Clostridium difficile toxin B gene. Nucl Acids Res. 1990;18:4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas A, Gagnon JN, Brouns SJJet al. CRISPRTarget. RNA Biol. 2013;10:817–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry P, Semenova E, Monot Met al. Function of the CRISPR-Cas system of the human pathogen Clostridium difficile. mBio. 2015;6:e01112–01115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Christopher RT, Smith JTet al. The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol Microbiol. 2014;93:98–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Raval IH. Chapter 32–pathogenic microbial genetic diversity with reference to health. In: Das S, Dash HR (eds.), Microbial Diversity in the Genomic Era. Cambridge, MA, USA: Academic Press, 2019, 559–77. [Google Scholar]
- Czepiel J, Dróżdż M, Pituch Het al. Clostridium difficile infection: review. Eur J Clin Microbiol Infect Dis. 2019;38:1211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FRet al. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deem MW. CRISPR recognizes as many phage types as possible without overwhelming the Cas machinery. P Natl Acad Sci USA. 2020;117:7550–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmée M. Laboratory diagnosis of Clostridium difficile disease. Clin Microbiol Infect. 2001;7:411–6. [DOI] [PubMed] [Google Scholar]
- Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Didelot X, Quan TPet al. Effects of control interventions on Clostridium difficile infection in England: an observational study. Lancet Infect Dis. 2017;17:411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Elliott B, Robinson Eet al. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evolut. 2014;6:36–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, Griffiths D, Didelot Xet al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One. 2011;6:e19993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez SR, Anderson LJ, Kotter CVet al. Comparison of whole-genome sequencing and molecular-epidemiological techniques for Clostridium difficile strain typing. J Ped Infect Dis. 2016;5:329–32. [DOI] [PubMed] [Google Scholar]
- Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinf. 2007;8:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekseth OK, Kuiper M, Mironov V. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics. 2013;30:734–6. [DOI] [PubMed] [Google Scholar]
- Ersoz Seyma S, Cosansu S. Prevalence of Clostridium difficile isolated from beef and chicken meat products in Turkey. Korean J Food Sci Anim Resour. 2018;38:759–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari Z, Jalali M, Ezzatpanah Het al. Prevalence and characterization of Clostridium difficile in beef and mutton meats of Isfahan region, Iran. Jundishapur J Microbiol. 2014;7:e16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier L-C, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster NF, Riley TV. Improved recovery of Clostridium difficile spores with the incorporation of synthetic taurocholate in cycloserine-cefoxitin-fructose agar (CCFA). Pathology (Phila). 2012;44:354–6. [DOI] [PubMed] [Google Scholar]
- Gawlik D, Slickers P, Engelmann Iet al. DNA-microarray-based genotyping of Clostridium difficile. BMC Microbiol. 2015;15:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenrod STE, Stoycheva M, Elphinstone Jet al. Global diversity and distribution of prophages are lineage-specific within the Ralstonia solanacearum species complex. BMC Genomics. 2022;23:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths D, Fawley W, Kachrimanidou Met al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol. 2010;48:770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort Vet al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. [DOI] [PubMed] [Google Scholar]
- Hargreaves KR, Clokie MRJ. Clostridium difficile phages: still difficult?. Front Microbiol. 2014;5:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves KR, Colvin HV, Patel KVet al. Genetically diverse Clostridium difficile strains harboring abundant prophages in an estuarine environment. Appl Environ Microb. 2013;79:6236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves KR, Flores CO, Lawley TDet al. Abundant and diverse clustered regularly interspaced short palindromic repeat spacers in Clostridium difficile strains and prophages target multiple phage types within this pathogen. mBio. 2014;5:e01045–01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves KR, Otieno JR, Thanki Aet al. As clear as mud? Determining the diversity and prevalence of prophages in the draft genomes of estuarine isolates of Clostridium difficile. Genome Biol Evol. 2015;7:1842–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves KR, Thanki AM, Jose BRet al. Use of single molecule sequencing for comparative genomics of an environmental and a clinical isolate of Clostridium difficile ribotype 078. BMC Genomics. 2016;17:1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan M, O'Sullivan O, Coffey Aet al. Genome analysis of the Clostridium difficile phage PhiCD6356, a temperate phage of the Siphoviridae family. Gene. 2010;462:34–43. [DOI] [PubMed] [Google Scholar]
- Imwattana K, Putsathit P, Leepattarakit Tet al. Molecular epidemiology and clinical characteristics of Clostridium difficile infection in Thailand. Int J Infect Dis. 2020;101:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra A, Huhulescu S, Schneeweis Met al. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol. 2008;57:1377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal WY, Mokaddas EM, Verghese TLet al. In vitro activity of 15 antimicrobial agents against clinical isolates of Clostridium difficile in Kuwait. Int J Antimicrob Agents. 2002;20:270–4. [DOI] [PubMed] [Google Scholar]
- Janezic S, Potocnik M, Zidaric Vet al. Highly divergent Clostridium difficile strains isolated from the environment. PLoS One. 2016;11:e0167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janezic S, Rupnik M. Development and implementation of whole genome sequencing-based typing schemes for Clostridioides difficile. Front Public Health. 2019;7:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janezic S, Rupnik M. Genomic diversity of Clostridium difficile strains. Res Microbiol. 2015;166:353–60. [DOI] [PubMed] [Google Scholar]
- Janezic S, Smrke J, Rupnik M. Isolation of Clostridioides difficile from different outdoor sites in the domestic environment. Anaerobe. 2020;62:102183. [DOI] [PubMed] [Google Scholar]
- Jolley K, Bray J, Maiden M. Open-access bacterial population genomics: bIGSdb software, the PubMLST.Org website and their applications [version 1; peer review: 2 approved]. Wellcome Open Res. 2018; 3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Kato N, Watanabe Ket al. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol. 1998;36:2178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A, Hendaus MA, Elmagboul Eet al. Incidence of Clostridium difficile infection and associated risk factors among hospitalized children in Qatar. Ther Clin Risk Manag. 2019;15:1343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight DR, Imwattana K, Kullin Bet al. Major genetic discontinuity and novel toxigenic species in Clostridioides difficile taxonomy. eLife. 2021;10:e64325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight DR, Squire MM, Collins DAet al. Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front Microbiol. 2017;7:2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Phil Trans R Soc B. 2019;374:20180087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SC, Knight D, Riley TV. Clostridium difficile and one health. Clin Microbiol Infect. 2020;26:857–63. [DOI] [PubMed] [Google Scholar]
- Liu Z, Dong H, Cui Yet al. Application of different types of CRISPR/Cas-based systems in bacteria. Microb Cell Fact. 2020;19:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maikova A, Kreis V, Boutserin Aet al. Using an endogenous CRISPR-Cas system for genome editing in the human pathogen Clostridium difficile. Appl Environ Microb. 2019;85:e01416–01419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maikova A, Severinov K, Soutourina O. New insights into functions and possible applications of Clostridium difficile CRISPR-Cas system. Front Microbiol. 2018;9:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OSet al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Micro. 2015;13:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008;190:6734–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meessen-Pinard M, Sekulovic O, Fortier LC. Evidence of in vivo prophage induction during Clostridium difficile infection. Appl Environ Microb. 2012;78:7662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AS, Giang TN, Dipali GS. CRISPR-Cas effector specificity and target mismatches determine phage escape outcomes. Biorxiv. 2022, 10.1101/2022.05.30.494023 [DOI] [Google Scholar]
- Mullany P, Allan E, Roberts AP. Mobile genetic elements in Clostridium difficile and their role in genome function. Res Microbiol. 2015;166:361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M, Ríos-Chaparro DI, Patarroyo MAet al. Determining Clostridium difficile intra-taxa diversity by mining multilocus sequence typing databases. BMC Microbiol. 2017;17:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nale JY, Shan J, Hickenbotham PTet al. Diverse temperate bacteriophage carriage in Clostridium difficile 027 strains. PLoS One. 2012;7:e37263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nale JY, Spencer J, Hargreaves KRet al. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2016;60:968–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. mBio. 2010;1:e00227–00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquale V, Romano VJ, Rupnik Met al. Isolation and characterization of Clostridium difficile from shellfish and marine environments. Folia Microbiol. 2011;56:431–7. [DOI] [PubMed] [Google Scholar]
- Payne LJ, Todeschini TC, Wu Yet al. Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types. Nucleic Acids Res. 2021;49:10868–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putsathit P, Maneerattanaporn M, Piewngam Pet al. Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand. New Microbes New Infect. 2017;15:27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyzocha NK, Chen S. Diverse class 2 CRISPR-Cas effector proteins for genome engineering applications. ACS Chem Biol. 2018;13:347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi E, Afzali ZS, Baghbadorani ZT. Clostridium difficile in ready-to-eat foods in Isfahan and Shahrekord, Iran. Asian Pac J Trop Biomed. 2015;5:128–31. [Google Scholar]
- Ramírez-Vargas G, Goh S, Rodríguez C. The novel phages phiCD5763 and phiCD2955 represent two groups of big plasmidial siphoviridae phages of Clostridium difficile. Front Microbiol. 2018;9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid SRJ, Clokie MRJ, Millard AD. Draft genome sequences of three novel Clostridium isolates from Northern Iraq. Genome Announc. 2016;4:e00033–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D, Amlinger L, Rath Aet al. The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–28. [DOI] [PubMed] [Google Scholar]
- Rinttilä T, Kassinen A, Malinen Eet al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–77. [DOI] [PubMed] [Google Scholar]
- Rodriguez C, Bouchafa L, Soumillion Ket al. Seasonality of Clostridium difficile in the natural environment. Transbound Emerg Dis. 2019;66:2440–9. [DOI] [PubMed] [Google Scholar]
- Rodriguez Diaz C, Seyboldt C, Rupnik M. Non-human C. difficile reservoirs and sources: animals, food, environment. In: Rupnik M, Mastrantonio P (eds.), Updates on Clostridium difficile in Europe. Cham: Springer, 2018, 227–43. [DOI] [PubMed] [Google Scholar]
- Rotimi VO, Jamal WY, Mokaddas EMet al. Prevalent PCR ribotypes of clinical and environmental strains of Clostridium difficile isolated from intensive-therapy unit patients in Kuwait. J Med Microbiol. 2003;52:705–9. [DOI] [PubMed] [Google Scholar]
- Sadeghifard N, Salari M, Ranjbar Ret al. The clinical and environmental spread and diversity of toxigenic Clostridium difficile diarrhea in the region of the Middle East. Rev Infect. 2010;1:180–7. [Google Scholar]
- Schunder E, Rydzewski K, Grunow Ret al. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303:51–60. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Sekulovic O, Garneau JR, Néron Aet al. Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins. Appl Environ Microb. 2014;80:2555–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith HMB, Biggel M, Roloff Tet al. Transition from PCR-ribotyping to whole genome sequencing based typing of Clostridioides difficile. Front Cell Infect Microbiol. 2021;11:681518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Patel KV, Hickenbotham PTet al. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Appl Environ Microb. 2012;78:6027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaei P, Shojaei H, Khorvash Fet al. Molecular epidemiology of Clostridium difficile infection in Iranian hospitals. Antimicrob Resist Infect Control. 2019;8:12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen Det al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Monot M, Boudry Pet al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire MM, Knight DR, Riley TV. Community-acquired Clostridium difficile infection and Australian food animals. Microbiol Aust. 2015;36:111–3. [Google Scholar]
- Stabler RA, Dawson LF, Valiente Eet al. Macro and micro diversity of Clostridium difficile isolates from diverse sources and geographical locations. PLoS One. 2012;7:e31559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs S, Rupnik M, Gibert Met al. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol Lett. 2000;186:307–12. [DOI] [PubMed] [Google Scholar]
- Tkalec V, Jamnikar-Ciglenecki U, Rupnik Met al. Clostridioides difficile in national food surveillance, Slovenia, 2015 to 2017. Euro Surveill. 2020;25:1900479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uelze L, Grützke J, Borowiak Met al. Typing methods based on whole genome sequencing data. One Health Outlook. 2020;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard G, Garrett RA, Shah SA. CRISPR adaptive immune systems of archaea. RNA Biol. 2014;11:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Holzbauer S, Pung Ket al. 473. Molecular typing of Clostridium difficile: concordance between PCR-ribotyping and multilocus sequence typing (MLST). Open Forum Infect Dis. 2018;5:S176. [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMAet al. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson CHD, Stone NE, Nunnally AEet al. Identification of novel, cryptic Clostridioides species isolates from environmental samples collected from diverse geographical locations. Microbial Genomics. 2022;8:000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Li Z, Wang Set al. Structural basis for the dimerization-dependent CRISPR-Cas12f nuclease. Biorxiv. 2020;49:4120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Weese JS, Flemming Cet al. Fate of Clostridium difficile during wastewater treatment and incidence in Southern Ontario watersheds. J Appl Microbiol. 2014;117:891–904. [DOI] [PubMed] [Google Scholar]
- Zhao H, Nickle DC, Zeng Zet al. Global landscape of Clostridioides difficile phylogeography, antibiotic susceptibility, and toxin polymorphisms by post-hoc whole-genome sequencing from the MODIFY I/II studies. Infect Dis Ther. 2021;10:853–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KHet al. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidaric V, Beigot S, Lapajne Set al. The occurrence and high diversity of Clostridium difficile genotypes in rivers. Anaerobe. 2010;16:371–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.