Abstract
Introduction
Since the introduction of pneumococcal conjugate vaccines, pneumococcal disease rates have declined for many vaccine-type serotypes. However, serotype 3 (SPN3) continues to cause significant disease and is identified in colonisation epidemiological studies as one of the top circulating serotypes in adults in the UK. Consequently, new vaccines that provide greater protection against SPN3 colonisation/carriage are urgently needed. The Experimental Human Pneumococcal Challenge (EHPC) model is a unique method of determining pneumococcal colonisation rates, understanding acquired immunity, and testing vaccines in a cost-effective manner. To enhance the development of effective pneumococcal vaccines against SPN3, we aim to develop a new relevant and safe SPN3 EHPC model with high attack rates which could be used to test vaccines using small sample size.
Methods and analysis
This is a human challenge study to establish a new SPN3 EHPC model, consisting of two parts. In the dose-ranging/safety study, cohorts of 10 healthy participants will be challenged with escalating doses of SPN3. If first challenge does not lead into colonisation, participants will receive a second challenge 2 weeks after. Experimental nasopharyngeal (NP) colonisation will be determined using nasal wash sampling. Using the dose that results in ≥50% of participants being colonised, with a high safety profile, we will complete the cohort with another 33 participants to check for reproducibility of the colonisation rate. The primary outcome of this study is to determine the optimal SPN3 dose and inoculation regime to establish the highest rates of NP colonisation in healthy adults. Secondary outcomes include determining density and duration of experimental SPN3 NP colonisation and characterising mucosal and systemic immune responses to SPN3 challenge.
Ethics and dissemination
This study is approved by the NHS Research and Ethics Committee (reference 22/NW/0051). Findings will be published in peer-reviewed journals and reports will be made available to participants.
Keywords: Respiratory infections, BACTERIOLOGY, Clinical Trial, Immunology, Adult thoracic medicine
STRENGTHS AND LIMITATIONS OF THIS STUDY.
The use of a novel inoculation regime of a second (targeted booster) inoculation to participants not colonised after first challenge could increase colonisation rates (unpublished data from ISRCTN12884329); higher colonisation rates will allow smaller sample sizes in future vaccine work, reducing study costs and time, resulting in improved, more globally accessible pneumococcal vaccines faster.
Additionally, the use of clinical globally relevant strains from the UK and Malawi not used before in Experimental Human Pneumococcal Challenge (EHPC) studies (serotype 3 10V CC700 and serotype 3 LIV014-S3 CC180) will allow the independent conduct of future pneumococcal vaccine studies in various settings and potentially a future global cochallenge collaborations network.
The evaluation of bacterial shedding of serotype 3 (SPN3) through hand swabs, cough plates and exhaled detection facemasks is novel and could provide greater understanding of how SPN3 is spread among individuals, eventually leading to improved infection and prevention control in hospitalised patients.
Follow-up beyond 14 days will allow evaluation of longer-term local immune response to SPN3 and as well as providing longer term data on colonisation rate, density and duration.
The main limitation of this study is the similarity to the EHPC Pneumo 1 study, which used different SPN3 isolates that are proprietary to a third party to establish an SPN3 model.
Introduction
Streptococcus pneumoniae (SPN) is a major cause of morbidity and mortality from a lower respiratory tract infection globally.1 Available pneumococcal conjugated vaccines (PCVs) confer protection by reducing vaccine-type (VT) SPN colonisation density and are effective against invasive pneumococcal disease (IPD).2 After the introduction of PCV-13 into national paediatric immunisation programmes, overall combined-VT IPD among all ages significantly reduced.3 However, surveillance data showed limited declines in serotype 3 (SPN3) IPD in all ages, despite this serotype being included in PCV-13.4 This is thought to be due to specific characteristics of the SPN3 capsule,5 which could theoretically overwhelm the protective capacity of antibodies that are produced in response to the vaccine. There could be also other evasion mechanisms apart from antibody responses that could play a role in why SPN3 is different and continues to circulate among vaccinated communities.
An observational study of IPD rates in England 2014–2018 showed an increase in SPN3 IPD cases, with SPN3 contributing the most to total IPD deaths.6 Furthermore, a randomised controlled trial (RCT) did not show significant difference in SPN3 colonisation post PCV-13 in infants when compared with PCV-7 (which does not include SPN3) as a control.7 Taken altogether, current data suggest that even if PCV-13 protects against SPN3 directly to some degree, it does not sufficiently produce the sustained indirect protective effects seen against other VTs. Therefore, novel vaccines providing higher levels of protection against SPN3 colonisation (and therefore disease) are needed.
The Experimental Human Pneumococcal Challenge (EHPC) model allows vaccines to be tested for their effect on experimental SPN colonisation/carriage, in a more cost-effective manner than field studies, with fewer participants and shorter follow-up.8–10 Participants are intranasally inoculated with SPN, inducing a stable colonisation episode for about 1–3 weeks, at a density typical of natural colonisation. Host samples including nasal washes, nasal cells and blood are taken to assess colonisation as well as the immune responses. The model has provided key insights into human immune mechanisms that are associated with protection and susceptibility to colonisation acquisition.11–13 The model is well developed for serotypes SPN6B and SPN15B with over 2000 challenges in over 15 independent studies, showing the model is safe and has reproducible attack rates. In recent studies, the model has been used to explore bacterial shedding and the transmission potential of SPN6B, demonstrating that hands can be a vehicle for transmission of SPN6B and lead to 18% colonisation when suspensions containing the bacteria are either sniffed from the hand or inserted into the nose via a finger.14 SPN transmission has been associated with living with a larger number of people, typically in prisons or nursing homes,15 therefore evaluating methods of shedding provides invaluable insight into how this transmission occurs and can be prevented.
More recently, 96 healthy participants were challenged with three different proprietary strains of SPN3 at various doses. Colonisation rates varied from 30%–70% and the model was shown to be safe and feasible.16 Interestingly, there was no increase in levels of nasal SPN3 anticapsular antibodies in colonised participants at day-14 post-inoculation, suggesting that there could be a lack of immunogenicity with SPN3, unlike has been demonstrated in previous studies.7 Additionally, 30.2% of participants reported symptoms when questioned at routine clinic visits, of whom the majority described a sore throat. SPN has not previously been commonly associated with pharyngitis17 and further investigation into this is required.
To ensure the EHPC model remains at the cutting edge of pneumococcal (current and future) vaccine assessment, we are proposing here to set up an EHPC model with carefully selected non-proprietary SPN3 strains and a second (targeted booster) inoculation to achieve maximum attack rates. In this study, in addition to determining the optimal dose and isolate of SPN3 to establish highest rates of colonisation in the human nasopharynx, we intend to improve the knowledge of both mucosal and serological immune responses to SPN3 colonisation. We will investigate longer-term immune response to SPN3 colonisation (beyond 14 days) and for the first time we will use a targeted booster SPN3 inoculation to improve colonisation rates and evaluate the impact that this has on immunogenicity. We hypothesise that this could better simulate natural SPN colonisation in high transmission settings, whereby individuals are likely to be repeatedly exposed.
Additional exploratory outcomes will be assessed, including evaluating SPN3 shedding and further investigating symptoms experienced post inoculation. The results from this study will be used to inform development of improved SPN3 vaccines and to inform design of future pneumococcal vaccine RCTs. We plan to transfer this SPN3 model to Malawi, where SPN3 is a dominant disease-causing and antimicrobial resistance-transmitting serotype. Our previous transfer of the SPN6B model has been safe and successful, revealing important differences in vaccine response in highly endemic low resource settings.18
The primary aim of this study is to determine the optimal SPN3 dose and isolate to establish experimental nasopharyngeal colonisation in healthy adults. Success in this project will result in:
Expansion of the EHPC model to demonstrate safe SPN3 colonisation with new non-proprietary isolates. This will allow better understanding of SPN3 colonisation dynamics and identification of correlates of protection.
Determination of the optimum isolate/dose and safety of SPN3 booster inoculation, to allow future testing of vaccines in double blind RCTs with even smaller numbers of participants than usual EHPC studies (and significantly less than are required for field studies).
Methods and analysis
Study overview
This protocol has been reported following the SPIRIT reporting guidelines.19
This is a human challenge study of healthy adult participants who will be nasally inoculated with well-characterised, fully sequenced-penicillin sensitive SPN3, for the assessment of acquisition of nasal pneumococcal colonisation and immune responses. We will conduct a dose-ranging study to determine the optimum SPN3 isolate and dose for safe colonisation acquisition and confirm the dose and safety in a subsequent larger cohort in a reproducibility study. This study will run from July 2022 to October 2023 at the Accelerator Research Building, Liverpool School of Tropical Medicine (LSTM), Liverpool, UK.
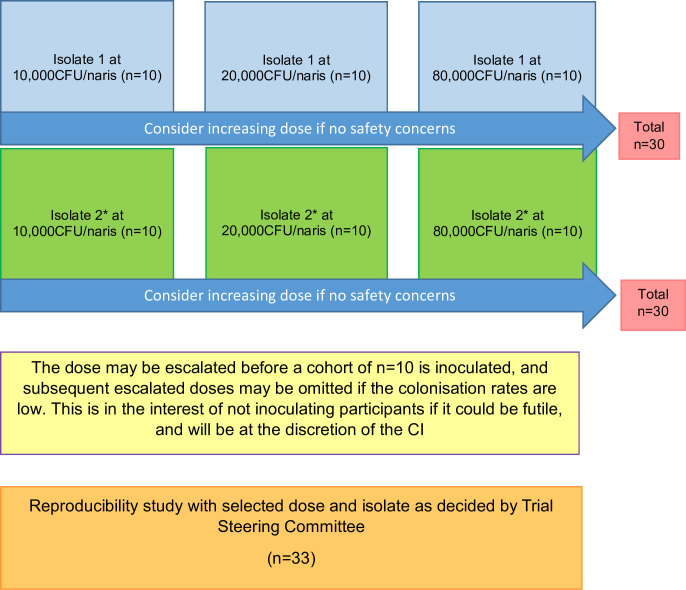
Figure 1 displays the study process. In the dose ranging study, sequential cohorts of 10 healthy participants will be challenged with escalating doses of SPN3. We will start at 10 000 colony-forming unit (CFU)/naris and after n=10, escalate to 20 000 CFU/naris (n=10) and then 80 000 CFU/naris (n=10) if safe to do so. The first challenge cohort at 10 000 CFU/naris will start slowly with a smaller group (n=1–6) inoculated per week, for safety, before completing the group (n=10). We may increase the dose further depending on colonisation rates after consultation with the Trial Monitoring Group (TMG). If optimum attack/colonisation acquisition rates are achieved (≥50%) in the lower doses, the higher dose escalation groups may not be completed—this will be discussed with the trial steering committee (TSC) before a decision to omit the higher doses is made. We may escalate the dose before a cohort of n=10 is inoculated, and subsequent escalated doses may be omitted if the colonisation rates are low. This is in the interest of not inoculating participants at a dose that could be futile. This will be at the discretion of the chief investigator (CI).
Figure 1.
Trial flow chart. *Isolate 2 may not be tested at the discretion of the trial steering committee. CFU, colony-forming unit; CI, chief investigator.
Up to 2 isolates will be tested in this manner, serotype 3 10V CC700 and serotype 3 LIV014-S3 CC180. In case of not achieving the desired attack rate with any of the isolates, the data obtained will be reviewed to either test an additional isolate or complete the cohort at a lower attack rate.
Using the dose, isolate and inoculation regime that results in ≥50% colonisation acquisition/attack rate, with a high safety profile, we will complete the cohort with another 33 participants to check for reproducibility of attack rate. In previous research conducted with SPN6B, colonisation rates of 58.5% were seen with single inoculation and of those who were negative after the first inoculation, a further 41.7% then became positive after a second inoculation, resulting in a combined colonisation rate following two inoculations of 70.7%.
Study objectives and outcomes
Study objectives and outcomes measures are demonstrated in table 1.
Table 1.
Objectives and outcome measures
| Objectives | Outcome measures | |
| Primary | To determine the optimal SPN3 dose and isolate to establish colonisation of the nasopharynx in healthy adults using the EHPC model. | The proportion of participants with experimental SPN3 colonisation of the nasopharynx, determined by SPN3 presence in classical microbiological culture in at least one nasal wash (NW) sample, at any time point following one or two inoculations (combined and individually). This will be assessed for each isolate and dose separately. |
| Secondary | To ascertain the rate of experimental colonisation acquisition of SPN3 in healthy adults post challenge, by classical and molecular methods. | The rate of occurrence of SPN3 experimental colonisation of the nasopharynx, determined by SPN3 presence in classical microbiological culture and qPCR (combined and individually) from at least one NW sample at any time point following one or two inoculations (combined and individually). |
| To determine the density of experimental SPN3 colonisation of the nasopharynx. | The bacterial density of experimental SPN3 colonisation of the nasopharynx in NW, at each and any time point following one or two inoculations (combined and individually), determined by classical microbiological culture and molecular methods. | |
| To determine the duration of experimental SPN3 colonisation of the nasopharynx. | The duration of experimental SPN3 colonisation of nasopharynx determined by the last NW sample following one or two inoculations (combined and individually) in which SPN3 is detected by classical microbiological culture or molecular methods. | |
| Exploratory | To determine grading score of symptoms during experimental colonisation (eg, sore throat, rhinitis, nasal congestion). | The presence of mild or moderate symptoms as recorded on a Likert scale in participants with SPN3 within the first 7 days after inoculations. Sore throat grading score will also be used if applicable. |
| To characterise mucosal immune cell populations and dynamics in response to SPN3 experimental inoculation in nasal cell samples. | Cell immunophenotyping using flow cytometry methods to identify and characterise cell populations such as neutrophils, monocytes, T cells and B cells in nasal cells samples at screening and 2, 7, 13, 16, 21 and 28 days after first inoculation. | |
| To determine the level of mucosal and systemic SPN3 polysaccharide-specific antibodies at baseline and after SPN3 experimental inoculation. | Measurement of anti-SPN3 polysaccharide specific immunoglobulin G (IgG) levels in serum and nasal wash samples using ELISAs. | |
| To determine the levels of polysaccharide specific SPN3 memory B cells at baseline and after SPN3 experimental inoculation. | Quantification and characterisation of the number of SPN3- polysaccharide specific memory B cell populations in PBMC samples using flow cytometry methods at screening and 13 and 28 days after first inoculation. | |
| To describe nasal inflammatory kinetics induced by SPN3 experimental inoculation. | Measurement of 30 cytokines and chemokines using multiplex Luminex in nasosorption samples at screening and 2, 7, 13, 16, 21 and 28 days after first inoculation. | |
| To assess bacterial shedding after pneumococcal colonisation. | The rate of pneumococcal bacterial shedding as defined by swabs of hand and cough-plate based assessment post inoculation (presence and density CFU/mL) at 2, 7, 13, 16, 21 and 28 days after first inoculation. (During COVID-19 pandemic cough sampling may not be performed.) | |
| To compare ‘exhaled detection facemask’ (EDF) with cough plate-based methods of assessing SPN3 bacterial shedding post pneumococcal inoculation. | The rate of pneumococcal bacterial shedding as defined by EDF and cough-plate based assessment post inoculation at 2 and 7 days post inoculation (presence and density (CFU/mL)) | |
| To determine the effect of natural SPN (non-SPN3) colonisation on SPN3 colonisation. | The rate of occurrence and density of SPN3 colonisation of the nasopharynx post inoculation in natural SPN carriers, determined by SPN3 presence and density in classical microbiology culture or qPCR from at least one NW sample at any timepoint post inoculation. | |
| To compare rates, density and duration of SPN3 colonisation in saliva versus nasopharyngeal samples. | The presence, density and duration of SPN3 colonisation in NW and saliva samples at any timepoint post inoculation, identified using classical microbiology culture or qPCR |
EHPC, Experimental Human Pneumococcal Challenge; SPN3, Streptococcus pneumoniae serotype 3.
Study participants
Inclusion/exclusion criteria
Healthy participants aged 18–50 (inclusive) who are fluent in English have access to a mobile telephone and have capacity to give informed consent will be allowed to participate. This age range minimises the risk of IPD and allows comparison with our previously published EHPC work.
Table 2 outlines the exclusion criteria for this study.
Table 2.
Exclusion criteria
| Research participant | Currently involved in another study unless observational or non-interventional, excluding the EHPC bronchoscopy study and COVID-19 observation and interventional trials |
| Participant in any previous EHPC trial in past 1 year | |
| Previous EHPC trial inoculated with SPN3 in last 3 years | |
| Vaccination | Previous pneumococcal vaccination PPV23 or PCV13 or PCV10. |
| Allergy | Allergy to penicillin |
| Comorbidities | Chronic respiratory, cardiac, kidney, liver or neurological disease |
| Connective tissue disease | |
| Diabetes | |
| Immunosuppressive disease | |
| Recurrent otitis media | |
| Asplenia or spleen dysfunction | |
| Cochlear Implants | |
| Major cerebrospinal fluid leak | |
| Uncontrolled medical/surgical conditions (at discretion of study doctor) | |
| Major pneumococcal illness requiring hospitalisation within the last 10 years | |
| Other conditions considered by clinical team as a concern for safety/integrity of the study | |
| Significant mental health problems (uncontrolled or previous admission to psychiatric unit) | |
| Medications | Immunosuppressive medication |
| Long-term antibiotic use or use of antibiotics in 28 days prior to inoculation | |
| Direct caring role or close contact with individuals at higher risk of infection (during the EHPC period) unless wearing personal protective equipment | Children aged under 5 |
| Chronic ill health or immunosuppressed adults | |
| People that are part of extremely vulnerable group as defined by Public Health England | |
| Smoking/drug/alcohol use | Current or ex-smoker (daily cigarettes/e-cigarettes/smoking of recreational drugs) in the last 6 months. Participants who smoke<5 cigarettes per week may be included |
| Previous significant smoking history (>20 pack years) | |
| History or current drug or alcohol abuse (frequently drinking alcohol): men and women should not regularly drink>3 units/day and >2 units/day, respectively) at discretion of the clinician | |
| Biologically female participants of childbearing potential who are currently pregnant/lactating, intending on becoming pregnant or not on effective birth control | |
| Overseas travel (involving air travel) planned in follow-up period of study visits | |
Participants who meet following criteria at time of screening:
| |
EHPC, Experimental Human Pneumococcal Challenge; SPN3, Streptococcus pneumoniae serotype 3.
Additionally, we will employ temporary exclusion criteria, including:
COVID-19 symptoms or confirmed current COVID-19 infection
Current/acute illness within 14 days prior to inoculation if COVID-19 negative
Positive COVID-19 swab within 10 days of inoculation. Participants will require negative lateral flow test prior to inoculation
COVID-19 vaccination 21 days prior to inoculation
Natural SPN3 colonisation identified in baseline nasal wash.
Participants who have been temporarily excluded at screening may be rescreened at a later date to assess their eligibility at this time for inclusion into the study.
Participant recruitment
Participants will be recruited from the general public, including through public engagement events, social media, generic research communication mailing lists, large local employers and local universities.
Participant timeline
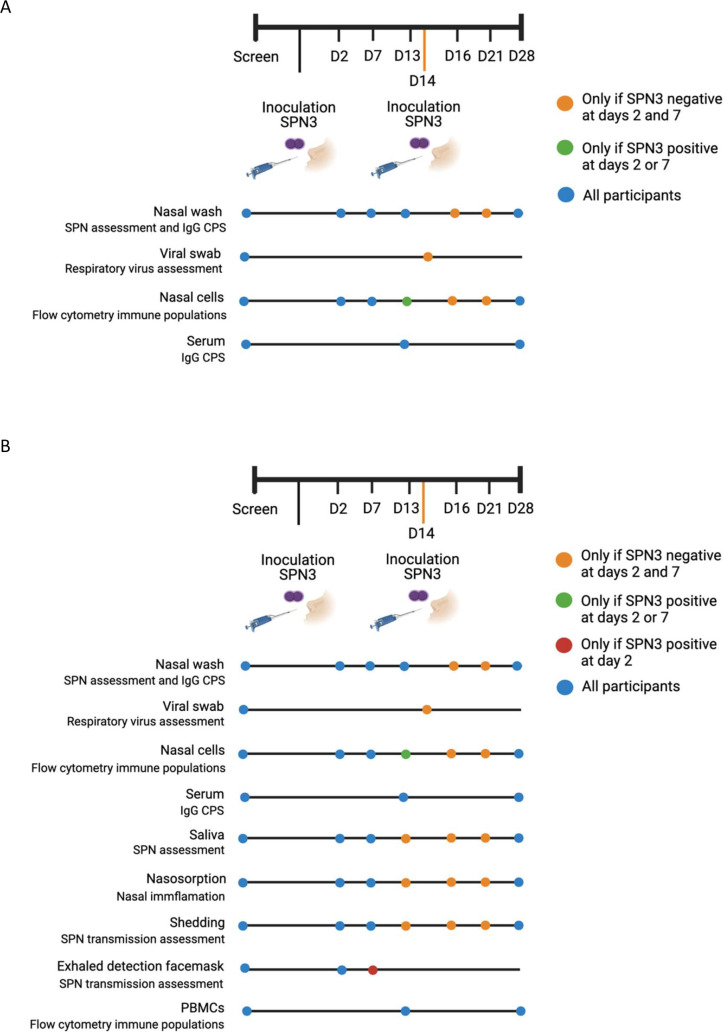
For both parts of the study, participants will attend an identical visit schedule; however, the samples taken at these visits will differ (figure 2). Participants will attend a screening visit, inoculation visit (day 0) and then follow-up visits at days 2, 7, 13 and 28. The screening visit should occur 5 days prior to inoculation but a window of −7/+4 days will be used. Figure 2 outlines the samples that are required at each visit.
Figure 2.
(A) Dose-ranging study sampling schedule. (B) Reproducibility study sampling schedule. CPS, capsular polysaccharide; IgG, immunoglobulin G; SPN3, Streptococcus pneumoniae serotype 3. Created with Biorender.com.
All will first attend a screening visit to confirm eligibility through medical history, clinical examination and acquisition of samples including a full blood count and nasal wash sampling. Baseline nasal washes will be evaluated for natural colonisation with SPN (and serotype if present) through classical microbiological culture and molecular methods.
All participants will then attend an inoculation visit (day 0), where fluid containing SPN3 will be instilled into their nose. At this visit, they will be given a safety pack containing a thermometer, safety information leaflet and a 5-day course of amoxicillin. They will be instructed to contact the research team daily for 3–5 days with their temperature recording and any symptoms. Participants who report symptoms consistent with pneumococcal disease will be reviewed in person by a clinician and may be instructed to take their antibiotic course. Participants have 24-hour access to research clinicians as well as access to hospital facilities and prompt treatment if required.
In the dose-ranging study, a second targeted booster inoculation will be given to participants on day 14 if they have tested negative for SPN3 on day 2 and 7 samples. In the reproducibility study, it will be at the discretion of the CI to decide if targeted booster inoculation should be applied, based on results of the dose-ranging study. If a second inoculation is included in the reproducibility study, it will be only given at day 14 to those participants who are negative for SPN3 at days 2 and 7 (these participants will be asked to attend additional follow-up visits at days 16 and 21 for further sampling).
During the dose-ranging phase, nasal wash samples preinoculation and post inoculation will be collected to assess colonisation acquisition and density, by classical microbiological culture and molecular methods. Additionally, serum samples will be taken at baseline, 13 and 28-days after inoculation, to measure levels of anticapsule polysaccharide immunoglobulin G (IgG). Nasal cell samples will be taken to characterise cellular populations and dynamics by flow cytometry.
During reproducibility phase, nasal wash and nasal cell samples will be collected as in the dose-ranging study. A viral swab will be collected preinoculation to test for viral coinfection. Nasal filters (nasosorption) will be collected preinoculation and post inoculation to assess mucosal inflammation using 30-plex Luminex. Saliva samples will be taken preinoculation and post inoculation to compare rates, density and duration of SPN3 colonisation in saliva and nasopharyngeal samples. Peripheral blood mononuclear cells (PBMCs) will be obtained at baseline, at 13-days and at 28-days post inoculation. PBMCs will be used to characterise memory B cell and other immune cell populations using flow-cytometry based methods. Serum samples will be taken at baseline, 13 and 28 days after inoculation to measure levels of anticapsule polysaccharide IgG. Exhaled detection facemasks (EDF) will be collected at day 2, to allow comparison against other shedding samples for detection of SPN3. A subgroup of participants who have demonstrated colonisation with SPN3 at day 2 will undergo EDF sampling at day 7.
At the end of the study, study participants who have been positive for SPN3 colonisation at any time point, and who have not subsequently had two consecutive negative nasal wash samples, will be asked to take oral amoxicillin 500 mg three times daily for 5 days with the aim to clear/assist with clearing of colonisation.
Participant retention
We will use an online booking system for appointments, to ensure that reminders are sent to participants for each appointment. There are windows of 2 days around all appointments to allow participants to move their appointments within window, if needed. Participants are remunerated on study completion.
Data collection methods
Samples will only be collected by staff members who are trained and delegated to do so. Table 3 describes the data collection methods.
Table 3.
Data collection methods
| Determination of colonisation | Colonisation will be defined as result of nasal washes taken at 2, 7, 13, 16, 21 and 28 days post inoculation. Nasal washes will be performed using the Naclerio method,14 which is a validated technique to collect nasal bacterial specimens. Nasal washes will be plated onto culture media. Colonies will be confirmed as SPN3 using classical microbiological techniques. Results from the cultured nasal wash will also be confirmed using PCR-based methods. |
| Molecular methods of determination of colonisation | DNA will be extracted from bacterial pellet post nasal wash sample centrifugation. SPN3 detection will be done by multiplex qPCR. This technique will enable us to detect individuals who are potential carriers with very low bacterial density. This multiplex qPCR is well validated in our laboratory.16 |
| Viral detection and quantification | Viral multiplex qPCR for detection and quantification will be performed on DNA and RNA of stored throat swab and/or nasal wash to detect all common respiratory viruses. |
| Mucosal and systemic immune responses | Serotype-specific responses and their association with both acquisition and clearance of colonisation (density and duration) will be measured. We will compare antibody levels and function between those colonised and those protected against colonisation. Levels of immunoglobulin to the capsular polysaccharides SPN3 in serum and nasal washes before and after inoculation will be determined. Levels of SPN3-specific memory B cells will be assessed using PBMCs collected preinoculation and post inoculation. Flow cytometry will be used to examine the induction of antigen-specific cellular responses in blood including B cell and T cells. Mucosal cellular responses will also be measured by flow cytometry on nasal cell samples. Additionally, the mucosal inflammatory response associated with inoculation will be evaluated using 30-plex Luminex method to detect cytokines and chemokines in nasal filters |
PBMCs, peripheral blood mononuclear cells; SPN3, Streptococcus pneumoniae serotype 3.
Sample size calculation
We have adopted a stepwise approach to escalating the inoculation dose. The protocol is designed to: (1) minimise the possibility that we try repeatedly to attain colonisation at a dose in which it is unlikely to happen; (2) maximise safety by inoculating small groups before continuing onto larger groups (in which we will have the statistical power to give reasonable precision of our estimate of colonisation rates).
Based on previous experience in studies of SPN6B and SPN3, we expect that with a colonisation rate of 45%, with 95% confidence level, and a margin of error of 15%, our study will be complete 43 participants with a single inoculation dose. Depending on colonisation rates at different inoculum doses of the SPN3 isolate, we need a minimum of 43 and a maximum of 93 participants to complete the study. This allows for up to two isolates to be taken through to the highest dose of 80 000 CFU/100 µL. For example, if two isolates were tested at each dose in 10 participants in the dose ranging study, this would equate to 60 participants. A further 33 participants would then take part in the reproducibility study, equating to a total of 93 participants. To ensure that we complete the correct number of participants, we will over recruit to allow for screen failures and exclusion from primary outcome analysis for natural colonisation and loss of participants due to drop out. Based on the assumption that two isolates will be tested and estimating a 20% rate of drop out/screening failure, we will recruit a maximum of 117 participants. Participants who are natural SPN carriers will be included in analysis of exploratory outcomes.
Statistical analysis plan
This is an open label, non-randomised, safety and dose escalation study in healthy participants. Analyses are descriptive.
Timing of analysis
Data will be reviewed at the middle and the end of follow-up for each cohort of 10 healthy participants from the dose ranging phase as soon as database completion and lock occurs for that cohort. Data reviewed after day 7 will be for targeted booster inoculations if participants have tested negative for SPN3 on both days 2 and 7. Safety and outcome results will be reported to the TSC and TMG. Interim analysis in the dose ranging phase is to consider addition of second inoculation dose. Analysis at the end of each cohort is to consider dose escalation (or cessation). Once the dose has been selected, we will complete then reproducibility study and final analysis will be done on completion of that cohort after database completion and lock.
Analysis populations
The intention-to-treat (ITT) population is all participants who have been enrolled and who have received at least one inoculation dose (ITT). The main analysis population for each cohort is the modified ITT population, consisting of all participants who have been enrolled and received at least one inoculation dose (D0) and have had at least one valid outcome assessment measure (nasal wash). The safety analysis population is the ITT population and will include all participants who have received at least one inoculation dose.
Analysis will use available data. No imputation for missing endpoints will be performed.
Covariates and subgroups
If targeted booster inoculations are given, the numbers and results of these will be described, including a summary of colonisation after the second dose. Depending on the number receiving second doses, and on the endpoint being analyses, the results after the second dose will either be included as a ‘new’ inoculation event, potentially requiring adjustment for repeated measures, or will be excluded. Immunological data will be described, using appropriate estimators for the characteristics of the variable (arithmetic or multiplicative means, for example) and appropriate transformations as required. Where appropriate, and in particular for the reproducibility cohort, regression models will be used to estimate adjusted associations between strain and number of doses with colonisation as an outcome, and with immunological response as an outcome.
Interim analysis and data monitoring
Data will be reviewed on an ongoing basis for any safety or adverse events (AEs) and will be reported per protocol to the TSC and data and safety monitoring committee (DSMC). Efficacy outcomes will be evaluated after day 7 to provide information on second dose and at the cohort completion to assist with decision-making for the next dose escalation in sequence. The reproducibility cohort will have a single interim look, to evaluate second inoculation dose and a final analysis at the end of cohort follow-up.
Efficacy analysis
Each dose cohort (dose selection and reproducibility) will be analysed separately. Binary endpoints will be summarised as frequency (%) with 95% confidence estimates at each time point and by participant level summary. Density will be summarised using number, mean, geometric mean, SD, geometric SD, minimum and maximum at each time point. 95% CIs will be estimated for geometric mean estimates. Duration of colonisation (days) will be analysed as a continuous variable and summarised using mean, SD, minimum and maximum in the event of no censored outcomes. In the event of censoring, product limit methods will be used to estimate endpoints. The duration will be defined as the time between inoculation and the time of the last NW sample that is positive for colonisation. Symptom and sore through grading scales will be summarised by frequency (%). Agreement between colonisation results from microbiological culture and qPCR will be estimated using Bland-Altman plots and estimates of agreement.
The reproducibility cohort alone will use generalised linear models for single measurement outcomes (colonisation at any time vs not) under a binomial model with logit link to estimate odds ratio (95% CI) for strain/procedure/one or two doses as independent variables. Censored endpoints (time to colonisation) will be analysed using product limit estimates of median (95% CI for the time). Exploratory immunological outcomes will be summarised as above.
Reporting conventions
P values>0.0001 will be reported to 4 decimal places; p values<0.0001 will be reported as‘< 0.0001’. Distribution estimates such as mean, geometric mean, SD, median and quartiles will be reported to 3 decimal places. Parameters estimates such as regression coefficients, CIs and HRs will be reported to three significant digits.
Safety reporting
AEs will be graded using the Division of AIDS Table for Grading the Severity of Adult and Paediatric Adverse Events.20 If the severity of an AE could fall in either one of two grades, the higher of the two grades should be selected.
Symptoms experienced and attendance to hospital/GP will be asked about at each visit. Serious adverse events (SAEs) will be reported from the time of consent until completion of day 28 visit, or until completion of antibiotic treatment if participants require this.
All AEs will be recorded in the electronic case report form (eCRF) and documented in a weekly safety report. Adverse events of special interest (AESI) such as (but not limited to) headache, cough, sore throat and earache will be specifically documented and reported to the TMG, TSC and DSMC in the safety report.
All SAEs will be recorded on an SAE form and reported to the DSMC, sponsor within 24 hours of discovery or notification of the event. All SAEs/AESIs will be followed until resolution/stabilisation or until the end of the participants last study visit. The DSMC will perform an independent review of SAEs.
Auditing
A Trial Monitoring Plan will be developed by the Sponsor and agreed by the TMG and CI based on the trial risk assessment. Following written standard operating procedures, the monitors will verify that the clinical trial is conducted, and data are generated, documented and reported in compliance with the protocol, good clinical practice and the applicable regulatory requirements.
Ethics and dissemination
Research ethics committee review
This protocol has been reviewed by the sponsor, funder and an external peer review process. Ethical approval has been obtained from Liverpool Central Research Ethics Committee (REC) with REC reference number 22/NW/0051. The protocol, informed consent form, participant information leaflet (PIL) and any proposed advertising material has been approved by REC as well. For any amendment to the study, the CI, in agreement with the sponsor, will submit information to REC and other appropriate bodies. Amendments will be discussed with participants.
Consent
Potential participants will be sent a copy of the PIL (online supplemental material 1) and invited to contact a member of the team if they would like to participate. They will then be invited to attend a presentation and to carry out a quiz to ensure they have understood the information given. If a participant has voluntarily agreed to take part in the research and the study team are satisfied that they meet the eligibility criteria, they will be invited to provide written informed consent with a delegated, trained member of staff (online supplemental material 2). In line with recommended practice (MRC tissue and biological samples for use in research), participants will be asked to consent to gift their anonymised samples for use in future studies and shared with research collaborators and stored for any future commercial respiratory partnerships. This is outlined in the PIL and consent form.
bmjopen-2023-075948supp001.pdf (1.8MB, pdf)
bmjopen-2023-075948supp002.pdf (31.2KB, pdf)
Data management and participant confidentiality
Study data will be recorded directly into REDcap, an Electronic Data Capture system.21 Any additional information that needs recording but is not relevant for the case report form (CRF) will be recorded on a separate paper source document. The eCRF must be completed by designated and trained study personnel. Quality control will be performed on each eCRF. The processing of eCRFs will include an audit trail, to include changes made, reason for change, date of change and person making change.
Each participant will be assigned a unique, non-identifiable study number at recruitment for anonymisation. Unlinked non-identifiable clinical data will be stored and analysed at the LSTM, MSD or collaborating laboratories. Only authorised members of the clinical research team will be able to access participant personal information which is directly relevant to the study. All electronic records containing personal information will be stored in a password-protected database on a password protected computer. Paper documentation containing personal information will be kept in a locked filing cabinet in a locked room. On completion of the study, the eCRF will be locked and source documents will be photocopied and archived on paper and electronically in a secure database. This data will be stored for a minimum of 25 years. See online supplemental material 2 for information on storage of biological specimens.
Dissemination policy
The findings from this study will be disseminated among the scientific community. We intend to publish our findings in peer-reviewed scientific journals and present data at appropriate local, national and international conferences. In addition, we will produce a report of our findings, which will be made available to all participants. Authorship of the final trial report and subsequent publications will include those who contribute to the design, delivery and analysis of the trial. Authorship will be defined on study completion in line with International Committee of Medical Journal Editors guidelines.22
Regarding data sharing, we will hold exclusivity of participant data for 2 years, until all analysis and publications are complete. Deidentified individual participant data that underlie the results reported at the end of the trial will then be made available in an open format with accompanying metadata to the LSTM archive team, who can make it available on request. Researchers who provide a methodologically sound proposal will be able to request access, to achieve aims in the agreed proposal. Proposals may be submitted up to 5 years following publication of the results of this protocol. After 5 years, the data will be available in LSTM archives but without investigator support.
Patient and public involvement and engagement
This study is run in conjunction with the EHPC studies, which have been studying pneumococcal colonisation over the last 11 years. There are numerous opportunities for public and patient involvement: newsletters are sent out to all participants to inform them of the study results and further work, previous participants assist with recruitment events and social media accounts update followers about current studies and our ongoing work. For this study, we have asked participants from previous studies to review the PIL and consent form, to ensure it is clear and easy to understand.
Supplementary Material
Acknowledgments
We would like to acknowledge the NIHR CRN North West Coast for providing clinical staff for this study. We would like to acknowledge Andrea Gori and Robert S Hyderman for providing the SPN3 strain from Malawi. We would also like to acknowledge members of the DSMC/TSC: Rob Read, Neil French, Marc Henrion and Chris Chiu. We would also like to acknowledge the Respiratory Consultants in the Liverpool University Hospitals NHS Foundation Trust who provide on-call cover for this study: Drs Kimberley Barber, Livingstone Chishimba, Paul Deegan, Justine Hadcroft, Gareth Jones, Patricia Yunger, Hassan Burhan and Seher Zaidi.
Footnotes
Twitter: @phoebehazenber1
Contributors: Study design set up: PH, RER, MF, CS, AH-W, DMF, AMC, KL, ML, AH, TKN, SBG. Statistics: ML. Ethics application: PH, KD, MF and AMC. Study coordination: PH, CS, AH-W, MF, KD, AMC, AB, JB, HF. Clinical cover including on-call responsibility: KL, RER, PH, AMC, JB, HF, AB. Writing the protocol: PH, MF, RER, CS, KL, KD, AH-W, EM, BU, DMF, AMC, ML. Bacterial selection, bacterial inoculum preparation: CS, AH, TK-N, DES, SBG. Manuscript writing: RH, RER, MF, CS, AMC, DMF. Manuscript review PH, RER, MF, CS, AMC, DMF, SBG, KL, AH-W, AH, ML, AB, HF, KD, TK-N, BU, SBG, DES.
Funding: The study is supported in part by a research grant from Investigator-Initiated Studies Programme of Merck Sharp and Dohme LLC (MISP 61493). The opinions expressed in this paper are those of the authors and do not necessarily represent those of Merck Sharp and Dohme LLC.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.GLRI C. Estimates of the global, regional, and national morbidity, mortality, and Aetiologies of lower respiratory infections in 195 countries, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet Infect Dis 2018;18:1191–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.German EL, Solórzano C, Sunny S, et al. Protective effect of PCV vaccine against experimental Pneumococcal challenge in adults is primarily mediated by controlling Colonisation density. Vaccine 2019;37:3953–6. 10.1016/j.vaccine.2019.05.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine OS, O’Brien KL, Knoll M, et al. Pneumococcal vaccination in developing countries. Lancet 2006;367:1880–2. 10.1016/S0140-6736(06)68703-5 [DOI] [PubMed] [Google Scholar]
- 4.Oligbu G, Collins S, Djennad A, et al. Effect of Pneumococcal conjugate vaccines on Pneumococcal meningitis. Emerg Infect Dis 2019:1708–18. 10.3201/eid2509.180747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Miguel S, Domenech M, González-Camacho F, et al. Nationwide trends of invasive Pneumococcal disease in Spain from 2009 through 2019 in children and adults during the Pneumococcal conjugate vaccine era. Clin Infect Dis 2021;73:e3778–87. 10.1093/cid/ciaa1483 [DOI] [PubMed] [Google Scholar]
- 6.Amin-Chowdhury Z, Collins S, Sheppard C, et al. Characteristics of invasive Pneumococcal disease caused by emerging Serotypes after the introduction of the 13-Valent Pneumococcal conjugate vaccine in England: A prospective observational cohort study, 2014-2018. Clin Infect Dis 2020;71:e235–43. 10.1093/cid/ciaa043 [DOI] [PubMed] [Google Scholar]
- 7.Dagan R, Patterson S, Juergens C, et al. Comparative Immunogenicity and efficacy of 13-Valent and 7-Valent Pneumococcal conjugate vaccines in reducing Nasopharyngeal Colonization: A randomized double-blind trial. Clin Infect Dis 2013;57:952–62. 10.1093/cid/cit428 [DOI] [PubMed] [Google Scholar]
- 8.Ferreira DM, Jambo KC, Gordon SB. Experimental human Pneumococcal carriage models for vaccine research. Trends Microbiol 2011;19:464–70. 10.1016/j.tim.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 9.Gritzfeld JF, Cremers AJH, Ferwerda G, et al. Density and duration of experimental human Pneumococcal carriage. Clin Microbiol Infect 2014;20:1145–51. 10.1111/1469-0691.12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gritzfeld JF, Wright AD, Collins AM, et al. Experimental human Pneumococcal carriage. J Vis Exp 2013. 10.3791/50115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jochems SP, de Ruiter K, Solórzano C, et al. Innate and adaptive nasal Mucosal immune responses following experimental human Pneumococcal Colonization. J Clin Invest 2019;129:4523–38. 10.1172/JCI128865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jochems SP, Marcon F, Carniel BF, et al. Inflammation induced by influenza virus impairs human innate immune control of Pneumococcus. Nat Immunol 2018;19:1299–308. 10.1038/s41590-018-0231-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitsi E, Carniel B, Reiné J, et al. Nasal Pneumococcal density is associated with Microaspiration and heightened human alveolar macrophage responsiveness to bacterial pathogens. Am J Respir Crit Care Med 2020;201:335–47. 10.1164/rccm.201903-0607OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor V, German E, Pojar S, et al. Hands are vehicles for transmission of Streptococcus Pneumoniae in novel controlled human infection study. Eur Respir J 2018;52. 10.1183/13993003.00599-2018 [DOI] [PubMed] [Google Scholar]
- 15.Musher DM. How contagious are common respiratory tract infections N Engl J Med 2003;348:1256–66. 10.1056/NEJMra021771 [DOI] [PubMed] [Google Scholar]
- 16.Robinson RE, Mitsi E, Nikolaou E, et al. Human infection challenge with Serotype 3 Pneumococcus. Am J Respir Crit Care Med 2022;206:1379–92. 10.1164/rccm.202112-2700OC [DOI] [PubMed] [Google Scholar]
- 17.Weiser JN. The Pneumococcus: why a Commensal Misbehaves. J Mol Med (Berl) 2010;88:97–102. 10.1007/s00109-009-0557-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton B, Burr S, Chikaonda T, et al. A feasibility study of controlled human infection with Streptococcus pneumoniae in Malawi. EBioMedicine 2021;72. 10.1016/j.ebiom.2021.103579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan A-W, Tetzlaff JM, Gøtzsche PC, et al. SPIRIT 2013 explanation and elaboration: guidance for protocols of clinical trials. BMJ 2013;346. 10.1136/bmj.e7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Institute of Allergy and Infectious Diseases . Division of AIDS table for grading the severity of adult and pediatric adverse events. 2021. Available: https://rsc.niaid.nih.gov/sites/default/files/daidsgradingcorrectedv21.pdf
- 21.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (Redcap) – A Metadata-driven methodology and Workflow process for providing Translational research Informatics support. J Biomed Inform 2009;42:377–81. 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International committee of medical Journal editors. recommendations for the conduct, reporting. Editing, and Publication of Scholarly Work in Medical Journals May 2022. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2023-075948supp001.pdf (1.8MB, pdf)
bmjopen-2023-075948supp002.pdf (31.2KB, pdf)