Abstract
Previous characterization of Pseudomonas aeruginosa clinical isolates has demonstrated an inverse correlation between cytotoxicity and internalization by epithelial cells. To further investigate this relationship, we tested PA103, a cytotoxic P. aeruginosa strain, and 33 isogenic noncytotoxic transposon mutants for internalization by Madin-Darby canine kidney cells. The majority of the mutants were not internalized, demonstrating that an inverse correlation between cytotoxicity and bacterial uptake by epithelial cells is not absolute. Six of the noncytotoxic mutants, however, demonstrated measurable levels of internalization by standard aminoglycoside exclusion assays even though internalization of wild-type strain PA103 was not detectable. All six had evidence of protein secretion defects involving two proteins, a 40-kDa protein and a 32-kDa protein. These proteins, designated PepB (for Pseudomonas exoprotein B) and PepD, respectively, each had characteristics of type III transported proteins. In addition, nucleotide sequencing studies demonstrated that PepB and PepD are homologs of YopB and YopD, respectively, type III secreted proteins of Yersinia spp. necessary for the translocation of effector molecules into the cytoplasmic compartment of eukaryotic cells. Thus, while many mutations in PA103 result in loss of cytotoxicity without an appreciable increase in internalization, defects in transport of type III secretion proteins PepB and PepD correlate with both loss of cytotoxicity and gain of internalization. These results are consistent with type III secretion of an inhibitor of internalization that requires PepB and PepD for translocation into the host cell.
Pseudomonas aeruginosa is a gram-negative bacterium which causes disease in compromised hosts (42). Infection may occur at a variety of sites, including the urinary tract in catheterized patients, the bloodstream in individuals suffering from neutropenia or burns, the cornea in contact lens wearers, and the lungs in people with cystic fibrosis or patients requiring mechanical ventilation. This organism accounts for a significant percentage of all hospital-acquired infections and is the leading cause of nosocomial pneumonia (18).
A remarkable number of bacterial secreted factors have been postulated to play roles in P. aeruginosa infections, including exoenzyme S (ExoS), exoenzyme T (ExoT), exotoxin A, phospholipase C, alkaline protease, and elastase (14, 32, 34, 49, 51). Recently, a type III secretion system in this organism has been identified and shown to be responsible for transport of two of these proteins, ExoS and ExoT (54). Type III secretion systems are sec-independent protein secretion pathways that are utilized by many gram-negative pathogens and are thought to be activated by host cell contact (29, 37, 41, 55). Effector proteins are secreted by these pathways without cleavage of a signal peptide, although the information necessary for secretion is located in the amino-terminal portion of the protein (44, 48, 54). Both intracellular pathogenic bacteria (e.g., Salmonella and Shigella spp.) and extracellular pathogenic bacteria (e.g., Yersinia spp. and enteropathogenic Escherichia coli) utilize conserved type III systems to secrete virulence factors (reviewed in reference 28).
Although P. aeruginosa has been traditionally viewed as an extracellular pathogen, recent reports indicate that this organism may be internalized by a variety of epithelial cells, including Madin-Darby canine kidney (MDCK) cells, corneal cells, and human respiratory epithelial cells (2, 6, 38–40, 56). In particular, respiratory epithelial cell shedding following P. aeruginosa internalization has been postulated to be a host defense mechanism (39). The mechanism of bacterial internalization remains poorly characterized, but it has been reported that the invasive potential of strains varies inversely with their ability to kill cells (cytotoxicity) (10). In addition, there is evidence to suggest that invasion is not a prerequisite for cytotoxicity (8). Finally, defects in expression of ExsA, a transcriptional activator of the Pseudomonas type III system (13, 19, 52, 54), result in uptake of strains not otherwise internalized (9). It has been postulated that ExsA is necessary for the secretion of a cytotoxic factor and that the absence of secretion of this factor by an ExsA mutant allows invasion to be detected in gentamicin exclusion assays (9). Examination of clinical isolates has also demonstrated that the secretion of ExoS correlates with an invasive phenotype, perhaps by subverting host cell signal transduction pathways through covalent modification of host proteins (9). Although these findings suggest that type III secretion modulates internalization, it remains unclear how this occurs and which components of this system are involved. We chose to further investigate the relationship between cytotoxicity and internalization by analyzing the uptake of PA103 mutants defective in cytotoxicity. Based on our results, we propose a model that explains how the type III secretion system of P. aeruginosa modulates both bacterial internalization by epithelial cells and bacterial killing of epithelial cells.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strains were routinely cultured in Luria-Bertani broth or in Vogel-Bonner minimal medium (50) with antibiotics as needed. For analysis of extracellular proteins, P. aeruginosa strains were grown in a defined minimal medium, MINS (25 mM KH2O4, 95 mM NH4Cl, 50 mM monosodium glutamate, 110 mM disodium succinate, 10 mM trisodium nitrilotriacetic acid, 2.5% glycerol, 5 mM MgSO4, and 18 μM FeSO4), that was originally optimized for ExoS production (33). The following antibiotic concentrations were used: ampicillin, 50 μg/ml; gentamicin, 100 μg/ml; and tetracycline, 100 μg/ml for Pseudomonas and 20 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain, phage, or plasmid | Relevant properties | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| PA103 | Wild-type cytotoxic respiratory clinical isolate | 1, 27 |
| Noncytotoxic mutants of PA103 | Summarized in Table 2 | 22 |
| E. coli strains | ||
| S17-1 | thi pro hsdR recA RP4-2(Tet::Mu) (Km::Tn7) | 47 |
| XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB hsdSMR mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene Corp. |
| Plasmids and cosmids | ||
| pBR322 | Cloning vector | Gibco BRL |
| pBluescriptII | Cloning vector | Stratagene Corp. |
| pLAFR5SK1 | Cosmid vector; pLAFR5 (23) with improved polylinker site | L. Shapiro |
| 4-7-H | pLAFR5SK1 cosmid with PA103 genomic DNA insert | This study |
| 4-11-H | pLAFR5SK1 cosmid with PA103 genomic DNA insert | This study |
| 5-9-E | pLAFR5SK1 cosmid with PA103 genomic DNA insert | This study |
| pAH830 | pBluescriptII containing the 2.0-kb SmaI-SmaI fragment of cosmid 4-7-H | This study |
| pAH831 | pBluescriptII containing the 8.0-kb EcoRI-EcoRI fragment of cosmid 4-7-H | This study |
Internalization assays.
Polarized monolayers were formed by adding 5 × 106 MDCK cells in minimal essential medium (MEM)-Eagle’s medium with Earle’s buffered saline solution (obtained from the University of California, San Francisco [UCSF], Cell Culture Facility) and 5% fetal calf serum (Gibco BRL, Gaithersburg, Md.) to 12-mm Transwell filters (Corning Costar Corp., Cambridge, Mass.). Cells were grown for 3 days at 37°C in 5% CO2. Bacteria were grown at 37°C with shaking for approximately 17 h in MINS medium. Immediately prior to use, bacteria were diluted to an appropriate concentration with MEM-Eagle’s medium containing Hanks buffered saline solution (Sigma Chemical Co., St. Louis, Mo.), 20 mM HEPES buffer (pH 8.0), 3.5% sodium bicarbonate, 0.6% bovine serum albumin and 5% fetal calf serum (referred to as MEM-Etc.). The MDCK monolayers were washed once with MEM-Etc., and approximately 1.5 × 107 CFU of bacteria were added to the apical surface of the MDCK cell monolayer. MEM-Etc. alone was added to the lower wells. The bacteria were incubated with the MDCK cells for 3 h at 37°C in room air to allow invasion. The MDCK cell monolayers were then washed once with MEM-Etc. and incubated for 2 h at 37°C in the presence of 400 μg of amikacin (Sigma Chemical Co.) per ml in MEM-Etc. to kill extracellular bacteria. Amikacin was added instead of gentamicin because the modified Tn5 transposon used to generate the mutants contains a gentamicin resistance marker (22). Monolayers were then washed four times with MEM-Etc., and the MDCK cells were lysed by vortexing with glass beads in the presence of 0.25% Triton X-100. Lysates were plated on Luria-Bertani agar and incubated at 37°C for 17 h to quantify the CFU of viable internalized bacteria.
Analysis of secreted proteins.
Secreted proteins were analyzed as follows. Bacteria were grown in 5 ml of MINS supplemented with appropriate antibiotics at 37°C for 17 h with shaking. Broths were centrifuged, and supernatants were partially purified and concentrated by ammonium sulfate precipitation and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (26) on 8% gels, as previously described (22). Proteins were electrotransferred to nitrocellulose for immunoblot analysis or visualized by silver staining.
Protein purification.
Extracellular aggregates were noted in cultures of PA103. Analysis indicated that these aggregates were composed of PepA, a 40-kDa protein, a 32-kDa protein, and several other proteins (see Fig. 2C). These aggregates were used to purify PepB and PepD as follows. Bacteria were grown in 25 ml of MINS at 37°C for 17 h with shaking. Extracellular aggregates were removed from broths with sterile streaking loops, washed once in 10 mM NaCl, and solubilized in SDS sample buffer. After boiling for 5 min, samples were electrophoresed on 8% SDS-polyacrylamide gels. For amino-terminal peptide sequencing studies, the electrophoresed aggregate proteins were electroblotted onto polyvinylidene fluoride membranes (Applied Biosystems, Foster City, Calif.). Peptide sequencing was performed on a model 470A protein sequencer (Applied Biosystems) by the Biomolecular Resource Center of UCSF.
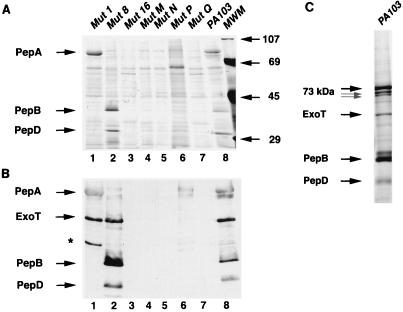
FIG. 2.
Analysis of secreted proteins of strain PA103 and mutants (Mut). Samples were prepared as described in the text and electrophoresed on SDS-polyacrylamide gels. The locations of bands corresponding to PepA, PepB, PepD, and ExoT are indicated. (A) Proteins present in concentrated culture supernatants as visualized by silver staining. Lane 6 is an example of a broth supernatant of mutant P with undetectable amounts of PepA, PepB, and PepD, but results varied as explained in the text. Molecular weight markers (MWM) are indicated on the right side of the panel. (B) Immunoblot analysis of proteins present in concentrated culture supernatants by using a combination of polyclonal antisera against PepA, PepB, PepD, and ExoT. The band in lane 1 indicated by an asterisk was noted occasionally in various samples and is thought to represent hybridization to a breakdown product of ExoT. (C) Electrophoretic analysis of broth protein aggregates of strain PA103. Samples were prepared and electrophoresed as described in the text. Bands were visualized by silver staining. The locations of bands corresponding to the PepA 73-, 71-, and 69-kDa triplet and to PepB, PepD, and ExoT are indicated.
Immunoblot analyses.
Polyclonal antiserum was prepared as described previously (4). Briefly, proteins were purified from PA103 broth aggregates by SDS-PAGE, excised from the acrylamide gels, and extracted by the method of Hager and Burgess (15). Proteins were then injected into rabbits, and sera were subsequently obtained by Animal Pharm Services, Inc. (Healdsburg, Calif.). Immunoblot analyses were performed by electrotransfer of ammonium sulfate-precipitated supernatant proteins from SDS-polyacrylamide gels to nitrocellulose followed by hybridization with the appropriate rabbit polyclonal antisera (43). Goat anti-rabbit immunoglobulin G horseradish peroxidase conjugate (Gibco BRL) diluted 1:5,000 was used as a secondary antibody. Detection was performed with the ECL system (Amersham Corp., Arlington Heights, Ill.).
Cosmid library preparation and manipulation.
A cosmid library of PA103 genomic DNA was constructed as follows. Chromosomal DNA from PA103 was purified by the freeze-thaw method (46) and partially digested with Sau3A. DNA fragments between 25 and 40 kb were isolated and purified by extraction from 0.4% agarose gels. The 3′ overhangs were partially filled in by incubation with the Klenow fragment (Gibco BRL) in the presence of dGTP and dATP. These fragments were then ligated with pLAFR5SK1, which had been previously digested with XhoI and incubated with the Klenow fragment in the presence of dTTP and dCTP. (The Klenow treatment prevented self-ligation of the genomic DNA fragments and the vector.) Ligated constructs were transduced into E. coli S17-1 (47) with a phage packaging kit (Stratagene Corp.). E. coli cells harboring cosmids were selected by growth on tetracycline-containing agar. A total of 1,536 individual cosmid clones were isolated.
DNA colony hybridization and Southern hybridization analyses.
Colony blots and hybridizations were performed by standard protocols (43). Probes were radiolabeled by nick translation (43). E. coli cells harboring cosmid clones were fixed to membranes in pools of eight and hybridized with radiolabeled probes. Members of the pools to which the probe bound were then fixed individually to membranes and again hybridized to the same probe to identify individual cosmid clones of interest.
Cosmid DNA used in Southern hybridization analyses was isolated by the alkaline lysis method (43). DNA was digested with the appropriate restriction endonucleases, electrophoresed on agarose gels (0.8%), and transferred to nylon membranes (Hybond-N; Amersham Corp.). Oligonucleotide probes were radioactively end labeled (43). Southern hybridizations, washes, and autoradiography were performed as described by Sambrook et al. (43).
Cloning of the genes encoding PepB and PepD.
Previously, 60 bp of nucleotide sequence adjacent to the insertion site of the transposon in mutant 1 had been determined by inverse PCR analysis (22). This short fragment, which did not show significant similarity to any sequences in the database, was radiolabeled and used to probe the cosmid library of PA103 genomic DNA. Three cosmid clones, 4-7-H, 4-11-H, and 5-9-E, contained sequences to which the probe hybridized.
These cosmids were analyzed further by Southern hybridization analysis. Degenerate oligonucleotide probes corresponding to the amino-terminal encoding portion of the PepB and PepD structural genes were synthesized, radioactively end labeled, and used as probes in these assays. Each probe hybridized to a 2.0-kb SmaI-SmaI fragment and an 8.0-kb EcoRI-EcoRI fragment present on both cosmids 4-7-H and 4-11-H, indicating that these cosmids may contain overlapping inserts. Endonuclease digestion analysis indicated that the 8.0-kb EcoRI-EcoRI fragment contained the entire 2.0-kb SmaI-SmaI fragment. Both fragments were purified by extraction from agarose gels, ligated into pBluescriptII, and transformed into E. coli XL1-Blue by standard protocols (43). Transformants were selected by growth on ampicillin plates. The plasmid containing the 2.0-kb SmaI-SmaI fragment was designated pAH830, and the plasmid containing the 8.0-kb EcoRI-EcoRI fragment was designated pAH831.
Nucleotide sequencing studies.
Both strands of the entire cloned 2.0-kb SmaI-SmaI insert of pAH830 were sequenced by a model 377 automated DNA sequencer (Applied Biosystems) at the Biomolecular Resource Center of UCSF. To complete sequencing of the 3′ open reading frame (ORF), which extended beyond the 3′ boundary of the 2.0-kb SmaI-SmaI insert, the 8.0-kb EcoRI-EcoRI insert of pAH831 was used as a template. Sequence analyses were performed with the Genetics Computer Group (Madison, Wis.) software package (3). Hydropathy analysis was performed by the method of Kyte and Doolittle (25).
Nucleotide sequence accession number.
The nucleotide sequences of the P. aeruginosa pepB and pepD genes have been submitted to GenBank and assigned accession no. AF035922.
RESULTS
Internalization of PA103 and isogenic mutants by MDCK cells.
To better assess the relationship between P. aeruginosa cytotoxicity and internalization, we examined the effects of mutations that eliminated cytotoxicity on the uptake of bacteria by epithelial cells. A variation (7) of the gentamicin exclusion assay (21) was used to quantify internalization by MDCK cells of parental strain PA103 and a group of 33 noncytotoxic isogenic mutants (Fig. 1). These mutants had been generated previously by transposon mutagenesis and partially characterized, as described elsewhere (22). Each harbors a single transposon insertion at a unique site. PA103 consistently lacked measurable internalization under the conditions of this assay, as did 27 of the 33 tested mutants (data not shown). Internalization, therefore, was not inversely correlated with cytotoxicity in a consistent manner, in that the majority of mutations decreased cytotoxicity without appreciably increasing bacterial uptake.
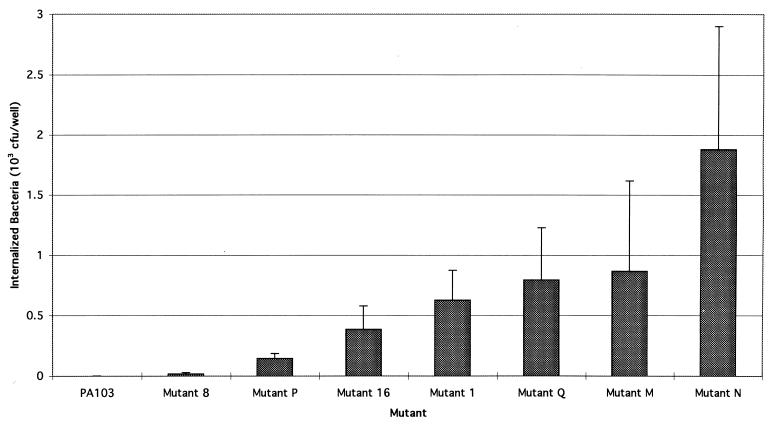
FIG. 1.
Internalization of strain PA103 and mutants. Assays were performed as described in the text. Each value represents the mean CFU of internalized bacteria per well. Although 27 of 33 tested mutants were not taken up by MDCK cells (e.g., mutant 8), six mutants, P, 16, 1, Q, M, and N, did demonstrate statistically significant internalization. Assays were performed a minimum of three times. Error bars represent standard errors of the means.
Six of the noncytotoxic mutants, however, demonstrated appreciable internalization (Fig. 1). These mutants were 1, 16, M, N, P, and Q. MICs and minimal bacteriocidal concentrations of amikacin, the aminoglycoside used in our internalization assays, were determined for each of these mutants and did not differ significantly from those of PA103 (data not shown), indicating that bacterial survival in the presence of antibiotic was due to internalization and not to increased resistance. Thus, transposon insertions in these six mutants had the dual effect of both decreasing the cytotoxic capacity of PA103 and increasing bacterial internalization by MDCK cells.
Protein secretion of the internalized mutants.
Since the loss of cytotoxicity alone did not correlate with bacterial internalization, we attempted to determine if mutants 1, 16, N, M, P, and Q shared any other properties that might account for their phenotype of internalization by MDCK epithelial cells. Interestingly, all six mutants had previously been reported to have potential defects in protein secretion (22). Two of these mutants, M and N, had transposon insertions in genes which had similarities to type III secretion genes of Yersinia spp. (22). We therefore further examined the secreted proteins of the six invasive mutants with protein gels. Culture supernatants were concentrated and partially purified by ammonium sulfate precipitation and analyzed by SDS-PAGE and silver staining (Fig. 2A and Table 2). Mutant 1 was defective in secretion of a 40-kDa protein, designated PepB for Pseudomonas exoprotein B), and a 32-kDa protein, designated PepD (Fig. 2A, lane 1). Mutants 16, M, N, and Q were defective in secretion of both of these proteins as well as PepA/ExoU, a 73-kDa protein necessary for cytotoxicity and virulence of PA103 (5, 17) (Fig. 2A, lanes 3, 4, 5, and 7). Repeated assays indicated that mutant P secreted variable amounts of the three proteins, ranging from undetectable to wild-type levels (Fig. 2A, lane 6). Immunoblot analysis with polyclonal antibodies against PepA, PepB, and PepD confirmed these findings (Fig. 2B). Together, these results verify those obtained previously by Kang et al. (22) and suggest that defects in secretion of PepA, PepB, or PepD by noncytotoxic mutants correlate with internalization.
TABLE 2.
Summary of PA103 and mutant phenotypes
| Strain | Invasiveness | Cytotoxicitya | Putative type III proteins secreted | Similarity of sequence at Tn5 insertion sitea | Postulated delivery to host cell of Putative AIFb |
|---|---|---|---|---|---|
| PA103 | No | Yes | PepA, PepB, PepD, ExoT | + | |
| Mutant 16 | Yes | No | Not tested | − | |
| Mutant M | Yes | No | yscN | − | |
| Mutant N | Yes | No | pscJ, yscJ | − | |
| Mutant P | Yes | No | PepA, PepB, PepD, ExoT (variable amounts) | Not tested | +/− |
| Mutant Q | Yes | No | Not tested | − | |
| Mutant 1 | Yes | No | PepA, ExoT | None | − |
| Mutant 8 | No | Minimal | PepB, PepD, ExoT | pepA/exoU | + |
| Mutant 13 | No | Minimal | PepB, PepD, ExoT, truncated PepA | pepA/exoU | + |
Further experiments, however, demonstrated that not all noncytotoxic mutants defective in secretion of one or more of these proteins were internalized. Mutant 8, which has previously been shown to be defective in production of PepA due to a transposon insertion in the gene encoding this protein (17), was not internalized (Fig. 1 and Fig. 2A and B, lane 2). Likewise, mutant 13, which secretes a truncated form of PepA (17), was not internalized (data not shown). Therefore, only a subset of secretion-defective noncytotoxic mutants were internalized—those that did not transport PepB and PepD. Furthermore, all noncytotoxic mutants that were not internalized did secrete PepB and PepD (data not shown).
PepB and PepD were further characterized in order that we might better understand why defective secretion of these proteins correlated with bacterial uptake. Interestingly, these proteins were noted to be present in insoluble macroscopic aggregates when PA103 was grown in MINS, a minimal medium known to enhance secretion of the type III effector protein ExoS (33). SDS-PAGE and immunoblot analyses demonstrated that the aggregates were composed primarily of PepA and its 71- and 69-kDa degradation products (17), PepB, PepD, and ExoT (Fig. 2C and data not shown). Interestingly, type III secreted proteins from other bacteria have been reported to form macroscopic aggregates in liquid cultures (20, 30, 36). Thus, the presence of PepB and PepD in aggregates containing ExoT, a protein secreted by a type III pathway (54), may indicate that PepB and PepD are also transported by this system.
Amino-terminal peptide sequences.
Following purification of PepB and PepD from these extracellular aggregates, amino-terminal peptide sequencing was performed. The amino terminus of PepB was found to be MNPITLERAGLPYGV and that of PepD was found to be MIDTQYSLAATQAAI. Of note, the amino-terminal residues of secreted PepB and PepD are methionines. These findings are consistent with both proteins being transported to the extracellular environment without cleavage of signal peptides, a hallmark of type III secreted proteins. Furthermore, they suggest that mutants 1, 16, M, N, P, and Q all have defects in type III secretion and that this transport pathway may be involved in modulating bacterial uptake by MDCK cells.
Type III secretion analyses.
To further investigate whether the mutants that demonstrated internalization upon contact with epithelial cells did indeed have defects in type III secretion, we chose to examine the transport of ExoT by these mutants. ExoT is secreted by a type III mechanism (54) and can therefore be used as a marker for an intact type III pathway. Immunoblot analysis of culture supernatants indicated that ExoT was secreted by PA103 (Fig. 2B, lane 8). (No cross-hybridization to the antigenically related protein ExoS was noted, because PA103 does not express ExoS.) In contrast, no hybridizing bands were noted in lanes corresponding to mutants 16, M, N, and Q (Fig. 2B, lanes 3, 4, 5, and 7). Thus, these mutants were defective in ExoT secretion as well as PepA, PepB, and PepD secretion. These findings are consistent with disruption of the type III secretion apparatus in mutants 16, M, N, and Q and with the premise that PepA, PepB, and PepD are type III transported proteins. Mutant P results were variable (Fig. 2B, lane 6). In contrast, mutants 1 and 8 secreted ExoT (Fig. 2B, lanes 1 and 2, respectively). The defect in mutant 8 is known to be limited to the gene encoding the effector molecule PepA (17); its type III secretion machinery is intact. Likewise, the defect in mutant 1 may be limited to secretion of PepB and PepD and may not involve the general type III secretion machinery, given the intact secretion of ExoT and PepA. Importantly, these results are consistent with each of the internalized mutants having defects in the type III secretion machinery or with one or more type III secreted proteins. In addition, bacterial internalization by epithelial cells correlates with loss of transport of PepB and PepD but not PepA (Table 2).
Cloning and sequencing of the genes encoding PepB and PepD.
Cloning of the genes encoding PepB and PepD, designated pepB and pepD, respectively, was performed to further characterize these proteins. As described in Materials and Methods, a PA103 genomic DNA cosmid library was screened by DNA hybridization analysis for the sequence into which the mutant 1 transposon had been inserted (obtained previously by inverse PCR [22]). Three cosmids, 4-7-H, 4-11-H, and 5-9-E, were identified as containing these sequences as well as the amino terminus-encoding portions of both pepB and pepD. Subsequent analysis demonstrated that an 8.0-kb EcoRI-EcoRI fragment of cosmid 4-7-H contained the amino terminus-encoding portions of both genes. Restriction mapping indicated that a 2.0-kb SmaI-SmaI fragment within the 8.0-kb EcoRI-EcoRI fragment contained the amino terminus-encoding portion of pepB and pepD. Both fragments were cloned and used in nucleotide sequencing studies.
Nucleotide sequence analysis of the 2.0-kb SmaI-SmaI fragment indicated the presence of two large ORFs separated by 11 nucleotides. One ORF was complete, while the second extended beyond the 3′ boundary of this fragment. Nucleotide sequencing of a portion of the 8.0-kb EcoRI-EcoRI fragment was performed to determine the sequence of the remaining portion of the incomplete ORF. Comparison of the inferred peptide sequences encoded by these ORFs to those obtained by amino-terminal sequencing studies indicated that the larger ORF (nucleotides 97 to 1269 of the submitted sequence) encoded PepB and that the second ORF (nucleotides 1281 to 2168 of the submitted sequence) encoded PepD. The molecular weight of PepB was predicted to be 39,961 and that of PepD was predicted to be 31,310. Shine-Dalgarno consensus sequences (45) were noted 5′ of the putative start codons of both pepB and pepD (data not shown). Since PepB and PepD purified from culture supernatants had the same amino-terminal peptide sequences as the deduced amino acid sequences, these proteins are likely not subject to amino-terminal signal cleavage upon secretion.
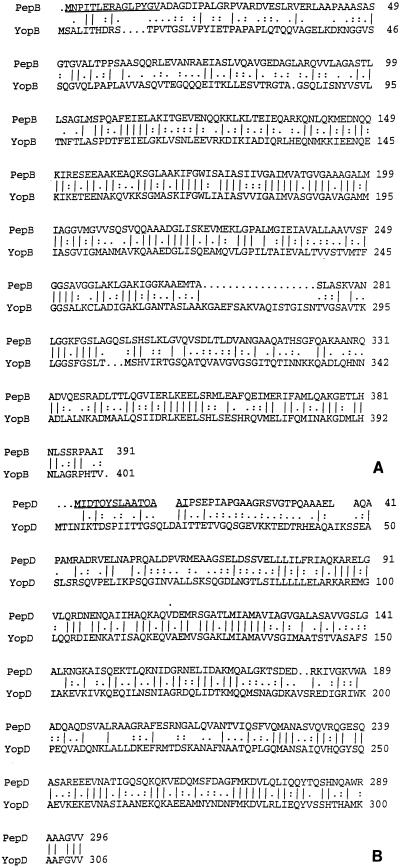
Comparison of the inferred peptide sequences encoded by these genes with the genetic database indicated that PepB and PepD have significant homology to YopB and YopD, respectively, type III secretion proteins of Yersinia spp. (16) (Fig. 3). Alignment of peptide sequences indicated that PepB was 41% identical to YopB of Yersinia enterocolitica. PepD was noted to be 38.8% identical to YopD of Y. enterocolitica. The spacing of only 11 nucleotides between the two genes suggested that they are part of an operon. PepB precedes PepD in this putative operon, as is the case with their Yersinia homologs (16), and the deduced proteins have similar sizes (YopB and YopD of Y. enterocolitica have molecular weights of 41,942 and 33,234, respectively). In addition, hydropathy analyses of the deduced amino acid sequences of PepB and PepD demonstrated patterns very similar to those of YopB and YopD, respectively (data not shown). For example, both PepB and YopB contain two hydrophobic regions in the central parts of the proteins consisting of approximately 44 and 35 amino acid residues (16). These portions of YopB are postulated to represent transmembrane domains, and their presence in PepB may indicate that this protein is also localized to the cytoplasmic membranes of eukaryotic cells. Hydropathy analyses also predicted a hydrophobic region consisting of approximately 31 amino acid residues in the central part of PepD, as has been shown for YopD (16). These findings suggested that PepB and PepD are P. aeruginosa homologs of YopB and YopD, respectively, and may be necessary for translocation of type III secretion effector molecules into host cells.
FIG. 3.
Deduced peptide sequence comparisons of PepB with YopB (A) and PepD with YopD (B). Underlining represents sequences obtained by amino-terminal peptide sequencing of secreted PepB and PepD. Alignments were performed by the method of Needleman and Wunsch (31). |, identity; :, residues with comparison values greater than or equal to the average positive nonidentical comparison value in the scoring matrix; ., residues with comparison values greater than or equal to 1.
The G+C content of the pepB coding sequence was found to be 65.6%, and that of the pepD coding sequence was found to be 68.0%. These values are in close agreement with those reported for the P. aeruginosa genome (67.2%) (35) and the pscB-L locus (67.5%) (54). The pscB-L locus encodes structural components of the P. aeruginosa type III secretion apparatus and is homologous to the yscB-L locus of Yersinia spp. (54).
Comparison of the nucleotide sequence of a partial ORF (nucleotides 2295 to 2688 of the submitted sequence) 3′ of pepD with sequences in the genetic database indicated that this ORF contains the coding sequence for ExsC, a part of the ExoS trans-regulatory locus (12). This regulatory locus is 5′ of the pscB-L operon (54). Thus, pepB, pepD, exsC, exsB, exsA, exsD, and pscB-L are part of a single pathogenicity island, and additional genes important in type III secretion may lie on either side of this locus.
DISCUSSION
We have examined the effects that mutations which decrease P. aeruginosa cytotoxicity have on bacterial internalization. An inverse correlation between cytotoxicity and internalization had been observed in previous studies with nonisogenic clinical isolates (9). In contrast, we were unable to detect internalization for 27 of 33 tested isogenic mutants with absent or minimal cytotoxicity. These findings provide further evidence that the absence of cytotoxicity does not necessarily imply a phenotype of internalization (8).
Unlike the majority of the examined noncytotoxic transposon insertion mutants, six such mutants were internalized by MDCK epithelial cells. This was surprising given that the parental strain, P. aeruginosa PA103, was not appreciably internalized. Each of the six mutants had properties suggesting defects in secretion. In particular, two or more of four distinct proteins were absent in culture supernatants of these mutants compared to the parental strain. These proteins were PepA, PepB, PepD, and ExoT. Of interest, all mutants with defects in transport of PepB and PepD were internalized, suggesting that secretion of these proteins may be essential for preventing bacterial uptake by MDCK epithelial cells.
Several properties of PepB and PepD were consistent with secretion by a type III pathway. Mutants M and N, which failed to secrete both PepB and PepD, have previously been shown to harbor transposon insertions in homologs of genes that encode structural components of the type III secretion machinery of Yersinia spp. (22) (Table 2). Both PepB and PepD are found in the supernatants of cultures during growth in media known to induce secretion of ExoS, a type III secreted protein. Both are secreted without amino-terminal processing. PepB and PepD form extracellular macroscopic aggregates with ExoT and PepA during growth in liquid cultures. Such aggregate formation has also been reported with regard to the type III secretion systems of other bacteria (20, 30, 36). Finally, PepB and PepD are themselves homologs of YopB and YopD, type III secreted proteins of Yersinia spp. Taken together, these findings suggest that defects in secretion of specific type III proteins correlate with internalization of P. aeruginosa by MDCK cells. Thus, in striking contrast to Salmonella typhimurium and Shigella spp., an intact type III secretion system in P. aeruginosa correlates with inhibition rather than augmentation of bacterial uptake by epithelial cells. This conclusion agrees with previously published results indicating that PA103exsA::Ω, a mutant of PA103 defective in expression of the type III system transcriptional activator ExsA, is internalized (9).
That PepB and PepD are homologs of YopB and YopD, respectively, may explain the noncytotoxic phenotype of mutant 1, which does not secrete these two proteins. While the lack of cytotoxicity of mutants 16, M, N, P, and Q was thought to be due to defects in secretion of PepA, the putative cytotoxin (5, 17), it was previously unclear why mutant 1 was noncytotoxic. Our results suggest that although mutant 1 secretes PepA, this protein is unable to reach the interior of epithelial cells, where it is functionally active, because of the absence of the PepB-PepD translocator.
A possible explanation for the internalization phenotype of the noncytotoxic mutants is as follows (Table 2). The lack of measurable internalization of the parental strain, PA103, is consistent with the secretion of a substance that inhibits bacterial uptake by epithelial cells. Mutants 16, M, N, and Q, which appear to be defective in secretion of multiple type III proteins, are internalized by epithelial cells. This suggests that the uptake-inhibiting molecule is secreted by a type III mechanism. Mutant 1, which secretes neither PepB nor PepD, is also internalized. This may indicate that these two proteins themselves function to inhibit bacterial internalization or that they are required for translocation of an internalization-inhibiting effector molecule into the host cell. Mutant 8, which is specifically defective in secretion of PepA (17), is informative in this regard. As is the case for the parental strain, PA103, mutant 8 does not exhibit appreciable internalization. Thus, PepA does not modulate bacterial uptake and is not the effector molecule responsible for inhibition of internalization. Our results are therefore consistent with different type III secretion effector molecules being responsible for cytotoxicity and for inhibiting bacterial uptake. It must be stressed, however, that our results demonstrate only correlations between protein secretion and bacterial uptake. Complementation studies will be necessary to conclusively demonstrate that these genetic defects are responsible for the lack of internalization of these mutants.
Although there are many possible explanations for the observations we describe, one model of the interaction between the P. aeruginosa type III secretion system and MDCK epithelial cells is shown in Fig. 4. This model assumes that in the absence of active inhibition of uptake, P. aeruginosa is internalized by MDCK epithelial cells. The type III system of PA103 secretes a cytotoxin(s) as well as a molecule, designated AIF (for anti-internalization factor), that functions to prevent the default bacterial internalization. (The former may be PepA, which is necessary for cytotoxicity in PA103 [5, 17].) This explains why the phenotype of wild-type PA103, which secretes both the cytotoxin and AIF, is cytotoxic but noninternalized. Anything that prevents the delivery of the cytotoxin to the cytoplasmic compartment of the host cell will result in loss of cytotoxicity. Likewise, the lack of delivery of AIF into the host cell yields a phenotype of bacterial internalization. Blocks in delivery of both of these factors can occur at several steps along the delivery pathway. For example, isogenic mutants of PA103 that are defective in expression or secretion of all type III proteins (e.g., PA103exsA::Ω or mutant N) secrete neither the cytotoxin nor AIF and are therefore noncytotoxic and internalized. Mutants which secrete both the cytotoxin and AIF but are unable to translocate them into the cytoplasmic compartment of eukaryotic cells (e.g., mutant 1, which secretes neither PepB nor PepD), where they are functionally active, are likewise noncytotoxic and internalized. Finally, mutants specifically defective in secretion of the putative cytotoxin PepA (e.g., mutant 8) are noncytotoxic. These mutants, however, still secrete and deliver AIF, and their uptake by epithelial cells is therefore inhibited; they are not internalized. Mutants that are specifically defective in secretion or delivery of AIF would be cytotoxic and internalized. The data presented here are compatible with any type III effector protein except PepA being AIF. ExoT is a candidate for such a factor in that it is secreted by the PA103 type III secretion system (54) and its function is currently unclear. It must be emphasized that PA103 does not secrete ExoS, and the role of this protein in cytotoxicity and inhibition of bacterial internalization (9) remains unclear. This model is highly speculative but is in agreement with what is currently known about Yersinia, which harbors the type III secretion system most closely related to that of P. aeruginosa (12, 24, 54). The Yersinia type III system secretes YopH, which blocks the invasion normally mediated by the bacterial surface protein invasin, and YopE, a potent cytotoxin (11). With regard to P. aeruginosa, the physiological relevance of bacterial internalization and secretion of factors that block it awaits further characterization.
FIG. 4.
Model of the interaction between the PA103 type III secretion system and epithelial cells. In wild-type bacteria, the transcriptional activator ExsA allows expression of type III secretion genes. These genes encode PepB, PepD, and effector proteins such as a cytotoxin(s) and a putative factor that inhibits bacterial internalization by epithelial cells, AIF. The cytotoxin and AIF are transported out of the bacterium by the secretion apparatus, which consists of PscJ and other proteins. The cytotoxin and AIF are then translocated by the PepB-PepD complex into the cytoplasmic compartments of epithelial cells, where they act to kill the cells and inhibit bacterial internalization, respectively. Thus, wild-type PA103 has a cytotoxic and noninternalized phenotype. Potential defects present in the mutants discussed in this study are numbered as follows. (1) An ExsA mutant is unable to transcribe type III secretion genes and therefore synthesizes neither the cytotoxin nor AIF (although our data is consistent also with AIF regulation being independent of ExsA). This mutant is noncytotoxic and internalized. (2) Mutant 8 is defective in production of the putative cytotoxin but does secrete AIF. It therefore does not kill epithelial cells, but delivery of AIF to the host cell cytoplasmic compartment prevents bacterial internalization. (3) Mutant N is defective in production of PscJ, a component of the bacterial type III secretion apparatus, and can secrete neither the cytotoxin nor AIF. It is thus noncytotoxic and internalized. (4) Mutant 1 is defective in secretion of PepB and PepD and is therefore unable to translocate the cytotoxin and AIF into epithelial cells. This mutant is also noncytotoxic and internalized by epithelial cells.
ACKNOWLEDGMENTS
We thank Jane Koehler and members of the Engel laboratory for reading the manuscript and for scientific advice. We thank Lucy Shapiro for the pLAFR5SK1 cosmid vector.
This work was supported by grants from the Cystic Fibrosis Research Institute (J.N.E.), the Lucille Markey Biomedical Foundation (J.N.E.), and the NIH (K.M. [RO1 HL55980] and S.F. [RO1 EY11221]); by a UC Berkeley Faculty Research Grant (S.F.); and by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship grant DRG-080 (A.R.H.). During a portion of this work, J.N.E. was a Lucille Markey Biomedical Scholar. K.M. is an Established Investigator of the American Heart Association.
ADDENDUM
Following submission of the manuscript, Yahr et al. (53) published the nucleotide sequences of the genes encoding PopB and PopD. Comparison of these sequences to those of pepB and pepD indicates that PepB and PopB are the same protein, as are PepD and PopD.
REFERENCES
- 1.Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D, Mostov K, Wiener-Kronish J. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi E, Mehl T, Nunn D, Lory S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect Immun. 1991;59:822–828. doi: 10.1128/iai.59.3.822-828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel J, Pollack J, Malik F, Ganem D. Cloning and characterization of RNA polymerase core subunits of Chlamydia trachomatis using the polymerase chain reaction. J Bacteriol. 1990;172:5732–5741. doi: 10.1128/jb.172.10.5732-5741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finck-Barbancon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M J, Wu C, Mende-Mueller L, Frank D. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 6.Fleiszig S M, Zaidi T S, Fletcher E L, Preston M J, Pier G B. Pseudomonas aeruginosa invades corneal epithelial cells during experimental infection. Infect Immun. 1994;62:3485–3493. doi: 10.1128/iai.62.8.3485-3493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleiszig S M, Zaidi T S, Pier G B. Pseudomonas aeruginosa invasion of and multiplication within corneal epithelial cells in vitro. Infect Immun. 1995;63:4072–4077. doi: 10.1128/iai.63.10.4072-4077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleiszig S M J, Evans D J, Vallas V, Frank D W. Abstracts of the 97th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1997. Epithelial cell invasion is not a prerequisite for cytotoxicity induced by Pseudomonas aeruginosa, abstr. B-354; p. 89. [Google Scholar]
- 9.Fleiszig S M J, Wiener-Kronish J P, Miyazaki H, Vallas V, Mostov K, Kanada D, Sawa T, Yen T S B, Frank D. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleiszig S M J, Zaidi T S, Preston M J, Grout M, Evans D J, Pier G B. Relationship between cytotoxicity and corneal epithelial cell invasion by clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:2288–2294. doi: 10.1128/iai.64.6.2288-2294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsberg A, Rosqvist R, Wolf-Watz H. Regulation and polarized transfer of the Yersinia outer proteins (Yops) involved in antiphagocytosis. Trends Microbiol. 1994;2:14–19. doi: 10.1016/0966-842x(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 12.Frank D, Iglewski B H. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frank D, Nair W G, Schweizer H P. Construction and characterization of chromosomal insertional mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect Immun. 1994;62:554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galloway D R. Pseudomonas aeruginosa elastase and elastolysis revisited: recent developments. Mol Microbiol. 1991;5:2315–2321. doi: 10.1111/j.1365-2958.1991.tb02076.x. [DOI] [PubMed] [Google Scholar]
- 15.Hager D A, Burgess R R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- 16.Hakansson S, Bergman T, Vanooteghem J-C, Cornelis G R, Wolf-Watz H. YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect Immun. 1993;61:71–80. doi: 10.1128/iai.61.1.71-80.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauser A R, Kang P J, Engel J. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 18.Horan T, Culver D, Jarvis W. Pathogens causing nosocomial infections. Antimicrob Newsl. 1988;5:65–67. [Google Scholar]
- 19.Hovey A K, Frank D W. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of te Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1995;177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 21.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature. 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 22.Kang P J, Hauser A R, Apodaca G, Fleiszig S, Wiener-Kronish J, Mostov K, Engel J N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol. 1997;24:1249–1262. doi: 10.1046/j.1365-2958.1997.4311793.x. [DOI] [PubMed] [Google Scholar]
- 23.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 24.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Liu P V. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. II. Effects of lecithinase and protease. J Infect Dis. 1966;116:112–116. doi: 10.1093/infdis/116.1.112. [DOI] [PubMed] [Google Scholar]
- 28.Mecsas J, Strauss E. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg Infect Dis. 1996;2:271–288. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michiels T, Wattlau P, Brasseur R, Ruysschaert J, Cornelis G. Secretion of Yop proteins by yersiniae. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 32.Nicas T, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 33.Nicas T I, Iglewski B H. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984;45:470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohman D E, Burns R P, Iglewski B H. Corneal infections in mice with toxin A and elastase mutants of Pseudomonas aeruginosa. J Infect Dis. 1980;142:547–555. doi: 10.1093/infdis/142.4.547. [DOI] [PubMed] [Google Scholar]
- 35.Palleroni N J. Pseudomonadaceae. In: Kreig N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. pp. 141–219. [Google Scholar]
- 36.Parsot C, Menard R, Gounon P, Sansonetti P. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 37.Persson C, Nodfelth R, Homstrom A, Hakansson S, Rosquist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 38.Pier G, Meluneni G, Neuger E. A murine model of chronic mucosal colonization by Pseudomonas aeruginosa. Infect Immun. 1992;60:4768–4776. doi: 10.1128/iai.60.11.4768-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pier G B, Grout M, Zaidi T S, Olsen J C, Johnson L G, Yankaskas J R, Goldberg J B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plotkowski M-C, Saliba A M, Pereira S H M, Cervante M P, Bajolet-Laudinat O. Pseudomonas aeruginosa selective adherence to and entry into human endothelial cells. Infect Immun. 1994;62:5456–5463. doi: 10.1128/iai.62.12.5456-5463.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosqvist R, Magnusson K-E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C: ASM Press; 1994. [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Schesser K, Frithz-Lindsten E, Wolf-Watz H. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J Bacteriol. 1996;178:7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silhavy T, Berman M, Enquist L. Experiments with gene fusion. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 47.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 48.Sory M-P, Lambermont B A I, Cornelis G. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasil M L, Graham L M, Ostroff R M, Shortridge V D, Vasil A I. Phospholipase C: molecular biology and contribution to the pathogenesis of Pseudomonas aeruginosa. In: Homma J Y, Tanimoto H, Holder I A, Hoiby N, Doring G, editors. Pseudomonas aeruginosa in human diseases. Vol. 44. Basel, Switzerland: Karger; 1991. pp. 34–47. [DOI] [PubMed] [Google Scholar]
- 50.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 51.Woods D E, Hwang W W, Shahrabade M S. Alteration of pulmonary structure by Pseudomonas aeruginosa exoenzyme S. J Med Microbiol. 1988;26:133–141. doi: 10.1099/00222615-26-2-133. [DOI] [PubMed] [Google Scholar]
- 52.Yahr T, Frank D. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yahr T, Mende-Mueller L M, Friese M B, Frank D W. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III secretion pathway. Mol Microbiol. 1996;22:991–1003. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 55.Zierler M, Galan J E. Contact with cultured epithelial cells induces the secretion of the Salmonella typhimurium invasion protein InvJ. Infect Immun. 1995;63:4024–4028. doi: 10.1128/iai.63.10.4024-4028.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoutman D E, Hulbert W, Pasloske B, Joffe A, Volpel K, Trebilcock M, Paranchych W. The role of polar pili in the adherence of Pseudomonas aeruginosa to injured canine tracheal cells: a semiquantitative morphological study. Scanning Microsc. 1991;5:109–126. [PubMed] [Google Scholar]