Abstract
Streptococcus pyogenes evades complement by binding the complement-regulatory protein factor H (fH) via the central conserved C-repeat region of M protein. However, the corresponding binding region within fH has not previously been precisely localized. fH is composed of 20 conserved modules called short consensus repeats (SCRs), each of which contains approximately 60 amino acids. A series of fH truncated and deletion mutants were prepared, and their interaction with M6 protein was examined. The M protein binding site was initially localized to SCRs 6 to 15 as demonstrated by ligand dot blotting, chemical cross-linking, and enzyme-linked immunosorbent assay. SCR 7 was then shown to contain the M protein binding site, as a construct consisting of the first seven SCRs bound M protein but a construct containing the first six SCRs did not bind. In addition, deletion of SCR 7 from full-length fH abolished binding to M protein. SCR 7 is known to contain a heparin binding domain, and binding of fH to M6 protein was almost totally inhibited in the presence of 400 U of heparin per ml. These results localize the M6 protein binding site of fH to SCR 7 and indicate that it is in close proximity to the heparin binding site.
The group A Streptococcus (Streptococcus pyogenes) is one of the most common and virulent human pathogens. It is responsible for a wide range of suppurative infections, ranging from skin infections and pharyngitis to necrotizing fasciitis, bacteremia, and overwhelming infection (4). Postinfectious sequelae of glomerulonephritis and rheumatic fever cause widespread morbidity and mortality, especially in developing countries (33).
Group A streptococci possess a wide variety of virulence factors, including M protein, hyaluronic acid capsule, serum opacity factor, C5a peptidase, and extracellular enzymes and toxins (8). M protein has been intensively studied since Lancefield showed that M protein-rich strains are resistant to phagocytic killing in nonimmune human blood (24). M protein appears on electron microscopy as multiple hairlike projections on the cell surface and consists of coiled dimers (30, 34). The N terminus of M protein, which is distal to the bacterial surface, contains a hypervariable region which defines more than 100 serotypes (19).
Strains of group A streptococci lacking M protein are efficiently opsonized by the alternative pathway of complement, but in the absence of type-specific antibody neither the alternative nor the classical pathway is activated by strains expressing M protein (3, 29). Horstmann et al. demonstrated that M6 protein and other M serotypes bind factor H (fH), a regulatory protein of the complement system, resulting in reduced deposition of C3b on the streptococcal surface (15). The fH binding site on M6 protein has subsequently been localized to the central conserved C-repeat region (11).
fH regulates complement activation by acting as a cofactor for factor I-dependent cleavage of C3b (27) and by disrupting the alternative pathway C3 convertase (35, 37). fH is a member of the genetically and structurally related regulators of complement activation family of proteins. These proteins all contain similar repetitive structural units of approximately 60 amino acids called short consensus repeats (SCRs) (16). Each SCR contains approximately 17 conserved amino acids involved in maintaining the tertiary structure of the module. The ligand binding specificity of each SCR is thought to reside within the remaining less well-conserved regions (2).
fH is composed of 20 SCR units, each of which is independent of its neighbor (2). This modularity makes it possible to delete individual or groups of SCRs without disrupting the overall structure of the protein. We and others have mapped the SCR domains required for cofactor activity, decay acceleration, C3b binding, and heparin and sialic acid binding (6, 13, 22, 23). During the course of these investigations, Sharma and Pangburn determined that the M protein binding site in fH is located within SCR 6 to SCR 10 (31). In this study we confirmed and extended this finding by localizing the M protein binding site to SCR 7 of fH. Moreover, we demonstrate that heparin inhibits the binding of fH to M6 protein, indicating that the two binding sites of fH are closely related in SCR 7.
MATERIALS AND METHODS
M6 protein.
M6 protein was purified from the periplasm of transformed Escherichia coli as previously described (12). M6 protein was biotinylated by incubating 500 μg of M6 protein per ml with 1,500 μg of NHS-biotin (Pierce, Rockford, Ill.) per ml in 50 mM bicarbonate buffer for 30 min at room temperature. Excess biotin was removed by ultrafiltration in a Microcon 10 microconcentrator (Amicon, Beverly, Mass.).
Cloning and expression of fH mutant proteins.
cDNA encoding full-length fH (H20), the first seven SCRs of fH (H7), and an SCR 7 deletion of fH (H20Δ7) were cloned into the mammalian expression vector BSRαEN as previously described (6, 13).
His-tagged proteins composed of the N-terminal 5 or 15 SCRs (H5 or H15, respectively) were expressed in CHO cells and purified by Ni2+ affinity chromatography. The construct BSRαEN-H5His was prepared by incorporating into the reverse primer an EcoRI site, a stop codon, six codons encoding His, and an XbaI site (reading 5′ to 3′). The forward primer was designed to anneal just 5′ to the 18-residue leader sequence and incorporated an XhoI restriction site. cDNA was amplified by PCR from a BSRαEN-H20 template by using Vent polymerase (New England Biolabs, Beverly, Mass.), and was cloned into the XhoI and EcoRI restriction sites of BSRαEN. This strategy introduced codons for Ser, Thr, and six His residues into the construct, and the introduced XbaI site was used to prepare cDNAs encoding other His-tagged proteins without the need for long reverse primers. BSRαEN-H15His was amplified by PCR in this manner, using a reverse primer incorporating an XbaI restriction site without a stop codon. Correct identity of the cloned products was shown by restriction analysis, partial sequencing of the cDNA, and Western blotting of the expressed protein.
CHO cells were stably transfected as previously described (6). Cells were grown in HyQ serum-free medium (HyClone, Logan, Utah) containing 250 μg of G418 (Life Technologies, Gaithersburg, Md.) per ml. Cell supernatants were harvested twice weekly, clarified by centrifugation, and stored at −70°C. H20 was purified by antibody affinity chromatography with anti-fH antibodies raised in rabbits. After washing of the column, bound protein was eluted in 3 M glycine acetate (pH 3), dialyzed against 50 mM phosphate buffer, and concentrated by placing the dialysis tubing in dry polyethylene glycol flakes (Mr 20,000; Sigma, St. Louis, Mo.). H5His and H15His were batch purified with Ni2+-nitrilotriacetic acid-agarose (Qiagen Inc., Chatsworth, Calif.). Bound proteins were eluted in 50 mM imidazole (Sigma), and the buffer was changed to 50 mM phosphate with Microcon spin concentrators.
Recombinant H7His and H6His were kindly provided by Jens Hellwage, Bernhard Nocht Institute, Hamburg, Germany. They were prepared in the pBSV-8His baculovirus expression system as previously described (21).
fH was purified from pooled human serum as previously described (1).
Binding of fH and mutant proteins to M6 protein. (i) Ligand dot blotting.
Five micrograms each of M6 protein and albumin was dried onto a nitrocellulose membrane (Hybond C+; Amersham, Buckinghamshire, United Kingdom) and dried at 37°C for 30 min. Nonspecific binding sites were blocked by incubation with 5% skim milk in 50 mM phosphate buffer for 60 min, and the membrane was then incubated with 0.5 μg of fH or mutant protein per ml for 3 h. Bound protein was then detected by immunoblotting, using polyclonal goat anti-fH antibody (Calbiochem, San Diego, Calif.) and horseradish peroxidase (HRP)-conjugated protein A (Pierce), and finally identified with the ECL chemiluminescence system (Amersham).
(ii) ELISA.
For the enzyme-linked immunosorbent assay (ELISA), 0.2 μg of M6 protein or albumin in 100 mM bicarbonate buffer (pH 9.5) was applied overnight to Maxisorb ELISA plates (Nunc, Copenhagen, Denmark). After blocking in 5% skim milk, test proteins were added and incubated for 3 h. After washing with 50 mM phosphate buffer–0.05% Tween 20, bound protein was detected by using polyclonal goat anti-fH antibody and HRP-conjugated protein A as described above. Substrate was added, and the optical density was determined at 490 nm.
The effects of heparin on the M protein-fH interaction were assessed by incubating H7 with immobilized M6 protein in the presence of 0 to 1,600 U of porcine heparin (David Bull Laboratories, Victoria, Australia) per ml. Binding was then assessed by using the same ELISA format as described above.
(iii) Chemical cross-linking.
Biotinylated M6 protein (0.7 μg) and 1 μg of fH, H15, or H5 were incubated in 50 mM bicarbonate buffer (pH 8.5) in a final volume of 10 μl. After 20 min, dithiobis(succinimidylpropionate) (DSP) (Pierce) was added to 1 mg/ml and incubated for another 30 min at room temperature. The reaction was quenched by the addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) nonreducing buffer, and the samples were separated with a 5 to 15% gradient gel. Duplicate reactions without addition of the cross-linker were included. After transfer to nitrocellulose, biotinylated M6 protein was detected with streptavidin-HRP (Vectastain; Vector Laboratories, Burlingame, Calif.) and chemiluminescence (Amersham).
RESULTS
Expression of fH and fH mutant proteins.
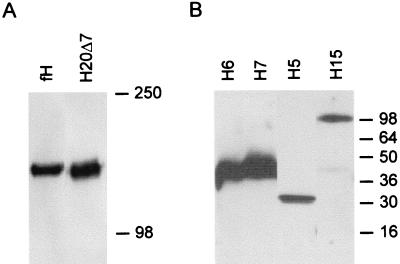
Figure 1 shows Western blots of the fH and mutant fH proteins used in these experiments. All proteins migrated on SDS-PAGE as single bands at the expected molecular weights.
FIG. 1.
Western blots of fH and fH mutant proteins. CHO cells were transfected with the appropriate cDNA, and supernatants were collected and separated by SDS-PAGE under nonreducing conditions. H20 and H20Δ7 were separated by SDS–6% PAGE (A). H5His, H6His, H7His, and H15 His were separated by SDS–12% PAGE (B). Recombinant proteins were detected by Western blotting, using polyclonal anti-fH antibodies as described in Materials and Methods. Apparent molecular masses (in kilodaltons) are indicated.
Binding of fH, H15, and H5 to M protein.
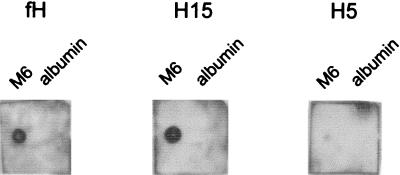
Preliminary localization of the M protein binding site of fH was obtained by ligand dot blotting. M6 protein or albumin, immobilized onto nitrocellulose, was incubated with either full-length fH or recombinant truncated proteins containing the first 15 (H15) or first 5 (H5) SCRs. Both fH and H15 bound to immobilized M6 protein, but not to albumin, while no binding of H5 occurred (Fig. 2). These results indicate that the M protein binding site is located somewhere between SCR 6 and SCR 15 inclusive.
FIG. 2.
Ligand dot blotting of binding of fH, H15, and H5 to immobilized M6 protein. M6 protein and albumin were dried onto nitrocellulose, blocked, and incubated with approximately 0.5 μg of the indicated fH-derived protein per ml. Bound protein was detected with anti-fH polyclonal antibodies and HRP-conjugated protein A followed by chemiluminescence.
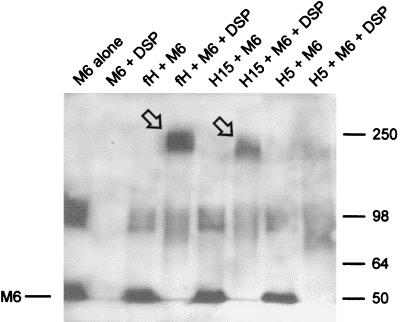
The interaction between M protein and fH, H15 and H5 was also investigated by chemical cross-linking experiments (Fig. 3). Biotinylated M protein was incubated with fH or truncated proteins in the presence or absence of DSP. The cross-linked proteins were analyzed by SDS-PAGE under nonreducing conditions and Western blotting. M6-containing proteins were then detected by streptavidin-HRP and chemiluminescence. In the absence of DSP, M6 protein migrated as two bands: as a monomer at the expected apparent molecular mass of ∼50 kDa and as a dimer of ∼100 kDa. An M6-containing cross-linked protein of ∼230 kDa appeared when M6 protein and fH were incubated with DSP, consistent with the combined molecular masses of dimeric M6 protein and fH. When H15 was substituted for fH, the cross-linked M6-containing protein migrated at around ∼200 kDa, consistent with the smaller size of H15. In contrast, no band corresponding to an M6-H5 cross-linked protein was detected (Fig. 3). Under reducing conditions, the higher-molecular-mass cross-linked proteins migrated as a ∼50-kDa single M6 protein band, indicating disruption of the disulfide bonds within the cross-linking agent (data not shown). Results from cross-linking experiments were thus consistent with those obtained by ligand dot blotting and confirmed that M6 protein binds to fH and H15 but not to H5.
FIG. 3.
Chemical cross-linking of fH, H15, and H5 to M6 protein. Biotinylated M6 protein and fH, H15, or H5 were incubated in 50 mM bicarbonate buffer (pH 7.4) with or without DSP cross-linker, as described in Materials and Methods. Proteins were separated by SDS–6% PAGE under nonreducing conditions, transferred to nitrocellulose, and then incubated in streptavidin-HRP. M6 protein and cross-linked M6 protein were detected by chemiluminescence. Cross-linking of M6 protein to fH and H15 is indicated by the arrows.
M protein interacts with SCR 7 of fH.
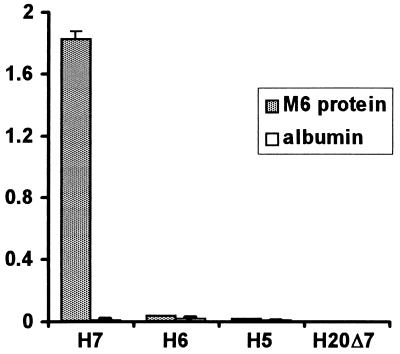
An ELISA test format was next used to further examine the binding of a series of fH constructs, including H7 and H6. H7, but neither H6 nor H5, bound to immobilized M6 protein (Fig. 4). No binding of any fH protein construct to albumin was detected. As expected, fH and H15 also bound to immobilized M6 protein (data not shown). These results indicate that SCR 7 is required for M protein binding. In addition, deletion of SCR 7 from H20 (H20Δ7) abolished all M6 protein binding (Fig. 4), confirming the requirement for SCR 7 and indicating that no other binding site is present in fH. Consistent results were obtained when His-tagged proteins or proteins expressed in different cell lines were used: for example, both H7 and H7His bound M6 protein, and H5His did not (data not shown).
FIG. 4.
Binding of C-terminal truncation mutants of fH to immobilized M6 protein. fH mutant proteins were added to ELISA wells on which M6 protein or albumin had been immobilized. After washing, bound protein was detected with polyclonal goat anti-fH antibodies followed by protein A-HRP and substrate. Amounts of fH mutant proteins equivalent to those shown in Fig. 1 were used. Results shown are the mean optical densities at 490 nm for four experiments, and error bars represent one standard deviation.
Binding of H7 to M protein is inhibited by heparin.
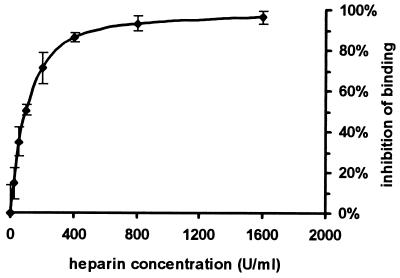
We have previously shown that the major heparin and sialic acid binding site of fH is located within SCR 7 (5). We therefore examined the effect of heparin on the H7-M protein interaction. Binding of H7 to M protein was almost totally inhibited by heparin concentrations of greater than 400 U/ml (Fig. 5).
FIG. 5.
Effect of heparin on binding of H7 to M6 protein. H7 in 50 mM phosphate buffer (pH 7.4) containing between 0 and 1,600 U of heparin per ml was incubated with immobilized M6 protein. Binding of H7 was measured by the same ELISA method described in the legend to Fig. 4. Results are expressed as [1 − (optical density at 490 nm of test wells/optical density at 490 nm of wells without heparin)] × 100. Error bars represent one standard deviation.
DISCUSSION
Binding of fH is one possible mechanism by which M protein exerts its antiphagocytic effects and contributes to the virulence of group A streptococci. fH binds to the C-repeat domain of M protein (11), but the corresponding M protein binding site on fH has not been precisely localized. The results of this study confirm and extend the results of Sharma and Pangburn (31), who showed that the M protein binding site of fH is located somewhere within SCRs 6 to 10. We have now more precisely localized the M protein binding site to SCR 7 of human fH.
Our initial results showed that fH binds to M protein via a site within SCRs 6 to 15 (Fig. 2 and 3). After the publication of Sharma and Pangburn’s results (31), we concentrated on SCR domains 6 to 10. By using an ELISA-based method, the M protein binding characteristics of proteins containing the N-terminal 5, 6, and 7 SCRs (H5, H6, and H7) were examined. H7 was the only one of these constructs to bind M protein, indicating the presence of the binding site in SCR 7. Moreover, a recombinant protein consisting of full-length fH from which SCR 7 had been deleted (H20Δ7) failed to bind M protein (Fig. 4). This result confirms that SCR 7 contains the M protein binding site and demonstrates that there are no other binding sites for M protein within fH. This is in contrast to heparin binding, which has been localized to two sites: one in SCR 7 (6) and another within or near SCR 20 (5). Therefore, deletion of SCR 7 from fH completely abrogates M protein binding yet leaves intact the second heparin binding site.
As we have previously shown that SCR 7 of fH contains an important heparin and sialic acid binding site (6), the influence of free heparin on the M protein-fH interaction was assessed. Heparin markedly inhibited binding of H7 to immobilized M protein, with almost complete inhibition obtained at concentrations above 400 U/ml (Fig. 5). This result demonstrates that the binding sites for M protein and heparin in SCR 7 are closely related or identical. However, binding of fH to heparin does not appear to be as stringent as the binding of fH to M6 protein: there are two binding sites for heparin (in SCRs 7 and 20), whereas there is just one for M6 protein (in SCR 7). Analysis of the linear amino acid sequences of SCRs 7 and 20 does not immediately identify putative M6 protein or heparin binding sites, but such analysis may be misleading because it does not take into account the complicated tertiary structure of the SCR. The most useful method to further define functional sites within an SCR is likely to be point mutation of individual amino acids. Another potentially useful method would be to analyze the binding of murine and bovine fH to M6 protein. SCR 7 of each has 57% amino acid identity with the human counterpart (32). Preparation and analysis of SCR 1 to 7 constructs of bovine and murine fH would thus assist in localizing binding sites within SCR 7.
fH is thought to play a key role in self- or non-self-recognition by the alternative pathway via its ability to bind to surfaces rich in sialic acid (10, 25). Similarly, in the absence of specific antibody, fH is thought to protect many pathogenic bacteria by binding to their sialic acid capsules (7, 9, 20). The observation that fH binds via SCR 7 to both sialic acid and streptococcal M protein suggests that the specificity of action of fH is mediated by SCR 7, while complement-regulatory activity resides in SCRs 1 to 4 (13, 22, 23). It is noteworthy that a 42-kDa fH-like protein-1 consisting of the N-terminal seven SCRs of fH and 4 additional amino acids exists in serum; this protein may be able to fulfill most of the crucial functions of fH.
Streptococcal M protein contains both a hypervariable and a conserved region. Only antibodies binding to the hypervariable region result in opsonization and phagocytosis (18). The streptococcus is thought to protect its conserved regions from complement by binding fH, thus controlling the amount of C3b deposited. In support of this hypothesis, C3 is deposited irregularly on M-positive streptococci (17), with an associated reduction in phagocytosis (36). M protein has also been reported to bind to a keratinocyte receptor identified as the complement-regulatory protein CD46 (membrane cofactor protein), and this binding has been implicated in streptococcal adherence (26).
After examining an S. pyogenes mutant in which most of the C repeats of the M6 protein were deleted, Perez-Casal et al. (28) concluded that bound fH may not be the only molecule to protect streptococci from phagocytosis, since the organisms were still resistant to killing in nonimmune blood. However, this deletion mutant still contained one half of a C repeat and was still able to bind fH and kerotinocytes, albeit weakly. Consistent with this, Sharma and Pangburn (31) observed that in contrast to an M-negative strain which bound no fH, the C-repeat deletion mutant prepared by Perez-Casal et al. (28) still bound fH but somewhat less than the strain containing an intact M protein molecule. While the pattern of C3b deposition was not examined by Perez-Casal et al., it is possible that there is sufficient binding of fH to the remaining C repeat to protect the mutant from phagocytosis. This is supported by a report of Fischetti et al. (11) showing that the fH binding site on the M protein is located in the spacer between the two repeats removed by Perez-Casal et al. Moreover, a spacer with 50% identity is still present in the mutant prepared by Perez-Casal et al. In addition, Horstmann et al. showed that fH is also able to bind to fibrinogen bound to the B-repeat region of the M molecule (14). Thus, in vivo, the combination of fH binding to both the B and C repeats (although low in the latter) could account for the retained resistance to phagocytosis in the C deletion mutant. The fact that removal of the complete M molecule results in an organism that is easily phagocytosed in normal human blood and does not bind fH supports the notion that M protein alone is the fH binding molecule on streptococci.
Comparison of the antiopsonic effects of fH with those of a mutant of fH (H20Δ7) lacking the binding domain to the M protein C repeat should further define the role of fH binding to other regions of M protein, particularly to the B-repeat-bound fibrinogen (14). Therefore, examination of the protective effects of fH, H20Δ7, and fH-like protein-1 on different M protein and antiphagocytic capsular mutants should help to clarify some of the complexities involved in the pathogenesis of infection by S. pyogenes.
ACKNOWLEDGMENTS
This research was supported by an Australian National Health and Medical Research Council Medical Postgraduate Research Scholarship and Project Grant.
We are grateful to Jens Hellwage and Peter Zipfel of the Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany, for providing H6His and H7His.
REFERENCES
- 1.Avery V M, Gordon D L. Characterization of factor H binding to human polymorphonuclear leukocytes. J Immunol. 1993;151:5545–5553. [PubMed] [Google Scholar]
- 2.Barlow P N, Campbell I D. Strategy for studying modular proteins: application to complement modules. Methods Enzymol. 1994;239:465–485. doi: 10.1016/s0076-6879(94)39018-5. [DOI] [PubMed] [Google Scholar]
- 3.Bisno A L. Alternate complement pathway activation by group A streptococci: role of M protein. Infect Immun. 1979;26:1172–1176. doi: 10.1128/iai.26.3.1172-1176.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisno A L, Stevens D L. Streptococcal infections of skin and soft tissues. N Engl J Med. 1996;334:240–245. doi: 10.1056/NEJM199601253340407. [DOI] [PubMed] [Google Scholar]
- 5.Blackmore T K, Hellwage J, Zipfel P F, Gordon D L. Identification of an additional heparin binding domain in the C-terminal region of human factor H. Exp Clin Immunogenet. 1997;14:30. [Google Scholar]
- 6.Blackmore T K, Sadlon T A, Ward H M, Lublin D M, Gordon D L. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J Immunol. 1996;157:5422–5427. [PubMed] [Google Scholar]
- 7.Brown E J, Joiner K A, Gaither T A, Hammer C, Frank M M. The interaction of C3b bound to pneumococci with factor H (β1H globulin), factor I (C3b/C4b inactivator) and properdin factor B of the human complement system. J Immunol. 1983;131:409–415. [PubMed] [Google Scholar]
- 8.Curtis N. Invasive group A streptococcal infection. Curr Opin Infect Dis. 1996;9:191–202. [Google Scholar]
- 9.Edwards M S, Kasper D L, Jennings H J, Baker C J, Nicholson-Weller A. Capsular sialic acid prevents activation of the alternative complement pathway by type III, group B streptococci. J Immunol. 1982;128:1278–1283. [PubMed] [Google Scholar]
- 10.Fearon D T. Regulation by membrane sialic acid of β1H-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc Natl Acad Sci USA. 1978;75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti V A, Horstmann R D, Pancholi V. Location of the complement factor H binding site on streptococcal M6 protein. Infect Immun. 1995;63:149–153. doi: 10.1128/iai.63.1.149-153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischetti V A, Jones K F, Manjula B N, Scott J R. Streptococcal M protein expressed in Escherichia coli. Localisation, purification, and comparison with streptococcal-derived M protein. J Exp Med. 1984;159:1083–1095. doi: 10.1084/jem.159.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon D L, Kaufman R M, Blackmore T K, Kwong J, Lublin D M. Identification of complement regulatory domains in human factor H. J Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 14.Horstmann R D, Sievertsen H J, Leippe M, Fischetti V A. Role of fibrinogen in complement inhibition by streptococcal M protein. Infect Immun. 1992;60:5036–5041. doi: 10.1128/iai.60.12.5036-5041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horstmann R D, Sievertsen H J, Knochbloch J, Fischetti V A. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci USA. 1988;85:1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hourcade D, Holers V M, Atkinson J P. The regulators of complement activation (RCA) gene cluster. Adv Immunol. 1989;45:381–416. doi: 10.1016/s0065-2776(08)60697-5. [DOI] [PubMed] [Google Scholar]
- 17.Jacks-Weis J, Kim Y, Cleary P P. Restricted deposition of C3 on M+ group A streptococci: correlation with resistance to phagocytosis. J Immunol. 1982;128:1897–1902. [PubMed] [Google Scholar]
- 18.Jones K F, Fischetti V A. The importance of the location of antibody binding on the M6 protein for opsonization and phagocytosis of group A M6 streptococci. J Exp Med. 1988;167:1114–1123. doi: 10.1084/jem.167.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones K F, Manjula B N, Johnston K H, Hollingshead S K, Scott J R, Fischetti V A. Location of variable and conserved epitopes among the multiple serotypes of M protein. J Exp Med. 1985;161:623–628. doi: 10.1084/jem.161.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasper D L. Bacterial capsule—old dogma and new tricks. J Infect Dis. 1986;153:407–415. doi: 10.1093/infdis/153.3.407. [DOI] [PubMed] [Google Scholar]
- 21.Kühn S, Zipfel P F. The baculovirus expression vector pBSV-8His directs secretion of histidine-tagged proteins. Gene. 1995;162:225–229. doi: 10.1016/0378-1119(95)00360-i. [DOI] [PubMed] [Google Scholar]
- 22.Kühn S, Skerka C, Zipfel P F. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H. J Immunol. 1995;155:5663–5670. [PubMed] [Google Scholar]
- 23.Kühn S, Zipfel P F. Mapping of the domains required for decay acceleration activity of the human factor H-like protein 1 and factor H. Eur J Immunol. 1996;26:2383–2387. doi: 10.1002/eji.1830261017. [DOI] [PubMed] [Google Scholar]
- 24.Lancefield R C. Current knowledge of type-specific M antigens of group A streptococci. J Exp Med. 1962;89:307–313. [PubMed] [Google Scholar]
- 25.Meri S, Pangburn M K. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci USA. 1990;87:3982–3986. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okada N, Liszewski M K, Atkinson J P, Caparon M G. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of the group A streptococcus. Proc Natl Acad Sci USA. 1995;92:2489–2493. doi: 10.1073/pnas.92.7.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pangburn M K, Schreiber R D, Müller-Eberhard H J. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J Exp Med. 1977;146:257–270. doi: 10.1084/jem.146.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Casal J, Okada N, Caparon M G, Scott J R. Role of the conserved C-repeat region of the M protein of Streptococcus pyogenes. Mol Microbiol. 1995;15:907–916. doi: 10.1111/j.1365-2958.1995.tb02360.x. [DOI] [PubMed] [Google Scholar]
- 29.Peterson P K, Schmeling D, Cleary P P, Wilkinson B J, Kim Y, Quie P G. Inhibition of alternative complement pathway opsonisation by group A streptococcal M protein. J Infect Dis. 1979;139:575–584. doi: 10.1093/infdis/139.5.575. [DOI] [PubMed] [Google Scholar]
- 30.Phillips G N, Flicker P F, Cohen C, Manjula B N, Fischetti V A. Streptococcal M proteins: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci USA. 1981;78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A K, Pangburn M K. Localization by site-directed mutagenesis of the site in human complement factor H that binds to Streptococcus pyogenes M protein. Infect Immun. 1997;65:484–487. doi: 10.1128/iai.65.2.484-487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soames C J, Day A J, Sim R B. Prediction from sequence comparisons of residues of factor H involved in the interaction with complement component C3b. Biochem J. 1996;315:523–531. doi: 10.1042/bj3150523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stollerman G H. Rheumatic fever. Lancet. 1997;349:935–942. doi: 10.1016/S0140-6736(96)06364-7. [DOI] [PubMed] [Google Scholar]
- 34.Swanson J, Hsu K C, Gotschlich E C. Electron microscopic studies on streptococci. I. M antigen. J Exp Med. 1969;130:1063–1091. doi: 10.1084/jem.130.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiler J M, Daha M R, Austen K F, Fearon D T. Control of the amplification convertase of complement by the plasma protein β1H. Proc Natl Acad Sci USA. 1976;73:3268–3272. doi: 10.1073/pnas.73.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis J J, Law S K, Levine R P, Cleary P P. Resistance to phagocytosis by group A streptococci: failure of deposited complement opsonins to interact with cellular receptors. J Immunol. 1985;134:500–505. [PubMed] [Google Scholar]
- 37.Whaley K, Ruddy S. Modulation of the alternative complement pathway by β1H globulin. J Exp Med. 1976;144:1147–1163. doi: 10.1084/jem.144.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]