Abstract
Cysteine protease of group A streptococci (GAS) is considered an important virulence factor. However, its role in invasiveness of GAS has not been investigated. We demonstrated in this study that two strains of protease-producing GAS had the ability to invade A-549 human respiratory epithelial cells. Isogenic protease mutants were constructed by using integrational plasmids to disrupt the speB gene and confirmed by Southern hybridization and Western immunoblot analyses. No extracellular protease activity was produced by the mutants. The mutants had growth rates similar to those of the wild-type strains and produced normal levels of other extracellular proteins. When invading A-549 cells, the mutants had a two- to threefold decrease in activity compared to that of the wild-type strains. The invasion activity increased when the A-549 cells were incubated with purified cysteine protease and the mutant. However, blockage of the cysteine protease with a specific cysteine protease inhibitor, E-64, decreased the invasion activity of GAS. Intracellular growth of GAS was not found in A-549 cells. The presence or absence of protease activity did not affect the adhesive ability of GAS. These results suggested that streptococcal cysteine protease can enhance the invasion ability of GAS in human respiratory epithelial cells.
Group A streptococci (GAS) (Streptococcus pyogenes) are commonly associated with pharyngitis, cellulitis, and impetigo, but the recent resurgence in blood infections has generated interest in their potential to invade deeper tissues (35). However, so far, their pathogenic factor is poorly defined. Streptococcal pyrogenic exotoxin B (SPE B) is among several extracellular products produced by GAS that may cause severe infections. The gene encoding SPE B is found in all isolates and is assumed to be chromosomal (43). SPE B is a cysteine protease produced extracellularly as a zymogen of about 40 kDa (13). The 28-kDa mature protease is formed by autoproteolytic truncation of the zymogen (42). In 1945, Elliott (10) first reported that streptococcal cysteine protease (SCP) had fibrinolytic activity, and later it was reported that intravenous injection of SCP into rabbits, guinea pigs, and mice caused myocardial necrosis (21). Patients with severe streptococcal infections had lower titers of antibody to SCP than patients with less severe infections (15, 25). Immunization with SCP protected mice from lethal challenge (3, 18). Inactivation of SCP significantly decreased lethality to mice (23). Moreover, purified SCP not only can cleave inactive human interleukin 1β precursor, monocytic cell urokinase receptor, and fibronectin but also can degrade vitronectin (19, 20, 41). SCP can release biologically active kinins and C5a peptidase as well as activate a 66-kDa human endothelial cell matrix metalloprotease (2, 4, 14). In addition, SCP acts synergistically with other S. pyogenes products to increase tissue injury (32). These pieces of evidence suggest that SCP plays multiple roles in GAS infection.
Recently, LaPenta et al. (22) have shown that GAS are efficiently internalized and can persist in cultured human respiratory epithelial cells, suggesting that the organisms actually enter the cell during the course of infection. In addition, Molinari et al. (24) and Cue and Cleary (7) demonstrated that the fibronectin-binding protein of GAS and fibrinogen are involved in the internalization of GAS. In this study, we found that cysteine protease of GAS also enhanced the ability of the microorganism to invade A-549 cells in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and tissue culture cells.
One GAS strain, A-20 (type M1, T1, opacity factor negative), was isolated from a blood culture and collected from National Cheng Kung University Hospital. S. pyogenes NZ131 (type M49, T14) was a gift from D. R. Martin, New Zealand Communicable Disease Center, Porirua. All S. pyogenes cultures were grown in tryptic soy broth supplemented with 0.5% yeast extract (TSBY). Escherichia coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was grown in Luria broth (LB). Plasmids pSF151 and pDL286 were kindly provided by L. Tao, University of Missouri, Kansas City (37). All strains were stored at −70°C in LB with 15% glycerol until testing. Human alveolar carcinoma epithelial cell monolayers (A-549; American Type Culture Collection CCL-185) were used for all invasion assays and were grown in Dulbecco’s modified Eagle medium (DMEM) (GIBCO Laboratories, Grand Island, N.Y.) supplemented with 10% fetal calf serum (GIBCO Laboratories), penicillin (100 μg/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml). The cells were subcultured every second day.
Protease assay.
Detection of protease activity was based on the method of Hynes and Tagg (16). GAS isolates were grown on Columbia agar base (Difco Laboratories, Detroit, Mich.) containing 3% skim milk for detection of protease production. After aerobic incubation at 37°C for 24 h, the zone of casein hydrolysis was measured. For the azocasein assay, bacterial supernatant was collected and assayed by a modified method of Ohara-Nemoto et al. (26). The reaction was initiated by addition of 200 μl of 24-h-growth supernatant of GAS to 400 μl of reaction mixture containing 2.7 mg of azocasein (Sigma Chemical Co., St. Louis, Mo.) per ml in 50 mM Tris-HCl (pH 8.0) which had been prewarmed at 37°C. After incubation at 37°C for 20 min, the reaction was stopped by addition of 100 μl of 15% ice-cold trichloroacetic acid. The reaction mixture was held on ice for 15 min and then centrifuged, and an equal volume of 0.5 M NaOH was added to the supernatant. The absorbance at 450 nm of the sample was measured with a kinetic microplate reader, V-max (Molecular Devices Corporation, Menlo Park, Calif.), to determine the amount of azopeptides not precipitated with trichloroacetic acid.
Adhesion assay.
Adhesion of 3H-labeled GAS to A-549 cells was measured by the method of Wang and Stinson (40). Bacteria were suspended in DMEM to a density of 4 × 108 CFU/ml. Aliquots of the bacterial suspension (250 μl) were added to prewashed confluent A-549 monolayer cultures containing about 5 × 105 cells per well. Bacteria were centrifuged onto the monolayer for 10 min at 183 × g at room temperature to accelerate the contact of bacteria and A-549 cells. After 90 min, nonadherent bacteria were removed by washing the monolayer three times with phosphate-buffered saline (PBS) (15 mM Na2HPO4, 145 mM NaCl [pH 7.2]). The monolayer with adherent bacteria was suspended in 200 μl of 1% Triton X-100, and radioactivity was determined by liquid scintillation spectrometry (Beckman Instruments, Somerset, N.J.).
Invasion assay.
The assay of streptococcal invasion of A-549 monolayers was based on the method of Isberg and Falkow (17). Briefly, A-549 cells (5 × 105) were seeded into each well of 24-well tissue culture plates (Costar Co., Cambridge, Mass.) containing DMEM with antibiotics. To remove the antibiotics, the cells were washed three times with PBS before inoculation. The bacterial cultures were washed three times with PBS and resuspended in the original volume. Before inoculation onto monolayers, bacterial suspensions were retained at room temperature and then diluted to the appropriate concentration in DMEM. Confluent monolayers were infected with approximately 5 × 107 CFU of streptococci (infection rate, 1:100). After 90 min of incubation at 37°C in 5% CO2, extracellular bacteria were removed by five washes with PBS. DMEM containing penicillin (0.8 μg/ml) was then added to the infected monolayer at 37°C in 5% CO2 and the monolayer was incubated for 90 min to eliminate extracellular bacteria. The infected monolayers were washed three times with PBS to remove the antibiotics. The cells were lysed by addition of 200 μl of 1% Triton X-100. The lysate was collected, and the number of intracellular bacteria was determined by plating on TSBY agar. All experiments were done at least three times for each strain. Wells without epithelial cells served as negative controls.
An invasion kinetics assay was done as described above except that the infected monolayers were incubated for 1.5, 2, 3, 4, 5, and 10 h before the addition of penicillin to eliminate extracellular bacteria (33). An intracellular multiplication assay was done as described above except that the infected monolayers were incubated for 1.5, 2, 3, and 4 h after the addition of penicillin to the infected monolayer (34).
Purification of SPE B from strain A-20.
The method for purification of SPE B was based on the methods of Kapur et al. (20) and Ohara-Nemoto et al. (26), with modifications. Bacteria were grown overnight at 35°C in TSBY medium. A 10-ml aliquot of the overnight growth was added to 500 ml of TSBY medium, which was then incubated at 35°C for 22 to 24 h. The cells were removed by centrifugation, and the supernatant was filtered through a 0.45-μm-pore-size membrane filter. The filtrate was diluted with 4 volumes of cold distilled water, the pH was adjusted to 8.0, and then a 1/20 volume of DEAE-Sephadex (Pharmacia Biotech, Sweden) equilibrated with 20 mM Tris-HCl (pH 8.0) was added. The suspension was left for 30 min with occasional mixing, and then unbound material was collected by filtration. The filtrate was concentrated to 100 ml by use of a 3-kDa-cutoff ultrafiltration cartridge (Amicon Division, W. R. Grace & Co., Beverly, Mass.). Buffer exchange by ultrafiltration was conducted with 1 liter of 20% ethanol–20 ml Tris-HCl (pH 7.0) (buffer A) at 4°C. The ultrafiltration solution was passed through a matrix gel Red A column (1.5 by 15 cm; Amicon Division) equilibrated with buffer A. The column was washed with buffer A until the absorbance (280 nm) returned to baseline, and the bound protein was eluted with buffer A containing 2 M NaCl. The eluted material was collected as one fraction and concentrated to 4 ml by ultrafiltration, and the buffer was exchanged with PBS (pH 7.2) by ultrafiltration. The proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The desired proteins contained in the gel slices were identified by amino acid sequencing (Applied Biosystems 477A autosequencer) after the proteins had been eluted and renatured.
Preparation and purification of anti-SPE B antibody.
Fifty micrograms of the purified SPE B was emulsified with complete Freund’s adjuvant and used to immunize rabbits. Four subsequent immunizations with 25 μg of SPE B emulsified with incomplete Freund’s adjuvant were given at 2-week intervals. The titers of the antisera were determined by an enzyme-linked immunosorbent assay using microdilution plates coated with SPE B. The immunoglobulin G fractions of the antisera were purified with a protein A column (Zymed Laboratories, Inc., South San Francisco, Calif.). The neutralizing activity of anti-SPE B immunoglobulin G was determined by azocasein assay according to the procedures described above.
Reconstitution and neutralization in the invasion assay.
For reconstitution studies, the purified SPE B was diluted to the appropriate concentration with 1× PBS buffer and added together with GAS to A-549 cells. Purified SPE B was also heated in a boiling water bath for 10 min and then subjected to the invasion assay. For neutralization, 8 and 16 μM E-64 (Sigma Chemical Co.) was used with purified SPE B and GAS at room temperature for 10 min. The rest of the invasion assay was carried out as described above. Control wells were preincubated with tissue culture medium without SPE B.
Cloning of speB.
The speB gene was amplified by PCR using S. pyogenes A-20 DNA as a template with the first primer (5′ GATCAAAACTTTGCTCGTAACG 3′) corresponding to bases 81 to 103 of the speB gene and the second primer (5′ CTAAGGTTTGATGCCTACAACAG 3′) designed to be complementary to speB nucleotides 1175 to 1195 in the published sequence (13). PCR was performed with a total volume of 50 μl containing 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, 100 ng of DNA, 50 pM primers, 10 mM deoxynucleoside triphosphates, and 1 U of Taq DNA polymerase (New England Biolabs Inc., Beverly, Mass.). Amplification was performed in a DNA thermal cycler (Perkin-Elmer Corporation, Norwalk, Conn.) that had been programmed for 4 cycles of 1 min at 94°C, 1 min at 37°C, and 2 min at 72°C followed by 35 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 72°C. Following amplification, the 1.1-kb PCR product was digested with either KpnI and PstI or HindIII and PstI to obtain 0.4- and 0.7-kb fragments of speB from nucleotides 578 to 1005 and 347 to 1005, respectively (13). The 0.4- and 0.7-kb fragments of speB were ligated into plasmids pSF151 and pDL286 to construct plasmids pMW152 and pMW153, respectively. The constructions were checked by analyzing digestion patterns obtained with appropriate endonucleases and DNA sequencing.
DNA preparation and sequencing.
Plasmid DNA was prepared from E. coli as described previously (29). Chromosomal DNA of GAS was prepared by the method of Cleary et al. (6). DNA sequencing was performed by the dideoxy-chain termination method of Sanger et al. (30). A Sequenase reagent kit (United States Biochemical Corp., Cleveland, Ohio) was used according to the manufacturer’s protocol.
Transformation.
E. coli was transformed by the method of Hanahan (11). S. pyogenes was electroporated by the method of Schalén et al. (31); selection for kanamycin and spectinomycin resistance was done with 50 and 100 μg/ml, respectively.
DNA-DNA hybridization.
Chromosomal DNA preparations from GAS were digested with restriction endonucleases and electrophoresed through a 0.8% agarose gel. DNA was transferred to nylon membranes (Amersham, Arlington Heights, Ill.) as previously described (29). The purified speB DNA fragment to be used as a probe was labeled with [α-32P]dCTP (Amersham) by use of a random-primer DNA labeling kit (Promega Corp., Madison, Wis.) according to the manufacturer’s instructions. Hybridizations were performed as follows: prehybridization and hybridization were carried out for 2 and 18 h, respectively, at 65°C in 5× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate)–0.02% SDS–1% (wt/vol) blocking reagent for nucleic acid hybridization (Boehringer, Mannheim, Germany)–0.1% N-lauryl-sarcosine, followed by two washings in 2× SSC–0.1% SDS at room temperature for 5 min and two washings in 0.2× SSC–0.1% SDS at 65°C for 15 min.
TEM.
Bacteria were prepared for the invasion assay as described above. Samples for electron microscopy were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.3) for 4 h, washed in sodium cacodylate buffer with 3 mM CaCl2, and postfixed in 1% osmium tetroxide at 4°C for 1 h. After being rinsed in cacodylate buffer, the cells were dehydrated in an ascending series of alcohol concentrations (50, 70, 85, 95, 100, 100, and 100%; each step for 15 min). After being rinsed in propylene oxide and then subjected to infiltration in propylene oxide-epoxy resin mixtures, the cells were embedded in epoxy resin and polymerized at 60 to 70°C for 48 h. Sections were cut 0.5 μm thick with a Reichert OM UIII ultramicrotome and stained with 1% toluidine blue in 1% borax. Areas suitable for ultrathin sectioning (approximately 90 nm) were selected. Sections were mounted on copper grids, stained with uranyl acetate and lead citrate, and then examined with a Hitachi H-7000 transmission electron microscope (TEM).
Susceptibility test.
The MIC of penicillin was determined by the Etest method (AB Biodisk, Piscataway, N.J.) according to the manufacturer’s instructions.
Statistical analysis.
The difference in invasion ability of the wild type and the isogenic mutant was analyzed by t test. A P value of <0.01 was considered statistically significant.
RESULTS
Disruption of speB.
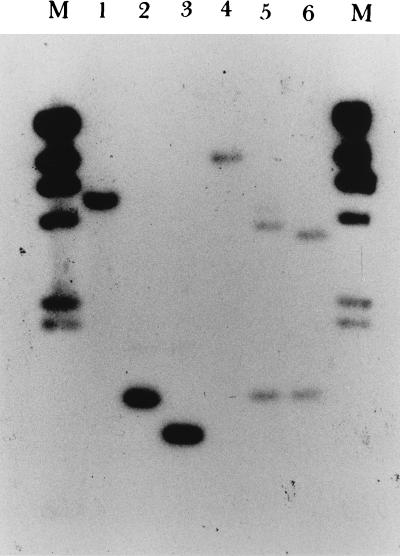
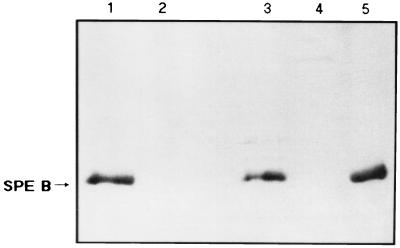
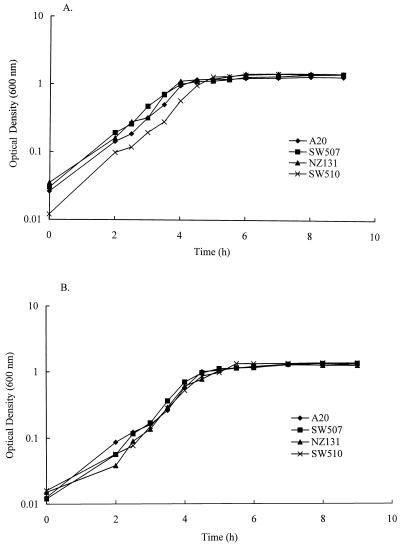
Integrational plasmids were used to disrupt the speB gene. Plasmids pMW152 and pMW153 were constructed by cloning into vectors pSF151 and pDL286 an insert of S. pyogenes DNA that contained part of the speB coding region. These plasmids replicated autonomously in E. coli but not in S. pyogenes when transformed into S. pyogenes with selection for the vector-determined resistance; transformants arose because of integration of the plasmid into the homologous region of the recipient chromosome by a Campbell-like mechanism. When the region cloned on an integrational plasmid was entirely internal to the speB transcriptional unit, it disrupted that transcriptional unit. Plasmid pMW152 integrated into A-20 to obtain an isogenic protease mutant was designated SW507; plasmid pMW153 integrated into NZ131 was designated SW510. Transformant colonies were initially screened on the basis of proteolytic activity on casein plates. Protease activity was present in the extracellular proteins secreted by strains A-20 and NZ131, while strains SW507 and SW510 had no extracellular protease activity (data not shown). As analyzed by Southern hybridization, 5.5- and 1.3-kb hybridization bands were obtained with NsiI and HindIII digestion, respectively, while a 1.0-kb fragment was seen in an NsiI-HindIII double digestion of wild-type strain A-20 (Fig. 1, lanes 1 to 3). For mutant SW507, hybridization bands of 1.3 and 3.9 kb were seen in the HindIII digestion, a 9.4-kb fragment was seen in the NsiI digestion, and both 3.6- and 1.3-kb fragments were seen in the NsiI-HindIII double digestions, confirming that integration occurred by a Campbell-like mechanism (Fig. 1, lanes 4 to 6). In the same way, integration into strains NZ131 and SW510 was also confirmed by Southern hybridization (data not shown). To demonstrate that strains SW507 and SW510 lost their ability to express SPE B protein, the supernatants from these strains were collected and the absence of SPE B protein was confirmed by Western immunoblot analysis using rabbit anti-SPE B antibody. As shown in Fig. 2, the antiserum recognized the mature form of SPE B (28 kDa) in supernatants of strains A-20 and NZ131, while no SPE B protein bands (either 40 or 28 kDa) were observed in the supernatants of SW507 and SW510. No difference in growth rates was observed between mutants and the wild-type strains in TSBY and DMEM (Fig. 3). No differences in streptokinase expression and hemolysis on blood agar were found (data not shown). In addition, the MIC of penicillin for strains A-20 and SW507 was 0.023 μg/ml, whereas that for strains NZ131 and SW510 was 0.016 μg/ml.
FIG. 1.
Southern blot analysis of genomic DNA extracted from GAS A-20 and SW507. The probes used were specific for the speB gene and λ DNA. The DNAs were digested with NsiI (lanes 1 and 4), HindIII (lanes 2 and 5), and HindIII-NsiI (lanes 3 and 6). Lanes 1 to 3, GAS A-20 genomic DNA; lanes 4 to 6, GAS SW507 genomic DNA; lanes M, λ HindIII marker used as a molecular size standard.
FIG. 2.
Western immunoblot analysis of SPE B present in the supernatants of wild-type GAS (A-20 and NZ131) and mutants (SW507 and SW510) after electrophoresis on an 8% polyacrylamide gel. The blot was developed with rabbit anti-SPE B polyclonal antibodies. Lane 1, A-20; lane 2, SW507; lane 3, NZ131; lane 4, SW510; lane 5, purified 28-kDa SPE B from A-20 used as a standard. The mature SPE B protein is indicated (arrow).
FIG. 3.
Growth curves for wild-type strains (A-20 and NZ131) and isogenic mutants (SW507 and SW510) in TSBY (A) and DMEM (B).
Adhesion assay.
The ability of GAS to adhere to monolayers of A-549 cells is shown in Table 1. All wild-type strains and isogenic mutants adhered to the A-549 monolayer, regardless of protease activity.
TABLE 1.
Adhesion abilities of protease-positive and -mutant strains of GAS
| Strain | Proteolytic activitya | Adhesionb (108 CFU/ml) |
|---|---|---|
| A-20 | + | 4.88 ± 0.13 |
| SW507 | − | 6.20 ± 0.21 |
| NZ131 | + | 3.13 ± 0.34 |
| SW510 | − | 5.39 ± 0.47 |
The proteolytic activity was determined by use of a casein plate. +, activity; −, no activity.
Adhesion ability was assayed as described in the text. Results are means ± standard errors of the means of three experiments.
Penicillin protection assay.
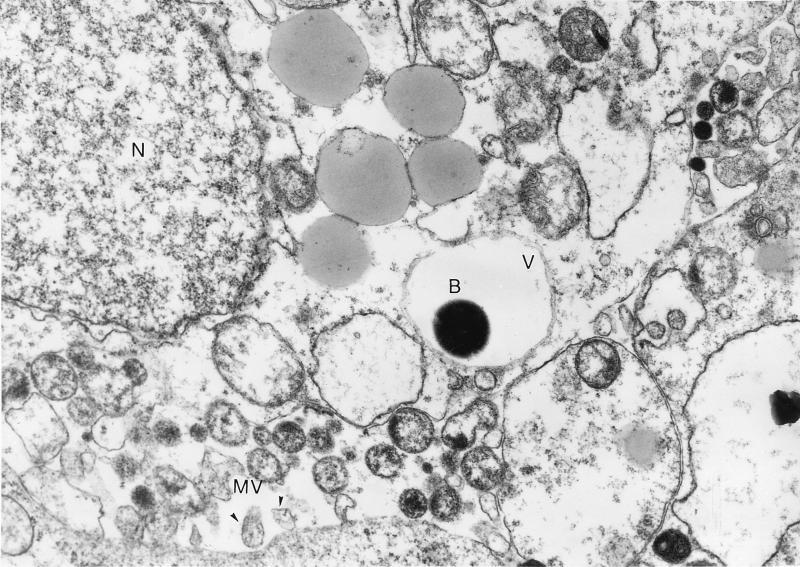
A penicillin-protected bacterium is shown in Fig. 4. A-549 monolayers were infected with GAS for 2 h before the addition of penicillin to eliminate extracellular bacteria. TEM demonstrated the intracellular presence of GAS in the vacuoles of A-549 cells.
FIG. 4.
Electron micrograph of GAS strain A-20 entry into cultured A-549 cells. Monolayers were infected with 5 × 107 CFU for 120 min before they were washed and exposed to penicillin. A bacterium (B) is enclosed in a vacuole (V). The nucleus (N) and microvilli (MV) are indicated. Magnification, ×10,000.
Effect of cysteine protease on GAS invasion.
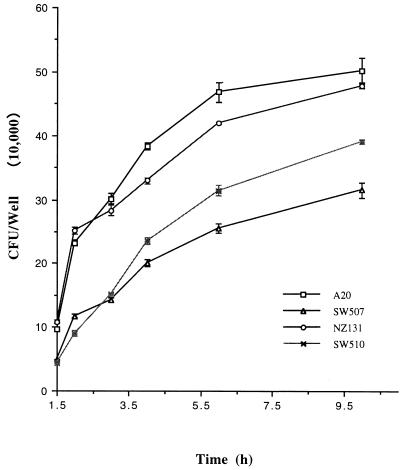
To examine the effect of cysteine protease on GAS invasion, the protease-positive strains and mutants were tested by an invasion assay based on the method of Isberg and Falkow (17). Strains A-20 and NZ131, both protease positive, had invasive activities two- to threefold greater than those of isogenic mutant strains SW507 and SW510 (Table 2 and Fig. 5). Statistically significant differences (P < 0.01) in invasion activities between wild-type strains and mutants were noted. The invasion kinetics assay showed that when the incubation time was extended from 1.5 to 10 h, the invasion activity for both protease-positive strains and mutants increased significantly (Fig. 5).
TABLE 2.
Invasion abilities of protease-positive and -mutant strains of GAS
| Strain | No. of streptococci (104 CFU/well) grown within A-549 cells reconstituted witha:
|
|||||
|---|---|---|---|---|---|---|
| None | SPE Bb | Heated SPE B | SPE B + E-64 (8 μM) | SPE B + E-64 (16 μM) | E-64 (16 μM) | |
| A-20 | 9.68 ± 0.14 | 12.6 ± 0.11 | 10.02 ± 0.37 | 10.2 ± 0.19 | 9.68 ± 0.30 | 9.88 ± 0.65 |
| SW507 | 4.87 ± 0.11 | 9.58 ± 0.29 | 5.32 ± 0.58 | 5.15 ± 0.17 | 4.38 ± 0.09 | 4.75 ± 0.39 |
| NZ131 | 10.79 ± 0.24 | 11.17 ± 0.39 | NDc | ND | ND | ND |
| SW510 | 4.43 ± 0.08 | 9.21 ± 0.29 | 5.40 ± 0.21 | 6.65 ± 0.81 | 6.40 ± 0.56 | 4.43 ± 0.05 |
Results are demonstrated as the number of intracellular bacteria ± standard error after 1.5 h of incubation.
The invasion assay was done with reconstituted purified SPE B.
ND, not done.
FIG. 5.
Invasion kinetics of the wild-type strains (A-20 and NZ131) and isogenic mutants (SW507 and SW510). Confluent cell monolayers were infected with 5 × 107 bacteria in 24-well plates. The cells were incubated for 1.5, 2, 3, 4, 5, and 10 h, washed with PBS, and incubated for another 1.5 h in medium containing 0.8 μg of penicillin per ml. GAS cell numbers were determined by serial dilutions of cell lysates and plating on agar. Results are means ± standard errors of the means (SEM) of the numbers of GAS per well from four separate experiments; at each time point, triplicate samples were analyzed.
To further examine the role of cysteine protease on the invasion of GAS, purified SPE B protein was tested in vitro. Identification of the purified protein as SPE B was confirmed via N-terminal sequence analysis of the protein from the Red A column; the first 10 amino acids (Gln-Pro-Val-Val-Lys-Ser-Leu-Leu-Asp-Ser) were confirmed exactly as predicted from the DNA sequence (13). The purified cysteine protease was mixed with GAS prior to the invasion assay. After a 1.5-h incubation, the invasion activities of the mutants increased to levels similar to those of protease-positive strains (P < 0.01) (Table 2). The SPE B-mediated enhancement of invasion activity was significantly inhibited by the addition of a specific cysteine protease inhibitor, E-64, for both SW507 and SW510 mutants (P < 0.01).
Intracellular growth of GAS.
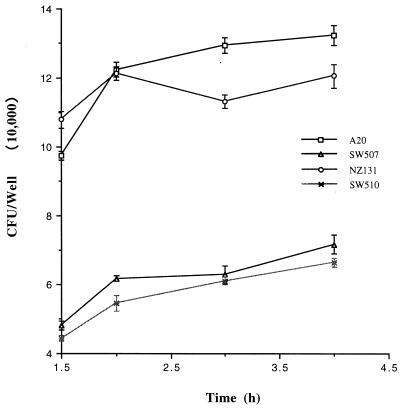
The intracellular multiplication of GAS was determined by addition of penicillin to the infected monolayers after GAS had invaded the cells for 1.5 h. Infected cells were collected for 4 h after the bacterial invasion, the cells were lysed, and the numbers of intracellular bacteria were determined by plating on TSBY agar. As shown in Fig. 6, there was no significant increase in the number of intracellular bacteria over the 4-h incubation period. Control experiments showed that no viable bacteria were recovered from wells without A-549 cells after incubation in culture medium containing 0.8 μg of penicillin per ml.
FIG. 6.
Intracellular growth of S. pyogenes in A-549 cells. Confluent cell monolayers were infected with 5 × 107 bacteria in 24-well plates. The cells were incubated for 90 min, washed, and incubated for another 1.5 to 4 h in medium containing 0.8 μg of penicillin per ml. GAS cell numbers were determined by serial dilution of cell lysates and plating on agar. Results are the means ± SEM of the numbers of GAS per well from four separate experiments; at each time point, triplicate samples were analyzed.
DISCUSSION
Recently, LaPenta et al. (22) have demonstrated for the first time that GAS, which represent typically extracellular pathogens of epithelial surfaces, can enter and persist in host cells. Invasion might play a role in the pathogenicity of GAS by facilitating tissue invasion or helping to protect the bacteria from extracellular defense mechanisms. Although evidence suggests that cysteine protease plays an important role in the pathogenesis of streptococcal infection, it is not known whether it plays a role in the invasion of epithelial cells. In this study, we used gene disruption of speB, protease reconstitution, and neutralization experiments to study the role of cysteine protease of GAS in the invasion of respiratory epithelial cells in vitro.
On the basis of the finding that penicillin-protected bacteria were inside the cells, we have provided data which demonstrate the ability of cysteine protease to enhance the entrance of GAS into eukaryotic cells. Strains A-20 and NZ131, both protease-positive GAS, displayed two- to threefold increases in invasive activity over isogenic protease mutants (SW507 and SW510). The effect was also observed when purified cysteine protease and the mutant were incubated together with A-549 cells prior to the invasion assay. Blockage of the cysteine protease with E-64 decreased the invasion activity (Table 2). These data suggest that cysteine protease not only enhanced bacterial uptake by the cells but also had a role in receptor-ligand interaction involved in the entry of GAS into the respiratory epithelial cells. However, we cannot rule out the possibility that a substance(s) other than cysteine protease present in the purified fraction may also have enhanced the invasive activity. No significant changes were observed if GAS were treated with E-64 alone prior to the invasion assay. The results suggest that cysteine protease was not present on the bacterial surfaces. How cysteine protease interacts with the host and the nature of the receptor recognized by cysteine protease require further study.
Factors other than cysteine protease may also have influenced the results of the invasion assay. The MICs of penicillin are the same for both mutants and wild-type strains, suggesting that the mutants were no more susceptible to penicillin than the wild-type strains. In addition, the growth rates of A-20 and NZ131 and their mutants in TSBY and DMEM appeared to be similar (Fig. 3). Therefore, the growth rates of the two isogenic GAS strains did not explain the differences in the invasion of A-549 cells. A previous report (22) showed that as GAS enter the stationary phase, they may invade cells more efficiently. Recently, Chaussee et al. (5) demonstrated that cysteine protease is produced to a greater extent during the stationary phase than during the other growth phases. Taken together, the differences in invasion ability observed reflected the differences in cysteine protease rather than growth characteristics.
As the incubation time increased, a drastic increase in the recovery of intracellular bacteria from cells was observed, suggesting that bacterial entry increased as the infection progressed. The increase in recovery rate could be due to greater production of a bacterial product that stimulated bacterial uptake or that allowed GAS to replicate inside the cell. However, our results have demonstrated that once internalized, wild-type strains did not grow inside A-549 epithelial cells; neither did the mutants (Fig. 6). A similar observation was made for group B streptococci (28). Whether this effect is due to intracellular starvation or is a result of culturing cells in a medium containing antibiotics is not clear.
Since mutants displayed a 0.1% invasive activity, it seems that factors other than the cysteine protease are important for invasion. Recently, Molinari et al. (24) showed that the binding of SfbI protein to eukaryotic cells via fibronectin can trigger bacterial internalization in HEp-2 cells. In addition, Cue and Cleary (7) demonstrated that fibrinogen and peptides containing the sequence Arg-Gly-Asp could promote bacterial invasion.
Studies of other pathogens have shown that adherence and invasion of host cells involve interactions between specific components on the surfaces of bacteria and the eukaryotic cells (1, 12, 17, 36, 39). S. pyogenes colonizes the epithelial surface of the nasopharynx and the skin of humans, often resulting in infections. Several investigators have shown that M+ or protein F-containing strains of S. pyogenes adhere more easily to human buccal and pharyngeal epithelial cells or HEp-2 cells than M− strains (8, 38, 40). Since SCP can enhance the invasion of GAS, it is of interest to determine the role of SCP in adhesion to epithelial cells. Our results indicated no striking differences in adhesion between protease-positive strains and isogenic mutants. Although there was an increase of adhesion in the mutants compared to that of the wild-type strain, it was not investigated further.
Although the protease isogenic mutant showed invasion activity lower than that of the wild-type strain, it was not known whether this was due to the polarity effect which was required for invasion. Based on the published data (13, 27), speB is transcribed via a monocistronic message. The transcript size is about 1.8 kb, which corresponds to the length predicted from the speB gene. In addition, there is no known gene either at least 600 bp upstream from the speB start codon (27) or at least 270 bp downstream from the speB termination codon (13). On the basis of this information, we believe that the decreased invasion activity was caused by a lack of speB gene product (cysteine protease). A similar protease isogenic mutant was constructed by Lukomski et al. (23).
In conclusion, cysteine protease is not the only factor responsible for invasion of respiratory epithelial cells by GAS; other bacterial factors may also be responsible. However, cysteine protease can enhance the ability of GAS to invade respiratory epithelial cells.
ACKNOWLEDGMENTS
We thank T.-Y. Chang and S.-C. Tsai for their technical assistance in electron microscopy and T.-C. Chang for his comments on the manuscript. We are also grateful to D. R. Martin, New Zealand Communicable Disease Center, Porirua, for typing of GAS.
This work was partly supported by grants NSC 85-2331-B-006-056, NSC 86-2314-B006-056, and NSC 86-2314-B006-054 from the National Science Council, Republic of China.
REFERENCES
- 1.Beachey E H. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surfaces. J Infect Dis. 1981;143:325–344. doi: 10.1093/infdis/143.3.325. [DOI] [PubMed] [Google Scholar]
- 2.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 3.Björck L, Akesson P, Bohus M, Trojnar J, Abrahamson M, Olafsson I, Grubb A. Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature (London) 1989;337:385–386. doi: 10.1038/337385a0. [DOI] [PubMed] [Google Scholar]
- 4.Burns E H, Jr, Marciel A M, Musser J M. Activation of a 66-kilodalton human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine protease. Infect Immun. 1996;64:4744–4750. doi: 10.1128/iai.64.11.4744-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee M S, Phillips E R, Ferretti J J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleary P P, Kaplan E L, Livdahl C, Skjold S. DNA fingerprints of Streptococcus pyogenes are M type specific. J Infect Dis. 1988;158:1317–1326. doi: 10.1093/infdis/158.6.1317. [DOI] [PubMed] [Google Scholar]
- 7.Cue D R, Cleary P P. High-frequency invasion of epithelial cells by Streptococcus pyogenes can be activated by fibrinogen and peptides containing the sequence RGD. Infect Immun. 1997;65:2759–2764. doi: 10.1128/iai.65.7.2759-2764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Ellen R P, Gibbons R J. M-protein-associated adherence of Streptococcus pyogenes to epithelial surfaces: prerequisite for virulence. Infect Immun. 1972;5:826–830. doi: 10.1128/iai.5.5.826-830.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellen R P, Gibbons R J. Parameters affecting the adherence and tissue tropisms of Streptococcus pyogenes. Infect Immun. 1974;9:85–91. doi: 10.1128/iai.9.1.85-91.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott S D. A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J Exp Med. 1945;81:573–592. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hanski E, Caparon M G. Protein F, a fibronectin-binding protein, is an adhesion of the group A streptococci. Proc Natl Acad Sci USA. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauser A R, Schlievert P M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J Bacteriol. 1990;172:4536–4542. doi: 10.1128/jb.172.8.4536-4542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herwald H, Collin M, Müller-Esterl W, Björck L. Streptococcal cysteine proteinase releases kinins: a novel virulence mechanism. J Exp Med. 1996;184:665–673. doi: 10.1084/jem.184.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm S E, Norrby A, Bergholm A M, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- 16.Hynes W L, Tagg J R. A simple plate assay for detection of group A streptococcus proteinase. J Microbiol Methods. 1985;4:25–31. [Google Scholar]
- 17.Isberg R R, Falkow S. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature (London) 1985;317:262–264. doi: 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 18.Kapur V, Maffei J T, Greer R S, Li L L, Adams G J, Musser J M. Vaccination with streptococcal extracellular cysteine protease (interleukin-1β convertase) protects mice against challenge with heterologous group A streptococci. Microb Pathog. 1994;16:443–450. doi: 10.1006/mpat.1994.1044. [DOI] [PubMed] [Google Scholar]
- 19.Kapur V, Majesky M W, Li L L, Black R A, Musser J M. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapur V, Topouzis S, Majesky M W, Li L L, Hamrick M R, Hamill R J, Patti J M, Musser J M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 21.Kellner A, Robertson T. Myocardial necrosis produced in animals by means of crystalline streptococcal proteinase. J Exp Med. 1954;99:495–504. doi: 10.1084/jem.99.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LaPenta D, Rubens C, Chi E, Cleary P P. Group A streptococci efficiently invade human respiratory epithelial cells. Proc Natl Acad Sci USA. 1994;91:12115–12119. doi: 10.1073/pnas.91.25.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molinari G, Talay S R, Valentin-Weigand P, Rohde M, Chhatwal G S. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun. 1997;65:1357–1363. doi: 10.1128/iai.65.4.1357-1363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norrby-Teglund A, Pauksens K, Holm S E, Norgren M. Relation between low capacity of human sera to inhibit streptococcal mitogens and serious manifestation of disease. J Infect Dis. 1994;170:585–591. doi: 10.1093/infdis/170.3.585. [DOI] [PubMed] [Google Scholar]
- 26.Ohara-Nemoto Y, Sasaki M, Kaneko M, Nemoto T, Ota M. Cysteine protease activity of streptococcal pyrogenic exotoxin B. Can J Microbiol. 1994;40:930–936. doi: 10.1139/m94-149. [DOI] [PubMed] [Google Scholar]
- 27.Podbielski A, Woischnik M, Pohl B, Schmidt K H. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol. 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 28.Rubens C E, Smith S, Hulse M, Chi E Y, van Belle G. Respiratory epithelial cell invasion by group B streptococci. Infect Immun. 1992;60:5157–5163. doi: 10.1128/iai.60.12.5157-5163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schalén C, Gebreselassie D, Ståhl S. Characterization of an erythromycin resistance (erm) plasmid in Streptococcus pyogenes. APMIS. 1995;103:59–68. doi: 10.1111/j.1699-0463.1995.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 32.Shanley T, Schrier D, Kapur V, Kehoe M, Musser J M, Ward P A. Streptococcal cysteine protease augments lung injury induced by products of group A streptococci. Infect Immun. 1996;64:870–877. doi: 10.1128/iai.64.3.870-877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw J H, Falkow S. Model for invasion of human tissue culture cells by Neisseria gonorrhoeae. Infect Immun. 1988;56:1625–1632. doi: 10.1128/iai.56.6.1625-1632.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Small P L C, Isberg R R, Falkow S. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocolitica to enter and replicate within HEp-2 cells. Infect Immun. 1987;55:1674–1679. doi: 10.1128/iai.55.7.1674-1679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens D L, Tanner M H, Winship J, Swarts R, Ries K M, Schlievert P M, Kaplan E. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N Engl J Med. 1989;321:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- 36.Talay S R, Valentin-Weigand P, Jerlström P G, Timmis K N, Chhatwal G S. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect Immun. 1992;60:3837–3844. doi: 10.1128/iai.60.9.3837-3844.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao L, LeBlanc D J, Ferretti J J. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene. 1992;120:105–110. doi: 10.1016/0378-1119(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 38.Tylewska S K, Fischetti V A, Gibbons R J. Binding selectivity gradients in studies of the adhesion of Streptococcus pyogenes and M protein to epithelial cells differ from that of lipoteichoic acid. Curr Microbiol. 1988;16:209–216. [Google Scholar]
- 39.Valentin-Weigand P, Gruhlich-Henn J, Chhatwal G S, Müller-Berghaus G, Blobel H, Preissner K T. Mediation of adherence of streptococci to human endothelial cells by complement S protein (vitronectin) Infect Immun. 1988;56:2851–2855. doi: 10.1128/iai.56.11.2851-2855.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J R, Stinson M W. M protein mediates streptococcal adhesion to HEp-2 cells. Infect Immun. 1994;62:442–448. doi: 10.1128/iai.62.2.442-448.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf B B, Gibson C A, Kapur V, Hussaini I M, Musser J M, Gonias S L. Proteolytically active streptococcal pyogenic exotoxin B cleaves monocytic cell urokinase receptor and releases an active fragment of the receptor from the cell surface. J Biol Chem. 1994;269:30682–30687. [PubMed] [Google Scholar]
- 42.Yonaha K, Elliott S D, Liu T Y. Primary structure of zymogen of streptococcal proteinase. J Protein Chem. 1982;1:317–334. [Google Scholar]
- 43.Yu C E, Ferretti J J. Frequency of the erythromycin toxin B and C genes (speB and speC) among clinical isolates of group A streptococci. Infect Immun. 1991;59:211–215. doi: 10.1128/iai.59.1.211-215.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]