Abstract
Precision tools for spatiotemporal control of cytoskeletal motor function are needed to dissect fundamental biological processes ranging from intracellular transport to cell migration and division. Direct optical control of motor speed and direction is one promising approach, but it remains a challenge to engineer controllable motors with desirable properties such as speed and processivity required for transport applications in living cells. Here we develop engineered myosin motors that combine large optical modulation depths with high velocities, and create processive myosin motors with optically controllable directionality. We characterize the performance of the motors using in vitro motility assays, single-molecule tracking, and live-cell imaging. Bidirectional processive motors move efficiently toward the tips of cellular protrusions in the presence of blue light, and can transport molecular cargos in cells. Robust gearshifting myosins will further enable programmable transport in contexts ranging from in vitro active matter reconstitutions to microfabricated systems that harness molecular propulsion.
Engineered cytoskeletal motors with dramatically altered properties and controllable functions1 have provided critical tests of motor mechanisms2,3 and tools for relating nanoscale biophysics to cellular and tissue scale functions4,5. An outstanding challenge is that engineered motors often have impaired parameters such as speed and processivity6,7,8 in comparison with natural counterparts. Eukaryotic cells rely on both processive and non-processive cytoskeletal motors with speeds ranging from the nm/s to the microns/s range9. Light-activated gearshifting10 provides direct control over cytoskeletal motor speed and direction in genetically encoded engineered proteins, and was proposed to be applicable for regulating cellular dynamics. This technology has been demonstrated in vitro, using gliding filament assays with purified proteins. However, existing designs for light-activated gearshifting10 are non-processive and suffer from either low overall speeds or modest velocity modulation, precluding their use in many areas of cell biology, devices, and reconstituted systems. Moreover, unlike complementary optogenetic approaches based on cargo recruitment11,12,13,14, gearshifting has not been shown to function in cellular contexts.
Here we demonstrate that engineered myosin motors can satisfy four critical design constraints for optically controllable transport: speeds that match the range of speeds of natural cytoskeletal motors, deep modulation in response to light, processive motility, and operability in living cells. Exploiting the principle of programmable changes in lever arm geometry6,7,10,15, we first introduce fast non-processive myosin motors that respond strongly to blue light in gliding filament assays. Collections of these motors show graded dose-dependent responses to optical stimulation. We then use in vitro single-molecule fluorescence imaging and gold nanoparticle tracking to characterize the motility of tetrameric engineered motor assemblies; we find that these designed motors walk processively along actin filaments and switch their predominant direction of stepping in response to blue light. Moving to live cell imaging, we observe light-dependent intracellular localization of the bidirectional tetramers in both fibroblasts and cultured neurons, and confirm high-speed motility of individual motors tagged with fluorescent protein arrays. Finally, we use co-expression in epithelial cells to show that the engineered myosins can be used to transport separate molecular cargos, demonstrating light-dependent localization of both cytoplasmic and transmembrane proteins to cellular protrusions.
Results
Engineering fast myosins with strong optical response
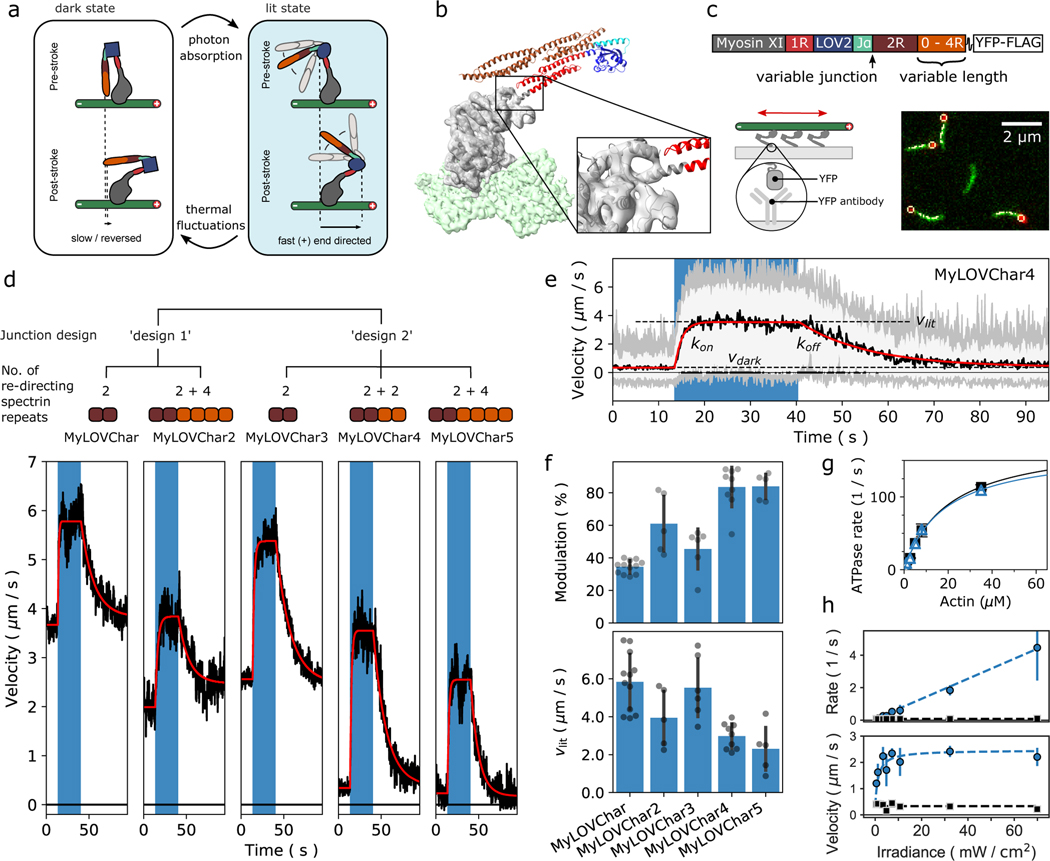
To enable cellular applications that require robust spatiotemporal control while matching or exceeding the transport velocities of natural motors, we first set out to design a gearshifting myosin that combines high velocity with deep optical modulation. The existing MyLOV family of engineered gearshifting myosins10 includes constructs (MyLOV1–4) that show deep light-dependent velocity modulation but low speeds (< 10 nm/s), and one construct (MyLOVChar) with high speed (microns/s) but only mild velocity modulation. We hypothesized that a suitable starting point for optimization could be MyLOVChar, a light-controlled motor incorporating the catalytic domain of Chara corallina myosin XI, the fastest known cytoskeletal motor16.
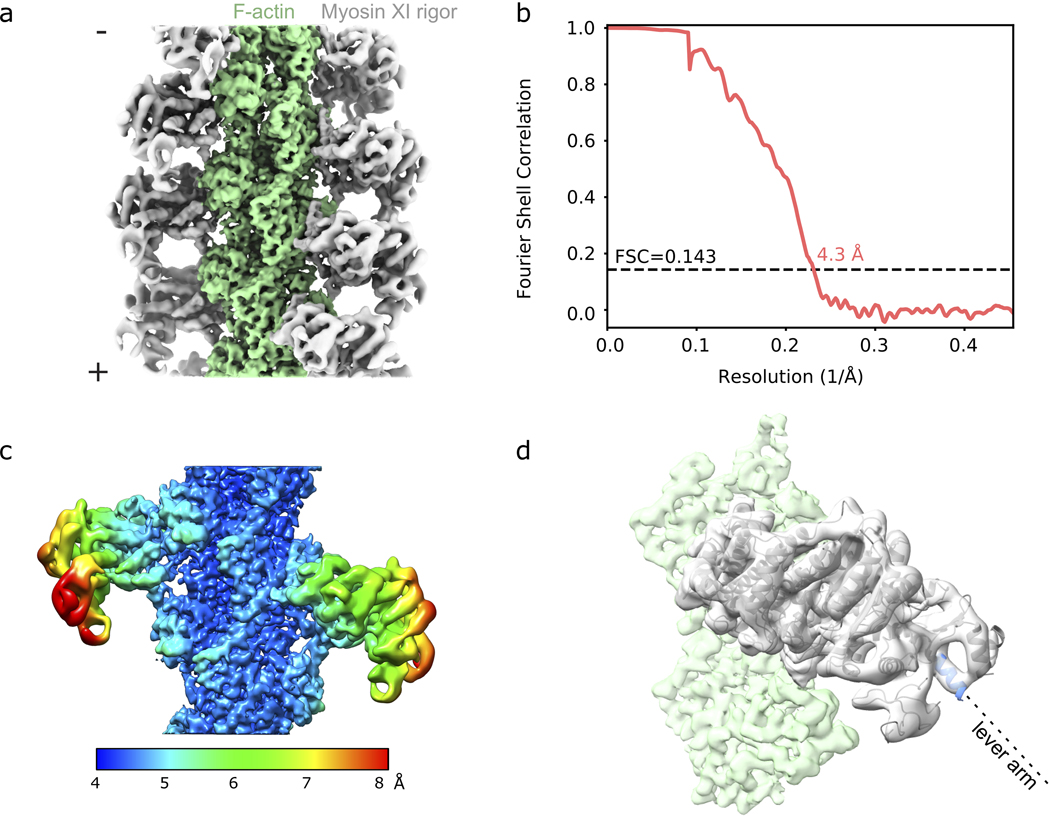
To identify possible strategies for achieving our design goals, we reviewed the switching mechanism and functional characteristics of MyLOVChar. The MyLOVChar design exploits a mechanical effect of light-induced unfolding in the N-terminal J-α helix of a LOV domain (Avena sativa LOV2)17 embedded in an engineered lever arm structure (Fig. 1a). The fully folded LOV domain is designed to act as a mechanical hairpin element that redirects the lever arm in the absence of blue light, resulting in a small stroke of the myosin towards the actin (+)-end. In the light, the J-α helix is undocked from the LOV domain and becomes disordered. The last rigid element of the LOV domain then becomes the effective end of the lever arm, predicted to produce a larger (+)-end directed stroke and an increased resulting velocity18,19. A coarse structural model of the MyLOVChar power stroke estimated that the velocity should increase by > 3-fold in the light10. We used cryo-EM (Extended Data Fig. 1a–c, Supplementary Table 1) to obtain a new experimental post-stroke structure of Chara corallina myosin XI bound to actin (Fig. 1b, Extended Data Fig. 1d and Supplementary Video 1) and generated updated models of MyLOVChar, producing a similar prediction with even larger velocity modulation (Extended Data Fig. 2a). However, as reported previously, the experimentally measured modulation is much smaller. In an in vitro gliding motility assay (Fig. 1c), the speeds of actin filaments driven by MyLOVChar increased from ~3–5 μm/s in the absence of blue light to ~5–7 μm/s when illuminated (Fig. 1d, Supplementary Video 2), for an optical modulation depth (defined as 1 – vdark/vlit, with vdark and vlit determined as shown in Fig. 1e) of ~35%. This discrepancy between prediction and experiment may be explained by unintended compliance or angular deviations introduced by junctions between structural modules in the engineered lever arm, leading to a power stroke that is suboptimally redirected in the dark. We pursued two complementary strategies for overcoming this deficiency: compensating for poor redirection by increasing the length of the redirected lever arm, and correcting suboptimal junctions by varying the junction sequences as in prior work10.
Figure 1. Engineering monomeric myosin XI motors for high speed and strong optical response.
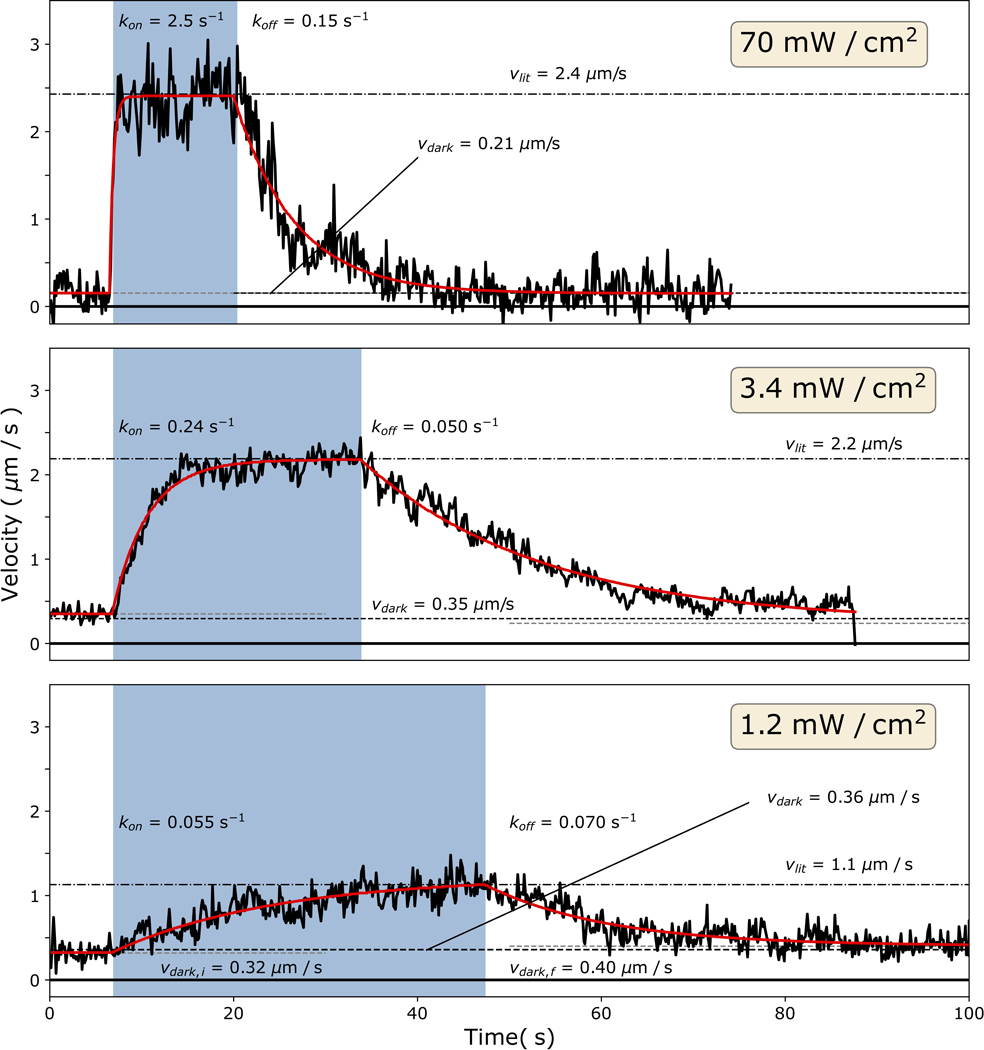
a, Cartoon model of the mechanism of optical switching. b, Cryo-EM reconstruction of Chara corallina myosin XI catalytic domain (apo, grey) bound to F-actin (green) and post-stroke model of MyLOVChar (ribbon diagram) based on the cryo-EM model, with fused lever arm structure built from crystal structure fragments using RosettaRemodel (see also Extended Data Fig. 1d, 2a). c, Block diagram of the molecular construct, and schematic and snapshot of the gliding filament assay with automated tracking overlay. d, Representative velocity traces for a series of MyLOVChar variant constructs with two different junction designs and variable lengths of the redirected lever arm. Plotted velocities are the mean directionally scored frame-to-frame velocities of all moving filaments in a video frame, averaged over 3 to 5 sequential acquisitions of the same illumination sequence on the same field of view in one assay chamber (blue box indicates period of blue light illumination, with irradiance of 11 mW/cm2). Gliding assays were performed at 2mM ATP. Red lines are fits to the exponential rise and decay of the velocity. e, Velocity trace of MyLOVChar4 from d in more detail. Gray and white colored bands display the envelopes of the center 96 and 72 percentiles of the distribution of frame-to-frame velocities, respectively. The model fit (red line) yields the annotated rate constants and and the steady state velocities in the dark, , and in the presence of blue light,. f, Velocity modulation and lit-state velocity for the series of constructs in d, with each grey dot representing the result of an independent experiment analyzed as in e, and each bar displaying the mean ± standard deviation across the results shown (between N=5 and N=12 experiments depending on the construct). The velocity modulation is defined as and displayed as a percentage. g, ATPase activity of MyLOVChar4 measured with an NADH enzyme-coupled assay, with (blue datapoints) and without (black datapoints) blue light at an irradiance of 10 mW/cm2. Lines are fits to a Michaelis-Menten expression, with and without blue light (black line), and with blue light (blue line). Data are plotted as mean of 1–5 enzyme velocity measurements at each actin concentration (see Supplementary Fig. 3 for details including individual measurements and N values). Error bars display standard deviations for datasets with 3 or more measurements. h, Characterization of MyLOVChar4 as function of blue light intensity (see also Extended Data Fig. 3). Blue datapoints: (top) and (bottom). Black datapoints: (top) and (bottom). The dotted lines are fits to a dose response model, with , , and the slope of the rate dependence m = 0.062 s−1 mW−1 cm2 (see Methods). Each data point represents the mean ± standard deviation from 3 to 5 independently prepared assays (see Supplementary Figure 4 for details); for each assay, parameters were obtained as in e from velocity traces averaged over 3 sequential acquisitions of the stimulation sequence.
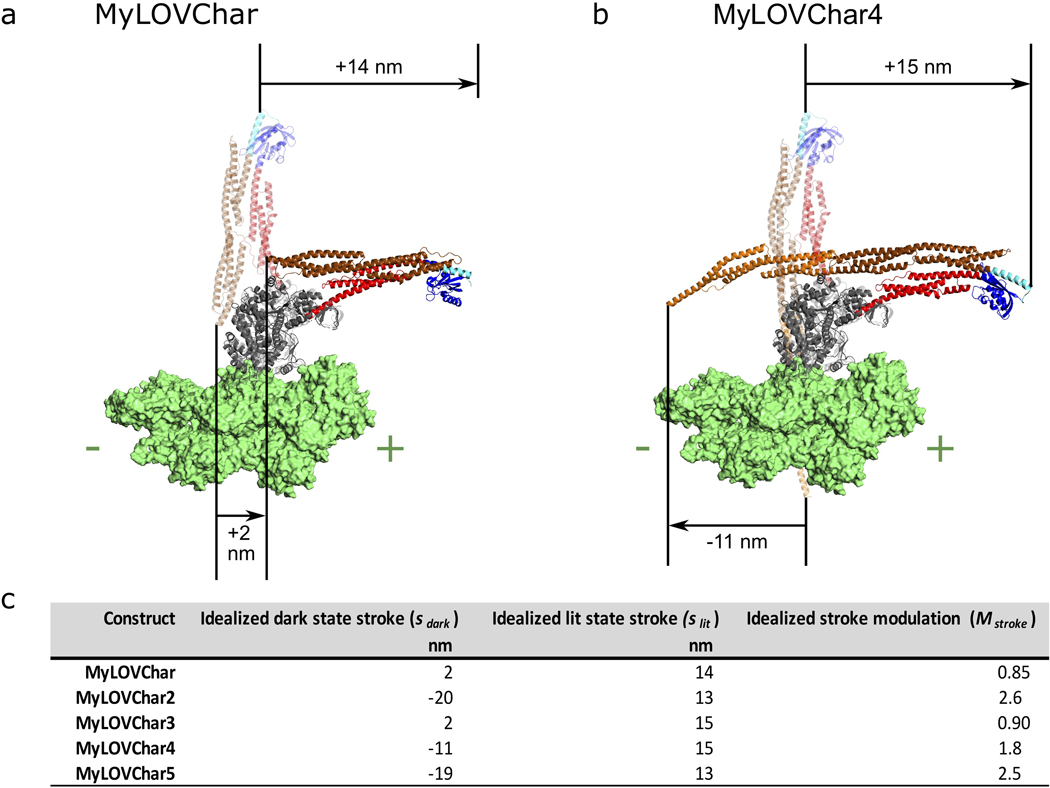
In the first strategy, we aimed to compensate for compliance in the MyLOVChar lever arm by adding spectrin repeats as rigid extensions that increase the length of the redirecting part of the lever arm. MyLOVChar2 incorporates 4 added spectrin repeats (Supplementary Fig. 2) and displays a reduced gliding velocity in the dark as expected for a more redirected lever arm (Fig. 1d, Supplementary Video 3). The lit velocity is also reduced relative to MyLOVChar, but to a smaller degree, so that the net optical modulation is increased to ~60% (Fig. 1f). Thus, building longer redirecting elements is one effective strategy to compensate for mechanical inefficiency in gearshifting myosins.
In the second strategy, we aimed to improve the mechanical efficiency of the design by optimizing the mechanical properties of the junction between the LOV domain and the flanking spectrin repeats. The resulting construct MyLOVChar3 (Fig. 1d, Supplementary Video 4) uses a junctional design adapted from MyLOV1, the junctional variant with the strongest redirected stroke from the MyLOV1–4 series of slow gearshifting motors based on porcine myosin VI10. MyLOVChar3 achieves an increased optical modulation of ~50%, showing that optimizing the junctions of engineered domain fusions can also improve the mechanical efficiency of gearshifting lever arm designs.
Next, to maximize optical modulation of velocity, we combined the two successful strategies, retaining the improved junction from MyLOVChar3 while adding either two or four additional redirecting spectrin repeats (Fig. 1d, Supplementary Videos 5 and 6, and Extended Data Fig. 2b–c). The resulting MyLOVChar4 and MyLOVChar5 designs both exhibit >80% optical modulation on average. Individual experiments can show close to 100% optical modulation, with close to zero average gliding velocity in the dark, increasing to ~3 μm/s when illuminated with blue light (Fig. 1e). As expected from our design, the actin-activated ATPase rate of MyLOVChar4 is not substantially affected by light (Fig. 1g and Supplementary Figure 3), indicative of a gearshifting mechanism in which enzyme kinetics remain constant while mechanical output is strongly modulated via redirection of the lever arm. While the lit velocities of the redesigned motors are reduced compared to MyLOVChar, they remain fast compared to most natural myosin classes20. Thus, we succeeded in our design goal to build a molecular motor that combines high velocity with deep optical modulation.
Directional switching of gliding filaments
As shown previously10, sufficient redirection of the lever arm should yield velocity modulation that exceeds 100%, crossing the origin to produce a change in direction. For MyLOVChar variants, this would give (−)-end directed motility in the dark switching to (+)-end directed motility in the light (Extended Data Fig. 2b–c). Close inspection of gliding filament results for MyLOVChar4 and MyLOVChar5 indeed reveals some bidirectional behavior. When signed velocities are averaged over many filaments, the mean velocities are close to zero in the dark. However, a fraction of the filaments displays clear (−)-end directed motion in the dark (Fig. 1e), and individual filaments can be seen to switch their direction of motion as the light is turned on (Supplementary Video 7). The stochastic behavior of individual filaments driven by small collections of these gearshifting motors thus yields distributions of velocities that extend into the (−)-end directed regime in the dark, switching to robust, rapid (+)-end directed motility in the light.
Dose-dependent response of gearshifting ensembles
A potentially useful feature of optically controlled motors is that the properties of motor ensembles may be tunable by altering the irradiance of the light signal. In isolated LOV domains the fractional population in the lit state can be varied by changing light intensity21, with steady-state populations resulting from a light-dependent photoproduct formation and a thermally driven recovery. In ensembles of optically controlled motors, the steady-state fraction of motors in the lit state thus depends on the irradiance, and the velocity of a filament or cargo driven by the ensemble might be continuously tuned to ensemble-averaged values that lie between the lit and dark extremes. We used gliding filament assays to analyze the dynamic and steady-state response of MyLOVChar4 ensembles to varying doses of blue light. As expected, at low irradiances, we found that the lit state gliding velocity depended on the blue light intensity (Fig. 1h, Extended Data Fig 3 and Supplementary Fig. 4). We also characterized the gliding velocity transients upon turning the blue light on and off. The dose-dependent rates of transition and steady-state velocities can be globally described by a simple two-state model for LOV photoswitching, in which the transition rate from the dark to the lit state depends linearly on the irradiance while the rate of thermal decay to the dark state is fixed21 (see Methods and Supplementary Note 1). Saturated velocities can be achieved with an irradiance of a few mW/cm2, 2–3 orders of magnitude smaller than typical intensities used in standard epifluorescence experiments on live cells22. To further test the dose response model, we directly probed the dose-dependent conformational change of the optically controllable lever arm of the motor, using Förster resonance energy transfer (FRET) between a pair of fluorescent dyes conjugated to a minimal LOV-containing lever arm construct (Extended Data Fig. 4a–b). Bulk fluorescence measurements (Extended Data Fig. 4c–f) show a light-dependent response that saturates at similar blue light intensities to the gliding filament velocities (Extended Data 4g), as expected if the dose-dependent characteristics of the motor are dominated by the conformational changes of the LOV domain.
Directional control of processive myosins in vitro
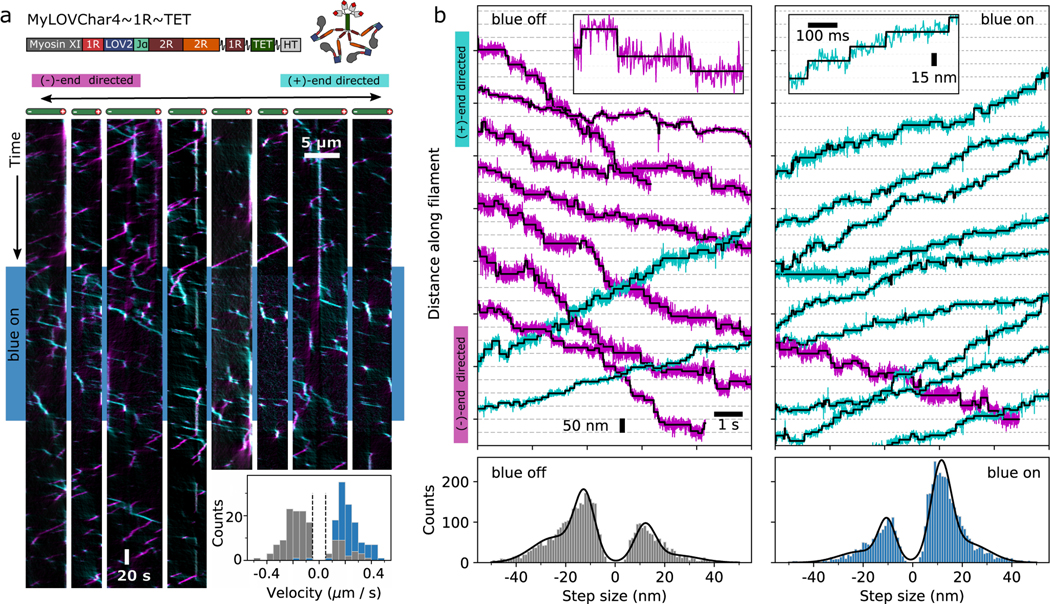
Transport functions carried out by a number of cellular cytoskeletal motors depend on processivity — the ability of a motor to take many steps along a filament before dissociating23. Light-activated gearshifting has not previously been combined with processive motion, although controllable bidirectional processive myosins have been engineered that respond to metal ions6,7 or oligonucleotide strands15. We designed a controllable processive motor based on MyLOVChar4 by adopting a strategy of forced tetramerization in combination with flexible linkers (Fig. 2a). In prior work, this strategy was found to be successful for building highly processive motors from engineered monomers, and was applied to a fixed-gear motor based on the Chara myosin XI catalytic domain7.
Figure 2. Directional control of processive myosins in vitro.
a, Characterization of an optically controllable tetrameric myosin motor MyLOVChar4~1R~TET using single-molecule fluorescence. Top: block diagram and cartoon of the molecular construct. 1R~TET: tetramerization domain with flexible linkages7. Main: Representative kymographs displaying processive runs on 4 actin filaments (left and right sets of 4 kymographs each show results of a 1st and 2nd stimulus sequence on the same filaments). The blue band in the kymograph indicates the period when the blue LED (irradiance: 11 mW/cm2) was turned on. Plus-end directed runs are colored cyan and minus-end directed runs are colored magenta, using an automated kymograph analysis42. Inset: Histograms of velocities of detected directional runs (blue: lit, gray: dark). Dashed lines indicate the minimum threshold speed used for scoring runs. The mean and standard deviation of minus-end directed velocities in the dark are (N = 138 runs), and in the light (N = 123 runs), with results collected over 4 assay chamber preparations and 2 different protein preparations. Data were collected at 10 ATP. b, Gold nanoparticle tracking motor stepping traces on MyLOVChar4L2(+4)~1R~TET, a variant that differs from the construct MyLOVChar4~1R~TET only by mutations to an actin binding loop in the catalytic domain of Chara corallina myosin XI43. Top, Representative stepping traces without blue light (left) and with blue light on (right). Raw trajectories (colored traces, acquired at 503 Hz) are shown together with idealizations (black) based on automated step-finding44. Predominantly (+)-end directed traces are colored in cyan, while predominantly (−)-end directed traces are colored in magenta. Insets show selected traces at high resolution. Scale bars (15 nm and 100 ms) apply to both insets. Data were collected at 5 ATP. Bottom, step size distributions at the annotated light condition. Distributions of scored step sizes (blue off: N= 4364, blue on: N=4809) are shown together with fits (black lines) of sums of Gaussian distributions. Peak locations ± standard errors are as follows. Blue off: −24.7 ± 0.4 nm, −12.4 ± 0.3 nm, +12.1 ± 0.4 nm, and +24.0 ± 0.9 nm. Blue on: −20.9 ± 0.6 nm, −10.4 ± 0.3 nm, +11.4 ± 0.2 nm, and +22.1 ± 0.4 nm.
We characterized the performance of MyLOVChar4~1R~TET (Fig. 2a, Supplementary Video 8) using in vitro single-molecule tracking, where single fluorescently labeled motor complexes are imaged translocating along actin filaments immobilized on a glass coverslip. In the absence of blue light (top and bottom part of the kymograph in Fig. 2a), the majority of motors move toward the actin (−)-end. A small fraction of runs are (+)-end directed, revealing the heterogeneity of motor function at the single-molecule level. When the blue light is turned on, almost all motors become (+)-end directed (Fig 2a kymograph, center region with blue band). This behavior is also seen in a histogram (Fig. 2a, inset) of several hundred motor runs, with almost exclusively (+)-end directed motion in the presence of blue light, and predominantly (−)-end directed motion in the absence of blue light. To probe the stepping behavior of bidirectional motors, we performed high-resolution gold nanoparticle tracking7 using a variant of MyLOVChar4~1R~TET (Fig. 2b, Supplementary Video 9). Nanoparticles also show processive light-dependent bidirectional motion, with discrete steps identified using automated changepoint detection (insets in Figure 2b; Supplementary Fig. 5). As seen previously for unidirectional tetrameric myosins incorporating flexible spacer elements7, the step size distributions are broad with frequent backsteps. Steps are predominantly (+)-end directed in the presence of blue light and predominantly (−)-end directed in the absence of blue light (Fig. 2b, bottom panel; Supplementary Fig. 6), with distributions peaked at similar magnitudes and opposite signs in the two conditions (dominants peaks in Fig. 2b: blue off, −11.4 nm; blue on: 12.4 nm). As expected, characteristic dwell times between detected steps do not substantially change upon illumination (Supplementary Fig. 6; blue off, 0.235 ± 0.004 s; blue on, 0.246 ± 0.004 s at 5 μM ATP). Using both fluorescence and gold nanoparticle tracking, we have thus shown that engineered processive myosins can switch their direction of motion under the influence of an optical signal in a reconstituted system with purified proteins.
Optical control of motor localization in living cells
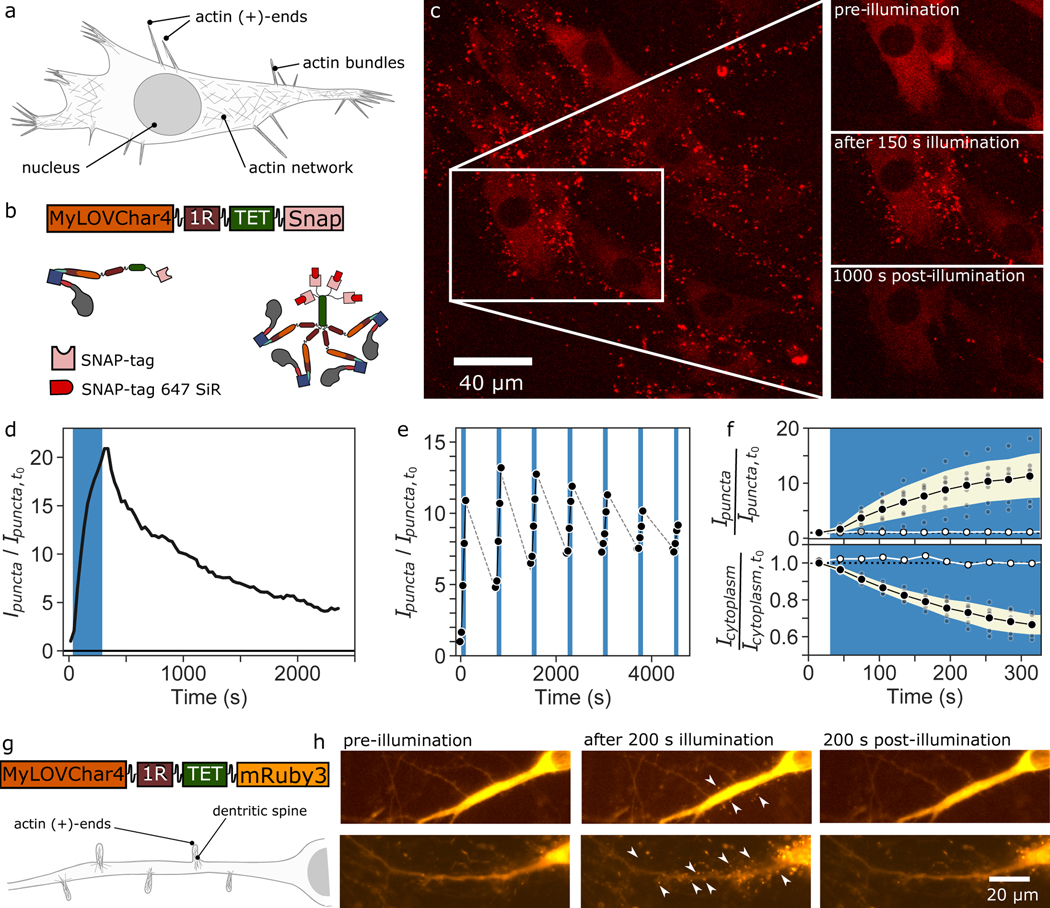
To satisfy our final performance criterion, we asked whether MyLOVChar4~1R~TET retained motility and optically controlled directionality in living cells. We characterized motor function in fibroblasts and in hippocampal neurons. Actin filament architectures within the cell include branched networks of cortical actin close to the cell membrane and polarized actin bundles in protrusions around the periphery of the cells, with actin (+)-ends at the protrusion tips24 (Fig. 3a). For experiments in fibroblasts, we expressed MyLOVChar4~1R~TET as a SNAP-tag fusion, and imaged the motors labeled with a red fluorescent dye (Fig. 3b, see Methods). Prior to optical stimulation, motors were distributed throughout the cytoplasm (Fig. 3c, Supplementary Movie 10). Within 30 seconds of illumination with blue light, distinct puncta became visible around the periphery of the cell, increasing in number and intensity over a 150 s stimulation period, and decaying after the removal of blue light on a longer timescale of 100–1000 s (Fig. 3c,d; Supplementary Fig. 7). We attribute these puncta to motors accumulating at the tips of cellular protrusions, after switching to (+)-end directed motion at the start of optical stimulation and translocating out to the (+)-ends of polarized actin structures. To further support this attribution, we simultaneously imaged the fluorescence of motors and the contours of the cell with differential interference contrast microscopy, and found that puncta colocalize with tips of protrusions (Supplementary Movie 11). In the absence of blue light, the strongly reduced number and intensity of puncta are consistent with directional motion of a dominant fraction of motors toward the actin (−)-end in the dark. Translocation can be repeated across multiple cycles of illumination and recovery (Fig. 3e), with residual persistence of some puncta in the dark, leading to reduced effects over successive cycles. The population of motors in puncta depends monotonically on blue light intensity at low irradiances (Supplementary Fig. 7), and strong effects are seen at irradiances on the order of 1 mW/cm2, comparable to levels commonly used for optogenetic experiments25. The cytoplasmic intensity decreases concomitantly with the appearance of puncta (Fig. 3f, Supplementary Movie 12), indicating that a substantial fraction of the total population of motors within the cell can be sequestered in cellular protrusions. Experiments in cultured neurons also revealed a rapid formation of fluorescent motor puncta around the periphery of neurites upon stimulation with blue light (Fig. 3g,h and Supplementary Movie 13), consistent with motors accumulating at tips of dendritic spines, where actin filaments are polarized with (+)-ends outward26. Taken together, our results show that we can optically control the localization of MyLOVChar4~1R~TET motors in cells.
Figure 3. Optically controlled localization of myosin motors in live cells.
a, Simplified schematic of a cell with different actin architectures at various locations in the cell. b, Block diagram and cartoon of the molecular construct imaged in live cells. c, Maximum intensity projection of a 3D confocal stack of fluorescence images (excitation at 639 nm) of a sheet of mouse fibroblasts in culture immediately after 150 s stimulation with blue light, displaying fluorescent puncta concentrated around the periphery of the cell. Insets show a region of the sheet before and after the blue light stimulation. Images are representative of a set of 7 independent experiments with comparable blue light stimulation protocols (Supplementary Figure 7). d-f, Fluorescence intensity contained in puncta, normalized to the total intensity in the image and plotted after further normalization to the value at the start of the series,. The blue bands indicate the period of blue light stimulation using a blue laser line in the scanning confocal microscope. d, Intensity in puncta calculated for the image series on the cells in panel c. e, Intensity in puncta over 7 stimulation cycles of 2 minutes each, separated by 40-minute wait times. In each stimulation cycle, cells were imaged for 5 frames, with one frame without blue light preceding 4 frames with blue light on. Grey dashed lines are visualization guides. f, Time traces of averaged responses (black datapoints) upon stimulation with blue light, from 7 independent sample preparations (grey datapoints). Black datapoints, mean of the 7 measurements. Error bands, standard deviation of the measurements. White datapoints, response in absence of blue light. Top, fluorescence intensity contained in puncta; the maximum level of after 300 s of stimulation was 11 ± 4. Bottom, fluorescence of cytoplasm. g-h, Characterization of MyLOVChar4~1R~TET~mRuby3 in hippocampal neurons in culture. g, Block diagram of molecular construct and illustration of actin architecture in neurons. h, Wide-field fluorescent images of MyLOVChar4~1R~TET~mRuby3 before, during and after stimulation with blue light (see also Supplementary Video 13). Arrows highlight puncta as expected for motor accumulation in dendritic spines. Data are representative of an experiment replicated twice on different days, with 13 cells.
Single-molecule tracking in living cells
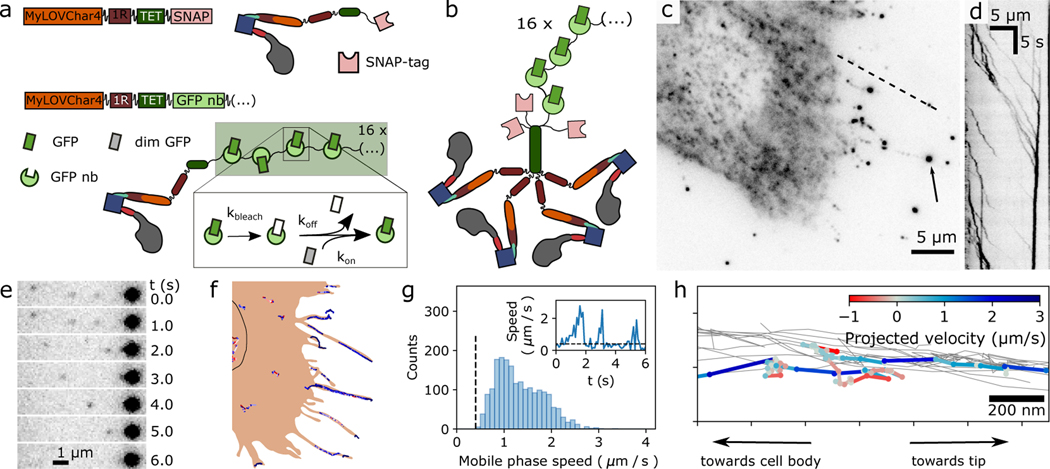
To directly measure the dynamics and motor properties of individual engineered myosins in cells, we performed high-resolution single-molecule tracking (Fig. 4, Supplementary Movie 14). Earlier reports of single-molecule tracking of motors in cells have used quantum dots27, individual fluorescent protein (GFP) tags28, GFP-tagged peroxisomes29, and scaffold arrays that tightly bind multiple fluorescent proteins30. Here we use a 16-copy version of the fluorogenic array tag ArrayG31, where up to 16 monomeric wild type GFPs (mwtGFPs) can bind to a scaffold of flexibly enchained camelid nanobodies. The mwtGFPs bind reversibly, so that the effect of bleaching can be diminished by exchange with a pool of unbleached mwtGFPs in the cytoplasm. mwtGFP fluorescence increases by approximately 26-fold upon binding to the array, which reduces imaging background (Fig. 4a). To favor robust tetramer (Fig. 4b) formation while keeping the concentration of the ArrayG16X tag low enough for single-molecule imaging, we co-expressed low levels of ArrayG16X -tagged motors with an excess of unlabeled SNAP-tagged motors (see Methods).
Figure 4. Single-molecule imaging of engineered motor constructs in live cells.
a, Design of molecular constructs for single-molecule tracking with high spatiotemporal resolution in cells. Block diagram and cartoons show two MyLOVChar4 constructs, C-terminally tagged with either a SNAP-tag (MyLOVChar4~1R~TET~SNAP) or a 16X ArrayG scaffold of GFP nanobodies (MyLOVChar4~1R~TET~ArrayG16X). Dim monomeric wildtype GFP (mwtGFP, grey rectangles) becomes brighter (green rectangles) upon binding to the scaffold; reversible association allows exchange of bleached mwtGFP (white rectangles) with unbleached protein from solution. b, Cartoon of the tetrameric motor complex imaged in cells overexpressing the non-visible species MyLOVChar4~1R~TET~SNAP, a minor population of MyLOVChar4~1R~TET~ArrayG16, and mwtGFP. c, Fluorescence image of a cell excited with 488 nm laser excitation. Arrow: bright fluorescence spot that is a terminal point of observed tracks. d, Kymograph along the dashed line in c. e-f, Single-molecule tracking of motors in cells. e, Image series from a region of the cell in panel c, with single motors moving toward, and merging with, a bright spot like the one indicated in panel c. f, Motor trajectories identified for the cell in panel c, overlaid on a schematic outline of the cell. Black outline, estimated location of the nucleus. Similar results to those presented in c-f were obtained on two independent biological replicates, across a total of 41 cells contained in 39 image acquisition series. g, Velocity histogram from all 41 cells, for all extracted directionally persistent tracks with a minimum length of 20 frames, tabulating mobile runs in which the speed exceeds 0.4 μm/s for 4 frames or longer (N= 1946 runs). The mean and mode of the distribution are 1.3 μm/s and 1.0 μm/s respectively. Inset: velocity trace with definition of mobile phases. h, Detail of motor trajectories in a cellular protrusion. Grey thin lines: set of all tracks associated with the protrusion. One selected trajectory is colored according to the projected velocity along the main axis of the protrusion.
Single-molecule imaging of ArrayG16X -tagged MyLOV4~1R~TET was performed under blue laser excitation, which both activates the LOV2 domain and excites the GFP fluorophore. Under these conditions, we expect the motors to be predominantly (+)-end directed. As expected, fluorescent spots around the periphery of the cell (Fig. 4c) are seen streaming away from the cell body along selected paths (Supplementary Video 14), as shown in a kymograph (Fig. 4d) and image series (Fig. 4e) along selected paths. Single-molecule trajectories often terminate in a brightly labeled endpoint that increases in intensity over time (Fig. 4e, Supplementary Videos 15). Supported by our observations of the cellular morphology and ensemble motor localization (Fig. 3, Supplementary Videos 10 and 11), we conclude these trajectories correspond to motors traveling toward the actin (+)-end on cellular protrusions containing polarized actin bundles.
To measure motor velocities in cells, we extracted high-resolution traces from the fluorescence imaging series using a tracking algorithm optimized for high spot densities32. Extracted traces for an example cell (Fig. 4c) show many traces around the cell periphery, a set of traces in the region below the nucleus, and a few isolated traces in between (Fig. 4f). The imaging conditions in our experiment are most favorable for detection of processive runs in thin cellular protrusions, because of the dense, polarized actin structure and lower fluorescence background. The motors predominantly travel away from the cell body, moving in intermittent bursts of high velocity separated by much slower phases (Fig. 4g, inset; Supplementary Fig. 9). We analyzed the frame-to-frame speeds of motors tracked in 53 cells, and found a broad distribution of mobile phase speeds with a peak at ~1 μm/s (Fig. 4g). Distinct trajectories can be resolved within sets of clustered traces around the cell periphery (Fig. 4h), revealing the spatial extent of the protrusions that motors are able to explore. Short directional reversals can be seen for some traces (Fig. 4h, Supplementary Fig. 9), consistent with motors that temporarily become (−)-end directed. Taken together, these results show that optically activated motors can efficiently traverse across various cellular actin structures, with velocities in the range of microns per second.
Controllable transport of molecular cargos
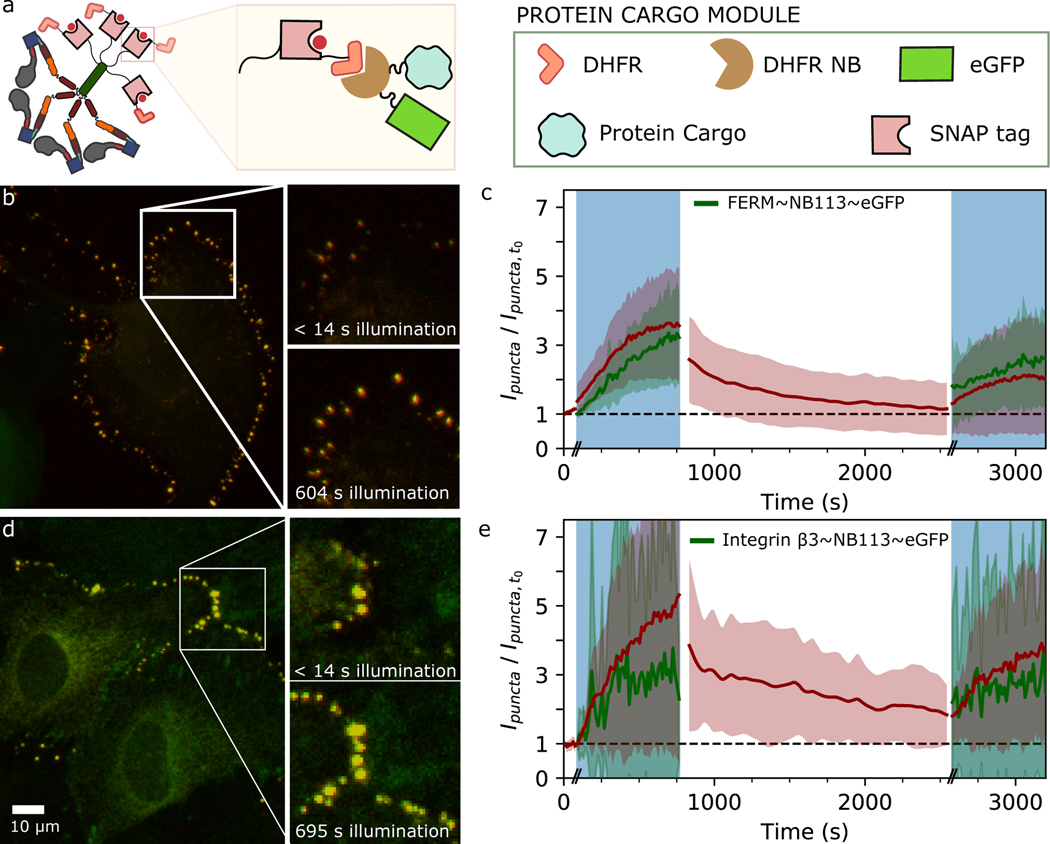
Having satisfied our performance criteria for controllable processive molecular motors that function in living cells, we set out to directly test whether the engineered motors could be harnessed for optically controlled cargo transport (Fig. 5). Native myosins are responsible for transporting diverse molecular and vesicular cargos33; we chose to test two distinct classes of molecular cargos: talin FERM domain34 (a cytosolic protein), and integrin β3 (an integral membrane protein). Prior work has shown that fusion of a myosin to a protein of interest or nanobody can be sufficient for localizing a binding partner into cellular protrusions35. We sought to couple controllable motors to protein cargos via a nanobody interaction31 (see Methods, and Fig. 5a), and co-expressed motor and cargo constructs in epithelial cells. In these experiments, the tetrameric motor can be visualized with a SNAP-tag fluorophore excited by a red laser, while blue excitation is used to both visualize the fluorescent cargo and switch the motor directionality. Upon stimulation with blue light, both the cytoplasmic FERM domain (Fig. 5b–c, Supplementary Fig. 10, Supplementary Videos 16 and 17 and the transmembrane integrin β3 (Fig 5d–e, Supplementary Fig 11, and Supplementary Videos 18 and 19) are enriched in puncta around the cell periphery, closely following motor enrichment. The localization of motors largely decays within several hundred seconds when blue light is removed, while a second stimulus reveals a more persistent localization of the cargos that increases again during the illumination period (Fig. 5c,e). Taken together, our observations show that an engineered myosin motor with optically controllable directionality can localize distinct classes of molecular cargos to cellular protrusions.
Figure 5. Optically controlled localization of motors and molecular cargos in live cells.
a, Schematic of MyLOVChar4~1R~TET~DFHR interacting with a protein cargo fused to a DFHR nanobody (NB113). b-c, Results from cells expressing MyLOVChar4~1R~TET~DFHR and FERM~NB113~eGFP. d-e, Results from cells expressing MyLOVChar4~1R~TET~DFHR and integrin β3~NB113~eGFP. b,d, Merged two-channel fluorescence images (maximum intensity projections of confocal z-stacks; red, motor; green, cargo) from sheets of MCF10A cells in culture, imaged with simultaneous 639 nm and 488 nm excitation. Images are representative of a set of 10 (in b) and 5 (in c) independent measurements, on 12 and 8 cells, respectively. c,e, Fluorescence intensity contained in puncta, normalized to values at the start of the imaging sequence, for motor (red) and cargo (green) channels. Results are averaged over individual time series (Supplementary Fig. 9 – 10) representing (c) 12 and (e) 8 cells, from 2 biological replicates in each case. Error bands are standard deviations of the measurements. Time axis includes approximate delays at transitions between the illumination phases (indicated by the double dashes), which for these experiments were collected as separate movies.
Discussion
We have presented a set of engineered myosins with optically controllable velocities and directionalities, with speeds matching the range of natural biological motors. Deviations in modulation depths from the predictions of idealized structural models (Extended Data Fig. 2a–c) can be overcome by extending redirected lever arms and optimizing engineered junctions. Monomeric motors can be assembled into controllable processive complexes and used for light-dependent intracellular transport of cargos. This strategy complements approaches based on light-dependent cargo recruitment11,12,13,14 while avoiding challenges associated with non-productive accumulation of the motor pool at target locations36 and allowing for the use of tight constitutive cargo binding interactions that can resist forces generated in cellular contexts. Light-activated gearshifting with photoswitchable cargo binding at orthogonal wavelengths37 could also provide additional levels of control. Fast monomeric non-processive gearshifting motors may be used to introduce programmability into devices that rely on gliding filament propulsion38, or spatial control in active fluids composed of propelled filaments39. The multimeric motors here could also be used for spatiotemporal control of active matter systems40 , and similar design principles may be used to control the mechanical output of filamentous myosins to enable control of cellular contractility. Further extension of the gearshifting mechanism to microtubule-based motors with swinging lever arm mechanisms such as kinesin-1410 and dynein41 would then complete a toolbox of optically controlled motors that can be used to probe the relationships between nanoscale motility and complex cellular functions.
Methods
Molecular constructs for in vitro experiments
Myosin constructs were assembled from DNA fragments encoding Chara corallina Myosin XI (codons 1–746), Nicotiana tabacum Myosin XI (codons 1–738), Dictyostelium discoideum alpha-actinin (codons 266–502), Avena sativa phot1 (codons 404–543), and Gallus gallus alpha-spectrin (codons 1663–2090) and cloned into the expression plasmid pBiEx-1 (Novagen). Constructs encoding the monomeric motors MyLOVChar and MyLOVChar1–5 include codons for a C-terminal flexible linker, enhanced YFP, and FLAG tag (DYKDDDDK) as in prior work10. Constructs for the tetrameric motors MyLOVChar4~1R~TET and NM11CD738_2R~1R~TET include codons for a C-terminal engineered leucine zipper variant pLI45, a spacer consisting of Dictyostelium discoideum alpha-actinin codons 266–388 flanked by flexible linkers, a Halotag protein sequence (Promega), and a FLAG tag (DYKDDDDK) as in prior work7. Motor proteins were expressed by direct transfection in Sf9 insect cells and purified using anti-FLAG resin (Sigma-Aldrich A2220), as previously described7,46. Prior to transfection, the cells were cultured in serum-free medium without antibiotics. Myosin proteins used for in vitro assays were eluted in a glycerol-containing storage buffer (25 mM KCl, 2 mM MgCl2, 5 mM EGTA, 25 mM Imidazole pH7.5, 0.1 mM ATP, 1 μg/mL Aprotinin, 1 μg/mL Leupeptin, 2 mM DTT, 55 % glycerol by volume), flash frozen in small aliquots within hours of the completed elution, and stored at −80° C until used in assays. The myosin construct characterized by electron microscopy (CM11L2+4CD746) was eluted in a glycerol-free storage buffer (25 mM KCl, 2 mM MgCl2, 5 mM EGTA, 25 mM Imidazole pH7.5, 0.1 mM ATP, 2 mM DTT), and concentrated using 10 kDa Amicon Ultra Millipore columns before flash freezing. For details of the constructs see Supplementary Fig. 2.
The minimal lever arm construct SNAP-1R-LOV-1R-HT for FRET measurements was assembled with DNA fragments coding for a section of the lever arm from MyLOVChar4 as described above and cloned into the bacterial expression vector pET24a. The construct also includes codons for a N-terminal SNAP tag and a C-terminal Halotag and His-Tag. The construct was expressed in Escherichia coli B21 (DE3) by induction with 0.5 mM Isopropyl-β-D-thiogalactoside (IPTG) at 16 °C in 2xYT broth for 20 hours. Cells were harvested by centrifugation and frozen. Bacterial pellets were thawed, lysed with hen egg lysozyme (1.7 mg/ml) and by mechanical lysis with a sonicator, and purified by metal ion affinity (Ni-NTA column, HisPur resin, Thermo Scientific) and size-exclusion chromatography (Superdex 200/10/ 300 GL). Lysis buffer consisted of 50 mM Tris (pH 7.5), 500 mM NaCl, and 1.7 mM PMSF. Wash buffer consisted of phosphate buffered saline (PBS) with 50 mM Imidazole (pH 7.5). Elution buffer consisted of PBS with 500 mM Imidazole (pH 7.5). Following the metal affinity columns, proteins were dialyzed overnight into PBS with 0.5 mM DTT and concentrated using 3kDa Amicon Ultra Millipore columns. Concentrations were measured by absorbance at 280 nm using a Nanodrop spectrophotometer (Thermo Scientific). For details of the construct see Supplementary Fig. 2.
Cryo-electron microcopy on Chara Myosin XI
Actin purification:
Chicken muscle actin was purified as previously described47 and stored in G-Mg buffer: 2mM Tris-HCl PH 8.0, 0.5mM DTT, 0.2mM MgCl2, 0.01% NaN3. F-actin was prepared by polymerizing 10μM monomeric actin in KMEI buffer (50mM KCl, 1mM MgCl2, 1mM EGTA, 10mM imidazole pH 7.0) overnight at 4 °C. 20 μM Phalloidin was added to F-actin 30 min before grid preparation.
Grid preparation and cryo-EM image acquisition:
Prior to vitrification, 10U/ml apyrase was added to 18.5 μM myosin XI to deplete nucleotide. 3 μl of phalloidin-bound F-actin, diluted to 0.6 μM, was applied to a glow discharged C-flat 1.2/1.3 holy carbon Au 300 mesh grid (Electron Microscopy Sciences) in a Leica GP plunge freezer at 25°C. 60 s later, 3 μl of myosin XI was applied and incubated for 60 s. After that, 3 μl of the mixture solution was removed and another 3 μl of myosin XI was applied. After an additional 60 s of incubation, 3 μl of the mixture solution was removed and the grid was blotted from the back with Whatman no. 5 filter paper for 5 s and flash frozen in liquid ethane. Data were collected on a FEI Titan Krios (New York Structural Biology Center) operating at 300 kV equipped with a Gatan K2-summit detector and a Cs-corrector using counting mode. Image stacks were recorded with the Leginon software at a nominal magnification of 105,000X corresponding to a pixel size of 1.096 Å. Each exposure was fractionated across 50 frames with a total electron dose of 67.12 e-/Å2 (1.34 e-/Å2 /frame) and a total exposure time of 10 s. Images were recorded with defocus values ranging from −1 to −4 μm underfocus. In total, 714 movie stacks were acquired.
Cryo-EM image processing:
Data processing was performed as described in Mei et al.48. Briefly, image processing was carried out using the Iterative Helical Real Space Refinement (IHRSR) protocol as implemented in the RELION 3.0 pipeline49, utilizing RELION functions unless otherwise noted. Movie stacks were motion corrected and dose weighted with MotionCor2 (https://emcore.ucsf.edu/ucsf-software) using 5 × 5 patches. CTF estimation was performed with CTFFIND4 (https://grigoriefflab.umassmed.edu/ctffind4). Filaments were auto-picked and split into overlapping segments with a step size of 81 Å, corresponding to 3 actin protomers. A total of 72,336 segments were extracted with a box size of 512 pixels. After two-dimensional (2D) classification and sorting, 45,779 segments were selected and subjected to three-dimensional (3D) classification initialized with the helical parameters of 27 Å rise and −166.7° twist. The cryo-EM map of myosin VI bound to F actin (EMD-7116) was low pass filtered to 30 Å to serve as an initial model. 3D auto-refinement and postprocessing using a 3D mask with a length of 30% of the box size resulted in a 5.7 Å map. CTF refinement, Bayesian polishing and another round of 3D refinement improved the resolution to 4.3 Å based on the Fourier shell correlation (FSC) 0.143 criterion. Final helical parameters converged to 27.45 Å rise and −167.11° rise. Local resolution was estimated using RELION 3.0. Figures were generated in UCSF Chimera and ChimeraX50.
Model building and structure refinement:
The initial model of myosin XI was generated with I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER). The myosin model and a published actin model (PDB: 6BNO) were docked into the EM density by rigid body fitting in UCSF Chimera. This initial actomyosin model was then rebuilt and refined as described in Mei et al.48 Briefly, 100 models were generated and the lowest energy model was manually adjusted in coot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot) and refined using Phenix.real_space.refine in the Phenix suite51. As the region of the cryo-EM map corresponding to myosin XI was of limited resolution (5–8 Å, Extended Data Fig. 1c), and largely lacked well-defined side chain density, the myosin chain of the atomic model was truncated to poly-alanine prior to deposition.
Structural modeling of optically controllable motor constructs
We generated idealized pre- and post- stroke structures for MyLOVChar-MyLOVChar5 using flexible backbone modeling with RosettaRemodel52 to model engineered junctions between protein domains. For modeling the catalytic domain, in post-stroke models we used the rigor structure of Chara corallina myosin XI obtained in this work (see section on cryo-electron microscopy), and in pre-stroke models we used a previously reported structure of human myosin Vc (PDB: 5HMP)53, a myosin isoform related to plant myosin XI. Engineered lever arms were constructed from available crystal structures for Dictyostelium discoideum alpha-actinin (PDB: 1g8xA residues 766–1002), Avena sativa phot1 (PDB: 2V0U) and (for MyLOVChar2, MyLOVChar4 and MyLOVChar5) Gallus gallus alpha-spectrin (PDB: 1U4Q). Models of the engineered constructs were obtained by remodeling 4 to 8 residues on either side of each engineered junction as alpha helical structures, followed by a relaxation protocol on the complete lever arm sequence. See Supplementary Fig. 1 for examples of the RosettaRemodel flags and blueprint files with the instructions for modeling of the junctions. We gathered 500 candidate structures for each model, and removed structures that clashed with the actin filament (which was not included in the initial computation) To obtain the idealized predicted power strokes of the motors in the light and the dark states, we aligned the 50 top-scoring structures to a subregion (corresponding approximately to the lower 50kDa domain L50 of myosin) of the cryo-electron microscopy reconstruction of rigor Chara corallina Myosin XI bound to actin, and extracted the mean projected stroke along the filament of the end of the lever arm (for the dark state) or the last rigid residue on the LOV2 domain, taken as G516 (for the lit state).
Gliding filament motility assays
Monomeric engineered myosin constructs were characterized using a dual-labelled gliding filament assay as described in Nakamura et al.10, with minor modifications to buffers and imaging conditions as described below. Flow channels were assembled with microscope coverslips attached to a cover slide with double-sided scotch tape. Channels were approximately 2 mm wide and had a volume of approximately 4–5 μl. Prior to assembly of the channel, the coverslips were cleaned by 5 minutes of plasma cleaning, and spin-coated with 0.1% nitrocellulose (Ladd Research #10800) for 30 seconds at 3000 rpm, following by 30 seconds at 6000 rpm, with 5 second ramps.
Assay buffer consisted of 25 mM Tris (pH 7.5), 25 mM KCl, 1 mM EGTA, 2 mM MgCl2 and 10 mM DTT. Blocking buffer consisted of assay buffer supplemented with 2 mg/ml BSA (Sigma Aldrich). Imaging buffer consisted of blocking buffer supplemented with an oxygen scavenging system (0.2 mg/ml glucose oxidase and 0.36 mg/ml catalase and 0.4% wt/vol glucose) and an ATP regeneration system (creatine phosphokinase at 0.95 μg/ml and 1 mM phosphocreatine). The assay conditions differ from Nakamura et al.10 due to the complete omission of imidazole, which even at the low levels present in the prior study can be sufficient to speed up the dark state recovery of the LOV domain by several fold54.
Flow channels were prepared in six steps: (1) Flow in 3.5 μl GFP antibody (Millipore Sigma MAB3580) diluted 3-fold from stock in phosphate buffered saline, followed by a 2-minute incubation period. (2) Flow in 30 μl of blocking buffer, followed by a 2-minute incubation period. (3) Flow in 10 μl of GFP-tagged monomeric myosin motor (diluted from stock in assay buffer to 100–400 nM), followed by a 2-minute incubation period, (4) Wash with 30 μl blocking buffer. (5) Flow in 30 μl of polarity-labeled actin by gravity flow and incubate for 3 minutes. Actin was freshly diluted 1000-fold to 4000-fold from stock into assay buffer for each channel, in a two-step dilution, gently tapping the Eppendorf-tube between steps to mix. (6) Flow in imaging buffer. Before imaging, the channels were sealed with vacuum grease.
Channels were imaged with total internal reflection fluorescence (TIRF) excitation, with a 1.49 NA 100X objective (Nikon), excited with a 532 nm laser (Coherent Sapphire, 30 μW at the back aperture) and a 633 nm laser (Blue Sky Research, 30–80 μW at the back aperture. Fluorescence from TMR/Cy5 labeled actin filaments was recorded on an EMCCD camera (Andor iXon DU-897E-CS0-#BV) at maximum gain. An LED light source (Thorlabs, M470L3, center wavelength 470 nm) provided continuous wave blue light stimulation via the TIRF illuminator arm (Nikon T-FL-TIRF2). The blue light intensity was controlled using a voltage driven modulating input on the LED current driver and synchronized to the camera acquisition with custom written LabVIEW software. For most assays (Fig. 1d–f) 800-frame movies were acquired at a 7.4 Hz frame rate (0.136 s/frame), with blue light on (irradiance 11 mW/cm2) in frames 100–300. For Fig. 1h we acquired movies of 550, 650, and 750 frames, with blue light turned on during frames 50–150, 50–250 and 50–350 respectively (see Extended Data Fig. 3), with the 100-frame blue light stimulation period for the highest irradiances (32 and 70 mW/cm2), the 200-frame stimulation period for intermediate irradiances (3.4, 4.8, 7.3, and 11 mW/cm2), and the 300-frame stimulation period for the lowest irradiances (0.5 and 1.2 mW/cm2).
Gliding filament motility analysis
Gliding filament assays were analyzed to extract frame-to-frame velocity of directionally scored filaments using a custom workflow building on the FAST software for gliding filament motility analysis by Aksel et al.55. Two-color movies with the registered images of the filaments in a green fluorescence channel and the filament ends in a red fluorescence channel were generated from the raw data using custom written scripts in Matlab, with image registration between the channels based on cross-correlation using a discrete Fourier Transform algorithm. We used custom written Jython scripts to automate the use of ImageJ plugins for filament detection (Ridge Detection plugin version 1.4 (Wagner, T., Hiner, M. & Raynaud, X., doi:10.5281/zenodo.845874 (2017), implementing the ridge detection algorithm of Steger) and tracking of polarity labels (TrackMate plugin, Tinevez et al.56). Motility was then analyzed using Python software modified from FAST55 and incorporating stuck filament rejection, selection of polarity-marked filaments, and directionality determination.
We collected sets of signed trajectories over multiple movies (3–5 repeats of the same light stimulus sequence acquired on the same field of view in a single sample chamber, with no additional waiting time between repeats) and computed frame-to-frame filament velocities that were averaged to obtain velocity traces as in Fig. 1d. The average number of filament velocities combined to obtain each timepoint in each velocity trace was <N>=51 for the dataset represented in Fig. 1d and compiled in Fig. 1f. The parameter value for the velocity in the dark before light stimulus, , was calculated as the mean of the datapoints before blue light was turned on. With fixed, we globally fitted a single exponential rise followed by an exponential decay using a non-linear least squares fitting procedure (Scipy.optimize curve_fit). This fit provides the rates and , the lit velocity , and the final velocity . In Fig. 1e and Fig. 1h the reported dark velocity is taken as .
Gliding filament dose response modeling
We modeled the dose response for MyLOVChar4 (Fig. 1h) as a two-state system with a population of motors in the dark state, and a population of motors in the lit state, , and corresponding characteristic velocities and . In this 2-state model, the lit state thermally decays into the dark state with a constant rate . Transition from the dark state into the lit state occurs with a rate that depends linearly on irradiance, as
with the irradiance of the light and a slope . The steady-state fraction of motors in the lit state is then
Assuming a simplified model for filament gliding that neglects strain-dependent effects (see Supplementary Note 1), this results in a dose-dependent steady-state velocity
The parameters of this two-state model may be related to the parameters fit to the velocity transients (see above) after noting that corresponds directly to the decay rate of the lit state population, , while the measured transient rate reflects the relaxation process of the two-state system, .
Steady-state ATPase assay
The steady-state ATPase activity was measured using the NADH coupled assay as described57, but instead of the imidazole buffer used by De la Cruz et al we used a Tris-based assay buffer as for the gliding motility assay (see above), supplemented with 1 mg/ml BSA. In addition, we implemented the capability to illuminate the sample with blue light. Briefly, assays were performed with 14 nM of MyLOVChar4 motors at 23 °C, monitoring NADH absorption at 340 nm wavelength in a spectrophotometer (Agilent Cary UV-Vis 6000). Assays were performed with 150 μl of sample in a 1 cm path length quartz cuvette with 100 μl nominal volume (Starna Cells 16.100-Q-10/Z15), using a microscope coverslip as a translucent lid. Samples could be illuminated from the top of the cuvette (perpendicular to the light path of the 340 nm absorption measurement) with a 12 mm diameter collimated beam (fiber collimator Thorlabs F950-FC-A) from a fiber-coupled light source (Thorlabs M470F3, controlled by voltage modulation with a function generator, Agilent 33220A). Blue light irradiance was 10 mW/cm2. In each measurement at a given actin concentration we measured slopes of decreasing absorbance during one or more cycles of a period without followed by a period with blue light. Illumination periods varied from 30 sec to 2 min, for the highest and lowest actin concentration respectively.
FRET assays on minimal lever-arm construct
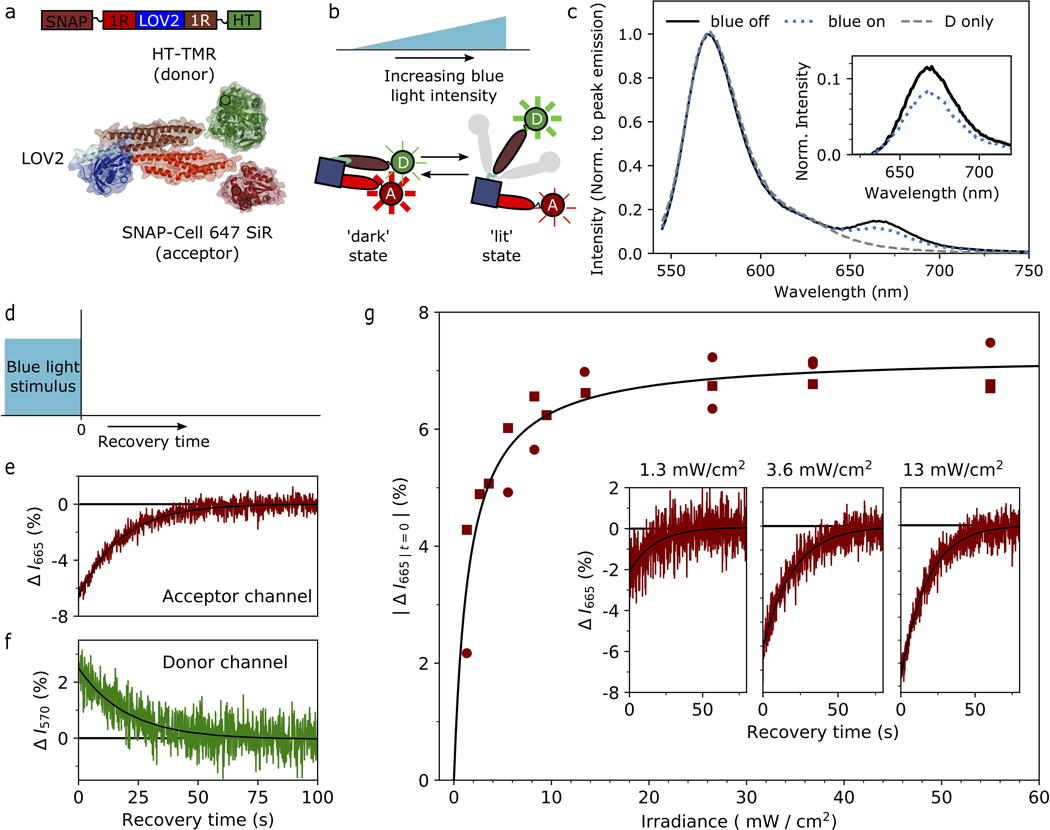
We performed Förster Resonance Energy Transfer (FRET) measurements in solution on the minimal lever arm construct Snap~1R-LOV-1R~HT at 100 nM, either singly labelled (with SnapAlexa 647 SiR or with Halotag TMR) or doubly labelled (Snap Alexa 647 SiR and Halotag TMR). Labeling was done on flash-frozen aliquots of the purified protein (see section “Constructs for in-vitro experiments”) in a single step rotating at 4 °C for 3.5 hours in phosphate buffered saline (PBS) buffer. We used 5-fold molar excess of the ligands, which were removed after the labeling by overnight dialysis into Tris assay buffer (25 mM Tris, 25 mM KCl, 1 mM EGTA, 1 mM DTT, 2 mM MgCl2), with 2 buffer exchanges and >500:1 sample-to-buffer volume ratio. Labeled proteins were flash frozen in aliquots for single-use experiments.
Fluorescence emission spectra and time-resolved measurements were collected on a fluorimeter (Horiba Fluorolog 3) with a 450W Xenon lamp light source. Samples were contained in a quartz cuvette with 40 μl nominal volume (Starna Cells 16.40F-Q-10/Z15). Samples could be stimulated with blue light during fluorescence emission experiments through the exit window in the cuvette, illuminated through a hole in the front panel of the fluorimeter by a fiber coupled LED light source as used in ATPase activity assays. The intensity of the 12 mm collimated beam was further concentrated with a 50 mm lens to slightly overfill the 2×2 mm exit window. Blue light irradiance was controlled by voltage modulation with a function generator as used in the ATPase activity assay, and insertion of appropriate optical density filters. The blue light was spectrally filtered with a bandpass filter (Semrock FF01–465/537/623–25) to diminish intensity of light with wavelengths 480 nm and longer, reducing direct excitation of the donor dye.
Fluorescence emission measurements were acquired with excitation centered at 532 nm or 632 nm (spectral width 10 nm). Emission spectra were collected with a 3 nm spectral width and 200 ms integration time. Time-resolved measurements collected emission in a 10 nm spectral band around 665 nm to record acceptor fluorescence or with a 3 nm band around 570 nm to record donor fluorescence, using a 100 ms integration time. Emission spectra and intensities in time traces were corrected for background from the buffer without sample.
In vitro processive single-molecule fluorescence tracking assays
In vitro assays on tetrameric motor constructs were performed as in Schindler et al.7 with some modifications. Filaments were immobilized on the glass surface using rabbit skeletal muscle myosin (Cytoskeletal Inc, Catalog no. MY02) that was treated with N-ethyl maleimide. Assay buffer consisted of 25 mM Tris (pH 7.5), 25 mM KCl, 1 mM EGTA, 2 mM MgCl2 and 10 mM DTT. Blocking buffer consisted of 25 mM Tris (pH 7.5), 25 mM KCl, 1 mM EGTA, 2 mM MgCl2, 10 mM DTT, and 2 mg/ml BSA (Sigma Aldrich). Imaging buffer consisted of blocking buffer supplemented with an oxygen scavenging system (0.2 mg/ml glucose oxidase and 0.36 mg/ml catalase and 0.4% wt/vol glucose) and an ATP regeneration system (creatine phosphokinase at 0.95 μg/ml and 1 mM phosphocreatine) and diluted motors (final dilution 1:300 to 1:2000 from the stock concentration of 80 nM). Unlike Schindler et al.7, we did not supplement any of the assay buffers with 0.1% Tween. The flow protocol was as follows: (1) Flow in 3.5 μl of N-ethyl maleimide treated muscle myosin followed by incubation for 2 minutes. (2) Flow in 30 μl of blocking buffer followed by incubation for 2 minutes. (3) Flow in 30 μl of diluted actin followed by incubation for 3 minutes. (4) Flow in imaging buffer.
Channels were imaged with total internal reflection fluorescence (TIRF) excitation. Fluorescence was excited with a solid state 633 nm laser (BSR). For each channel, we first performed a quick scan across several fields of view with 532 nm laser excitation to verify that there was a good distribution of filaments across the channel. We then performed the following image acquisition protocol on a suitable field of view: (1) Turn on red laser, acquire ~20 frame movie (typically 100 ms exposure time) of red labelled filament heads. (2) Turn on green laser, switch to green imaging channel on Optosplit. Image filaments with short movie (10–20 frames, 1 second exposure). (3) Switch back to red laser, at high power (1 mW at objective back aperture). Wait for filament heads to bleach to a dim level compatible with motor imaging. (4) Start image acquisition on motor channel (red), using a 1 s exposure time.
Motor trajectories were analyzed using the kymograph generation and analysis software KymographClear and KymographDirect42. We manually selected only clearly polarity labelled filaments, analyzing each of the three phases of the acquisition sequence (dark pre, lit, and dark post) as a separate movie. In KymographDirect we used the “noise rejection” detection mode and processed results for forward and backward trajectories separately. For each run, we recorded the average velocity of the detected trajectory, contributing one measurement to the velocity histogram. Multiple separate runs were recorded on a single motor displaying bidirectional behavior when the directional phases were picked up as separate segments by the detection algorithm.
In vitro processive single-molecule gold-nanoparticle tracking assays
Gold nanoparticle tracking assays were performed as described in Schindler et al.7 with some modifications to channel preparation and imaging conditions. Streptavidin-conjugated 50 nm gold nanoparticles (Cytodiagnostics) were washed twice in binding buffer (500 mM NaCl, 40 mM Tris pH 7.5, 0.2% (vol/vol) Tween-20 (Sigma) and 1 mg/ml BSA) by centrifugation and then resuspended by sonication. Gold nanoparticles were combined with PEG-biotin labeled proteins at an approximately 1:1 protein:nanoparticle molar ratio and then incubated on ice for 15 minutes before dilution into imaging buffer. Channels were prepared as described for the in vitro processive single-molecule tracking assays with the addition of 0.2% (vol/vol) Tween-20 in the blocking buffer and imaging buffer. Polarity of Alexa 633 phalloidin-labeled actin filaments was determined using single-molecule tracking of engineered Nicotiana tabacum myosin XI tetramers NM11CD738_2R~1R~TET (Supplementary Fig. 1–2) labeled with TMR (Supplementary Movie 9).
Gold nanoparticles were imaged using objective-side evanescent dark-field microscopy on a custom-built inverted microscope, with a 1.49 NA 60X objective (Nikon) and a 552 nm laser (Coherent OBIS 552 LS) at an irradiance of 0.8 W/cm2 at the sample plane. Scattered light was imaged on a sCMOS camera (Andor Zyla) at 503 Hz. Actin filaments and fluorescently labeled myosins were imaged with TIRF excitation at 637 nm (Coherent OBIS 637 LX) and 552 nm, respectively, and fluorescence imaging videos were collected on an EMCCD camera (Andor iXon Ultra DU-897U). Blue illumination was provided by an LED (Thorlabs M470L2-C3) mounted above the objective. The blue LED was manually switched on to an irradiance of 17 mW/cm2. For each channel, first a movie of the actin was acquired in TIRF with 637 nm excitation. The TMR-labeled Nicotiana tetramers were imaged in TIRF at 552 nm excitation for directionality scoring of actin. The fluorescent tetramers were either included in the channel at the same time as the nanoparticles and imaged before imaging the nanoparticles, or else separately flowed in and imaged after the movies of the nanoparticles were collected. To acquire movies of the nanoparticles, the imaging was switched to dark field scattering at 552 nm excitation. Five to six movies of gold nanoparticles were acquired for each channel. The blue illumination was alternated between on and off for each subsequent movie within a channel, remaining either on or off for the duration of each movie.
Analysis of gold nanoparticle movies was performed as described7, with some modifications. Nanoparticles were tracked using 2D gaussian fitting with a custom routine written in C58, and the 2D trajectory was fit with a polynomial representing the path along an actin filament. The XY positions of the nanoparticle centers were projected onto the polynomial to obtain a 1D trajectory of distance along the actin filament. Each trajectory was examined manually, and trajectories were rejected from further analysis if they displayed tracking errors (such as missed frames or tracking of nearby diffusing particles) or if the amplitude of the high frequency noise was too large to distinguish state changes within the trajectory by eye. Steps were fit to the 1D trajectory using the Steppi changepoint detection algorithm44. The following settings were used for Steppi: global unknown κ, global unknown ɛ, local unknown μ, ⍺ = 0, protected state radius = 10 frames, penalty factor = 1. For step size distributions, maximum likelihood estimates of the parameters for a 4-component gaussian mixture model were obtained using the fitgmdist function in Matlab. Standard errors for the maximum likelihood estimates were calculated using bootstrapping with 10000 samples. Dwell time distributions were fitted to an exponential decay, omitting dwells shorter than 0.1 s which may be undercounted due to limited experimental resolution. The standard errors of estimated decay times were calculated using the mlecov function in Matlab.
Molecular constructs and cell lines for motor localization
ArrayG16X was synthesized from monomeric codon optimized ArrayG units as described in Ghosh et al31. Using a three-piece ligation strategy, MyLOVChar4~1R~TET and ArrayG16X were ligated in sequence into the multiple cloning site of a PiggyBac Cumate switch system (PBQM812A-1, SBI) plasmid. This generated a MyLOVChar4~1R~TET~ArrayG16X fusion plasmid, where the cumate repressor was expressed under a separate EF1 promoter as a CymR repressor-T2A-Puro cassette. To generate a binder FP plasmid, wildtype monomeric GFP (wtmGFP)31 and HaloTag were ligated in sequence into a modified PiggyBac Cumate switch plasmid where the cumate operator sequence (CuO) was replaced with a Tet-On 3G Tetracycline inducible promoter and the EF1-CymR repressor-T2A-Puromycin cassette was replaced with an EF1-Tet-Activator-P2A-Hygromycin cassette. MyLOVChar4~1R~TET~SNAP plasmid was generated by ligating MyLOVChar4~1R~TET and a SNAP-tag in sequence into the same modified PiggyBac Tet-On 3G plasmid, except that the EF1-CymR repressor-T2A-Puromycin cassette was replaced with an EF1-Tet-Activator-P2A-Blasticidin cassette. This was done so that if needed, all three plasmids could be stably integrated into the same cell line using three different antibiotic resistances as selection markers. MyLOVChar4~1R~TET~mRuby3 for expression in neurons was constructed using sequence coding for the fluorescent protein mRuby359 and cloned into the vector pcDNA3.1/Puro-CAG.
The transformed mouse embryonic fibroblast cell line BALB/3T3 was purchased from ATCC (ATCC® CCL-163™) and cultured in standard DMEM media supplemented with 10% fetal bovine serum and penicillin-streptomycin antibiotics.
For generating stable cell lines as well as for transient transfection, cells were transfected with appropriate amounts of plasmids using the Neon Transfection system (Thermo Fisher Scientific) and the following pulse program (Pulse voltage: 1350V, Pulse Width: 20ms, 2 pulses). To generate the MyLOVChar4~1R~TET~SNAP stable cell line, BALB/3T3 cells were transfected with PiggyBAC-Tet-MyLOVChar4~1R~TET~SNAP plasmid and a Super PiggyBac transposase plasmid at 2.5: 1 ratio. Three days after transfection, the growth medium was replaced with fresh growth medium supplemented with 10 μg/ml Blasticidin. Fresh growth medium with Blastidicin was added on alternate days throughout the selection process.
Molecular constructs and cell lines for controllable cargo localization
To design an optogenetic platform for measuring directional and reversible protein-cargo delivery, we generated a two-component recruitment system. The first component, MyLOVChar4~1R~TET~SNAP~DHFR, was generated by appending a dihydrofolate reductase (DFHR) coding sequence in frame to the end of SNAP-tag from the plasmid PBTH MyLOVChar4~1R~TET~SNAP. (PBTH: PiggyBac Tet-On 3G plasmid with an EF1-Tet-Activator-P2A-Hygromycin cassette). For the second component, either (1) the F3 subdomain of the FERM domain of mouse Talin protein, or (2) Integrin beta 3 was fused in frame to NB113 DHFR nanobody60 followed by a chemically inducible dimerizer sequence and eGFP, and the entire cassette was inserted at the MCS of a PBTB plasmid. Future applications might use the dimerizer to induce clustering using a chemical signal (TakaraBio A/C Heterodimerizer, Catalog # 635055, a Rapamycin analog), but this functionality was not tested or used here.
To generate dual stable cell lines, MCF10A mammary epithelial cell line (ATCC® CRL-10317™) was transfected with the appropriate plasmids at a 4 (Super PiggyBac transposase): 1 (component 1 plasmid): 1 (component 2 plasmid) ratio using a Neon electroporator (30ms pulse duration: 1100V: 2 pulse electroporation program). Three days after transfection, the growth medium was replaced with fresh growth medium (DMEM/F12 + 5% Horse serum supplemented with EGF (20ng/ml), Insulin (10μg/ml), Cholera toxin (100ng/ml), Hydrocortisone (0.5μg/ml) and Pen/Strep) supplemented with 10 μg/ml Blasticidin and 250ug/ml Hygromycin). Fresh growth medium with Blastidicin and Hygromycin was added on alternate days throughout the selection process.
Live cell bulk localization assays
For bulk motor imaging in fibroblasts the BALB/3T3 cell line expressing-MyLOVChar4~1R~TET~SNAP was induced with doxycycline overnight and then live stained with SNAP-Cell® 647-SiR (New England Biolabs) as per manufacturer’s instruction. For simultaneous two-color imaging of cargo and motor, MCF10A cell lines co-expressing MyLOVChar4~1R~TET~SNAP~DHFR and NB113~Integrin Beta 3~eGFP or NB113~FERM~eGFP were induced with doxycycline overnight and then live stained with SNAP-Cell® 647-SiR (New England Biolabs).
Cells were imaged with a Zeiss LSM 700 scanning confocal microscope. All SNAP-Cell® 647-SiR labeled cells were imaged with a 639 nm laser, 128–180 μm pinhole, pixel dwell time of 2.55 μs and a 63x plan-apochromat 1.40 NA objective.
For experiments with motors in fibroblasts, blue light stimulation was performed with either the blue 488 nm laser line of the confocal microscope at 1% of nominal power, or an LED light source (Thorlabs M470L3) with a center wavelength of 470 nm. The LED was driven by a direct current source (Thorlabs LEDD1B). For most dose response experiments the LED intensity was controlled by means of pulse width modulation at 500 Hz with output of an Arduino Uno board.
For simultaneous blue light stimulation of motors and imaging of cargo in MCF10A cells, we used the 488 nm laser line of the confocal microscope at 2% laser power. Data for the four phases of the stimulus assays (pre-illumination, illumination phase 1, post-illumination, and illumination phase 2) were collected in 4 separate movies. Three of the four phases were acquired with a typical frame rate of 14 s per frame, and a lower frame rate (60 s per frame) was used for the post-illumination phase to reduce photo-bleaching. Movie acquisition of an individual phase within a stimulus assay was started manually, approximately 14 s after the last frame of a previous acquisition. See Supplementary Table 2 for an overview of light stimulation conditions for the complete set of experiments.
Live cell bulk localization analysis
We quantified the accumulation of motors in puncta with the TrackMate plugin in ImageJ and subsequent analysis with custom written scripts in Python. Tracking and puncta quantification was performed on a maximum intensity projection of the confocal z-stack.
To detect puncta for the experiments with motors in fibroblasts, we used the Find Maxima detector (https://imagej.net/Find_maxima_(Trackmate_module), with a spot diameter of 1 micrometer and a noise tolerance of 100. Spots were tracked using the linear assignment problem tracker with a frame-to-frame linking distance of 2 microns, a maximum frame gap of 2 frames, and maximum track segment closing distance and track segment merging distance of 4 microns.
We quantified the population of motors in the cytoplasm by recording the mean intensity from 4–6 cells in each analyzed movie. For each cell, the cytoplasmic intensity was computed as the mean value in a rectangular region of interest. The size and position of each static ROI was manually defined in ImageJ and optimized to fully remain in the cytoplasmic region of the cell for the full duration of the analysis interval. For each selected cell, we also defined a region of interest in the nucleus of the cell to apply a background subtraction. Analysis was performed on a single plane chosen from the confocal z-stack. To prevent loss of intensity due to focal drift, we selected the plane of the z-stack with maximum average intensity within the set of ROIs for each frame of the movie. For the dataset presented in Fig. 3f, the selection plane was constant for the duration of the experiment.
We used custom Python scripts to tabulate the tracked spots at each time point in the movie and compute the average intensity of spots as a function of time, normalized to the total intensity contained in each image. For the dose response data, we extracted the saturated value and characteristic rise time from fitting a single exponential rise to the trace (Supplementary Fig. 8).
We analyzed accumulation of puncta in experiments on the MCF10a cells as for the experiments in fibroblasts described above, with a few modifications. We selected regions in the movies that primarily contained cells that clearly expressed both the motor and the cargo. Collections of puncta in both channels were extracted from these movie crops using a Laplacian of a Gaussian (LoG) spot detection algorithm, with a spot radius of 2 pixels, a noise tolerance of 1000 and selecting spots with a minimum contrast of 0.2. Spots were tracked using the linear assignment problem tracker with a frame-to-frame linking distance of 2 pixels, a maximum frame gap of 2 frames, and maximum track segment closing distance and track segment merging distance of 4 pixels. We filtered the set of found trajectories to preserve tracks of at least 4 frames in length, with a minimum track velocity of 1 pixel/frame; we found this empirical additional filtering useful in the epithelial cell movies to select for the puncta in protrusions, which are mobile cellular structures, excluding stuck spots. We used custom python scripts to tabulate the tracked spots at each time point in each phase of the stimulus assay and compute the average intensity, normalized to the total intensity contained in each image. We then combined the time traces for the individual phases into a single sequence, and further normalized the traces to the value in the first frame in the trace. The resulting trajectories for each acquisition sequence (Supplementary Fig. 10 – 11) include some variable time gaps between individually acquired movies for each of the four phases of the stimulus experiment. To average the plots and place them on a common time axis in Fig. 5c,e, we used the start times of each of the two blue light stimulus phases as common reference points, and resampled the data during the post-illumination phase at common timepoints by interpolation before computing averages and standard deviations.
Neuron culture imaging and light stimulation
All cell culture reagents were purchased from Thermo Fisher Scientific unless otherwise specified. Glass-bottom cell culture dishes were pre-coated with poly-D-lysine (10μg/mL, Sigma-Aldrich) overnight at 4oC and washed with PBS. Hippocampal neurons were dissected from embryonic day 18 rats and plated at a density of 30,000 per cm2 in the plating medium (Neurobasal medium supplemented with B27, 2mM GlutaMAX and 10% FBS). The entire medium was replaced with the growth medium (Neurobasal medium supplemented with B27, 2mM GlutaMAX and 1% FBS) on the following day, and half of the media was refreshed every 3–4 days. Neurons were transfected with MyLOVChar4~1R~TET~mRuby3 on 9DIV using Lipofectamine following the manufacturer’s instruction. Cultures were maintained at 37oC in 5% CO2 and 100% humidity during the entire culturing period. All animal procedures were approved by the Stanford Institutional Animal Care and Use Committee. Dynamics of MyLOVChar4~1R~TET~mRuby3 were imaged in 12DIV transfected neurons using an inverted wide-field microscope (Zeiss, Axiovert 200). Prior to the experiment, the cell culture media was replaced with the imaging solution (Hank’s Balanced Buffered Solution supplemented with 2mM GlutaMAX,10mM HEPES and 1mM D-glucose). The culture dish was equilibrated for 30min in a 37˚C heated chamber supplied with 5% CO2. Cells were excited with Xenon Arc lamp filtered at 568/20nm under a 20 × 0.75 NA objective lens, and images were detected with an emission filter at 620/60 nm. Time lapse images were acquired every 10s for 80 time points. Light stimulation (~10mW/cm2) was applied with a blue LED lamp (Thorlabs M470L3, 470 nm center wavelength) placed on top of the cell culture dish between time points 21–50 for each time lapse series.
Cell staining and imaging for single-molecule assays
For single-molecule imaging of MyLOVChar4~1R~TET~ArrayG16X, the BALB/3T3-Tet-MyLOVChar4~1R~TET~SNAP cell line was transiently transfected with the Cumate-inducible MyLOVChar4~1R~TET~ArrayG16X and Tet-inducible mwtGFP-Halo plasmids. The cells were then induced overnight with Doxycycline to ensure robust expression of MyLOVChar4~1R~TET~SNAP and the binder protein mwtGFP-Halo. Cumate was not added, as the leaky expression of MyLOVChar4~1R~TET~ArrayG16X under zero cumate induction was ideal for single-molecule imaging. Overexpression of MyLOVChar4~1R~TET~SNAP ensured that the MyLOVChar4~1R~TET~ArrayG16X were tetramers. Single molecules were imaged in HILO using a TIRFM setup on an Olympus CellTIRF system through a 1.49 NA 100x objective and 1.6x Optovar magnifier. For imaging MyLOVChar4~1R~TET~ArrayG16X / mwtGFP-Halo we used a BrightLine 482/18 excitation filter and a BrightLine 525/45 emission filter. Images were acquired at 10 Hz on an Andor iXon Plus EMCCD camera.
Live cell single-molecule analysis
We tracked single particles using the MATLAB software package μ-track, implementing tracking as a linear assignment problem algorithm32. For detection, we used an initial gaussian standard deviation of 2 pixels, rolling-window averaging of 3 frames, iterative estimation of gaussian standard deviation and iterative gaussian mixture modeling with residuals, distance and amplitude parameters set to 0.05. For tracking, we used a problem dimensionality of 2, maximum gap to close of 4 frames, minimum length of track from the first step of 1 frame, and the options for segment splitting and segment merging turned on. We used Brownian and directed motion models as the cost functions for frame-to-frame linking, gap closing, merging and splitting, and a Kalman filter function. For initial motion analysis in μ-track, we used a problem dimensionality of 2 and an alpha value of 0.1 to analyze asymmetric tracks.
Extracted tracks were then analyzed with custom written MATLAB scripts, to extract parameters of all tracks classified as directed motion, with a minimum length of 20 frames. Within a track, we designated mobile and stalled time points by simple thresholding. A histogram of velocities of the mobile phases was populated with the average velocities of all contiguous mobile sections of the tracks that were longer than 4 frames, recording the average velocity of that each section as one datapoint.
Replication and Reproducibility
All in vitro assay data were collected on two or more independent preparations of the assays. Additionally, for direct comparisons of properties between different molecular motor constructs (Figure 1f), data were collected using at least 2 independent protein preparations for each construct. All data in live cells were collected over at least two independent biological replicates, on separate days.
Data availability
Data for all results reported in this study are freely available as follows. The cryo-EM map and atomic model for the myosin XI-F-actin complex have been deposited in the EMDB (22808) and PDB (7KCH) respectively. Source data are available on the journal website as spreadsheet files linked to figures 1–5 and Extended Data figures 3–4. Source data for supplementary figures 3–11 are available from the Stanford Digital Repository (https://doi.org/10.25740/65j8-6114). All unique biological materials generated for this study, including plasmids for purification of engineered motors and for live cell experiments, are available from the authors on request.
Code availability
Custom code used in this study is freely available from the Stanford Digital Repository (https://doi.org/10.25740/65j8–6114).
Extended Data
Extended Data Fig. 1. Cryo-EM analysis of F-actin decorated with rigor state myosin XI.
a, Cryo-EM map of the rigor state of the Chara myosin XI catalytic domain CM11L2+4 CD746 bound to F-actin, reconstructed at 4.3 Å. b, The gold-standard Fourier shell correlation (FSC) curve for the 3D reconstruction. The average resolution is estimated at the FSC value of 0.143. c, Local resolution estimation of the 3D reconstruction. d, Cryo-EM map of two actin and one myosin subunits with the ribbon model of myosin fitted. The terminal helix of the myosin converter domain (residues 738–746) is highlighted in cornflower blue; this helix can be extended by a fused lever arm as indicated by the dashed line.
Extended Data Fig. 2. Structural models and predicted stroke vectors.
a-b,Structural models of MyLOVChar and MyLOVChar4, displaying representative pre-stoke (transparent) and post-stroke (solid) models from a set of 50 top-scoring structures modelled using RosettaRemodel (Methods and Supplementary Table 1), with annotated actin polarity. Post-stroke models are based on an actin-bound rigor stroke structure of Chara corallina myosin XI structure (Fig. 1b and Extended Data Fig. 1). Pre-stroke structure models built from myosin V pre-stroke crystal structure were aligned to the actin-bound myosin XI rigor. Idealized power stroke sizes and directions in the dark state (arrows below structures) and in the lit state (arrows above structures) were calculated by projecting the motion of the effective tip of the lever arm along the actin filament for the 50 top-scoring structures, and recording the mean. The effective tip of the lever arm was taken as the position of the C-terminal residues in the dark state, and as the position of the last rigid element on the LOV2 domain in the lite state (G516) c, Tabulation of calculated stroke sizes and optical stroke modulation for structural models of MyLOVChar-MyLOVChar5. Negative values indicate a stroke directed to the actin minus end. The stroke modulation was calculated as. A modulation larger than 1 indicates a directional reversal of the stroke.
Extended Data Fig. 3. Velocity traces of gliding filament assays on MyLOVChar4 at variable blue light intensity.
The traces are selected from the dataset used to compose Fig. 1h in the main text. Each trace displays the frame-to-frame velocity at the annotated blue light irradiance (box in the top right corner), averaged over all tracked polarity labelled filaments in 2–3 consecutive repeats of the stimulation sequence. The initial dark velocity was taken as the mean velocity of the time before the start of the blue light stimulus, indicated by the left grey dashed lines, and annotated for the bottom plot. The subsequent parts of the trace were globally fit to an exponential rise and decay, extracting the characteristic rates and , the steady-state lit velocity (black dash-dotted lines), and the final velocity (right grey dashed lines, annotated for bottom plot). In Fig. 1e and Fig. 1h the dark velocity (black dashed lines) was reported, taken as the mean of and.
Extended Data Fig. 4. Dose-dependent conformational changes in a minimal LOV2-containing lever arm construct.
a, Block diagram and idealized molecular cartoon of a minimal lever-arm construct with N-terminal SNAP-tag and C-terminal HaloTag fusions (approximate positions shown). HaloTag was labeled with TMR (donor) and SNAP-tag with SNAP-Cell 647-SiR (acceptor) for fluorescence measurements. b, Schematic of light-dose dependent population of conformational states of the construct. Transparent elements in the lit state indicate increased flexibility. Positions of the fluorescent dyes are annotated (D: donor, A: acceptor). c, Fluorescence emission spectra normalized to the peak donor emission. Inset: extracted acceptor emission, obtained by subtracting the normalized emission from a donor-only sample from the normalized emission of the FRET sample. d, Schematic of the light stimulus experiment in bulk in solution. e-f,Time traces of fluorescence intensity immediately after a removal of a blue light stimulus (irradiance: 10 mW/cm2), with continued excitation of the donor dye. e, Fluorescence intensity collected at the emission maximum of the acceptor (665 nm). f, Fluorescence intensity collected at the emission maximum of the donor (570 nm), recorded on an independent run of the stimulus sequence, on the same sample as d using the same blue light irradiance. g, Amplitude of the normalized intensity difference of acceptor channel after recovery from a light stimulus as function of irradiance of the blue light, showing two replicate datasets (circular and square datapoints) taken on different days. Insets: time-dependent traces for measurements at the annotated irradiances. Black lines in e-f and insets in g are single exponential decays with amplitude A and characteristic time t. Listed in pairs (A, t), parameter values are e: (−7 %, 18 s), f: (+3 %, 20 s), g (left): (−2 %, 20 s), g (center): (−5 %, 20 s), g (right): (−7%, 18 s) s). Black line in g is a saturation curve with A=7 % at saturation, with half-saturation occurring at a dose of 1.6 mW/cm2 . The average decay time <t> over all n=20 experiments was 19 ± 1 s (mean ± s.e.m.).
Supplementary Material
Acknowledgements
This work was supported by a Human Frontiers Science Program (HFSP) long-term fellowship to P.V.R., an NSF graduate research fellowship to S.Z., a W.M. Keck Foundation grant to Z.B. and M. Prakash, HFSP grant RPG0023/2014, NIH R01 GM114627 to Z.B., and Stanford Bio-X seed grants to Z.B. and M.Z.L. and to P.-S.H. and Z.B. Work performed at the Stanford Nano Shared Facilities (SNSF) was supported by the National Science Foundation under award ECCS-1542152. Cryo-EM work was conducted at the Simons Electron Microscopy Center (SEMC) and the National Resource for Automated Molecular Microscopy (NRAMM) located at the New York Structural Biology Center, supported by NIH grant P41 GM103310, NYSTAR, and Simons Foundation grant SF349247. We thank N. Denans, M. Barna, and Z. Zhang for useful discussions and for conducting and sharing results on early tests of engineered motors in live cells, T. Aksel and C. Espenel for advice on gliding filament analysis, P. Mangeol for providing a custom version of the KymographClear software to enable processing ROI sets, S. Sutton for providing purified F-actin, and members of the Bryant lab for discussions and assistance.
Footnotes
Competing interests
The authors declare they have no conflicts of interest relating to this work.
Methods-Only References
References
- 1.Furuta K. y. & Furuta A Re-engineering of protein motors to understand mechanisms biasing random motion and generating collective dynamics. Current Opinion in Biotechnology 51, 39–46 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Anson M, Geeves MA, Kurzawa SE & Manstein DJ Myosin motors with artificial lever arms. The EMBO Journal 15, 6069–6074 (1996). [PMC free article] [PubMed] [Google Scholar]
- 3.Can S, Lacey S, Gur M, Carter AP & Yildiz A. Directionality of dynein is controlled by the angle and length of its stalk. Nature 566, 407–410 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelke MF et al. Engineered kinesin motor proteins amenable to small-molecule inhibition. Nature Communications 7, 11159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner W, Brenowitz SD & Hammer Iii JA Myosin-Va transports the endoplasmic reticulum into the dendritic spines of Purkinje neurons. Nature Cell Biology 13, 40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Nakamura M, Schindler TD, Parker D. & Bryant Z. Engineering controllable bidirectional molecular motors based on myosin. Nature Nanotechnology 7, 252 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindler TD, Chen L, Lebel P, Nakamura M. & Bryant Z. Engineering myosins for long-range transport on actin filaments. Nat Nanotechnol 9, 33–38 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuta A. et al. Creating biomolecular motors based on dynein and actin-binding proteins. Nat Nanotechnol 12, 233–237 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Howard J. Molecular motors: structural adaptations to cellular functions. Nature 389, 561–567 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Nakamura M. et al. Remote control of myosin and kinesin motors using light-activated gearshifting. Nat Nanotechnol 9, 693–697 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Bergeijk P, Adrian M, Hoogenraad CC & Kapitein LC Optogenetic control of organelle transport and positioning. Nature 518, 111–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan L. et al. Optogenetic Control of Molecular Motors and Organelle Distributions in Cells. Chemistry & Biology 22, 671–682 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ballister ER, Ayloo S, Chenoweth DM, Lampson MA & Holzbaur ELF Optogenetic control of organelle transport using a photocaged chemical inducer of dimerization. Current Biology 25, R407–R408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French AR, Sosnick TR & Rock RS Investigations of human myosin VI targeting using optogenetically controlled cargo loading. Proc Natl Acad Sci U S A 114, E1607–E1616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omabegho T. et al. Controllable molecular motors engineered from myosin and RNA. Nature Nanotechnology 13, 34–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higashi-Fujime S. et al. The fastest-actin-based motor protein from the green algae, Chara, and its distinct mode of interaction with actin. FEBS Letters 375, 151–154 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Crosson S, Rajagopal S. & Moffat K. The LOV Domain Family: Photoresponsive Signaling Modules Coupled to Diverse Output Domains. Biochemistry 42, 2–10 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Uyeda TQ, Abramson PD & Spudich JA The neck region of the myosin motor domain acts as a lever arm to generate movement. Proceedings of the National Academy of Sciences 93, 4459 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsiavaliaris G, Fujita-Becker S. & Manstein DJ Molecular engineering of a backwards-moving myosin motor. Nature 427, 558–561 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Mooseker MS & Cheney RE Unconventional Myosins. Annual Review of Cell and Developmental Biology 11, 633–675 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Salomon M, Christie JM, Knieb E, Lempert U. & Briggs WR Photochemical and Mutational Analysis of the FMN-Binding Domains of the Plant Blue Light Receptor, Phototropin. Biochemistry 39, 9401–9410 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Schilling Z. et al. Predictive-focus illumination for reducing photodamage in live-cell microscopy. Journal of Microscopy 246, 160–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vale RD & Milligan RA The Way Things Move: Looking Under the Hood of Molecular Motor Proteins. Science 288, 88 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Taylor MP, Koyuncu OO & Enquist LW Subversion of the actin cytoskeleton during viral infection. Nature Reviews Microbiology 9, 427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W. et al. A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nature Biotechnology 35, 864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirokawa N, Niwa S. & Tanaka Y. Molecular Motors in Neurons: Transport Mechanisms and Roles in Brain Function, Development, and Disease. Neuron 68, 610–638 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Nan X, Sims PA, Chen P. & Xie XS Observation of Individual Microtubule Motor Steps in Living Cells with Endocytosed Quantum Dots. The Journal of Physical Chemistry B 109, 24220–24224 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Kerber ML et al. A Novel Form of Motility in Filopodia Revealed by Imaging Myosin-X at the Single-Molecule Level. Current Biology 19, 967–973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kural C. et al. Kinesin and Dynein Move a Peroxisome in Vivo: A Tug-of-War or Coordinated Movement? Science 308, 1469 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Tanenbaum Marvin E., Gilbert Luke A., Qi Lei S., Weissman Jonathan S. & Vale, Ronald D. A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh RP et al. A fluorogenic array for temporally unlimited single-molecule tracking. Nature Chemical Biology 15, 401–409 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Jaqaman K. et al. Robust single-particle tracking in live-cell time-lapse sequences. Nature Methods 5, 695 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartman MA & Spudich JA The myosin superfamily at a glance. Journal of Cell Science 125, 1627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bugaj LJ et al. Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nature Communications 6, 6898 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bird JE et al. Harnessing molecular motors for nanoscale pulldown in live cells. Molecular Biology of the Cell 28, 463–475 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harterink M. et al. Light-controlled intracellular transport in Caenorhabditis elegans. Current Biology 26, R153–R154 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Adrian M, Nijenhuis W, Hoogstraaten RI, Willems J. & Kapitein LC A Phytochrome-Derived Photoswitch for Intracellular Transport. ACS Synthetic Biology 6, 1248–1256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicolau DV et al. Parallel computation with molecular-motor-propelled agents in nanofabricated networks. Proceedings of the National Academy of Sciences 113, 2591–2596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaller V, Weber C, Semmrich C, Frey E. & Bausch AR Polar patterns of driven filaments. Nature 467, 73–77 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Ross TD et al. Controlling organization and forces in active matter through optically defined boundaries. Nature 572, 224–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt H, Zalyte R, Urnavicius L. & Carter A. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518, 435–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangeol P, Prevo B. & Peterman EJG KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Molecular Biology of the Cell 27, 1948–1957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito K, Yamaguchi Y, Yanase K, Ichikawa Y. & Yamamoto K. Unique charge distribution in surface loops confers high velocity on the fast motor protein Chara myosin. Proceedings of the National Academy of Sciences 106, 21585–21590, doi: 10.1073/pnas.0910787106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiggins Paul A. An Information-Based Approach to Change-Point Analysis with Applications to Biophysics and Cell Biology. Biophysical Journal 109, 346–354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harbury PB, Zhang T, Kim PS & Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262, 1401 (1993). [DOI] [PubMed] [Google Scholar]
- 46.Liao J-C, Elting MW, Delp SL, Spudich JA & Bryant Z. Engineered Myosin VI Motors Reveal Minimal Structural Determinants of Directionality and Processivity. Journal of Molecular Biology 392, 862–867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLean-Fletcher S. & Pollard TD Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochemical and Biophysical Research Communications 96, 18–27 (1980). [DOI] [PubMed] [Google Scholar]
- 48.Mei L. et al. Molecular mechanism for direct actin force-sensing by α-catenin. eLife 9, e62514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Leiro R. & Scheres SHW A pipeline approach to single-particle processing in RELION. Acta Crystallographica Section D 73, 496–502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallographica Section D 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang PS et al. RosettaRemodel : A Generalized Framework for Flexible Backbone Protein Design. Plos One 6, e24109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ropars V. et al. The myosin X motor is optimized for movement on actin bundles. Nature Communications 7, 12456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alexandre MTA, Arents JC, van Grondelle R, Hellingwerf KJ & Kennis JTM A Base-Catalyzed Mechanism for Dark State Recovery in the Avena sativa Phototropin-1 LOV2 Domain. Biochemistry 46, 3129–3137 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Aksel T, Choe Yu E, Sutton S, Ruppel KM & Spudich JA Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep 11, 910–920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tinevez J-Y et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 57.De La Cruz EM, Sweeney HL & Ostap EM ADP Inhibition of Myosin V ATPase Activity. Biophysical Journal 79, 1524–1529 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebel P, Basu A, Oberstrass FC, Tretter EM & Bryant Z. Gold rotor bead tracking for high-speed measurements of DNA twist, torque and extension. Nature Methods 11, 456–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bajar BT et al. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Scientific Reports 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oyen D, Wechselberger R, Srinivasan V, Steyaert J. & Barlow JN Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihydrofolate reductase of Escherichia coli. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1834, 2147–2157 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for all results reported in this study are freely available as follows. The cryo-EM map and atomic model for the myosin XI-F-actin complex have been deposited in the EMDB (22808) and PDB (7KCH) respectively. Source data are available on the journal website as spreadsheet files linked to figures 1–5 and Extended Data figures 3–4. Source data for supplementary figures 3–11 are available from the Stanford Digital Repository (https://doi.org/10.25740/65j8-6114). All unique biological materials generated for this study, including plasmids for purification of engineered motors and for live cell experiments, are available from the authors on request.
Code availability
Custom code used in this study is freely available from the Stanford Digital Repository (https://doi.org/10.25740/65j8–6114).