Abstract
Western blot analysis was used to assess the reactivity of convalescent-phase sera from patients who were associated with an outbreak of hemolytic-uremic syndrome (HUS) caused by fermented sausage contaminated with Shiga toxin-producing Escherichia coli (STEC). The predominant STEC isolated from HUS patients belonged to serotype O111:H−, and reactivity to O111:H− whole-cell lysates, treated or untreated with proteinase K, was examined. As expected, all five serum samples demonstrated a marked anti-lipopolysaccharide response, but several protein bands were also immunoreactive, particularly one with an apparent size of 94 kDa. One convalescent-phase serum sample was subsequently used to screen an O111:H− cosmid bank and 2 of 900 cosmid clones were found to be positive, both of which contained a similar DNA insert. Western blot analysis of one of these clones identified three major immunoreactive protein bands of approximately 94, 70, and 50 kDa. An immune response to the three proteins was detectable with all five convalescent-phase serum samples but not with healthy human serum. Immunoreactive 94- and 50-kDa species were produced by a deletion derivative of the cosmid containing a 7-kb STEC DNA insert. Sequence analysis of this region indicated that it is part of the locus for enterocyte effacement, including the eaeA gene which encodes intimin. The deduced amino acid sequence of the O111:H− intimin was 88.6% identical to intimin from O157:H7 STEC, and the most divergent region was the 200 residues at the carboxyl terminus, which were only 75% identical. Such variation may be antigenically significant as serum from a HUS patient infected only with the O111:H− STEC reacted with intimin from an enteropathogenic E. coli O111 strain, as well as several other eaeA-positive STEC isolates, but not with an eaeA-positive STEC belonging to serotype O157:H−. Sera from two of the other HUS patients also failed to react with intimin from this latter strain. However, intimin from O157:H− STEC did react with serum from a patient infected with both O111:H− and O157:H− STEC.
Shiga toxin-producing Escherichia coli (STEC) strains are increasingly being recognized as causes of diarrhea and hemorrhagic colitis in humans, and these infections can result in potentially life-threatening systemic sequelae such as hemolytic-uremic syndrome (HUS) (12, 21). STEC strains are commonly found in the intestines of livestock, and human infections are usually a consequence of consumption of contaminated meat or dairy products which have been improperly cooked or processed or uncooked vegetable products which have come into contact with manure (12). However, the serotype distribution of STEC strains present in the gastrointestinal tracts of domestic animals is very broad, and a limited range of serotypes (particularly O157, O111, and O26) are responsible for the majority of serious human infections (7, 12). Previous studies have compared properties of STEC from human and animal sources in order to identify traits which distinguish human pathogenic strains from those of lesser clinical significance. Capacity to produce attaching and effacing (A/E) lesions on intestinal mucosa (mediated by intimin, the eaeA gene product), production of a plasmid-encoded enterohemolysin, and production of Shiga toxin type 2 (Stx2) rather than Shiga toxin type 1 (Stx1) have all been associated preferentially with human STEC isolates or with more severe cases of infection (2, 4, 13, 22, 32). STEC isolates associated with HUS cases also have an enhanced capacity to adhere to intestinal epithelial cells in vitro compared with STEC isolates from cases of uncomplicated diarrhea or nonhuman sources (25).
We have recently described a large food-borne outbreak of STEC disease caused by contaminated fermented sausage (mettwurst) (24). There were a total of 21 cases of HUS, one of which was fatal, as well as one case of thrombotic thrombocytopenic purpura and over 100 reports of noncomplicated gastrointestinal disease. STEC strains (producing Stx1 and Stx2) belonging to serotype O111:H− were isolated from the patient with thrombotic thrombocytopenic purpura and from 16 of the patients with HUS (the remaining HUS patients had either PCR or serological evidence of O111 infection). One of the HUS patients was infected with O157:H− and O111:H− STEC strains, as was an additional patient with bloody diarrhea and microangiopathic hemolytic anemia. STEC isolates belonging to other serogroups (e.g., O26, O91, O123, and O128) were also isolated from some of the patients with diarrhea linked to the contaminated food source. The mettwurst itself yielded O111:H− STEC but also many other STEC isolates which were not found in any of the patients with diarrhea or HUS. Thus, only a subset of the STEC strains to which the community was exposed appeared capable of colonizing the human gut and causing disease, and among these, the O111:H− STEC appeared to have an enhanced capacity to cause serious complications such as HUS. Presumably, some product of the O111:H− STEC contributes to its apparently enhanced human virulence.
In the present study we performed Western immunoblot analysis using convalescent-phase sera from patients with HUS to determine whether patients respond to specific STEC proteins and whether such proteins are associated with more virulent STEC strains. Convalescent-phase serum was also used to isolate and characterize genes encoding immunoreactive proteins from a cosmid library of O111:H− DNA constructed in E. coli K-12.
MATERIALS AND METHODS
Bacterial strains and cloning vectors.
The STEC strains associated with the mettwurst outbreak, including 95NR1 (O111:H−) and 95SF2 (O157:H−), were isolated at the Women’s and Children’s Hospital (WCH), South Australia, Australia, as previously described (24). STEC strains from other HUS cases as well as the O111 enteropathogenic E. coli (EPEC) strain 87A were also isolated at WCH and have been described previously (25). The O157:H7 STEC strain EDL933 and its 60-MDa plasmid-cured derivative, EDL933-Cu, were provided by R. Robins-Browne. E. coli K-12 strain DH5α was obtained from Gibco-BRL, Gaithersburg, Md. The cosmid vector pPM2101, a derivative of pHC79 containing the RP4 mobilization region, has been described previously (27). The phagemid pBC SK, which encodes chloramphenicol resistance, was obtained from Stratagene, La Jolla, Calif. All E. coli strains were routinely grown in Luria-Bertani (LB) medium (19) with or without 1.5% Bacto-Agar (Difco Laboratories, Detroit, Mich.). Where appropriate, ampicillin or chloramphenicol was added to the growth medium at a concentration of 50 or 25 μg/ml, respectively.
Western blot analysis.
Crude lysates of STEC or other E. coli strains (with or without predigestion with proteinase K) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, as described by Laemmli (15), and antigens were electrophoretically transferred onto nitrocellulose filters, as described by Towbin et al. (29). Filters were probed with convalescent-phase sera from various HUS patients (kindly provided by K. F. Jureidini, Renal Unit, WCH) or with serum from a healthy donor (used at a dilution of 1:5,000), followed by goat anti-human immunoglobulin G conjugated to horseradish peroxidase. Immunoreactive bands were visualized with an enhanced chemiluminescence substrate system, as recommended by the supplier (Boehringer GmbH, Mannheim, Germany).
Construction of cosmid gene bank.
High-molecular-weight chromosomal DNA was extracted as described previously (23) from STEC 95NR1 and was digested partially with Sau3A1 so as to optimize the yield of fragments in the size range of 35 to 40 kb. This DNA was ligated with a fivefold molar excess of pPM2101 DNA, which had been digested with BamHI. Ligated DNA was packaged into lambda heads with a Packagene kit (Promega Biotec, Madison, Wis.) and transfected into E. coli DH5α, which had been grown in LB plus 2% maltose. The cells were then plated onto LB agar supplemented with ampicillin, and after incubation, clones were stored in LB plus ampicillin plus 15% glycerol in microtiter plates at −70°C.
Screening of cosmid clones by immunoblotting.
Cosmid clones were grown overnight at 37°C in 200 μl of LB plus ampicillin in microtiter plates and then were spotted onto a nitrocellulose filter. Filters were then blocked, reacted with serum from HUS patient 1 (diluted 1:5,000), and then developed as described above for Western blots.
DNA sequencing.
Nested deletions of STEC DNA cloned into pBC SK were constructed by the method of Henikoff (8) by using an Erase-a-base kit (Promega Biotec). This DNA was transformed into E. coli DH5α, and the resulting plasmid DNA was characterized by restriction analysis. Double-stranded template DNA for sequencing was prepared as recommended in the Applied Biosystems sequencing manual. The sequences of both strands were then determined with dye-labelled primers on an Applied Biosystems model 373A automated DNA sequencer. The sequence was analyzed using DNASIS and PROSIS Version 7.0 software (Hitachi Software Engineering, San Bruno, Calif.). Comparison with sequence databases was carried out with the program BLASTX (1).
Nucleotide sequence accession number.
The nucleotide sequence of the segment of 95NR1 DNA described in this study has been deposited with GenBank under accession no. AF025311.
RESULTS
Western blot analysis of STEC isolates using convalescent-phase sera from HUS patients.
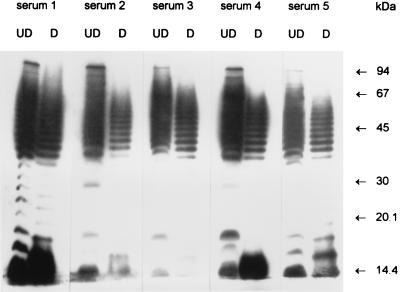
Convalescent-phase sera from five HUS patients (used at a dilution of 1:5,000) were used to probe Western blots of STEC 95NR1 (one of the O111:H− STEC isolates responsible for the outbreak), with or without predigestion of antigen preparations with proteinase K (Fig. 1). Each of the serum samples tested reacted with a ladder-like pattern of proteinase K-resistant bands typical of lipopolysaccharide (LPS), as previously reported (26). However, examination of the undigested tracks indicated that each of the serum samples also reacted with a proteinase K-sensitive species with an apparent size of 94 kDa. Smaller immunoreactive protein species were also present, although these were largely obscured by the heavily labelled LPS.
FIG. 1.
Western immunoblot analysis of convalescent-phase sera from HUS patients. Convalescent-phase sera from five HUS patients were reacted with electrophoresed undigested extracts of O111:H− STEC 95NR1 (UD) or with extracts which had been digested with proteinase K (D) as described in Materials and Methods. The positions of the protein size markers are indicated at right.
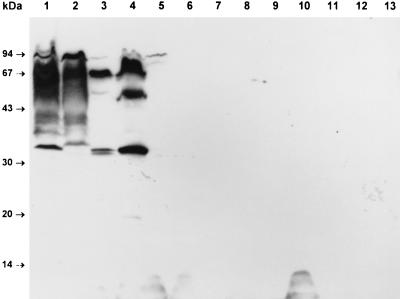
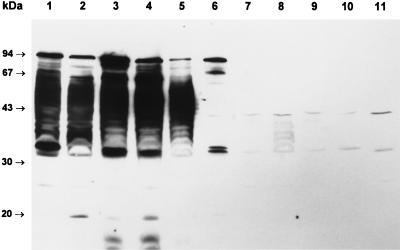
Serum from patient 1 was then used to probe a Western blot of a variety of STEC and reference E. coli strains (Fig. 2). This serum reacted with a number of species, including LPS and the 94-kDa protein, in 95NR1 as well as in O111 EPEC 87A. LPS reactivity was somewhat weaker than that seen for the same serum in Fig. 1; the reason for this is unknown but may be a consequence of batch to batch variation in the nitrocellulose membranes. O157:H7 STEC EDL933 contained several immunoreactive species with approximate sizes of 94, 70, 50, and 34 kDa, all of which were also present in a derivative (EDL933-Cu) which had been cured of its 60-MDa plasmid. The extract of STEC 95ZG1 (O26, eaeA positive) contained a 94-kDa immunoreactive species, but none of the remaining STEC isolates reacted with the serum. The nonreactive STEC were all eaeA negative, with the exception of 95SF2, an O157:H− isolate known to be eaeA positive (25). E. coli K-12 DH5α also did not contain any immunoreactive species.
FIG. 2.
Reactivity of HUS patient serum with various STEC and reference E. coli strains. Serum from HUS patient 1 was used to probe Western blots of undigested E. coli lysates as described in Materials and Methods. Lanes: 1, 95NR1 (O111:H−, eaeA positive); 2, EPEC 87A (O111, eaeA positive); 3, EDL933 (O157:H7, eaeA positive); 4, EDL933-Cu (O157:H7, eaeA positive, 60-MDa plasmid negative); 5, 95ZG1 (O26, eaeA positive); 6, 95SF2 (O157:H−, eaeA positive); 7, 94CR (O48:H21); 8, MW13 (O98); 9, MW10 (O113); 10, 95AS1 (O128); 11, 95PM2 (O123); 12, 95HE4 (O91); 13, E. coli DH5α. The positions of the protein size markers are indicated at left.
Cloning and characterization of genes encoding immunoreactive O111:H− STEC proteins.
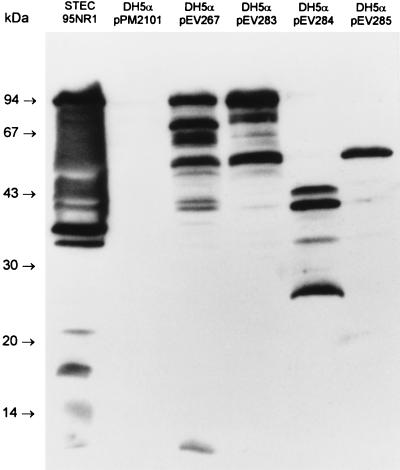
Serum from patient 1 was then used to screen an O111:H− STEC gene bank constructed in E. coli K-12 DH5α with cosmid pPM2101, as described in Materials and Methods. Two of 900 clones were found to be immunoreactive. Cosmids extracted from these clones appeared to contain similar DNA inserts (as judged by restriction analysis), and so one (designated pEV267) was selected for further study. Western blot analysis of E. coli DH5α[pEV267] showed that the recombinant cosmid directed the production of three major immunoreactive protein species with approximate sizes of 94, 70, and 50 kDa (Fig. 3). Similar labelling patterns were seen with all five convalescent-phase serum samples, but no proteins reacted with a similar dilution of normal human serum (result not shown). A deletion derivative of pEV267 was then constructed by digestion of the cosmid with HindIII, followed by religation. This derivative, designated pEV283, contained a 7-kb 95NR1 DNA insert, and E. coli DH5α[pEV283] produced 94- and 50-kDa immunoreactive species. Two further subclones (in pBC SK), designated pEV284 and pEV285, were also constructed, as shown in Fig. 4. The latter directed the production of only a 50-kDa immunoreactive species, while pEV284 directed the production of a series of smaller immunoreactive proteins (Fig. 3).
FIG. 3.
Reactivity of HUS patient serum with E. coli DH5α clones. Serum from HUS patient 1 was used to probe Western blots of 95NR1 and of E. coli DH5α carrying the cosmid vector (pPM2101), the immunoreactive cosmid pEV267, its deletion derivative pEV283, and two pBC SK subclones of pEV283 (pEV284 and pEV285; see Fig. 4 for map). The positions of the protein size markers are indicated at left.
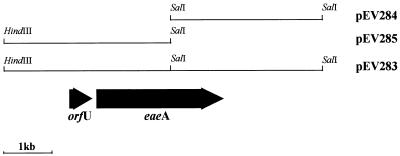
FIG. 4.
Locations of ORFs within the region of 95NR1 DNA cloned in pEV283 and in subclones pEV284 and pEV285. The cosmid deletion derivative pEV283 was constructed by digestion of pEV267 with HindIII followed by religation. This deleted approximately 30 kb of 95NR1 insert DNA 5′ to the HindIII site shown. Subclones pEV284 and pEV285 are in pBC SK.
The DNA inserts in pEV284 and pEV285 were then subjected to sequence analysis to identify genes encoding the immunoreactive proteins. The positions of open reading frames (ORFs) within the compiled 6,956-bp sequence are shown in Fig. 4. Examination of the DNA sequence suggested that the 50-kDa immunoreactive species encoded by pEV285 could be a truncated derivative of the 94-kDa species. Examination of the sequence at the junction of vector and insert DNA in pEV284 indicated that the larger of the immunoreactive species could be a fusion protein, as there is an in-frame TTG eight codons 5′ to the distal portion of the major ORF. The TTG codon is also preceded by a possible ribosome binding site 5 to 8 nucleotides upstream. Such a fusion protein would have a predicted size of 40.6 kDa. The smaller species are presumably degradation products of this polypeptide or may result from translational initiation at internal ATG codons.
Comparison with sequences deposited with GenBank indicated that the cloned 95NR1 DNA was part of the locus for enterocyte effacement (LEE), a chromosomal virulence island containing genes necessary for generation of A/E lesions on enterocytes (5). Moreover, the major ORF within pEV283 is a member of the intimin gene family (eaeA). At the deduced amino acid sequence level there is 88.6% identity between 95NR1 intimin and that reported for intimin from O157:H7 STEC (3, 33). There is a lesser degree of amino acid identity (81.3%) with intimin from enteropathogenic E. coli (EPEC) (11). The predicted size of 95NR1 intimin is 101.6 kDa (935 amino acids), and it is one residue longer than the homolog from O157:H7 STEC. The alignment of these two proteins is shown in Fig. 5, along with a previously reported sequence for a 254-amino-acid C-terminal portion of intimin from an O111:H8 STEC (18). The region of greatest divergence between the intimins from 95NR1 and O157:H7 STEC is the C-terminal 200 amino acids, which exhibited 75% identity. In contrast, there was 92.5% identity over the proximal 735 residues. As might be expected, the C-terminal fragment of O111:H8 intimin was more closely related to 95NR1 intimin, exhibiting 97.6% identity over the 254 amino acids for which sequence is available. Both these proteins have the additional residue N713 compared with O157:H7 intimin.
FIG. 5.
Alignment of the deduced amino acid sequence of intimin from O111:H− STEC 95NR1 with that previously published for intimin from an O157:H7 STEC isolate (33) and that of a partial sequence for intimin from an O111:H8 STEC isolate (18). Identical residues are represented by dots, while the dash indicates the absence of an amino acid.
The only other significant ORF in pEV283 is 156 amino acids long and is located 60 bp 5′ to eaeA (Fig. 4). Comparison with sequence databases indicated that it is 99.4 and 96.2% identical to the 156-amino-acid OrfUs of the LEE loci of EPEC and O157:H7 STEC, respectively (11, 34). However, none of the immunoreactive species found in lysates of E. coli DH5α carrying pEV283 or pEV285 corresponded to the expected size of OrfU.
Additional Western blot analysis.
Identification of the major protein species encoded by pEV283 which reacts with convalescent-phase serum from HUS patient 1 as intimin is largely consistent with the Western blot analysis of other STEC strains presented in Fig. 2. All the E. coli strains with a 94-kDa immunoreactive species (Fig. 2, lanes 1 to 5) are known to be eaeA positive (25). Conversely, known eaeA-negative STEC strains (lanes 7 to 12) and E. coli DH5α did not react with the patient serum. However, the negative immunoblot result for O157:H− STEC strain 95SF2 (lane 6) was unexpected, as this strain is eaeA positive (25). This finding could be explained either by lack of expression of eaeA in 95SF2 or by antigenic variation between intimins such that convalescent-phase serum from a patient infected with O111:H− STEC 95NR1 does not cross-react with intimin from the O157:H− strain. To examine this, we conducted additional Western blot analysis of lysates from various STEC strains by using convalescent-phase serum from HUS patient 2 (Fig. 6). This patient had a dual O111-O157 infection, as both an O111:H− STEC strain indistinguishable from 95NR1 and 95SF2 itself were isolated from feces during the acute phase of illness. The serum reacted with LPS from both O111 (Fig. 6, lanes 1 and 2) and O157 (lanes 3 to 5) strains. It also reacted with a 94-kDa species in all eaeA-positive lysates (lanes 1 to 6), including 95SF2. Thus, intimin is indeed produced by this O157:H− STEC strain. Additional Western blot analysis indicated that sera from patients 3 and 4 (from whom only O111 STEC strains were isolated) did not react with intimin from 95SF2 (result not shown). However, serum from patient 5 reacted weakly with a 94-kDa species in 95SF2 lysates (result not shown). Although this HUS patient showed a serological response to O111 LPS and was epidemiologically linked to the same outbreak, no STEC strains were isolated from feces. Thus, the possibility that patient 5 had a dual infection (as did patient 2) cannot be eliminated.
FIG. 6.
Additional Western blot analysis. Serum from HUS patient 2, who was infected with both O111:H− and O157:H− STEC, was used to probe Western blots of undigested E. coli lysates as described in Materials and Methods. Lanes: 1, 95NR1 (O111:H−, eaeA positive); 2, EPEC 87A (O111, eaeA positive); 3, EDL933 (O157:H7, eaeA positive); 4, EDL933-Cu (O157:H7, eaeA positive, 60-MDa plasmid negative); 5, 95SF2 (O157:H−, eaeA positive); 6, 95ZG1 (O26, eaeA positive); 7, MW13 (O98); 8, 94CR (O48:H21); 9, MW10 (O113); 10, 95PM2(O123); 11, 95AS1 (O128). The positions of the protein size markers are indicated at left.
DISCUSSION
It has been known for more than a decade that certain strains of STEC are capable of causing A/E lesions on enterocytes (6, 28). These lesions are characterized by ultrastructural changes including loss of enterocyte microvilli, and there is intimate attachment of the bacterium to the cell surface. Beneath the adherent bacteria, there is accumulation of cytoskeletal components, resulting in the formation of pedestals, and this is recognizable by electron microscopy and by fluorescence microscopy after staining with phalloidin-fluorescein isothiocyanate (14). Capacity to produce A/E lesions was initially recognized in EPEC strains, and recent studies have elucidated the molecular events involved in their generation (reviewed by Donnenberg et al. [5]). All of the genes necessary for generation of A/E lesions in EPEC are located on a 35.5-kb chromosomal virulence island termed LEE. LEE contains the eaeA gene, which encodes intimin, an outer membrane protein which mediates intimate attachment to the enterocyte. LEE also includes a cluster of genes which encode a type III secretion system responsible for export of other LEE-encoded proteins, including EspA, EspB, and EspD, which are necessary for initiation of signal transduction events involved in the generation of enterocyte cytoskeletal rearrangements (5, 16). The mechanism whereby STEC strains generate A/E lesions is less well characterized but is essentially analogous to that for EPEC. STEC strains displaying the A/E phenotype have a LEE homolog (20), which although not yet fully characterized, encodes intimin as well as various secreted proteins (including an EspB homolog) and a type III secretion system (3, 9, 33).
There is no doubt that there is a strong link between carriage of eaeA and STEC strains associated with severe human disease such as hemorrhagic colitis and HUS (2, 4, 18, 32). In the present study we have shown that convalescent-phase sera from five HUS patients associated with an outbreak of O111:H− STEC infection recognize a protein in STEC lysates that comigrates with intimin (EaeA). A serological response to EPEC intimin has previously been reported for patients with EPEC diarrhea (10, 17). We have also shown that the only clones among a large O111:H− STEC cosmid gene bank that reacted with HUS patient serum contained part of the LEE locus. The 7-kb subclone pEV283 was strongly immunoreactive and contained a copy of eaeA. There were no other ORFs within pEV283 capable of encoding proteins with sizes similar to those of the other immunoreactive species, and so these are presumed to be degradation products of intimin. No immunoreactive species of the expected size of OrfU was observed. OrfU is thought to be a cytoplasmic chaperone protein (5) and so may not be exposed to the immune system during infection, or alternatively, it may be poorly immunogenic.
Assessment of the contribution of intimin to STEC pathogenesis, however, is complicated by sequence heterogeneity, particularly in the C-terminal portion of the protein, which is thought to be involved in binding to the epithelial cell (30). EPEC intimin and the homolog from O157:H7 STEC exhibit a high degree of amino acid identity for the first 700 or so amino acids, but the C-terminal portions (about 25% of the total length) are quite divergent, displaying only about 50% homology (3, 33). This may account for marked differences in tissue tropism observed in gnotobiotic piglets challenged with O157:H7 STEC eaeA-negative mutants complemented with either EPEC or O157:H7 eaeA (30). Heterogeneity also occurs within STEC strains, as shown in the present study; the deduced amino acid sequence of intimin from 95NR1 is only 88.6% identical to that of O157:H7 intimin. Again, the most divergent region is the C-terminal portion, the last 200 residues of which exhibit only 75% identity. These findings are consistent with an earlier report which showed a similar divergence between the last 254 amino acids of intimins from O157:H7 and O111:H8 STEC (18). This partial O111:H8 intimin sequence exhibits 97.6% identity to the respective portion of the 95NR1 protein. Given the apparent involvement of the C terminus in receptor binding, it is possible that differences in intimin sequence may be functionally significant. This might account for the observation of Wieler et al. (31) who found that only 65% of eaeA probe-positive bovine STEC isolates were capable of forming A/E lesions, as judged by fluorescent actin staining of infected HEp-2 cells.
The heterogeneity of intimin contrasts markedly with the high degree of conservation of the gene (orfU) which is located immediately 5′ to eaeA. The amino acid identities between the OrfU from 95NR1 and homologs from O157:H7 STEC and EPEC strains were 96.2 and 99.4%, respectively. This implies that selective pressure in favor of heterogeneity is stronger for intimin than for OrfU. Unlike OrfU, intimin is exposed on the bacterial cell surface, and systemic or local immune responses to this protein may be capable of blocking adherence of the STEC to the intestinal epithelium. Thus, antigenic variation, particularly in the exposed intimin domains, would be a significant advantage to the bacterium, assuming such variation does not compromise receptor interactions. The present study provides the first direct evidence for antigenic diversity of intimin within STEC strains as the eaeA-positive O157:H− STEC 95SF2 failed to react with sera from three of five patients who were infected with O111:H− STEC. The fact that a 94-kDa species in the O157:H7 STEC strain (EDL933) reacted strongly with one of these serum samples is also intriguing. The difference in reactivity between 95SF2 and EDL933 is not related to expression of eaeA, as 95SF2 did react with serum from patient 2 who had a dual O111:H−-O157:H− infection. This suggests that immune responses to intimin may even be specific to strains within an O serogroup, a possibility which may compromise the efficacy of intimin as a STEC vaccine antigen.
ACKNOWLEDGMENTS
We are grateful to K. F. Jureidini for providing the convalescent-phase sera from HUS patients used in this study. We also thank Monica Ogierman for assistance with Western blot analyses.
This work was supported by a grant from the National Health and Medical Research Council of Australia. A.W.P. holds an NHMRC Australian Postdoctoral Fellowship.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barrett T J, Kaper J B, Jerse A E, Wachsmuth I K. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165:979–980. doi: 10.1093/infdis/165.5.979. [DOI] [PubMed] [Google Scholar]
- 3.Beebakhee G, Louie M, De Azavedo J, Brunton J. Cloning and nucleotide sequence of the eae gene homologue from enterohemorrhagic Escherichia coli serotype O157:H7. FEMS Microbiol Lett. 1992;91:63–68. doi: 10.1016/0378-1097(92)90563-4. [DOI] [PubMed] [Google Scholar]
- 4.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 6.Francis D H, Collins J E, Duimstra J R. Infection of gnotobiotic pigs with an Escherichia coli O157:H7 strain associated with an outbreak of hemorrhagic colitis. Infect Immun. 1986;51:953–956. doi: 10.1128/iai.51.3.953-956.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R I, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–761. [Google Scholar]
- 8.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 9.Jarvis K G, Kaper J B. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun. 1996;64:4826–4829. doi: 10.1128/iai.64.11.4826-4829.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jerse A E, Kaper J B. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton protein, the expression of which is influenced by the EAF plasmid. Infect Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karmali M A. Infection by verotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleanthous H, Smith H R, Scotland S M, Gross R J, Rowe B, Taylor C M, Milford D V. Haemolytic uraemic syndromes in the British Isles, 1985-8: association with Verocytotoxin producing Escherichia coli. Part 2. Microbiological aspects. Arch Dis Child. 1990;65:722–727. doi: 10.1136/adc.65.7.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knutton S, Baldwin T, Williams P H, McNeisch A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lai L-C, Wainwright L A, Stone K D, Donnenberg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine M M, Nataro J P, Karch H, Baldini M M, Kaper J B, Black R E, Clements M L, O’Brien A D. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding the enteroadhesiveness factor. J Infect Dis. 1985;152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 18.Louie M, De-Azavedo J, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 20.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien A D, Holmes R K. Shiga and shiga-like toxins. Microbiol Rev. 1987;51:206–220. doi: 10.1128/mr.51.2.206-220.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargrett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–999. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 23.Paton A W, Paton J C, Heuzenroeder M W, Goldwater P N, Manning P A. Cloning and nucleotide sequence of a variant Shiga-like toxin II gene from Escherichia coli OX3:H21 isolated from a case of Sudden Infant Death Syndrome. Microb Pathog. 1992;13:225–236. doi: 10.1016/0882-4010(92)90023-h. [DOI] [PubMed] [Google Scholar]
- 24.Paton A W, Ratcliff R, Doyle R M, Seymour-Murray J, Davos D, Lanser J A, Paton J C. Molecular microbiological investigation of an outbreak of hemolytic uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J Clin Microbiol. 1996;34:1622–1627. doi: 10.1128/jcm.34.7.1622-1627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paton A W, Voss E, Manning P A, Paton J C. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun. 1997;65:3799–3805. doi: 10.1128/iai.65.9.3799-3805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton, A. W., E. Voss, P. A. Manning, and J. C. Paton. Antibodies to lipopolysaccharide block adherence of Shiga toxin-producing Escherichia coli to human intestinal epithelial (Henle 407) cells. Microbial Pathog., in press. [DOI] [PubMed]
- 27.Sharma D P, Stroeher U H, Thomas C J, Manning P A, Attridge S R. The toxin-coregulated pilus (TCP) of Vibrio cholerae: molecular cloning of genes involved in pilus biosynthesis and evaluation of TCP as a protective antigen in the infant mouse model. Microb Pathog. 1989;7:437–448. doi: 10.1016/0882-4010(89)90024-7. [DOI] [PubMed] [Google Scholar]
- 28.Sherman P, Soni R, Karmali M. Attaching and effacing adherence of Vero cytotoxin-producing Escherichia coli to rabbit intestinal epithelium in vivo. Infect Immun. 1988;56:756–761. doi: 10.1128/iai.56.4.756-761.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzipori S, Gunzer F, Donnenberg M S, de-Montigny L, Kaper J B, Donohue-Rolfe A. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect Immun. 1995;63:3621–3627. doi: 10.1128/iai.63.9.3621-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wieler L H, Vieler E, Erpenstein C, Schlapp T, Steinruck H, Bauerfeind R, Byomi A, Baljer G. Shiga toxin-producing Escherichia coli strains from bovines: association of adhesion with carriage of eae and other genes. J Clin Microbiol. 1996;34:2980–2984. doi: 10.1128/jcm.34.12.2980-2984.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willshaw G A, Scotland S M, Smith H R, Cheasty T, Thomas A, Rowe B. Hybridization of strains of Escherichia coli O157 with probes derived from the eaeA gene of enteropathogenic E. coli and the eaeA homolog from a Vero cytotoxin-producing strain of E. coli O157. J Clin Microbiol. 1994;32:897–902. doi: 10.1128/jcm.32.4.897-902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Kaper J B. Cloning and characterization of the eae gene of enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1992;6:411–417. doi: 10.1111/j.1365-2958.1992.tb01484.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao S, Mitchell S E, Meng J, Doyle M P, Kresovich S. Cloning and nucleotide sequence of a gene upstream of the eaeA gene of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett. 1995;133:35–39. doi: 10.1111/j.1574-6968.1995.tb07856.x. [DOI] [PubMed] [Google Scholar]