Abstract
Adeno-associated viruses (AAVs) hold tremendous promise as delivery vectors for gene therapies. AAVs have been successfully engineered—for instance, for more efficient and/or cell-specific delivery to numerous tissues—by creating large, diverse starting libraries and selecting for desired properties. However, these starting libraries often contain a high proportion of variants unable to assemble or package their genomes, a prerequisite for any gene delivery goal. Here, we present and showcase a machine learning (ML) method for designing AAV peptide insertion libraries that achieve fivefold higher packaging fitness than the standard NNK library with negligible reduction in diversity. To demonstrate our ML-designed library’s utility for downstream engineering goals, we show that it yields approximately 10-fold more successful variants than the NNK library after selection for infection of human brain tissue, leading to a promising glial-specific variant. Moreover, our design approach can be applied to other types of libraries for AAV and beyond.
Computational, data-driven redesign of a state-of-the-art AAV initial library improves downstream selection for therapeutic uses.
INTRODUCTION
Adeno-associated viruses (AAVs) hold major promise as delivery vectors for gene therapy. While naturally occurring AAVs can be clinically administered safely and in some cases efficaciously, they have a number of shortcomings that limit their use in many human therapeutic applications. For example, naturally occurring AAVs do not target delivery to specific organs or cells, their delivery efficiency is limited, and they are susceptible to preexisting neutralizing antibodies (1–3). Consequently, directed evolution of the AAV capsid protein has emerged as a powerful strategy for engineering therapeutically suitable or optimal AAV variants. In directed evolution, a diversified library of AAV capsid sequences is subjected to multiple rounds of selection for a specific property of interest, with the aim of identifying and enriching the most effective variants (1, 4). Primary techniques for constructing AAV starting libraries include error-prone polymerase chain reaction (PCR) (1, 5), DNA shuffling (6, 7), structurally guided recombination (8), peptide insertions (9), and phylogenetic reconstruction (10). Recent studies have also explored computational strategies for setting the parameters that control the construction of these libraries. For example, genomic junctions that minimize AAV structure disruptions, suitable for recombination libraries, were computationally identified (9). For mutagenesis libraries, genomic locations and their mutation probabilities were identified using single-substitution variant data or by way of ancestral imputation from phylogenetic analysis (10, 11).
Although successes have been achieved with directed evolution (4, 5, 8, 9, 12), several challenges are slowing progress (13). For instance, a substantial fraction of the variants in the starting libraries for these selections are unable to assemble properly or package their payload efficiently—a basic requirement for any functional selection (11, 14, 15). Consequently, much of the library is wasted, thereby decreasing the chance of successfully achieving any desired engineering goal in the downstream selections. Next-generation sequencing (NGS) technologies enable analysis of properties for individual variants within a library, such as packaging fitness and infectivity, and the large quantity of data resulting from such assays suggests that machine learning (ML) could be a useful tool to help design more effective starting libraries for directed evolution. Here, we propose a method to design such an ML-guided library that balances the requirements of packaging and diversity, to improve the probability of success in any general AAV directed evolution goal.
Recent studies have applied ML models trained on experimental data to generate previously unidentified AAV variants (16, 17); however, these studies examined diversity post hoc and provided no way to systematically navigate an optimal trade-off between diversity and packaging. In earlier work, Parker et al. (18) balanced “quality” and “novelty” in their library design. Quality was estimated from a statistical model evaluated on each sequence [specifically, a Potts model (19) trained on a fixed set of natural sequences], whereas novelty captured how different the library sequences were from naturally occurring sequences but provided no indication of diversity within the library. Extensions of this work considered multiple fitness, or quality, scores (20). In contrast to these, our approach will (i) allow for the use of any predictive model of fitness, (ii) explicitly address and control the diversity within the designed library, and (iii) be broadly applicable to different kinds of library construction.
We instantiated and evaluated our library design approach by designing a 7-mer peptide insertion library for AAV serotype 5 (AAV5) to optimally balance diversity and overall packaging fitness. Among the natural AAV serotypes, AAV5 has been suggested as a promising candidate for clinical gene delivery because of the low prevalence of preexisting neutralizing antibodies and successful clinical development for hemophilia B (21–24). We focus, specifically, on peptide insertion libraries because they are both simple and highly practical, having already been translated to the clinic (e.g., NCT03748784, NCT04645212, NCT04483440, NCT04517149, NCT04519749, NCT03326336, and NCT05197270) (25).
Briefly, our approach is as follows: First, we assess the packaging fitness of variants in an NNK 7-mer insertion library, a standard library type that is used as a starting point for experimental selections of insertions to AAV capsids. We then use these estimated packaging efficiencies as labels to build a predictive model from peptide insertion sequence to packaging fitness. Last, we develop a design approach that can systematically trade off library diversity with packaging fitness, enabling us to choose an optimal trade-off. Our approach to ML-guided library design biases library construction toward variants that package well, thereby reducing the amount of wasted sequences and space in screening tasks. We show that our design approach yields a library with fivefold higher packaging fitness than the NNK library, with negligible sacrifice to diversity, suggesting that our library will be more generally useful. As further evidence, when we subjected the NNK library to one round of packaging selection, the resulting pool of variants still had a lower packaging fitness than that of our initially designed library while also being substantially less diverse. Last, to demonstrate the general downstream utility of our designed library on an engineering task for which it was not designed, we showed in a primary human brain tissue selection that the ML-guided library yielded a 10-fold higher number of infectious variants compared to the NNK library, and these variants can be further selected for efficient and cell-specific infectivity. This ML-guided AAV capsid library design is thus highly useful for selection in human tissue. While we focus on a therapeutically relevant capsid 7-mer peptide insertion library, our methods are general and can be applied to other AAV library types and to proteins beyond AAV.
RESULTS
AAV5–7-mer peptide insertion library preparation and packaging selection
We used libraries with a variable seven–amino acid (7-mer) NNK sequence inserted at position 575–577 in the viral protein monomer, within a loop at the threefold symmetry axis associated with receptor binding and cell-specific entry (26, 27). The “NNK” moniker refers to a broadly used strategy (28–30) involving a uniform distribution over all four nucleotides (N) in the first two positions of a codon, and equal probability on nucleotides G and T (K) in the third position, where the K in the third position was chosen to reduce the chance of stop codons that typically render the protein nonfunctional. Each of the seven amino acids in the insertion is sampled at random from this distribution during library construction. Although NNK libraries are among the most promising AAV libraries (2), a substantial fraction (>50%) of the variants in these libraries fail to package (i.e., do not assemble into viable capsids), and many more have lower packaging fitness than the parental virus (14, 15). For example, placing a large hydrophobic residue in the 7-mer (solvent-exposed) region is likely destabilizing. Much of the experimental library is thus effectively wasted on poor fitness variants.
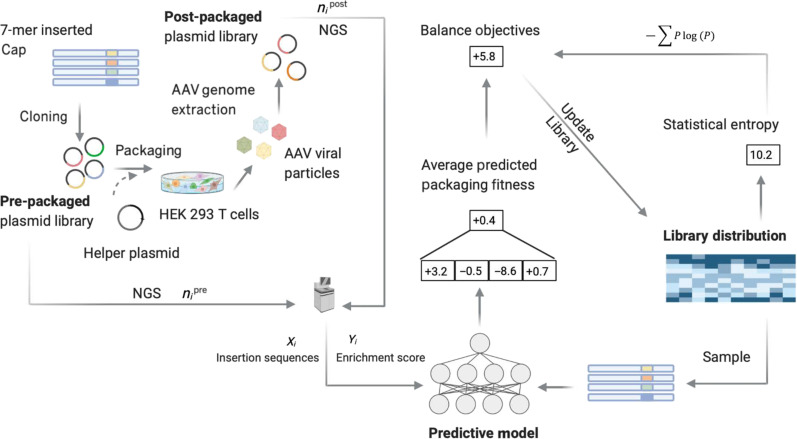
Our goal was to improve upon the commonly used NNK library and implicitly uncover a broad set of rules, as yet unknown, for insertion sequences that confer higher packaging fitness and then encode them in our library design so as to avoid such problems. In particular, our design approach will specify probabilities for each nucleotide in each position of the codon, at each position in the 7-mer, in a manner that achieves better overall packaging than NNK while maintaining high diversity. For example, we might specify for the first codon that the first nucleotide in the codon should be chosen with 20% chance as an A, 40% chance as a C, and 35% chance as a T and 5% G and then specify four other such probabilities for the other two positions in the codon, for a total of 12 specified values. A designed library will specify these 84 (=7 × 12) probabilities, which, in turn, will dictate the mean packaging fitness—through a complicated relationship that will be approximated with our ML predictive model—and library sequence diversity. We refer to designed libraries specified in this way as position-wise nucleotide specified. First, we experimentally synthesized roughly 107 variants from the NNK library to yield the NNK pre-packaged library. This plasmid library was then packaged, and the resulting viral particles were harvested and purified, and their genomes were extracted, yielding the NNK post-packaged library (Fig. 1) (31). The sequences from both pre- and post-packaged libraries were then PCR-amplified and deep-sequenced (Materials and Methods).
Fig. 1. Experimental workflow for generating pre- and post-packaged AAV5–7-mer library data for ML-based library design.
Ni, number of reads for each unique insertion sequence i. Experimental data were used to build a supervised regression model where the target variable reflects the packaging success of each insertion sequence. The predictive model was then systematically inverted to design libraries that trace out an optimal trade-off curve between diversity and packaging fitness. Schematic illustration created with BioRender.com.
These experiments yielded 49,619,716 pre-packaged and 55,135,155 post-packaged sequencing reads, which collectively yielded read counts for 8,552,729 unique peptide sequences. For each unique sequence, we used the pre and post read counts to calculate a log enrichment score (Materials and Methods) (11, 16, 32, 33), a measure of its packaging fitness. Note, however, that a variant that appeared in 10 pre- and 100 post-packaged sequencing reads would have the same log enrichment score as one that appeared in 1 and 10 sequencing reads, although the former has more data to support its value (i.e., more stably statistically estimated). Consequently, we derived a procedure to take this into account in a statistically principled manner when estimating our regression model parameters. Our procedure assigns a weight to each unique sequence that is higher when the statistical estimate is more stable, and higher weighted sequences have more influence on the regression model (Materials and Methods). In the previous example, the variant with a read count ratio of 10:1 would get a smaller weight than the one with a ratio 100:10, as the former provides weaker evidence of enrichment.
Training and evaluation of predictive models
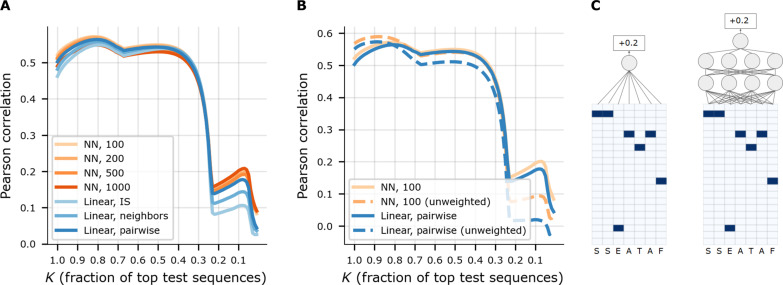
To find the best model type to use for our ML-guided library design, we compared seven classes of ML regression models: three linear models and four feed-forward neural networks (NNs) (Fig. 2). Each model was trained using the log enrichment scores as the target variable and the sequence-specific weights described above (Materials and Methods). The three linear models differed in the set of input features used. One used the “independent site” (IS) representation wherein individual amino acids in each 7-mer insertion sequence were one-hot–encoded. Another used a “neighbors” representation composed of IS features and, additionally, pairwise interactions between all positions that are directly adjacent in the amino acid sequence. The third used a “pairwise” representation composed of the IS features and, additionally, all pairwise interactions among all positions in the sequence. All NN models used the IS features alone, as these models have the capacity to construct higher-order interaction features from the IS features. Each NN architecture comprised exactly two densely connected hidden layers with tanh activation functions. The four NN models differed in the size of the hidden layers, with each using either 100, 200, 500, or 1000 nodes in both hidden layers.
Fig. 2. Predictive model architectures and results.
(A) Comparison of models for predicting AAV5–7-mer packaging log enrichment scores, using Pearson correlation, and “top-K” Pearson correlation, where K denotes what fraction of top-ranked observed log enrichment test sequences were used. The correlation is between predicted and true log enrichment scores. Seven different models including four neural network (NN) architectures, distinguished by the number of nodes in the hidden layers (100, 200, 500, and 1000). (B) Similar plot to (A) except comparing the use of weighted versus unweighted sequences during training, for the final selected model, NN, 100, and a baseline, linear, pairwise. (C) Schematic illustrations of “linear, IS” (left) and “NN, 4” (right) predictive models. Each model predicts packaging log enrichment from peptide insertion sequence.
We compared the performance of these seven models using the standard (unweighted) Pearson correlation between model predictions and true log enrichment scores on a held-out test set (training with weighted samples as described earlier). We randomly split the data into a training set containing 80% of the data points and a test set containing the remaining 20% of the points. Because our ultimate aim was to design a library of sequences that package well, we also studied how the models’ predictive accuracy changed when restricted to sequences in the test set with observed high packaging log enrichment. Specifically, we computed the Pearson correlation on subsets of the test set restricted to the fraction K of sequences with the highest observed log enrichment. By varying K, we traced out a performance curve where for lower K, the evaluation is more focused on accurate prediction of higher log enrichment scores rather than lower ones (Fig. 2A and fig. S1). Overall, we found that the NN models performed better than the linear models, presumably owing to their capacity to construct more complex functions, particularly to capture higher-order epistatic interactions in the fitness function. We selected “NN, 100” as our final model, as it performed similarly to the overall best-performing model, “NN, 1000,” but with many fewer parameters. The comparison between observed log enrichment scores and scores predicted by the (NN, 100) model for all sequences in the test set is shown in fig. S2.
Next, we assessed the effect of training with our sequence-specific weights by retraining two of the models—the final model, (NN, 100) model, and the “linear, pairwise” model—this time with all weights set to 1.0 (i.e., unweighted), again using Pearson correlation to evaluate (Fig. 2B). Training in this unweighted manner, rather than weighted, resulted in a performance benefit for K near 1.0 but degraded the performance near K < 0.25, a regime of particular interest because it focuses on variants with high log enrichment, and we ultimately aim to design a library that packages well (i.e., with high enrichment). These results further supported our choice of the weight trained (NN, 100) model with which to do library design.
Experimental validation of the predictive models
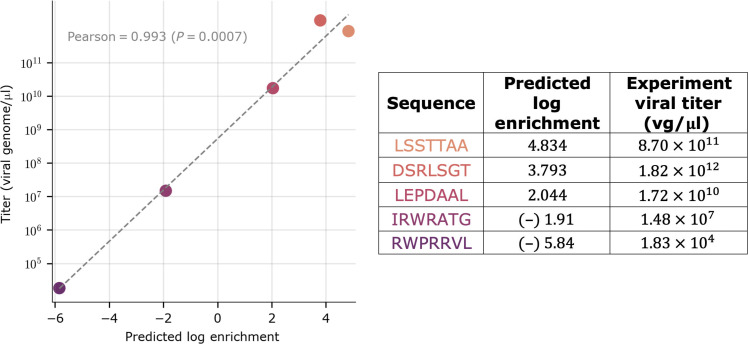
Before proceeding to using our predictive model for library design, we first validated the (NN, 100) model by identifying and synthesizing five individual 7-mer insertion sequences that were not present in our original experiment dataset. These five sequences were chosen to span a broad range of predicted log enrichment scores (−5.84 to 4.83; see Fig. 3 for correspondence with viral titers). The five variants were packaged individually into viral particles, harvested, and titered by quantifying the resulting number of genome-containing particles using digital-droplet PCR (ddPCR; Materials and Methods). High titer values indicated the variant was capable of packaging its genome properly in the assembled capsid. To differentiate whether our capsid variants failed to assemble in the first place or whether there is a defect with loading the genome following capsid assembly, we applied single-ion charge detection mass spectrometry (CDMS; Materials and Methods) and showed that our selected capsid variants exhibited similar levels of full-to-empty capsids in the packaged pools (fig. S3). The agreement between model predictions and corresponding experimental measurement of viral titers [1.83 × 104 to 8.70 × 1011 viral genomes (vg)/μl] (Fig. 3) demonstrates that the predictive model was sufficiently accurate to be used for library design. The accuracy of model predictions reported in Fig. 3 is higher than that reported in Fig. 2. This can be largely attributed to the choice of five sequences that spanned a large range of predicted log enrichment scores to produce the results of Fig. 3 (fig. S4).
Fig. 3. Experimental titers versus predicted log enrichment scores.
The five variants were selected to span a broad range of predicted log enrichment scores. Log enrichment scales are computed using natural logarithm. Experimental titers are measured on three biological replicates.
Model-guided library design
Having validated our final model for use with our library design task, we next aimed to design a library that packages better than the NNK library while maintaining good diversity. Inherent in this challenge is a trade-off between library diversity and mean predicted packaging fitness of the library. For example, note that mean predicted packaging fitness is maximized with a library that contains only a single variant with the highest predicted fitness, while diversity is maximized with a library uniformly distributed across sequence space, irrespective of packaging fitness. The library that is most effective for downstream selections will lie between these two extremes, balancing mean packaging fitness with diversity. Because the best trade-off between these two extremes is not clear a priori, our approach to library design was to provide the tools to trace out an optimal trade-off curve, also known as a Pareto frontier (e.g., Fig. 4A). Each point lying on this optimal frontier represents a library for which it is not possible to improve one desiderata (packaging or diversity), without hurting the other. Our Pareto optimal frontier, therefore, allows us to assess what mean library packaging fitness can be achieved for any given level of diversity. To generate each point (library) that lies on the optimal frontier curve, we define a library optimization objective that seeks to maximize mean predicted fitness subject to a library diversity constraint controlled by the value λ. This knob, λ, controls the trade-off between library diversity and packaging ability; we set it to different values to trace out the Pareto frontier. We quantified the diversity of each theoretical library by computing the statistical entropy of the probabilistic distribution that it corresponds to (Materials and Methods). We refer to this overall methodology enabling tracing out the optimal curve as diversity-constrained optimal library design. We note that the optimization problem is challenging to solve exactly (i.e., it is non-convex). Consequently, libraries computed as we trace out λ may not lie exactly on the optimal frontier. However, the frontier can nevertheless be inferred approximately, providing useful insights, as we shall see next.
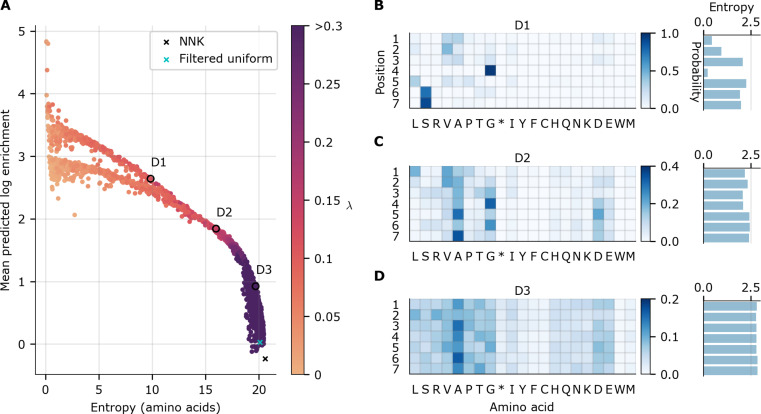
Fig. 4. Designed AAV5-based 7-mer insertion libraries.
Each point in (A) represents a theoretical library designed with our diversity-constrained optimal library approach, with one particular diversity constraint, λ (higher values yields more diverse libraries). Entropy indicates diversity of the library distribution, while MPLE indicates overall library fitness; both quantities were computed from the theoretical library distribution. The baseline NNK library is denoted with a black “×”, while a cyan “×” denotes the filtered uniform library that is uniform over all 21-mer nucleotide sequences except for those containing stop codons. Three designed libraries have been circled and labeled D1 to D3 for reference. Because of the non-convex optimization problem, some dots are suboptimal (i.e., lie strictly below or to the left of other dots) and are therefore further from the optimal frontier but are displayed for completeness. (B to D) Designed library parameters (probability of each amino acid at each position) for the three designed libraries D1 to D3, respectively, highlighted in (A).
We applied this diversity-constrained optimal library design methodology to the design of an improved AAV5–7-mer peptide insertion library, yielding some notable implications (Fig. 4A). We call out three designed libraries in particular—D1, D2, and D3—as representative of three important areas of the curve and also show the NNK library overlayed. The NNK library has a markedly poor mean predicted log enrichment (MPLE), much lower than any designed library. In contrast, library D3 had nearly identical diversity but substantially higher mean packaging fitness (top 50% of all designed libraries). This observation implies that D3 effectively dominates NNK in the sense that we increased the predicted packaging fitness without taking much loss to the diversity. Such concrete conclusions can be drawn from a Pareto frontier whenever one point on the frontier lies vertically above another. In addition, we see that, compared to D3, D2 is less diverse but is predicted to package better (2.0-fold higher MPLE). Similarly, D1 is less diverse than D2 but, again, is predicted to package better (1.4-fold higher MPLE).
Although the original motivation for creating the NNK library was to reduce the number of stop codons, it does not eliminate them entirely. Therefore, for further comparison, we computed the mean packaging fitness and diversity of the theoretical library containing all possible sequences, except for any containing a stop codon. In practice, such a library is not physically realizable using this position-wise nucleotide specification strategy but serves as a useful comparator. We call this the “filtered uniform” library and find that, although it does have slightly higher mean packaging fitness than NNK and correspondingly less diversity, these differences are negligible compared to the differences between NNK and D3, suggesting that the removal of stop codons is not the primary mechanism by which our ML-designed libraries achieve higher predicted packaging fitness.
Richer library generation mechanisms
As mentioned earlier, each designed library specifies the 84 marginal probabilities of individual nucleotides at each position in the 21–base pair (bp) insertion (tables S1 and S2). Our diversity-constrained optimal library design approach can, however, be used for any library construction method, such as one where we specify and synthesize individual 21-bp nucleotide sequences to create a library. We use the term “unconstrained” to refer to libraries that are designed with this construction method because individual synthesis offers the most control over sequences in the library. In contrast, a position-wise nucleotide specification strategy, such as the one we have used, cannot guarantee the inclusion of any particular sequence; we thus refer to libraries constructed in this manner as “constrained” libraries. We have focused our experiments on these constrained libraries because they are now more cost-effective and thus most widely used. Weinstein et al. (34) showed that, for a fixed cost, the use of a constrained library construction can yield orders of magnitude more promising leads in protein engineering than an unconstrained (individual synthesis) approach. As the cost of individual synthesis declines, it will become increasingly useful to use our design approach to specify unconstrained libraries that are both diverse and fit. With this future in mind, we also estimated the Pareto frontier for an unconstrained library (fig. S5), which shows that, if cost were no concern, then it would be advantageous to use an individual synthesis library construction approach, as its frontier substantially dominates that of our constrained library.
Experimental validation of designed libraries
We synthesized two designed libraries (D2 and D3) from our optimality curve (Fig. 4A) to assess the accuracy of the designed libraries’ trade-off between diversity and mean packaging fitness. Later, we also tested D2 in a downstream selection task of infecting brain tissue. The library D2 was chosen for being at the “elbow” of the curve, suggestive of a library making a good trade-off. The library D3 was chosen because, as discussed earlier, it dominates NNK by achieving much higher predicted packaging fitness with a negligible drop in diversity.
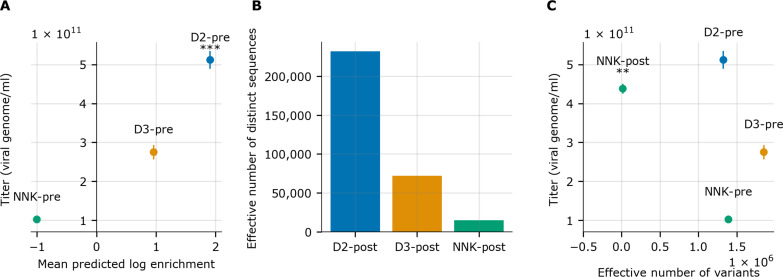
After experimentally constructing and deep sequencing these two designed libraries, we first checked that the physically realized library matched the statistics of the theoretical designed library distribution. We found that the empirically observed position-wise probabilities for each amino acid in each of the designed libraries were within 5% of the designed specification (tables S1 and S2). Furthermore, these sequencing data demonstrate that the reduction in the diversity between the NNK and designed libraries is relatively small, with approximately 2.7 and 4.4 million unique variants observed in the D2 and D3 libraries, respectively (table S3 and fig. S6). Having validated that the constructed libraries were as specified, we packaged and harvested each library using the same methods as for the NNK library, yielding a pre- and post-packaged version of each. Deep sequencing data for each pre- and post-packaged library confirmed that these designed libraries, D2 and D3, are substantially different from the standard NNK library: Only roughly 0.2 to 0.5% of variants are shared with the NNK library (table S4). Next, we assessed to what degree the MPLE of each library reflected the measured library titers and found a strong positive Pearson correlation between them (r = 0.959; Fig. 5A).
Fig. 5. Comparison of ML-designed libraries D2 and D3 to the NNK library.
(A) Experimental titers (viral genome/ml) plotted against the MPLE. ***P < 0.001 compared to NNK. (B) Comparison of the effective number of variants present in each library after packaging and post-packaged libraries are labeled with the “-post” suffix. (C) Experimental titers and effective number of variants for D2, D3, and NNK DNA libraries (pre-packaging selection), and the NNK-post library (post-packaging selection). The NNK-post library represents the NNK library after one round of packaging selection. One-way ANOVA followed by Tukey test (**P < 0.01 compared to D2). In all cases, experimental titers are measured on three biological replicates. Graphs show means ± SD.
As discussed earlier, D3 dominates the NNK library in fitness (one lies vertically above the other) and is thus predicted to be the better library. The choice between D3 and D2 is less clear, as they trade off packaging fitness and diversity. To assess such trade-offs, we subjected each of D2, D3, and NNK to one round of packaging selection and analyzed the diversity of each library post-packaging. When analyzing packaged libraries, the true underlying probability distributions corresponding to each library are not known, and, thus, we cannot exactly compute entropies. Instead, we estimate the effective sample size—specifically, the effective number of variants—of each packaged library from the observed deep sequencing data (see the “Comparison of constructed libraries” section). Effective sample size is commonly used to estimate phylogenetic diversity (35, 36), the number of nonredundant homologous sequences in multiple sequence alignments (37), cell-type specificity of transcription factor expression (38), and population sizes in population genetics (39) because it measures the uniformity of the distribution (i.e., relative abundances) of the unique observations rather than just the number of unique observations. In our context, the effective number of variants for a given library is defined, mathematically, as
where pempirical(s) corresponds to the empirical read frequency of the variant with sequence s in the sequencing data, and, therefore, for a given library, the effective number of variants reflects not just the number of unique variants but also relative read frequencies among unique variants. For example, if 100 sequencing reads are distributed among five unique variants with read counts (25, 25, 25, 20, and 5), then the estimated effective number of variants is
whereas the estimated effective number of variants if the 100 reads are instead distributed as (90, 3, 3, 2, and 2) is
A larger effective number of variants after packaging selection indicate that the post-packaged library is less likely to be dominated by a small number of variants and thus that the library contains more variants able to be packaged. We were also able to confirm, via supplementary experiments that artificially equalized the total number of reads across libraries, that the effective number of variants is not sensitive to the small observed differences in read coverage between libraries in our study (table S5 and fig. S7). An analysis of the effective number of variants revealed D2 to be more promising than D3 (Fig. 5B). Consequently, we continued our comparison to NNK with only the D2 library.
Looking back at our measured titers, designed library D2 (MPLE ~ 2.0) showed a fivefold higher packaging titer than that of the NNK library (MPLE ~ −0.9) with titers of 5.12 × 1011 and 1.02 × 1011 vg/ml, respectively. Next, we also measured the packaging titer of the NNK library after one round of packaging selection (NNK-post), finding that its titer (4.38 × 1011 vg/ml) was lower than that of D2 (5.12 × 1011 vg/ml) (Fig. 5C). This result suggests that the additional round of packaging was not enough to lift the NNK library’s titer level to that of library D2. Note also that (i) the NNK-post library contains only 1.48 × 104 effective variants compared to the 1.33 × 106 effective variants in D2 and (ii) the designed libraries differ substantially from the observed amino acid frequencies at each position in the NNK-post library (fig. S8). Therefore, the designed library D2 is different from and preferable to the library resulting from subjecting the NNK library to a round of packaging in both packaging titer and library diversity. Collectively, these experimental results suggest that our ML-guided library design procedure yielded a more useful library than the NNK library, the current standard peptide insertion library for AAV directed evolution experiments.
ML-designed AAV library for primary brain tissue infection
Having demonstrated our ability to design and construct libraries with better packaging and good diversity, we next investigated how these gains would translate into performance on a downstream selection task for which the library had not been tailored. After all, our goal was to design a generally useful library, agnostic to the downstream selection goal. Thus, we moved forward with two of the libraries, the NNK as a baseline and our designed library D2 and used each to infect primary adult brain tissue. Infecting such tissue with AAV can be a first step toward numerous clinical applications in the central nervous system. AAV selection using directed evolution is sensitive to the choice of experimental system, where evolved variants display high specificity in the context of cell types (40), species, and even strain within the same species (41). Given transcriptional differences between mouse and human cell types within the brain (42, 43) and evolutionary emergence of new cell types in the human brain that are absent in rodents, such as outer radial glia (44), it is crucial to select the starting biological material and model system. To make our work relevant for therapeutic interventions in humans across different disease states, we used fresh, surgically resected adult cortical tissues from epilepsy patients to develop and select the AAV variants, which would efficiently infect and drive gene expression in the human-specific context.
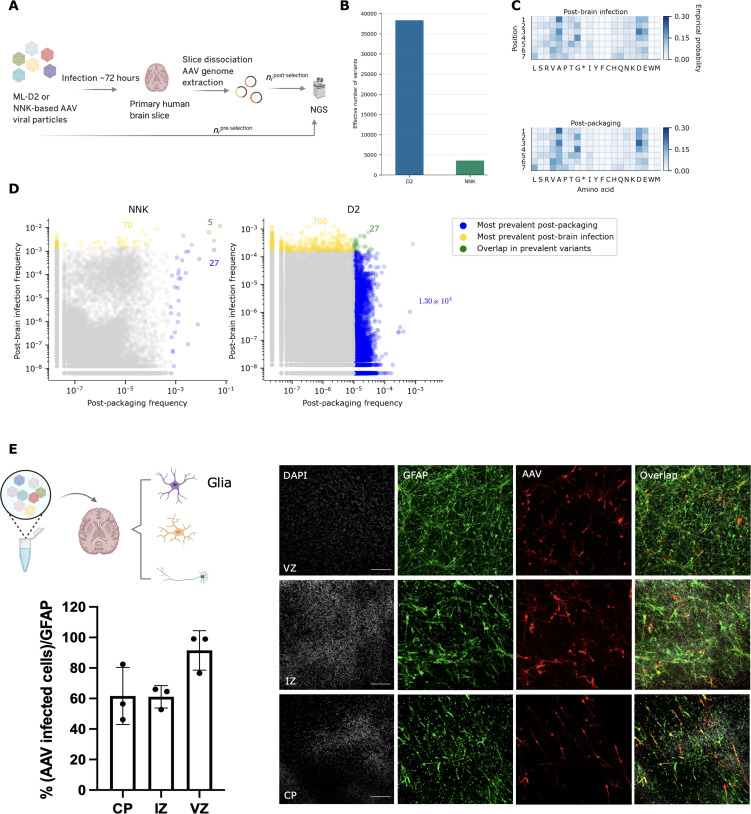
We applied each library onto human adult brain slices (fig. S9 and Materials and Methods) and harvested the tissues after 72 hours of infection (Fig. 6A). We evaluated the success of each library on this task by comparing the effective number of variants in each pool after infectivity selection. A higher effective number of variants post-brain infection would suggest that the starting library contained more variants that were able to successfully infect human brain tissue, indicating a more useful starting library and larger set of promising variants.
Fig. 6. ML-designed AAV library for primary brain tissue infection.
(A) General workflow of the primary adult brain infection study. (B) Effective number of variants (calculated from entropy) in NNK post-brain infection versus D2 post-brain infection; D2 post-brain infection exhibits a ~10-fold increase in effective number of variants compared to that of NNK post-brain infection. (C) Empirical probabilities of each amino acid at each position for D2 post-packaging and post-brain infection. (D) Scatterplots illustrating the behavior of individual variants over packaging and primary brain selection. Each axis shows the (log) prevalence of the variant in each library, as a fraction of reads in the library. For each library, variants in the top 20% are determined by first sorting unique variants by read count in descending order and then counting the number of unique variants comprising 20% of the total sequencing reads. Variants in the top 20% after packaging are colored blue, while those in the top 20% after brain selection are colored yellow. Those variants in the top 20% of both packaging and selection are colored green. The annotated colored numbers indicate the number of variants of each colored pool. A pseudo-count of 1 was added to each variant in each library before plotting. See fig. S11 for additional versions of (D) displaying variants in the top 50 and 80% of each library. (E) Cell-specific AAV validation (VVKQRGD) selected from the post-brain infection pool [green, glial fibrillary acidic protein (GFAP) marker; red, AAV infected cells; scale bars, 100 μm; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; DAPI, 4′,6-diamidino-2-phenylindole). Schematic illustration created with BioRender.com.
We found that designed library D2 had a 10-fold higher post-brain infection effective number of variants than the NNK library (Fig. 6B and fig. S10): 38,350 versus 3541 effective variants. In terms of diversity, which can be achieved in different ways, we were interested to know whether diversity of a given library was spread over the length of the 7-mer insertion or was more concentrated on particular positions. Thus, for each post-packaging and post-brain infection library, we examined the diversity at each position, finding that, for both selections, no amino acid has higher that 0.3 frequency in any position Fig. 6C and fig. S11), revealing a diversity that arose from across the 7-mer.
Other noteworthy observations include that our D2 library design showed a modest depletion of threonine (T) and serine (S) compared to the NNK library (Fig. 6C and fig. S11). Previous research has shown that the removal of several critical residues of S, tyrosine (Y), and T on the AAV capsid can significantly increase transduction efficiency compared to the wild-type vectors. Reduced S, Y, and T residues can result in lower capsid phosphorylation, thereby avoiding intracellular degradation and enabling higher nuclear translocation (45–47). Such mutations have also been explored previously to demonstrate enhanced transduction within various cell types (45–49). A modest depletion of S and T residues on capsids from ML-designed library D2 could thus potentially improve the transduction of target cells.
We next compared the post-packaging and post-brain infection libraries at the level of individual variants to assess some practical implications of the difference in diversity between the NNK and D2 libraries (Fig. 6D). We found a small set of variants dominated the post-packaging NNK library: The 32 most prevalent variants post-packaging (blue and green points in Fig. 6E) accounted for 20% of the total sequencing reads. There were roughly 100-fold more unique variants in the top 20% of the D2 library post-packaging (1.32 × 104 blue and green points), meaning that there is a much larger set of variants in the D2 library that package well when compared to the NNK library. In terms of downstream selection, the post-brain infection NNK library is dominated by a much smaller set of variants (~10-fold fewer) compared to D2 (75 yellow and green points for NNK compared to 727 for D2). This suggests that the chances of discovering individual variants that successfully package and pass downstream selection are increased by using D2 instead of NNK as the starting library. In practice, then, the higher entropy for D2 post-packaging and post-brain infection translates to a much larger set of promising individual variants after each type of selection. We also considered the top 50 and 80% of the post-packaging and post-brain infection libraries (fig. S12) and found these conclusions to be consistent. Collectively, the results shown in Fig. 6 demonstrate that our designed library D2 provided more useful diversity over the widely used NNK library, thereby making it an effective, general starting library for downstream selections for which it was not specifically designed.
Last, we validated that individual AAV variants from the ML-designed library D2 can not only package well but also successfully mediate cell-specific infection, which is a significant challenge in AAV engineering. For example, glial cells are important regulators of many aspects of human brain functions and diseases; however, true glial cell–specific targeting AAVs remain elusive (50).
To identify top variants for cell-specific expression validation, we applied the D2 library to human brain tissue, dissociated and isolated glial cells using magnetic-activated cell sorting (MACS), extracted the AAV genomes that successfully entered the glial cells, and applied NGS (Materials and Methods). We then ranked the glia-infectious variants in the D2 post-glia infection library by enrichment score and selected three top variants for individual validation. We see clearly that these three variants ranked top (in the yellow zone) of the D2 library, whereas they would not have been identified from the NNK (fig. S13). In general, we can see that, although these glial-specific variants were not highly enriched in the post-packaging libraries for either NNK or D2, because the distribution of variants post-packaging is less skewed in D2 compared to NNK (i.e., the few most prevalent variants in NNK post-packaging are an order of magnitude more abundant than the most prevalent variants in D2 post-packaging), these three variants appeared more enriched in D2 after both the whole brain-selection and the glial-specific selection. In contrast, in NNK, the variants are unlikely to be selected for the downstream application. Each of these top selected glial specific AAV variants showed high titers (~1012 vg /μl) when packaged with a green fluorescent protein (GFP)–encoding genome (table S6). Furthermore, AAV variant VVKQRGD insertion selected for glia population showed high levels of glial infection across multiple regions of the primary brain tissue in immunostaining (Fig. 6E). Future work can extend our library design and selections to other cell types in brain or other tissues for a variety of therapeutic applications.
DISCUSSION
We developed an ML-based method for systematically designing diverse AAV libraries with good packaging capabilities, so that they can be used as starting libraries in directed evolution for engineering-specific and enhanced AAV properties. A brief summary of our overall workflow was to (i) synthesize and sequence a baseline NNK library, the pre-packaged library; (ii) transfect the library into packaging cells [i.e., human embryonic kidney (HEK) 293 T] to produce AAV viral vectors, harvest the successfully packaged capsids, extract viral genomes, and sequence to obtain the post-packaging library; (iii) build a supervised regression model where the target variable reflects the packaging success of each insertion sequence found; (iv) systematically invert the predictive model to design libraries that trace out an optimal trade-off curve between diversity and fitness; and (v) select a library design with a suitable trade-off. We then validated both the predictive model and the designed library by experimentally measuring library packaging success and sequence diversity. Last, we demonstrated that our ML-designed library is better able to infect primary human brain tissues as compared to the baseline NNK library.
In doing so, we have shown that (i) we can build accurate predictive models for AAV packaging fitness for 7-mer insertion libraries; (ii) we can leverage these predictive models to design libraries that optimally trade off diversity with packaging fitness; and (iii) these designed libraries can be better starting libraries for downstream selection than standard libraries used today, despite not being tailored to the downstream task. This work develops and uses a generalizable and impactful ML-based design to systematically identify a suite of optimal libraries along a trade-off curve of diversity and fitness. In addition, it provides an end-to-end set of ML-based library design solutions, realized through experiments, in a therapeutically relevant system. We plan to generalize and apply this approach to further downstream selection tasks, including those relevant to gene replacement in the nervous system and evasion of preexisting antibodies. Our approach can, in principle, be used for other library construction techniques, such as individual gene sequence specification and synthesis (fig. S5). Our framework can also be extended to design libraries with multiple desired properties beyond diversity, by replacing the predictive model with one trained to simultaneously predict multiple functions or fitness combining such models when the properties are independent. This could be particularly useful to design libraries with improved cell sensitivity and specificity, which is particularly challenging using conventional experiment approaches.
Although we used replication during experimental preparation of our training dataset to mitigate noise, potential biases could still exist, such as those arising from uneven amplification from PCR or biased cloning errors. It could be possible to address some of these biases through further experimental and computational work; however, as our results demonstrate, the models as are have already proved useful in moving the field forward.
Beyond introducing a new approach for ML library design, a major contribution of our work has been to perform AAV directed evolution selected on primary human brain tissue, a clinically relevant physiological system for downstream therapeutic applications. Gene therapies have been explored as promising treatments for multiple brain diseases, including but not limited to Parkinson’s disease, Huntington’s disease, and lysosomal storage disorders. Specifically, there is a consensus that dysfunction of nonneuronal populations of brain cells (e.g., astrocytes, a subtype of glial cells) contributes to the progression of multiple pathologies such as neurodegenerative diseases (51–52), and gene delivery to these cells could help reinforce their normal functional roles. However, a lack of robust and specific targeting tools for accessing and manipulating these human brain cells remains a challenge in developing effective therapies. Through application of our ML-designed libraries, we succeeded in developing glial-targeting capsid variants (Fig. 6E), which we had previously been unable to do.
MATERIALS AND METHODS
Construction of the NNK-based 7-mer insertion library
We used libraries with a variable seven–amino acid (7-mer) insertion region flanked by amino acid linkers [Threonine-Glycine-Glycine-Leucine-Serine (TGGLS)] introduced at position 575–577 in the viral protein monomer. (NNK)7 oligo was first synthesized (Elim) and introduced to the 5′ end of the right fragment by a primer overhang (7mer_F). For ML-designed libraries, instead of using NNK, we specified position-specific nucleotide probabilities (table S1) at the time of synthesis (GeneWiz) to be incorporated at the 5′ end of the 7mer_F primer. Left and right fragments were each PCR-amplified by primer pairs Seq_F/Seq_R and 7mer_F/7mer_R, respectively (table S7). PCR products of the two fragments were then purified individually and subjected to overlap extension PCR (using HindIII_F and NotI_R primers) with Vent DNA polymerase (Thermo Fisher Scientific) with equimolar amounts of the left and right fragments for a total of 250-ng DNA templates. The resulting library was then digested with Hind III and Not I (New England Biolabs Inc.) and ligated into replication incompetent AAV packaging plasmid pSub2repKO (8) for library construction. The resulting ligation reaction was electroporated (Bio-Rad) into electrocompetent Escherichia coli (Thermo Fisher Scientific, catalog no. 18290015) for plasmid production and purification. HEK 293 T cells were originally obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium (Gibco) with 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin (Gibco) at 37°C and 5% CO2. The passage number of 293 T for packaging AAV libraries was between 10 and 15.
We note that experimental noise and bias could arise from several procedures, including the PCR amplification, electroporation in transformation, and plasmid purification. To mitigate experimental noise, we used biological replicates (n = 3) in each of these steps that averaged the replicates at the biological level, before computational analysis.
Vector packaging and production
AAV library was packaged with transfection of HEK 293 T cells where each biological replicate (n = 3) was separately transfected in each round. Specifically, in a ~75 to 80% confluent density of 15-cm dish of HEK 293 T cells, 13.5 μg of pHelper, 9 μg of pBluescript (Addgene), 70 ng of the capsid plasmid library, and 5 μg of pRepHelper were cotransfected by the polyethyleneimine (PEI) method. This ratio was calculated to minimize occurrences of cross-packaging as previously reported (1, 26, 53). Seventy-two hours later, cells were harvested, and the supernatant was collected. The cell pellet was resuspended in a lysis buffer [50 mM tris and 150 mM NaCl (pH 8.5)] and freeze/thawed for three times at dry ice/ethanol. The lysate was then incubated at 37°C for 30 min with an addition of Benzonase (10 U/ml; Invitrogen). Then, the lysate was first spun at 2000 rpm for 2 min, followed by a 10,000-rpm spin for 10 min, before the supernatant was all collected for purification. Collected virus was then purified via iodixanol density centrifugation and buffer-exchanged into phosphate-buffered saline (PBS) by Amicon (Ultra-15, Merck Millipore) filtration.
This packaging process has the potential to be confounded by cross-packaging, in which viral particles are composed of viral genomes and capsid proteins derived from different library variants. To minimize cross-packaging, we diluted the plasmid library according to previously determined concentration that minimizes the event of multiple members of the capsid plasmid library entering into the same cell (1, 54). To quantify capsid cross-packaging in different libraries, we used GFP plasmid mixed with capsid libraries in 1:7 molar ratio and determined correctly packaged versus cross-packaged viral particles using either Cap-specific or GFP-specific primers, respectively (table S8). These findings quantitatively characterized cross-packaging and provided experimental evidence of a similar but minimal level (less than 2%) of cross-packaging in all libraries.
Each individual sequence plasmid (in Fig. 3) was packaged separately with biological replicates (n = 3), and its titer was measured with technical replicates (n = 3) for each run. Specifically, in a ~75 to 80% confluent density of 15-cm dish of HEK 293 T cells, 12 μg of pHelper, 10 μg of the pRepCap (AAV capsid variant), and 6 μg of GFP-encoding AAV vector plasmid were cotransfected by the PEI method. Seventy-two hours later, collected virus was purified and buffer-exchanged into PBS. We then measured the packaged viral titers using ddPCR with GFP probe (CGCGATCACATGGTCCTGCTGG).
AAV viral genome extraction and titer
Packaged AAV vectors were first combined with equal volume of 10× deoxyribonuclease (DNase) buffer (New England Biolabs, B0303S) and 0.5 μl DNase I (10 U/μl; New England Biolabs, M0303L) incubated for 30 min at 37°C. Then, equal volume of 2× proteinase K buffer was added with sample to break open capsid. After heat inactivating for 20 min at 95°C, the sample was further diluted at 1:1000 and 1:10,000 and use as templates for titer. DNase-resistant viral genomic titers were measured using ddPCR (Bio-Rad) using with Hex-ITR probes (CACTCCCTCTCTGCGCGCTCG) tagging the conserved regions of encapsidated viral genome of AAV. After primary tissue infection, capsid sequences were recovered by PCR from harvested cells using primers HindIII_F and NotI_R (table S7). A ~75- to 85-bp region containing the 7-mer insertion was PCR-amplified from harvested DNA. Primers included the Illumina adapter sequences containing unique barcodes to allow for multiplexing of amplicons from multiple libraries. PCR amplicons were purified and sequenced with a single-read run-on Illumina NovaSeq 6000.
Single-ion CDMS
Experiments were performed using an in-house built charge detection mass spectrometer at University of California, Berkeley that has been described previously (55–56). AAV5–7-mer variants were packaged and harvested at 72 hours after transfection followed by the purification through iodixanol gradients. Purified AAVs were washed six times using an Amicon Ultra Centrifugal Filter Unit (MilliporeSigma, St. Louis, MO): three times each with 1× PBS + 0.001% Tween, followed by 500 mM ammonium acetate. These AAVs were subsequently analyzed with CDMS as previously described (57), with a minimum of ~2200 ions measured to determine the capsid mass. By fitting the mass spectrum to a sum of three Gaussians corresponding to each component, we computed the ratio of full to empty capsid by dividing the integral of the full capsid mass peak by that of the empty capsid mass peak.
Data filtering and processing
The raw sequencing data consisted of 49,619,716 and 55,135,155 sequencing reads corresponding to the pre- and post-selection libraries, respectively. Each read contained (i) a 5-bp unique molecular identifier, (ii) a fixed 21-bp primer sequence, (iii) a 6-bp sequence representing the pre-insertion linker (two fixed amino acids that connect the insertion sequence to the capsid sequence at position 575), (iv) a variable 21-bp sequence containing the nucleotide insertion sequence, and (v) a 9-bp representing the post-insertion linker (three fixed amino acids that connect the insertion sequence to the capsid sequence at position 577). We filtered the reads, removing those that either contained more than two mismatches in the primer sequences or contained ambiguous nucleotides. After this filtering, the pre- and post- libraries contained 46,046,268 and 45,303,374 reads, respectively. The insertion sequences were then extracted from each read and translated to amino acid sequences. Analysis of overlap variants between pools of sequences can be found in table S4.
Log enrichment score and variance
We calculated the log enrichment scores (Eq. 1) for each insertion sequence using the (filtered) sequencing data to quantify each sequence’s effect on packaging. Note that only 218,942 of the 8,552,729 unique sequences appear in both the pre- and post-selection libraries. A pseudo-count of 1 was added to each count so that the log enrichment score could still be calculated when the sequence appeared in only one of the libraries. In all cases, the natural log was used.
| (1) |
We estimated a variance associated with each log enrichment score using Eq. 2, which follows by noting that each of the raw counts associated with a log enrichment score is a random variable. Specifically, the count associated with a sequence can be modeled as a Binomial random variable (32). The log enrichment score (Eq. 1) is then the log ratio of two Binomial random variables; it can be shown with the Delta method (58) that, in the limit of infinite samples, the log ratio of two Binomial random variables converges in distribution to a Normal random variable with mean and variance approximated by Eqs. 1 and 2, respectively (32, 33)
| (2) |
Model training and evaluation
Our data processing yields a dataset of the form where the xi are unique insertion sequences, yi are log enrichment scores associated with the insertion sequences, are the estimated variances of the log enrichment scores, and M = 8,555,729 is the number of unique insertion sequences in the data. We randomly split this dataset into a training set containing 80% of the data and a test set containing the remaining 20% of the data.
We assume that the distribution of a log enrichment score given the associated insertion sequence is
where fθ is a function with parameters θ that parameterizes the mean of the distribution and represents a predictive model for log enrichment scores. We determined suitable settings of the parameters θ with maximum likelihood estimation (MLE). The log likelihood of the parameters of this model given the training set of M′ ≤ M data points is given by
Performing MLE by optimizing this likelihood with respect to the model parameters, θ, results in the weighted least squares loss function in Eq. 3.
| (3) |
We tested both linear and NN forms for the function fθ; linear models are standard baseline models due to their simplicity, while NNs have been shown to produce state-of-the-art performance in sequence-to-fitness modeling tasks (59). For the linear forms of fθ, the loss (Eq. 3) is a convex function that can be solved exactly for the minimizing ML parameters. To stabilize training, we used a small amount of l2 regularization for the neighbors and pairwise representations (with regularization coefficients 0.001 and 0.0025, respectively, chosen by cross-validation). For the NN forms of fθ, the objective (Eq. 3) is non-convex, and we use stochastic optimization techniques to solve for suitable parameters. We implemented these models in TensorFlow (60) and used the built-in implementation of the Adam algorithm (61) to approximately solve Eq. 3.
All NNs had two hidden layers, each with tanh activation functions. A two hidden layer architecture was chosen so as to allow hierarchical representations while reducing the total number of parameters in the model. As we did not observe an increase in model performance with an increased number of parameters (Fig. 2A), we did not explore deeper model architectures. The tanh activation function is a common choice for regression models, and we did not find the model performance to be sensitive to the choice of activation function (fig. S1).
To assess the prediction quality of each model, we calculated the Pearson correlation between the model predictions and observed log enrichment scores for different subsets of the sequences in the test set. Our aim is to use these models to design a library of sequences that package well (i.e., would be highly enriched in the post-selection library). We, therefore, assess how well the models perform for highly enriched sequences by progressively culling the test set to only include sequences with the largest observed log enrichment scores (Fig. 2).
Diversity-constrained optimal library design
We developed a general framework for sequence library design that (i) can be used with any predictive model of fitness, (ii) is broadly applicable to different library construction mechanisms (e.g., error prone PCR, site-specific marginal probability specification, and individual synthesized sequences), and (iii) is simple to implement and extend. This framework balances mean predicted packaging fitness with entropy, a measure of diversity for probability distributions, which has been used extensively in ecology to describe the diversity of populations (62). Our approach is based on a maximum entropy formalism: We represent libraries as probability distributions and aim to find maximum entropy distributions that maximize entropy while also satisfying a constraint on the mean fitness, which is predicted by a user-specific model such as a NN.
Let χ be the space of all sequences that may be included in a library (e.g., all amino acid sequences of length 7). We consider a library to be an abstract quantity represented by a probability distribution with support on. Let ℘ represent all such libraries and one particular library. In other words, p represents a library, and p(x) refers to the probability of a sequence x in the library (63)
Now, let f(x) be a predictive model of fitness (e.g., from a trained NN). Our goal is to find a diverse library, p, where the mean predicted fitness in the library, , is as high as possible. Formally, we want to find the library with the largest entropy such that the mean predicted fitness is above some cutoff. This objective is written
where a is the cutoff on the mean predicted fitness. It is straightforward to show that the solution to this optimization problem is given by (64)
| (4) |
where λ > 0 is a Lagrange multiplier that is a monotonic function of the cutoff a and Z(λ) = ∑x∈χ exp[f(x)/λ] is a normalizing constant. Equation 4 gives the probability mass of what is known as the maximum entropy distribution. The parameter λ controls the balance between diversity and mean fitness in the library (higher λ corresponds to more diversity). Each library, pλ, represents a point on a Pareto optimal frontier of libraries, which balances diversity and mean predicted fitness; these distributions cannot be perturbed in such a manner as to increase both the entropy and the mean fitness. Theoretically, the entire Pareto frontier could be traced out by calculating the mean predicted fitness and entropy of pλ for every possible setting of λ. In practice, we pick a discrete set of λ that traces out a practically useful curve.
As written so far, this framework can be used to select a particular library distribution, pλ(x), with value λ, from the Pareto optimal curve. Then, if designing libraries composed of individually specified sequences, one can sample individual sequences from this distribution, thereby designing a realizable, synthesizable library. However, for many cases of practical interest, it will not be cost-effective to synthesize individual sequences. We will, therefore, consider a more affordable library construction mechanism: A library of oligonucleotides is generated in a stochastic manner based on specified position-wise nucleotide probabilities. Because this position-wise nucleotide specification strategy does not allow one to specify individual sequences, we refer to libraries constructed in this way as constrained. In the next section, we describe how we use our design framework to set the parameters of these constrained libraries.
Maximum entropy design for constrained libraries
In this section, we describe the design of libraries that are not specified at the level of individual sequences, but rather at the (less precise) level of position-specific distributions. In particular, we controlled the marginal probability of each nucleotide at each position. The probability mass function of the distribution representing a library specified by position-wise probabilities is given by
where L is the sequence length, K is the alphabet size (i.e., K = 4 for nucleotide libraries), ϕ ∈ ℝL×K is a matrix of distribution parameters, ϕj is the jth row of ϕ, δk(xj) = 1 if xj = k and zero otherwise, and
| (5) |
In words, qϕj(k) refers to the probability of observing the kth alphabet element at the jth position in the sequence.
For an arbitrary predictive model (such as a NN to predict log enrichment scores from sequence), the maximum entropy distribution (Eq. 4) will generally not have the form of Eq. 5. To apply the maximum entropy formulation to the design of libraries that are constrained to take a particular form, what we refer to as constrained library design, we take a variational approach: For a single, fixed value of λ, we find the constrained library distribution, qθ, that is the best approximation to the maximum entropy library distribution, pλ, in terms of the KL divergence
| (6) |
Our objective (Eq. 6) is a non-convex function of the library parameters. The stochastic gradient descent (SGD) algorithm has been shown to consistently find optimal or near-optimal solutions to a variety of non-convex problems, particularly in ML (65). We use a variant of SGD on the basis of the score function estimator (66) to solve Eq. 6. We randomly initialize a parameter matrix, ϕ(0), with independent Normal samples and then update the parameters according to
| (7) |
for t = 1, …, T, where we define to be the objective function in Eq. 6. The number of iterations, T, was set such that we observed convergence of the objective function values in most runs of the optimization. After T iterations, we assumed that we had reached a near-optimal solution [i.e., ϕ(T) can be used as an approximation of ϕλ]. The components of the gradient in Eq. 7 are given by
| (8) |
where we define the weights w(x) ≔ f(x) − λ[1 + log qϕ(x)] (Supplementary Materials). The expectation in Eq. 8 cannot be solved exactly, so we use a Monte Carlo approximation
where M is the number of samples used for the MC approximation. We applied this maximum entropy framework to design site-specific marginal probability libraries of the 21 nucleotides corresponding to the seven–amino acid insertion using the (NN, 100) predictive model of fitness. Figure 3 shows the near-optimal Pareto frontier resulting from 2238 such library optimizations with α = 0.01, T = 2000, and M = 1000 and a range of settings of λ.
In practice, the nucleotide sequences sampled from qϕ must be translated to amino acid sequences before being passed to f(x), which is a model trained to predict log enrichment scores from amino acid sequences. We have omitted this translation step from the above equations for notational simplicity.
Comparison of constructed libraries
Entropy is closely related to another notion of diversity known as effective sample size. The effective sample size of a library with entropy H is defined as Ne = eH and corresponds to how many unique variants one would need to obtain entropy H, if each variant was constrained to have equal probability mass. This can be seen by noting that . This interpretation of entropy is commonly used in the population genetics literature, first introduced by Wright in 1931 (39).
When comparing designed theoretical libraries, we were able to compute the statistical entropy of each library distribution exactly in terms of its position-wise probabilities. However, when analyzing post-selection libraries, there is no known underlying probability distribution with which we can exactly compute entropy. Consequently, we instead estimated and compared the effective sample size of the empirically observed distribution in each library. Specifically, we estimated the effective number of samples in a library using the sequencing observations
where pempirical(s) corresponds to the empirical frequency of sequence s appearing in the post-selection sequencing data.
Consent statement UCSF
De-identified tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Sample use was approved by the Institutional Review Board at University of California San Francisco (UCSF) and experiments conform to the principles set out in the World Medical Association (WMA) Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Primary human adult brain slices culture and library infection
Adult surgical specimens from epilepsy cases were obtained from the UCSF medical center in collaboration with neurosurgeons with previous patient consent. Surgically excised specimens were immediately placed in a sterile container filled with N-methyl-d-glucamine (NMDG)–substituted artificial cerebrospinal fluid (aCSF) of the following composition: 92 mM NMDG, 2.5 mM KCl, 1.25 mM NaH2PO4, 30 mM NaHCO3, 20 mM Hepes, 25 mM glucose, 2 mM thiourea, 5 mM Na-ascorbate, 3 mM Na-pyruvate, 0.5 mM CaCl2·4H2O, and 10 mM MgSO4·7H2O. The pH of the NMDG aCSF was titrated from pH 7.3 to pH 7.4 with 1 M tris-base at pH 8, and the osmolality was 300 to 305 mosmol/kg. The solution was prechilled to 2° to 4°C and thoroughly bubbled with carbogen (95% O2/5% CO2) gas before collection. The tissue was transported from the operating room to the laboratory for processing within 40 to 60 min. Blood vessels and meninges were removed from the cortical tissue, and, then, the tissue block was secured for cutting using superglue and sectioned perpendicular to the cortical plate to 300 μm using a Leica VT1200S vibrating blade microtome in aCSF. The slices were then transferred into a container of sterile-filtered NMDG aCSF that was prewarmed to 32° to 34°C and continuously bubbled with carbogen gas. After 12 min of recovery incubation, slices were transferred to slice culture inserts (Millicell, PICM03050) on six-well culture plates (Corning) and cultured in adult brain slice culture medium containing 840 mg of MEM Eagle medium with Hanks’ salts and 2 mM l-glutamine (Sigma-Aldrich, M4642), 18 mg of ascorbic acid (Sigma-Aldrich, A7506), 3 ml of Hepes (1 M stock) (Sigma-Aldrich, H3537), 1.68 ml of NaHCO3 (892.75 mM solution, Gibco, 25080-094), 1.126 ml of d-glucose (1.11 M solution; Gibco, A24940-01), 0.5 ml of penicillin/streptomycin, 0.25 ml of GlutaMAX (at 400×; Gibco, 35050-061), 100 μl of 2 M stock MgSO4·7H2O (Sigma-Aldrich, M1880), 50 μl of 2 M stock CaCl2·2H2O (Sigma-Aldrich, C7902), 50 μl of insulin from bovine pancreas (10 mg/ml; Sigma-Aldrich, I0516), 20 ml of horse serum heat-inactivated, and 95 ml of MilliQ H2O [as previously described (67)]. The following day after plating, adult human brain slices were infected with the viral library. Specifically, purified AAV library suspension in 0.001% Tween PBS was gently mixed by pipetting with tissue culture medium and directly added on top of the slices (n = 3 per group). The volume of each library added to the slice is calculated as follows
Slices were cultured at the liquid-air interface created by the cell culture insert in a 37°C incubator at 5% CO2 for 72 hours after infection.
Slice culture dissociation, cell purification, and Hirt extraction
Seventy-two hours after infection with the viral library, cultured brain tissue slices were first rinsed twice with Dulbecco's PBS (DPBS; Gibco, 14190250) and detached from the filters, then mechanically minced to 1-mm2 pieces, and enzymatically digested with a papain digestion kit (Worthington, LK003163) with the addition of DNase for 1 hour at 37°C. After the enzymatic digestion, tissue was mechanically triturated using fire-polished glass pipettes (Thermo Fisher Scientific, catalog no. 13-678-6A), filtered through a 40-μm cell strainer (Corning, 352340), pelleted at 300g for 5 min, and washed twice with DBPS. Following mechanical digestion, the slices were first treated with lysis buffer [10% SDS, 1 M tris-HCl (pH 7.4 to 8.0), and 0.5 M EDTA (pH 8.0)] with the addition of ribonuclease (RNase) A (Thermo Fisher Scientific, EN0531) for 60 min at 37°C and proteinase K (New England Biolabs, P8107S) for 3 hours at 55°C. The enzymatically digested tissue homogenate was then proceeded to the Hirt column protocol as previously published (68).
Primary prenatal brain slices
Deidentified primary tissue samples were collected with previous patient consent in strict observance of the legal and institutional ethical regulations. Cortical brain tissue was immediately placed in a sterile conical tube filled with oxygenated aCSF containing 125 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 1.25 mM NaH2PO4 bubbled with carbogen (95% O2/5% CO2). Blood vessels and meninges were removed from the cortical tissue, and, then, the tissue block was embedded in 3.5% low–melting point agarose (Thermo Fisher Scientific, BP165-25) and sectioned perpendicular to the ventricle to 300 μm using a Leica VT1200S vibrating blade microtome in a sucrose protective aCSF containing 185 mM sucrose, 2.5 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 1.25 mM NaH2PO4, 25 mM NaHCO3, and 25 mM d-(+)-glucose. Slices were transferred to slice culture inserts (Millicell, PICM03050) on six-well culture plates (Corning) and cultured in prenatal brain slice culture medium containing 66% (v/v) Eagle’s basal medium, 25% (v/v) Hanks’ balanced salt solution, 2% (v/v) B27, 1% N2 supplement, 1% penicillin/streptomycin, and GlutaMAX (Thermo Fisher Scientific). Slices were cultured in a 37°C incubator at 5% CO2, 8% O2, at the liquid-air interface created by the cell culture insert.
Slice dissociation and cell purification
Cultured brain slices were washed twice with DPBS (Gibco, 14190250), detached from the filters, and enzymatically digested with a papain digestion kit (Worthington, LK003163) with the addition of DNase for 30 min at 37°C. Following enzymatic digestion, slices were mechanically triturated using a fire-polished glass pipette, filtered through a 40-μm cell strainer test tube (Corning 352235), pelleted at 300g for 5 min, and washed twice with DBPS.
Dissociated cells were resuspended in MACS buffer (DPBS with 1 mM EGTA and 0.5% bovine serum albumin) with addition of DNAse and incubated with CD11b antibody for 15 min on ice. After the incubation, cells were washed in 10 ml of MACS buffer and loaded on LS columns (Miltenyi Biotec, 130-042-401) on the magnetic stand. Cells were washed three times with 3 ml of MACS buffer, then the column was removed from the magnetic field, and microglia cells were eluted using 5 ml of MACS buffer. The flow-through cells were then gently prepared to separate out neurons using polysialylated-neural cell adhesion molecule, and the flow-through cell population was used as glial cell type. Cells were pelleted, resuspended in 1 ml of culture medium, and counted.
Immunofluorescence and antibodies
Primary human brain slices were fixed on the filters in 4% paraformaldehyde for 1 hour at room temperature and washed three times with PBS for 5 min each wash. Slices were carefully detached from the culture filter inserts and places into 12-well plates. Blocking and permeabilization were performed in a blocking solution consisting of 10% normal donkey serum, 1% Triton X-100, and 0.2% gelatin for 1 hour. Primary and secondary antibodies were diluted and incubated in the blocking solution. Prenatal brain slices were incubated with primary antibodies at 4°C overnight and washed three times with washing buffer (1% Triton X-100 in PBS). Adult brain slices were incubated with primary antibodies for 2 days and washed three times with washing buffer (1% Triton X-100 in PBS). Slices were incubated with secondary antibodies in the blocking buffer at 4°C overnight and washed with washing buffer five times for 10 min each. Images were collected using Leica SP8 confocal system with 10× and 20× air objective and processed using ImageJ/Fiji and Affinity Designer software. Primary antibodies used in this study included chicken glial fibrillary acidic protein (1:1000; Abcam, ab4674), rabbit dsRed (1:250; Takara, 632496), and 4′,6-diamidino-2-phenylindole. Secondary antibodies were species-specific AlexaFluor secondary antibodies (1:2000; Thermo Fisher Scientific).
Statistical analysis
All comparisons were performed using Prism 8 (GraphPad Software). AAV viral titers were compared using a one-way analysis of variance (ANOVA), and comparisons between different groups were doing using a Tukey’s comparison test.
Acknowledgments
Funding: D.Z. was supported by Siebel Stem Cell Fellowship. D.Z., T.J.N., and J.L. were supported by the Chan Zuckerberg Biohub. A.B., C.F., and A.C. were supported by the National Science Foundation Graduate Research Fellowship Program under grant nos. DGE 1752814, DGE 2146752, and DGE 2146752; any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. G.P. was supported by NIH NRSA F32 1F32MH118785. This work was supported, in part, by National Institutes of Health 5R01GM139338 (E.R.W.) and NIH 1 U01 MH130700-01, a grant from The Shurl and Kay Curci Foundation (T.J.N.), and gifts from the Schmidt Futures and the William K. Bowes Jr Foundation (T.J.N.).
Author contributions: D.Z. and D.H.B. designed and performed the experiments. D.Z., D.H.B., and A.B. analyzed, plotted, and interpreted the data and wrote the manuscript. A.C. and C.F. contributed to data interpretations. G.P., D.S., and K.C.D. performed brain tissue processing. E.F.C. and T.J.N. contributed to brain tissue access. L.F.L., Z.M.M., and E.R.W. contributed to additional experiments during manuscript revision. J.L. and D.V.S. supervised the project, providing insights in experimental design and data interpretation, and revised and edited the manuscript.
Competing interests: D.Z., D.H.B., J.L., and D.V.S. are inventors on a patent application related to improving packaging and diversity of AAV libraries with ML. D.H.B. and A.B. are now an employee for Dyno Therapeutics. The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Source code for data preparation, model training and evaluation, library design, analysis, and designed library specifications are available DOI: 10.5281/zenodo.10064777 (https://zenodo.org/records/10064777). The specifications for the designed libraries synthesized in our study (i.e., the position-wise nucleotide probabilities required for synthesis) are also available in the Supplementary Materials (table S1). The designed plasmid libraries D2 and D3 can be provided by D.V.S.’s group pending scientific review and a completed material transfer agreement upon reasonable request. Requests for the plasmid libraries should be submitted to schaffer@berkeleye.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S8
REFERENCES AND NOTES
- 1.Maheshri N., Koerber J. T., Kaspar B. K., Schaffer D. V., Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 24, 198–204 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Dalkara D., Byrne L. C., Klimczak R. R., Visel M., Yin L., Merigan W. H., Flannery J. G., Schaffer D. V., In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 5, 189ra76 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Tse L. V., Klinc K. A., Madigan V. J., Castellanos Rivera R. M., Wells L. F., Havlik L. P., Smith J. K., Agbandje-McKenna M., Asokan A., Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. U.S.A. 114, E4812–E4821 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel M. A., Weinstein J. R., Schaffer D. V., Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Ther. 19, 694–700 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Jang J. H., Koerber J. T., Kim J. S., Asuri P., Vazin T., Bartel M., Keung A., Kwon I., Park K. I., Schaffer D. V., An evolved adeno-associated viral variant enhances gene delivery and gene targeting in neural stem cells. Mol. Ther. 19, 667–675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimm D., Lee J. S., Wang L., Desai T., Akache B., Storm T. A., Kay M. A., In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koerber J. T., Jang J.-H., Schaffer D. V., DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol. Ther. 16, 1703–1709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojala D. S., Sun S., Santiago-Ortiz J. L., Shapiro M. G., Romero P. A., Schaffer D. V., In vivo selection of a computationally designed SCHEMA AAV library yields a novel variant for infection of adult neural stem cells in the SVZ. Mol. Ther. 26, 304–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deverman B. E., Pravdo P. L., Simpson B. P., Kumar S. R., Chan K. Y., Banerjee A., Wu W. L., Yang B., Huber N., Pasca S. P., Gradinaru V., Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santiago-Ortiz J., Ojala D. S., Westesson O., Weinstein J. R., Wong S. Y., Steinsapir A., Kumar S., Holmes I., Schaffer D. V., AAV ancestral reconstruction library enables selection of broadly infectious viral variants. Gene Ther. 22, 934–946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogden P. J., Kelsic E. D., Sinai S., Church G. M., Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science 366, 1139–1143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koerber J. T., Klimczak R., Jang J.-H., Dalkara D., Flannery J. G., Schaffer D. V., Molecular evolution of adeno-associated virus for enhanced glial gene delivery. Mol. Ther. 17, 2088–2095 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero P. A., Arnold F. H., Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne L. C., Day T. P., Visel M., Strazzeri J. A., Fortuny C., Dalkara D., Merigan W. H., Schaffer D. V., Flannery J. G., In vivo-directed evolution of adeno-associated virus in the primate retina. JCI Insight 5, e135112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adachi K., Enoki T., Kawano Y., Veraz M., Nakai H., Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun. 5, 3075 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marques A. D., Kummer M., Kondratow O., Banerjee A., Moskalenko O., Zolotukhin S., Applying machine learning to predict viral assembly for adeno-associated virus capsid libraries. Mol. Ther. Methods Clin. Dev. 20, 276–286 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant D. H., Bashir A., Sinai S., Jain N. K., Ogden P. J., Riley P. F., Church G. M., Colwell L. J., Kelsic E. D., Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 39, 691–696 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Parker A. S., Griswold K. E., Bailey-Kellogg C., Optimization of combinatorial mutagenesis. J. Comput. Biol. 18, 1743–1756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopf T. A., Ingraham J. B., Poelwijk F. J., Schärfe C. P. I., Springer M., Sander C., Marks D. S., Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma D., Grigoryan G., Bailey-Kellogg C., Pareto optimization of combinatorial mutagenesis libraries. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 1143–1153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Yang C., Hu H., Chen C., Yan M., Ling F., Wang K. C., Wang X., Deng Z., Zhou X., Zhang F., Lin S., Du Z., Zhao K., Xiao X., Directed evolution of adeno-associated virus 5 capsid enables specific liver tropism. Mol. Ther. Nucleic Acids 28, 293–306 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M. F., Masurier C., Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: Implications for gene therapy using AAV vectors. Hum. Gene Ther. 21, 704–712 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Von Drygalski A., Giermasz A., Castaman G., Key N. S., Lattimore S., Leebeek F. W. G., Miesbach W., Recht M., Long A., Gut R., Sawyer E. K., Pipe S. W., Etranacogene dezaparvovec (AMT-061 phase 2b): Normal/near normal FIX activity and bleed cessation in hemophilia B. Blood Adv. 3, 3241–3247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calcedo R., Vandenberghe L. H., Gao G., Lin J., Wilson J. M., Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J Infect Dis 199, 381–390 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ClinicalTrials. 2020, U.S. National Library of Medicine ClinicalTrials.gov.
- 26.Müller O. J., Kaul F., Weitzman M. D., Pasqualini R., Arap W., Kleinschmidt J. A., Trepel M., Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 21, 1040–1046 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Perabo L., Büning H., Kofler D. M., Ried M. U., Girod A., Wendtner C. M., Enssle J., Hallek M., In vitro selection of viral vectors with modified tropism: The adeno-associated virus display. Mol. Ther. 8, 151–157 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Zheng L., Baumann U., Reymond J.-L., An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kille S., Acevedo-Rocha C. G., Parra L. P., Zhang Z.-G., Opperman D. J., Reetz M. T., Acevedo J. P., Reducing codon redundancy and screening effort of combinatorial protein libraries created by saturation mutagenesis. ACS Synth. Biol. 2, 83–92 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Li A., Acevedo-Rocha C. G., Reetz M. T., Boosting the efficiency of site-saturation mutagenesis for a difficult-to-randomize gene by a two-step PCR strategy. Appl. Microbiol. Biotechnol. 102, 6095–6103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zolotukhin S., Byrne B. J., Mason E., Zolotukhin I., Potter M., Chestnut K., Summerford C., Samulski R. J., N., Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6, 973–985 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Matuszewski S., Hildebrandt M. E., Ghenu A.-H., Jensen J. E., Bank C., A statistical guide to the design of deep mutational scanning experiments. Genetics 204, 77–87 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz D., Baptista J., Azen S. P., Obtaining confidence intervals for the risk ratio in cohort studies. Biometrics 34, 469–474 (1978). [Google Scholar]
- 34.E. N. Weinstein, A. N.Amin, W. S. Grathwohl, D. Kassler, J. Disset, D. Marks, “Optimal design of stochastic DNA synthesis protocols based on generative sequence models” in Proceedings of the 25th International Conference on Artificial Intelligence and Statistics, vol. 151 (PMLR, 2022), pp. 7450–7452. [Google Scholar]
- 35.Tucker C. M., Cadotte M. W., Carvalho S. B., Davies T. J., Ferrier S., Fritz S. A., Grenyer R., Helmus M. R., Jin L. S., Mooers A. O., Pavoine S., Purschke O., Redding D. W., Rosauer D. F., Winter M., Mazel F., A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. Camb. Philos. Soc. 92, 698–715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Chao, C.-H. Chiu, L. Jost, “Phylogenetic diversity measures and their decomposition: A framework based on Hill numbers” in Biodiversity Conservation and Phylogenetic Systematics: Preserving Our Evolutionary Heritage in an Extinction Crisis, R. Pellens, P. Grandcolas, Eds. (Springer International Publishing, 2016), pp. 141–172. [Google Scholar]
- 37.Peng J., Xu J., Low-homology protein threading. Bioinformatics 26, i294–i300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng Y. R., Shekhar K., Yan W., Herrmann D., Sappington A., Bryman G. S., van Zyl T., Tri M., Do H., Regev A., Sanes J. R., Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell 176, 1222–1237.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wright S., Evolution in mendelian populations. Genetics 16, 97–159 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin R., Zhou Y., Yan T., Wang R., Li H., Wu Z., Zhang X., Zhou X., Zhao F., Zhang L., Li Y., Luo M., Directed evolution of adeno-associated virus for efficient gene delivery to microglia. Nat. Methods 19, 976–985 (2022). [DOI] [PubMed] [Google Scholar]
- 41.He T., Itano M. S., Earley L. F., Hall N. E., Riddick N., Samulski R. J., Li C., The influence of murine genetic background in adeno-associated virus transduction of the mouse brain. Hum. Gene. Ther. Clin. Dev. 30, 169–181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.La Manno G., Gyllborg D., Codeluppi S., Nishimura K., Salto C., Zeisel A., Borm L. E., Stott S. R. W., Toledo E. M., Villaescusa J. C., Lönnerberg P., Ryge J., Barker R. A., Arenas E., Linnarsson S., Molecular diversity of midbrain development in mouse, human, and stem cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathiesen S. N., Lock J. L., Schoderboeck L., Abraham W. C., Hughes S. M., CNS transduction benefits of AAV-PHP.eB over AAV9 are dependent on administration route and mouse strain. Mol. Ther. Methods Clin. Dev. 19, 447–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lui J. H., Hansen D. V., Kriegstein A. R., Development and evolution of the human neocortex. Cell 146, 18–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong L., Li B., Mah C. S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R. W., Zolotukhin I., Warrington K. H. Jr., Weigel-Van Aken K. A., Hobbs J. A., Zolotukhin S., Muzyczka N., Srivastava A., Next generation of adeno-associated virus 2 vectors: Point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. U.S.A. 105, 7827–7832 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markusic D. M., Herzog R. W., Aslanidi G. V., Hoffman B. E., Li B., Li M., Jayandharan G. R., Ling C., Zolotukhin I., Ma W., Zolotukhin S., Srivastava A., Zhong L., High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 18, 2048–2056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aslanidi G. V., Rivers A. E., Ortiz L., Govindasamy L., Ling C., Jayandharan G. R., Zolotukhin S., Agbandje-McKenna M., Srivastava A., High-efficiency transduction of human monocyte-derived dendritic cells by capsid-modified recombinant AAV2 vectors. Vaccine 30, 3908–3917 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrs-Silva H., Dinculescu A., Li Q., Min S.-H., Chiodo V., Pang J.-J., Zhong L., Zolotukhin S., Srivastava A., Lewin A. S., Hauswirth W. W., High-efficiency transduction of the mouse retina by tyrosine-mutant AAV serotype vectors. Mol. Ther. 17, 463–471 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aslanidi G. V., Rivers A. E., Ortiz L., Song L., Ling C., Govindasamy L., Vliet K. V., Tan M., Agbandje-McKenna M., Srivastava A., Optimization of the capsid of recombinant adeno-associated virus 2 (AAV2) vectors: The final threshold? PLoS One 8, e59142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Carroll S. J., Cook W. H., Young D., AAV targeting of glial cell types in the central and peripheral nervous system and relevance to human gene therapy. Front. Mol. Neurosci. 13, 618020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halliday G. M., Holton J. L., Revesz T., Dickson D. W., Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol. 122, 187–204 (2011). [DOI] [PubMed] [Google Scholar]
- 52.Miyazaki I., Asanuma M., Neuron-astrocyte interactions in Parkinson’s disease. Cells 9, 2623 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Batard P., Jordan M., Wurm F., Transfer of high copy number plasmid into mammalian cells by calcium phosphate transfection. Gene 270, 61–68 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Schmit P. F., Pacouret S., Zinn E., Telford E., Nicolaou F., Broucque F., Andres-Mateos E., Xiao R., Penaud-Budloo M., Bouzelha M., Jaulin N., Adjali O., Ayuso E., Vandenberghe L. H., Cross-packaging and capsid mosaic formation in multiplexed AAV libraries. Mol. Ther. Methods Clin. Dev. 17, 107–121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elliott A. G., Merenbloom S. I., Chakrabarty S., Williams E. R., Single particle analyzer of mass: A charge detection mass spectrometer with a multi-detector electrostatic ion trap. Int. J. Mass Spectrom. 414, 45–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harper C. C., Miller Z. M., McPartlan M. S., Jordan J. S., Pedder R. E., Williams E. R., Accurate sizing of nanoparticles using a high-throughput charge detection mass spectrometer without energy selection. ACS Nano 17, 7765–7774 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller Z. M., Harper C. C., Lee H., Bischoff A. J., Francis M. B., Schaffer D. V., Williams E. R., Apodization specific fitting for improved resolution, charge measurement, and data analysis speed in charge detection mass spectrometry. J. Am. Soc. Mass Spectrom. 33, 2129–2137 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R. W. Keener, Theoretical Statistics: Topics for a Core Course (Springer, 2010). [Google Scholar]
- 59.Gelman S., Fahlberg S. A., Heinzelman P., Romero P. A., Gitter A., Neural Networks to learn protein sequence-function relationships from deep mutational scanning data. Proc. Natl. Acad. Sci. U.S.A. 118, e2104878118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. Abadi, P. Barham, J. Chen, Z. Chen, A. Davis, J. Dean, M. Devin, S. Ghemawat, G. Irving, M. Isard, M. Kudlur, J. Levenberg, R. Monga, S. Moore, D. G. Murray, B. Steiner, P. Tucker, V. Vasudevan, P. Warden, M. Wicke, Y. Yu, X. Zheng, “TensorFlow: A system for large-scale machine learning” in Proceedings of the 12th USENIX Conference on Operating Systems Design and Implementation (USENIX Association, 2016), pp. 265–283. [Google Scholar]
- 61.D. P. Kingma, J. Ba, Adam: A method for stochastic optimization. arXiv:1412.6980; https://arxiv.org/pdf/1412.6980.pdf [accessed 18 September 2022].
- 62.Tuomisto H., A diversity of beta diversities: Straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography 33, 2–22 (2010). [Google Scholar]
- 63.D. J. C. MacKay, Information Theory, Inference and Learning Algorithms (Cambridge Univ. Press, 2003). [Google Scholar]
- 64.Jaynes E. T., Information theory and statistical mechanics. Phys. Rev. 106, 620–630 (1957). [Google Scholar]
- 65.K. P. Murphy, Machine Learning: A Probabilistic Perspective (MIT Press, 2012). [Google Scholar]
- 66.J. Kleijnen, R. Y. Rubinstein, Optimization and sensitivity analysis of computer simulation models by the score function method (Tilburg University, Center for Economic Research, 1995).
- 67.Ting J. T., Kalmbach B., Chong P., de Frates R., Keene C. D., Gwinn R. P., Cobbs C., Ko A. L., Ojemann J. G., Ellenbogen R. G., Koch C., Lein E., A robust ex vivo experimental platform for molecular-genetic dissection of adult human neocortical cell types and circuits. Sci. Rep. 8, 8407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arad U., Modified Hirt procedure for rapid purification of extrachromosomal DNA from mammalian cells. Biotechniques 24, 760–762 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S8