Abstract
In neurological and neuropsychiatric diseases, different brain regions are affected, and differences in gene expression patterns could potentially explain this mechanism. However, limited studies have precisely explored gene expression in different regions of the human brain. In this study, we performed long-read RNA sequencing on three different brain regions of the same individuals: the cerebellum, hypothalamus, and temporal cortex. Despite stringent filtering criteria excluding isoforms predicted to be artifacts, over half of the isoforms expressed in multiple samples across multiple regions were found to be unregistered in the GENCODE reference. We then especially focused on genes with different major isoforms in each brain region, even with similar overall expression levels, and identified that many of such genes including GAS7 might have distinct roles in dendritic spine and neuronal formation in each region. We also found that DNA methylation might, in part, drive different isoform expressions in different regions. These findings highlight the significance of analyzing isoforms expressed in disease-relevant sites.
Using long reads, over half the detected brain isoforms are unregistered, and region-specific isoforms influence neurite formation.
INTRODUCTION
Brain disorders such as neurodegenerative diseases and psychiatric disorders are multifactorial diseases with complex etiologies that contribute significantly to human morbidity and mortality. In neuropsychiatric disorders, the brain regions associated with each disease vary and different neurodegenerative diseases and their clinical symptoms are associated with distinctive neurodegenerative systems (1). Although pathology studies suggested the involvement of several factors in this site selectivity, such as changes in neurotransmitters, protein homeostasis, and energy demand, the detailed mechanism is still unclear, and other changes that are not measurable by histological techniques, such as synaptic connections or gene expression patterns, may also be involved (1). Previous studies on psychiatric disorders examined gene expression in postmortem brain samples of patients with schizophrenia and bipolar disorder. These studies identified altered expression of γ-aminobutyric acid–related genes in the superior temporal gyrus and hippocampus in both disorders (2–4) and anterior cingulate cortex–specific expression changes in schizophrenia (5). Identifying and understanding the specific genes and gene expression patterns in each brain region may provide crucial insights into the underlying mechanisms of various neurological and psychiatric diseases.
Previous studies on gene expression analysis in the brain have revealed not only genes involved in brain disorders but also transcripts that are altered by factors such as sleep status and aging (6–8). However, most of these analyses were based on short-read sequence or microarray data, and their isoforms were only inferred from sequences with short read lengths. Recent advances in transcriptome technology using long-read sequencing allow us to read the entire length of the transcript from the 5′ to the 3′ untranslated region (UTR) and polyA tail at once, enabling us to capture more complex and precise transcript features without phasing problems (9). Previous studies have reported that tissue-specific transcripts are more abundant in the brain compared to other organs (10) and that their expression is also elevated overall (11), suggesting that the brain transcriptome is considered to be relatively specific. A recent study analyzed the single-cell long reads of cells from various regions of the postmortem mouse brain and reported region-specific expression of isoforms, which is very important in unraveling the complex integration of cells in the brain (12). While studies using animal models and cells are invaluable, many studies have investigated human brain site–specific isoforms (6, 11, 13–17), and numerous unregistered isoforms and tissue- and age-specific gene expression patterns have been detected in studies that have analyzed postmortem human brain samples using long-read methods (18, 19). However, the findings of these studies do not provide a lot of information on the regional specificity of the brain.
Here, we performed long-read sequencing of RNA extracted from three regions of the postmortem human brain—the temporal cortex, hypothalamus, and cerebellum (cerebellar cortex)—with a focus on characteristics of region-specific transcriptomes. The temporal cortex is involved in language, memory, and hearing (20–22) and has been reported to be associated with schizophrenia (23),Alzheimer’s disease (24), and temporal lobe epilepsy (25). The hypothalamus is a critical brain region that regulates endocrine and autonomic functions (26, 27). It is composed of numerous neuronal nuclei and coordinates and manages diverse physiological functions, including thermoregulation, stress response, feeding behavior, and sleep arousal. The cerebellum is primarily involved in the integration of sensory and motor functions and is associated with several developmental disorders (28, 29). The purpose of this study was to elucidate the region-dependent gene and isoform expression patterns in the human brain and to obtain data that can be used as a control for future disease research. We analyzed three regions of the postmortem brains of four male subjects who had no brain lesions or infections and died in their 50s or later. Our analysis is not single-cell analysis, so when differences are observed between regions, it is challenging to determine whether it is due to differences in the composition of the cells or if it is due to region-specific effects; however, to accurately capture the features that reflect distinct cell compositions between at least different brain regions, we attempted to minimize the influence of genomic sequences and other environmental factors by making comparisons among different regions from the same samples. In addition, epigenetics may play a significant role in intraindividual expression differences. It is well known that the methylation of CpG islands in the promoter region typically results in silencing of gene expression and that gene-body methylation is associated with higher levels of gene expression (30–32). DNA methylation has been also reported to be involved in alternative splicing either by acting as an indicator of an alternative intragenic promoter (33) or by recruiting methyl CpG binding protein 2 (MeCP2) to promote exon recognition (34, 35). In this study, we performed a combined analysis with long-read RNA sequencing (Iso-Seq) and DNA methylation data obtained from the same regions in the same samples to examine the effect of DNA methylation on alternative splicing.
RESULTS
Overview of the full-length transcriptome of three brain regions
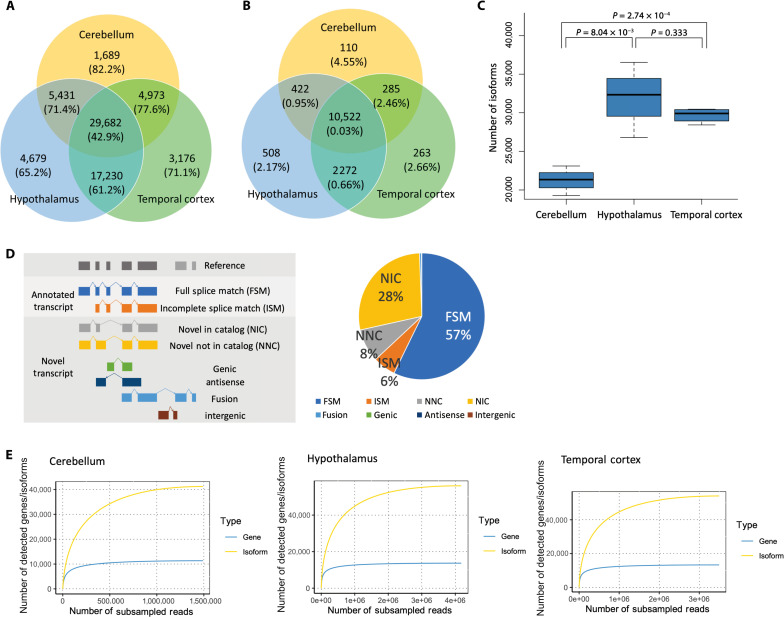
To characterize the profiles of full-length isoforms in different human brain regions, we extracted RNA from the cerebellum, hypothalamus, and temporal cortex and performed Iso-Seq using PacBio’s Sequel IIe system (fig. S1A). Quality control (QC) results for raw sequencing data are described in table S1. In all samples, more than 80% of the zero-mode waveguides, the small pores through which each library is sequenced, met all requirements of the Sequel IIe system (table S1). For each sample, we generated 225 to 401 Gb of raw sequencing data and 7 to 14 Gb of HiFi data satisfying >3 full-pass sub-reads and quality value (QV) ≥20 (table S2). Reads supported by at least two full-length non-concatemer (FLNC) reads with accuracy >99% were mapped to the human genome (hg38) and unique isoforms were clustered using IsoSeq v3 software (fig. S1B). We checked the number of FL, FLNC, and FLNC with polyA reads and confirmed that all sample reads were ≥2,000,000 and that the read number was not related to the RNA quality number (RQN) (fig. S2). After additional QC and filtering out isoforms like nonsense-mediated decay and false cDNA molecules resulting from reverse transcriptase template switching using SQANTI3, an average of 27,643 isoforms per sample and 66,860 nonredundant isoforms in which each isoform is unique, with no duplicates, across all samples were identified (Fig. 1A and table S3). These isoforms were annotated to 14,382 known and 52 unregistered genes (Fig. 1B). When compared by brain region, the number of isoforms was significantly lower in the cerebellum than in the hypothalamus and temporal cortex (Ce versus Hy: P = 8.04 × 10−3, Ce versus Tc: P = 2.74 × 10−4) (Fig. 1C). The proportion of unregistered isoforms commonly expressed in the three regions was 42.9%, whereas the proportion of unregistered isoforms expressed at only one or two of the regions was higher (61.2 to 82.2%) (Fig. 1A).
Fig. 1. Isoforms detected in the human cerebellum, hypothalamus, and temporal cortex using Iso-Seq.
(A) Number of isoforms detected in the cerebellum, hypothalamus, and temporal cortex and the percentage of unregistered isoforms indicated in parentheses. (B) Number of genes detected in the three brain regions and the percentage of unregistered genes indicated in parentheses. (C) Number of detected isoforms by brain region. The P value was calculated using a two-tailed t test. (D) Definition of isoform categories and breakdown of detected isoforms. FSM: isoform with all splice junctions matching the reference; ISM: isoform with fewer exons than existing splice junctions, but matching the known splice junctions, NIC: isoform with combinations of known donor-acceptor sites; NNC: isoform with at least one unregistered donor-acceptor site; genic: within the known genes; antisense: reverse stranding of known gene regions; fusion: fusion gene; intergenic: outside of known genetic regions. (E) Rarefaction curves of genes and transcripts detected in each brain region.
We then compared the detected isoforms to those in the comprehensive human gene annotation database GENCODE v.41 and categorized them into the following eight categories: full splice match (FSM), incomplete splice match (ISM), novel not in catalog (NNC), novel in catalog (NIC), fusion, genic, antisense, and intergenic (Fig. 1D). Since the number of fusion, genic, antisense, and intergenic isoforms was very small, we focused mainly on FSM, ISM, NIC, and NNC isoforms. In the data of this study, approximately 57.3% of the isoforms were FSM, 27.9% were NIC isoforms, and 8.5% were NNC isoforms. ISM did not account for a large proportion (5.8%) of the identified isoforms (Fig. 1D), which may be due to the strict filtering applied to ISM isoforms, as these are difficult to distinguish from degradation products. QC of SQANTI3 with these final isoforms showed that only ISM isoforms had a “Coverage Cage” of around 30 to 60% (fig. S3). “Coverage Cage” is one of the indicators used for good quality isoforms and a measure of whether a peak is detected by the CAGE method within 50 base pairs (bp). All good quality indicators for the other categories generally exceeded 75%, which assured the quality of the filtered isoform (fig. S3).
Next, we performed rarefaction analysis to examine whether isoforms and genes were adequately detected in the sequencing analyses performed in this study. The analysis performed for each gene and isoform showed that the gene rarefaction curves were well saturated in all three brain regions (Fig. 1E). Since the isoform rarefaction curves were also mostly saturated in all three brain regions, but the degree of saturation was not as high as that of the genes, it is possible that some previously unidentified isoforms could be identified if the depth of sequencing is increased further (Fig. 1E). When analyzed by isoform category, the curves were generally saturated in all regions and in all categories (fig. S4).
Furthermore, we performed cluster analysis and principal components analysis (PCA) to examine the relationship between the isoform profiles of each sample. In terms of transcription profiles, the cerebellum differed from the hypothalamus and the temporal cortex, while the hypothalamus and the temporal cortex did not differ much (fig. S5, A and B). Since the transcriptome profile was expected to be markedly affected by the proportion of cells constituting the sample, we estimated the proportion of cells. On the basis of our current understanding, there has not been a deconvolution method that has undergone verification for its estimation accuracy specifically for long-read RNA sequencing data. Consequently, we estimated cell components using MeDeCom, a reference-free estimation method (36), using genome-wide DNA methylation data obtained from the same samples used for the Iso-Seq analysis. The estimation results were not homogeneous across samples in the hypothalamus, and some samples were found to have a similar cellular composition to the temporal cortex (fig. S5C).
Comparison of isoform features between categories and brain regions
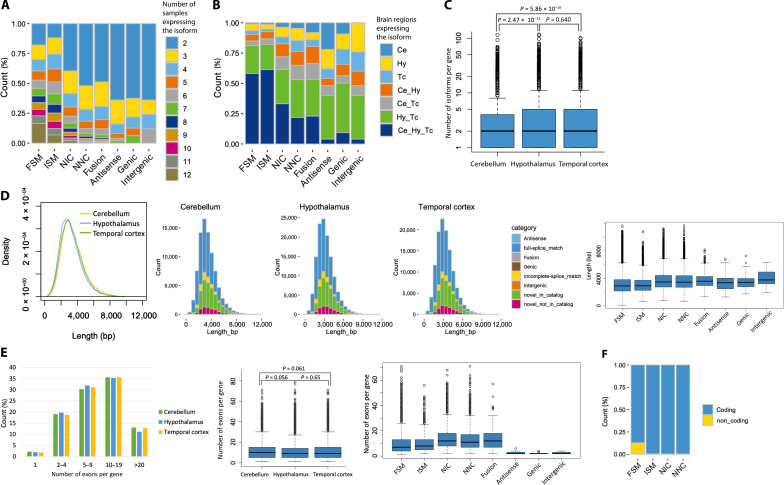
We investigated the characteristics of isoforms and compared them between isoform categories and brain regions. First, we examined how many samples expressed isoforms in each of the eight categories. Since the isoforms that were expressed in only one sample were removed to consider more plausible isoforms, those in the analysis were expressed in at least two samples. Compared to unregistered isoforms, the proportion of FSM isoforms was higher in many samples (average 6.51), and the number of samples detected decreased with increasing degree of novelty, ISM (average 6.12, P = 4.80 × 10−16), NIC (average 4.05, P < 2.2 × 10−16), and NNC (average 3.29, P < 2.2 × 10−16) (Fig. 2A). As for brain regions, isoforms expressed in all three regions were most abundant in ISM, followed by FSM; compared to FSM, NIC, and NNC had significantly higher percentages of isoforms expressed in only one or two regions [NIC: P < 2.2 × 10−16, odds ratio (OR) = 2.79, NNC: P < 2.2 × 10−16, OR = 4.97] (Fig. 2B). Expression levels [transcripts per million (TPM)] tended to be higher for FSM (fig. S6A) and for isoforms that were expressed in more brain regions (fig. S6B).
Fig. 2. Overview of the detected transcripts by category or brain region.
(A) Distribution of the number of samples in which the detected isoforms were found. Only isoforms that were expressed in at least two samples were included in subsequent analyses. (B) Distribution of the brain regions in which the detected isoforms were found. Ce, cerebellum; Hy, hypothalamus; Tc, temporal cortex. (C) Number of isoforms per gene in each brain region. The P value was calculated using a two-tailed t test. (D) Distribution of the read lengths of all detected isoforms by brain region. In the major four categories, NIC and NNC showed a tendency to have longer lengths. (E) Distribution of the number of exons per gene. The P value was calculated using a two-tailed t test. (F) Percentage of estimated protein-coding isoforms.
Next, the number of isoforms per gene was examined and an average of 2.74 isoforms were identified (Fig. 2C). When compared by region, the number of isoforms per gene was significantly lower in the cerebellum than in the other regions (Ce versus Hy: P = 2.47 × 10−11, Ce versus Tc: P = 5.86 × 10−10) (Fig. 2C). We then examined the distribution of isoform lengths. Isoform read lengths peaked at around 3 kb and tended to be slightly shorter in the hypothalamus (Fig. 2D and fig. S7A). When examining the length of each isoform by category, unregistered isoforms were longer in all brain regions (P < 2.2 × 10−16), as reported in previous studies (Fig. 2D and fig. S7, B and C) (18, 37). When examining the number of exons per gene in each brain region, the number was higher in the cerebellum (average 11.1 per gene) and significantly lower in the hypothalamus (average 10.6 per gene, P < 2.2 × 10−16) (Fig. 2E). When compared by category, the distribution of the number of exons per gene was significantly different between FSM (average 9.32 per gene) and unregistered isoforms (Fig. 2E), especially NIC (average 13.5 per gene, P < 2.2 × 10−16) and NNC (average 12.8 per gene, P < 2.2 × 10−16), which was consistent between regions (fig. S8). When we examined the proportion of protein-coding isoforms, the results showed that 13.0% of the FSMs were noncoding, while noncoding isoforms of the other unregistered isoforms were ~0.5%, indicating that most of the unregistered isoforms were predicted to be protein-coding (Fig. 2F).
To validate the unregistered isoforms detected in this study, we then used proteome analysis data of the postmortem human brain cortex reported in a previous study (38). Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) results were used to detect false discovery rate (FDR) < 0.01 proteins with both unregistered isoform sequences and known protein sequences from GENCODE as a reference. Among the amino acid sequences used to detect unregistered isoforms, those were not detected when using known protein sequences as a reference and not identical to any known sequences were searched. We identified 37 amino acid sequences derived from 34 proteins (table S4), which is evidence of translated unregistered isoforms. Alternative splicing events such as exon skipping (fig. S9A) and intron retention (fig. S9B) of NIC and unregistered splice junction of NNC (fig. S9C) were validated by the identified amino acid sequences.
Comparison of genes and isoforms between brain regions
We first examined genes that were highly expressed throughout the brain. The TPM for each gene is shown in table S5. There were 201 genes with TPM >200, of which myelin basic protein (MBP) was the most highly expressed gene throughout the brain, and pathway analysis using these genes detected numerous pathways for nervous and brain development and neurogenesis (fig. S10A and table S6). Similar trends were observed when we examined brain regions separately and many of the genes that were highly expressed in each region were related to brain and nervous system pathways (tables S7 to S9).
Next, we searched for genes that were only expressed in each respective region. Very few genes were expressed in only one of the three brain regions, and only 42 genes met our criteria for the cerebellum (table S10), 25 in the hypothalamus (table S11), and 44 in the temporal cortex (table S12). Pathway analysis using these region-specific genes detected no significant pathways in the analyses of the cerebellum and hypothalamus, while synaptic signaling-related pathways were detected in the analysis of the temporal cortex (table S13).
Since Iso-Seq was considered to be less suitable for differential expression analysis than short read analysis (39), we searched for genes that were differentially expressed in each brain region based not only on statistical significance but also on the strict criterion of fold change (FC) >5 (fig. S11 and tables S14 to S16). The number of genes up-regulated in the cerebellum according to our criteria was 58 (fig. S11A and table S14), including zinc finger protein of the cerebellum (ZIC) family members ZIC1, ZIC3, and ZIC4, and pathway analysis showed that the top pathways included “neuronal pathways,” “ion transport,” and “brain development” (fig. S10B and table S17). Similar pathways involved in neurogenesis were detected in the analysis using 77 up-regulated genes in the temporal cortex (figs. S10D and S11C and tables S16 and S19). In the hypothalamus, on the other hand, 53 genes with elevated expression were detected (table S15), and many of these genes were involved in the “immune response” and “cell migration” pathways (figs. S10C and S11B and table S18). As for the site-specific down-regulated genes, only 25 were detected in the hypothalamus and 4 in the temporal cortex, whereas 314 were detected in the cerebellum (tables S14 to S16). Pathway analysis showed that many of the cerebellum-dependent genes with decreased expression were involved in “actin filament-based process,” “anatomical structure,” and “cellular component size” (fig. S10E and table S17).
We then focused on genes with different numbers of isoforms per gene. First, we searched for genes expressing more than 20 isoforms per gene in the whole brain and detected 488 genes (table S20). Pathway analysis using these genes identified “macromolecule and protein localization” and “cell projection organization” pathways in addition to brain-specific pathways such as “neurogenesis” (fig. S12A and table S21). To investigate whether this trend was brain-specific, we used long-read data from the GTEx (19) to detect genes with high isoform counts in cells and tissues other than the brain and performed a pathway analysis using these genes. The results showed that five of the top 10 pathways, including “macromolecule and protein localization,” were detected in the analysis of tissues other than the brain and that the genes involved in these pathways generally expressed numerous isoforms (fig. S12A and table S21). Next, we searched for genes with a particularly high number of isoforms per gene in the regions of interest compared to other regions and found 34 genes in the cerebellum, 136 genes in the hypothalamus, and 77 genes in the temporal cortex (tables S22 to S24). No significant pathways were detected in the analysis of genes that had a higher number of isoforms per gene in the cerebellum, but these genes included ZIC3 and ZIC4, which were also detected as cerebellum-dependent expressed genes (table S22). Pathway analyses of the hypothalamus and temporal cortex detected such pathways as “membrane and cytoskeleton organization” and “immune response” in the hypothalamus and “neuron differentiation” and “cell projection organization” in the temporal cortex, respectively (fig. S12, B and C, and tables S25 and S26). The correlation between the number of isoforms per gene and expression levels was not strong (r2 = 0.127; fig. S13); however, pathways related to the immune response in the hypothalamus and neurogenesis in the temporal cortex were detected not only for genes that were highly expressed in a region-dependent manner but also for genes with a high number of isoforms.
Genes with different major isoforms in different brain regions
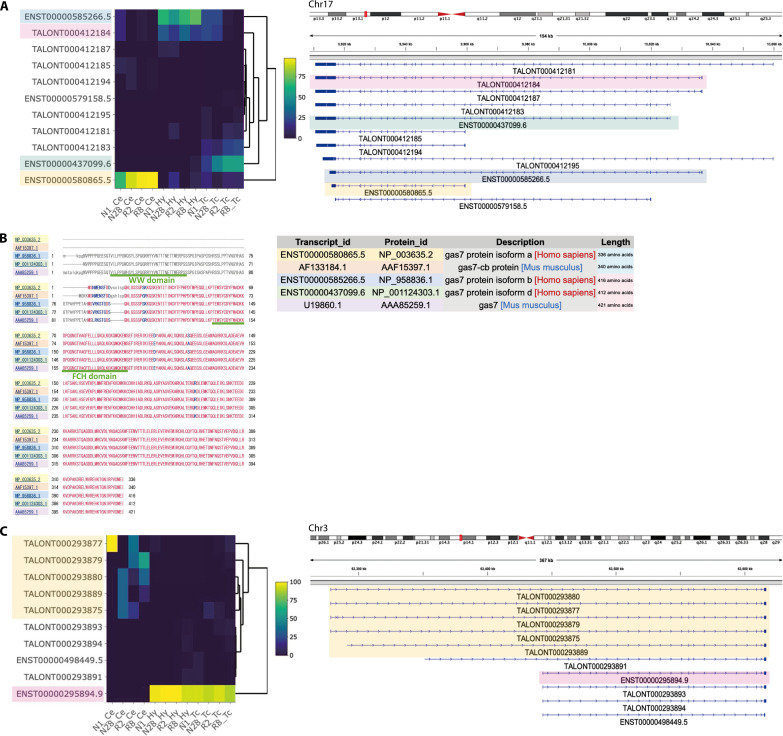
Next, we searched for genes with different major isoforms in different regions. For each sample, the ratio (%) of the expression of individual isoforms to total gene expression was calculated, and the ratios and their ranks were compared among regions. We used both parametric and nonparametric tests to search for isoforms with ratios that differed by more than 20% (P < 0.05), and with ranks of expression that differed between regions. As a result, 193 isoforms of 145 genes were identified in the cerebellum versus the hypothalamus, 147 isoforms from 114 genes were identified in the cerebellum versus the temporal cortex, and 55 isoforms from 47 genes were identified in the hypothalamus versus the temporal cortex (Table 1 and tables S27 to S29). Among the genes for which isoforms were detected in all three comparisons, growth arrest–specific 7 (GAS7) was the gene with the lowest P value (tables S27 to S29). The transcripts ENST00000580865.5 and ENST00000437099.6 were predominantly expressed in the cerebellum and temporal cortex, respectively (Fig. 3A). In the hypothalamus, both ENST00000585266.5 and an unregistered isoform, TALONT000412184, were expressed (Fig. 3A). The predominance of the shorter isoform, ENST00000580865.5, in the cerebellum was replicated in the GTEx brain samples (fig. S14) (19). In a previous study using Western blotting in mouse brains, different isoforms of GAS7 that were predominantly expressed in the cerebellum and cerebral cortex were reported (40). Therefore, we compared the predicted amino acid sequence encoded by the isoforms detected in this study with those of mice. As a result, the longer isoform, Gas7, expressed in the mouse cerebrum showed 97.33% identity with ENST00000585266.5 detected in the hypothalamus and 97.35% identity with ENST00000437099.6 detected in the temporal cortex. On the other hand, the shorter isoform, Gas7-cb, expressed in the mouse cerebellum showed 97.9% identity with ENST00000580865.5 detected in the cerebellum (Fig. 3B).
Table 1. Top 10 isoforms for which the proportion of gene expression differs depending on the brain region.
Ce, cerebellum; Hy, hypothalamus; Tc, temporal cortex.
| Ce versus Hy | Ce versus Tc | Hy versus Tc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Average TPM (%)* |
t test | Gene | Average TPM (%)* |
t test | Gene | Average TPM (%)* |
t test | ||||||
| Isoform name | Symbol | Ce | Hy | P value | Isoform name | Symbol | Ce | Tc | P value | Isoform name | Symbol | Hy | Tc | P value |
| ENST00000295894.9 | SYNPR | 0.0 | 97.7 | 9.7 × 10−06 |
ENST00000645548.2 | KIFC2 | 11.7 | 51.5 | 4.5 × 10−06 | TALONT000372117 | SCN4B | 3.2 | 33.4 | 2.0 × 10−05 |
| ENST00000448602.5 | ELMO1 | 9.5 | 44.8 | 4.8 × 10−05 |
ENST00000295894.9 | SYNPR | 0.0 | 91.8 | 7.9 × 10−06 | ENST00000585266.5 | GAS7 | 64.0 | 12.3 | 9.1 × 10−04 |
| ENST00000585266.5 | GAS7 | 4.0 | 64.0 | 6.4 × 10−05 |
TALONT000280002 | UBXN4 | 54.5 | 14.4 | 5.4 × 10−05 | TALONT000434444 | CPNE1 | 27.0 | 3.0 | 9.6 × 10−04 |
| TALONT000280002 | UBXN4 | 54.5 | 20.3 | 1.4 × 10−04 |
ENST00000272638.14 | UBXN4 | 32.3 | 80.8 | 7.4 × 10−05 | TALONT000265350 | IGSF8 | 26.6 | 0.0 | 1.1 × 10−03 |
| ENST00000418337.6 | PRKAG2 | 10.0 | 64.5 | 1.4 × 10−04 |
ENST00000496484.3 | MRPS25 | 48.1 | 16.8 | 1.0 × 10−04 | TALONT000384069 | ZNF10 | 67.4 | 45.3 | 1.6 × 10−03 |
| ENST00000272638.14 | UBXN4 | 32.3 | 73.9 | 1.6 × 10−04 |
ENST00000222792.11 | CHN2 | 71.1 | 17.1 | 1.0 × 10−04 | TALONT000333397 | KRIT1 | 3.1 | 25.6 | 1.8 × 10−03 |
| ENST00000412711.6 | CHN2 | 7.5 | 60.6 | 1.8 × 10−04 |
TALONT000346238 | PHF24 | 0.0 | 59.3 | 1.4 × 10−04 | TALONT000306327 | CAMK2D | 0.7 | 34.0 | 2.3 × 10−03 |
| ENST00000222792.11 | CHN2 | 71.1 | 22.9 | 2.1 × 10−04 |
ENST00000615255.1 | DIXDC1 | 9.7 | 74.6 | 5.7 × 10−04 | ENST00000351424.8 | TPD52L2 | 5.4 | 42.9 | 2.6 × 10−03 |
| ENST00000580865.5 | GAS7 | 87.5 | 5.5 | 2.5 × 10−04 |
TALONT000306327 | CAMK2D | 2.7 | 34.0 | 6.1 × 10−04 | ENST00000264312.12 | OCIAD1 | 42.6 | 22.2 | 3.5 × 10−03 |
| ENST00000370558.8 | SH3GLB1 | 21.6 | 77.8 | 2.6 × 10−04 |
ENST00000249786.9 | SERF2 | 20.6 | 60.4 | 6.3 × 10−04 | ENST00000261168.9 | ATF7IP | 56.9 | 16.4 | 3.9 × 10−03 |
*Average percentage of expression (TMP) accounted for by isoform within a gene.
Fig. 3. Examples of a gene that uses different major isoforms depending on the brain region.
(A) GAS7 is an example of a gene with different major isoforms at each region. The heatmap represents the proportion of expression levels of each isoform relative to the overall expression levels of the gene. The colors on the transcript IDs correspond to the colors of the transcript structures displayed in IGV. (B) Comparison of predicted amino acid sequences between transcripts detected in this study and those reported to be expressed in the mouse cerebellum (AF133184.1/AAF15397.1) and cerebral cortex (U19860.1/AAA85259.1). (C) An example of SYNPR found to have isoforms that are the most significantly differentially expressed between Ce and Hy and were also the second most significant when comparing Ce and Tc.
The isoform with the lowest P value in the cerebellar and hypothalamic comparison was that of synaptoporin (SYNPR), which was one of the top two isoforms in the cerebellar and temporal cortex analysis (tables S27 and S28). ENST00000295894.9 was expressed in the hypothalamus and the temporal cortex, whereas the cerebellum expressed five new, longer isoforms with transcription start sites (TSSs) more than 150 kb upstream of ENST00000295894.9 (Fig. 3C).
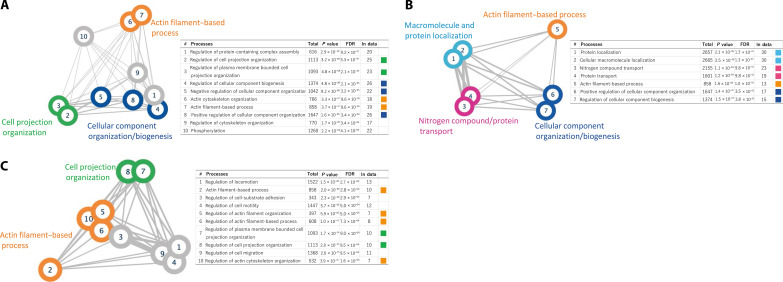
By analyzing the genes that expressed different major isoforms between the brain regions, we identified pathways such as “actin filament-based process” and “cell projection organization” consistently across multiple comparisons (Fig. 4 and tables S30 to 32).
Fig. 4. Pathway analysis of genes expressing different major isoforms in different regions of the brain.
When more than 10 pathways are detected, the top 10 pathways with the smallest FDR are shown. Edge thickness is proportional to the number of related genes shared among pathways. (A) Pathway analysis results for genes with different major isoforms expressed in the cerebellum and hypothalamus. (B) Pathway analysis results for genes with different major isoforms expressed in the cerebellum and temporal cortex. (C) Pathway analysis results for genes with different major isoforms expressed in the hypothalamus and temporal cortex.
Integrative analysis of transcriptome and methylation data
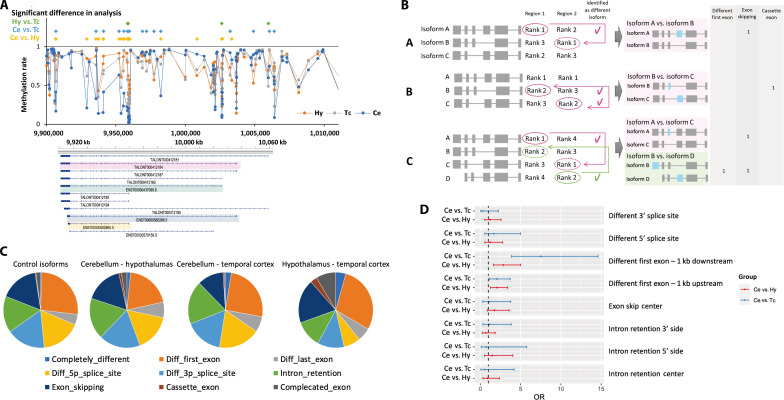
We identified several genes that expressed different isoforms in different brain regions. The samples used in this study were composed of different brain regions of the same samples and there were no differences in genomic sequences (e.g., single-nucleotide polymorphisms) among regions, and the effects of environmental factors were also expected to be similar between regions. Therefore, considering the possibility that epigenetics may play an important role as a mechanism for the expression of different isoforms within a gene, we focused on DNA methylation. Using data from a previous study (41) in which we performed array-based whole-genome methylation analyses using DNA extracted from the same samples used in this study, we first searched for differentially methylated positions (DMPs) between regions and identified 66,908, 32,841, and 8752 DMPs in comparisons between the cerebellum and the hypothalamus, the cerebellum and the temporal cortex, and the hypothalamus and the temporal cortex, respectively. We examined whether these DMPs were abundant in the vicinity of genes with different major isoforms in the different regions. The results showed that DMPs were significantly more abundant around genes with different major isoforms between the regions than around all of the expressed genes in the brain in this study (Ce versus Hy; P = 3.87 × 10−07, Ce versus Tc; P = 4.48 × 10−05, Hy versus Tc; P = 4.51 × 10−02) (Table 2).
Table 2. DMPs around genes with different major isoforms expressed in each region.
| Comparison | Genes with different major isoforms |
All genes expressed in the brain |
Fisher’s exact test |
Odds ratio (OR) | ||||
|---|---|---|---|---|---|---|---|---|
| 95% CI | ||||||||
| annotated | not_annotated | annotated | not_anno tated |
P value | Lower | Upper | ||
| Ce versus Hy | 99 | 46 | 6,785 | 7,599 | 3.87 × 10−07 | 2.41 | 1.70 | 3.43 |
| Ce versus Tc | 62 | 52 | 5,074 | 9,308 | 4.48 × 10−05 | 2.19 | 1.51 | 3.17 |
| Hy versus Tc | 11 | 36 | 1,841 | 12,541 | 4.51 × 10−02 | 2.08 | 1.06 | 4.10 |
When we examined the areas where these DMPs were present, we found that DMPs were more common in the region 200 to 1500 bp upstream of TSS, 5′ UTR, and first exon (Table 3). We also found that DMPs were abundant in the enhancer regions and transcription factor (TF) binding sites; however, the relationship between DMPs and CpG islands was not strong (Table 3). Given the existence of many DMPs around genes with different major isoforms between regions, we examined methylation around GAS7 and found that there were many DMPs, especially around TSSs where isoforms of different lengths were expressed (Fig. 5A). Since our methylation analysis was based on an array, for the GAS7 region, we used data from a previous study (42) that performed bisulfite sequencing on neurons derived from the cortex and the cerebellum to examine more densely populated methylation sites. As a result, there was a strong correlation between the methylation rate in our samples and the methylation rate based on bisulfite sequencing (our cerebellum samples versus neurons derived from the cerebellum r2 = 0.92, our temporal cortex samples versus neurons derived from the cortex r2 = 0.73) (fig. S15, A and B). When we examined the relationship between the sequence-based methylation rate and isoforms, similar to the results from the array-based method, sites with large methylation differences between neurons derived from the cerebellum and those from the cortex were often found near the TSSs of isoforms that have different expressions between the regions (fig. S15C).
Table 3. Characteristics of areas with DMPs around genes with different major isoforms in each region.
TFBS, transcription factor binding site; DNase, deoxyribonuclease.
| Group | Ce versus Hy | Ce versus Tc | Hy versus Tc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rate 1* | Rate 2† | P value | OR | Rate 1* | Rate 2† | P value | OR | Rate 1* | Rate 2† | P value | OR | ||
| Relation to gene |
TSS1500 | 0.448 | 0.241 | 7.84 × 10−08 | 2.56 | 0.272 | 0.160 | 2.88 × 10−03 | 1.96 | 0.128 | 0.041 | 1.20 × 10−02 | 3.45 |
| TSS200 | 0.172 | 0.107 | 1.50 × 10−02 | 1.74 | 0.114 | 0.072 | 9.94 × 10−02 | 1.66 | 0.106 | 0.023 | 4.57 × 10−03 | 5.05 | |
| 5’ UTR | 0.455 | 0.266 | 1.12 × 10−06 | 2.31 | 0.333 | 0.195 | 5.05 × 10−04 | 2.07 | 0.192 | 0.070 | 5.12 × 10−03 | 3.13 | |
| First exon | 0.172 | 0.074 | 8.60 × 10−05 | 2.60 | 0.105 | 0.048 | 1.28 × 10−02 | 2.32 | 0.106 | 0.016 | 8.76 × 10−04 | 7.49 | |
| Exon boundary | 0.200 | 0.056 | 2.95 × 10−09 | 4.21 | 0.044 | 0.027 | 0.242 | 1.65 | 0.000 | 0.005 | 1.000 | 0.00 | |
| 3’ UTR | 0.372 | 0.179 | 4.71 × 10−08 | 2.72 | 0.237 | 0.115 | 2.97 × 10−04 | 2.40 | 0.000 | 0.035 | 0.414 | 0.00 | |
| Genomic region features |
Enhancer | 0.345 | 0.160 | 7.72 × 10−08 | 2.76 | 0.263 | 0.111 | 6.97 × 10−06 | 2.86 | 0.128 | 0.037 | 7.53 × 10−03 | 3.83 |
| TFBS | 0.324 | 0.167 | 3.36 × 10−06 | 2.40 | 0.175 | 0.100 | 1.22 × 10−02 | 1.91 | 0.085 | 0.019 | 1.23 × 10−02 | 4.83 | |
| DNase | 0.559 | 0.383 | 2.26 × 10−05 | 2.04 | 0.430 | 0.275 | 4.56 × 10−04 | 1.99 | 0.128 | 0.108 | 0.636 | 1.21 | |
| Open chromatin | 0.255 | 0.150 | 9.69 × 10−04 | 1.95 | 0.149 | 0.088 | 2.99 × 10−02 | 1.82 | 0.021 | 0.017 | 0.546 | 1.29 | |
| Relation to CpG island |
N_Shelf | 0.117 | 0.049 | 8.01 × 10−04 | 2.60 | 0.026 | 0.029 | 1.000 | 0.91 | 0.021 | 0.006 | 0.262 | 3.38 |
| N_Shore | 0.297 | 0.156 | 2.68 × 10−05 | 2.27 | 0.140 | 0.101 | 0.159 | 1.46 | 0.043 | 0.025 | 0.324 | 1.76 | |
| Island | 0.069 | 0.056 | 4.66 × 10−01 | 1.25 | 0.035 | 0.037 | 1.000 | 0.96 | 0.021 | 0.014 | 0.478 | 1.57 | |
| S_Shore | 0.214 | 0.140 | 1.58 × 10−02 | 1.67 | 0.132 | 0.089 | 0.134 | 1.55 | 0.043 | 0.021 | 0.269 | 2.02 | |
| S_Shelf | 0.117 | 0.043 | 2.11 × 10−04 | 2.94 | 0.044 | 0.027 | 0.246 | 1.63 | 0.000 | 0.004 | 1.000 | 0.00 | |
*Rate of DMPs around a gene with different major isoforms per brain region.
†Rate of DMPs around all genes expressed in the brain and identified in this study.
Fig. 5. Relationship between genes that have different isoforms in different brain regions and DNA methylation status.
(A) The DNA methylation status in the vicinity of GAS7 in each brain region. (B) Strategies for selecting isoforms to compare. I. If isoform A with the highest expression level (rank 1) in region 1 is detected, then the isoform that ranks first in region 2 is selected as the comparison target. II. If isoform A is ranked first in both regions and isoform B with the second lowest expression (rank 2) in region 1 is detected, then isoform C with a rank of 2 in region 2 is selected as the comparative isoform. In some cases, isoform C is also detected as having a significantly different expression ratio between regions. III. Isoforms A and D have been detected, and isoforms corresponding to the lower ranks, rank 1 and rank 2, are selected as comparative isoforms. (C) Proportion of structural differences in isoforms with different expression ratios between brain regions. The control isoform was defined as the isoform with one of the top two most highly expressed isoforms in at least one region of the brain and was consistent across all regions without any significant differences in expression levels between them. (D) Forest plot of ORs in the test for whether DMPs are more common in areas associated with the structure of isoforms that are expressed differentially between brain regions.
To examine methylation associations in more detail for the differences in alternative splicing patterns, we defined the pair of isoforms to compare (Fig. 5B). Besides, the structural differences between isoforms were categorized into isoforms with different first and/or last exons, the same exon with different 5′ and/or 3′ splice sites, intron retention, exon skipping, cassette exons, or other complicated exon structures (fig. S16). In each case, we explored the DMPs of the associated regions. To compare the degree of DMP accumulation, we performed the same analysis for control isoforms, the expression patterns of which did not differ among regions. The distribution of the isoform structure patterns did not differ markedly between genes with different major isoforms in the different regions and those of the control isoforms (Fig. 5C). Next, we examined the DMPs in the vicinity of related regions for isoform structural differences. Although the number of identified structural patterns, such as cassette exons and exon skipping were too small to evaluate, especially in the hypothalamus versus the temporal cortex comparisons (table S33), in the comparisons of the cerebellum versus the hypothalamus and the cerebellum versus the temporal cortex, we found that there were significantly more DMPs within regions 1 kb downstream from the TSSs of the isoforms that were differently expressed between regions and had different first exons (Ce versus Hy; P = 6.75 × 10−04, Ce versus Tc; P = 1.46 × 10−07) (Fig. 5D and table S34). There were also more DMPs of nominal significance in areas 1 kb upstream of the isoform TSSs that varied in expression between regions, but the degree of association was not as strong as in 1-kb downstream area (Ce versus Hy; P = 5.66 × 10−03, Ce versus Tc; P = 0.023) (Fig. 5D and table S34). Genes with DMPs within 1 kb downstream of the TSSs of isoforms with different first exons are shown in table S35. When we examined the directionality of hyper- and hypomethylation at the associated methylation sites, we found that for isoforms with different first exons, the methylation rate of DMPs located within 1 kb downstream from the TSS showed a negative correlation with gene expression in 78.8% of the sites (table S35). Furthermore, we investigated whether DMRs (differentially methylated regions) accumulate in regions related to the differences between each isoform. Similar to the DMP analysis, we found that there were significantly more DMRs both upstream and downstream within 1 kb from the TSSs of isoforms with different first exons (downstream; Ce versus Hy; P = 3.42 × 10−03, Ce versus Tc; P = 2.93 × 10−07, upstream; Ce versus Hy; P = 9.79 × 10−04, Ce versus Tc; P = 5.33 × 10−06) (tables S36 and S37).
DISCUSSION
In this study, we conducted a full-length transcriptome analysis of mRNA extracted from postmortem human brains. Long-read sequencing using human brain samples is very rare, and this is the first study to use samples from the hypothalamus. As a result, more than half of the isoforms identified in this study were unregistered in the GENCODE reference. The unregistered isoforms NIC and NNC, which accounted for most of the identified unregistered isoforms, were longer and contained more exons compared to the existing isoforms, which was consistent with the results of previous studies (18, 37). Most of the unregistered isoforms were presumed to be protein-coding according to the SQANTI3 prediction, suggesting that they may have some function in the brain. The detection of peptides derived from the unregistered isoforms by LC-MS/MS of the human postmortem brain samples further supports the possibility that the unregistered isoforms are translated and functional.
We focused on genes and isoforms that are differentially expressed in different regions of the brain. The samples used in this study were all postmortem brains from men in their 50s or older, and the comparison of different regions of the same samples minimized the effects of genetic variations and environmental factors on the analysis. When we compared gene expression levels in different regions of the brain, we found that many of the genes involved in neural pathways were highly expressed throughout the brain, and this trend was also observed in each brain region. We also found some genes that showed differences in expression levels between different regions. In the cerebellum, we found the ZIC family genes including ZIC1, ZIC3, and ZIC4. ZIC genes encode a group of evolutionarily conserved proteins with zinc finger motifs that are known to regulate cell differentiation and are involved in neural and early development (43–45). In adults, ZIC is expressed only in cerebellar granule cells and a few other nuclei (46). ZIC3 and ZIC4 were also detected as genes with a higher number of isoforms in the cerebellum than in other regions in this study, again suggesting that these genes play an important role, especially in the cerebellum.
Analysis of genes that were specifically up-regulated in the temporal cortex also revealed pathways related to neurogenesis, as in the cerebellum. However, in the hypothalamus, immune response pathways were detected exclusively. The reason is not certain; however, hypotheses include the possibility that the infiltration of immune cells is greater in the hypothalamus than in other brain regions and that the immune process and metabolism regulation by cytokines are specific to the hypothalamus (47, 48). In some regions of the brain, the blood-brain barrier has been modified to form a more permeable barrier to allow certain substances from systemic circulation to access the central nervous system (49). One such region, the median eminence, is in physical proximity to the hypothalamus, making the hypothalamus more accessible to immune cells and other substances than other brain regions (47, 49). Our results appear to corroborate the existence of such immune processes in the hypothalamus, but further studies are necessary.
Next, we searched for genes that express more than 20 isoforms. Analysis of the entire brain identified numerous genes that are involved in pathways such as protein localization. Our analysis of the public data of long-read transcriptome sequencing in tissues other than the brain found that the genes involved in these pathways have a high number of expressed isoforms not only in the brain but also in many other tissues. After searching for genes with a higher number of isoforms in the cerebellum and temporal cortex, we identified genes related to pathways involved in the nervous system and cell projection organization. On the other hand, in the hypothalamus, immune response pathways were detected. The correlation between the number of isoforms and expression levels per gene was not strong (r2 = 0.127), and therefore, genes with higher expression levels in the hypothalamus did not always have a higher number of isoforms. These findings suggest that, as with genes with high expression levels, genes with a high number of isoforms may also play important functions in pathways such as neurogenesis in the cerebellum and temporal cortex, as well as in pathways of the immune system in the hypothalamus.
We then performed an exploration of genes that express different major isoforms in each brain region, which is the main focus of our research. As a result, the gene GAS7 was found to show the most significant difference in the major expressed isoforms among the three brain regions. The result showing that the shorter isoform is predominantly expressed in the cerebellum was also replicated in the analysis using the long-read data from the GTEx Consortium (19). On the other hand, in our data, the longer isoform was hardly expressed in the cerebellum, whereas it was expressed in the GTEx data. This discrepancy is likely attributed to the distinct sources of the cerebellum samples: ours being derived from the cerebellar cortex, in contrast to the GTEx Consortium, which classifies the entire cerebellar hemisphere as the cerebellum. It has been reported that GAS7 not only has important functions in neurite outgrowth (50, 51) but is also involved in neuronal mitochondrial dynamics and is required for normal neuronal function (52). In a previous study, the expression of two murine isoforms of GAS7, Gas7 and Gas7-cb, were examined by Western blotting (40). The findings showed that Gas7 is expressed in the cerebral cortex and growth-arrested NIH3T3 fibroblasts, while Gas7-cb is predominantly expressed in the cerebellum. The shorter isoform, Gas7-cb, which consists of 340 amino acids, was found to have an almost identical amino acid sequence to ENST00000580865.5, which was predominantly expressed in the human cerebellum in this study. The longer isoform consisting of 421 amino acids had a high sequence identity with those of ENST00000585266.5 and ENST00000437099.6, which were predominantly expressed in the hypothalamus and temporal cortex, respectively. Furthermore, another study has reported that transfection of cells with cDNAs of the longer and shorter human isoforms yielded the 38- or 50-kDa protein and differentially induced neurite outgrowth (53). These results support the differences in isoform expression found in our analysis for each brain region and showed that distinct isoforms predominantly expressed in specific brain regions play crucial roles in biological processes respectively.
Pathway analysis using genes with different major isoforms in the different regions identified pathways such as “actin filament-based process” and “cell projection organization”. The actin cytoskeleton is a fundamental regulator of cell morphology, including that of neuronal cells, and in neurons, actin is concentrated in dendritic spines. Since cell projections are structures such as axons and dendrites in brain cells, these findings suggest the possibility that some of the genes involved in determining the morphology of axons and dendrites in neurons and glial cells use different isoforms in the different brain regions. The expression profile of guanine-nucleotide-exchange factors that activate calcium/calmodulin-dependent kinases and small guanosine triphosphatase Rac, which are involved in actin reorganization and spine morphogenesis, has been reported to be regionally controlled in the brain (54, 55), supporting the results of this study.
Last, we considered the possibility that epigenetics may play an important role in the background of the different usage of isoforms in different brain regions and showed that there were significantly more DMPs and DMRs in the vicinity of genes with different isoforms in different regions. The methylation sites were particularly abundant in the upstream regions and enhancer of genes, and when the structural differences between isoforms were considered in detail, it became clear that DMPs are particularly abundant in the 1-kb region downstream of the TSS between isoforms with different first exons. Several studies have reported that DNA methylation is not only related to gene expression (31, 56–58) but also to alternative splicing (33–35). In particular, the regulation of exon inclusion is considered to be one mechanism that contributes to alternative splicing (33, 34). Our results did not show a clear correlation between methylation and the structural differences in isoforms related to exon inclusion, such as cassette exons or exon skipping. However, this may be due to the limited coverage of the methylation probes in the gene regions, which may have prevented the detection of sufficient effects in this study. Further analysis using sequencing-based methylation analysis could reveal a more detailed picture of methylation sites and their associations with alternative splicing.
Our study has several limitations. First, the sample size was small. Matching sample characteristics such as age and sex as much as possible may have compensated for this limitation to some extent, and strict criteria were used to minimize false positives. However, an analysis with more samples would reveal a more detailed profile of isoforms in the brain. Second, some of the hypothalamus-derived RNA used in this study was of poor quality, and it is possible that some of the mRNA included was degraded. In this study, we applied very strict filtering for ISMs and excluded many ISMs from the analysis. By using better-quality RNA and relaxing the stringency of filtering conditions, it may be possible to detect truly functionally important ISMs more accurately. Third, our brain region–specific analyses were performed on bulk tissue fragments containing a variety of cells, so it is difficult to determine whether the results are specific to the region or to a specific cell type within that region. In the future, single-cell sequencing techniques could be used to obtain isoform profiles of individual cells in specific brain regions. Last, our genome-wide methylation data, which we used to investigate the association with alternative splicing, were based on an array-based analysis. While the probe density is high in the upstream region of genes, the density is not high in the gene body, which prevents adequate detection of associations. Sequence-based methylation data with sufficient coverage would be more helpful for more detailed analysis. Furthermore, in this study, we have only been able to investigate DNA methylation among the mechanisms of epigenetics. There is a possibility that by examining region-specific TF binding and histone marks, we may discover important factors that play a role in the expression of different isoforms.
In summary, we analyzed the full-length transcriptomes of the human cerebellum, hypothalamus, and temporal cortex from the same samples and showed that several genes, including GAS7, use region-dependent isoforms, and that DNA methylation might be involved in this mechanism. These genes with different major isoforms across regions are particularly involved in dendritic morphology and neuronal pathways in the nervous system, and they are likely to play important roles in their respective regions of the brain. Our results including a list of isoforms currently unregistered in the brain and a list of genes that use different isoforms for each brain region provide potentially valuable findings for future research on brain disorders and shed light on the mechanisms underlying isoform diversity in the human brain.
MATERIALS AND METHODS
Brain samples and RNA extraction
Three regions—the temporal cortex, hypothalamus, and cerebellum (cerebellar cortex)—were excised from postmortem brain samples from unrelated Japanese individuals (N = 4 × 3). All of the tissues were from male subjects without brain lesions or infections and who died in their 50s or later. The demographic data of these brain samples are summarized in table S38. All of the samples were autopsied within a postmortem interval of 6 hours and promptly stored at −80°C at the Niigata University Brain Research Institute Brain Bank, Japan.
RNA was extracted using RNeasy Plus Universal Kit (QIAGEN, Netherlands) after 10 to 35 mg of tissues was placed in RNAlater-ICE (Thermo Fisher Scientific, MA, USA) for a minimum of 16 hours at −20°C to stabilize the RNA. RNA quality was assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and Femto Pulse System (Agilent Technologies, CA, USA), and RQN was calculated according to the Agilent Technical Overview, “Comparison of RIN and RQN for the Agilent 2100 Bioanalyzer and the Fragment Analyzer Systems”; RQN is generally compatible with RNA integrity number (RIN). Samples with RQN greater than 5.0 were used for sequencing.
Iso-Seq library preparation and single molecule, real-time (SMRT) sequencing
First-strand cDNA synthesis and cDNA amplification were performed with a NEBNext Single Cell/Low Input cDNA Synthesis and Amplification Module (New England Biolabs, MA, USA) and Iso-Seq Express Oligo Kit (Pacific Biosciences (PacBio), CA, USA) according to the procedure and checklist (i.e., Iso-Seq Express Template Preparation for Sequel and Sequel II Systems) provided by PacBio. Amplified cDNA samples were purified with ProNex beads (Promega, WI, USA) using the standard workflow and assuming that most transcripts are ~2 kb. Quantification was performed using a Qubit Fluorometer (Thermo Fisher Scientific) and size distribution was confirmed by a Femto Pulse System.
The Iso-Seq library was prepared by going through the steps of DNA damage repair, end repair and A-tailing, overhang adopter ligation, and cleanup using a SMRTbell Express Template Prep Kit 2.0 (PacBio). Sequencing primer was annealed and polymerase binding, complex cleanup, and sample loading were performed with a Sequel II binding kit 2.1 and internal control 1.0 (PacBio) following the manufacturer’s protocol, and sequencing was performed using the Sequel IIe system. Using one SMRTcell per sample, movie runtime was set to 24 hours for each SMRTcell.
Data processing
The overview of the data processing used in this study is shown in fig. S1B. The list of links to the GitHub repositories used is provided in the Supplementary Materials. Using the circular consensus sequence reads generated from each of the sub-reads, HiFi reads, which are reads that satisfy the conditions of >3 full-pass sub-reads and QV ≥ 20, were automatically generated in the Sequel IIe system using SMRT Link v11.0. The GitHub repository, IsoSeq v3, was used to process the HiFi reads: The 3′ and 5′ primers, concatemer, and polyA tail were removed and only reads supported by at least two FLNC reads with an accuracy ≥99% were used for subsequent analysis. These reads were mapped to the human genome (hg38) using the GitHub repository, pbmm2, and the isoforms that were considered to be identical were combined into one according to the default value of the collapse command in IsoSeq v3.
QC assessment and filtering were performed using SQANTI3 (59). The human genome (hg38) and comprehensive gene annotation data from GENCODE v.41 were used as the reference for the QC. For the reference files of CAGE Peak data, polyA motif list, and polyA site data, we used the files provided in SQANTI3. Each isoform was judged against the reference to determine which of the following isoform categories it fell into FSM, ISM, NIC, NNC, genic, intron, antisense, fusion, and intergenic. Since the isoforms in each category have different probabilities of not being an artifact, a rule-based filter was applied with different conditions for each category, as shown in filtering.json in the GitHub repository, Brain_Iso-Seq. In addition, since partial RNA degradation was a concern for some of the samples in this study, additional filtering was performed for ISM isoforms to carefully detect ISM isoforms, i.e., only isoforms that were either determined not to be artifacts by the machine learning filter or that were detected by other long-read studies with brain samples were retained [data provided by GTEx (19) and PacBio (www.pacb.com/general/data-release-alzheimer-brain-isoform-sequencing-iso-seq-dataset/)]. QC was performed again on only those isoforms remaining after application of the filter.
TALON (v5.0) (60) was used for chaining multiple samples together. We initialized a TALON database using comprehensive gene annotation data from GENCODE v.41. The default settings were used as the condition for considering isoforms to be identical, except that differences in the length of the 5′ and 3′ ends of the first and last exon were allowed. We used minimap2 and SAMtools (61) to prepare MD-tagged SAM files for each sample and each sample was added to the database with the conditions --cov 0.95 and --identity 0.95. Last, only isoforms expressed in two or more samples were retained for analysis. After chaining, we focused our analysis on isoforms that were expressed in at least two samples and summarized isoforms, i.e., in this study, differences only at both ends of the same first/last exons were not detected. We carried out all the data processing using the script ‘Data_Processing1–4.py’. All command lines are described in the Supplementary Materials.
Rarefaction curve analysis
Rarefaction curve analysis of isoform diversity was performed to confirm whether genes or isoforms were sufficiently detected in this study. Programs subsample.py and subsample_with_category.py in the GitHub repository, cDNA_Cupcake, were used for the analysis.
PCA and cluster analysis
PCA and cluster analysis were performed using information on the presence or absence of all isoforms after filtering. The analysis was performed in R (62) and the code is described in the Supplementary Materials. Euclidean distance was used for PCA and clustering was performed using the Ward method.
Gene-based comparison between brain regions
We first examined genes with high expression levels in each of the brain regions (median TPM ≥ 200) and genes expressed only in specific brain regions using the following condition: genes with TPM >2 in three out of four samples from the region of interest and TPM <1 in all other samples. Next, we examined genes with differential expression levels in different brain regions under the conditions of Wilcoxon test P < 0.01, t test P < 0.05, and FC >5. Considering the number of samples (four samples for the region of interest versus eight samples for the other regions), a threshold of P < 0.01 was the most stringent criterion for the Wilcoxon test.
We further examined genes that have a number of isoforms (≥20) in each brain region and genes with a higher number of isoforms only in the region of interest compared to other regions with Wilcoxon test P < 0.01 (four samples for the region of interest versus eight samples for the other sites). Furthermore, genes that met the following conditions were excluded: (i) genes that are not expressed except in the region of interest and (ii) genes that express only one type of isoform in the site of interest.
To explore whether isoform-rich genes were brain-specific, we performed an analysis using long-read data from GTEx (https://gtexportal.org/home/datasets). We targeted isoforms expressed in at least two of the non-brain samples, and pathway analysis was performed using genes expressing more than 20 isoforms per gene.
Isoform-based comparison between brain regions
We examined genes with different major isoforms expressed in different brain regions. First, we determined the percentage of each isoform’s TPM relative to the total expression of the gene from which it was derived using TPM_table.py and Isoform_Proportion.py. The percentages were used to compare the cerebellum versus the hypothalamus, the cerebellum versus the temporal cortex, the hypothalamus versus the temporal cortex with the condition of the median TPM of the higher expression region >20, and the median TPM of the lower expression region >5. Genes that had isoforms with Wilcoxon test P < 0.05 and t test P < 0.05 and a difference in the average TPM percentage between the two compared regions of >20% were selected. Here, a threshold of P < 0.05 was the most stringent criterion used for the Wilcoxon test, considering the number of samples (four samples for the region of interest versus four samples for the other region). Isoforms were imaged using IGV software (v2.15.2) (63).
Pathway analysis
Pathway analysis was performed to explore the characteristics of the genes detected in each analysis or genes to which detected isoforms belong. We used MetaCore software (version 6.24 build 67895, Thomson Reuters, NY, USA) with high-quality, manually curated content and the PANTHER Classification System (PANTHER 17.0) (64) to validate the results of MetaCore software. The Gene Ontology database (65, 66) was used to search for significant gene sets. We searched a maximum of the top 500 gene sets in the MetaCore analysis that met the following conditions: (i) FDR < 0.05 in MetaCore and replicated in PANTHER with P < 0.05; (ii) if a pathway contains fewer than 100 network objects, at least 10% of them are included in the test gene set; (iii) if a pathway has more than 100 network objects, at least 10 objects are included in the test gene set; and (iv) the total number of objects in the pathway is less than 3000. When more than 10 pathways were detected, Cytoscape (67) was used to show the relationships among the top 10 pathways. The coordinates, which indicated the positional relationship between pathways, used the first and second principal components calculated based on the presence or absence of the included network objects.
DNA methylation data
The methylation rate data obtained for the DNA derived from three brain regions of the four samples used in this study are described in detail elsewhere (41). Briefly, DNA samples were extracted with a QIAamp DNA mini Kit (Qiagen), and DNA methylation was examined with an Infinium Methylation EPIC BeadChip kit (Illumina Inc., CA, USA). Data filtering was performed excluding underperforming probes (68) and data were normalized for probe design bias with the Beta Mixture Quantile dilation normalization method (69) and batch effects with ComBat software (70). For a more detailed procedure, it is described elsewhere (41). Reference-free estimation of the percentage of cell components was performed using MeDeCom (36).
Integrative analysis of transcriptome and methylation data
We examined whether there were any DMPs between brain regions in the vicinity of genes that expressed different major isoforms between regions. We first detected DMPs between brain regions. For each comparison of the cerebellum versus the hypothalamus, the cerebellum versus the temporal cortex, and the hypothalamus versus the temporal cortex, methylation sites with Wilcoxon’s test P < 0.05 and t test P < 0.05, and with >20% difference in methylation rate between groups, were identified as DMPs. We then compared the presence of the DMPs in the vicinity of genes, between genes expressing different major isoforms, and genes expressed in all brains using CpG_annotation.py. Annotation information such as whether the methylation site is upstream or downstream of the gene was obtained from the annotation file provided by Illumina Inc. (infinium-methylationepic-v-1-0-b5-manifest-file.csv). Since our data are based on array-based methylation, we conducted a more detailed examination of the GAS7 region. We used methylation rate data from bisulfite sequencing of neurons derived from the cortex and cerebellum obtained from publicly available data (42). We acquired the bigwig files registered in the NCBI’s GEO dataset GSE186458 (cortex N = 8, cerebellum N = 1).
Furthermore, we determined alternative splicing patterns shown in fig. S16 between isoform pairs that were differentially expressed between regions. We have shown three cases as examples in Fig. 5B. For isoforms detected as having differential expression between the regions being compared (Region1 and Region2), we calculated min(rank_Region1, rank_Region2). We then selected isoforms ranked in the other region based on this min(rank_Region1, rank_Region2) as the comparison target. Next, we identified areas in the vicinity of isoforms that differed from each other according to their alternative splicing patterns (fig. S16). Detailed definitions of the areas are shown in fig. S16. Comparison of isoforms and extraction of relevant areas was performed with AS_investigation.py. We then searched for DMPs of brain regions in these areas. As a control for comparison, we performed the same analysis between isoforms that showed no differences in expression patterns between brain regions to search for the extent to which DMPs were present in areas associated with differences in isoforms. Genes with TPM >10 were targeted, and of the isoforms for which P > 0.2 in any of the statistical tests of comparisons between brain regions, isoforms that were ranked second or higher in terms of their expression level within a gene at any region were included. Comparisons of these selected isoforms within the same gene were used as controls for statistical analysis.
To conduct a similar analysis for DMR, we detected DMR using the bump hunting method (71) under the following conditions: (i) A minimum number of probes to detect a DMR was set at three, (ii) the maximum gap between probes inside a DMR was set at 1000 bp, and (iii) bump hunter DMR P values computed through permutation procedures <0.05. We then searched for the DMRs of brain regions in the areas relevant to each differentially expressed isoform following the same analysis as DMP.
Utilization of proteome analysis for the validation of unregistered isoforms
To confirm whether the newly identified isoform is translated and exists as a protein in the brain, we used the results of LC-MS/MS analysis of the human postmortem cortex obtained in a previous study (38). The raw data were downloaded from the ProteomeXchange Consortium (www.proteomexchange.org) and processed as an 11-plex tandem mass tag using MaxQuant (72). TMT parameter files (lot nos. SI258088 and SJ258847) were obtained from the Thermo Fisher Scientific website. The ‘Type’ for the group-specific parameters was set to ‘11plex TMT’, and the charge for each was set to the value in the TMT parameter files. In addition to Trypsin, LysC was also selected for ‘Digestion’. For the ‘Sequence’ in the global parameters, we used the GENCODE protein-coding transcript translation sequences Release41, which is the same version of the transcript information used for identifying isoforms. All other settings were left as default values. Only the peptides used for identifying proteins detected with a criterion of 1% FDR were analyzed. We also conducted an analysis with the same settings but changed the amino acid sequences of the predicted ORF for the unregistered isoforms to the reference only. We then searched for peptides that did not match the amino acid sequences registered in GENCODE and those estimated from the known transcripts identified in our study, specifically targeting those that were detected for the amino acid sequences derived from the unregistered isoforms.
Ethics approval and consent to participate
Written informed consent was obtained from all participants or their families, and the protocols were approved by the Institutional Review Board of the National Center for Global Health and Medicine (NCGM-A-003267), Tokyo Metropolitan Institute of Medical Science (20–15), and other collaborative institutes.
Acknowledgments
We extend our gratitude to all the donors who provided samples for this study. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 08/15/2022.
Funding: This study was supported by Grants-in-Aid for Scientific Research (grant numbers 18 K15053 and 21 K15360) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Furthermore, this study was endorsed by the joint usage/research program of the Brain Research Institute of Niigata University. This study was also supported by a grant (grant number JP22tm0424222) from the Japan Agency for Medical Research and Development (AMED).
Author contributions: Conceptualization: M.S. and A.F. Methodology: M.S., Y.O., and Y.H. Validation: Y.O., M.S., and T.M. Formal analysis: M.S. and Y.O. Investigation: M.S. and Y.O. Resources: A.K., R.G., and Y.O. Data curation: M.S. and Y.O. Writing—original draft: M.S. and Y.O. Writing—review and editing: M.S., Y.O., and Y.H. Visualization: M.S. and Y.O. Project administration: M.S. Supervision: A.F., Y.H., T.M., K.T., and M.H. Funding acquisition: A.K., K.T., and M.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper other than the raw sequence data and GitHub dataset are present in the paper and/or the Supplementary Materials. Iso-Seq analysis raw data have been deposited into DDBJ (www.ddbj.nig.ac.jp/) with the BioProject accession number: PRJDB15555. The GitHub repository, containing all the code used for analysis along with the complete set of simulated datasets assumed by the code, is accessible here: https://github.com/mihshimada/Brain_Iso-Seq and has been archived in Zenodo with doi:10.5281/zenodo.10398061.
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Command lines used in this study
Legends for tables S1 to S38
The list of links to the GitHub repositories
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S38
REFERENCES AND NOTES
- 1.Fu H., Hardy J., Duff K. E., Selective vulnerability in neurodegenerative diseases. Nat. Neurosci. 21, 1350–1358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang Y., Kim J., Shin J. Y., Kim J. I., Seo J. S., Webster M. J., Lee D., Kim S., Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl. Psychiatry 3, e321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohen R., Dobra A., Tracy J. H., Haugen E., Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl. Psychiatry 4, e366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darby M. M., Yolken R. H., Sabunciyan S., Consistently altered expression of gene sets in postmortem brains of individuals with major psychiatric disorders. Transl. Psychiatry 6, e890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramaker R. C., Bowling K. M., Lasseigne B. N., Hagenauer M. H., Hardigan A. A., Davis N. S., Gertz J., Cartagena P. M., Walsh D. M., Vawter M. P., Jones E. G., Schatzberg A. F., Barchas J. D., Watson S. J., Bunney B. G., Akil H., Bunney W. E., Li J. Z., Cooper S. J., Myers R. M., Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 9, 72 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egervari G., Kozlenkov A., Dracheva S., Hurd Y. L., Molecular windows into the human brain for psychiatric disorders. Mol. Psychiatry 24, 653–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ham S., Lee S. V., Advances in transcriptome analysis of human brain aging. Exp. Mol. Med. 52, 1787–1797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishir M., Bhat A., Essa M. M., Ekpo O., Ihunwo A. O., Veeraraghavan V. P., Mohan S. K., Mahalakshmi A. M., Ray B., Tuladhar S., Chang S., Chidambaram S. B., Sakharkar M. K., Guillemin G. J., Qoronfleh M. W., Ojcius D. M., Sleep deprivation and neurological disorders. Biomed. Res. Int. 2020, 5764017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilgner H., Grubert F., Sharon D., Snyder M. P., Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. U.S.A. 111, 9869–9874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu J., Chen G., Zhu S., Li S., Wen Z., Li B., Zheng Y., Shi L., Identification of tissue-specific protein-coding and noncoding transcripts across 14 human tissues using RNA-seq. Sci. Rep. 6, 28400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cáceres M., Lachuer J., Zapala M. A., Redmond J. C., Kudo L., Geschwind D. H., Lockhart D. J., Preuss T. M., Barlow C., Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. U.S.A. 100, 13030–13035 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joglekar A., Prjibelski A., Mahfouz A., Collier P., Lin S., Schlusche A. K., Marrocco J., Williams S. R., Haase B., Hayes A., Chew J. G., Weisenfeld N. I., Wong M. Y., Stein A. N., Hardwick S. A., Hunt T., Wang Q., Dieterich C., Bent Z., Fedrigo O., Sloan S. A., Risso D., Jarvis E. D., Flicek P., Luo W., Pitt G. S., Frankish A., Smit A. B., Ross M. E., Tilgner H. U., A spatially resolved brain region- and cell type-specific isoform atlas of the postnatal mouse brain. Nat. Commun. 12, 463 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enard W., Khaitovich P., Klose J., Zöllner S., Heissig F., Giavalisco P., Nieselt-Struwe K., Muchmore E., Varki A., Ravid R., Doxiadis G. M., Bontrop R. E., Pääbo S., Intra- and interspecific variation in primate gene expression patterns. Science 296, 340–343 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Khaitovich P., Muetzel B., She X., Lachmann M., Hellmann I., Dietzsch J., Steigele S., Do H. H., Weiss G., Enard W., Heissig F., Arendt T., Nieselt-Struwe K., Eichler E. E., Pääbo S., Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14, 1462–1473 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konopka G., Bomar J. M., Winden K., Coppola G., Jonsson Z. O., Gao F., Peng S., Preuss T. M., Wohlschlegel J. A., Geschwind D. H., Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature 462, 213–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konopka G., Friedrich T., Davis-Turak J., Winden K., Oldham M. C., Gao F., Chen L., Wang G. Z., Luo R., Preuss T. M., Geschwind D. H., Human-specific transcriptional networks in the brain. Neuron 75, 601–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oldham M. C., Konopka G., Iwamoto K., Langfelder P., Kato T., Horvath S., Geschwind D. H., Functional organization of the transcriptome in human brain. Nat. Neurosci. 11, 1271–1282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung S. K., Jeffries A. R., Castanho I., Jordan B. T., Moore K., Davies J. P., Dempster E. L., Bray N. J., O'Neill P., Tseng E., Ahmed Z., Collier D. A., Jeffery E. D., Prabhakar S., Schalkwyk L., Jops C., Gandal M. J., Sheynkman G. M., Hannon E., Mill J., Full-length transcript sequencing of human and mouse cerebral cortex identifies widespread isoform diversity and alternative splicing. Cell Rep. 37, 110022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glinos D. A., Garborcauskas G., Hoffman P., Ehsan N., Jiang L., Gokden A., Dai X., Aguet F., Brown K. L., Garimella K., Bowers T., Costello M., Ardlie K., Jian R., Tucker N. R., Ellinor P. T., Harrington E. D., Tang H., Snyder M., Juul S., Mohammadi P., MacArthur D. G., Lappalainen T., Cummings B. B., Transcriptome variation in human tissues revealed by long-read sequencing. Nature 608, 353–359 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramezanpour H., Fallah M., The role of temporal cortex in the control of attention. Curr Res Neurobiol 3, 100038 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickok G., Poeppel D., The cortical organization of speech processing. Nat. Rev. Neurosci. 8, 393–402 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Wu Z., Buckley M. J., Prefrontal and medial temporal lobe cortical contributions to visual short-term memory. J. Cogn. Neurosci. 35, 27–43 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Kaur A., Basavanagowda D. M., Rathod B., Mishra N., Fuad S., Nosher S., Alrashid Z. A., Mohan D., Heindl S. E., Structural and functional alterations of the temporal lobe in Schizophrenia: A literature review. Cureus 12, e11177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migliaccio R., Cacciamani F., The temporal lobe in typical and atypical Alzheimer disease. Handb. Clin. Neurol. 187, 449–466 (2022). [DOI] [PubMed] [Google Scholar]
- 25.Mathon B., Clemenceau S., Surgery procedures in temporal lobe epilepsies. Handb. Clin. Neurol. 187, 531–556 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Barbosa D. A. N., de Oliveira-Souza R., Monte Santo F., de Oliveira Faria A. C., Gorgulho A. A., De Salles A. A. F., The hypothalamus at the crossroads of psychopathology and neurosurgery. Neurosurg. Focus 43, E15 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Saper C. B., Lowell B. B., The hypothalamus. Curr. Biol. 24, R1111–R1116 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Delgado-García J. M., Structure and function of the cerebellum. Rev. Neurol. 33, 635–642 (2001). [PubMed] [Google Scholar]
- 29.Stoodley C. J., The cerebellum and neurodevelopmental disorders. Cerebellum 15, 34–37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holliday R., Pugh J. E., DNA modification mechanisms and gene activity during development. Science 187, 226–232 (1975). [PubMed] [Google Scholar]
- 31.Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., Thomson J. A., Ren B., Ecker J. R., Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellman A., Chess A., Gene body-specific methylation on the active X chromosome. Science 315, 1141–1143 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Maunakea A. K., Nagarajan R. P., Bilenky M., Ballinger T. J., D'Souza C., Fouse S. D., Johnson B. E., Hong C., Nielsen C., Zhao Y., Turecki G., Delaney A., Varhol R., Thiessen N., Shchors K., Heine V. M., Rowitch D. H., Xing X., Fiore C., Schillebeeckx M., Jones S. J., Haussler D., Marra M. A., Hirst M., Wang T., Costello J. F., Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maunakea A. K., Chepelev I., Cui K., Zhao K., Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 23, 1256–1269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierrez-Arcelus M., Ongen H., Lappalainen T., Montgomery S. B., Buil A., Yurovsky A., Bryois J., Padioleau I., Romano L., Planchon A., Falconnet E., Bielser D., Gagnebin M., Giger T., Borel C., Letourneau A., Makrythanasis P., Guipponi M., Gehrig C., Antonarakis S. E., Dermitzakis E. T., Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing. PLoS Genet. 11, e1004958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lutsik P., Slawski M., Gasparoni G., Vedeneev N., Hein M., Walter J., MeDeCom: Discovery and quantification of latent components of heterogeneous methylomes. Genome Biol. 18, 55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang K. K., Huang J., Wu J. K. L., Lee M., Tay S. T., Kumar V., Ramnarayanan K., Padmanabhan N., Xu C., Tan A. L. K., Chan C., Kappei D., Göke J., Tan P., Long-read transcriptome sequencing reveals abundant promoter diversity in distinct molecular subtypes of gastric cancer. Genome Biol. 22, 44 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ping L., Kundinger S. R., Duong D. M., Yin L., Gearing M., Lah J. J., Levey A. I., Seyfried N. T., Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer's disease. Sci Data 7, 315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amarasinghe S. L., Su S., Dong X., Zappia L., Ritchie M. E., Gouil Q., Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazakovitch E. M., She B. R., Lien C. L., Woo W. M., Ju Y. T., Lin-Chao S., The Gas7 gene encodes two protein isoforms differentially expressed within the brain. Genomics 61, 298–306 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Shimada M., Miyagawa T., Takeshima A., Kakita A., Toyoda H., Niizato K., Oshima K., Tokunaga K., Honda M., Epigenome-wide association study of narcolepsy-affected lateral hypothalamic brains, and overlapping DNA methylation profiles between narcolepsy and multiple sclerosis. Sleep 43, (2020). [DOI] [PubMed] [Google Scholar]
- 42.Loyfer N., Magenheim J., Peretz A., Cann G., Bredno J., Klochendler A., Fox-Fisher I., Shabi-Porat S., Hecht M., Pelet T., Moss J., Drawshy Z., Amini H., Moradi P., Nagaraju S., Bauman D., Shveiky D., Porat S., Dior U., Rivkin G., Or O., Hirshoren N., Carmon E., Pikarsky A., Khalaileh A., Zamir G., Grinbaum R., Abu Gazala M., Mizrahi I., Shussman N., Korach A., Wald O., Izhar U., Erez E., Yutkin V., Samet Y., Rotnemer Golinkin D., Spalding K. L., Druid H., Arner P., Shapiro A. M. J., Grompe M., Aravanis A., Venn O., Jamshidi A., Shemer R., Dor Y., Glaser B., Kaplan T., A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aruga J., Minowa O., Yaginuma H., Kuno J., Nagai T., Noda T., Mikoshiba K., Mouse Zic1 is involved in cerebellar development. J. Neurosci. 18, 284–293 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahmane N., Altaba A. R., Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126, 3089–3100 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Merzdorf C. S., Emerging roles for zic genes in early development. Dev. Dyn. 236, 922–940 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Aruga J., Yozu A., Hayashizaki Y., Okazaki Y., Chapman V. M., Mikoshiba K., Identification and characterization of Zic4, a new member of the mouse Zic gene family. Gene 172, 291–294 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Haddad-Tóvolli R., Dragano N. R. V., Ramalho A. F. S., Velloso L. A., Development and function of the blood-brain barrier in the context of metabolic control. Front. Neurosci. 11, 224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banks W. A., The blood-brain barrier in neuroimmunology: Tales of separation and assimilation. Brain Behav. Immun. 44, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett L., Yang M., Enikolopov G., Iacovitti L., Circumventricular organs: A novel site of neural stem cells in the adult brain. Mol. Cell. Neurosci. 41, 337–347 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.She B. R., Liou G. G., Lin-Chao S., Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp. Cell Res. 273, 34–44 (2002). [DOI] [PubMed] [Google Scholar]
- 51.You J. J., Lin-Chao S., Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J. Biol. Chem. 285, 11652–11666 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhupana J. N., Huang B. T., Liou G. G., Calkins M. J., Lin-Chao S., Gas7 knockout affects PINK1 expression and mitochondrial dynamics in mouse cortical neurons. FASEB Bioadv. 2, 166–181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chao C. C., Chang P. Y., Lu H. H., Human Gas7 isoforms homologous to mouse transcripts differentially induce neurite outgrowth. J. Neurosci. Res. 81, 153–162 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Penzes P., Cahill M. E., Jones K. A., Srivastava D. P., Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 18, 405–413 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Penzes P., Cahill M. E., Deconstructing signal transduction pathways that regulate the actin cytoskeleton in dendritic spines. Cytoskeleton (Hoboken) 69, 426–441 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Razin A., Riggs A. D., DNA methylation and gene function. Science 210, 604–610 (1980). [DOI] [PubMed] [Google Scholar]
- 57.Klose R. J., Bird A. P., Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 31, 89–97 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Aran D., Toperoff G., Rosenberg M., Hellman A., Replication timing-related and gene body-specific methylation of active human genes. Hum. Mol. Genet. 20, 670–680 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Tardaguila M., de la Fuente L., Marti C., Pereira C., Pardo-Palacios F. J., Del Risco H., Ferrell M., Mellado M., Macchietto M., Verheggen K., Edelmann M., Ezkurdia I., Vazquez J., Tress M., Mortazavi A., Martens L., Rodriguez-Navarro S., Moreno-Manzano V., Conesa A., SQANTI: Extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res. 28, 396–411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D. Wyman, G. Balderrama-Gutierrez, F. Reese, S. Jiang, S. Rahmanian, W. Zeng, B. Williams, D. Trout, W. England, S. Chu, R. C. Spitale, A. Tenner, B. Wold, A. Mortazavi, A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification. bioRxiv 672931 (2020). 10.1101/672931. [DOI]
- 61.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Core Team, R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2022); www.R-project.org/.
- 63.Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P. D., PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G., Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gene Ontology Consortium , The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou W., Laird P. W., Shen H., Comprehensive characterization, annotation and innovative use of Infinium DNA methylation BeadChip probes. Nucleic Acids Res. 45, e22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teschendorff A. E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S., A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29, 189–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson W. E., Li C., Rabinovic A., Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Jaffe A. E., Murakami P., Lee H., Leek J. T., Fallin M. D., Feinberg A. P., Irizarry R. A., Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int. J. Epidemiol. 41, 200–209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Command lines used in this study
Legends for tables S1 to S38
The list of links to the GitHub repositories
Tables S1 to S38