Abstract
The nucleotide sequence of the Actinomyces naeslundii T14V type 2 fimbrial structural subunit gene, fimA, and the 3′ flanking DNA region was determined. The fimA gene encoded a 535-amino-acid precursor subunit protein (FimA) which included both N-terminal leader and C-terminal cell wall sorting sequences. A second gene, designated orf365, that encoded a 365-amino-acid protein which contained a putative transmembrane segment was identified immediately 3′ to fimA. Mutants in which either fimA or orf365 was replaced with a kanamycin resistance gene did not participate in type 2 fimbriae-mediated coaggregation with Streptococcus oralis 34. Type 2 fimbrial antigen was not detected in cell extracts of the fimA mutant by Western blotting with anti-A. naeslundii type 2 fimbrial antibody, but the subunit protein was detected in extracts of the orf365 mutant. The subunit protein detected in this mutant also was immunostained by an antibody raised against a synthetic peptide representing the C-terminal 20 amino acid residues of the predicted FimA. The antipeptide antibody reacted with FimA isolated from the recombinant Escherichia coli clone containing fimA but did not react with purified type 2 fimbriae in extracts of the wild-type strain. These results indicate that synthesis of type 2 fimbriae in A. naeslundii T14V may involve posttranslational cleavage of both the N-terminal and C-terminal peptides of the precursor subunit and also the expression of orf365.
Two major fimbrial types have been identified in strains of Actinomyces naeslundii that colonize the oral cavity. Fimbriae designated type 1 mediate bacterial adherence to salivary proline-rich proteins that coat the tooth enamel (9, 18). In contrast, those designated type 2 exhibit a lectin activity (3) that was initially detected by the lactose-sensitive coaggregation of A. naeslundii strains with several streptococcal strains, such as Streptococcus oralis 34 (27), that also colonize teeth. Type 2 fimbriae also mediate bacterial adhesion to various host cells (3), including erythrocytes, epithelial cells, and polymorphonuclear leukocytes. Activation of the latter cell type by type 2 fimbriated Actinomyces strains results in phagocytosis and bacterial killing (32) and the release of mediators such as superoxide (33) that may contribute to the initiation of gingival inflammation. Consequently, the identification of the fimbrial lectin(s) would provide an improved understanding of bacterium-host cell interactions. However, the nature of the type 2 fimbria-associated lectin activity, whether it is a part of the major fimbrial subunit or a minor fimbrial component, remains unknown. A major obstacle in distinguishing between these alternatives is the inability to dissociate A. naeslundii fimbriae to monomer subunits.
The lectin-like adhesins of several gram-negative bacteria have been identified in studies of fimbria biogenesis at the genetic level (22, 39). However, little is known concerning bacterial adhesins and assembly of fimbriae in gram-positive bacteria. The expression of both type 1 and type 2 fimbriae by A. naeslundii T14V (8) makes this strain a model system for studies of biogenesis of fimbriae in gram-positive bacteria.
The genes that encode the structural subunits of A. naeslundii T14V type 1 and type 2 fimbriae and A. naeslundii WVU45 type 2 fimbriae have been cloned previously, and results indicate that these genes encode proteins of approximately 54 to 59 kDa (13, 45–47). Nucleotide sequencing of the type 1 subunit of strain T14V and the type 2 subunit of strain WVU45 (47) revealed significant similarity between the encoded proteins. These studies also showed the presence in each subunit of an N-terminal leader and a C-terminal cell wall sorting signal, which is common among gram-positive cell surface proteins (37). The detection of a cell wall sorting signal in the fimbrial subunits is of interest since individual subunits are not expected to become covalently anchored to the cell wall peptidoglycan. The possible role of this sorting signal in fimbrial processing and polymerization in A. naeslundii has not been examined. Interestingly, results from a recent study showed that mutant strains generated by insertional inactivation of a fimbria-associated gene, orf4, 3′ to the A. naeslundii T14V type 1 fimbrial subunit gene expressed subunits that were not assembled into functional type 1 fimbriae (49). A comparison of unassembled to polymerized subunits would provide insights into assembly of fimbriae.
The A. naeslundii T14V type 2 fimbrial subunit gene, fimA, was cloned and expressed previously in Escherichia coli from a recombinant cosmid, pAV1402 (13). This clone expressed a protein of approximately 59 kDa that was detected with an antibody raised against type 2 fimbriae (5). In this report, we present the nucleotide sequence of fimA and an additional gene, designated orf365, 3′ to fimA. Mutants generated by allelic replacement of either fimA or orf365 were examined for type 2 fimbria expression and fimbria-mediated adherence. The immunoreactions of fimbrial antigens from wild-type and isogenic mutants were compared with those of antibodies against either type 2 fimbriae from A. naeslundii T14V or a 20-amino-acid synthetic peptide prepared from the predicted C-terminal sequence of the fimbrial subunit. The results demonstrate clearly that expression of both fimA and orf365 was required for the synthesis of type 2 fimbriae. Moreover, the carboxyl-terminal peptide of the precursor fimbrial subunit appeared to have been cleaved during assembly. To our knowledge, the proposed posttranslational modification is a novel step in biogenesis of fimbriae.
MATERIALS AND METHODS
Bacteria and plasmids.
Bacterial strains and plasmids used in this study are described in Table 1. A complex medium (7) or Lactobacillus-carrying medium (15) supplemented with 20 mM d,l-threonine was used to prepare cultures of Actinomyces strains, and Luria-Bertani (LB) (31) was used for E. coli strains. The antibiotics (Sigma Chemical Co., St. Louis, Mo.) used in this study were kanamycin sulfate, streptomycin, and ampicillin, at 40, 50, and 100 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used
| Bacterial strain or plasmid | Relevant phenotype and/or genotypea | Reference or source |
|---|---|---|
| Strain | ||
| A. naeslundii | ||
| T14Vb | Wild-type strain, expresses type 1 and 2 fimbriae, Smr Kms | 8 |
| 5951 | Spontaneous mutant of strain T14V, lacks type 1 fimbriae, expresses type 2 fimbriae, Smr Kms | 8 |
| 147 | Spontaneous mutant of strain 5951, lacks type 1 and 2 fimbriae, Smr Kms | 8 |
| MYT2-DC7c | Lacks type 2 fimbriae, ΔfimA::kan Smr Kmr | This study |
| MYT2-SC8d | Lacks type 2 fimbriae, ΔfimA::kan, contains a copy of pMY2201, Smr Kmr | This study |
| MYT2-SC3d | Expresses FimA, contains a copy of pMY2201, ΔfimA::kan Smr Kmr | This study |
| MY2366-DC2c | Expresses FimA, Δorf365::kan Smr Kmr | This study |
| MY2366-SC1d | Expresses type 2 fimbriae, Δorf365::kan, contains a copy of pMY2366, Smr Kmr | This study |
| S. oralis 34 | Synthesizes cell wall polysaccharide that serves as the receptor for Actinomyces type 2 fimbriae | 26, 27 |
| E. coli | ||
| DH5α | Aps | Gibco-BRL |
| TG1 | Aps | Amersham |
| JM109 | Aps | Gibco-BRL |
| AV3502 | TG1 carrying pGP1-2 and pAV3502, Apr Kmr | This study |
| Plasmids | ||
| pAV1402 | Contains a 48-kb A. naeslundii T14V chromosomal DNA subcloned onto pHC79 | 13 |
| pAV3022 | Contains a 9.0-kb HindIII DNA fragment from pAV1402 subcloned onto pUC13, Apr | This study |
| pAV3502 | Contains a 2.4-kb SmaI DNA fragment from pAV3022 subcloned onto pGEM3Z for high-level expression of FimA, Apr | This study |
| pAV2606 | Contains a 1.85-kb BamHI DNA fragment from pAV3022 that encoded a 22-kDa truncated amino-terminal portion of FimA, Apr | This study |
| pAV2621 | Contains a 4.4-kb BamHI DNA fragment from pAV3022 that encoded a 35-kDa truncated carboxyl-terminal portion of FimA, Apr | This study |
| pMY300 | Contains the kan gene from pJRD215 subcloned onto pGEM7Zf(+), Apr Kmr | This study |
| pMY221 | Contains a 2.4-kb SmaI DNA fragment from pAV3022 subcloned onto pUC13 that harbors A. naeslundii T14V fimA, Apr | This study |
| pMY2201 | The 975-bp KpnI DNA fragment internal to fimA is deleted and replaced with the kan gene from pJRD215; transcription of kan is opposite that of fimA; ΔfimA::kan Apr Kmr | This study |
| pMY2366 | The 600-bp BstXI DNA fragment internal to orf365 is substituted with the kan gene; transcription of kan is opposite that of orf365; Δorf365::kan Apr Kmr | This study |
| pJRD215 | Kmr Smr | 10 |
| pUC13 | Apr | Gibco-BRL |
| pGEM7Zf(+) | Apr | Promega |
| pGEM3Z | Apr | Promega |
| pGP1-2 | Contains the T7 RNA polymerase gene, Kmr | 40 |
Km, kanamycin; Sm, streptomycin; Ap, ampicillin.
Made resistant to streptomycin by selection of colonies from a growth medium containing the antibiotic.
Mutant strain generated by a double-crossover recombinational event.
Mutant strain generated by a single-crossover recombinational event predicted by the mechanism of Campbell.
Preparation of A. naeslundii T14V type 2 fimbrial antigens.
Type 2 fimbriae were isolated from A. naeslundii 5951, a spontaneous mutant that expresses only type 2 fimbriae (Table 1). Bacteria from the stationary phase of growth were washed with Tris HCl-buffered saline (TBS; 0.15 M NaCl, 0.02 M Tris-HCl [pH 7.8], 0.1 mM CaCl2, 0.1 mM MgCl2, 0.02% sodium azide) and subjected to sonication (4). Fimbriae collected in fractions at or near the void volume of a Sephacryl S400 (Pharmacia Biotech, Inc., Piscataway, N.J.) column were purified by fractional ammonium sulfate precipitation at 4°C as described previously (5, 29). Edman degradation of purified type 2 fimbriae was performed as described previously (47).
The precursor subunit protein (FimA) from E. coli AV3502 was purified by a procedure similar to that described previously (45). A sonicated extract was applied to a DEAE-Sephacel (Pharmacia Biotech) column and eluted with a gradient of 0.05 to 0.2 M NaCl in TBS. FimA was monitored by solid-phase immunoassay with anti-A. naeslundii T14V type 2 fimbrial antibody and further purified by Sephacryl S-300 (Pharmacia Biotech) gel filtration column chromatography. Final purification of FimA was by immunoaffinity chromatography with a column prepared with an anti-A. naeslundii T14V type 2 fimbrial monoclonal antibody and by elution with 3 M sodium thiocyanate. The concentration of antigens was determined by a micro-bicinchoninic acid protein assay (Pierce, Rockford, Il.), using bovine serum albumin as the standard.
A synthetic peptide consisting of an amino-terminal cysteine followed by the carboxyl-terminal 20 amino acid residues (VGSVLVARYRERKQNANLAL) of A. naeslundii T14V FimA was synthesized on a 430A automated peptide synthesizer (Applied Biosystems, Inc., Foster City, Calif.). The amino acid composition of the peptide was determined by amino acid analysis as described previously (47). For immunization, the peptide was conjugated to keyhole limpet hemocyanin (KLH), using sulfo-m-maleimidobenzoyl-N-hydroxysulfo-succinimide ester (Pierce) as the cross-linker.
Antisera and immunological methods.
A rabbit was immunized with peptide-KLH conjugate (1 mg) in Freund’s complete adjuvant on day 1 and the same amount of conjugate in incomplete adjuvant on days 21, 42, and 63. The antipeptide antiserum (JC8) was obtained 1 week after the last injection. The production of rabbit antiserum (R55) against purified A. naeslundii T14V type 2 fimbriae and the preparation of monospecific immunoglobulin G (IgG) fractions from these antisera have been described previously (5, 9).
Enzyme-linked immunosorbent assay (ELISA) was performed with flat-bottom wells of Immulon I plates (Dynatech Laboratories, Inc., Alexandria, Va.) that were coated overnight at 4°C with purified type 2 fimbriae (3 μg/ml), purified recombinant subunit (3 μg/ml), or synthetic peptide (100 μg/ml). The amount of antibody bound to adsorbed antigen was detected with a secondary horseradish peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad Laboratories, Hercules, Calif.). Western blotting was performed as described previously (44), using either rabbit immune IgG (1 μg/ml) or antiserum diluted to at least 1:500. Horseradish peroxidase-conjugated goat anti-rabbit IgG was the secondary antibody, and blots were developed with reagents supplied in the immunoassay kit (Bio-Rad Laboratories).
Molecular DNA manipulations.
Restriction endonuclease maps of plasmid DNA were determined by standard methods (31). Subcloning and construction of integration vectors were performed with DNA fragments eluted from agarose gels with reagents from an Elu Quik kit (Schleicher & Schuell, Keene, N.H.). Ligations and transformations of E. coli with various constructs were performed by procedures described previously (14, 31). The host strain for plasmids pAV3022, pAV2621, and pAV2606 was E. coli JM109. For plasmids pMY221, pMY2366, and pMY2201, E. coli DH5α was the host strain, and E. coli TG1 carrying the resident plasmid pGP1-2 (Amersham Life Science Inc., Arlington Heights, Ill.) (40) was the host strain for plasmid pAV3502. Transformants were selected on LB agar containing antibiotics, and plasmid DNA was isolated by the alkaline lysis method and purified by CsCl-ethidium bromide density gradient centrifugation (31). The nucleotide sequence of A. naeslundii T14V chromosomal DNA in plasmids pAV3502 and pAV2621 was determined by the Sanger dideoxy-chain termination procedure (34), using a Sequenase kit (version 7.0; United States Biochemical Corp., Cleveland, Ohio) and [35S]dATP (12.5 mCi/ml; DuPont England Nuclear, Boston, Mass.). Primers for DNA sequencing and for amplification of DNA fragments by PCR were prepared on an Applied Biosystems model 391 DNA synthesizer. PCR was performed with either Pfu DNA polymerase (Stratagene, La Jolla, Calif.) or Taq DNA polymerase (Life Technologies, Inc.), using conditions similar to those described previously (44). Nucleotide sequences were analyzed by using the software package of the Genetics Computer Group (version 9.0; University of Wisconsin Biotechnology Center) (12). Sequence homology searches to other bacterial proteins in public databases were performed with the program BLAST (1).
Isolation and characterization of mutants.
Purified integration plasmid DNA (100 ng) was used to transform A. naeslundii T14V by electroporation (48), and transformants were selected on brain heart infusion agar containing kanamycin and streptomycin. Chromosomal DNA from mutants was digested with restriction endonucleases, separated by agarose gel electrophoresis, and analyzed by Southern blot hybridization, under conditions of high stringency, to various 32P-labeled DNA probes (43). Briefly, DNA on filters was prehybridized at 42°C for 2 to 4 h in a solution containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50% formamide, 10% dextran sulfate (Pharmacia Biotech), 1× Denhardt’s solution, 1% sodium dodecyl sulfate (SDS), 1 M NaCl, 0.5% sodium pyrosphosphate, and 200 μg of denatured herring sperm DNA (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) per ml. Hybridizations were at the same temperature for 18 to 20 h. Posthybridization washes were with 0.1× SSC–0.5% SDS at 65°C for 1 h with one change of buffer. Sonicated bacterial cell extracts from A. naeslundii strains were prepared as described previously (44). A cell extract enriched for cytoplasmic proteins also was obtained by disruption of washed bacteria which had been passed twice at 500 lb/in2 (equivalent to cell pressure of 8,000 lb/in2) through a French pressure cell (American Instrument Co., Silver Spring, Md.). Cell debris was removed by centrifugation at 16,000 rpm in a SW40Ti rotor (Beckman Instruments, Inc., Fullerton, Calif.) for 1 h at 4°C, and the supernatant fluid was brought to 40% saturation with solid ammonium sulfate at 4°C. The precipitated proteins were dissolved in TBS and dialyzed extensively against TBS prior to SDS-polyacrylamide gel electrophoresis (PAGE) (25) on 10% polyacrylamide gels and transfer to nitrocellulose.
The adherence properties of each mutant strain were assessed by the coaggregation assay using S. oralis 34 as the partner strain (26). Cell suspensions (5 × 108 in a final volume of 50 μl) of A. naeslundii parent or mutant strains and S. oralis 34 were mixed in wells of microtiter plates, and results were scored as described previously (7). Reversibility of coaggregation was determined in the presence of 125 mM lactose.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was assigned GenBank accession no. AF019629.
RESULTS
Restriction site mapping, subcloning, and expression.
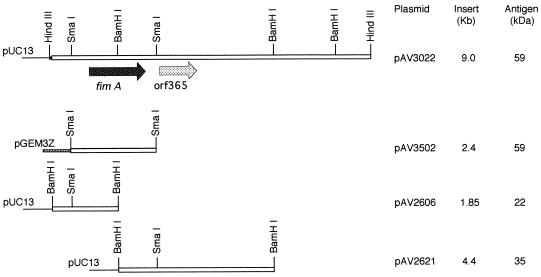
A restriction endonuclease map was determined for the recombinant cosmid pAV1402 (13), and fimA was localized on the inserted DNA fragment by immunological screening of various subclones with anti-A. naeslundii T14V type 2 fimbrial antibody. The 59-kDa type 2 fimbrial subunit protein was detected by Western blot analysis in a subclone carrying the plasmid pAV3022, which contained a 9.0-kb HindIII DNA fragment from pAV1402 (Fig. 1). Shotgun subcloning of pAV3022 after SmaI digestion resulted in the isolation of plasmids pAV3502 (Fig. 1) and pMY221 (Table 1), which had an insertion of a 2.4-kb SmaI DNA fragment that encoded the 59-kDa subunit protein. Two additional derivatives, designated pAV2606 and pAV2621, which contained BamHI DNA fragments from pAV3022 were obtained. These subclones encoded proteins of approximately 22 and 35 kDa, respectively, that were immunostained with anti-A. naeslundii T14V type 2 fimbrial antibody. From the physical maps of the plasmids, these proteins represented the truncated N- and C-terminal portions, respectively, of the structural subunit (Fig. 1). Expression of these truncated proteins was directed by the promoter of the vector, and the A. naeslundii T14V DNA in pAV2621 was fused in frame to the lacZ′ sequence of pUC13.
FIG. 1.
Restriction endonuclease maps of recombinant cosmid pAV1402 and its derivatives. The size of the inserted A. naeslundii T14V DNA in each plasmid and the apparent molecular weights of the plasmid-encoded proteins detected by immunostaining with the anti-A. naeslundii T14V type 2 fimbriae antibody are indicated. Selected restriction endonuclease recognition sites are included for reference. Symbols: ▪, pHC79 DNA; _____, pUC13 DNA; ▨, pGEM3Z DNA; and □, A. naeslundii T14V DNA.
Sequence analysis of fimA and a putative gene 3′ to fimA.
The nucleotide sequences of the 2.4-kb SmaI and the 4.4-kb BamHI DNA fragments from pAV3502 and pAV2621, respectively, were determined. The DNA sequence containing fimA and an open reading frame, designated orf365, is presented in Fig. 2. Both fimA and orf365 started with an ATG initiation codon that was preceded by a putative ribosomal binding site (Fig. 2) (38). The fimA gene (nucleotides 482 to 2086) encoded a predicted protein of 535 amino acids, and the amino-terminal 30+ residues of the deduced protein had properties characteristic of a leader sequence (41). Results of Edman degradation of type 2 fimbriae purified from A. naeslundii T14V identified glutamate at position 34 of the predicted protein sequence as the N-terminal amino acid. The next 29 amino acid residues determined by amino acid sequencing were identical to the predicted protein sequence (Fig. 2). Thus, the putative leader sequence cleavage site of the precursor protein encoded by fimA is between threonine and glutamate at positions 33 and 34, respectively (Fig. 2).
FIG. 2.
Nucleotide sequence of a 3.67-kb A. naeslundii T14V chromosomal DNA region containing the type 2 fimbrial subunit gene, fimA, and a putative gene, orf365, involved in fimbrial biogenesis. The presumptive ribosomal binding site (rbs; underline), two inverted repeats (arrow and dotted underline) downstream of the termination codon, TGA (∗), of fimA, the amino-terminal amino acid sequence of type 2 fimbriae (dotted underline) determined by Edman degradation, the leader peptide processing site (upward arrow) of the subunit precursor, the conserved cell wall anchoring motif (LPXTG; boxed), the 20-amino-acid carboxyl-terminal sequence (thick underline) of FimA used to prepare a rabbit antipeptide antibody, and the putative transmembrane segment in ORF365 (open bar) are indicated. Selected restriction endonuclease recognition sites are included for reference.
The predicted type 2 fimbrial subunit of A. naeslundii T14V, obtained following cleavage of the leader, consisted of 502 amino acid residues and had a calculated molecular weight of 52,847. The hydropathy of the predicted protein, plotted by the Kyte-Doolittle method (24), suggested a molecule that was predominately hydrophilic except for the presence of a hydrophobic region at the C-terminal end. The sequence of the C-terminal 43 amino acid residues resembled that of a cell wall sorting signal (28, 36, 37) which has been noted in many gram-positive cell surface proteins and other Actinomyces fimbrial subunits (46, 47). This signal comprises the consensus cell wall anchoring motif, LPXTG (Fig. 2), followed, in sequence, by a hydrophobic domain and a positively charged hydrophilic domain. To detect the C-terminal end of the fimbrial subunit protein, rabbit antibody was prepared against a synthetic peptide consisting of the C-terminal 20 amino acid residues (Fig. 2). This antiserum reacted strongly in ELISA with the recombinant subunit protein (FimA) purified from E. coli AV3502 and with the unconjugated synthetic peptide. Significantly, the antibody did not react above the level of preimmune serum with type 2 fimbriae purified from strain T14V (Table 2). In contrast, rabbit antiserum against type 2 fimbriae from strain T14V did not react above the level of preimmune serum with the unconjugated peptide but reacted strongly both with the recombinant fimbrial subunit and with type 2 fimbriae. Comparable results were obtained with purified type 2 fimbriae and recombinant subunit protein that were subjected to SDS-PAGE and transferred to nitrocellulose. The patterns from Western blotting with anti-A. naeslundii T14V type 2 fimbriae antibody were similar to those observed previously (13) and consisted of a characteristic ladder of high-molecular-weight proteins in addition to a relatively weak band of monomeric subunit for type 2 fimbriae and a single band at 59 kDa for the recombinant protein. In contrast, the antipeptide antibody did not detect bands from transferred fimbriae but reacted strongly with the recombinant FimA (profile not shown). Thus, epitopes associated with the C-terminal 20 amino acids predicted by the nucleotide sequence of fimA were detected in the recombinant fimbrial subunit but not in fimbriae isolated from A. naeslundii T14V cell surface.
TABLE 2.
Reactions of rabbit antisera with A. naeslundii T14V type 2 fimbriae and related antigens measured by ELISA
| Antigen | Dilution−1 of antiserum for
optical density of 0.5 in ELISA
|
|
|---|---|---|
| Anti-carboxyl-terminal peptide | Anti-A. naeslundii T14V type 2 fimbriae | |
| C-terminal peptidea | 6 × 104 | <1 × 102 |
| FimAb | 2 × 104 | 5 × 105 |
| Fimbriaec | <1 × 102 | 1 × 106 |
Synthetic peptide representing the 20 amino acids at the carboxyl terminus of the fimA-encoded protein.
Type 2 subunit protein purified from E. coli AV3502 (FimA).
Type 2 fimbriae purified from A. naeslundii 5951 (fimbriae).
Two inverted repeats were located immediately 3′ of the termination codon of fimA (nucleotides 2097 to 2134 and 2217 to 2762) (Fig. 2). The calculated free energies (17) of these potential hairpin structures were −35 and −49 kcal, respectively. A predicted RNA secondary structure encompassing these repeat sequences generated by the program FOLD (50) had an overall calculated free energy of −137.7 kcal. This region of dyad symmetry was followed by a putative gene, orf365 (nucleotides 2468 to 3562), that encoded a predicted protein of 365 amino acids with a calculated molecular weight of 39,425. Similar to the type 2 fimbrial structural subunit, ORF365 also was predominately hydrophilic (24). However, unlike the subunit protein, no detectable leader sequence or cell wall sorting signal motifs were observed in ORF365. Further analysis of ORF365 suggested that this protein contained one membrane helix (between amino acids 238 and 255) (Fig. 2) (42). Results of a topology prediction (with a reliability of 7 on a scale of 0 to 9, with 9 being most reliable) (30) indicated that the N-terminal two-thirds and the C-terminal one-third of ORF365 were located outside and inside the cytoplasmic membrane, respectively.
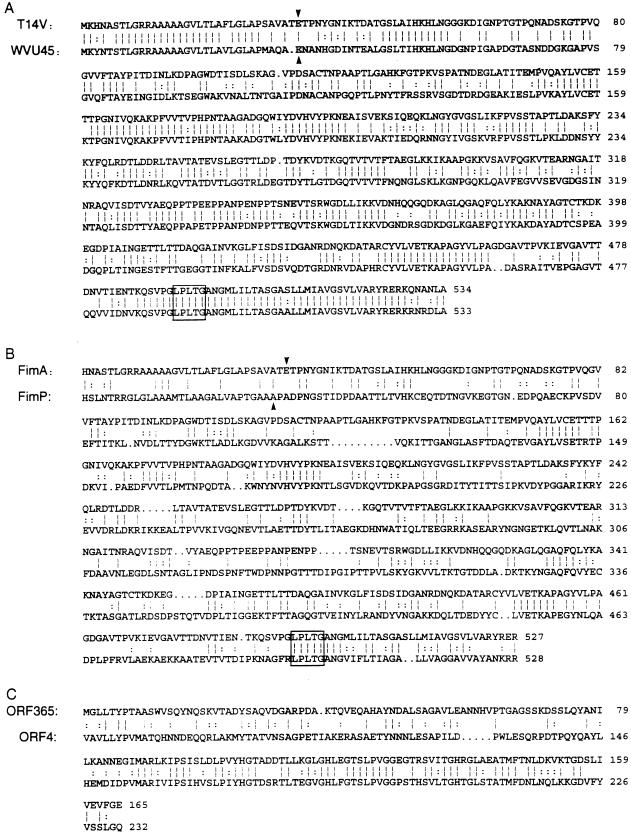
Sequence homology between FimA or ORF365 and other bacterial proteins was noted only with fimbria-associated proteins from A. naeslundii T14V or WVU45 (46, 47, 49). Sequence alignments by the program Bestfit (Genetics Computer Group) showed significantly greater sequence similarity between FimA of strain T14V and the type 2 fimbrial subunit of strain WVU45 (65% sequence identity and 77% similarity) (Fig. 3A) (46) than between FimA and the type 1 fimbrial subunit (FimP) of strain T14V (31% sequence identity and 38% similarity) (Fig. 3B). Significant homology (40% identity and 47.5% similarity) also was noted between the amino-terminal half of ORF365 and the central region of the predicted protein encoded by orf4, a type 1 fimbria-associated gene of A. naeslundii T14V (49).
FIG. 3.
Sequence homology between the deduced amino acid sequences of type 2 fimbrial subunits of A. naeslundii T14V and A. naeslundii WVU45 (A), type 2 (FimA) and type 1 (FimP) fimbrial subunits of A. naeslundii T14V (B), and the N-terminal portion of the protein encoded by orf365 flanking fimA and the central portion of the protein encoded by orf4 flanking fimP of the type 1 fimbrial gene cluster (C). Identical (|) and conserved substituted (:) amino acid residues are indicated. The N-terminal amino acid (downward arrow) determined by Edman degradation of purified fimbriae and the consensus cell wall anchoring motif (LPXTG; boxed) are indicated.
Construction of fimA and orf365 mutants.
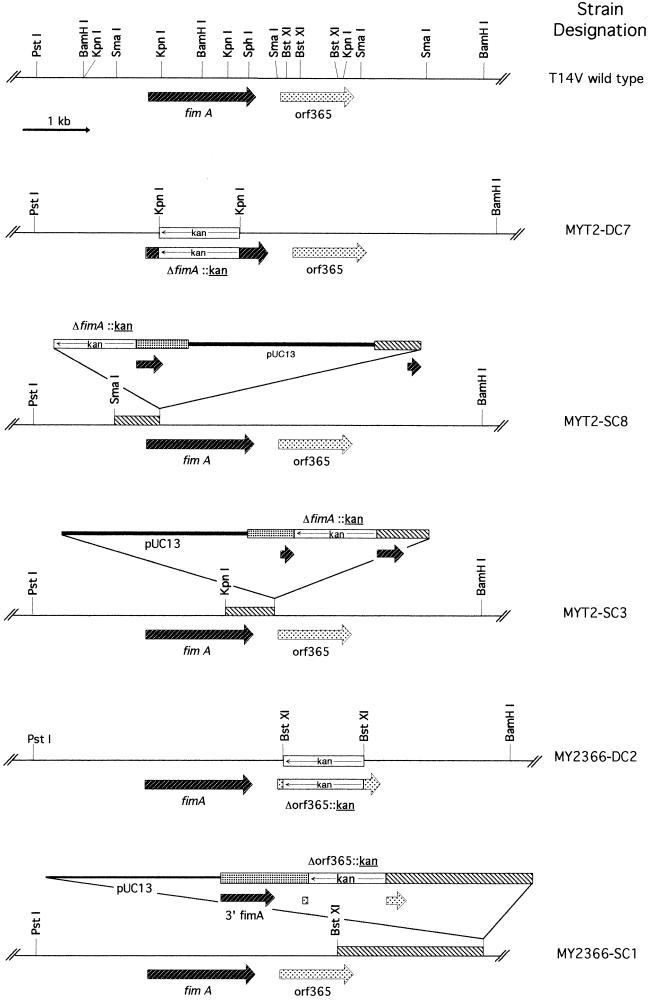
Integration plasmids pMY2201 and pMY2366 were constructed by substituting the kanamycin resistance (kan) gene from pJRD215 (10) for the 975-bp KpnI and 600-bp BstXI DNA fragments internal to fimA and orf365, respectively (Table 2 and Fig. 2). Kanamycin-resistant transformants were obtained by transformation of A. naeslundii T14V with these plasmids. The physical maps of representative mutants (Fig. 4) were determined by Southern blot analysis of genomic DNA digested with various restriction endonucleases and hybridized to various DNA probes, including pUC13 DNA, kan, fimA, and orf365. Results of these analyses showed that strains MY2T-DC7 and MY2366-DC2 were generated by allelic replacement of fimA and orf365, respectively, with the kan gene. The lack of hybridization signal between strain MYT2-DC7 and the 975-bp KpnI DNA internal to fimA, and between strain MY2366-DC2 and the 600-bp BstXI DNA internal to orf365, confirmed that each specific DNA sequence was deleted from the respective mutant. As expected from the insertion-and-duplication mechanism predicted by Campbell (2), two types of single-crossover mutants were obtained with pMY2201. Those like strains MYT2-SC8 and MYT2-SC3 each contained a copy of the intact fimA sequence (Fig. 4). The only single-crossover mutants obtained with pMY2366 were those like MY2366-SC1 (Fig. 4); mutants with insertions of pMY2366 between the BamHI site in fimA and the 5′ BstXI site of orf365 were not isolated.
FIG. 4.
Restriction endonuclease maps of A. naeslundii wild-type strain T14V and isogenic mutants generated by allelic replacement of fimA (strain MYT2-DC7) or orf365 (strain MY2366-DC2) and those generated by single crossover with the integration plasmid pMY2201 (strains MYT2-SC8 and MYT2-SC3) or pMY2366 (strain MY2366-SC1). Only the chromosomal DNA region flanking fimA and orf365 and selected restriction endonucleases are included. Symbols: _____, A. naeslundii T14V DNA; ——, pUC13 DNA; □, kan gene; , fimA; , orf365. The chromosomal region where plasmid integration occurred as mediated by the Campbell insertion-duplication mechanism is also indicated (▧).
Roles of fimA and orf365 in biogenesis of fimbriae.
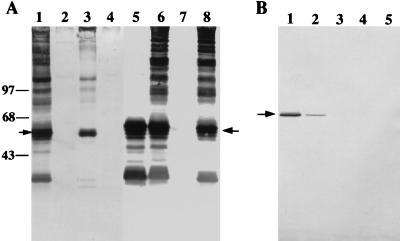
Type 2 fimbria-mediated adherence, as determined by the lactose-sensitive coaggregation of Actinomyces strains with S. oralis 34, was completely abolished by allelic replacement of fimA in strain MYT2-DC7 and by integration of pMY2201 in fimA of strain MYT2-SC8 or MYT2-SC3. No type 2 fimbrial antigens were detected in strains MYT2-DC7 and MYT2-SC8 (Fig. 5A, lanes 2 and 4, respectively), as shown by Western blot analysis of sonicated cell extracts with anti-type 2 fimbrial antibody. Thus, although strain MYT2-SC8 contained a copy of fimA (Fig. 4), the lack of FimA production suggested that fimA might be part or an operon or that the expression of genes 5′ to fimA was required for fimA expression. Some high-molecular-weight protein bands along with the fimbrial subunit were present in strain MYT2-SC3 (Fig. 5A, lane 3). However, minor differences were noted between the immunostained protein profile of this strain and that of the wild-type strain (Fig. 5A, lane 1), suggesting the possibility of truncated type 2 fimbriae produced by this strain.
FIG. 5.
(A) Composite of Western blots of sonicated cell extracts of A. naeslundii T14V (wild type), mutant strains MYT2-DC7, MYT2-SC3, MYT2-SC8, MY2366-DC2, and MY2366-SC1, strain 147, and strain 5951 with anti-A. naeslundii T14V type 2 fimbrial antibody (lanes 1 through 8, respectively). Proteins were separated by SDS-PAGE, and transferred proteins on nitrocellulose were immunostained with anti-A. naeslundii T14V type 2 fimbrial antibody. The apparent molecular sizes (in kilodaltons) are indicated on the left. (B) Western blot of cell extracts of A. naeslundii mutant strains MY2366-DC2, MYT2-SC3, 147, and 5951 and wild-type strain T14V (lanes 1 through 5, respectively). Transferred proteins on nitrocellulose were immunostained with rabbit anti-peptide antibody prepared against the predicted C-terminal sequence of FimA. Arrows indicate the 59-kDa type 2 fimbrial subunit.
Lactose-sensitive coaggregation activity was also abolished by allelic replacement of orf365 in mutant strain MY2366-DC2 but was not affected by insertion of pMY2366 3′ to orf365 in strain MY2366-SC1. Only the fimbrial subunit protein was detected in sonicated extract of strain MY2366-DC2 along with immunoreactive bands of lower molecular weight (Fig. 5A, lane 5). The latter bands were degradative products of the subunit, as suggested previously (13). In contrast, the immunoblot profile of sonicated extract of strain MY2366-SC1 was similar to that observed in the wild-type strain T14V or strain 5951, which expresses only type 2 fimbriae (Fig. 5A; compare lane 6 to lanes 1 and 8). The subunit expressed by strains MY2366-DC2 and MYT2-SC3 was also detected by Western blotting with the antipeptide antibody prepared against the C-terminal end of FimA. The subunit protein appeared as a sharp narrow immunostained band with this antibody but as a broadly stained band with the anti-type 2 fimbrial antibody, even though equal amounts of sonicated extract were used in both analyses. A difference in the amount of antigen detected by the antipeptide antibody in strain MY2366-DC2 and MYT2-SC3 also was indicated by the greater intensity of the subunit band observed with 35 and 70 μg, respectively, of sonicated extract from these strains (Fig. 5B, lanes 1 and 2). Significantly, the antipeptide antibody did not react with 150 μg of sonicated extracts or French press extracts from strains 147 and 5951 and wild-type strain T14V (Fig. 5B, lanes 3, 4, and 5, respectively). However, the subunit and higher-molecular-weight type 2 fimbrial antigens were readily detected by Western blotting with anti-type 2 fimbrial antibody in the extracts of strains T14V and 5951 (Fig. 5A, lanes 1 and 8, respectively). The specificity of the anti-type 2 fimbrial antibody used in these experiments was indicated by the absence of any reaction with strain 147, which lacks both type 1 and 2 fimbriae (Fig. 5A, lane 7; Fig. 5B, lane 3). Thus, the unassembled subunit expressed in the mutant strain MY2366-DC2 or MYT2-SC3 also contained the C-terminal peptide of the precursor subunit FimA.
DISCUSSION
Results from this study demonstrate that at least two genes, namely, the fimbrial subunit gene, fimA, and the 3′ adjacent gene orf365, are necessary for the synthesis of functional type 2 fimbriae in A. naeslundii T14V. The conclusion is supported by the lack of expression of type 2 fimbrial antigen by the fimA mutant MYT2-DC7 and of assembled fimbriae by the orf365 mutant MY2366-DC2. The mutant strains, MYT2-DC7 and MYT2-SC8, that lacked type 2 fimbriae were unable to coaggregate with S. oralis 34, which has a receptor polysaccharide for the type 2 fimbrial lectin (26). However, expression of fimA alone was not sufficient for the adherence properties observed in the wild-type strain, since mutant strains MYT2-SC3 and MY2366-DC2, which synthesized only the fimbrial subunit, also failed to coaggregate with S. oralis 34. In a previous study of strain T14V type 1 fimbriae (44, 49), mutants that produced the unassembled type 1 subunits but not type 1 fimbriae did not adhere to proline-rich proteins that are specific receptors of these fimbriae (18). Further studies to identify and characterize the genes involved in biogenesis of type 1 and type 2 fimbriae should provide a firm basis for associating the receptor binding sites of these structures either with the structural subunits, FimP and FimA, respectively, or with minor fimbrial proteins.
The hypothesis that orf365 is involved in the assembly of type 2 fimbriae was supported by the following observations. First, mutants generated by allelic replacement of orf365 coding sequence with the kan cassette expressed only monomeric fimbrial subunit. Second, significant sequence similarity was observed between the predicted protein encoded by orf365 and that encoded by orf4, a gene which is located immediately 3′ to the A. naeslundii T14V type 1 fimbrial subunit gene, fimP (49). Third, an isogenic mutant of orf4 created by allelic replacement also expressed only unassembled fimbrial subunits (49). These similarities suggested that orf365 and orf4 may play similar roles in the synthesis of type 2 and type 1 fimbriae, respectively. It is of interest that the mutant strain, MY2366-SC1, which contained pMY2366 integrated beyond the 3′ end of orf365 produced functional type 2 fimbriae. Thus, orf365 may be the last member of the type 2 fimbrial gene cluster(s). Indeed, analysis of the nucleotide sequence of the chromosomal DNA region between the SmaI site 3′ to orf365 and the downstream BamHI site (Fig. 5; note restriction endonuclease map of strain T14V [Fig. 4]) revealed the presence of genes encoding ribosomal proteins (data not shown). Further analysis of the DNA sequence 5′ of fimA may reveal additional fimbria-associated genes, as is the case with the type 1 fimbrial gene cluster in A. naeslundii T14V (49), which consists of seven genes, including fimP.
Striking similarities have been observed among different fimbrial types from E. coli and related gram-negative bacteria (11, 16, 19, 23, 39). The similarities include significant homologies between structural subunit and fimbria-associated proteins. In addition, the organizations of genes that encode major and minor fimbrial components, chaperone proteins, or other proteins involved in subunit transport and control of fimbrial assembly are similar (11, 16, 19, 23). Information gained from the sequence analysis of fimA and its gene product in this study extends previous observations that common characteristics also exist among various A. naeslundii fimbrial genes. Thus, each of three fimbrial subunit genes (type 1 of A. naeslundii T14V and type 2 of strains T14V and WVU45) encodes a precursor subunit that has a typical leader sequence of approximately 30 amino acids. The fimbrial subunits of A. naeslundii are generally more hydrophilic than the subunits of gram-negative bacterial fimbriae (21). At the protein level, significant sequence homology (33%) was observed between the type 1 fimbrial subunit of A. naeslundii T14V and the type 2 fimbrial subunit of strain WVU45 (47). A similar level of sequence homology also was observed between the fimP- and fimA-encoded proteins (Fig. 3B), suggesting that an overall sequence identity of approximately 30% may be expected between the structural subunits of A. naeslundii type 1 and type 2 fimbriae. Greater homology would be anticipated between the subunits of functionally similar fimbriae. Indeed, an overall sequence similarity of 77% was found between the A. naeslundii T14V and WVU45 type 2 fimbrial subunits (Fig. 3A) even though the type 2 fimbriae of these strains are only weakly cross-reactive (6). Finally, the relative locations of orf365 and orf4 with respect to the structural subunit genes would favor the hypothesis that the type 1 and type 2 fimbrial gene clusters are organized similarly.
An epitope(s) associated with the predicted carboxyl terminus of the type 2 fimbrial subunit was detected in recombinant FimA from E. coli and unassembled FimA synthesized by the orf365 knockout mutant but not in fimbriae from A. naeslundii T14V. Thus, the C terminus of FimA must either become inaccessible to antibody or, alternatively, be removed during assembly of fimbriae. The latter possibility is favored by the results of Western blotting of cell extracts of strains 5951 and T14V in which FimA monomer was not detected by antibody against the predicted C-terminal peptide of FimA (Fig. 5B, lanes 4 and 5, respectively) but was readily detected by antibody against type 2 fimbriae (Fig. 5A, lanes 8 and 1, respectively). In addition to these findings, the removal of the peptide at the C-terminal end of FimA during assembly of fimbriae would be consistent with the presence of a cell wall sorting signal in the subunit (Fig. 2). Based on the general model for trafficking of surface proteins in gram-positive bacteria (28, 35), it is likely that the carboxyl terminus of the precursor fimbrial subunit may be cleaved between threonine and glycine of the LPLTG sequence and that the C-terminal threonine, instead of being anchored to cell wall peptidoglycan, is linked to another subunit either directly or through a peptidoglycan fragment that has yet to be detected in mature fimbriae. Alternatively, the processed subunit may be transiently associated with a cell wall protein that functions to initiate subunit assembly. In this regard, a cell-bound nucleator protein that primes the polymerization of E. coli curlins during pilus assembly has been described (20). The possible existence of covalent linkages between subunits of A. naeslundii fimbriae would account for the inability of techniques such as SDS-PAGE to dissociate these structures to subunits (3). Consistent with the predicted primary sequence and experimental data, the A. naeslundii fimbrial subunit precursor would be expected to undergo two posttranslational modifications: removal of the amino-terminal leader sequence during export of the precursor through the cytoplasmic membrane, and removal of the carboxyl-terminal peptide at the cell wall anchoring sequence during subunit assembly. However, the possibility that carboxyl-terminal peptide processing and fimbrial assembly are independent events cannot be excluded. Clearly, further studies are needed to define the mechanism(s) of fimbrial subunit polymerization. The results of such studies should advance our knowledge of fimbrial biosynthesis in this gram-positive species.
ACKNOWLEDGMENTS
We thank Frank Robey (NIDR) for preparation of the synthetic peptide and KLH conjugate, Bob Harr (NIDR) for technical assistance, and Linda Lee (UTHSC at San Antonio) and Donald J. LeBlanc (Indiana University School of Dentistry) for critical review of the manuscript.
This study was supported by grant DE11102 awarded to M.K.Y. from the National Institute of Dental Research.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Campbell A M. Episomes. Adv Genet. 1962;11:101–145. [Google Scholar]
- 3.Cisar J O. Fimbrial lectins of the oral actinomyces. In: Mirelman D, editor. Microbial lectins and agglutinins: properties and biological activity. New York, N.Y: John Wiley & Sons, Inc.; 1986. pp. 183–196. [Google Scholar]
- 4.Cisar J O, Barsumian E L, Curl S H, Vatter A E, Sandberg A L, Siraganian R P. Detection and localization of a lectin on Actinomyces viscosusT14V by monoclonal antibodies. J Immunol. 1981;127:1318–1322. [PubMed] [Google Scholar]
- 5.Cisar J O, Curl S H, Kolenbrander P E, Vatter A E. Specific absence of type 2 fimbriae on a coaggregation-defective mutant of Actinomyces viscosusT14V. Infect Immun. 1983;40:759–769. doi: 10.1128/iai.40.2.759-765.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cisar J O, David V A, Curl S H, Vatter A E. Exclusive presence of lactose-sensitive fimbriae on a typical strain (WVU45) of Actinomyces naeslundii. Infect Immun. 1984;46:453–458. doi: 10.1128/iai.46.2.453-458.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cisar J O, Kolenbrander P E, McIntire F C. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun. 1979;24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cisar J O, Vatter A E, Clark W B, Curl S H, Hurst-Calderone S, Sandberg A L. Mutants of Actinomyces viscosusT14V lacking type 1, type 2 or both types of fimbriae. Infect Immun. 1988;56:2984–2989. doi: 10.1128/iai.56.11.2984-2989.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark W B, Wheeler T T, Cisar J O. Specific inhibition of adsorption of Actinomyces viscosusT14V to saliva-treated hydroxyapatite by antibody against type 1 fimbriae. Infect Immun. 1984;43:397–501. doi: 10.1128/iai.43.2.497-501.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 11.de Graaf F K. Genetics of adhesive fimbriae of intestinal Escherichia coli. Curr Top Microbiol Immunol. 1990;151:29–53. doi: 10.1007/978-3-642-74703-8_2. [DOI] [PubMed] [Google Scholar]
- 12.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donkersloot J A, Cisar J O, Wax M E, Harr R J, Chassy B M. Expression of Actinomyces viscosus antigens in Escherichia coli: cloning of a structural gene (fimA) for type 2 fimbriae. J Bacteriol. 1985;162:1075–1078. doi: 10.1128/jb.162.3.1075-1078.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coliby high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efthymiou C, Hansen C A. An antigenic analysis of Lactobacillus acidophilus. J Infect Dis. 1962;110:258–267. doi: 10.1093/infdis/110.3.258. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich M J, Kinsey N E, Vila J, Kadner R J. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- 17.Freier S M, Kierzek R, Jaeger J A, Sugimoto N, Caruthers M H, Neilson T, Turner D H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbons R J, Hay D I, Cisar J O, Clark W B. Adsorbed salivary proline-rich protein I and statherin: receptors of type 1 fimbriae of Actinomyces viscosusT14V-J1 on apatitic surfaces. Infect Immun. 1988;56:2990–2993. doi: 10.1128/iai.56.11.2990-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hacker J. Genetic determinants coding for fimbriae and adhesins of extraintestinal Escherichia coli. Curr Top Microbiol Immunol. 1990;151:1–27. doi: 10.1007/978-3-642-74703-8_1. [DOI] [PubMed] [Google Scholar]
- 20.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinson G, Williams P H. Adhesins of pathogenic Escherichia coli. In: Hopwood D A, Chater K C, editors. Genetics of bacterial diversity. New York, N.Y: Academic Press; 1989. pp. 287–307. [Google Scholar]
- 22.Hultgren S J, Jones C H, Normark S. Bacterial adhesins and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 2730–2756. [Google Scholar]
- 23.Hultgren S J, Normark S, Abraham S N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:383–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 24.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.McIntire F C, Crosby L K, Vatter A E, Cisar J O, McNeil M R, Bush C A, Tjoa S S, Fennessey P V. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosusT14V. J Bacteriol. 1988;170:2229–2235. doi: 10.1128/jb.170.5.2229-2235.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIntire F C, Vatter A E, Baros J, Arnold J. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis34. Infect Immun. 1978;21:978–988. doi: 10.1128/iai.21.3.978-988.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 29.Revis G J, Vatter A E, Crowle A J, Cisar J O. Antibodies against the Ag2 fimbriae of Actinomyces viscosusT14V inhibit lactose-sensitive bacterial adherence. Infect Immun. 1982;36:1217–1222. doi: 10.1128/iai.36.3.1217-1222.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Sandberg A L, Mudrick L L, Cisar J O, Brennan M J, Mergenhagen S E, Vatter A E. Type 2 fimbrial lectin-mediated phagocytosis of oral Actinomycesspp. by polymorphonuclear leukocytes. Infect Immun. 1986;54:472–476. doi: 10.1128/iai.54.2.472-476.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandberg A L, Mudrick L L, Cisar J O, Metcalf J A, Malech H L. Stimulation of superoxide and lactoferrin release from polymorphonuclear leukocytes by the type 2 fimbrial lectin of Actinomyces viscosusT14V. Infect Immun. 1988;56:267–269. doi: 10.1128/iai.56.1.267-269.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 36.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 38.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli16S ribosomal RNA: complementarily to nonsense triplets and ribosome-binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 40.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 43.Yeung M K. Conservation of an Actinomyces viscosus T14V type 1 fimbrial subunit homolog among divergent groups of Actinomycesspp. Infect Immun. 1992;60:1047–1054. doi: 10.1128/iai.60.3.1047-1054.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung M K. Construction and use of integration plasmids to generate site-specific mutations in the Actinomyces viscosusT14V chromosome. Infect Immun. 1995;63:2924–2930. doi: 10.1128/iai.63.8.2924-2930.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeung M K, Chassy B M, Cisar J O. Cloning and expression of a type 1 fimbrial subunit of Actinomyces viscosusT14V. J Bacteriol. 1987;169:1678–1683. doi: 10.1128/jb.169.4.1678-1683.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeung M K, Cisar J O. Cloning and nucleotide sequence of a gene for Actinomyces naeslundiiWVU45 type 2 fimbriae. J Bacteriol. 1988;170:3803–3809. doi: 10.1128/jb.170.9.3803-3809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung M K, Cisar J O. Sequence homology between the subunits of two immunologically and functionally distinct types of fimbriae of Actinomycesspp. J Bacteriol. 1990;172:2462–2468. doi: 10.1128/jb.172.5.2462-2468.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeung M K, Kozelsky C S. Transformation of Actinomycesspp. by a gram-negative broad-host-range plasmid. J Bacteriol. 1994;176:4173–4176. doi: 10.1128/jb.176.13.4173-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung M K, Ragsdale P A. Synthesis and function of Actinomyces naeslundiiT14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun. 1997;65:2629–2639. doi: 10.1128/iai.65.7.2629-2639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]