Abstract
Early-life stress experiences can produce lasting impacts on organismal adaptation and fitness. How transient stress elicits memory-like physiological effects is largely unknown. Here, we show that early-life thermal stress strongly up-regulates tsp-1, a gene encoding the conserved transmembrane tetraspanin in C. elegans. TSP-1 forms prominent multimers and stable web-like structures critical for membrane barrier functions in adults and during aging. Increased TSP-1 abundance persists even after transient early-life heat stress. Such regulation requires CBP-1, a histone acetyltransferase that facilitates initial tsp-1 transcription. Tetraspanin webs form regular membrane structures and mediate resilience-promoting effects of early-life thermal stress. Gain-of-function TSP-1 confers marked C. elegans longevity extension and thermal resilience in human cells. Together, our results reveal a cellular mechanism by which early-life thermal stress produces long-lasting memory-like impact on organismal resilience and longevity.
Early-life heat exposure produces long-lasting benefits on longevity in the nematode C. elegans.
INTRODUCTION
Epidemiological and clinical studies in humans show that life stress of various forms can exert profound lasting impacts on mental and physical health outcomes and life spans (1–4). For example, severe nutritional stress early in life can incur life-span costs, while psychosocial early-life stress increases vulnerability to psychiatric disorders (5–7). By contrast, milder physiological stresses, such as fasting with adequate nutrition or thermal stimuli via sauna exposure, are associated with long-lasting health benefits (8, 9). Transient periods of stress can induce persistent changes in the endocrine response, epigenetic regulation of gene expression, and plasticity changes in various organs (10–12). However, the underlying molecular and cellular mechanisms by which transient early-life stress can produce memory-like physiological effects remain poorly understood. In addition, it is challenging to establish causal relationship between putative cellular mechanisms and long-term health outcomes in human studies.
The free-living nematode Caenorhabditis elegans has emerged as a tractable model system to study how early-life stress may affect adult phenotypes. Early-life exposure to starvation, anoxia or osmotic stresses in C. elegans induces the transition into a larval stage 1 (L1) arrested developmental stage or stress-resistant dauer stage, which can persist until environmental conditions become favorable (13–15). Adults that have undergone the dauer stage preserve a memory of their early-life starvation experience, resulting in alterations in gene expression, extended life span, and decreased reproductive capacity (16, 17). Early-life starvation experience can also reorganize neuronal modulation of stage-specific behavioral responses to stresses (18). In addition, a 1-day shift from 20° to 25°C during early adulthood in C. elegans appears to improve stress resistance and extend life span through known stress-responding transcription factors: Forkhead box transcription factor (DAF-16), heat shock transcription factor (HSF-1), and hypoxia inducible transcription factor (HIF-1) (19, 20). It remains unclear how specific effectors of these transcription factors, or other epigenetic mechanisms independent of these factors, may elicit long-lasting impacts on adult stress resilience and longevity.
In this study, we use a robust thermal stress paradigm in C. elegans to uncover causal mechanisms by which transient stress may exert lasting impacts on organismal resilience and longevity. We show that transient heat exposure at 28°C during late larval development activates the gene tsp-1, which encodes a C. elegans homolog of the evolutionarily conserved tetraspanin protein family. Tetraspanin 1 (TSP-1) proteins form tetraspanin web-like structures and are essential for maintaining membrane permeability, barrier functions, and heat-induced organismal resilience and longevity. Initial induction of tsp-1 by heat requires the histone acetyltransferase CBP/p300 homolog (CPB-1); however, unexpectedly, this results in sustained up-regulation of TSP-1 protein without a corresponding increase in mRNA abundance. We propose that the stability of the tetraspanin web-like structure, achieved through TSP-1 multimerization, mediates the long-lasting organismal impact triggered by transient mild thermal stress in early life.
RESULTS
Using RNA sequencing, we previously identified tsp-1 as one of the genes highly up-regulated by cold-warming (CW) stress (transient exposure to 4 or −20°C followed by recovery at 20°C) in C. elegans (21–23). We generated translational reporters by fusing green fluorescent protein (GFP) with endogenous regulatory DNA sequences (promoter and coding regions) for many of the CW stress-inducible genes to monitor their induction kinetics and protein localization in live C. elegans during development and adult responses to thermal stresses. We focus on tsp-1 in this study as the constructed tsp-1p::tsp-1::GFP translational reporter shows robust and notable induction at L4 stages by not only CW stresses but also mild heat stress at 25°C, more so at 28°C, less so at 20°C or above 32°C (Fig. 1, A and B, and fig. S1). Induction by 28°C is heat-duration dependent (fig. S1) and does not require HSF-1 (heat shock factor) (fig. S2) that mediates the canonical heat shock response in C. elegans (24–28). By contrast, the heat shock chaperon gene hsp-16 is also induced by 28°C and requires HSF-1 for heat induction (fig. S2). These results highlight a previously unknown, HSF-independent thermal induction of TSP-1 in C. elegans.
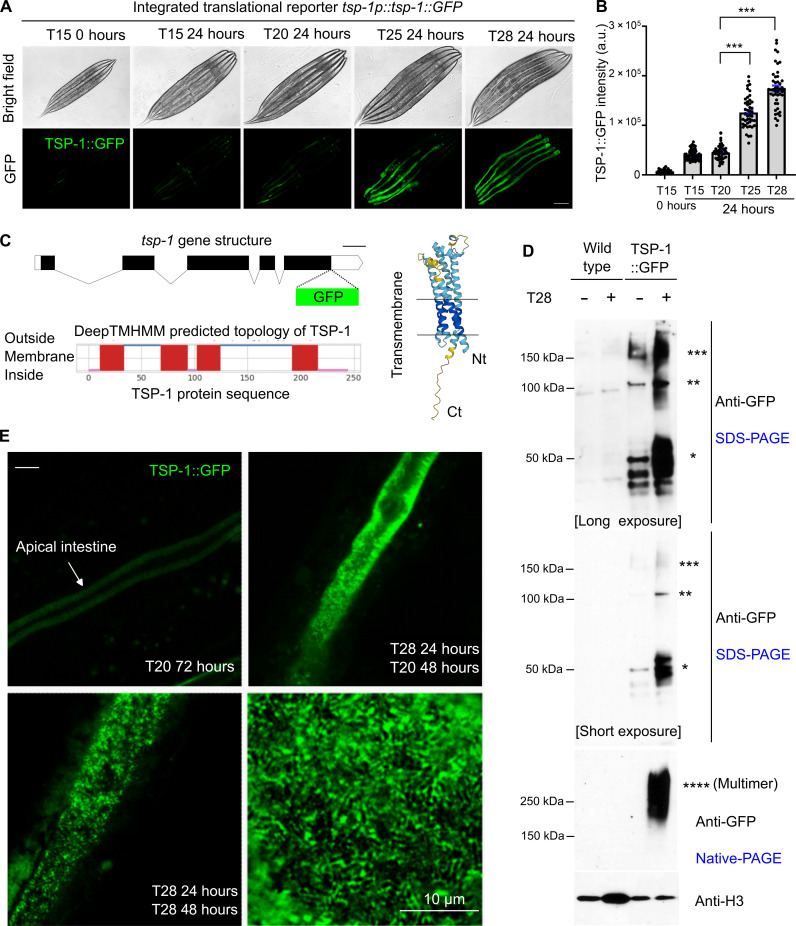
Fig. 1. Thermal stress induces TSP-1 expression and tetraspanin web-like structure formation.
(A) Representative bright field and epifluorescence images showing expression of an integrated transgene tsp-1p::tsp-1::GFP under temperatures and durations indicated. Scale bar, 100 μm. (B) Quantification of TSP-1::GFP fluorescence intensities under conditions indicated. ***P < 0.001 (n > 40 animals per condition). (C) Schematic of the tsp-1 gene structure, predicted membrane topology (by DeepTMHMM), and structure (by AlphaFold2) of TSP-1. (D) SDS-PAGE and native-PAGE Western blots showing heat-induced increase in abundance and formation of dimers and multimers (asterisk) by TSP-1::GFP. (E) Representative confocal fluorescence images showing high-resolution Z-stack views of tetraspanin web structure formed by TSP-1::GFP, with enlarged view at right bottom. Scale bar, 10 μm. a.u., arbitrary units.
TSP-1 is a C. elegans homolog of the evolutionarily conserved tetraspanin protein family in eukaryotes (29–31), predicted to form four-transmembrane α-helical segments and two extracellular loop domains (Fig. 1C). We fused GFP to the C terminus of TSP-1, leaving the N terminus of TSP-1 intact, which contains the first transmembrane helix targeting TSP-1 to plasma membranes (Fig. 1C). Tetraspanins are versatile scaffolding transmembrane proteins, specific members of which can interact with themselves to form multimers and control the spatial organization of membrane lipids and proteins in networks called tetraspanin webs or tetraspanin-enriched microdomains (29–31). We used SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and native PAGE to analyze the thermal induction and multimerization of TSP-1::GFP (Fig. 1D). Under baseline conditions at 20°C, TSP-1::GFP formed protein species corresponding to predicted monomers, SDS-resistant dimers and trimers (in molecular weights). Heat at 28°C for 24 hours drastically increased all TSP-1::GFP species in SDS-PAGE, and native PAGE further revealed the formation of high–molecular weight species larger than dimers and trimers (Fig. 1D). Confocal microscopy revealed that TSP-1::GFP was highly enriched along the apical membrane of intestinal cells (Fig. 1E). Heat at 28°C for 72 hours or a transient period of 24 hours starting from L4 markedly increased the abundance of TSP-1::GFP, forming tetraspanin web-like structures discernable at high resolution (Fig. 1E). Such structures were not caused by temperature effect on the tag GFP per se since intestinal apical membrane-tethered GFP alone did not show such a pattern (fig. S3). In addition, tsp-1 endogenously tagged with a worm codon–optimized fluorescent protein wrmScarlet by CRISPR-Cas showed similar up-regulation by heat at 28°C and formation of tetraspanin web-like structures (fig. S4). Compared with multicopy transgenic TSP-1::GFP, endogenously tagged TSP-1::wrmScarlet exhibited overall weaker fluorescent intensity and enabled us to better resolve finer tetraspanin webs with regular lattice-like structures (fig. S4, F to I).
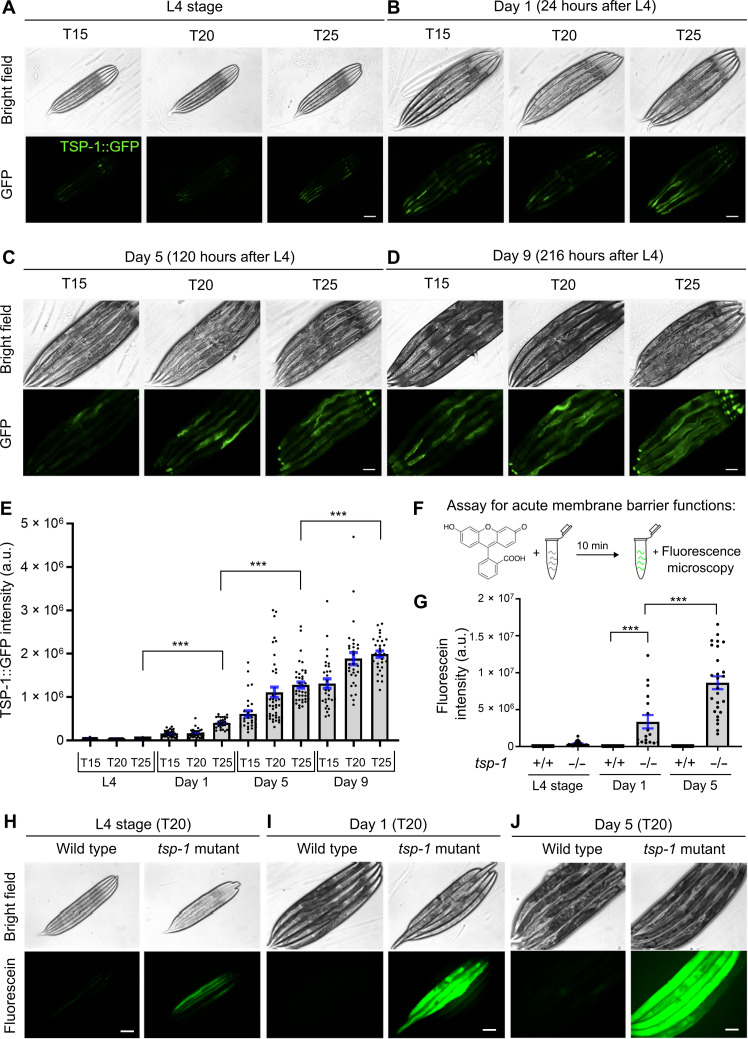
Tetraspanin has been implicated in numerous biological processes, yet its precise cellular functions remain largely unknown. We next sought to determine normal TSP-1 expression pattern under nonstress conditions and its adult physiological function during aging in C. elegans. At day 1 (young adult), 5 (adult), and 9 (old adult) post-L4 stages, TSP-1::GFP exhibited a progressive increase in abundance (Fig. 2, A to E). Cultivation at 25°C accelerated the time-dependent increase of TSP-1::GFP abundance, albeit to a lesser degree than a transient 24-hour period at 28°C (Figs. 1E and 2, A to E) (28°C represents harsh stress precluding chronic analyses of TSP-1::GFP at days 5 and 9 because of organismal death by prolonged exposure, see below). The notable tetraspanin web-like structure formed by TSP-1::GFP (Fig. 1E) prompted us to test the role of TSP-1 in maintaining membrane integrity. We developed a fluorescein-based assay modified from our previous studies (21) to measure the acute barrier function or permeability of intestinal membranes in C. elegans (Fig. 2F). In wild-type animals, a short (10 min) incubation with fluorescein did not result in detectable fluorescence signals in the intestine (Fig. 2G). By contrast, the same procedure led to a marked accumulation of fluorescein in the intestine of tsp-1–deficient animals (Fig. 2, G to J). This difference was notable at the L4 stage and particularly prominent in adults at day 5 after L4 (Fig. 2J). We observed similar defects in membrane barrier functions using RNA interference (RNAi) against two different coding regions of tsp-1 (fig. S5). These results indicate that TSP-1 up-regulation during aging or by thermal stress may physiologically maintain intestinal barrier functions.
Fig. 2. Expression and roles of TSP-1 in membrane barrier integrity in adults and during aging.
(A to D) Representative epifluorescence images showing TSP-1::GFP up-regulation from L4 (A) to young adults (B) and during aging (C) and (D). (E) Quantification of fluorescence intensities of TSP-1::GFP under conditions indicated. ***P < 0.001 (n > 30 animals per condition). (F) Schematic of the assay for acute membrane barrier functions. (G) Quantification of intensities of fluorescein accumulated by live animals under conditions indicated. ***P < 0.001 (n > 20 animals per condition). (H to J) Representative epifluorescence images showing fluorescein accumulation in animals of indicated stages, temperature and genotypes (wild-type and tsp-1 deletion mutant allele ok3594). Scale bars, 100 μm.
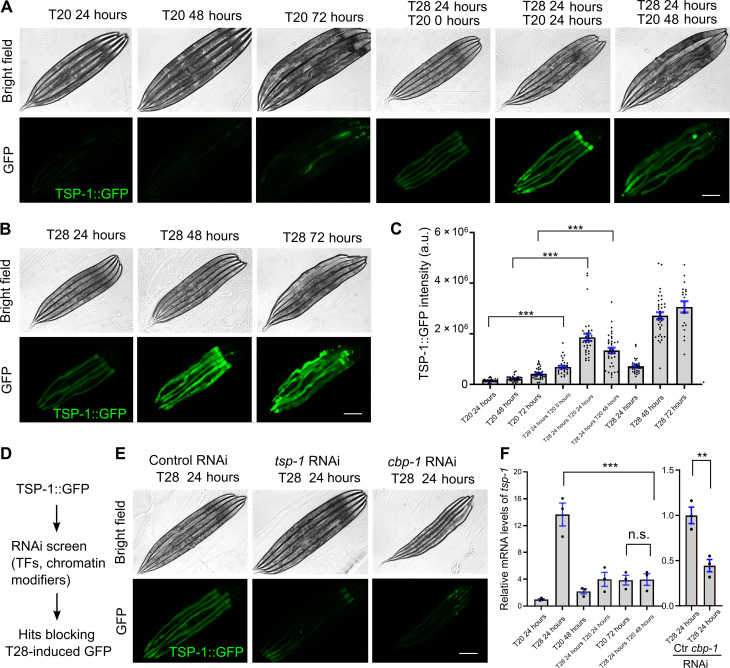
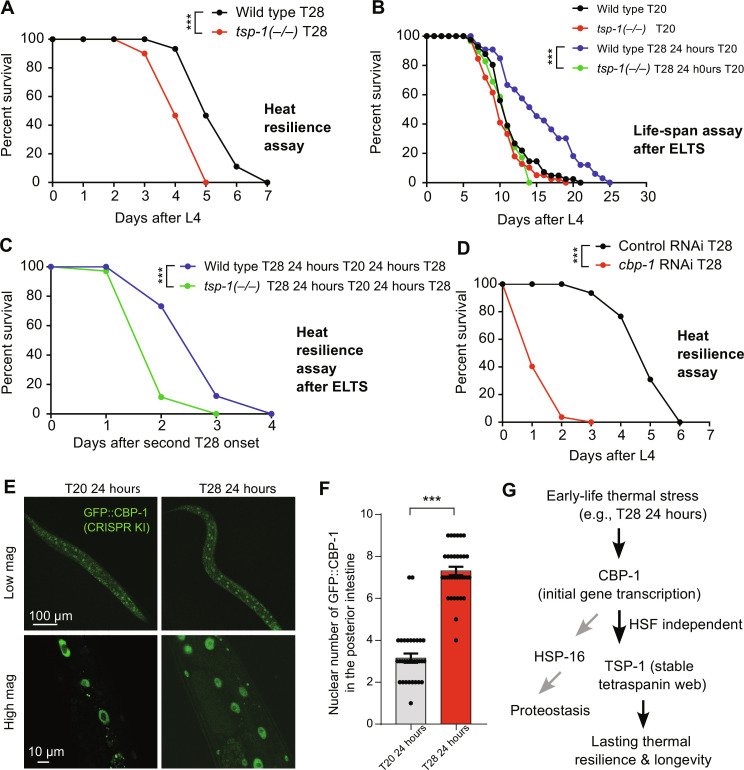
We characterized how early-life thermal stress (ELTS; 28°C for 24 hours starting from L4 stages) may trigger lasting TSP-1::GFP expression. In control animals at 20°C, TSP-1::GFP remained at low baseline levels largely unaltered for the first 48 hours and became slightly elevated at 72 hours (Fig. 3A). By contrast, ELTS (28°C for 24 hours at L4) triggered robust up-regulation of TSP-1::GFP abundance at 24 hours after L4, which remained still high at 20°C even for another 24 and 48 hours after the initial ELTS (Fig. 3A). Chronic exposure to 28°C starting at L4 induced stronger expression of TSP-1::GFP for 48 hours than 24 hours but reached peak levels at 72 hours (Fig. 3, B and C). We took advantage of the robust TSP-1::GFP activation by ELTS and performed RNAi screens for genes that are required for TSP-1::GFP up-regulation. Given the importance of transcription factors and histone-modifying enzymes in epigenetic gene regulation (32–34), we assembled a customized library of RNAi clones targeting >100 genes with adequate expression in the intestine (transcript per million, >2) and that encode proteins including stress-responding transcription factors and chromatin/epigenetic regulators (Fig. 3D and table S1). From such candidate screens, using RNAi vector only as negative control and tsp-1 RNAi as a positive control, we found that RNAi against only one hit, cbp-1, robustly prevented TSP-1::GFP up-regulation by ELTS (Fig. 3, E and F). cbp-1 encodes the C. elegans ortholog of histone acetyltransferase p300/CBP that promotes gene transcription (35–43). RNAi against many of the known genes encoding heat-responding transcription factor, including hsf-1, hsf-2, hif-1, and nhr-49, did not block TSP-1::GFP up-regulation (table S1). Although TSP-1::GFP remained high for 24 and 48 hours at 20°C after initial ELTS (Fig. 3C), quantitative reverse transcription polymerase chain reaction (qRT-PCR) measurements revealed that the up-regulation of tsp-1 mRNA transcripts by 28°C was transient but not sustained and required CBP-1 (Fig. 3F). These results indicate that transient ELTS can trigger lasting up-regulation of TSP-1 abundance, which requires initial CBP-1–dependent tsp-1 transcription but remains stable even after tsp-1 expression has normalized after ELTS.
Fig. 3. ELTS-triggered TSP-1::GFP is long-lasting and requires CBP-1.
(A) Representative epifluorescence images showing long-lasting up-regulation of TSP-1::GFP upon transient ELTS (T28 for 24 hours at L4). (B) Representative epifluorescence images showing up-regulation of TSP-1::GFP upon sustained thermal stress (T28 for 24, 48, or 72 hours starting at L4). (C) Quantification of fluorescence intensities of TSP-1::GFP under conditions indicated. ***P < 0.001 (n > 30 animals per condition). (D) Schematic of experimental flow using RNAi screen to identify genes required for ELTS induction of TSP-1::GFP. TFs, transcription factors. (E) Representative bright-field and epifluorescence images showing that RNAi against tsp-1 or cbp-1 blocks up-regulation of TSP-1::GFP by ELTS. (F) Quantitative RT-PCR measurements of tsp-1 expression levels under conditions indicated, showing transient tsp-1 up-regulation by ELTS in a CBP-1–dependent manner. **P < 0.01 and ***P < 0.001 (three independent biological replicates). Scale bars, 100 μm. n.s., not significant.
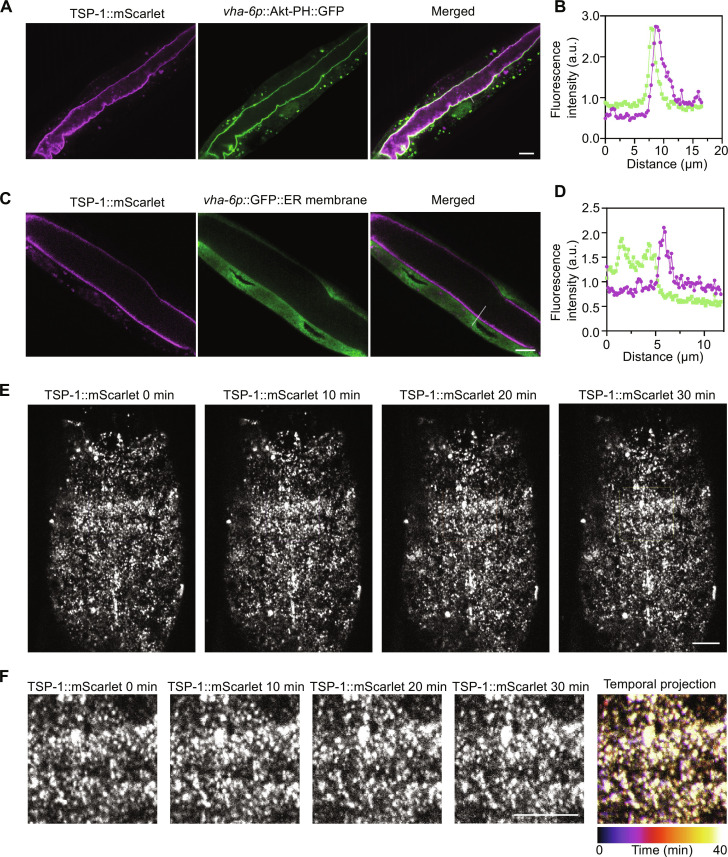
We aimed to determine the subcellular localization and stability of the heat-induced TSP-1 tetraspanin web formation. We generated strains with CRISPR-mediated wrmScarlet knock-in at the endogenous tsp-1 locus that was genetically crossed with previously validated GFP reporters for intestinal subcellular structures, including Akt-PH::GFP that binds to phosphatidylinositol 4,5-bisphosphate (PIP2)/phosphatidylinositol 3,4,5-trisphosphate (PIP3) at the inner leaflet of plasma membranes or GFP::C34B2.10(SP12) that labels the endoplasmic reticulum membrane (ERm) (21, 44, 45). Confocal microscopic imaging revealed the proximity of Akt-PH::GFP and TSP-1::wrmScarlet throughout intestinal apical membranes (Fig. 4A), corroborated by quantitative fluorescent intensity correlation analysis (Fig. 4B). By contrast, ERm::GFP and TSP-1::wrmScarlet did not exhibit apparent colocalization (Fig. 4, C and D). We further assessed the temporal dynamics of tetraspanin webs by imaging endogenous TSP-1::wrmScarlet (Fig. 4E) and found that the tetraspanin web structure exhibited markedly consistent stability across the entire field and timescale of imaging (Fig. 4F and movie S1). These results indicate that TSP-1–dependent tetraspanin webs, once induced by heat, form rather stable structures closely associated with the intestinal apical membrane.
Fig. 4. Subcellular localization and stability of tetraspanin web formed by endogenous TSP-1::wrmScarlet.
(A) Representative confocal images showing colocalization of the marker Akt-PH::GFP, which binds to intestinal apical plasma membrane inner leaflet PIP2/PIP3, with endogenous TSP-1 tagged with wrmScarlet by CRISPR. Scale bars, 10 μm. (B) Fluorescent intensity correlation analysis showing close proximity of Akt-PH::GFP and TSP-1::wrmScarlet. (C) Representative confocal images showing non-colocalization of the marker ERm::GFP, which labels intestinal ER membrane, with endogenous TSP-1 tagged with wrmScarlet by CRISPR. Scale bars, 10 μm. (D) Fluorescent intensity correlation analysis for ERm::GFP and TSP-1::wrmScarlet. (E) Representative confocal time-series images showing stability of tetraspanin webs formed by endogenous TSP-1::wrmScarlet, with enlarged views in (F). Scale bars, 10 μm.
Next, we investigated the consequences of TSP-1 up-regulation triggered by ELTS at the organismal level. To address this, we measured the population life span and survival rates of wild-type and tsp-1 deletion mutants under various heat stress conditions. Under constant 28°C starting from L4, deficiency of tsp-1 caused shortened life span and premature population death compared with wild-type animals (Fig. 5A). Transient ELTS (28°C for 24 hours at L4) extended the longevity of wild-type animals at subsequent normal cultivation temperature (20°C). By contrast, the ELTS effect on longevity extension was abolished in tsp-1 mutants (Fig. 5B). We applied 28°C exposure to animals growing constantly for 48 hours at 20°C after L4 and found that transient ELTS (28°C for 24 hours at L4) enhanced survival rates in wild-type but not tsp-1–deficient animals (Fig. 5C). We used RNAi against cbp-1 (null mutations are lethal) and observed that reducing cbp-1 expression caused even more shortened population life spans (Fig. 5D). Given the more severe phenotype of cbp-1 deficiency, CBP-1 likely regulates additional genes beyond tsp-1. We found that the heat shock chaperon-encoding gene hsp-16 was transiently induced by ELTS, and its induction required both HSF-1 and CBP-1 (fig. S6). Unlike hsp-16, ELTS induction of tsp-1 was HSF-1 independent (fig. S2), yet its similarly transient induction led to stable tetraspanin multimerization and web-like structural formation (Fig. 4). Last, we used a CRISPR-mediated GFP knock-in allele of cbp-1 to show that ELTS markedly increased the probability of nuclear entry by endogenous CBP-1 in the intestine, suggesting that CBP-1 plays an instructive (actively mediating the effect of heat) rather than permissive (gate-keeping to allow heat effect) role in tsp-1 up-regulation by heat (Fig. 5, E and F). Like induction of tsp-1 mRNAs but not TSP-1 proteins, increased nuclear entry of CBP-1 also followed a similarly transient pattern upon ELTS (fig. S7 and Fig. 3). Together, these results demonstrate that TSP-1 promotes survival under 28°C heat stress, and its CBP-1–dependent induction mediates the effects of transient ELTS on lasting benefits to longevity and organismal resilience to subsequent heat stress conditions (Fig. 5G).
Fig. 5. ELTS-triggered thermal resilience and longevity requires TSP-1.
(A) Life-span curves of wild-type and tsp-1 mutants at 28°C starting at L4. A total of 20% median and 28% maximal survival decrease in tsp-1 mutants. ***P < 0.001 (n > 40 animals per condition). (B) Life-span curves of wild-type and tsp-1 mutants at constant 20°C or after transient 28°C for 24 hours at L4 (ELTS). A total of 36% median and 20% maximal life-span extension by ELTS in wild type. *** P < 0.001 (n > 35 animals per condition). (C) Life-span curves of wild-type and tsp-1 mutants at 28°C after transient 28°C for 24 hours at L4 (ELTS). A total of 33% median and 33% maximal survival decrease in tsp-1 mutants. ***P < 0.001 (n > 35 animals per condition). (D) Life-span curves of wild-type animals with RNAi against cbp-1 or control at 28°C starting at L4. A total of 80% median and 50% maximal survival decrease with RNAi against cbp-1. ***P < 0.001 (n > 40 animals per condition). (E) Representative confocal fluorescence images showing ELTS-induced increase in numbers of nuclei with GFP::CBP-1 in the posterior intestine. (F) Quantification of numbers of nuclei with GFP::CBP-1 in the posterior intestine under conditions indicated. (G) Model of how ELTS induces lasting organismal thermal resilience through HSF-independent epigenetic regulation of TSP-1 and stable tetraspanin web structure formation.
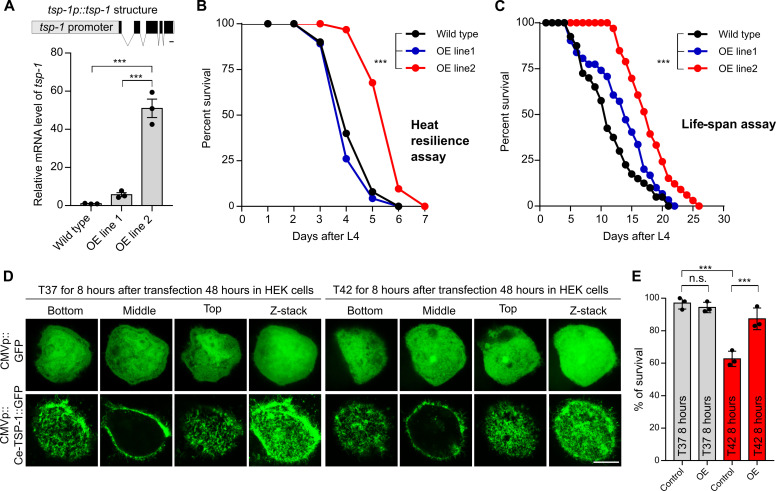
To assess the sufficiency of TSP-1 in conferring stress resilience, we determined the gain-of-function effects of TSP-1 in C. elegans and ectopically in human cells. In C. elegans, we generated two transgenic strains carrying genomic tsp-1 regions as extrachromosomal arrays at either low or high copy numbers (overexpression or OE, line 1 and 2, respectively) (Fig. 6A). qRT-PCR confirmed that tsp-1 mRNA expression levels are differentially up-regulated in the two strains with array-positive animals (Fig. 6A). In thermal resilience assays, we found that OE line 2 with high-copy tsp-1 arrays exhibited markedly enhanced survival rates upon continuous 28°C heat stress, compared with array-negative control or OE line 1 with low-copy tsp-1 arrays (Fig. 6B). In life-span assays, OE line 2 exhibited similarly extended survival and longevity under 20°C cultivation conditions, with a marked increase in both median (63%) and maximal (24%) life spans in animals carrying high-copy expression of tsp-1 (Fig. 6C). Furthermore, to assess the gain-of-function effect of TSP-1 in a heterologous cellular context, we overexpressed C. elegans tsp-1 cDNA with a tagged GFP driven by the CMV promoter in human embryonic kidney cell lines [human embryonic kidney (HEK) 293] (Fig. 6D). Such ectopic expression of tsp-1 resulted in prominent tetraspanin web-like structures (Fig. 6E) and functionally protected HEK293 cells against thermal stress conditions (42°C). These results demonstrate notable gain-of-function effects of tetraspanin-encoding tsp-1 in both C. elegans and heterologous mammalian cells.
Fig. 6. Gain of function of TSP-1 confers longevity extension in C. elegans and thermal resilience in human cells.
(A) Schematic of the transgene that produces tsp-1 gain-of-function OE lines (OE line 1 at low level, OE line 2 at high level as measured by qRT-PCR). ***P < 0.001. (B) Life-span curves of wild type and two representative tsp-1 OE lines at 28°C starting at L4. A total of 50% median and 16% maximal survival extension by overexpression of tsp-1 in wild type, ***P < 0.001 (n > 40 animals per condition). (C) Life-span curves of wild-type and tsp-1 OE lines at constant 20°C. A total of 63% median and 24% maximal life-span extension by overexpression of tsp-1 in wild type, ***P < 0.001 (n > 40 animals per condition). (D) Serial confocal fluorescence images showing tetraspanin web-like structures formed in the human cell line HEK293 by CMV promoter-driven expression of C. elegans tsp-1::GFP (bottom) but not GFP alone (top). Scale bar, 10 μm. (E) Quantification of survival rates after heat shock in HEK293 cells showing enhanced thermal resilience conferred by ectopic gain-of-function of C. elegans TSP-1::GFP. ***P < 0.001 (three independent biological replicates).
DISCUSSION
Previous studies have shown that mild and transient metabolic or environmental stress in early life can produce beneficial effects, extending longevity in C. elegans (38, 46–51). While heat exposure activates numerous genes involved in proteostasis and defense responses, specific HSF-independent target genes with causal effects on longevity remain unidentified. In this work, we identify tsp-1 as a heat-inducible gene that mediates effects of ELTS on organismal resilience and longevity. Regulatory mechanisms governing how heat stress activates CBP-1 in coordination with other unidentified factors await further exploration. Our data suggest that tsp-1 expression leads to TSP-1 protein multimerization and the formation of stable tetraspanin web structures, which persist even in the absence of initial stimuli and tsp-1 transcript up-regulation. This tetraspanin web-based stable protein structure formation represents an intriguing mechanism of cellular memory, distinct from previously known modes of epigenetic regulation primarily occurring in the nucleus, such as DNA and histone modifications.
We demonstrate that TSP-1 is crucial for maintaining intestinal membrane barrier functions and enhancing animal adaptation and survival under heat stress conditions and during aging. The C. elegans genome encodes 21 tetraspanin family proteins, including tsp-1 and tsp-2 located in the same operon, which may be similarly regulated by heat. Other tetraspanin family members have been implicated in plasma membrane repair, extracellular vesicle formation, and oxidative stress responses (52–55). It is plausible that different tetraspanins may be regulated in a tissue-specific, developmental stage-specific, and stress-specific manner to confer distinct biological functions. In addition to heat that affects membrane barrier and integrity, other types of specific membrane-perturbing stress can also induce tsp-1 expression through CBP-1, which likely regulates additional transcriptional targets that together safeguard intestinal barrier functions (figs. S8 and S9). Several mammalian tetraspanins have been shown to play important roles in maintaining blood-brain or retina-blood barriers in endothelial cells and attenuating inflammation, cell senescence, and organismal aging (56–58). Thus, we propose that the functional roles of tetraspanins are evolutionarily conserved, likely mediating long-lasting effects of transient cellular and organismal stresses in mammals, including humans.
MATERIALS AND METHODS
C. elegans strains
C. elegans strains were grown on nematode growth media (NGM) plates seeded with Escherichia coli OP50 at 20°C with laboratory standard procedures unless otherwise specified. The N2 Bristol strain was used as the reference wild type (59). Mutants and integrated transgenes were back-crossed at least five times. Genotypes of strains used are as follows: dmaIs125 [tsp-1p::tsp-1::GFP; unc-54p::mCherry], dmaIs8 [hsp-16p::GFP; unc-54p::mCherry]; him-5(e1490), pwIs890[Pvha-6::AKT(PH)::GFP],hjIs14 [vha-6p::GFP::C34B2.10(SP12 ER membrane) + unc-119(+)], etEx2 [glo-1p::GFP::ras-2 CAAX + rol-6], tsp-1(ok3594), hsf-1(sy441), tsp-1(syb7456[tsp-1::wrmScarlet]), cbp-1(ust475[GFP::cbp-1]).
For CRISPR knock-in of cbp-1 with 3xFLAG::GFP::cbp-1, a cbp-1 promoter region was PCR-amplified with the primers 5′-gggtaacgccagCACGTGtgggctgactcgtgctg-3′ and 5′-ccgtcatggtctttgtagtctggtggttcatccatcaattagta-3′ from N2 genomic DNA. A cbp-1 coding sequence region was PCR-amplified with the primers 5′-AAGGAGGTGGAGGTGGAGCTatggatgaaccaccatcaa-3′ and 5′-cagcggataacaatttcacaatgcatcattggatatccacc-3′ from N2 genomic DNA. A 3xFLAG::GFP region was PCR-amplified with the primers 5′-gactacaaagaccatgacgg-3′ and 5′-AGCTCCACCTCCACC-3′ from SHG1890 genomic DNA. The ClonExpress MultiS One Step Cloning Kit (Vazyme C113-02, Nanjing) was used to connect these fragments with vector amplified with 5′-tgtgaaattgttatccgctgg-3′ and 5′-caCACGTGctggcgttacc-3′ from L4440. The injection mix contained pDD162 (50 ng/μl), cbp-1 repair plasmid (50 ng/μl), pCFJ90 (5 ng/μl), and three single guide RNAs (30 ng/μl). The mix was injected into young adult N2 animals, and the coding sequence of 3xFLAG::GFP was inserted after the start codon using the CRISPR-Cas9 system. The tsp-1::wrmScarlet knock-in allele was generated similarly (SunyBiotech), with wrmScarlet sequences inserted before the termination codon of tsp-1 in the N2 background.
For overexpression of tsp-1 (3.26 kb), the tsp-1 promotor (1.98 kb) and 1283-bp genomic DNA fragment of tsp-1 coding region was PCR-amplified with the primers 5′-aaatataatttcagggatgtggtctcaaat-3′ and 5′-tgacaagggtactgtagttcgtct-3′ from N2 genomic DNA. The injection mix contained tsp-1p::tsp-1 (10 to 50 ng/μl) and unc-54p::GFP (25 ng/μl) to establish transgenic strains carrying extrachromosomal arrays of varying copy numbers.
Compound and confocal imaging
Epifluorescence compound microscopes (Leica DM5000 B Automated Upright Microscope System) were used to capture fluorescence images (with a 10× objective lens). Animals of different genotypes and different stages (L4, day 1, day 5, and day 9) and different heat treatments were randomly picked and treated with 10 mM sodium azide solution (71290-100MG, Sigma-Aldrich) in M9, aligned on an 2% agarose pad on slides for imaging. The same settings (for bright field: exposure time of 1 s, for GFP: exposure time of 10 s) were maintained for the images of all samples. The integrated density (IntDen) of TSP-1::GFP was measured by National Institutes of Health (NIH) Image program (Fiji ImageJ), and average of mean gray value (three background areas of each image randomly selected) was used to normalized the TSP-1::GFP. For confocal images, the worms were randomly chosen and treated with 10 mM sodium azide in M9 solution and aligned on a 2% agarose pad on slides, and images were acquired using a confocal microscope (Leica TCS SPE) with a 63× objective lens, the same settings were maintained for the images of all samples. Imaging in Fig. 4 was conducted under room temperature conditions using a Nikon Eclipse Ti inverted microscope equipped with a Borealis beam conditioning unit (Andor), a CSU-W1 Yokogawa spinning disk (Andor, Belfast, Northern Ireland), a 100× PlanApo total internal reflection fluorescence 1.49 numerical aperture (NA) objective (Nikon, Toyko, Japan), an iXon Ultra EMCCD camera (Andor), and a laser merge module (LMM5, Spectral Applied Research; Exton, PA) containing 405-, 440-, 488-, and 561-nm laser lines. Micro-Manager (UCSF) was used to control all the hardware. Fiji (NIH) (60) and Prism (GraphPad Software Inc.) were used for image display and colocalization analysis.
SDS-page and native-page Western blotting
For SDS-PAGE samples, stage-synchronized animals for control and experiment groups were picked (n = 50) in 60 μl of M9 buffer and lysed directly by adding 20 μl of 4× Laemmli sample buffer (1610747, Bio-Rad) containing 10% of 2-mercaptoethanol [M6250-100ML, Sigma-Aldrich (v/v)]. Protein extracts were denatured at 95°C for 10 min and separated on 10% SDS-PAGE gels (1610156, Bio-Rad) at 80 V for ∼40 min followed by 110 V for ∼70 min. The proteins were transferred to a nitrocellulose membrane (1620094, Bio-Rad,) at 25 V for 40 min by the Trans-Blot Turbo Transfer System (Bio-Rad). The nitrocellulose membrane was initially blocked with 5% nonfat milk and 2% bovine serum albumin [BSA; A4503, Sigma-Aldrich (v/v)] in tris-buffered saline with 0.1% Tween 20 (TBS-T; 93773, Sigma-Aldrich) at room temperature for 1 hour. Proteins of interest were detected using antibodies against GFP (A6455, Invitrogen) and histone H3 (ab1791, Abcam) in cold room for overnight. After three washes of 5 min each with TBST, anti-rabbit immunoglobulin G (IgG) and horseradish peroxidase (HRP)–linked antibody (7074S, Cell Signaling Technology) were added at a dilution of 1:5000.
For native-PAGE samples, stage-synchronized animals were washed down from NGM plates using M9 solution and subjected to 850g for 60 s, and the pellet animals were resuspended in precooled 300-μl lysis buffer (M9 buffer + protein inhibitors cocktails, A32965, Thermo Fisher Scientific), then lysed by TissueRuptor (Motor unit “6”) for 10 s, and taken out, repeated three to five times, followed by diluting the samples with native sample buffer (161-0738, Bio-Rad). Proteins were resolved by 4 to 15% Mini-PROTEAN TGX Precast Protein Gels (4561086, Bio-Rad) and transferred to a nitrocellulose membrane (1620094, Bio-Rad). The NC membrane was initially blocked with 5% nonfat milk and 2% BSA [A4503, Sigma-Aldrich (v/v)] in TBST (93773, Sigma-Aldrich) at room temperature for 1 hour. Native TSP-1::GFP were detected using antibodies against GFP (1:1000, 66002-1-Ig, Proteintech) at cold room overnight. After three washes of 5 min each with TBST, goat anti-mouse IgG (H + L) and secondary antibody (HRP) (NB7539, Novus) were added at a dilution of 1:5000 as the secondary antibody.
Fluorescein assay
Stage-synchronized animals (L4, day 1, and day 5) of different genotypes at 20°C were randomly picked to 20 μl of M9 solution in a 1.5-ml tube, followed by treatment with 100 μl of fluorescein (F0095-25G, TCI America, 10 mg/ml in M9 buffer) at room temperature for 10 min. Worms were transferred to a NGM plate (without OP50), collected to new NGM plate with 200 μl of M9 buffer to wash for three times, followed by randomly picking and treatment with 10 mM sodium azide in M9 buffer and aligned on an 2% agarose pad on slides for compound microscope imaging. The IntDen of fluorescein was measured by NIH image program (Fiji ImageJ), and average of mean gray value (three background areas of each image randomly selected) was used to normalize the fluorescein signals.
RNAi cloning and screen
To clone the tsp-1 RNAi sequences, tsp-1 exon 1-3 (Chr. III, C02F5.8, 1124-bp size, primer forward: GGCGGCCGCTCTAGAACTAGTACAGATTTCCTCCACCTCTTCC, primer reverse: TCCACGCGTCACGTGGCTAGCATTCCTAATTTTTCAGAGCCCACC) and exon 1-5 (Chr. III, C02F5.8, 1472-bp size, primer forward: GGCGGCCGCTCTAGAACTAGTTCCACCTCTTCCACCTTCATTAC, primer reverse: TCCACGCGTCACGTGGCTAGCTGACAAGGGTACTGTAGTTCGTCT) were PCR-amplified from wild-type N2 gDNA and subcloned into the Spe I and Nhe I sites of a pL4440 expression vector with the NEBuilder HiFi DNA Assembly Cloning Kit (E2621L, New England Biolabs). Briefly, PCR was performed with the following protocol on a MyCyclerTM Thermal Cycler (Bio-Rad): 98°C for 30 s, 98°C for 10 s, 55°C for 30 s, 72°C for 2 min (35 cycles); 72°C for 5 min and a final hold at 12°C. The PCR products were analyzed on a 1.5% agarose gel. Twenty-five nanograms of precut pL4440 vector with twofold excess of tsp-1 PCR fragment (50 ng) was used for assembly in a thermocycler at 50°C for 30 min, and 25 ng of precut pL4440 vector with the same water instead of PCR fragment was used as native assembly control. Five microliters of assembly preparations was transformed to NEB 5-alpha Competent E. coli (C2987H, NEB) by heat shock at exactly 42°C for exactly 30 s. Positive clones were verified by bacteria PCR by 2x Thermo Scientific DreamTaq Green PCR Master Mix (pL4440 forward: CTTATCGAAATTAATACG, pL4440 reverse: AGGGCGAATTGGGTACCG). Positive pL4440-tsp-1 RNAi plasmids were transformed to competent HT115 by electroporation, and the positive HT115 clones were verified by bacteria PCR.
RNAi and screen for hits blocking TSP-1::GFP were performed by feeding worms with E. coli strain HT115 (DE3) expressing double-strand RNA (dsRNA) targeting endogenous genes. Briefly, dsRNA-expressing bacteria were replicated from the Ahringer library to LB plates containing ampicillin (100 μg/ml; BP1760-25, Thermo Fisher Scientific) at 37°C for 16 hours. Single clone was picked to LB medium containing ampicillin (100 μg/ml) at 37°C for 16 hours, and positive clones (verified by bacteria PCR with pL4440 forward and pL4440 reverse primers) were spread onto NGM plates containing ampicillin (100 μg/ml) and 1 mM isopropyl-β-d-thiogalactopyranoside (420322, Millopore) for 24 hours (namely RNAi plates). Developmentally synchronized embryos from bleaching of gravid adult hermaphrodites were plated on RNAi plates and grown at 20°C to L4 stage followed by transfer to 28°C incubator for 24 hours. Randomly selected population was observed under the a epifluorescence microscope (SMZ18, Nikon) with hits considered to block TSP-1::GFP when observing GFP levels comparable to that by tsp-1 RNAi (positive control).
Quantitative reverse transcription polymerase chain reaction
Animals were washed down from NGM plates using M9 solution and subjected to RNA extraction using TissueDisruptor and RNA lysis buffer (Motor unit “6” for 10 s and take it out, repeat three to five times on ice), total RNA was extracted following the instructions of the Quick-RNA MiniPrep kit (Zymo Research, R1055), and reverse transcription was performed by SuperScript III (18080093, Thermo Fisher Scientific). Real-time PCR was performed by using ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme) on the Roche LightCycler96 (Roche, 05815916001) system. Ct values of tsp-1 were normalized to measurements of rps-23 (C. elegans) levels. Primers for qRT-PCR were as follows: tsp-1 (forward, CTTTGATTGCCGTTGGATTT; reverse, CCCAAAGAAAGGCCGATAAT), hsp-16.2 (forward: ACGTTCCGTTTTTGGTGATCTTAT; reverse, TCTGGTTTAAACTGTGAGACGTTG), and rps-23 (forward, CGCAAGCTCAAGACTCATCG; reverse, AAGAACGATTCCCTTGGCGT).
Thermal resilience and life-span assays
Animals were cultured under nonstarved conditions for at least two generations before heat stress assays. For treatment of “early-life thermal stress” (ELTS), bleach-synchronized eggs were growth to the L4 stage at 20°C, and populations were kept at 28°C for 24 hours and then recovered for 0 to 72 hours at 20°C. For thermal resilience assays, stage-synchronized L4 stage worms (n = 50) were picked to new NGM plates seeded with OP50 and transferred to a 28°C incubator. Animals were scored for survival per 24 hours. Worms failing to respond to repeated touch of a platinum wire were scored as dead. For life-span assays, stage-synchronized L4 stage worms (n = 50) were picked to new NGM plates seeded with OP50 containing 50 μM 5-fluoro-2′-deoxyuridine to prevent embryo growth at a 20°C incubator. Animals were scored for survival per 24 hours. Worms failing to respond to repeated touch of a platinum wire were scored as dead.
Mammalian expression constructs for C. elegans TSP-1
The C. elegans tsp-1 open reading frame was PCR-amplified with the primers 5′-GCGGCCTTAATTAAACCTCTAGAATGGCAACTTGGAAATTTATCATACG-3′ and 5′-AGCTCGAGATCTGAGTCCGGcAAAACGAGTGTCTTCGGTGATG-3′ from cDNA prepared from heat-induced (T28 24 hours) wild-type animals. GFP fragment was PCR-amplified with 5′-gCCGGACTCAGATCTCGAGCTATGGTGAGCAAGGGCGCCG-3′ and 5′-CGGATCTTACTTACTTAGCGGCCGCTTACTTGTACAGCTCATCCATGCC-3′ from the pHAGE2-gfp plasmid. The tsp-1–GFP fragment was PCR-amplified using overlapping PCR and subcloned by T4 DNA ligase to the pHAGE2-gfp plasmid at the Xba I and Not I sites.
HEK293T cells and thermal resilience assay
HEK 293T cells were maintained in Dulbecco’s modified Eagle’s medium with 10% inactive fetal bovine serum and penicillin-streptomycin (Gibco, Grand Island, 15140122) at 37°C supplied with 5% CO2 in an incubator (Thermo Fisher Scientific) with a humidified atmosphere. Cells were washed once using phosphate-buffered saline (PBS) and digested with 0.05% trypsin-EDTA (Gibco) at 37°C for routine passage of the cells. All HEK 293T cells were transiently transfected with indicated constructions using the Lipo2000 (1 mg/ml; Life Technologies) reagent. The Lipo2000/DNA mixture with the ratio of Lipo2000 to DNA at 3:1 was incubated for 30 min at room temperature before being added to the HEK 293T cell cultures dropwise. For thermal resilience assay, mock control and transfected HEK293T cells (48 hours) in a 24-well plate were placed in a culture incubator with an ambient temperature at 42°C and humidified 5% CO2 for 8 hours followed by cell death assay or imaging with 4% paraformaldehyde treatment for 12 min at room temperature. For cell death assay, the collected cells were resuspended with 100-μl buffer with addition of 0.1 μl of Sytox blue (Thermo Fisher Scientific) for an additional 15 min at room temperature. Twenty-five microliters of incubated cells was loaded into Arthur cell analysis slide (NanoEntek, AC0050). The fluorescence intensity was measured for individual cells using automated cytometry (Arthure image–based cytometer, NanoEntek, AT1000) by viability assay. The 226 relative fluorescence unit (RFU) threshold and a cell size minimum of 5 to maximum of 25 were used for cell death analysis and quantification.
Statistics
Data were analyzed using GraphPad Prism 9.2.0 Software (GraphPad, San Diego, CA). Numeric results are presented as means ± SD unless otherwise specified, with P values calculated by unpaired two-tailed t tests (comparisons between two groups), one-way analysis of variance (ANOVA) (comparisons across more than two groups), and two-way ANOVA (interaction between genotype and treatment), with post hoc Tukey and Bonferroni’s corrections. The life-span assay was quantified using Kaplan-Meier life-span analysis, and P values were calculated using the log-rank test.
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding: The work was supported by NIH grants (R35GM139618 to D.K.M. and R35GM118167 to O.D.W.), BARI Investigator Award (D.K.M.), and UCSF PBBR New Frontier Research Award (D.K.M.), and UCSF CIRM Scholars Training Program EDUC4-12812 (W.J).
Author contributions: Conceptualization: W.I.J., O.D.W., and D.K.M. Methodology: W.I.J., H.D.B., B.W., A.W., M.K., F.O., J.D., X.H., and S.G. Investigation: W.I.J., H.D.B., B.W., A.W., M.K., F.O., J.D., X.H., S.G., O.D.W., and D.K.M. Visualization: W.I.J., H.D.B., O.D.W., and D.K.M. Supervision: O.D.W. and D.K.M. Writing—original draft: W.I.J. and D.K.M. Writing—review and editing: W.I.J., H.D.B., B.W., A.W., M.K., F.O., J.D., X.H., S.G., O.D.W., and D.K.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs S1 to S9
Legends for tables S1 and S2
Legend for movie S1
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
Movie S1
REFERENCES AND NOTES
- 1.de Kloet E. R., Joëls M., Holsboer F., Stress and the brain: From adaptation to disease. Nat. Rev. Neurosci. 6, 463–475 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Taylor S. E., Way B. M., Seeman T. E., Early adversity and adult health outcomes. Dev. Psychopathol. 23, 939–954 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Fogelman N., Canli T., Early life stress, physiology, and genetics: A review. Front. Psychol. 10, 1668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felitti V. J., Anda R. F., Nordenberg D., Williamson D. F., Spitz A. M., Edwards V., Koss M. P., Marks J. S., Relationship of childhood abuse and household dysfunction to many of the leading causes of death in adults. The Adverse Childhood Experiences (ACE) Study. Am. J. Prev. Med. 14, 245–258 (1998). [DOI] [PubMed] [Google Scholar]
- 5.English S., Uller T., Does early-life diet affect longevity? A meta-analysis across experimental studies. Biol. Lett. 12, 20160291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLaughlin K. A., Koenen K. C., Bromet E. J., Karam E. G., Liu H., Petukhova M., Ruscio A. M., Sampson N. A., Stein D. J., Aguilar-Gaxiola S., Alonso J., Borges G., Demyttenaere K., Dinolova R. V., Ferry F., Florescu S., de Girolamo G., Gureje O., Kawakami N., Lee S., Navarro-Mateu F., Piazza M., Pennell B.-E., Posada-Villa J., Ten Have M., Viana M. C., Kessler R. C., Childhood adversities and post-traumatic stress disorder: Evidence for stress sensitisation in the World Mental Health Surveys. Br. J. Psychiatry 211, 280–288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown D. W., Anda R. F., Tiemeier H., Felitti V. J., Edwards V. J., Croft J. B., Giles W. H., Adverse childhood experiences and the risk of premature mortality. Am. J. Prev. Med. 37, 389–396 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Longo V. D., Mattson M. P., Fasting: Molecular mechanisms and clinical applications. Cell Metab. 19, 181–192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laukkanen T., Kunutsor S. K., Khan H., Willeit P., Zaccardi F., Laukkanen J. A., Sauna bathing is associated with reduced cardiovascular mortality and improves risk prediction in men and women: A prospective cohort study. BMC Med. 16, 219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupien S. J., McEwen B. S., Gunnar M. R., Heim C., Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat. Rev. Neurosci. 10, 434–445 (2009). [DOI] [PubMed] [Google Scholar]
- 11.McEwen B. S., In pursuit of resilience: Stress, epigenetics, and brain plasticity. Ann. N. Y. Acad. Sci. 1373, 56–64 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Ma D. K., Jang M.-H., Guo J. U., Kitabatake Y., Chang M.-L., Pow-Anpongkul N., Flavell R. A., Lu B., Ming G.-L., Song H., Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padilla P. A., Ladage M. L., Suspended animation, diapause and quiescence. Cell Cycle 11, 1672–1679 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton N. O., Furuta T., Webster A. K., Kaplan R. E. W., Baugh L. R., Arur S., Horvitz H. R., Insulin-like signalling to the maternal germline controls progeny response to osmotic stress. Nat. Cell Biol. 19, 252–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassada R. C., Russell R. L., The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46, 326–342 (1975). [DOI] [PubMed] [Google Scholar]
- 16.Hall S. E., Beverly M., Russ C., Nusbaum C., Sengupta P., A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr. Biol. 20, 149–155 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ow M. C., Borziak K., Nichitean A. M., Dorus S., Hall S. E., Early experiences mediate distinct adult gene expression and reproductive programs in Caenorhabditis elegans. PLOS Genet. 14, e1007219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali Nasser R., Harel Y., Stern S., Early-life experience reorganizes neuromodulatory regulation of stage-specific behavioral responses and individuality dimensions during development. eLife 12, e84312 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J., Wang K., Wang M., Wu Z., Xie G., Peng Y., Zhang Y., Zhang X., Shao Z., One-day thermal regime extends the lifespan in Caenorhabditis elegans. Comput. Struct. Biotechnol. J. 21, 495–505 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim B., Lee J., Kim Y., Lee S.-J. V., Regulatory systems that mediate the effects of temperature on the lifespan of Caenorhabditis elegans. J. Neurogenet. 34, 518–526 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Wang C., Wang B., Pandey T., Long Y., Zhang J., Oh F., Sima J., Guo R., Liu Y., Zhang C., Mukherjee S., Bassik M., Lin W., Deng H., Vale G., McDonald J. G., Shen K., Ma D. K., A conserved megaprotein-based molecular bridge critical for lipid trafficking and cold resilience. Nat. Commun. 13, 6805 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C., Long Y., Wang B., Zhang C., Ma D. K., GPCR signaling regulates severe stress-induced organismic death in Caenorhabditis elegans. Aging Cell 22, e13735 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W., Wei Y., Long Y., Owen A., Wang B., Wu X., Luo S., Dang Y., Ma D. K., A genetic program mediates cold-warming response and promotes stress-induced phenoptosis in C. elegans. eLife 7, e35037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunquell J., Morris S., Lu Y., Cheng F., Westerheide S. D., The genome-wide role of HSF-1 in the regulation of gene expression in Caenorhabditis elegans. BMC Genomics 17, 559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baird N. A., Douglas P. M., Simic M. S., Grant A. R., Moresco J. J., Wolff S. C., Yates J. R., Manning G., Dillin A., HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science 346, 360–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Pastor R., Burchfiel E. T., Thiele D. J., Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morimoto R. I., The heat shock response: Systems biology of proteotoxic stress in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 76, 91–99 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Prahlad V., Cornelius T., Morimoto R. I., Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320, 811–814 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charrin S., Jouannet S., Boucheix C., Rubinstein E., Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Hemler M. E., Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 (2005). [DOI] [PubMed] [Google Scholar]
- 31.van Deventer S. J., Dunlock V.-M. E., van Spriel A. B., Molecular interactions shaping the tetraspanin web. Biochem. Soc. Trans. 45, 741–750 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Cui M., Han M., Roles of chromatin factors in C. elegans development. WormBook, 1–16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González-Aguilera C., Palladino F., Askjaer P., C. elegans epigenetic regulation in development and aging. Genomics 13, 223–234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodhouse R. M., Ashe A., How do histone modifications contribute to transgenerational epigenetic inheritance in C. elegans? Biochem. Soc. Trans. 48, 1019–1034 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Shi Y., Mello C., A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12, 943–955 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eastburn D. J., Han M., A gain-of-function allele of cbp-1, the Caenorhabditis elegans ortholog of the mammalian CBP/p300 gene, causes an increase in histone acetyltransferase activity and antagonism of activated Ras. Mol. Cell. Biol. 25, 9427–9434 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vora M., Shah M., Ostafi S., Onken B., Xue J., Ni J. Z., Gu S., Driscoll M., Deletion of microRNA-80 activates dietary restriction to extend C. elegans healthspan and lifespan. PLOS Genet. 9, e1003737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou L., He B., Deng J., Pang S., Tang H., Histone acetylation promotes long-lasting defense responses and longevity following early life heat stress. PLOS Genet. 15, e1008122 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabianca D. S., Muñoz-Jiménez C., Kalck V., Gaidatzis D., Padeken J., Seeber A., Askjaer P., Gasser S. M., Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature 569, 734–739 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Ganner A., Gerber J., Ziegler A.-K., Li Y., Kandzia J., Matulenski T., Kreis S., Breves G., Klein M., Walz G., Neumann-Haefelin E., CBP-1/p300 acetyltransferase regulates SKN-1/Nrf cellular levels, nuclear localization, and activity in C. elegans. Exp. Gerontol. 126, 110690 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Mathies L. D., Lindsay J. H., Handal A. P., Blackwell G. G., Davies A. G., Bettinger J. C., SWI/SNF complexes act through CBP-1 histone acetyltransferase to regulate acute functional tolerance to alcohol. BMC Genomics 21, 646 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T. Y., Sleiman M. B., Li H., Gao A. W., Mottis A., Bachmann A. M., El Alam G., Li X., Goeminne L. J. E., Schoonjans K., Auwerx J., The transcriptional coactivator CBP/p300 is an evolutionarily conserved node that promotes longevity in response to mitochondrial stress. Nat. Aging 1, 165–178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barrett L. N., Westerheide S. D., The CBP-1/p300 lysine acetyltransferase regulates the heat shock response in C. elegans. Front. Aging 3, 861761 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi A., Liu O., Koenig S., Banerjee R., Chen C. C.-H., Eimer S., Grant B. D., RAB-10-GTPase-mediated regulation of endosomal phosphatidylinositol-4,5-bisphosphate. Proc. Natl. Acad. Sci. U.S.A. 109, E2306–E2315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu N., Zhang S. O., Cole R. A., McKinney S. A., Guo F., Haas J. T., Bobba S., Farese R. V., Mak H. Y., The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J. Cell Biol. 198, 895–911 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao R., Zhang B., Dong Y., Gong J., Xu T., Liu J., Xu X. Z. S., A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell 152, 806–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lithgow G. J., White T. M., Melov S., Johnson T. E., Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci. U.S.A. 92, 7540–7544 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu D., Wu X., Zhou J., Li X., Huang X., Li J., Wu J., Bian Q., Wang Y., Tian Y., NuRD mediates mitochondrial stress-induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci. Adv. 6, eabb2529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian Y., Garcia G., Bian Q., Steffen K. K., Joe L., Wolff S., Meyer B. J., Dillin A., Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt). Cell 165, 1197–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oleson B. J., Bhattrai J., Zalubas S. L., Kravchenko T. R., Ji Y., Jiang E. L., Lu C. C., Madden C. R., Coffman J. G., Bazopoulou D., Jones J. W., Jakob U., Early life changes in histone landscape protect against age-associated amyloid toxicities through HSF-1-dependent regulation of lipid metabolism. Nat. Aging 10.1038/s43587-023-00537-4, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen A., Vantipalli M. C., Lithgow G. J., Lifespan extension of Caenorhabditis elegans following repeated mild hormetic heat treatments. Biogerontology 7, 221–230 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Xu Z., Hu Y., Deng Y., Chen Y., Hua H., Huang S., Nie Q., Pan Q., Ma D. K., Ma L., WDR-23 and SKN-1/Nrf2 coordinate with the BLI-3 dual oxidase in response to iodide-triggered oxidative stress. G3 8, 3515–3527 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z., Shi H., Liu J., The C. elegans TspanC8 tetraspanin TSP-14 exhibits isoform-specific localization and function. PLOS Genet. 18, e1009936 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikonorova I. A., Wang J., Cope A. L., Tilton P. E., Power K. M., Walsh J. D., Akella J. S., Krauchunas A. R., Shah P., Barr M. M., Isolation, profiling, and tracking of extracellular vesicle cargo in Caenorhabditis elegans. Curr. Biol. 32, 1924–1936.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y., Yang Q., Meng X., Wijaya C. S., Ren X., Xu S., Recruitment of tetraspanin TSP-15 to epidermal wounds promotes plasma membrane repair in C. elegans. Dev. Cell 57, 1630–1642.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Jin Y., Takeda Y., Kondo Y., Tripathi L. P., Kang S., Takeshita H., Kuhara H., Maeda Y., Higashiguchi M., Miyake K., Morimura O., Koba T., Hayama Y., Koyama S., Nakanishi K., Iwasaki T., Tetsumoto S., Tsujino K., Kuroyama M., Iwahori K., Hirata H., Takimoto T., Suzuki M., Nagatomo I., Sugimoto K., Fujii Y., Kida H., Mizuguchi K., Ito M., Kijima T., Rakugi H., Mekada E., Tachibana I., Kumanogoh A., Double deletion of tetraspanins CD9 and CD81 in mice leads to a syndrome resembling accelerated aging. Sci. Rep. 8, 5145 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C., Lai M. B., Pedler M. G., Johnson V., Adams R. H., Petrash J. M., Chen Z., Junge H. J., Endothelial cell-specific inactivation of TSPAN12 (Tetraspanin 12) reveals pathological consequences of barrier defects in an otherwise intact vasculature. Arterioscler. Thromb. Vasc. Biol. 38, 2691–2705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franz J., Tarantola M., Riethmüller C., How tetraspanins shape endothelial and leukocyte nano-architecture during inflammation. Biochem. Soc. Trans. 45, 999–1006 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Brenner S., The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs S1 to S9
Legends for tables S1 and S2
Legend for movie S1
Tables S1 and S2
Movie S1