Abstract
To understand the basis of pathogenesis by Legionella longbeachae serogroup 1, the importance of the Mip protein in this species was examined. Amino-terminal analysis of the purified, cloned L. longbeachae serogroup 1 ATCC 33462 Mip protein confirmed that the cloned gene protein was expressed and processed in an Escherichia coli background. DNA sequence analysis of plasmid pIMVS27, containing the entire L. longbeachae serogroup 1 mip gene, revealed a high degree of homology to the mip gene of Legionella pneumophila serogroup 1, 76% homology at the DNA level and 87% identity at the amino acid level. Primer extension analysis determined that the start site of transcription was the same for both species, with some differences observed for the −10 and −35 promoter regions. Primers designed from the mip gene sequence obtained for L. longbeachae serogroup 1 ATCC 33462 were used to amplify the mip genes from L. longbeachae serogroup 2 ATCC 33484 and an Australian clinical isolate of L. longbeachae serogroup 1 A5H5. The mip gene from A5H5 was 100% identical to the type strain sequence. The serogroup 2 strain of L. longbeachae differed by 2 base pairs in third-codon positions. Allelic exchange mutagenesis was used to generate an isogenic mip mutant in ATCC 33462 and strain A5H5. The ATCC mip mutant was unable to infect a strain of Acanthamoebae sp. both in liquid and in a potting mix coculture system, while the A5H5 mip mutant behaved in a manner siilar to that of L. pneumophila serogroup 1, i.e., it displayed a reduced capacity to infect and multiply within Acanthamoebae. To determine if this mutation resulted in reduced virulence in the guinea pig animal model, the A5H5 mip mutant and its parent strain were assessed for their abilities to establish an infection after aerosol exposure. Unlike the virulent parent strain, the mutant strain did not kill any animals under two different dose regimes. The data indicate that the Mip protein plays an important role in the intracellular life cycle of L. longbeachae serogroup 1 species and is required for full virulence.
Legionella longbeachae serogroup 1 was first recognized as a cause of pneumonia in 1981 (30). In May 1987, L. longbeachae serogroup 1 was isolated for the first time from a patient in Australia (25). Since then, numerous cases of infection caused by this species have been reported (8, 24), and presently approximately 50% of all pneumonia cases in South Australia are attributable to this species (8, 39a, 45), a statistic which reflects the national trend. Subsequent studies showed that L. longbeachae serogroup 1 was present in commercial potting mix and in the soil of potted plants of patients and that it survived for long periods in these environments, indicating that soil, rather than water, may be the natural habitat of this species and a possible source of infection in the community (40). Restriction fragment-length polymorphism and allozyme studies performed to compare L. longbeachae serogroup 1 isolates from clinical and environmental origins demonstrated that they were all closely related and similar to isolates from L. longbeachae serogroup 1 ATCC 33462, indicating a close relationship between organisms isolated from countries as far apart as Australia and the United States (24).
No virulence studies of L. longbeachae serogroup 1 have been done, although L. longbeachae serogroup 2 has been examined by intraperitoneal injection into guinea pigs and for the ability to infect and multiply in a protozoan model of infection with Tetrahymena pyriformis and Hartmannella verformis (17, 44). Recent publications detailing in vitro models for intracellular growth of L. longbeachae serogroup 1 have shown that it can replicate in U937 cells (35) but is unable to replicate in Mac 6 cells or in Acanthamoebae castellanii (33). Little is known about the intracellular life cycle of this species, the factors which may contribute to pathogenesis, and whether these factors are shared with Legionella pneumophila serogroup 1.
L. longbeachae serogroup 1 pathogenesis studies have focused on the Mip protein and have examined the significance of this protein in pathogenesis by the organism. The Mip protein of L. pneumophila serogroup 1 has been established as a virulence factor of the organism, playing an important role in the intracellular life cycle, as mutant strains which lack the protein are significantly impaired in their ability to infect alveolar macrophages and protozoa (9, 12). They are also attenuated in their ability to cause disease in experimentally infected guinea pigs (11). The L. pneumophila serogroup 1 Mip protein displays homology to the FK506 binding protein (FKBP) class of immunophilins and shows characteristic peptidyl prolyl cis-trans isomerase (PPIase) activity (18). A homolog of the Mip protein also occurs in Legionella micdadei (2), a species of Legionella associated with disease in humans, and a mip mutant in this species also shows reduced intracellular infection (34). Mip analogs have been detected in all species of Legionella examined so far, including L. longbeachae serogroup 1 (10, 37, 38). Mip-like analogs which also display homology to the FKBP class of proteins have been reported in other intracellular pathogens such as Chlamydia trachomatis (27) and Coxiella burnetti (32), with PPIase activity having been demonstrated for both organisms (28, 32). Hence, Mip-like proteins with homology to the FKBP class of immunophilins may play a critical role in the life cycles of these organisms (19).
In this report, we document the cloning and sequence analysis of the mip gene from L. longbeachae serogroup 1 ATCC 33462 and compare the results with those from L. pneumophila serogroup 1 (16), L. longbeachae serogroup 2 ATCC 33484, L. micdadei (2), and an Australian clinical isolate of L. longbeachae serogroup 1, strain A5H5. To understand the significance of Mip in L. longbeachae serogroup 1, we constructed and characterized isogenic mip mutants in L. longbeachae serogroup 1 ATCC 33462 and the Australian clinical isolate of this species, strain A5H5. The mutants, which represent the first reported genetic manipulation of this species, were tested for their abilities to infect a strain of Acanthamoebae and to establish infection in guinea pigs. There were apparent differences between the two isolates of L. longbeachae serogroup 1 in both of these models.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial isolates of Legionella, E. coli strains, and plasmids used or constructed in this study are listed in Table 1. Legionella strains were routinely cultured on charcoal yeast extract α-ketoglutarate (CYE) plates (24) at 35°C. Legionella broth was used as a liquid growth medium (41). When required, selective agents were used at the following concentrations: chloramphenicol (CM), 5 μg/ml; kanamycin (KM), 25 μg/ml; and aztreonam, 4 μg/ml. For the amoeba coculture experiments, Legionella organisms were plated onto CYE plates containing pimafucin (250 mg/liter), polymixin B (80,000 IU/liter), and vancomycin (2 mg/liter) (CYE-VPP). E. coli strains were grown in Luria broth or on Columbia agar, and where appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; CM, 25 μg/ml, and KM, 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| L. pneumophila serogroup 1 (Philadelphia) | ATCC 33152; type strain | CDCa |
| L. longbeachae serogroup 1 | ATCC 33462; type strain | CDC |
| L. longbeachae serogroup 2 | ATCC 33484; type strain | CDC |
| A5H5 (L. longbeachae serogroup 1) | Australian clinical isolate | This study |
| B10 | ATCC 33462 L. longbeachae serogroup 1 strain with a mip deletion mutation | This study |
| B8 | L. longbeachae serogroup 1 A5H5 with a mip deletion mutation | This study |
| B8.22 | Strain B8 complemented with plasmid SKW27 | This study |
| E. coli DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rk− mk+) deoR thi-1 supE44 λ− gyrA96 relA1 | 20 |
| E. coli S17-1 | recA derivative of E. coli 294 (hsdR Pro) with RP4-2Tc::Mu (Ap Km Nm Tc::Mu) Km::Tn7 in the chromosome | 39 |
| Plasmids | ||
| pGEM-7Zf(−) | Apr cloning vector | Promega |
| pUC18K | pUC18 with the aphA-3 Kmr resistance cassette | P. Sansonetti, reference 31 |
| pCACTUS | Cmr cloning vector containing sacB and a temperature-sensitive replicon | C. A. Clark |
| pIMVS26 | pGEM with ca. 8-kb fragment of L. longbeachae serogroup 1 ATCC 33462 genomic DNA | This study |
| pIMVS27 | pGEM carrying a SacI fragment of pIMVS26 | This study |
| pIMVS28 | pGEM containing deleted mip gene generated in pIMVS27 | This study |
| pCACTUS49 | pCACTUS containing deleted mip gene fragment from pIMVS28 | This study |
| pCACTUS50 | Derivative of pCACTUS49 containing Kmr from pUC18K and mob deletion | This study |
| pWKS130 | Kmr cloning, sequencing vector | 46 |
| pIMVS29 | pWKS130 containing entire mip gene on SacI fragment from pIMVS27 | This study |
CDC, Centers for Disease Control and Prevention, Atlanta, Ga.
Antisera and antibodies.
L. pneumophila serogroup 1 polyclonal monospecific anti-Mip antisera, used initially to screen the L. longbeachae serogroup 1 plasmid bank, were a kind gift from N. P. Cianciotto (Department of Microbiology and Immunology, Northwestern University, Chicago, Ill.). Polyclonal antiserum was prepared specifically against L. longbeachae serogroup 1 Mip, excised from a 15% polyacrylamide gel, and emulsified in phosphate-buffered saline (PBS), pH 7.2. The acrylamide mix was injected subcutaneously into two New Zealand White rabbits. The injection was repeated after 2 weeks, and the serum was harvested at 6 weeks. The antiserum was extensively absorbed with E. coli DH5α(pGEM-7Zf[−]) prior to use.
Western immunoblot.
Total cell protein extractions were prepared by the method of Pearlman et al. (36). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by the method of Lugtenberg et al. (26) with a 15% running gel. Western immunoblot was performed per the procedure of Towbin et al. (43), using the staining procedure of Hawkes et al. (21), with 4-chloro-1-napthol.
Construction and screening of the plasmid bank.
Whole chromosomal DNA was extracted from L. longbeachae serogroup 1 ATCC 33462, by the method of Manning et al. (29), and digested with BamHI-EcoRI. The fragments were cloned into pGEM-7Zf(−) and transformed into DH5α. A clone carrying an 8-kb fragment was identified by colony immunoblot with L. pneumophila serogroup 1 anti-Mip serum. This clone, designated DH5α(pIMVS26), expressed a protein of approximately 27 kDa, as demonstrated by Western immunoblot. A subclone expressing the protein was generated by SacI digestion of pIMVS26 and recloning into pGEM-7Zf(−). A clone containing a 1.3-kb SacI fragment was identified, and the plasmid was designated pIMVS27.
DNA sequencing.
Sequencing was performed with the Applied Biosystems model 373A DNA sequencer. Plasmid pIMVS27 was sequenced in the forward direction with Dye Primer kits (ABI, Foster City, Calif.). The protocol was applied to templates generated by nested deletion of pIMVS27 with the Erase-a-Base kit (Promega, Madison, Wis.), according to the manufacturer’s instructions. The complementary strand of the clone was determined by using the Dye Terminator kit (ABI), with primers designed from the forward-sequence data, with double-stranded pIMVS27 as the template. The entire mip gene sequence was analyzed by DNASIS and PROSIS (Hitachi Software). Two primers, 844 (5′-GAGTATGATGAGAAAGAA-3′) and 845 (5′-ACAATTAATCTGATTTAAGG-3′), were designed from the completed sequence to amplify the entire mip gene from ATCC 33484 and strain A5H5. The expected 850-bp PCR product was purified with the QIAquick PCR purification kit (Qiagen) and sequenced with the Dye Terminator kit (ABI).
Primer extension from total bacterial RNA.
Primer extension analysis was used to map the 5′ end of the mip mRNA with a synthetic oligonucleotide primer (5′-GGCTGCAACTGATGCTACATCGCTT-3′). Total bacterial RNA was extracted from L. longbeachae serogroup 1 ATCC 33462, DH5α(pIMVS27), and DH5α(pGEM-7Zf[−]) by the hot-phenol method of Aiba et al. (1) and treated with RNase-free DNase I (Boehringer Mannheim). The oligonucleotide primer was radioactively labeled with [γ-32P]ATP by using T4 polynucleotide kinase (Boehringer Mannheim). The primer was hybridized to 20 μg of total RNA, and the mix was extended per the method of Williams et al. (47), with Moloney murine leukemia virus reverse transcriptase (Boehringer Mannheim). The reaction was loaded onto a 6% acrylamide-urea sequencing gel and visualized by autoradiography. Plasmid pIMVS27 was sequenced with the DNA sequencing kit version 2 (Amersham, Buckinghamshire, United Kingdom).
Allelic exchange mutagenesis, construction, and complementation of mip mutants.
Allelic exchange was carried out to generate mutations in the mip gene with the suicide vector pCACTUS. Vector pCACTUS is a derivative of plasmids containing the sacB gene of Bacillus subtilis (pIB279) and pIB307, containing a temperature-sensitive pSC101 replicon (6). pCACTUS also contained a mob region and a chloramphenicol resistance gene. Plasmid pCACTUS49 was introduced into L. longbeachae serogroup 1 ATCC 33462 by conjugation with the modified method from Bradley et al. (7) from a 48-h plate subculture of Legionella growth. The mating was incubated for 6 h at 30°C on CYE plates, serially diluted in PBS, and plated onto CYE plates containing 5 μg of CM per ml and 4 μg of aztreonam per ml. The natural resistance of L. longbeachae to aztreonam was used to select against the donor. All plates were incubated at 30°C. Electroporation was used to introduce pCACTUS50 into L. longbeachae serogroup 1 A5H5. Electrocompetent A5H5 cells were prepared according to the method of Dower et al. (13), except that PBS was used in the initial washes. Glycerol-treated A5H5 cells and plasmid DNA (approximately 1 μg) were subjected to an electric pulse of 2.3 kV in a 0.2-cm cuvette (Bio-Rad) with a Bio-Rad gene pulser at 100 Ω. The cells were incubated in broth at 30°C for 5 to 6 h and plated onto CYE plates containing KM.
The resulting Kmr or Cmr L. longbeachae colony was cultured in broth at 30°C with the appropriate antibiotic. Subsequent culture on CYE plates containing CM or KM at the nonpermissive temperature for pCACTUS replication in L. longbeachae (39°C) resulted in the cointegration of the plasmid into the chromosome via homologous recombination. One resultant antibiotic-resistant colony was incubated in broth with antibiotic selection at 30°C and plated onto CYE containing 6% sucrose to select for resolved cointegrates. Colonies from the sucrose plates were patched onto CYE plates and screened by PCR to assess allelic exchange, and potential mutants were confirmed by Southern blot hybridization and immunoblot. Mutant strains were complemented with plasmid pIMVS29, which was introduced by electroporation.
Southern blot hybridization.
DNA was transferred to nylon membranes (Hybond-N+; Amersham) by the method of Southern (42) and hybridized with digoxigenin (DIG)-labeled probe at 42°C overnight. Probes were labeled with DIG and hybridized with the filter under conditions described previously (24). The filters were developed according to the manufacturer’s protocol (Boehringer Mannheim).
Infection of Acanthamoebae with Legionella strains.
Acanthamoebae group 2 spp. used in coculture experiments were originally isolated from potting mix. Their identities were confirmed by Brett Robinson, South Australian Water Corporation, Bolivar, South Australia, Australia, and one strain, designated ACO97, was chosen for all experimental work.
Liquid cocultures of Acanthamoebae ACO97 and Legionella species were set up essentially as described by other workers (12). Duplicate cocultures containing approximately 103 Legionella organisms per ml and 104 Acanthamoebae cysts per ml were set up in 4 ml of amoeba saline (2 mM NaCl, 0.016 mM MgSO4, 0.027 mM CaCl2, 1 mM Na2HPO4, 1 mM KH2PO4). Legionella organisms were prepared by suspending growth from a 72-h CYE plate in sterile tap water to give approximately 109 organisms/ml by comparison with a turbidity standard (McFarland standard number 4); this was confirmed spectrophotometrically by using an optical density of 1.0 at 550 nm. These organisms were serially diluted and plated onto CYE agar to determine numbers of viable bacteria. Cocultures were incubated at 30°C. Samples were taken at days 1, 3, and 7, diluted in 0.2 M HCl-KCl buffer (pH 2.2), and plated onto CYE plates. Potting mix coculture samples were set up essentially as for liquid coculture, except that Legionella and Acanthamoebae were added to presteamed potting mix (Nu-Erth, Meadows, South Australia, Australia). Twenty grams of steamed soil seeded with Legionella and amoebae was incubated at 30°C, and samples were taken at days 3, 7, 11, and 15. At each interval, a 1-g aliquot of soil was removed, diluted in sterile tap water, mixed thoroughly, allowed to settle for 15 min, and then diluted in 0.2 M HCl-KCl acid buffer to reduce the number of unwanted soil microorganisms. Aliquots were plated onto CYE-VPP.
Animal studies. (i) Intraperitoneal inoculation.
Outbred guinea pigs (IMVS colored stock; Institute of Medical and Veterinary Science-Veterinary Services, Gilles Plains, South Australia, Australia), weighing between 300 and 600 grams, were inoculated intraperitoneally with a suspension of Legionella prepared in sterile tap water, as outlined for coculture experiments, and enumerated initially in a counting chamber (Hausser Scientific Partnership, Horsham, Pa.). The actual dose administered was accurately determined retrospectively by serial dilution and plating on CYE plates.
(ii) Aerosol inoculation.
Guinea pigs were infected by exposure to an aerosolized dose of Legionella within a closed chamber. The test strain dose was prepared as outlined for the amoeba coculture, except that strain B8.22 was grown on CYE plates containing KM and was enumerated retrospectively. The chamber was constructed of Perspex (Lucite) and measured 220 by 220 by 240 mm, with a removable top. A nebulizer pump therapy kit (Ventalair Forte II; Allersearch, Granville, Australia) was used to generate the aerosol, which had an average particle size of 3.9 microns, as specified by the manufacturer. An inlet was constructed on one side of the chamber, through which the nebulizer hose was inserted; this hose was sealed in place. The hose connected the nebulizer bowl on the inside of the chamber to the nebulizer pump unit on the outside. The nebulizer pump generated positive pressure in the chamber which was vented through a small outlet valve on the opposite side of the box. The chamber was placed in a laminar flow hood during aerosolization as a safety measure. Guinea pigs were placed in the chamber, and a 3-ml test suspension (containing approximately 109 or 1010 Legionella organisms total) was aerosolized into the chamber over a 15-min interval. The guinea pigs were held in the chamber for a further 5 min. One animal in each test group was killed immediately after exposure to enumerate the Legionella organisms introduced into the lungs. Lungs were homogenized in 100 ml of sterile tap water by using a Waring commercial blender (Waring Products, New Hartford, Conn.), and viable counts were determined by plating the homogenate, in duplicate, onto CYE and CYE-VPP plates.
Animals were checked three times daily for signs of illness, and their weights were recorded. Lungs were taken from animals that died to confirm experimental pneumonia.
Nucleotide sequence accession numbers.
The mip gene sequence data obtained for L. longbeachae serogroup 1 ATCC 33462 and L. longbeachae serogroup 2 ATCC 33484 in this study are available under GenBank and EMBL accession numbers X83036 and AF000958, respectively.
RESULTS
Analysis of L. longbeachae serogroup 1 Mip.
Amino-terminal analysis of clone DH5α(pIMVS26) was performed to ensure the identity of the Mip protein from L. longbeachae serogroup 1 and that it was processed in E. coli. N-terminal sequencing of the purified Mip protein from clone DH5α(pIMVS26) was performed at Macquarie University Centre for Analytical Biotechnology (Macquarie University, School of Biological Sciences, New South Wales, Australia) on a 470A Applied Biosystems protein sequencer. The protein was homologous to the first 16 amino acids in the processed form of the Mip outer membrane protein from L. pneumophila serogroup 1 (16), except for a threonine residue at position 8 in place of an alanine residue (Ala-Thr-Asp-Ala-Thr-Ser-Leu-Thr-Thr-Asp-Lys- Asp-Lys-Leu-Ser-Tyr). Subsequent sequencing of plasmid pIMVS27 showed one potential open reading frame (ORF) of 699 bp. A strong ribosome binding site was also found in close proximity to the putative ATG start site for translation. Downstream of the ORF, a stop codon was seen in conjunction with a region of dyad symmetry, corresponding to a putative transcriptional terminator, similar to that seen for the L. pneumophila serogroup 1 mip gene (16). This most likely represents a factor-independent transcription termination signal and has also been proposed for the L. micdadei mip gene (2).
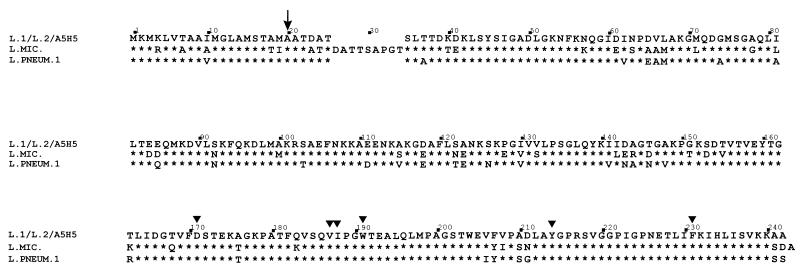
The inferred translated mip gene product was a polypeptide of 233 amino acids with a predicted molecular mass of 24,661 Da. The inferred amino acid sequences of the Mip proteins from L. longbeachae serogroup 1 A5H5 and the ATCC 33484 serogroup 2 isolate were identical with that of the ATCC 33462 L. longbeachae serogroup 1 Mip protein (Fig. 1) and were very similar to that of the L. pneumophila serogroup 1 Mip protein (16), displaying approximately 87% identity, and also to that of the L. micdadei Mip protein (2) (Fig. 1). The first 20 amino acids suggested a signal sequence and in conjunction with the N-terminal analysis suggest that Mip is processed in E. coli in a manner similar to the way it is processed in L. pneumophila serogroup 1. The key sites proposed to be involved in the PPIase activity (19) determined for the L. pneumophila serogroup 1 (Philadelphia) Mip protein (18) are conserved in L. longbeachae serogroup 1 Mip (Fig. 1), suggesting a similar function and role in pathogenesis.
FIG. 1.
Amino acid comparison of the Mip proteins of L. longbeachae serogroup 1 ATCC 33462 (L.1), L. longbeachae serogroup 1 A5H5 (A5H5), L. longbeachae serogroup 2 ATCC 33484 (L.2), L. pneumophila serogroup 1 (L. PNEUM. 1), and L. micdadei (L. MIC.). Asterisks indicate amino acids identical to those of L. longbeachae; triangles indicate amino acids predicted to form part of the active site for PPIase activity of Mip. The arrow indicates the site of signal peptidase cleavage.
Analysis of L. longbeachae serogroup 1 mip transcriptional signals.
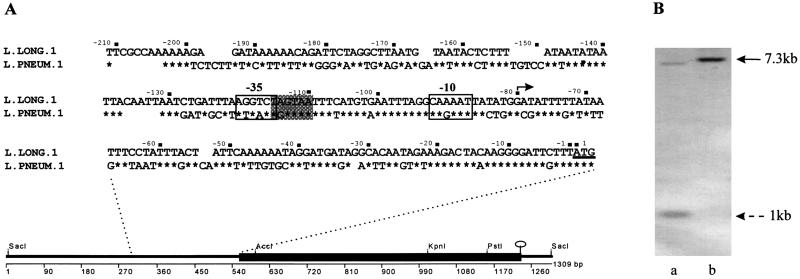
To confirm the start site for transcription of mip, and to compare this with the case for L. pneumophila serogroup 1, primer extension analysis was performed. Identification of the 5′ ends of the mip mRNA isolated from L. longbeachae serogroup 1 and the E. coli clones was determined by synthesis of cDNA with an oligonucleotide primer that was complementary to a region of DNA 54 bp downstream from the putative ATG start site on the mip mRNA. Identical cDNA bands were synthesized from RNA from L. longbeachae serogroup 1 ATCC 33462 and DH5α(pIMVS27), with no band detected in the control track where DH5α(pGEM7Zf[−]) was used as a template (data not shown). By comparing these bands with the sequencing reaction of pIMVS27, primed with the same oligonucleotide, the 5′ end of the mip mRNA was mapped to the G residue at nucleotide position 473 of the L. longbeachae serogroup 1 mip gene sequence. This result confirmed that the start sites for transcription in L. longbeachae serogroup 1 and L. pneumophila serogroup 1 (16) were identical in both species (Fig. 2A). The probable −10 and −35 promoter consensus sequences were identified and compared with those for L. pneumophila serogroup 1 (Fig. 2A). The −10 region was the same for L. longbeachae serogroup 1 and L. pneumophila serogroup 1; however, a −35 region was identified (Fig. 2A) in L. longbeachae serogroup 1 that is a more likely part of the promoter sequence than that suggested for L. pneumophila serogroup 1 (16). The spacing of the −10 and −35 regions for L. longbeachae serogroup 1 is a closer match with the consensus sequences determined for E. coli, and the spacing is optimal (17 ± 1 nucleotide).
FIG. 2.
(A) Line diagram depicts the DNA sequence determined from sequencing pIMVS27. The solid box shows the mip gene from L. longbeachae serogroup 1 ATCC 33462, selected restriction sites in the mip gene, and the stem loop structure at the end of the ORF. The inset sequence is the DNA sequence upstream of the ATG start site for translation in L. longbeachae serogroup 1 and L. pneumophila serogroup 1, showing the −10 and −35 promoter regions determined by primer extension analysis. The shaded box indicates the −35 promoter region proposed for L. longbeachae. The nonshaded box is the −35 region proposed in reference 16. The start site for transcription is shown with a solid arrow. (B) Southern hybridization demonstrating mutagenesis by allelic exchange of the L. longbeachae serogroup 1 mip gene. DNA was digested with KpnI and probed with DIG-labeled pIMVS27. Lanes: a, L. longbeachae serogroup 1 A5H5; b, B8. The solid arrow indicates the 7.3-kb fragment generated in B8 due to the loss of an internal KpnI site which generates 1- and 7-kb fragments in the parent strain. A similar pattern was observed for L. longbeachae serogroup 1 ATCC 33462 (data not shown).
Construction and complementation of L. longbeachae serogroup 1 mip mutants.
Isogenic mip mutants were generated in L. longbeachae serogroup 1 ATCC 33462 and strain A5H5 by allelic exchange with a plasmid construct, pCACTUS49, constructed in several stages. First, the mip gene in pIMVS27 was mutated by digestion with AccI and PstI to delete a 650-bp fragment within the coding region (Fig. 2A). The resulting construct, designated pIMVS28, was transformed into DH5α, and transformants were screened by Western blot to confirm the loss of production of Mip (data not shown). The residual 850-bp SacI fragment of pIMVS28 was cloned into pCACTUS to yield pCACTUS49. Plasmid pCACTUS49 was introduced into S17-1, which then served as a donor strain in subsequent conjugation experiments.
Due to difficulties in conjugation with strain A5H5, construct pCACTUS50 was made and delivered by electroporation. Plasmid pCACTUS50 was constructed from pCACTUS49 by removing the mob site by restriction digestion and inserting the Kmr marker from pUC18K into the SmaI site of the polylinker in order to use KM in addition to CM to select for transformants.
To identify colonies that had undergone complete allelic exchange, PCR analysis was performed with primers 844 and 845. A fragment of approximately 650 bp amplified in the case of the mutant strain was in contrast to an 850-bp fragment for the wild type (data not shown). Chromosomal DNA from the putative mip mutants and the parent was then digested with KpnI, a restriction site internal to the mip gene sequence (Fig. 2A). Duplicate Southern blots were probed with DIG-labeled pIMVS27 and pCACTUS. The mutant strains had only one hybridizing fragment (Fig. 2B, lane b) lacking the internal KpnI site, which had been removed by restriction deletion, while the parent strain had two hybridizing fragments (Fig. 2B, lane a). No bands were detected with the pCACTUS probe, indicating the vector sequence had been completely resolved from the chromosome (data not shown). In addition, Western immunoblot showed loss of Mip production in the mutant strains (data not shown).
To ensure that the mutation process had not affected genes other than mip, complemented mutant strains were constructed with vector pIMVS29. This construct was derived by cloning the SacI fragment from pIMVS27, containing the entire L. longbeachae serogroup 1 mip gene, into vector pWKS130. Only strain B8 was complemented, as B10 and the parent strain were both avirulent. The complemented mip mutant in A5H5 was screened by Western immunoblot to confirm the production of Mip (data not shown).
Effect of mip on intracellular infection.
To determine whether Mip promotes infection of amoebae in L. longbeachae serogroup 1, we assessed the abilities of both mip mutants to infect Acanthamoebae, a common soil amoeba. Two systems were used to assess the levels of multiplication of Legionella strains, with potting mix considered a more natural, nonaquatic environment for L. longbeachae serogroup 1. The potting mix was steamed for approximately 1 h to kill any preexisting Legionella spp.; however, the steaming process did not sterilize the mix, as spore-forming organisms were not killed. The same multiplicity of infection was used for both systems, and samples were taken frequently during the experiment to determine the level of multiplication of Legionella (Fig. 3). The mean number of CFU (± standard deviations) was determined for each time point, and the Student-Newman-Keuls comparison of means (P < 0.05) was used to determine statistical significance.
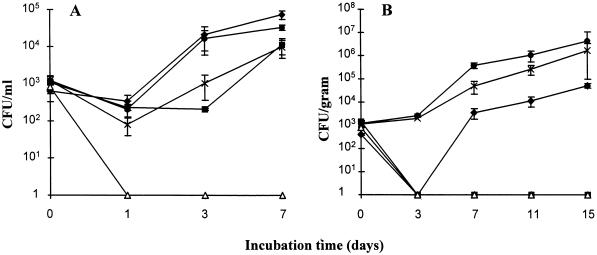
FIG. 3.
Coculture of Acanthomoebae with strains of Legionella. (A) Amoeba liquid cocultures were set up in saline with approximately 104 amoebae/ml and 103 CFU (each) of L. pneumophila serogroup 1 (Philadelphia) (⧫), L. longbeachae serogroup 1 ATCC 33462 (▪), B10 (▵), A5H5 (•), and B8 (×) per ml. Samples were taken at various time intervals, and the number of Legionella organisms was determined by plating on selective media. Each time point represents the mean number of CFU recovered, and the vertical bars indicate standard deviations. (B) Amoebae were cocultured in an artificial potting mix system with strains of Legionella as indicated above. Numbers of viable Legionella organisms were determined at various time points by treatment of the soil sample with acid and plating on selective media. The experiments shown are representative of two independent experiments.
L. pneumophila serogroup 1 (Philadelphia), L. longbeachae serogroup 1 ATCC 33462, and strain A5H5 multiplied in liquid coculture similarly to other Legionella organisms (12, 34) (Fig. 3A), showing an initial lag period with a steady increase in bacterial numbers during the experiment. The mip mutants of the two strains of L. longbeachae serogroup 1 showed growth patterns different from that of their parent strain and also from those of each other. Mutant B8 increased in numbers at a lower rate than A5H5, with a statistically significant difference in recovery observed at day 7. Statistically significant differences were not seen at days 1 and 3, most likely due to large variations in the counts and the low sample numbers. However, the expected growth trend was observed, and the end result was similar to that determined for the L. pneumophila serogroup 1 mip mutant (12). Complemented mip mutant B8.22 also grew in amoebae (data not shown) and was recovered at day 7 in numbers that were not statistically different from those of the wild-type strain A5H5. Strain B10 was unable to replicate in this system and in several repeat experiments.
L. longbeachae serogroup 1 A5H5 and B8 were both able to replicate in potting mix, showing similar growth trends, as seen with the liquid coculture (Fig. 3B). Statistically significant differences in strain recovery were observed at days 7 and 11. The numbers of organisms observed at day 15 were not statistically different, most likely due to reasons stated above; however, the expected growth trend was observed. L. pneumophila serogroup 1 (Philadelphia) replicated in this system, with an initial lag phase observed at day 3, where numbers dropped to undetectable levels; this may reflect the low number of organisms added initially to the coculture or the inappropriate levels of sampling for that time point. In all experiments, however, L. pneumophila serogroup 1 (Philadelphia) multiplied. L. longbeachae serogroup 1 ATCC 33462 and mutant B10 were unable to replicate in this system and in several repeat experiments.
Animal model of infection.
Two models were established in the laboratory and assessed for their ability to allow a comparison of the virulences of experimental Legionella strains. The intraperitoneal model allowed accurate doses to be administered and test strains to be compared (Table 2). L. pneumophila serogroup 1 (Philadelphia) was virulent in this model, with death occurring in all animals within 30 h. The L. longbeachae serogroup 1 strains, however, rarely caused death by this mode of transmission. The L. longbeachae serogroup 1 and serogroup 2 ATCC type strains were completely avirulent in this model, while L. longbeachae serogroup 1 A5H5 did produce symptoms and death in 1 of 3 animals after 4 days. The data indicated that this was not a suitable model to assess the mutant strains, given the relative avirulence of L. longbeachae in this model.
TABLE 2.
Intraperitoneal inoculation of Legionella strains
| Strain | Dose (total CFU) | No. of guinea pigs killed/no. tested | Comments |
|---|---|---|---|
| L. pneumophila serogroup 1 (Philadelphia) | 5 × 108 | 3/3 | Spleens contained 107 to 108 organisms |
| L. longbeachae serogroup 1 ATCC 33462 | 2 × 109 | 0/3 | |
| L. longbeachae serogroup 2 ATCC 33484 | 5 × 108 | 0/3 | |
| L. longbeachae serogroup 1 A5H5 | 1 × 109 | 1/3 | Death at 4 days |
The aerosol model allowed doses of Legionella to be administered by the respiratory route of entry. The symptoms produced and the time course of the disease were similar to those predicted by other workers (11). L. pneumophila serogroup 1 and strain A5H5 were both virulent by this model and caused death in 3 of 5 animals tested. Examination of the lungs taken from a guinea pig that died due to exposure to A5H5 revealed that the air spaces of the lung parenchyma were filled with a dense cellular infiltrate consisting of neutrophils and monocyte cells, and the histological appearance was consistent with a severe acute pneumonia similar to that seen with L. pneumophila serogroup 1 (3, 4, 15). Animals exposed to aerosols of L. longbeachae serogroup 1 ATCC 33462 and L. longbeachae serogroup 2 ATCC 33484 developed no symptoms and were avirulent, although the numbers of Legionella organisms retained in the lung with each test strain were comparable (Table 3). For this reason, only the A5H5 mip mutant (B8) was assessed in the aerosol model, along with the complemented strain B8.22. Guinea pigs were tested with two doses of the mutant strain, and the percentage weight gain or loss was plotted for each animal and compared with that for animals inoculated with the isogenic parent strain as well as with that for animals inoculated with the complemented strain (Fig. 4). The parent strain killed 3 of 5 animals (all symptomatic) within 5 days after exposure (Fig. 4A). Guinea pigs exposed to 109 CFU (approximately 105 retained organisms) of B8 showed no evidence of the disease (Fig. 4B). Some evidence of disease, predominantly weight loss, was observed in some of the animals exposed to a 1010-CFU dose (approximately 106 retained organisms) of the same mutant strain (Fig. 4C). The mip mutant in strain A5H5 did not cause death with either dose. Reintroduction of the intact wild-type mip gene from L. longbeachae serogroup 1 was able to fully complement the mutation in strain B8, leading to restored virulence (Fig. 4D).
TABLE 3.
Aerosol inoculation of Legionella strains
| Strain | Dose (total CFU) | No. of guinea pigs killed/no. tested | Comments | Retained dose |
|---|---|---|---|---|
| L. pneumophila serogroup 1 (Philadelphia) | 1 × 109 | 2/3 | Death within 5 days | ∼2 × 105 |
| L. longbeachae serogroup 1 ATCC 33462 | 1 × 109 | 0/3 | No symptoms | 2 × 105 |
| L. longbeachae serogroup 2 ATCC 33484 | 1 × 109 | 0/3 | No symptoms | ∼2 × 105 |
| L. longbeachae serogroup 1 A5H5 | 1 × 109 | 3/5 | Death within 5 days (days 2 and 4) | 3.5 × 105 |
| B8 | 1 × 109 | 0/5 | No symptoms | 1 × 105 |
| B8 | 1 × 1010 | 0/5 | Slight symptoms in most animals | 1.6 × 106 |
FIG. 4.
Percentage weight gain or loss in guinea pigs exposed to an aerosol of different strains of Legionella longbeachae serogroup 1. (A) Animals exposed to a dose of 109 L. longbeachae serogroup 1 A5H5 organisms; (B) animals exposed to a dose of 109 B8 organisms; (C) animals exposed to a dose of 1010 B8 organisms; (D) animals exposed to a dose of 109 B8.22 organisms. Guinea pig death is indicated by the termination of the ribbon graph prior to the end of the experiment on day 7.
DISCUSSION
In this study, the mip gene from L. longbeachae serogroup 1 was sequenced, and the role played by this protein in facilitating infection of guinea pigs and Acanthamoebae was examined. The mip gene sequences for L. longbeachae ATCC 33462 and A5H5 were identical, while the sequence for L. longbeachae ATCC 33484 differed from the former by two bases (positions 517 and 523). The translated protein sequences were identical and highly conserved in comparison to those from L. pneumophila serogroup 1 and L. micdadei. The start sites of transcription for L. longbeachae serogroup 1 and L. pneumophila were identical, and this confirms the high degree of conservation of mip genes and hence the probability that the proteins have similar functions. PPIase activity was not determined for L. longbeachae serogroup 1 Mip, but conserved amino acids critical to this enzymatic activity suggest the protein has a similar mechanism of action.
The role of the Mip protein as a potentiator of intracellular infection in L. longbeachae is further suggested by the behavior of the mip mutants in the Acanthamoebae coculture models. The mutant in strain A5H5 showed a growth pattern similar to those of the mip mutants of L. pneumophila serogroup 1 and L. micdadei (12, 34). The mip mutant in L. longbeachae serogroup 1 ATCC 33462 was unable to multiply in the amoeba models and warrants further analysis, but these results may simply reflect a greater level of attenuation of the ATCC parent strain. Differences were observed between the two parent strains, in both models, with the type strain ATCC L. longbeachae showing a lower level of infectivity in comparison to strain A5H5. This strain difference was most significant in the animal model, where L. longbeachae serogroup 1 ATCC 33462 was unable to establish infection in either model, while strain A5H5 was virulent.
The results obtained in the animal model for the mip mutant in L. longbeachae serogroup 1 A5H5 are of interest, as no other mip mutant, other than those of L. pneumophila serogroup 1, has been assessed in an aerosol animal model. The results are consistent with those seen for the mip mutant of L. pneumophila serogroup 1 (11). The mutant was unable to cause death in guinea pigs under two test dose conditions. However, the test doses trialed in this study resulted in lower numbers of bacteria being deposited into the lungs than those achieved by intratracheal inoculation in the study by Cianciotto et al. (11). The aerosol model of infection makes it difficult to achieve higher numbers of deposited bacteria, and hence we cannot say whether the mip mutation in L. longbeachae serogroup 1 would have yielded different results at higher doses. It is tempting to speculate that this would be the case, as L. longbeachae serogroup 1 differs from L. pneumophila serogroup 1 in that it does not possess the major outer membrane protein (references 14, 22, and 23 and unpublished observations). The major outer membrane protein is believed to play a role in uptake of L. pneumophila serogroup 1 into macrophages through its ability to bind complement component C3b (5). Therefore, L. longbeachae serogroup 1 may be more susceptible to changes in outer membrane proteins. The difference between the wild-type parent and the mip mutant in L. longbeachae serogroup 1 on the severity of the symptoms shown indicates a significant effect on the organism. The Mip protein is likely to have a significant role in pathogenesis of the organism or in survival in protozoa and the environment.
Given the close clonal nature of L. longbeachae serogroup 1 (24), why is the American ATCC L. longbeachae serogroup 1 isolate less virulent than the Australian clinical isolate? Are there fundamental differences between the two strains that may account for these discrepancies? Work is currently under way to investigate these questions.
ACKNOWLEDGMENTS
This project was partly funded by the Horticultural Research and Development Corporation. We thank the Clive and Vera Ramaciotti Foundations for the purchase of laboratory equipment.
We are also grateful to N. Cianciotto for the kind gift of the L. pneumophila serogroup 1 anti-Mip serum and thank C. A. Clark for helpful discussions regarding pCACTUS.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 2.Bangsborg J M, Cianciotto N P, Hindersson P. Nucleotide sequence analysis of the Legionella micdadei mip gene, encoding a 30-kilodalton analog of the Legionella pneumophila Mip protein. Infect Immun. 1991;59:3836–3840. doi: 10.1128/iai.59.10.3836-3840.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baskerville A, Fitzgeorge R B, Broster M, Hambleton P. Histopathology of experimental Legionnaires’ disease in guinea pigs, rhesus monkeys and marmosets. J Pathol. 1983;139:349–362. doi: 10.1002/path.1711390310. [DOI] [PubMed] [Google Scholar]
- 4.Baskerville A, Dowsett A B, Fitzgeorge R B, Hambleton P, Broster M. Ultrastructure of pulmonary alveoli and macrophages in experimental Legionnaires’ disease. J Pathol. 1983;140:77–90. doi: 10.1002/path.1711400202. [DOI] [PubMed] [Google Scholar]
- 5.Bellinger-Kawahara C, Horwitz M A. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J Exp Med. 1990;172:1201–1210. doi: 10.1084/jem.172.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomfield I C, Vaughn V, Rest R F, Eisenstein B I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol Microbiol. 1991;5:1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 7.Bradley D E, Taylor D E, Cohen D R. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980;143:1466–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron S, Roder D, Walker C, Feldheim J. Epidemiological characteristics of Legionella infection in South Australia: implications for disease control. Aust N Z J Med. 1991;21:65–70. doi: 10.1111/j.1445-5994.1991.tb03007.x. [DOI] [PubMed] [Google Scholar]
- 9.Cianciotto N P, Eisenstein B I, Mody C H, Toews G B, Engleberg N C. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianciotto N P, Bangsborg J M, Eisenstein B I, Engleberg N C. Identification of mip-like genes in the genus Legionella. Infect Immun. 1990;58:2912–2918. doi: 10.1128/iai.58.9.2912-2918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cianciotto N P, Eisenstein B I, Mody C H, Engleberg N C. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J Infect Dis. 1990;162:121–126. doi: 10.1093/infdis/162.1.121. [DOI] [PubMed] [Google Scholar]
- 12.Cianciotto N P, Fields B S. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehret W, Ruckdeschel G. Species specific membrane proteins of Legionellaceae. Zentralbl Bakteriol Hyg Abt Orig A. 1983;255:33–38. [PubMed] [Google Scholar]
- 15.Eisenstein T B, Tamada R, Meissler J, Flesher A, Oels H C. Vaccination against Legionella pneumophila: serum antibody correlates with protection induced by heat-killed or acetone-killed cells against intraperitoneal but not aerosol infection in guinea pigs. Infect Immun. 1984;45:685–691. doi: 10.1128/iai.45.3.685-691.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engleberg N C, Carter C, Weber D R, Cianciotto N P, Eisenstein B I. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect Immun. 1989;57:1263–1270. doi: 10.1128/iai.57.4.1263-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fields B S, Barbaree J M, Shotts E B, Jr, Feeley J C, Morrill W E, Sanden G N, Dykstra M J. Comparison of guinea pig and protozoan models for determining virulence of Legionella species. Infect Immun. 1986;53:553–559. doi: 10.1128/iai.53.3.553-559.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer G, Bang H, Ludwig B, Mann K, Hacker J. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPIase) activity. Mol Microbiol. 1992;10:1375–1383. doi: 10.1111/j.1365-2958.1992.tb00858.x. [DOI] [PubMed] [Google Scholar]
- 19.Hacker J, Fischer G. Immunophilins: structure-function relationship and possible role in microbial pathogenicity. Mol Microbiol. 1993;10:445–456. doi: 10.1111/j.1365-2958.1993.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D. Bacterial transformation. In: Glover D M, editor. DNA cloning. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 21.Hawkes R, Niday E, Gordon J. A dot-immunoblotting assay for monoclonal and other antibodies. Anal Biochem. 1982;119:142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- 22.Hindahl M S, Iglewski B H. Cloning and expression of a common Legionella outer membrane antigen in Escherichia coli. Microb Pathog. 1987;2:91–99. doi: 10.1016/0882-4010(87)90101-x. [DOI] [PubMed] [Google Scholar]
- 23.Hindahl M S, Iglewski B H. Outer membrane proteins from Legionella pneumophila serogroups and other Legionella species. Infect Immun. 1986;51:94–101. doi: 10.1128/iai.51.1.94-101.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanser J A, Adams M, Doyle R, Sangster N, Steele T W. Genetic relatedness of Legionella longbeachae isolates from human and environmental sources in Australia. Appl Environ Microbiol. 1990;56:2784–2790. doi: 10.1128/aem.56.9.2784-2790.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim I L, Sangster N, Reid D P, Lanser J A. L. longbeachae pneumonia: report of two cases. Med J Aust. 1989;150:599–601. doi: 10.5694/j.1326-5377.1989.tb136700.x. [DOI] [PubMed] [Google Scholar]
- 26.Lugtenberg B J, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K-12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 27.Lundemose A G, Rouch D A, Birkelund S, Christiansen G, Pearce J H. Chlamydia trachomatis Mip-like protein. Mol Microbiol. 1992;6:2539–2548. doi: 10.1111/j.1365-2958.1992.tb01430.x. [DOI] [PubMed] [Google Scholar]
- 28.Lundemose A G, Kay J E, Pearce J H. Chlamydia trachomatis Mip-like protein has peptidyl-prolyl cis/trans isomerase activity that is inhibited by FK506 and rapamycin and is implicated in initiation of chlamydial infection. Mol Microbiol. 1993;7:777–783. doi: 10.1111/j.1365-2958.1993.tb01168.x. [DOI] [PubMed] [Google Scholar]
- 29.Manning P M, Heuzenroeder M W, Yeadon J, Leavesley D I, Reeves P R, Rowley D. Molecular cloning and expression in Escherichia coli K-12 of the O antigens of the Inaba and Ogawa serotypes of the Vibrio cholerae O1 lipopolysaccharides and their potential for vaccine development. Infect Immun. 1986;53:272–277. doi: 10.1128/iai.53.2.272-277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney R M, Porschen R K, Edelstein P H, Bissett M J, Harris P P, Bondell S P, Steigerwalt A G, Weaver R E, Ein M E, Lindquist D S, Kops R S, Brenner D J. Legionella longbeachae species nova, another etiological agent of human pneumonia. Ann Intern Med. 1981;94:739–741. doi: 10.7326/0003-4819-94-6-739. [DOI] [PubMed] [Google Scholar]
- 31.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo Y-Y, Cianciotto N P, Mallavia L P. Molecular cloning of a Coxiella burnetti gene encoding a macrophage infectivity potentiator (Mip) analogue. Microbiol. 1995;141:2861–2871. doi: 10.1099/13500872-141-11-2861. [DOI] [PubMed] [Google Scholar]
- 33.Neumeister B, Schöniger S, Faigle M, Eichner M, Deitz K. Multiplication of different Legionella species in Mac 6 cells and in Acanthamoebae castellanii. Appl Environ Microbiol. 1997;63:1219–1224. doi: 10.1128/aem.63.4.1219-1224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Connell W A, Bangsborg J M, Cianciotto N P. Characterization of a Legionella micdadei mip mutant. Infect Immun. 1995;63:2840–2845. doi: 10.1128/iai.63.8.2840-2845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell W A, Dhand L, Cianciotto N P. Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect Immun. 1996;64:4381–4384. doi: 10.1128/iai.64.10.4381-4384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearlman E, Engleberg N C, Eisenstein B I. Identification of protein antigens of Legionella pneumophila serogroup 1. Infect Immun. 1985;47:74–79. doi: 10.1128/iai.47.1.74-79.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratcliff R M, Donnellan S C, Lanser J A, Manning P A, Heuzenroeder M W. Interspecies sequence differences in the Mip protein from the genus Legionella: implications for function and evolutionary relatedness. Mol Microbiol. 1997;25:1149–1158. doi: 10.1046/j.1365-2958.1997.5471908.x. [DOI] [PubMed] [Google Scholar]
- 38.Riffard S, Vandenesch F, Reyrolle M, Etienne J. Distribution of mip-related sequences in 39 species (48 serogroups) of Legionellaceae. Epidemiol Infect. 1996;117:501–506. doi: 10.1017/s0950268800059173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilisation system for in vivo genetic engineering: transposon metagenesis in gram-negative bacteria. Bio/Technology. 1982;1:784–791. [Google Scholar]
- 39a.South Australian Health Commission. Unpublished data.
- 40.Steele T W, Lanser J, Sangster N. Isolation of Legionella longbeachae serogroup 1 from potting mixes. Appl Environ Microbiol. 1990;56:49–53. doi: 10.1128/aem.56.1.49-53.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinmetz I, Rheinheimer C, Hübner I, Bitter-Suermann D. Genus-specific epitope on the 60-kilodalton Legionella heat shock protein recognized by a monoclonal antibody. J Clin Microbiol. 1991;29:346–354. doi: 10.1128/jcm.29.2.346-354.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Souther E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 43.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wadowsky R M, Wilson T M, Kapp N K, West A J, Kuchta J M, States S J, Dowling J N, Yee R B. Multiplication of Legionella spp. in tap water containing Hartmannella vermiformis. Appl Environ Microbiol. 1991;57:1950–1955. doi: 10.1128/aem.57.7.1950-1955.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker C, Weinstein P. Communicable Diseases Intelligence. 16:70–71. Canberra, Australia: Department of Health, Housing and Community Services; 1992. [Google Scholar]
- 46.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 47.Williams S G, Manning P A. Transcription of the Vibrio cholerae haemolysin gene hlyA, and cloning of a positive regulatory locus, hlyU. Mol Microbiol. 1991;5:2031–2038. doi: 10.1111/j.1365-2958.1991.tb00825.x. [DOI] [PubMed] [Google Scholar]