Abstract
Background and Aims
Several lines of evidence indicate that carbohydrate storage in plant below-ground organs might be positively related to genome size because both these plant properties represent resource sinks and can affect cell size, cell cycle time, water-use efficiency and plant growth. However, plants adapted to disturbance, such as root sprouters, could be an exception because their strategy would require higher carbohydrate reserves to fuel biomass production but small genomes to complete their cell cycles faster.
Methods
We used data from a field survey to test the relationship between genome size and the probability of root sprouting ability in 172 Central European herbaceous species. Additionally, we conducted a pot experiment with 19 herbaceous species with different sprouting ability (nine congeneric pairs plus one species), and measured root non-structural carbohydrate concentrations and pools at the end of a growing season.
Key Results
In the Central European flora, the probability of root sprouting ability was lower in large-genome species but this pattern was weak. In the pot experiment, both total non-structural and water-soluble carbohydrates (mainly fructans) were positively and non-linearly related to genome size, regardless of sprouting strategy. The concentrations of mono- and disaccharides and all carbohydrate pools showed no link to genome size, and starch was absent in large-genome species. The link between genome size and carbohydrate storage was less apparent at a small phylogenetic scale because we only observed a higher carbohydrate concentration in species with larger genomes for four of the species pairs.
Conclusions
Root sprouters may have smaller genomes because of their frequent occurrence in dry and open habitats. Large-genome species with presumably large cells and vacuoles could accumulate more water-soluble carbohydrates at the end of the growing season to fuel their growth and perhaps protect vulnerable organs from freezing early in the next season.
Keywords: Below-ground organ, carbon storage, cell size, fructan, genome size, root sprouting
INTRODUCTION
Genome size (DNA content in the nucleus) is a plant trait that is increasingly being used to inform ecological models, thanks to its huge between-species variability, relative stability within species and independence from environmental conditions. At the cellular level, genome size is recognized as a fundamental constraint of minimum cell size (Bennett, 1987; Beaulieu et al., 2008; Šímová and Herben, 2012; Greilhuber and Leitch, 2013) and minimum cell cycle length (Francis et al., 2008; Greilhuber and Leitch, 2013), especially the S phase (Šímová and Herben, 2012). Genome size, through its effect on cell size and cell cycle length, can also be correlated with various whole-plant properties, such as water-use efficiency, CO2 assimilation or metabolic rates (Faizullah et al., 2021), and, ultimately, plant ecological strategies (Bennett, 1987; Greilhuber and Leitch, 2013). Small-genome species are often fast-growing ruderals (Bennett, 1987; Bennett et al., 1998; Greilhuber and Leitch, 2013), while large-genome size species are often spring geophytes (Veselý et al., 2012). Many spring geophytes can have high concentrations of water-soluble carbohydrates, namely fructans (Hendry, 1987; Brocklebank and Hendry, 1989), and this high concentration of fructans has been interpreted as an adaptation to cold weather at the beginning of the growing season when early flowers emerge (Hendry, 1987; Orthen and Wehrmeyer, 2004). Nevertheless, water-soluble carbohydrates are part of carbohydrate storage together with other compounds, such as starch, that is also crucial for the regeneration of fast-growing ruderals. However, the relationship between regeneration capacity, carbohydrate storage and genome size has not yet been explored.
Genome size can be negatively correlated with plant growth because larger genomes might take more time to replicate during the S phase (Šímová and Herben, 2012), and thus slow the relative growth rate (Gruner et al., 2010) and prolong generation time (although evidence is currently highly mixed; Knight et al., 2005 and references therein). Therefore, early-spring plant species are not restricted by the speed of cell division because they fully preform organs such as flowers prior to winter, and in spring they only need to fill up the existing cells with water (Schnáblová et al., 2021). In contrast, quickly regenerating weedy plants are dependent on fast cell division, and thus small genomes are beneficial for them (Bennett, 1987). Some weeds are also perennial and resprout after disturbance from below-ground organs, often roots (Bartušková et al., 2021; Klimešová and Martínková, 2022). This strategy requires sufficient carbohydrate storage to boost their fast regrowth (Martínková et al., 2023).
Non-structural carbohydrates are formed during the growing season and stored in the below-ground storage organs of herbs before mobilization for seasonal regrowth or response to damage and stress (Janeček et al., 2011). These are a diverse group of molecules that play a variety of roles in plant life and growth strategy. They can be more generally organized by their size and mobility, with starch as the best recognized storage carbohydrate, which is a large and immobile polysaccharide (Almeida et al., 2021). The smallest and thus most mobile non-structural carbohydrates are the mono- and disaccharides (e.g. fructose, glucose and sucrose); these are present in all plant species and play particularly important roles in signalling and transport (Liu et al., 2012; Jensen et al., 2016), but can represent a large component of carbohydrate storage in select species (Lubbe et al., 2021b; Chlumská et al., 2022). Somewhere in-between are the water-soluble reserves, including disaccharides such as melibiose and trehalose, sugar alcohols, and the fructans and raffinose family oligosaccharides (RFOs). Fructans and RFOs range from very small oligosaccharides to more mid-sized polysaccharides and usually form an abundant part of carbohydrate storage for a wide variety of herbaceous species, especially those in Asteraceae, Amaryllidaceae, Lamiaceae and the Pooideae subfamily of Poaceae (Hendry, 1987; Van den Ende, 2013). All these compounds can have some role in physiological responses (especially to frost and drought; Hincha et al., 2002; Dias-Tagliacozzo et al., 2004; Patton et al., 2007).
The mechanistic link between carbohydrate storage and genome size could be a positive scaling between genome and cell size, which effectively sets the storing capacity of cells. This would suggest a positive link between genome size and the amount of stored carbohydrates, especially the water-soluble carbohydrates stored in cell vacuoles, such as fructans (fructose oligo- and polysaccharides) or vacuolar sucrose (Hendry, 1987; Chapin et al., 1990). The vacuole is typically the largest organelle in plant cells and its size is generally positively associated with cell size (Owens and Poole, 1979). Starch, another important storage carbohydrate, is not stored in vacuoles but in amyloplasts, and its concentration may be higher in small-genome species (Hendry, 1987) and therefore might be needed to speed up tissue regeneration of ruderals. We can see two contrasting outcomes for the relationship between genome size and carbohydrate storage that is dependent on carbohydrate type (large-genome fructan-producing species vs. small-genome starch-producing species; Hendry 1987). In this respect, plants that are able to produce adventitious buds on roots (root sprouters) could be an important group of presumably small-genome species (with fast cell cycles) that store higher amounts of carbohydrates to fuel resprouting (Klimešová and Martínková, 2022; Martínková et al., 2023).
Root sprouting is known as an alternative strategy in severely disturbed habitats otherwise dominated by annuals (Herben et al., 2018; Klimešová and Martínková, 2022). Root sprouters are frequent in open and dry habitats (Bartušková et al., 2021), i.e. in conditions that are expected to favour species with smaller genomes (Faizullah et al., 2021). When disturbed, root sprouters have an advantage in comparison to other plants because of their ability to resprout even from small root fragments after fragmentation of the root system and therefore are successful even on arable land. In a pot experiment, root sprouters tended to store slightly more mono- and disaccharides and starch than fructans compared to non-root sprouting congeners but these differences were not consistent throughout a growing season (Martínková et al., 2023). In terms of genome size, to our knowledge no study has tested whether root sprouters have smaller genomes than non-root sprouters because previous studies of ruderal plants were focused mainly on annual and not perennial weeds (Bennett et al., 1998).
To understand the link between genome size and carbohydrate storage and how this link is affected by species sprouting ability, we combined two alternative approaches: a field survey and a glasshouse pot experiment. The former allowed us to capture a large range of environmental factors and a sufficient taxon sampling, while the latter allowed us to minimize variation in non-structural carbohydrates (e.g. because of seasonality or environmental factors, Martinez-Vilalta et al., 2016) in a smaller set of species. We addressed the following questions: (1) Is the probability of root sprouting (RS) ability associated with genome size in a set of Central European herbaceous species? (2) Is there a relationship between root non-structural carbohydrate concentrations and pools (concentration × dry root biomass) and genome size in a set of congeneric herbaceous species with different sprouting strategies? (2a) Is the nature of the genome–carbohydrate storage relationship affected by RS ability? (2b) Is the genome–carbohydrate storage relationship similar in different carbohydrate functional groups? For the first question, we expected lower genome size in root sprouters because faster cell cycles can be advantageous for strategies that rely on fast regeneration from roots in disturbed habitats. For the second question, we expected total carbohydrate concentrations and pools to be independent of genome size but water-soluble carbohydrates to be positively and starch to be negatively associated with genome size.
MATERIALS AND METHODS
Survey of RS ability and genome size
To examine the relationship between RS ability and genome size, we used data from Bartušková et al. (2021) that included 172 Central European herbaceous species that occur along major environmental gradients, such as moisture, light availability and disturbance. Approximately one-quarter of the species in this list were confirmed root sprouters. Holoploid genome size (1C value), i.e. the DNA content of one non-replicated chromosome set, and monoploid genome size (1Cx value), i.e. the total mass of DNA in the nucleus (2C value) divided by ploidy level, were obtained from www.pladias.cz (Šmarda et al., 2019; Chytrý et al., 2021).
Glasshouse experiment
To analyse carbohydrate storage and biomass production of root-sprouting versus non-root-sprouting herbs, we set up a pot glasshouse experiment in 2019. For the experiment, we acquired nine congeneric species pairs (and standalone Silene vulgaris), with one root-sprouting and one non-root-sprouting species from the same genus (Table 1; CLO-PLA database, Klimešová and de Bello, 2009), to minimize the effects of phylogeny on the observed plant behaviour (see ‘Species congeners’). Seeds were obtained from a commercial supplier (Planta Naturalis, Markvartice u Sobotky, Czech Republic).
Table 1.
Basic information on species used in the glasshouse experiment. Family, carbohydrate concentration ranges (%) and genome size category (following Leitch et al., 1998) are shown. Genome size categories: very small = 1C < 1.4 pg; small = 1.4 < 1C < 3.5 pg; intermediate = 3.5 < 1C < 14 pg; 1 pg = 978 Mb. TNC = total non-structural carbohydrates
| Species | Family | TNC | Mono- and disaccharides | Starch | Water-soluble reserves | Genome size |
|---|---|---|---|---|---|---|
| Achillea millefolium | Asteraceae | 49.4, 62.1 | 0.9, 1.4 | 0.1, 0.5 | 47.7, 60.8 | Intermediate |
| Achillea nobilis | Asteraceae | 28.5, 39.5 | 1.4, 1.9 | 0.1, 0.3 | 26.8, 37.9 | Small |
| Artemisia absinthium | Asteraceae | 41.6, 52.4 | 1.6, 1.8 | 0.2, 2.8 | 39.8, 50.3 | Intermediate |
| Artemisia campestris | Asteraceae | 44.7, 52.8 | 1.5, 2.3 | 0.1, 0.8 | 42.5, 51.1 | Intermediate |
| Centaurea jacea | Asteraceae | 46.0, 57.3 | 1.6, 2.5 | 0.1, 0.8 | 44.0, 55.4 | Small |
| Centaurea pseudophrygia | Asteraceae | 37.5, 51.9 | 1.5, 2.5 | 0.08, 0.1 | 35.3, 49.6 | Very small |
| Hypericum maculatum | Hypericaceae | 10.2, 14.9 | 3.3, 4.1 | 4.8, 8.8 | 1.8, 2.5 | Very small |
| Hypericum perforatum | Hypericaceae | 7.4, 10.7 | 2.3, 4.1 | 2.3, 5.4 | 1.6, 2.1 | Very small |
| Inula britannnica | Asteraceae | 50.5, 61.0 | 1.5, 3.4 | 0.2, 0.3 | 48.7, 59.2 | Small |
| Inula salicina | Asteraceae | 42.9, 54.3 | 1.0, 2.1 | 0.3, 0.4 | 41.3, 52.7 | Small |
| Pilosella lactucella | Asteraceae | 39.0, 43.2 | 2.1, 2.5 | 0.1, 0.7 | 36.6, 40.5 | Small |
| Pilosella officinarum | Asteraceae | 31.0, 44.1 | 2.2, 3.7 | 0.1, 0.3 | 28.5, 41.0 | Small |
| Plantago maritima | Plantaginaceae | 41.1, 50.1 | 4.3, 5.5 | 2.0, 4.1 | 33.9, 42.8 | Very small |
| Plantago media | Plantaginaceae | 39.8, 49.1 | 3.8, 8.7 | 3.1, 10.8 | 27.2, 39.7 | Small |
| Senecio erraticus | Asteraceae | 53.6, 71.1 | 2.1, 4.3 | 0.02, 0.1 | 51.4, 68.6 | Small |
| Senecio jacobaea | Asteraceae | 62.5, 72.7 | 3.1, 5.3 | 0.04, 0.8 | 58.8, 68.8 | Intermediate |
| Silene vulgaris | Caryophyllaceae | 8.2, 20.2 | 1.0, 3.6 | 0.02, 0.8 | 6.7, 15.9 | Very small |
| Trifolium pratense | Fabaceae | 29.4, 38.3 | 1.6, 2.9 | 27.1, 34.9 | 0.7, 1.2 | Very small |
| Trifolium repens | Fabaceae | 10.6, 28.3 | 1.7, 3.1 | 7.8, 24.0 | 1.1, 1.4 | Very small |
Seeds were sown separately by species on sterilized wet sand in Petri dishes and were kept under wet–cold stratification in a refrigerator (dark, 3 °C) in March 2019. After 1 month of stratification, the Petri dishes were transferred to a growth chamber (day: 23 °C for 15 h, night: 16 °C for 9 h). In mid-April 2019, 5-d-old seedlings were transplanted from the Petri dishes to 2.2-L pots filled with sand and garden loam substrate at a 3:2 volume ratio. Immediately after transplantation, the pots were placed in the unheated glasshouse without artificial light at the Institute of Botany, Třeboň, Czech Republic. A standard liquid NPK nutrition solution (0.5/0.1/0.07 g N, P, K per litre of substrate; KristalonTM by AGRO CS a.s., Říkov, Czech Republic) was added every 3 weeks, and the plants were watered with tap water throughout the experiment. Plants were harvested between the end of September and the beginning of October 2019. Above- and below-ground biomass were separated, dried and weighed. Plants in three pots died during the experiment (one of Pilosella officinarum and two of Pilosella lactucella). Genome size values for the species growing in the experiment were also obtained from Šmarda et al. (2019).
Species congeners
Nine congeneric pairs and one standalone species, Silene vulgaris, were used in the pot experiment (Table 1). The production of certain carbohydrates and their accumulation is often associated with phylogeny (Hendry, 1987; Van den Ende, 2013; Lubbe et al., 2021b) and it might be necessary to account for evolutionary history when relating carbohydrate storage to other plant traits. However, comparative phylogenetic methods typically require one value per species (e.g. average). Because of high genetic, environmental or temporal variability of carbohydrate storage within species (Chapin et al., 1990; Hartmann and Trumbore, 2016; Blumstein et al., 2022), average species values might be less representative. To overcome this impediment, we used pairs of congeneric species and thus also included intraspecific variability in carbohydrate storage.
Carbohydrate measurements
To measure root non-structural carbohydrates, we followed the methodology described in further detail in Klimešová et al. (2019) and Martínková et al. (2023). After washing off the substrate, the samples were placed in cryovials, deep-frozen in liquid nitrogen and placed a freezer at −80 °C. After several days, samples were lyophilized and ground by an oscillating mill (Retsch MM 400). Approximately 100 mg of the sample was extracted for 12 min in boiling ethanol (80:20, v/v) and centrifuged for 10 min at 3000 r.p.m. (repeated three times). The supernatant was dried, redissolved in distilled water and filtered through a nylon microfilter (0.45 µm). The ethanol-soluble carbohydrates (mainly glucose, fructose, saccharose, raffinose and stachyose) and sugar alcohols (myo-inositol, sorbitol, mannitol) were assessed using high-performance anion exchange chromatography coupled with pulsed amperometric detection (HPAE-PAD, Dionex ICS-3000 system). For separation, we used a Dionex CarboPac PA1 column (4 × 250 mm, 10 µm) and guard column CarboPac PA1 (4 × 50 mm, 10 µm) with gradient elution composed of 16 mm and 200 mm NaOH.
Starch content was determined using the remaining sediment from ethanol-soluble carbohydrate extractions. We followed the total starch assay procedure AOAC (Association of Official Agriculture Chemists) Method 996.11 and AACC (American Association of Cereal Chemists) Method 76-13.01 developed by Megazyme Ltd (www.megazyme.com). Starch in the sediment was hydrolysed using thermostable α-amylase at 100 °C to maltodextrins, which were further hydrolysed by amyloglucosidase at 50 °C to glucose. Glucose was finally coloured using a glucose oxidase/peroxidase (GOPOD) reagent and the amount of glucose was measured by spectrophotometry at 510 nm (Shimadzu UV-1800 spectrophotometer).
For fructan determination we used the fructan assay procedure based on AOAC Method 999.03 and AACC Method 32-32.01 (www.megazyme.com). Approximately 100 mg of the sample (below-ground biomass) was extracted with boiling water. For the removal of sucrose, starch and reducing sugars, specific enzymes were applied, and alkaline borohydride was added. Fructo-oligosaccharides (FOS), fructans and reduced FOS were hydrolysed by exo- and endo-inulinase and endo-levanase to glucose and fructose. Released fructose and glucose were finally coloured using PAHBAH (p-hydroxybenzoic acid hydrazide) reagent. The amount of fructose and glucose was measured by spectrophotometry at 410 nm.
All non-structural carbohydrates in this study were expressed as percentage of dry root biomass. Carbohydrates were grouped into four categories based on their size or functional properties: (1) total non-structural carbohydrates (TNC, the sum of all carbohydrates measured); (2) mono- and disaccharides (higher quantity sugars: glucose, fructose and saccharose), i.e. carbohydrates often associated with a transport function, (3) starch (storage polysaccharide); and (4) water-soluble reserves, consisting of fructans (dominant carbohydrate of this group), RFOs (raffinose, stachyose and galactose), sugar alcohols (myo-inositol, sorbitol and mannitol), and small but lower-quantity sugars (melibiose and trehalose). We estimated carbohydrate concentration and pools (dry biomass × concentration) in roots as two aspects of carbohydrate storage (Klimešová et al., 2019).
Statistical analysis
To assess whether the probability of RS ability is related to genome size in the field survey of herbaceous species, we used a binomial generalized linear model (GLM) with a logit link function. RS ability was considered as a binary response variable (0/1), and genome size (both 1C and 1Cx values) as an explanatory continuous variable that was also log10-transformed. To compare the binomial GLM with a phylogenetically informed alternative, we simultaneously performed phylogenetic logistic regression using penalized log likelihood with Firth’s penalty as the estimation method (Ho and Ané, 2014). Fits of both models were compared based on Akaike’s information criterion (AIC). For this analysis, the dataset included 172 species (Supplementary Data Fig. S1).
For the glasshouse experiment, we tested the relationships between carbohydrate concentrations and pools, and genome size (1C and 1Cx value) using generalized least squares (GLS) models. Models were fitted using restricted maximum likelihood (REML). We used the compound symmetry correlation structure to account for autocorrelation within species measurements. We tested the significance of the effects of treatment, genome size, RS ability and their interactions. Because the relationships appeared to be non-linear we fitted the data using either logarithmic growth (y = a + b lnx) or exponential decay [y = a(1 − b)x] formulas. The model fitting starch concentrations accounted for decreasing heteroscedasticity using the exponential variance function structure. To account for a positively skewed distribution of residuals, heteroscedasticity and potential non-linearities in carbohydrate pools, we also fitted generalized additive models (GAMs; Wood, 2017) assuming a Gamma error distribution using a thin plate regression spline as a smoothing term. GAMs were fitted by REML as well. The dataset consisted of 111 harvested plants from 19 species (nine pairs of congeners and one standalone species, Silene vulgaris, Table 1).
To assess the importance of genome size as a predictor of carbohydrate concentrations and pools, we conducted commonality analysis (Ray-Mukherjee et al., 2014) and hierarchical partitioning (Chevan and Sutherland, 1991), which have been merged recently into a single quantitative framework (Lai et al., 2022). The aim of this analysis was to differentiate the effect of genome size from the general effect of species because genome size is a species trait and both predictors were essentially multicollinear in this case. Model coefficients only consider unique contributions of predictors, which might be incorrectly interpreted when strong multicollinearity is involved. By contrasting unique and common fractions of variation, the model interpretability can be improved (Lai et al., 2022). In the context of this study, the unique contribution of genome size to predict carbohydrate concentrations and pools is expected to be zero (because it is fully masked by species), whereas the contribution common to genome size and species is expected to reflect the variation in carbohydrate concentrations and pools attributed to genome size.
To examine the relationship between carbohydrate storage (concentrations and pools) and genome size at a small phylogenetic scale, we conducted a comparison between congeners. Standalone Silene vulgaris (Table 1) was omitted from this congener comparison. To test whether the congener with a larger genome has higher carbohydrate concentrations and pools, we used multivariate linear models (Wang et al., 2012). Significance testing was based on a likelihood ratio test and 999 permutations. Univariate P-values were adjusted for multiple testing, and ridge regularization (Warton, 2008) was used to account for correlations between carbohydrate concentrations and pools (response variables).
We made all analyses and graphics in R 4.2.1 (R Core Team 2022) using the functions: gls (nlme 3.1-157; Pinheiro et al., 2022) to fit GLS models, nls (nlme 3.1-157; Pinheiro et al., 2022) to fit exponential decay models, gam (mgcv 1.8-40; Wood, 2017) to fit GAMs, rdacca.hp (rdacca.hp 1.0-8; Lai et al., 2022) to conduct hierarchical partitioning, manylm and summary.manylm (mvabund 4.2.1; Wang et al., 2012) to fit and test multivariate linear models, and phyloglm (phylolm 2.6.2; Ho and Ané, 2014) to fit phylogenetic logistic regression. The species phylogenies for both datasets (the field survey and the glasshouse experiment) were obtained using the V.PhyloMaker2 package (Jin and Qian, 2022). Phylogenetic visualization was done with the ggtree 3.4.2 package (Yu et al., 2017) and some figures were drawn in ggplot2 3.3.6 (Wickham, 2016). All data is available in the Mendeley data depository (Bitomský et al., 2023).
RESULTS
For the field survey, we found a slight tendency of the probability of RS ability to decrease with increasing genome size (Fig. 1). For 1C value (Fig. 1A), the results of the binomial GLM (b = −1.0009, s.e. = 0.45, P = 0.025) and phylogenetic logistic regression (b = −1.1229, s.e. = 0.50, P = 0.026; both b coefficients on a logit scale) were relatively similar, but the phylogenetic model showed a better fit (an improvement of AIC by four units). For 1Cx value (Fig. 1B), the slopes were slightly steeper and also significant in both binomial GLM (b = −1.0535, s.e. = 0.40, P = 0.009) and phylogenetic logistic regression (b = −1.1345, s.e. = 0.46, P = 0.014). The binomial GLM and phylogenetic model showed similar fit accuracy as their AICs differed only by 1.47 units. Importantly, the goodness of fit of genome size was quite low: McFadden pseudo-R2 was 2.7 % for 1C value and 3.7 % for 1Cx value.
Fig. 1.
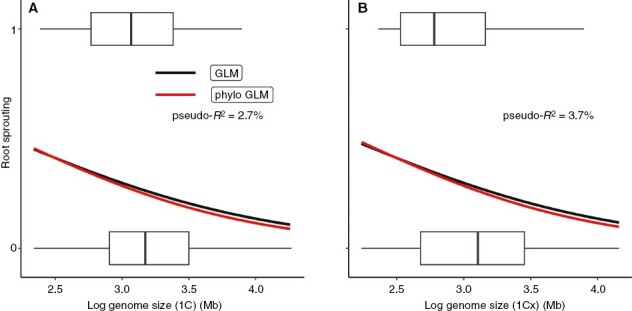
Probability of root sprouting ability plotted against species genome size: (A) 1C value and (B) 1Cx value. Predicted values for both the binomial generalized linear model (GLM) and phylogenetic logistic regression are shown. Boxplots indicate the distributions of genome size in non-root sprouters (0) and root sprouters (1). Models and significance (n = 172 in all cases): GLM for 1C: y = 2.1446 − 1.0009x (P = 0.025), phylo GLM for 1C: y = 2.44749 − 1.12289x (P = 0.026), GLM for 1Cx: y = 2.1462 − 1.0535x (P = 0.009) and phylo GLM for 1Cx: y = 2.29966 − 1.13452x (P = 0.014). Note that all coefficients are on a logit scale and that genome size was log-transformed; pseudo-R2 = goodness of fit.
For the glasshouse experiment, the concentrations of TNC and water-soluble reserves were positively and non-linearly related to genome size (Fig. 2). The interaction between root sprouting ability and genome size was not significant for all carbohydrate groups (F ≤ 0.4 and lower, P ≥ 0.523), and therefore the relationships between TNC, water-soluble reserves and genome size were consistent in both root sprouters and non-root sprouters (Fig. 2). Models based on both 1C (Table 2) and 1Cx values (Supplementary Data Table S1) showed similar results. For TNC and water-soluble reserves, the rate of increase with increasing genome size was relatively high in small-genome species but lower for species with larger genomes (Fig. 2A, D; Table S2). For mono- and disaccharides, we did not observe any relationship between them and genome size in any treatment (Table 2b; Fig. 2B). Starch concentrations were relatively high for only a few samples of some small-genome species (both Trifolium species or Plantago media, Table 1) and then exponentially decreased towards species with larger genomes (Fig. 2C; Table S2). However, starch concentrations in these starch-rich species were highly variable and the coefficient of genome size was not significant when accounting for decreasing heteroscedasticity in residuals (Table 2c). As expected, the fraction of the variation in carbohydrate concentrations explained exclusively by genome size (its unique contribution) was zero for all carbohydrates. In contrast, unique contributions common to species and genome size were 0.43 for TNC, 0.05 for mono- and disaccharides, 0.30 for starch, and 0.55 for water-soluble reserves.
Fig. 2.
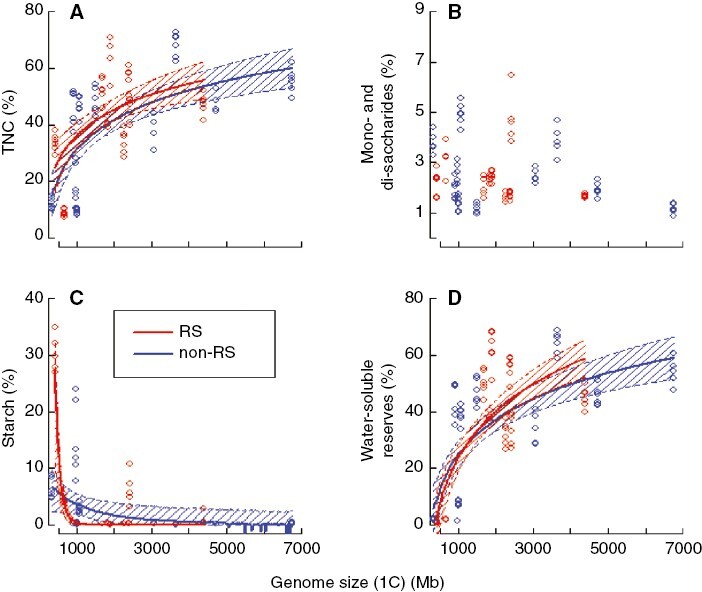
Carbohydrate concentrations plotted against species genome size (1C value). Fitted models and their 95 % confidence intervals for root (RS) and non-root (non-RS) sprouters are indicated (n = 111). (A) Total non-structural carbohydrates (TNC), (B) mono- and disaccharides (glucose, fructose and saccharose), (C) starch and (D) water-soluble reserves, i.e. mainly fructans + raffinose family oligosaccharides (RFOs) and sugar alcohols. Estimated models (merged for both non-root and root sprouters): TNC = −64.8 + 14.2 ln(GS), Starch = 35.3 (1 − 0.002)GS, WSR = −112.4 + 19.8 ln(GS), where ln = natural logarithm, GS = genome size and WSR = water-soluble reserves. Fitted lines are not plotted for the relationships that were not significant (P > 0.05); all significant relationships displayed P < 0.001.
Table 2.
Significance testing of the relationships between carbohydrate concentrations and species holoploid genome size (GS, 1C value). Generalized least squares models were used (d.f. = 4/107 in all cases). For each model term, F- and P-values are indicated. For each model, Cox and Snell pseudo-coefficient of determination (R2) and estimated compound symmetry correlation (Rho) for within-species measurements are indicated. (a) Total non-structural carbohydrates (TNC), (b) mono- and disaccharides (glucose, fructose and saccharose), (c) starch and (d) water-soluble reserves, i.e. mainly fructans + raffinose family oligosaccharides (RFOs) and sugar alcohols.
| Carbohydrate concentration | F-value | P-value | R 2 | Rho |
|---|---|---|---|---|
| (a) TNC | 0.25 | 0.92 | ||
| Sprouting ability | 0.4 | 0.552 | ||
| GS | 11.3 | 0.001 | ||
| Sprouting × GS | 0.04 | 0.847 | ||
| (b) Mono- and disaccharides | 0.08 | 0.89 | ||
| Sprouting ability | 0.02 | 0.893 | ||
| GS | 1.0 | 0.320 | ||
| Sprouting × GS | 0.1 | 0.740 | ||
| (c) Starch | 0.02 | 0.85 | ||
| Sprouting ability | 0.02 | 0.886 | ||
| GS | 2.7 | 0.105 | ||
| Sprouting × GS | 0.05 | 0.828 | ||
| (d) Water-soluble reserves | 0.28 | 0.94 | ||
| Sprouting ability | 0.1 | 0.782 | ||
| GS | 19.5 | <0.001 | ||
| Sprouting × GS | 0.4 | 0.523 |
In contrast to concentrations, carbohydrate pools had no link to genome size (Supplementary Data Fig. S2). Although linear models suggested significant relationships of the pools of TNC and water-soluble reserves to genome size, these models fitted the data poorly and did not pass the model diagnostics. GAMs fitted the data better but did not suggest any clear patterns between genome size and carbohydrate pools.
Carbohydrate storage and genome size were generally associated with phylogeny in our 20 species (Table 1; Fig. 3). In five out of nine congeneric pairs (Achillea, Artemisia, Pilosella, Senecio and Trifolium) the non-root sprouting congener had a larger genome. In four congeneric pairs, the congener with the larger genome also had higher concentrations of TNC or water-soluble reserves than the small-genome congener (Fig. 4). Interestingly, we found a contradictory pattern, i.e. higher TNC concentrations in a small-genome congener, in Trifolium (Fig. 4). In Achillea, TNC concentrations were also lower in the large-genome congener but the concentrations of starch and water-soluble reserves increased (Fig. 4). Finally, Inula was the only genus for which we observed a difference in carbohydrate pool between congeners; the large-genome congener stored more mono- and disaccharides (Fig. 4).
Fig. 3.
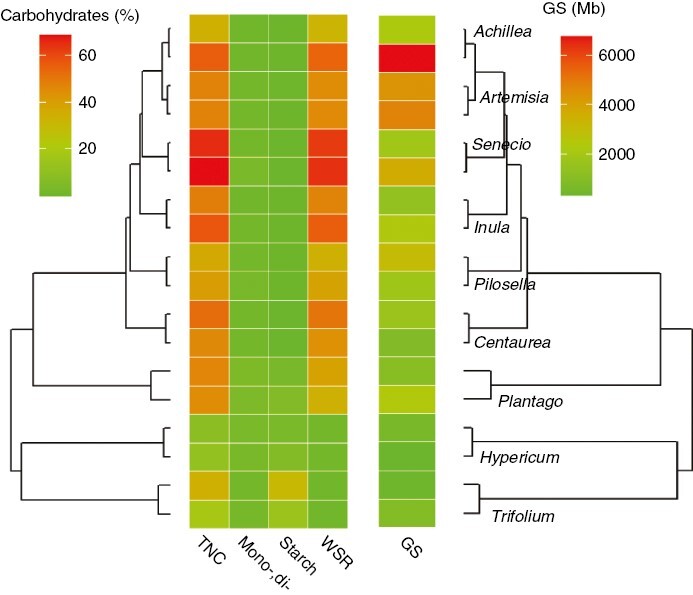
Carbohydrate concentrations (average species values) and genome size (1C value) mapped on the species phylogeny. Average carbohydrate pools were highly correlated with concentrations at the species level (r > 0.82 in all cases), and therefore they are not shown to reduce overplotting. TNC = total non-structural carbohydrates, mono-, di- = mono- and disaccharides (glucose, fructose and saccharose), WSR = water-soluble reserves, i.e. mainly fructans + raffinose family oligosaccharides (RFOs) and sugar alcohols, and GS = genome size.
Fig. 4.
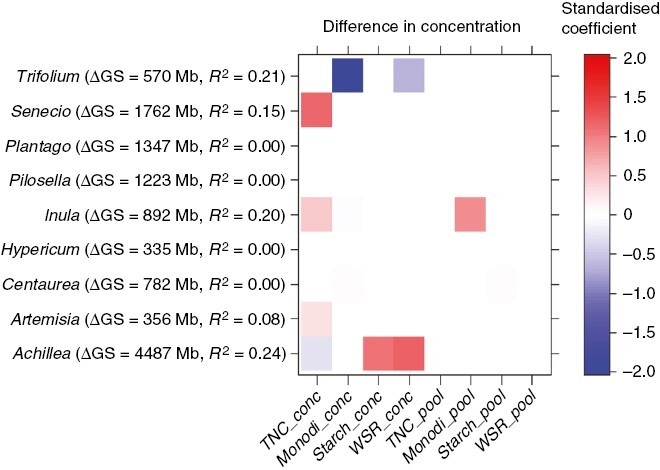
Heatmap showing differences in carbohydrate concentrations and pools between pairs of congeners. Standardized coefficients indicate significant differences (based on multivariate linear models, P < 0.05) of the congener with larger genome from the congener with smaller genome. ∆GS = genome size difference between congeners (Mb), R2 = Hooper’s coefficient of determination of the multivariate linear model, TNC = total non-structural carbohydrates, Monodi = mono- and disaccharides (glucose, fructose and saccharose), WSR = water-soluble reserves, i.e. mainly fructans + raffinose family oligosaccharides (RFOs) and sugar alcohols.
DISCUSSION
Here, we report two major results: (1) a weak negative relationship between the probability of root sprouting ability and genome size in a set of temperate herbs and (2) a positive and non-linear relationship between genome size and the concentrations of TNC and water-soluble reserves (predominantly fructans) accumulated at the end of the growing season in a set of congeneric herbs, independently of species sprouting strategy. This link seems to be stronger at larger phylogenetic scales because at the genus level we only observed higher carbohydrate concentrations in species with larger genomes in four out of nine congeneric pairs. Interestingly, carbohydrate pools varied independently of genome size.
Root sprouting ability and genome size
In the field survey, we observed that large-genome species were less likely to be able to resprout from roots. This suggests that genome size might be another constraint limiting the prevalence of RS ability in plants besides phytohormonal balance (Martínková et al., 2023). Root sprouters might take advantage of their smaller genomes to complete their cell cycles faster in order to quickly regenerate after biomass removal. Slightly smaller genome size in root sprouters could also explain their higher occurrence in dry and open habitats (Bartušková et al., 2021). Small-genome species have more efficient control of water balance thanks to smaller and denser stomata (Beaulieu et al., 2008; Faizullah et al., 2021). However, the goodness of fit of genome size was very low (≤3.7 %), and thus these results do not allow us to draw strong conclusions; and potential connections between genome traits (size, base content, etc.) and sprouting ability clearly require further research.
Carbohydrate storage and genome size
Our results from the pot experiment are in accord with the observations of Hendry (1987) who found a positive linear correlation with shoot fructan concentrations (measured in February) and 2C DNA content in a set of 15 temperate herbs. Nevertheless, a logarithmic growth model better explained our data, because the rate of increase of TNC and fructan concentrations with increasing genome size was not constant but was slower in species with larger genomes. This phenomenon can be attributed to the storage capacity of root cells (the highest TNC concentration in our dataset was 72.7 %). Clearly, there is a limit in carbohydrate storage potential for root cells because there must be adequate space to accommodate other organelles within these cells. It is worth noting that there is a rather indirect link between carbohydrate storage and genome size. The genome–cell–vacuole size scaling (Owens and Poole, 1979; Bennett, 1987; Beaulieu et al., 2008; Šímová and Herben, 2012; Greilhuber and Leitch, 2013) can be a driving mechanism of a relatively high accumulation of carbohydrates stored in the vacuole in species with large genomes. In terms of carbon source–sink interactions, large root cells are expected to have better carbon sink efficiency (White et al., 2016), while higher fructan concentrations could be needed for the osmoregulation of large vacuoles in these large cells (Hendry, 1987).
Pairwise congeneric comparisons from the pot experiment revealed that large-genome species had higher TNC and fructan concentrations in their roots than small-genome species, but this was only confirmed in Senecio, Inula, Artemisia and Achillea (all from the fructan-storing family Asteraceae). This suggests that a positive relationship between genome size and carbohydrate concentrations can be sometimes observed even at small phylogenetic scales but its consistency needs to be further studied. There was a contradictory pattern in Trifolium where the congener with the larger genome had lower concentrations of mono- and disaccharides, RFOs, and sugar alcohols. Species from this genus and family (Fabaceae) primarily store starch, but other genera from this family can have very high content of water-soluble carbohydrates, especially in cold climates (Chlumská et al., 2022). We may expect a stronger relationship between genome size and carbohydrate concentrations at larger rather than smaller phylogenetic scales.
Because the production of starch vs. fructans as storage carbohydrates is often associated with certain phylogenetic groups (Hendry, 1987; Vijn and Smeekens, 1999; Lubbe et al., 2021b), it is also important to consider possible phylogenetic effects at various phylogenetic scales. Taxon sampling of our pot experiment primarily enabled us to address small phylogenetic scales (especially at the genus level), yet most of our species belong to the family Asteraceae and it is difficult to further extrapolate regarding the trends for starch, RFOs or even fructans across other fructan-storing families. Plants probably store fructans exclusively in the vacuole (Vijn and Smeekens, 1999) but starch is typically stored in amyloplasts. It is therefore not clear whether the genome–cell–vacuole size scaling rationale to interpret links between carbohydrate storage and genome size can be applied to starch as well. Starch granule morphology can further alter responses under physiological stresses, such as drought (Almeida et al., 2021). Starch-producing species, such as Hypericum and Trifolium, had smaller genomes within our dataset, but greater taxon sampling is needed to examine whether the production of starch or fructans is associated with genome size.
In contrast to concentrations, carbohydrate pools were independent of genome size in the pot experiment. A potential explanation for this is that genome size may affect the storing capacity in roots primarily at the cellular level but its influence does not scale-up to the organ level. Such a scenario occurs when two species differing in genome size and minimum cell size produce the same amount of below-ground biomass. Because fewer large cells are needed for a certain amount of biomass than small cells, higher carbohydrate concentrations in large cells may be effectively blurred when converted to pools (per storage organ). Plants with larger genomes and larger storage of carbohydrates therefore do not profit from having larger overall carbohydrate storage for regrowth after disturbance or seasonal dormancy, suggesting that carbohydrates in large-celled plants could more probably be involved in stress tolerance.
Ecological role of carbohydrates and genome size
To explain the link between high genome size and high carbohydrate concentrations in some plants, the role of stress tolerance, osmoregulation or phenology is often considered (Hendry, 1987; Brocklebank and Hendry, 1989). To assess the role of stress, Cseri et al. (2020) compared salt tolerance of tetraploids and diploids of three Salix species and found that one tetraploid variant accumulated more starch in leaves and coincidently exhibited higher photosynthetic assimilation rates under salt stress than diploids. Nevertheless, our dataset consisted mainly of Asteraceae species with fructans as the primary storage carbohydrate. The ecological significance of water-soluble fructans is often attributed to cold resistance (Hendry, 1987; Brocklebank and Hendry, 1989; Martinez-Vilalta et al., 2016) and a higher fructan concentration is associated with the potential reduction or avoidance of tissue frost damage (Livingston et al., 2009) including within the below-ground stems and buds of plants (Livingston et al., 2006). Similarly to a high fructan concentration, large genome size is often found in species with early spring growth and strong ability to survive lower temperatures (Grime and Mowforth, 1982; Knight et al., 2005). However, the evidence for a relationship between genome size and temperature resistance is often inconsistent (Knight et al., 2005; Sklenář et al., 2022), and thus integrating both carbohydrate storage and genome size into a single framework might be needed to overcome the obscuring effects of unmeasured variables.
Early spring growth at low temperatures can be facilitated by large genome size and the associated larger cell size thanks to cell expansion driven by turgor pressure and not cell division (Grime and Mowforth, 1982; Hendry, 1987). The importance of osmoregulation was shown for the early spring geophyte Lachenalia minima (Orthen, 2001). This species uses both fructans and starch, but starch reserves are primarily broken down and used for sprouting whereas total fructan content within the bulb did not change while the concentration increased within the innermost leaves, probably to facilitate osmoregulation and cell expansion (Orthen, 2001). A similar trend may occur within Galanthus nivalis although there is greater evidence that these plants also use fructans as a cryoprotectant, thus taking advantage of the dual roles of this carbohydrate (Orthen and Wehrmeyer, 2004). In contrast to the unpredictable disturbances that root sprouters survive, both species above are responding to the regular pattern of spring regrowth for which they prepare before winter.
Although many fructan-storing species begin growth in early spring, many others have very long growing seasons and can experience hot and dry conditions that are also eased by fructans and their ability to serve in osmoregulation (Dias-Tagliacozzo et al., 2004; de Moraes et al., 2016; Lubbe et al., 2021b). Plants with small genomes are better in coping with drought stress thanks to smaller and denser stomata (Beaulieu et al., 2008; Faizullah et al., 2021), and even when they store starch, they are able to cope with drought stress (Lubbe et al., 2021a). Whether greater storage of starch in small genome species that can regenerate from roots is used preferentially for resprouting will need further study.
CONCLUSIONS
We observed a tendency of large-genome herbaceous species to store high concentrations of water-soluble carbohydrates and a weak link between genome size and root sprouting ability. We speculate that genome–cell size scaling, the association of both genome size and carbohydrate concentrations with plant strategies (phenology), stress tolerance (cold, drought), water-use efficiency or metabolism (photosynthesis, respiration, growth rate) can explain why genome size and carbohydrate storage need to be coordinated. To fully understand the role of genome and cell size in plant carbon dynamics, further research needs to include various time scales of carbohydrate storage (daily, weekly and seasonal) and concomitantly quantify several sinks of non-structural carbohydrates, such as growth, respiration, defence, reproduction, mycorrhizal symbionts or storage per se.
SUPPLEMENTARY DATA
Supplementary data are available at Annals of Botany online and consist of the following.
Figure S1: Phylogeny of 172 Central European herbaceous species. Figure S2: Carbohydrate pools plotted against species genome size.
ACKNOWLEDGEMENTS
We thank Šárka Haumerová, Lenka Leštinová and Michael Bartoš for technical assistance with the experiment. M.B., J.M. and J.K. conceived the ideas. J.M. and J.K. designed the study. M.B. analysed the data and led the writing of the manuscript. F.C.L. cowrote the manuscript. J.M. and I.M. collected the data. All authors discussed the results, contributed critically to the drafts and gave final approval for publication.
Contributor Information
Martin Bitomský, Institute of Botany of the Czech Academy of Sciences, Dukelská 135, 379 01 Třeboň, Czech Republic.
Jana Martínková, Institute of Botany of the Czech Academy of Sciences, Dukelská 135, 379 01 Třeboň, Czech Republic.
F Curtis Lubbe, Institute of Botany of the Czech Academy of Sciences, Dukelská 135, 379 01 Třeboň, Czech Republic.
Iveta Marešová, Institute of Botany of the Czech Academy of Sciences, Dukelská 135, 379 01 Třeboň, Czech Republic.
Jitka Klimešová, Institute of Botany of the Czech Academy of Sciences, Dukelská 135, 379 01 Třeboň, Czech Republic; Department of Botany, Charles University, Benátská 2, 128 01 Prague, Czech Republic.
FUNDING
This research was supported by the Czech Science Foundation GAČR [nos. 22-10897S and 19-13103S], Premium Academiae awarded by the Czech Academy of Sciences to J.K., and long-term research development project of the Czech Academy of Sciences (RVO 67985939). M.B. was supported by the Programme for the Promotion of Prospective Human Resources – Postdocs (PPLZ) of the Czech Academy of Sciences.
LITERATURE CITED
- Almeida VO, Di-Medeiros MC, Batista KA, Moraes MG, Fernandes KF.. 2021. Morphological and physicochemical characterization of starches from underground stems of Trimezia juncifolia collected in different phenological stages. International Journal of Biological Macromolecules 166: 127–137. doi: 10.1016/j.ijbiomac.2020.10.109. [DOI] [PubMed] [Google Scholar]
- Bartušková A, Filartiga AL, Herben T, Qian J, Klimešová J.. 2021. Comparative analysis of root sprouting and its vigour in temperate herbs: anatomical correlates and environmental predictors. Annals of Botany 127: 931–941. doi: 10.1093/aob/mcab030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA.. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist 179: 975–986. doi: 10.1111/j.1469-8137.2008.02528.x. [DOI] [PubMed] [Google Scholar]
- Bennett MD. 1987. Variation in genomic form in plants and its ecological implications. New Phytologist 106: 177–200. doi: 10.1111/j.1469-8137.1987.tb04689.x. [DOI] [Google Scholar]
- Bennett MD, Leitch IJ, Hanson L.. 1998. DNA amounts in two samples of angiosperm weeds. Annals of Botany 82: 121–134. doi:10.1006/anbo.1998.0785 [Google Scholar]
- [dataset] Bitomský M, Martínková J, Lubbe FC, Marešová I, Klimešová J.. 2023. Genome size is strongly linked to carbohydrate storage and weakly linked to root sprouting ability in herbs. doi: 10.17632/t8zwb7dxyb.2 [DOI] [PMC free article] [PubMed]
- Blumstein M, Sala A, Weston DJ, Holbrook NM, Hopkins R.. 2022. Plant carbohydrate storage: intra- and inter-specific trade-offs reveal a major life history trait. New Phytologist 235: 2211–2222. doi: 10.1111/nph.18213. [DOI] [PubMed] [Google Scholar]
- Brocklebank KJ, Hendry GAF.. 1989. Characteristics of plant species which store different types of reserve carbohydrates. New Phytologist 112: 255–260. doi: 10.1111/j.1469-8137.1989.tb02381.x. [DOI] [Google Scholar]
- Chapin FS III, Schulze ED, Mooney HA.. 1990. The ecology and economics of storage in plants. Annual Review of Ecology and Systematics 21: 423–447. [Google Scholar]
- Chevan A, Sutherland M.. 1991. Hierarchical partitioning. American Statistician 45: 90–96. doi: 10.2307/2684366. [DOI] [Google Scholar]
- Chlumská Z, Liancourt P, Hartmann H, et al. 2022. Species- and compound-specific dynamics of nonstructural carbohydrates toward the world’s upper distribution of vascular plants. Environmental and Experimental Botany 201: 104985. doi: 10.1016/j.envexpbot.2022.104985. [DOI] [Google Scholar]
- Chytrý M, Danihelka J, Kaplan Z, et al. 2021. Pladias database of the Czech flora and vegetation. Preslia 93: 1–87. [Google Scholar]
- Cseri A, Borbély P, Poór P, et al. 2020. Increased adaptation of an energy willow cultivar to soil salinity by duplication of its genome size. Biomass and Bioenergy 140: 105655. doi: 10.1016/j.biombioe.2020.105655. [DOI] [Google Scholar]
- de Moraes MG, de Carvalho MAM, Franco AC, Pollock CJ, Figueiredo-Ribeiro RCL.. 2016. Fire and drought: soluble carbohydrate storage and survival mechanisms in herbaceous plants from the Cerrado. BioScience 66: 107–117. [Google Scholar]
- Dias-Tagliacozzo GM, Itaya NM, de Carvalho MAM, Figueiredo-Ribeiro RDL, Dietrich SMC.. 2004. Fructans and water suppression on intact and fragmented rhizophores of Vernonia herbacea. Brazilian Archives of Biology and Technology 47: 363–373. [Google Scholar]
- Faizullah L, Morton JA, Hersch-Green EI, Walczyk AM, Leitch AR, Leitch IJ.. 2021. Exploring environmental selection on genome size in angiosperms. Trends in Plant Science 26: 1039–1049. doi: 10.1016/j.tplants.2021.06.001. [DOI] [PubMed] [Google Scholar]
- Francis D, Davies MS, Barlow PW.. 2008. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Annals of Botany 101: 747–757. doi: 10.1093/aob/mcn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Leitch IJ. 2013. Genome size and the phenotype. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF. eds. Plant genome diversity, Vol. 2. Vienna: Springer, 323–344. [Google Scholar]
- Grime JP, Mowforth MA.. 1982. Variation in genome size—an ecological interpretation. Nature 299: 151–153. doi: 10.1038/299151a0. [DOI] [Google Scholar]
- Gruner A, Hoverter N, Smith T, Knight CA.. 2010. Genome size is a strong predictor of root meristem growth rate. Journal of Botany 2010: 1–4. doi: 10.1155/2010/390414. [DOI] [Google Scholar]
- Hartmann H, Trumbore S.. 2016. Understanding the roles of nonstructural carbohydrates in forest trees – from what we can measure to what we want to know. New Phytologist 211: 386–403. doi: 10.1111/nph.13955. [DOI] [PubMed] [Google Scholar]
- Hendry G. 1987. The ecological significance of fructan in a contemporary flora. New Phytologist 106: 201–216. doi: 10.1111/j.1469-8137.1987.tb04690.x. [DOI] [Google Scholar]
- Herben T, Klimešová J, Chytrý M.. 2018. Effects of disturbance frequency and severity on plant traits: An assessment across a temperate flora. Functional Ecology 32: 799–808. doi: 10.1111/1365-2435.13011. [DOI] [Google Scholar]
- Hincha DK, Zuther E, Hellwege EM, Heyer AG.. 2002. Specific effects of fructo- and gluco- oligosaccharides in the preservation of liposomes during drying. Glycobiology 12: 103–110. doi: 10.1093/glycob/12.2.103. [DOI] [PubMed] [Google Scholar]
- Ho LST, Ané C.. 2014. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Systematic Biology 63: 397–408. doi: 10.1093/sysbio/syu005. [DOI] [PubMed] [Google Scholar]
- Janeček S, Lanta V, Klimešová J, Doležal J.. 2011. Effect of abandonment and plant classification on carbohydrate reserves of meadow plants. Plant Biology 13: 243–251. doi: 10.1111/j.1438-8677.2010.00352.x. [DOI] [PubMed] [Google Scholar]
- Jensen KH, Berg-Sørensen K, Bruus H, et al. 2016. Sap flow and sugar transport in plants. Reviews of Modern Physics 88: 035007. [Google Scholar]
- Jin Y, Qian H.. 2022. VPhyloMaker2: An updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Diversity 44: 335–339. doi: 10.1016/j.pld.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimešová J, de Bello F.. 2009. CLO-PLA: the database of clonal and bud bank traits of Central European flora. Journal of Vegetation Science 20: 511–516. doi: 10.1111/j.1654-1103.2009.01050.x. [DOI] [Google Scholar]
- Klimešová J, Martínková J.. 2022. Clonal growth, resprouting and vegetative propagation of weeds. In: Upadhyaya MK, Clements DR, Shrestha A. eds. Persistence strategies of weeds. Oxford: Wiley-Blackwell, 200–218. [Google Scholar]
- Klimešová J, Martínková J, Pausas JG, et al. 2019. Handbook of standardized protocols for collecting plant modularity traits. Perspectives in Plant Ecology, Evolution and Systematics 40: 125485. doi: 10.1016/j.ppees.2019.125485. [DOI] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA.. 2005. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany 95: 177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J, Zou Y, Zhang J, Peres-Neto P.. 2022. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdaccahp R package. Methods in Ecology and Evolution 13: 782–788. doi:10.1111/2041-210X.13800 [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD.. 1998. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany 82: 85–94. doi:10.1006/anbo.1998.0783 [Google Scholar]
- Liu DD, Chao WM, Turgeon R.. 2012. Transport of sucrose, not hexose, in the phloem. Journal of Experimental Botany 63: 4315–4320. doi: 10.1093/jxb/ers127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston DP, Premakumar R, Tallury SP.. 2006. Carbohydrate partitioning between upper and lower regions of the crown in oat and rye during cold acclimation and freezing. Cryobiology 52: 200–208. doi: 10.1016/j.cryobiol.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Livingston DP, Hincha DK, Heyer AG.. 2009. Fructan and its relationship to abiotic stress tolerance in plants. Cellular and Molecular Life Sciences 66: 2007–2023. doi: 10.1007/s00018-009-0002-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbe FC, Bitomský M, Hájek T, et al. 2021a. A tale of two grasslands: how belowground storage organs coordinate their traits with water-use traits. Plant and Soil 465: 533–548. doi: 10.1007/s11104-021-05031-7. [DOI] [Google Scholar]
- Lubbe FC, Klimeš A, Doležal J, et al. 2021b. Carbohydrate storage in herbs: the forgotten functional dimension of the plant economic spectrum. Annals of Botany 127: 813–825. doi: 10.1093/aob/mcab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vilalta J, Sala A, Asensio D, et al. 2016. Dynamics of nonstructural carbohydrates in terrestrial plants: a global synthesis. Ecological Monographs 86: 495–516. doi:10.1002/ecm.1231 [Google Scholar]
- Martínková J, Klimeš A, Motyka V, et al. 2023. Why is root sprouting not more common among plants? Phytohormonal clues and ecological correlates. Environmental and Experimental Botany 205: 105147. doi: 10.1016/j.envexpbot.2022.105147. [DOI] [Google Scholar]
- Orthen B. 2001. Sprouting of the fructan- and starch-storing geophyte Lachenalia minima: effects on carbohydrate and water content within the bulbs. Physiologia Plantarum 113: 308–314. doi: 10.1034/j.1399-3054.2001.1130302.x. [DOI] [PubMed] [Google Scholar]
- Orthen B, Wehrmeyer A.. 2004. Seasonal dynamics of non-structural carbohydrates in bulbs and shoots of the geophyte Galanthus nivalis. Physiologia Plantarum 120: 529–536. doi: 10.1111/j.0031-9317.2004.0284.x. [DOI] [PubMed] [Google Scholar]
- Owens T, Poole RJ.. 1979. Regulation of cytoplasmic and vacuolar volumes by plant cells in suspension culture. Plant Physiology 64: 900–904. doi: 10.1104/pp.64.5.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton AJ, Cunningham SM, Volenec JJ, Reicher ZJ.. 2007. Differences in freeze tolerance of Zoysiagrasses: II Carbohydrate and proline accumulation. Crop Science 47: 2170–2181. doi: 10.2135/cropsci2006.12.0784. [DOI] [Google Scholar]
- Pinheiro J, Bates D, R Core Team. 2022. nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-157. https://CRAN.R-project.org/package=nlme
- R Core Team. 2022. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.r-project.org [Google Scholar]
- Ray-Mukherjee J, Nimon K, Mukherjee S, Morris DW, Slotow R, Hamer M.. 2014. Using commonality analysis in multiple regressions: a tool to decompose regression effects in the face of multicollinearity. Methods in Ecology and Evolution 5: 320–328. doi:10.1111/2041-210X.12166 [Google Scholar]
- Schnáblová R, Huang L, Klimešová J, Šmarda P, Herben T.. 2021. Inflorescence preformation prior to winter: a surprisingly widespread strategy that drives phenology of temperate perennial herbs. New Phytologist 229: 620–630. doi: 10.1111/nph.16880. [DOI] [PubMed] [Google Scholar]
- Šímová I, Herben T.. 2012. Geometrical constraints in the scaling relationships between genome size, cell size and cell cycle length in herbaceous plants. Proceedings of the Royal Society B: Biological Sciences 279: 867–875. doi: 10.1098/rspb.2011.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenář P, Ptáček J, Klimeš A.. 2022. Genome size of alpine plants does not predict temperature resistance. Planta 256: 18. doi: 10.1007/s00425-022-03935-x. [DOI] [PubMed] [Google Scholar]
- Šmarda P, Knápek O, Březinová A, et al. 2019. Genome sizes and genomic guanine+cytosine (GC) contents of the Czech vascular flora with new estimates for 1700 species. Preslia 91: 117–142. doi: 10.23855/preslia.2019.117. [DOI] [Google Scholar]
- Van den Ende W. 2013. Multifunctional fructans and raffinose family oligosaccharides. Frontiers in Plant Science 4: 247. doi: 10.3389/fpls.2013.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T.. 2012. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany 109: 65–75. doi: 10.1093/aob/mcr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijn I, Smeekens S.. 1999. Fructan: more than a reserve carbohydrate? Plant Physiology 120: 351–360. doi: 10.1104/pp.120.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Naumann U, Wright ST, Warton DI.. 2012. mvabund – an R package for model-based analysis of multivariate abundance data. Methods in Ecology and Evolution 3: 471–474. doi: 10.1111/j.2041-210x.2012.00190.x. [DOI] [Google Scholar]
- Warton DI. 2008. Penalized normal likelihood and ridge regularization of correlation and covariance matrices. Journal of the American Statistical Association 103: 340–349. doi: 10.1198/016214508000000021. [DOI] [Google Scholar]
- White AC, Rogers A, Rees M, Osborne CP.. 2016. How can we make plants grow faster? A source–sink perspective on growth rate. Journal of Experimental Botany 67: 31–45. doi: 10.1093/jxb/erv447. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: Elegant graphics for data analysis. New York: Springer. [Google Scholar]
- Wood SN 2017. Generalized additive models: an introduction with R. New York: Chapman & Hall/CRC. [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TT.. 2017. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution 8: 28–36. doi: 10.1111/2041-210x.12628. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


