Abstract
Background and Aims
Cactaceae are succulent plants, quasi-endemic to the American continent, and one of the most endangered plant groups in the world. Molecular phylogenies have been key to unravelling phylogenetic relationships among major cactus groups, previously hampered by high levels of morphological convergence. Phylogenetic studies using plastid markers have not provided adequate resolution for determining generic relationships within cactus groups. This is the case for the tribe Cereeae s.l., a highly diverse group from tropical America. Here we aimed to reconstruct a well-resolved phylogenetic tree of tribe Cereeae and update the circumscription of suprageneric and generic groups in this tribe.
Methods
We integrated sequence data from public gene and genomic databases with new target sequences (generated using the customized Cactaceae591 probe set) across representatives of this tribe, with a denser taxon sampling of the subtribe Cereinae. We inferred concatenated and coalescent phylogenetic trees and compared the performance of both approaches.
Key Results
Six well-supported suprageneric clades were identified using different datasets. However, only genomic datasets, especially the Cactaceae591, were able to resolve the contentious relationships within the subtribe Cereinae.
Conclusions
We propose a new taxonomic classification within Cereeae based on well-resolved clades, including new subtribes (Aylosterinae subtr. nov., Uebelmanniinae subtr. nov. and Gymnocalyciinae subtr. nov.) and revised subtribes (Trichocereinae, Rebutiinae and Cereinae). We emphasize the importance of using genomic datasets allied with coalescent inference to investigate evolutionary patterns within the tribe Cereeae.
Keywords: Cactaceae, succulents, systematics, phylogenomics, Angiosperm353, Cactaceae591, target sequencing
INTRODUCTION
Cactus species are mostly succulent, popular plants found primarily in dry and arid regions in the Neotropics. This family is one of the most diverse groups of succulent plants in the Caryophyllales (Hernández-Ledesma et al., 2015), and one of the most endangered plant families in the world (Goettsch et al., 2015; Amaral et al., 2022; Pillet et al., 2022). Over the last two decades, molecular phylogenetics have been fundamental to elucidating phylogenetic relationships among major lineages of this family (Nyffeler, 2002; Bárcenas et al., 2011; Hernández-Hernández et al., 2011), previously hindered by the high level of morphological convergence. Traditionally, this family has been subdivided into four subfamilies, namely ‘Pereskioideae’ (possibly non-monophyletic; Edwards et al., 2005; Walker et al., 2018), Maihuenioideae, Opuntioideae and Cactoideae (Guerrero et al., 2019), the last exhibiting the greatest species diversity and morphological variation (Anderson, 2001; Applequist and Wallace, 2002; Hunt et al., 2006; Nyffeler and Eggli, 2010; Hernández-Hernández et al., 2011). Regardless of these efforts, the delimitation of suprageneric lineages within the subfamily Cactoideae has long been a subject of debate, particularly for the core Cactoideae II (Guerrero et al., 2019), a group of tropical American cacti with unclear phylogenetic placement.
The main systematic controversy within the core Cactoideae II is a well-supported clade composed of globose and columnar cacti from the former tribes Browningieae, Cereeae and Trichocereeae (also known as the BCT clade, Nyffeler, 2002), which exhibited a recent radiation in the late Miocene (~5 Mya; Arakaki et al., 2011; Hernández-Hernández et al., 2014). The relationships among these tribes remained unknown due to the non-monophyly at the tribal and generic levels (Nyffeler, 2002; Ritz et al., 2007, 2016; Hernández-Hernández et al., 2011; Schlumpberger and Renner, 2012; Franck et al., 2013; Lendel, 2013; Calvente et al., 2017; Franco et al., 2017; Fantinati et al., 2021; Romeiro-Brito et al., 2022). The most recent systematic classification proposed for the family broadened the circumscription of the tribe Cereeae, including all representatives from clade BCT (tribe Cereeae s.l.), and divided it into three subtribes: Cereinae, Trichocereinae and Rebutiinae (Fig. 1; Nyffeler and Eggli, 2010). Despite this taxonomic rearrangement, non-monophyletic groups continued to exist in the tribe Cereeae s.l. (Schlumpberger and Renner, 2012; Romeiro-Brito et al., 2022).
Fig. 1.

Representatives of subtribe Rebutiinae (A, C, G, K), Trichocereinae (B1, B2, D1, D2, E, F) and Cereinae (H, I, J, L, M), sensuNyffeler and Eggli (2010), showing different growth forms. (A) Rebutia minuscula K.Schum, (B1 and B2) Harrisia adscendens (Gürke) Britton & Rose, (C) Uebelmannia pectinifera Buining, (D1 and D2) Arthrocereus rondonianus Backeb. & Voll, (E) Echinopsis sp., (F) Cleistocactus winteri D.R.Hunt, (G) Aylostera fiebrigii (Gürke) Backeb., (H) Melocactus glaucescens Buining & Brederoo, (I) Discocactus bahiensis Britton & Rose, (J) Cereus jamacaru DC., (K) Gymnocalycium denudatum (Link & Otto) Pfeiff. ex Mittler, (L) Micranthocereus auriazureus Buining & Brederoo and (M) Facheiroa squamosa (Gürke) P.J.Braun & Esteves. Photo credits: A and G: M. Lowry; B, D, H, I, L and M: M. C. Telhe; C: E. M. Moraes; E and K: M. Kohler; F and J: M. Romeiro-Brito.
Few studies have investigated phylogenetic relationships using morphological or molecular data at the suprageneric level in the tribe Cereeae s.l. (Taylor and Zappi, 1989; Soffiatti, 2003; Lendel, 2013; Fantinati et al., 2021). The circumscription of the subtribes within Cereeae lacks apomorphic characteristics capable of distinguishing them, particularly between the subtribes Trichocereinae and Cereinae (treated at tribal level in Taylor and Zappi, 2004). While the subtribe Trichocereinae traditionally included taxa with flowers and fruits with trichomes, bristles and scales, the subtribe Cereinae is characterized by species with naked flowers (or with minute scales; Barthlott and Hunt, 1993). Thought to be the result of convergent phenotypic evolution, these reproductive morphological characters have not been sufficient for delimiting subtribes and genera in either group (Taylor and Zappi, 1989; Schlumpberger and Renner, 2012; Franck et al., 2013). Moreover, molecular systematic studies in this group have shown low phylogenetic support to delineate groups at generic levels (Lendel, 2013; Fantinati et al., 2021). The lack of phylogenetic resolution might be due to the recent and rapid diversification of this group during the late Miocene (Hernández-Hernández et al., 2014), but also insufficient phylogenetic signal when using only a few traditional molecular markers.
The use of next-generation sequencing has been emerging as an alternative to traditional molecular markers to study phylogenetic relationships in Cactaceae (Franco et al., 2022). To date, most of the phylogenomic studies conducted in the family have focused on examining relationships at generic and specific levels using different high-throughput sequencing approaches (RAD-seq: Bombonato et al., 2020; Amaral et al., 2021b; GBS: Merklinger et al., 2021; Breslin et al., 2021; genome skimming: Majure et al., 2019, 2021, 2022). The recent availability of genome and transcriptome sequencing of Cactaceae representatives (Copetti et al., 2017; Walker et al., 2018; Wang et al., 2019; Amaral et al., 2021a; Chen et al., 2022) has enabled the development of lineage-specific target sequencing by hybridization capture in Cactaceae (Acha and Majure, 2022; Romeiro-Brito et al., 2022). The Cactaceae591, for instance, is a probe set targeting both non-coding and coding orthologue regions, mostly nuclear, which has proven to be an effective resource for unravelling relationships at shallow and deep levels in Cactaceae (Romeiro-Brito et al., 2022) as well as for resolving the contentious relationship of a recently diverged genus in this group (N. P. Taylor et al., unpubl. res.). Therefore, the Cactaceae591 probe set is a potential source of data for establishing a phylogenetic hypothesis and proposing taxonomic adjustments within Cereeae.
In this study, our main goal was to infer suprageneric relationships within the tribe Cereeae s.l. and generic relationships in the subtribe Cereinae, proposing a new taxonomic classification. To this end, we integrated newly generated genome-scale data gathered with the Cactaceae591 target sequencing probe set and with sequence data from public databases. Considering the long-lasting controversies in the systematics of Cactoideae (Nyffeler and Eggli, 2010), we assume as hypotheses the non-monophyly of the current circumscription of subtribes Cereinae, Rebutiinae and Trichocereinae, as well as of the current generic delimitation within the subtribe Cereinae. Furthermore, we explored the potential of concatenated and coalescent inference approaches in resolving contentious phylogenetic relationships in the subtribe Cereinae.
MATERIALS AND METHODS
Datasets and taxon sampling
To explore distinct gene and genome-scale datasets for estimating the relationships within the tribe Cereeae, we used different datasets compiled from newly generated sequences as well as publicly available data. Our datasets were named as follows: ‘Cactaceae591’ was composed of newly generated data of more than 500 nuclear regions obtained with the Cactaceae591 probe set (Romeiro-Brito et al., 2022); ‘Angiosperm353’ was composed of data of more than 300 nuclear regions generated with the Angiosperm353 universal probe set (Johnson et al., 2016), retrieved from the Kew Tree Of Life Explorer Platform (Baker et al., 2022, https://treeoflife.kew.org/tree-of-life); and ‘gene-scale’ consisted of sequences of eight plastid (rbcl, atpB-rbcL, trnK-matK, rpL16, petL-psbE, trnT-trnL, trnL-trnF, trnS-trnG) and two nuclear (phyC and ppc) regions, which were recovered from the Cactaceae591 raw data and from GenBank (National Center for Biotechnology Information, NCBI). The Cactaceae591 and Angiosperm353 probe sets have 13 shared loci (Romeiro-Brito et al., 2022). However, only a limited subset of these loci was common to both datasets. Given the relatively small number of shared base pairs that could be recovered (Table 1), it was not feasible to conduct a phylogenetic analysis using both datasets simultaneously.
Table 1.
Genetic statistics of nucleotide variation calculated for the datasets used in the present study.
| Dataset | No. of loci | No. of taxa | Alignment length (bp) | Percentage missing data | S (%) | PIS ( %) | Supported nodes (%) |
|---|---|---|---|---|---|---|---|
| Cactaceae591 | 459 | 146 | 511 090 | 15.75 | 287 596 (0.563) | 171 109 (0.335) | 94.1/93.5 |
| Angiosperm353 | 318 | 84 | 136 589 | 12.63 | 59 975 (0.439) | 27 508 (0.201) | 82.3/87.1 |
| Shared loci Cactaceae591 and Angiosperm353 | 5 | 105 | 2873 | 25.53 | 1222 (0.425) | 601 (0.209) | – |
| Gene-scale | 10 | 209 | 9923 | 34.36 | 3502 (0.353) | 1901 (0.191) | 63.4 |
S: variable sites, PIS (%): parsimony-informative sites (proportion of PIS). Supported nodes: percentage of supported nodes from concatenated inferences for the gene-scale (ultrafast bootstrap > 95) and from concatenated and coalescent inferences for the genome-scale datasets (ultrafast bootstrap > 95/local posterior probability > 0.8). No. of taxa includes both ingroup and outgroup sampled taxa in each dataset.
The Cactaceae591 dataset consisted of 29 of 48 recognized genera and 122 of 561 recognized species from the tribe Cereeae s.l. (according to Korotkova et al., 2021). This sampling comprised all genera from the subtribe Cereinae (14 genera), three of seven genera from the subtribe Rebutiinae, and 12 of 27 genera from the subtribe Trichocereinae (Supplementary Data Table S1). To ascertain the phylogenetic position of monotypic genera (e.g. Espostoopsis Buxb. and Leocereus Britton & Rose) or lineages with controversial taxonomy (e.g. species from the genus Micranthocereus Backeb.), we sampled two specimens of each from different geographical records or different collectors.
The Angiosperm353 dataset included 43 of 48 recognized genera and 53 of 561 recognized species from the tribe Cereeae s.l. This sampling comprised all genera from the subtribe Cereinae, six of seven genera from the subtribe Rebutiinae, and 23 of 27 genera from the subtribe Trichocereinae (Supplementary Data Table S1).
The gene-scale sampling dataset was obtained using the SuperCRUNCH v.1.3 pipeline (Portik and Wiens, 2020) to parse, edit and generate the dataset gathered from public repositories. First, we updated the recognized species name according to synonyms from the latest checklist in Korotkova et al. (2021) and then selected all sequences from the tribe Cereeae s.l., allowing up to 60 % of missing data for each locus. Whenever a taxon presented multiple entries for a given locus, we prioritized selecting the longest sequence or, if available, the one obtained from our Cactaceae591 dataset (details of the final dataset in Supplementary Data Table S2). This dataset included 47 of 48 recognized genera and 177 of 561 recognized species from the tribe Cereeae s.l. The final sampling comprised all genera from the subtribe Cereinae and Rebutiinae, and only one genus (Weberbauerocereus Backeb.) from the subtribe Trichocereinae is missing (Table S2).
We attempted to select the same species and/or genus for our outgroup sampling in the three datasets (Supplementary Data Tables S1 and S2). The outgroup taxa included representatives of major clades of the subfamily Cactoideae (tribe Notocacteae, tribe Rhipsalideae, tribe Phyllocacteae and tribe Cacteae; sensuNyffeler and Eggli, 2010), representatives of the subfamilies Opuntioideae and Pereskioideae, and one representative of Portulacaceae (Portulaca hirsutissima Cambess.).
DNA extraction and target capture sequencing library of the Cactaceae591 dataset
Genomic DNA from the Cactaceae591 dataset was extracted from the root and epidermis of preserved tissues or herbarium material using a high-salt CTAB protocol [modified from Martínez-González et al. (2017) and Inglis et al. (2018) and detailed in Supplementary Data Appendix S1]. We isolated and enriched 591 regions using the Cactaceae591 probe set (Romeiro-Brito et al., 2022). This probe set includes 587 genic (exonic and intronic regions) and intergenic nuclear regions and also four regions commonly used in Cactaceae studies (plastidial regions: trnK-matK and rbcl; nuclear regions: ppc and phyc). Library preparation and sequencing were performed by RAPiD Genomics LLC (Gainesville, FL, USA) using their high-throughput workflow with proprietary chemistry. Samples were pooled in equimolar concentrations and sequenced on a NovaSeq 6000 (2 × 150 and 2 × 250 bp).
Processing raw reads of the Cactaceae591 dataset
Raw reads from the Cactaceae591 dataset were trimmed using AdapterRemoval v.2 (Schubert et al., 2016), removing poor quality reads (phred < 20), adapters and short reads (<60 bp). The trimmed reads were mapped and assembled using the HybPiper v.2.0.3 pipeline (Johnson et al., 2016, https://github.com/mossmatters/HybPiper). We obtained the 591 on-target regions using the DNA sequence references available in Romeiro-Brito et al. (2022), setting eight reads as the minimum coverage. The putative paralogues identified in Romeiro-Brito et al. (2022) were removed from the final dataset. We skimmed additional plastid regions commonly used for Cereeae s.l. phylogenies (atpB-rbcl, rpL16, petL-psbE, trnT-trnL, trnL-trnF, trnS-trnG; Supplementary Data Appendix S1) as off-target regions by using Cactaceae reference sequences and setting the minimum coverage reads to 4.
Alignment and trimming genome-scale datasets
We used MAFFT v.7 (Katoh and Standley, 2013) for sequence alignment and trimAL v.1.3 (Capella-Gutiérrez et al., 2009) for removing gaps found in at least 40 % of the sequences and poorly aligned regions.
We removed from the Cactaceae591 and Angiosperm353 datasets the nuclear regions with more than 60 % of missing data and the outlier regions using the genesortR script (Mongiardino Koch, 2021). Here the outlier loci were represented by loci with phylogenetic metrics that deviated significantly from all other loci. This script uses phylogenetic signal (e.g. average bootstrap support) and phylogenetic bias metrics (e.g. level of saturation, and root-to-tip variance) and resumes it using a principal component analysis, accounting for the increase of phylogenetic usefulness of each locus against major phylogenetic biases. This script requires concatenated alignment, a partition file, gene trees of each locus and a coalescent-based species tree. The gene trees were estimated using IQ-TREE v.2 (Minh et al., 2020) with 1000 ultrafast bootstrap replicates (Hoang et al., 2018) and rooted using the pxrr program available in phyx (Brown et al., 2017). The species tree was summarized using ASTRAL v.5.7 (Sayyari and Mirarab, 2016). Finally, we removed 5 % of outlier genes according to the genesortR script. We estimated genetic diversity statistics from all datasets using AMAS (Borowiec, 2016).
Concatenated and coalescent phylogenetic inference
We used the genome-scale datasets Cactaceae591 and Angiosperm353 to estimate phylogenetic trees employing both the concatenated and coalescent approaches. For the gene-scale dataset, we implemented only the concatenated approach. The concatenated inferences were implemented in IQ-TREE v.2 (Minh et al., 2020) with 10 000 ultrafast bootstraps (UfBoot) replicates (Hoang et al., 2018) and 10 000 SH-like approximate likelihood ratio test (SH-aLRT) replicates (Guindon et al., 2010). The best substitution model for each partition was estimated by ModelFinder (Kalyaanamoorthy et al., 2017), using the -m command available on IQ-TREE. We inferred coalescent species trees using the summary approach implemented in ASTRAL-hybrid (Zhang and Mirarab, 2022). Maximum-likelihood (ML) gene trees were generated in IQ-TREE v.2 (Minh et al., 2020) with 10 000 ultrafast bootstrap replicates (Hoang et al., 2018). We set the minimum bootstrap values of the gene tree branch support to be considered for species tree estimation as 40. Branch supports of species trees were accessed by local posterior probabilities (LPPs).
We assessed gene tree conflict against species trees inferred from the Cactaceae591 dataset using PhyParts (Smith et al., 2015) and checked for gene tree incongruences using PhyPartsPieCharts (https://github.com/mossmatters/phyloscripts/tree/master/phypartspiecharts).
RESULTS
Variability of genetic and genomic datasets
The efficiency of capture of Cactaceae591 regions was higher within representatives of Cactoideae (ranging from 480 to 567 regions) compared to representatives of Opuntioideae (ranging from 383 to 404 regions), Pereskioideae (ranging from 485 to 521 regions) and Portulacaceae (137 regions) (Supplementary Data Fig. S1). After removing regions with more than 60 % of missing data, paralogues and outlier regions, the Cactaceae591 dataset included 459 nuclear regions and the Angiosperm353 dataset included 318 nuclear regions (Table 1). The Cactaceae591 dataset presented the highest proportion of variable sites, parsimony-informative sites and branch support among the three datasets delimited in this study (Table 1). The Angiosperm353 presented the smallest percentage of missing data and the gene-scale dataset displayed the most complete taxon sampling regarding genera and species sampling (Table 1).
Phylogenetic relationships among major clades of tribe Cereeae
All phylogenetic inferences recovered the tribe Cereeae as a well-supported clade and sister to the tribe Notocacteae (Fig. 2). None of the Cereeae subtribes circumscribed by Nyffeler and Eggli (2010) was recovered as monophyletic, regardless of the dataset used in the present study. The genome-scale datasets (Cactaceae591 and Angiosperm353) provided a higher phylogenetic resolution along all nodes of the tribe Cereeae (Table 1; Fig. 2A, B, respectively). The gene-scale dataset exhibited insufficient node support for both the backbone and shallow relationships of the Cereeae phylogeny (Fig. 2C; Supplementary Data Fig. S2). The major clades within tribe Cereeae were recovered with high resolution regardless of the datasets and phylogenetic inferences used: Uebelmannia Buining clade, Aylostera Speg clade, Browningia Britton & Rose clade (including Rebutia K.Schum. and Weingartia Werderm.), Trichocereinae clade (excluding Espostoopsis dybowskii), Gymnocalycium Mittler clade and Cereinae clade [including Stetsonia coryne (Salm-Dyck) Britton & Rose and Espostoopsis dybowskii]. Due to low phylogenetic resolution (Fig. 2C; UfBoot = 49) or inadequate taxon representation in each dataset (Fig. 2), we could not determine whether Uebelmannia or Aylostera was the earliest diverging group within the Cereeae tribe. In other way, both genome-scale datasets were consistent in recovering the Browningia clade as an early divergent lineage sister to the remaining representatives from the tribe Cereeae s.l.: subtribe Trichocereinae, Gymnocalycium clade and subtribe Cereinae (Figs 3 and 5).
Fig. 2.
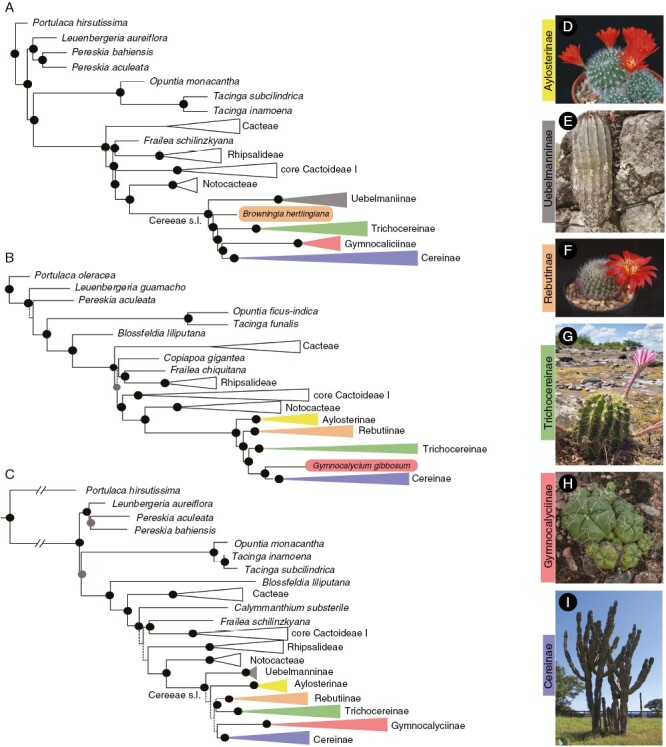
Phylogenetic tree reconstructions showing major clades of subfamily Cactoideae and tribe Cereeae from coalescent-based inference of the Cactaceae591 dataset (A), coalescent-based inference of the Angiosperm353 dataset (B) and maximum-likelihood inference of the gene-scale dataset (C). Highly supported branches (LPP > 0.9 for coalescent-based inference and BS/SH-aLRT > 95/80 for maximum-likelihood inference) are depicted with black circles; moderately supported branches (0.9 > LPP > 0.7 for coalescent-based inference and 95 > BS and 80 < SH-aLRT for maximum-likelihood) are depicted with grey circles. Low supported nodes (LPP < 0.7 in coalescent-based inference and BS/SH-aLRT < 95/80 in maximum-likelihood inference) are represented by dashed branch lines. Representatives of Cereeae clades: (D) Aylostera fiebrigii, (E) Uebelmannia pectinifera Buining, (F) Rebutia minuscula K.Schum, (G) Echinopsis sp., (H) Gymnocalycium denudatum (Link & Otto) Pfeiff. ex Mittler, (I) Cereus jamacaru DC. Photo credits: D and F: M. Lowry; E: E. M. Moraes; G and H: M. Kohler; and J: M. Romeiro-Brito.
Fig. 3.
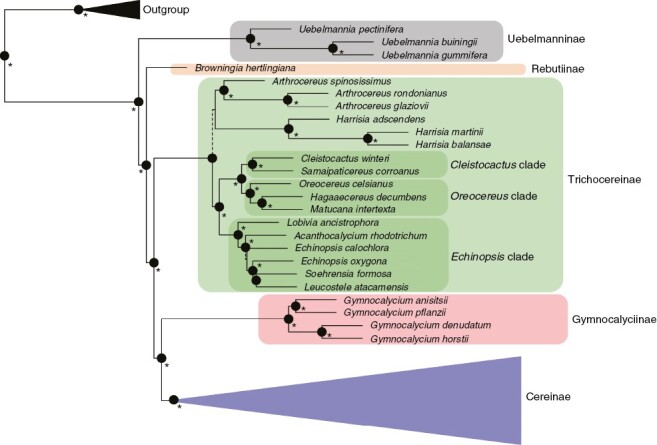
Phylogenetic tree reconstruction of coalescent-based inference using the Angiosperm353 dataset, showing the relationships within major clades of tribe Cereeae. Highly supported branches (LPP > 0.95) are depicted by black circles at nodes. Nodes with LPP = 1 are highlighted with an asterisk. Moderately supported branches are depicted with dark grey (0.95 > LPP > 0.8) and light grey (0.8 > LPP > 0.75) circles. Low supported nodes (LPP < 0.7) are shown with dashed lines in respective branches.
Fig. 5.
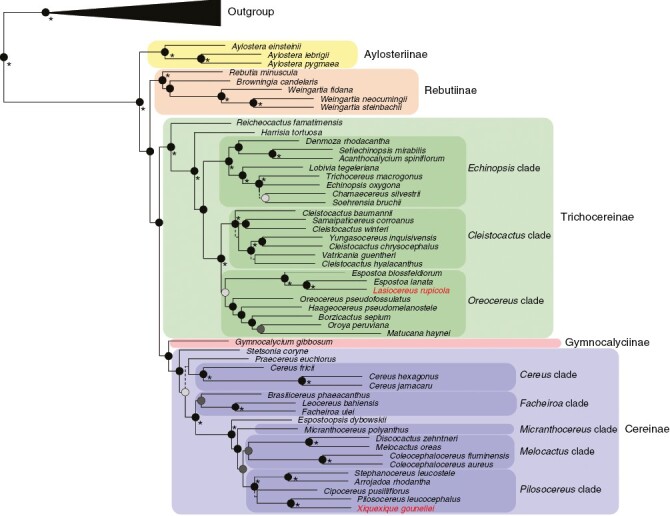
Phylogenetic tree reconstruction of coalescent-based inference using the Cactaceae591 dataset, highlighting the relationships within subtribe Cereinae. Highly supported branches (LPP > 0.95) are depicted by black circles at nodes. Nodes with LPP = 1 are highlighted with an asterisk. Moderately supported branches (0.95 > LPP > 0.8) are depicted by dark grey circles. Low supported nodes (LPP < 0.7) are shown with dashed lines in respective branches.
Phylogenetic relationships within major clades of tribe Cereeae s.l.
The Cactaceae591 dataset recovered Arthrocereus A.Berger and Harrisia as the early divergent lineages of subtribe Trichocereinae (Fig. 3), whereas the Angiosperm353 dataset recovered Reicheocactus Backeb. and Harrisia Britton as the early divergent lineages of this group (Fig. 5). The phylogenetic inferences of all datasets supported the Cleistocactus Lem. s.s. clade as sister to the Oreocereus (A.Berger) Riccob clade, although the genus Cleistocactus was recovered as a non-monophyletic group in either genomic-scale or gene-scale datasets (Figs 3 and 4; Supplementary Data Fig. S2, respectively). Both Cleistocactus and Oreocereus clades were sisters to the Echinopsis s.l. clade using the genome-scale datasets, considering that in the Angiosperm353 dataset the Echinopsis Zucc. clade included the genus Denmoza Britton & Rose (Fig. 4). The gene-scale dataset did not support Echinopsis s.l. as monophyletic, recovering some representatives closely related to the genus Harrisia and the remaining Echinopsis representatives in a clade including the genus Denmoza (Fig. S2). Another incongruence among datasets is related to the genus Lasiocereus F.Ritter, which is allied to the Browningia and Rebutia clade in the gene-scale dataset (Fig. S2), but closely related to Espostoa in the Angiosperm353 dataset (Fig. 5).
Fig. 4.
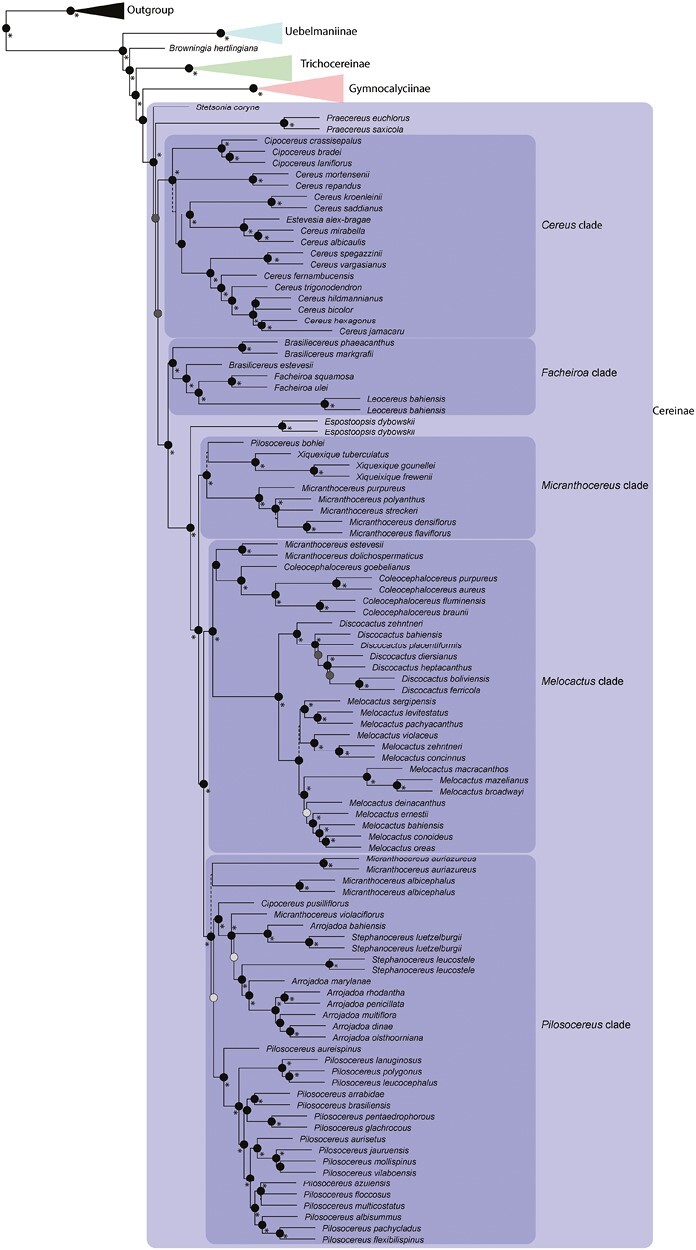
Phylogenetic tree reconstruction of coalescent-based inference using the Cactaceae591 dataset, highlighting the relationships of early-diverging lineages of tribe Cereeae and within subtribe Trichocereinae. Highly supported branches (LPP > 0.95) are depicted by black circles at nodes. Nodes with LPP = 1 are highlighted with an asterisk. Moderately supported branches (0.95 > LPP > 0.8) are depicted by dark grey circles. Low supported nodes (LPP < 0.7) are shown with dashed lines in respective branches.
Regardless of the dataset used, all phylogenetic inferences consistently resolve the genus Gymnocalycium as a sister group to the subtribe Cereinae s.l., which includes Stetsonia coryne and Espostoopsis dybowskii. The genomic dataset resolved the generic relationships within the subtribe Cereinae (especially the Cactaceae591 dataset, Fig. 4), while the gene-scale dataset did not present enough phylogenetic support in the deep and shallow nodes of subtribe Cereinae (Fig. S2).
The backbone of the subtribe Cereinae showed several short internal branches (Fig. 4). The first group to diverge in this subtribe was the genus Stetsonia, followed by the genus Praecereus Buxb., the Cereus clade (including Cereus Mill. Cipocereus F.Ritter), the Facheiroa clade (including Facheiroa Britton & Rose, Brasilicereus Backeb. and Leocereus), and the monotypic genus Espostoopsis. The next well-supported group comprised most of the diversity of the subtribe Cereinae.
Melocactus Link & Otto, Discocactus Pfeiff., Coleocephalocereus Backeb. and Xiquexique Lavor et al. represent the few monophyletic genera within subtribe Cereinae. We highlight the polyphyly of the genus Micranthocereus, whose representatives are scattered within the three main clades assigned as: the Micranthocereus s.s. clade [which includes Micranthocereus polyanthus (Werderm.) Backeb. and its allies, Xiquexique and Pilosocereus bohlei Hofacker], the Melocactus clade [which includes Coleocephalocereus, Micranthocereus subgen. Siccobaccatus (P.J.Braun & Esteves) N.P.Taylor, Discocactus and Melocactus], and the Pilosocereus clade [including Pilosocereus Byles & G.D.Rowley, Arrojadoa Britton & Rose, Stephanocereus A.Berger, Cipocereus pusilliflorus (F.Ritter) Zappi & N.P.Taylor, Micranthocereus violaciflorus Buining., Micranthocereus albicephalus (Buining & Brederoo) F.Ritter and Micranthocereus auriazureus Buining & Brederoo]. The position of one representative from the genus Micranthocereus [Micranthocereus purpureus (Gürke) F.Ritter] was nested within the outgroup taxa in the Angiosperm353 phylogenetic tree (Fig. 4), although this same species is recovered as closely related to the Micranthocereus s.s. clade in the Cactaceae591 dataset (Fig. 5).
Incongruences among concatenated and coalescent inferences using the genomic dataset
The backbone of the phylogenies obtained from the genome-scale datasets was congruent across different phylogenetic approaches (Supplementary Data Fig. S3 and S4), differing only in the presence of the lineages Uebelmannia and Aylostera in each genomic dataset. At shallower levels, incongruences among phylogenetic approaches occurred within the genus Melocactus, and within the Cereus, Micranthocereus and Pilosocereus clade in the Cactaceae591 dataset (Fig. S3), and within the Brasilicereus and Cleistocactus clade in the Angiosperm353 dataset (Fig. S4). Most incongruences presented high phylogenetic support in both phylogenetic approaches (represented by red lines in Figs S3 and S4). Incongruences involving the generic level are related to the placement of Cipocereus pusilliflorus (whether close to the Arrojadoa s.l. clade or to Micranthocereus albicephalus) and the relationship between Cereus and Cipocereus in the Cactaceae591 dataset (Fig. S3).
Gene tree discordances have been shown to be widespread throughout the evolutionary history of the tribe Cereeae, with no dominant alternative topology among the gene trees in the Cactaceae591 dataset (Supplementary Data Fig. S5). The genera that display a higher proportion of gene tree–species tree concordant topology are Uebelmannia, Harrisia, Gymnocalycium, Praecereus and Xiquexique (Fig. S5).
DISCUSSION
Target sequencing strategies have demonstrated their effectiveness in unravelling contentious phylogenetic relationships in plant groups that experienced recent radiation, despite extensive patterns of morphological convergence and gene tree–species tree discordance (Lagomorsino et al., 2022). These strategies may contribute to the progress toward a robust taxonomic classification at multiple levels in Cactaceae, particularly using the customized Cactaceae591 probe set (Romeiro-Brito et al., 2022). Although both genomic datasets used in this study presented similar topologies, the Cactaceae591 dataset recovered higher genetic variability and higher node support than the Angiosperm353 dataset. For instance, only the Cactaceae591 dataset successfully elucidated the relationship at the backbone of subtribe Cereinae. Hence, we agree with previous studies indicating that lineage-specific panels may outperform universal panels when conducting phylogenetic studies at the interspecific level or within challenging plant groups (Christe et al., 2021; Eserman et al., 2021; Yardeni et al., 2021).
On the other hand, the combination of lineage-specific and universal probe sets has been a useful strategy for recovering well-supported topologies in large-scale plant groups and resolving major clades at lineage-specific groups (e.g. Ogutcen et al., 2021; Siniscalchi et al., 2021; Ufimov et al., 2021). For instance, both genomic datasets revealed a surprisingly close and well-supported relationship between Frailea Britton & Rose (an orphan genus of tiny globose cacti) and tribe Rhipsalideae (a group of epiphytic cacti) (Fig. 2A, B). This example shows that robust relationships provided by the genomic datasets can also shed light on relationships of orphan lineages within the subfamily Cactoideae and may help to understand the evolutionary trend of convergent growth forms across Cactaceae.
This study explored comprehensive genetic dataset information, from a few molecular markers to hundreds of orthologous loci, and across contrasting phylogenetic inferences. In the meantime, this is the first phylogenomic analysis comprising most representatives of the tribe Cereeae, resulting in a higher-resolution topology. The presence of the same major clades in our findings as well as in early phylogenetic studies (Ritz et al., 2007; Schlumpberger and Renner, 2012; Lendel, 2013) provides further support for the need to delimit these subtribes and genera in Cereeae. Moreover, the well-supported topology obtained here will provide a solid basis for exploring the evolutionary and biogeographical patterns within this group.
Major groups of the tribe Cereeae s.l.
The monophyly of the tribe Cereeae s.l. and the close relationship with the tribe Notocacteae, as evidenced by the present study, agree with previous taxonomic (Nyffeler and Eggli, 2010) and molecular phylogenetic studies in this group (Nyffeler, 2002; Hernández-Hernández et al., 2011). Likewise, the non-monophyly of the Cereeae subtribes was previously observed in molecular phylogenetic studies within this group (Ritz et al., 2007; Bárcenas et al., 2011; Schlumpberger and Renner, 2012; Bombonato et al., 2020; Romeiro-Brito et al., 2022). While previous studies (Lendel, 2013; Fantinati et al., 2021) have proposed the monophyly of the subtribes Cereinae and Trichocereinae, the genera Espostoopsis and Stetsonia (representatives of the subtribes Trichocereinae and Rebutiinae, respectively) were not previously included for testing. Here, both genera were recovered with high support within the subtribe Cereinae (Figs 4 and 5), suggesting the inclusion of these taxa in an expanded delimitation of this subtribe. Therefore, the circumscription of the subtribe Trichocereinae should exclude the genus Espostoopsis, while the circumscription of the subtribe Rebutiinae should exclude the genus Stetsonia.
We identified six well-supported major clades across all phylogenetic inferences, indicating the existence of six subtribes within Cereeae. These subtribes consist of three monogeneric lineages and three clades with high support: Uebelmannia, Aylostera, Gymnocalycium, Rebutiinae s.s. (excluding Uebelmannia, Aylostera, Gymnocalycium and Stetsonia), Trichocereinae s.s. (excluding Espostoopsis) and Cereinae s.l. (including Espostoopsis and Stetsonia). As observed in previous phylogenetic studies (Ritz et al., 2007; Demaio et al., 2011; Mosti et al., 2011), the monogeneric clades were recovered as well-supported monophyletic groups in all inferences of the present study. Considering that the relationship among the major groups of the tribe Cereeae is now clarified using genome-scale datasets, these subgroups should be finally recognized as independent lineages within Cereeae s.l.
The earliest divergent lineage within Cereeae s.l. remains unknown. Previous phylogenetic studies have pointed to Uebelmannia as the first divergent lineage (Ritz et al., 2007; Hernández-Hernández et al., 2011; Lendel, 2013; Romeiro-Brito et al., 2022), although these studies did not include representatives from Aylostera, or they lacked phylogenetic support. Both issues were also the main concern in the present study. To properly address this matter within the tribe Cereeae, it is crucial to incorporate both lineages into future phylogenomic studies.
Generic relationships of subtribe Cereinae
To date, the Cactaceae591 dataset includes the most comprehensive sampling of the subtribe Cereinae in phylogenetic studies, including all genera and nearly half of its species diversity. To identify generic relationships and revise taxonomic classifications within this subtribe, it is essential to sample this subtribe broadly in light of its diversity and taxonomic complexity.
The present study corroborates the inclusion of two genera first placed in the subtribes Trichocereinae (Espostoopsis dybowskii) and Rebutiinae (Stetsonia coryne) in the subtribe Cereinae. All phylogenetic inferences in this study are in line with previous phylogenetic studies of the tribe Cereeae s.l., which grouped all taxa of the subtribe Cereinae in the same clade (Ritz et al., 2007; Schlumpberger and Renner, 2012; Lendel, 2013; Bombonato et al., 2020; Romeiro-Brito et al., 2022).
The early divergent lineages of the subtribe Cereinae consist of genera with few species, such as Stetsonia and Praecereus, followed by a clade that contains most of the subtribe’s diversity. The relationships of its major clade are partially congruent with the findings of Fantinati et al. (2021). Both studies confirm the non-monophyly of Arrojadoa, Micranthocereus, Pilosocereus and Cipocereus, and the close relationship between the following genera: (1) Cereus and Cipocereus s.s., (2) Arrojadoa and Stephanocereus, and (3) Micranthocereus s.s. and Xiquexique. On the other hand, our phylogenomic inferences disagreed with the close relationship between Facheiroa and Arrojadoa, and the non-monophyly of Cereus, Discocactus and Coleocephalocereus, as recovered by Fantinati et al. (2021).
The controversial relationship between Cereus and Cipocereus was first documented by phylogenetic inferences with few molecular markers (Franco et al., 2017). Later, the use of genome-scale datasets allied with coalescent phylogenetic inferences was decisive in untangling the relationship between these genera (Fig. 2; Bombonato et al., 2020; N. P. Taylor et al., unpubl. res.). Our phylogenetic analysis based on the Cactaceae591 dataset supported the monophyly of Cereus and Cipocereus. Moreover, we observed a significant proportion of incongruences among gene trees and species trees associated with these genera, particularly at the ancestral node of Cereus (Supplementary Data Fig. S5). This supports the hypothesis of Bombonato et al. (2020) that Cipocereus and Cereus are sister taxa that diverged rapidly during a radiation event, resulting in extensive incomplete lineage sorting and creating an ‘anomaly zone’ (Degnan and Rosenberg, 2006) near their ancestral node.
The Micranthocereus s.s. clade comprises Xiquexique and Pilosocereus bohlei. Although P. bohlei shares many characteristics with Pilosocereus species, it has long been recovered outside the Pilosocereus s.s. group (Calvente et al., 2017; Lavor et al., 2018; Fantinati et al., 2021; Romeiro‐Brito et al., 2023). The close relationship of these taxa within the Micranthocereus s.s. clade gives insight into common characteristics shared among them, such as the branching pattern occurring only at the base, hairy areoles and curved hypanthium (Hunt et al., 2006). Pilosocereus bohlei is similar to Xiquexique gounellei with regard to the broader and hairy areoles, but its branching pattern is similar to that of Micranthocereus species. The interesting discovery of the proximity of Micranthocereus s.s., Xiquexique and P. bohlei, with the possibility of the last of these being a hybrid taxon within this clade, highlights the need for additional scrutiny in future phylogenomic studies.
Three closely related genera (Facheiroa, Brasilicereus and Leocereus) were previously placed within the tribes Trichocereeae and Browningieae (Taylor and Zappi, 2004) due to the presence of scales and hair-spines on the pericarpel of the flower. Early molecular phylogeny inferences including these genera (Soffiatti, 2003) suggested a position within the tribe Cereeae (Soffiatti, 2003). Here, we find these genera compose a monophyletic group regardless of the dataset and phylogenetic inference. These results reinforce that the similarity of floral traits previously used to delineate major groups in Cactoideae may at least partially result from convergent evolution, particularly within the BCT clade (Guerrero et al., 2019).
The Melocactus clade clustered genera into two distinct groups, one including Melocactus and Discocactus, and the other comprising Coleocephalocereus and Micranthocereus subgen. Siccobaccatus. The former group consists of globose plants with apical cephalia, while the latter includes columnar plants with unilateral cephalia. The close relationship between the genera Melocactus and Discocactus has been consistently established by molecular phylogenetic studies (Hernández-Hernández et al., 2011; Santos, 2013; Silva et al., 2017). However, the present study is the first to mention the close relationship between M. subgen. Siccobaccatus and Colecephalocereus. Indeed, the species from M. subgen. Siccobaccatus share characteristics with most Colecephalocereus species, including a columnar, erect, growth habit, sunken lateral cephalium and association with rock outcrops. Based on these findings, we suggest the inclusion of the former to be recognized as a subgenus of Coleocephalocereus.
The Pilosocereus clade was composed of the genera Arrojadoa, Stephanocereus and Pilosocereus s.s., and two incertae sedis: Micranthocereus albicephalus and Micranthocereus auriazureus. This clade comprises columnar and shrubby species inhabiting the Caatinga, rock outcrops in the Cerrado, and campos rupestres from Minas Gerais to Bahia (Brazil). The inclusion of two representatives of M. albicephalus and M. auriazureus corroborates its position in the Pilosocereus clade, even though the low support could not delineate its placement within the Pilosocereus or Arrojadoa clades. Further studies investigating the source of low phylogenetic resolution among these taxa should be carried out.
The Pilosocereus clade was divided into two distinct groups with strong support in our study: one consisting of the Pilosocereus species, and the other comprising the genus Arrojadoa and its allies. Our findings support the monophyly of Pilosocereus, excluding Pilosocereus bohlei, a taxon previously positioned outside the genus Pilosocereus (Calvente et al., 2017; Lavor et al., 2018; Romeiro‐Brito et al., 2023). Although our analysis detected inconsistencies in the species relationships across various phylogenetic inference methods in the genus Pilosocereus, we obtained a topology comparable to those of previous phylogenetic studies (Lavor et al., 2018; Romeiro‐Brito et al., 2023), but with stronger node support (Fig. 5). These findings underline the significance of using a genome-wide dataset to gain a thorough understanding of the intricate evolutionary past of Pilosocereus.
The well-supported Arrojadoa clade is composed of Arrojadoa, Stephanocereus, Cipocereus pusilliflorus and Micranthocereus violaciflorus. The early divergent lineages of Arrojadoa consist of Cipocereus pusilliflorus and Micranthocereus violaciflorus, both species lacking a true cephalia. The following clade comprises Arrojadoa and Stephanocereus species, which split into two groups: one composed of species bearing terminal/ring-cephalia (most Arrojadoa species and Stephanocereus leucostele) and the other lacking or with a primitive chlorophyllous cephalium (Arrojadoa bahiensis and Stephanocereus luetzelburgii). Previous phylogenetic studies on this group (Soffiatti, 2003; Fantinati et al., 2021) have suggested a close relationship between Cipocereus pusilliflorus and Stephanocereus to Arrojadoa species, but the positioning of Micranthocereus violaciflorus near Arrojadoa is a novelty. These results highlight the need to recognize an expanded Arrojadoa that includes Cipocereus pusilliflorus, Micranthocereus violaciflorus and Stephanocereus species.
Generic relationships of subtribe Trichocereinae
Though the non-monophyly of subtribes is widespread in Cereeae s.l. (Guerrero et al., 2019), many phylogenetic studies using few molecular markers have recovered Trichocereinae as a monophyletic subtribe (Hernández-Hernández et al., 2011; Schlumpberger and Renner, 2012; Lendel, 2013). Here we highlight an updated circumscription of the subtribe Trichocereinae, excluding Espostoopsis dyboswkii. Although this taxon resembles Espostoa, suggesting a close relationship with the subtribe Trichocereinae, its naked flower tube corroborates its inclusion in the subtribe Cereinae (Taylor and Zappi, 2004). Considering this aspect, flower tube characteristics are also variable within the subtribes of Cereeae and should be treated with caution in taxonomic classifications of these groups.
According to gene-scale dataset phylogenetic inferences, the genus Reicheocactus was the earliest lineage to diverge within the subtribe Trichocereinae, followed by the genus Arthrocereus. We were unable to confirm this relationship using the genome-scale datasets because one of those genera was missing in each dataset. Nonetheless, the phylogenetic inferences with the Angiosperm353 and Cactaceae591 datasets recovered Reicheocactus and Arthrocereus as early divergent lineages in subtribe Trichocereinae, respectively, somehow agreeing with the gene-scale results. The next divergent clade is commonly recovered by the genomic dataset, placing the genus Harrisia as a sister group to all remaining representatives of Trichocereinae. The genus Harrisia was consistently identified as closely related to either the Cleistocactus clade (Franck et al., 2013) or the Echinopsis s.s. clade (Schlumpberger and Renner, 2012; Lendel, 2013). However, the present study suggests that Harrisia may not be closely related to either of these clades. The inclusion of Reicheocactus, Arthrocereus and Harrisia in future phylogenomic studies may shed light on Reicheocactus as the first divergent group of Trichocereinae and a closer relationship between Arthrocereus and Harrisia. The latter two genera are the only members of the subtribe Trichocereinae occurring outside the Andean region, spanning across Central-western and Eastern Brazil (Arthrocereus) to the Caribbean region (Harrisia).
Echinopsis was recovered as a polyphyletic group in our gene-scale phylogenetic tree, corroborating previous phylogenetic studies (Schlumpberger and Renner, 2012; Lendel, 2013). Indeed, the recent taxonomic checklist from Korotkova et al. (2021) segregated Echinopsis into multiple genera. However, all trees inferred from the Cactaceae591 and Angiosperm353 datasets recovered the monophyly of this genus, including Denmoza and excluding Reicheocatus. Thus, to properly address whether Echinopsis is or is not monophyletic, as once defined by Hunt et al. (2006), we recommend the use of genome-scale datasets with comprehensive taxonomic sampling in future phylogenetic studies.
The remaining complex groups of Trichocereinae mostly agree with previous plastid phylogenetic inferences, consisting of the Cleistocactus clade and Oreocereus clade (Schlumpberger and Renner, 2012; Lendel, 2013). The Cleistocactus clade includes species pollinated by bats (Samaipaticereus, Yungasocereus and Vatricania guentheri) or hummingbirds (Cleistocactus and Cephalocleistocactus). The Oreocereus clade, as recovered from genomic datasets, comprises the genera Borzicactus, Espostoa, Mila, Oreocereus, Oroya, Haageocereus, Matucana and Lasiocereus. The position of Lasiocereus among Trichocereinae in the Angiosperm353 tree should be taken with caution, considering that previous phylogenetic studies placed Lasiocereus species within the subtribe Rebutiinae (Supplementary Data Fig. S3; Ritz et al., 2007; Schlumpberger and Renner, 2012; Lendel, 2013).
Potential causes of extensive gene tree and species tree discordance
Resolving the phylogenetic relationships among representatives of the subtribe Cereinae has been a persistent challenge for many decades (Taylor and Zappi, 1989), due primarily to the absence of apomorphic characteristics (Taylor and Zappi, 2004) and low phylogenetic resolution (e.g. Calvente et al., 2017; Franco et al., 2017; Fantinati et al., 2021). The use of genome-scale datasets to resolve relationships at multiple levels is not only a trending practice for studying Cactaceae groups that underwent rapid and recent divergence (Franco et al., 2022), but it is an essential approach for investigating the evolutionary and diversification histories of its tribes. Moreover, the extensive level of gene-tree/species-tree discordance in Cactaceae also highlighted the usefulness of coalescent inference approaches in this group, which may outperform concatenated inference approaches at contentious nodes (Bombonato et al., 2020; Romeiro-Brito et al., 2022, 2023).
Despite the extensive gene tree discordance, the Cereeae topology is stable using different genomic resources and phylogenetic approaches. Some minor discordances at terminal branches were found among concatenated and coalescent inferences, including the Cereus, Pilosocereus and Melocactus clades in subtribe Cereinae, and Cleistocactus clade in subtribe Trichocereinae. Incomplete lineage sorting has been discussed as the main source of gene-tree and species-tree discordance in cacti (Copetti et al., 2017; Walker et al., 2018; Wang et al., 2019; Romeiro-Brito et al., 2022, 2023), especially if we consider that the rapid diversification experienced by cactus lineages (Arakaki et al., 2011) may also lead to short internal branches generating an ‘anomaly zone’ in the tree (Bombonato et al., 2020). However, other sources of phylogenetic conflict have been poorly investigated in these groups, such as hybridization/introgression events.
Hybridization may be an important source of phylogenetic discordance in the complex diversification of tribe Cereeae, given the large number of natural intergeneric and congeneric hybrids described within this group in the past years (Rowley, 1994; Machado, 2008; Khan et al., 2020; Taylor and Albuquerque-Lima, 2020; Arakaki et al., 2021). However, the effects of recent and ancient introgression on diversification within this group remain unknown. Previous studies with Melocactus have demonstrated that despite the widespread occurrence of hybridization events within these genera, there are low levels of introgression within different species (Khan et al., 2020). Hence, this pattern results in the maintenance of species boundaries due to the prevalence of genetic integrity of the parental lineages.
Considering the substantial evidence of hybridization observed in many rapid plant radiations (Schley et al., 2022), we hypothesize that the intricate diversification of tribe Cereeae may be attributed to deep hybridization events. These events, when combined with incomplete lineage sorting, probably contribute to the significant levels of gene tree incongruence observed. The expansion of arid conditions during the Miocene and Pliocene (Arakaki et al., 2011) may have enabled the rapid diversification within this group and promoted the contact and subsequent hybridization among related lineages. So far, the extensive phylogenomic discordance of North American cacti has primarily been linked to incomplete lineage sorting rather than introgression (Copetti et al., 2017). In addition to different methods available for detecting introgression events (e.g. Hibbins and Hahn, 2022), we now have the capability to discern the contributions of different sources of phylogenetic discordance (e.g. Cai et al., 2021; Morales-Briones et al., 2021). Investigating the roles played by different sources of phylogenetic conflict would provide valuable insights into the diversification of Cereeae and shed light on the persistently contentious and poorly resolved relationships within this group.
Taxonomic synopsis of Cereeae
Species are listed in cases where the circumscription of the genus or subgenus is being changed significantly from that in the standard works by Hunt et al. (2006, 2013). Newly published names are indicated in bold type.
Tribe Cereeae Salm-Dyck (as ‘Cereastreae’)
Superficially like Echinocereeae (Pachycereeae), but pericarpel, hypanthial tube and pericarp of unripe fruit usually lacking areoles and stiff spines (if spiny, then plant very slender, not pachycaul). Type: Cereus Mill. Comprising the following six subtribes:
1. Subtribe Uebelmanniinae N.P.Taylor, subtr. nov.
Globular to shortly columnar, many ribbed/tuberculate-ribbed cacti. Stems unbranched, with internal mucilage ducts; epidermis roughened, bearing waxy scales (cf. Copiapoa), grey-green to reddish; areoles on mature stems with long hairs and short straight spines. Cephalium lacking. Flowers small, apical, diurnal, pericarpel and very short hypanthial tube with bract-scales woolly in their axils, perianth-segments expanding, green to yellow, stigma-lobes few. Fruit scarcely fleshy, more or less naked but with the hairy perianth remains attached at apex, reddish. Seeds few, medium-sized, testa smooth.
Type and only genus: Uebelmannia Buining (3 spp.).
Distribution: Narrowly endemic to the central-northern part of Minas Gerais state, Brazil.
2. Subtribe Aylosterinae N.P.Taylor, subtr. nov.
Dwarf globular, simple or caespitose, sometimes semi-geophytic; stems with ±spiralled tubercles or indistinct ribs; spines short, but often dense; cephalium absent. Flowers diurnal, from the sides or base of the stem, shortly funnelform, pericarpel and tube with bract-scales, the style and tube ±fused in the lower half or more. Fruits and seeds small.
Type and only genus: Aylostera Spegazzini (incl. Mediolobivia Backeb., Digitorebutia Donald) (~11 spp.).
Distribution: Eastern Andes of Bolivia and Argentina.
3. Subtribe Rebutiinae Donald [incl. Browningieae F.Buxb., Krainz, Die Kakteen, Lfg 33: CIV/1 (1966)].
Tall columnar, ribbed pachycaul cacti, branched and treelike, rarely single-stemmed, or dwarf globular-stemmed and simple or clustering (caespitose). Stems 7–many-ribbed or with spiralled tubercles, when ribbed these mostly low and rounded. Areoles spiny on juvenile plants, later sometimes spineless on fertile stems. Cephalium lacking (but cf. Browningia columnaris F.Ritter). Flowers borne laterally to basally, nocturnal and whitish or diurnal and brightly coloured, shortly tubular to infundibuliform, pericarpel and hypanthial tube bearing small to large and often overlapping bract-scales, sometimes these shortly hairy, persisting on the fleshy indehiscent fruit. Seeds small, very numerous, testa variously ornamented.
Type: Rebutia K.Schum. (3 spp.)
Distribution: Andes.
Browningia Britt. & Rose (8 spp.)
Weingartia Werderm. (incl. Sulcorebutia Backeb., Cintia Knize & Riha) (~16 spp.).
4. Subtribe Gymnocalyciinae N.P.Taylor, subtr. nov.
Dwarf to medium-sized globular to discoid cacti; ribs few to many, mostly low, often tuberculate. Areoles spiny or sometimes almost spineless. Cephalium lacking. Flowers diurnal, from near the stem apex or from the ‘shoulder’ of the stem, pericarpel and short-to-long hypanthial tube scaly but otherwise naked, perianth variously coloured. Fruit dehiscent, revealing the funicular pulp in which the seeds of diverse testa morphology are embedded.
Type and only genus: Gymnocalycium Mittler (~65 spp.)
Distribution: southeastern South America from the eastern Andes to southern Brazil and central Argentina.
5. Subtribe Trichocereinae F.Buxb.
Stems of diverse size and form, from depressed globose to tall columnar, unbranched to treelike, many-ribbed. Areoles usually spiny. Cephalium lacking or occasionally present, then lateral. Flowers of diverse size and shape, from small to very large, nocturnal or diurnal, pericarpel and hypanthial tube clothed in discrete scales bearing abundant hairy spines (woolly hairs) in their axils, perianth expanded or segments remaining erect to incurved; stamens often inserted in the tube in two series; stigma-lobes many. Fruits scaly-hairy, mostly dehiscent to reveal the funicular pulp. Seeds medium-sized, testa smooth to tuberculate.
Type: Trichocereus Britt. & Rose (= Echinopsis Zucc.)
Distribution: Caribbean and Andes to southeastern South America.
Genera:
Mila Britt. & Rose
Pygmaeocereus Johns. & Backeb.
Haageocereus Backeb.
Espostoa Britt. & Rose (incl. Thrixanthocereus Backeb.)
Rauhocereus Backeb.
Weberbauerocereus Backeb.
Cleistocactus Lem. (incl. Samaipaticereus Cárd., Yungasocereus F.Ritter, Vatricania Backeb.)
The following new name combinations are required: Cleistocactus corroanus (Cárd.) N.P.Taylor, comb. nov. Basionym: Samaipaticereus corroanus Cárd., Cact. Succ. J. (US) 24: 141 (1952). Cleistocactus inquisivensis (Cárd.) N.P.Taylor, comb. nov. Basionym: Samaipaticereus inquisivensis Cárd., Cactus (Paris) 12(57): 246–247 (1957). Cleistocactus guentheri (Kupper) N.P.Taylor, comb. nov. Basionym: Cephalocereus guentheri Kupper, Monatss. Deuts. Kakteen-Gesels. 3: 159 (1931).
Borzicactus Riccob.
Oreocereus (A.Berger) Riccob.
Matucana Britt. & Rose
Oroya Britt. & Rose
Harrisia Britt.
Arthrocereus A.Berger
Echinopsis Zucc. (incl. Denmoza Britt. & Rose)
Reicheocactus Backeb.
Incertae sedis: Lasiocereus F.Ritter (cf. Espostoa)
6. Subtribe Cereinae Britt. & Rose
Habit diverse as in Trichocereinae. Lateral or terminal cephalia developed in many taxa. Flowers of diverse size and shape, from small to very large, but sometimes very small (e.g. Melocactus), mostly lacking areoles and hairs, bract-scales often inconspicuous or widely spaced. Fruits scaly or more often naked, dehiscent or indehiscent; seeds mostly small.
Type: Cereus Mill.
Distribution: Mexico and the Caribbean to eastern Andes and southeastern South America.
Genera:
Stetsonia Britt. & Rose (1 sp.)
Praecereus F.Buxb. (2 spp.)
Cipocereus F.Ritter (5 spp.)
Type:
1. C. pleurocarpus F.Ritter
2. C. minensis (Werderm.) F.Ritter
3. C. bradei (Backeb. & Voll) Zappi & N.P.Taylor
4. C. laniflorus N.P.Taylor & Zappi
5. C. crassisepalus (Buin. & Brederoo) Zappi & N.P.Taylor
Cereus Mill. Type: C. hexagonus (L.) Mill. (~33 spp.)
C. subg. Oblongicarpi (Croizat) D.R.Hunt & N.P.Taylor. (5 spp.). Type:
1. C. repandus (L.) Mill.;
2. C. fricii Backeb.
3. C. horrispinus Backeb.
4. C. mortensenii Croizat.
5. Cereus serruliflorus Haw. (C. haitiensis A.R.Franck, nom. illeg.).
C. subg. Mirabella (F.Ritter) N.P.Taylor (incl. Mirabella F.Ritter, Estevesia P.J.Braun).
Type:
6. C. mirabella N.P.Taylor (Mirabella minensis F.Ritter)
7. C. albicaulis (Britt. & Rose) Luetzelb.
Incertae sedis:
8. C. saddianus (Rizzini & Matos-F.) P.J.Braun
9. C. phatnospermus K.Schum. (incl. C. kroenleinii N.P.Taylor)
10. C. aethiops Haw.
11. C. adelmarii (Rizzini & Mattos-F.) P.J.Braun
12. C. estevesii P.J.Braun (cf. C. albicaulis).
C. subg. Cereus [incl. C. subg. Ebneria (Backeb.) D.Hunt]
13. C. spegazzinii F.A.C.Weber
14. C. vargasianus Cárd.
15. C. trigonodendron Ule
16. C. pierre-braunianus E.Esteves-Pereira
17. C. calcirupicola F.Ritter
18. C. lepidotus Salm-Dyck
19. C. gerardi N.P.Taylor
20. C. jamacaru DC.
21. C. ingens N.P.Taylor & M.C.Machado
22. C. sericifer (F.Ritter) P.J.Braun
23. C. fernambucensis Lem.
24. C. insularis Hemsl.
25. C. bicolor Rizz. & Mattos-F.
26. C. hildmannianus K.Schum.
27. C. stenogonus K.Schum.
Incertae sedis:
28. C. cochabambensis Cárd.
29. C. huilunchu Cárd.
30. C. hankeanus K.Schum.
31. C. lamprospermus K.Schum.
32. C. lanosus (F.Ritter) P.J.Braun
33. C. braunii Cárd.
Micranthocereus Backeb. (6 spp.). Type:
1. M. polyanthus (Werderm.) Backeb.
2. M. flaviflorus Buin. & Brederoo
3. M. alvinii (Hofacker & M.C.Machado) N.P.Taylor & M.Lowry
4. M. streckeri Van Heek. & Van Criek.
5. M. hofackerianus (P.J.Braun & E.Esteves-Pereira) M.C.Machado
6. M. purpureus (Gürke) F.Ritter.
Xiquexique Lavor & Calvente (Pilosocereus subg. Gounellea Zappi; incl. Caerulocereus Guiggi). (4 spp.). Type:
1. X. gounellei (F.A.C.Weber) Lavor & Calvente
2. Xiquexique bohlei (Hofacker) N.P.Taylor, comb. nov. Basionym: Pilosocereus bohlei Hofacker, Kakt. and. Sukk. 52: 253–257 (2001).
3. X. tuberculatus (Werderm.) Lavor & Calvente
4. X. frewenii (Zappi & N.P.Taylor) Lavor & Calvente.
Arrojadoa Britt. & Rose (incl. Stephanocereus A.Berger, Floribunda F.Ritter, Pierrebraunia E.Esteves-Pereira, Arrojadoopsis Guiggi) (13 spp.).
Revised description: Dwarf to medium-tall (to 4 m) cylindrical-stemmed cacti, erect to decumbent, shrub-like or sometimes solitary columnar; subterranean stem base and rootstock often tuberous, vascular cylinder woody. Stems 7–many ribbed, ribs low, rounded, never acute, axes sometimes segmented, and then interrupted by ring cephalia. Areoles usually bearing long hairs, at least when young, always spiny. Flowers mostly from or from near the apex of stem-segments, often from bristly terminal or encircling ring cephalia, small to medium-sized (2–10 cm), shortly tubular, pericarpel very small, it and hypanthial tube almost naked or with few inconspicuous bract-scales, with relatively small, mostly scarcely expanded perianth-segments, diurnal or nocturnal, hummingbird or bat syndrome, reddish pink to magenta, or bicoloured with paler to whitish inner segments, or greenish white. Fruits mostly indehiscent or opening by a basal pore (A. leucostele), fleshy, never dry when ripe, variously coloured, perianth remains persistent, blackish. Seeds mostly small, 1–2 mm. Seedlings, where known, globular at first, only later becoming elongate-cylindrical. Type:
1. A. rhodantha (Gürke) Britt. & Rose
2. Arrojadoa pusilliflora (F.Ritter) N.P.Taylor, comb. nov. Basionym: Floribunda pusilliflora F.Ritter, Kakt. Südamer. 1: 58–60 (1979).
3. Arrojadoa violaciflora (Buining & Brederoo) N.P.Taylor, comb. nov. Basionym: Micranthocereus violaciflorus Buin., Kakt. and. Sukk. 20: 129–130 (1969).
4. A. bahiensis (P.J.Braun & E.Esteves-Pereira) N.P.Taylor & Eggli
5. Arrojadoa luetzelburgii (Vaupel) N.P.Taylor, comb. nov. Basionym: Cereus luetzelburgii Vaupel, Zeitschr. Sukkulentenk. 1: 57 (1923).
6. A. marylaniae Soares-F. & M.C.Machado
7. Arrojadoa leucostele (Guerke) N.P.Taylor, comb. nov. Basionym: Cereus leucostele Gürke, Monatsschr. Kakt.-Kunde 18: 53 (1908).
8. A. dinae Buin. & Brederoo
9. A. albiflora Buin. & Brederoo
10. A. eriocaulis Buin. & Brederoo
11. A. multiflora F.Ritter
12. A. olsthoorniana Hofacker & M.C.Machado
13. A. penicillata (Gürke) Britt. & Rose.
Facheiroa Britt. & Rose (incl. Zehntnerella Britt. & Rose, Leocereus Britt. & Rose, Brasilicereus Backeb., Bragaia P.J.Braun) (7 spp.).
Revised description: Medium to tall cylindrical cacti, erect and self-supporting or slender and sometimes leaning on surrounding vegetation, shrubby to treelike, sparsely to many-branched, vascular cylinder woody. Stems unsegmented, 7–many-ribbed, ribs low, rounded, never acute. Areoles always spiny, mostly lacking long-hairs. Flowers lateral, never terminal, sometimes borne from a bristly/woolly unilateral cephalium, shortly tubular, to 7.5 × 7.5 cm, pericarpel and hypanthial tube scaly, woolly or spiny, never naked, perianth small and hardly expanded or as broad as the flower is long, diurnal to nocturnal, bird, bat or moth syndrome, reddish, green or white. Fruit indehiscent or disintegrating when mature, scaly or with deciduous spiny areoles, fleshy. Seeds small to medium-sized, 1–2.5 mm. Seedlings, where known, globular at first, later elongating. Type:
1. F. ulei (Gürke) Werderm.
2. F. cephaliomelana Buining & Brederoo
3. F. squamosa (Gürke) P.J.Braun & E.Esteves-Pereira
4. Facheiroa phaeacantha (Gürke) N.P.Taylor, comb. nov. Basionym: Cereus phaeacanthus Gürke, Monatsschr. Kakt.-Kunde 18: 57 (1908).
5. Facheiroa markgrafii (Backeb. & Voll) N.P.Taylor, comb. nov. Basionym: Brasilicereus markgrafii Backeb. & Voll, Arch. Jard. Bot. Rio de Janeiro 9: 155 (1949, publ. 1950).
6. Facheiroa bragaia N.P.Taylor, nom. nov. Replaced synonym: Bragaia estevesii Hofacker & P.J.Braun, Kakt. and. Sukk. 60(12): 328 (2009), non Facheiroa estevesii P.J.Braun (= F. cephaliomena subsp. estevesii (P.J.Braun) N.P.Taylor & Zappi).
7. Facheiroa bahiensis (Britt. & Rose) N.P.Taylor, comb. nov. Basionym: Leocereus bahiensis Britt. & Rose, Cact. 2: 108 (1920).
Melocactus Link & Otto, nom. cons. (~40 spp.). Type: Cactus melocactus L., typ. cons.
Discocactus Pfeiff. (13 spp.). Type: D. insignis Pfeiff. [=D. placentiformis (Lehm.) K.Schum.].
Coleocephalocereus Backeb. (incl. Buiningia F.Buxb., Siccobaccatus P.J.Braun & E.Esteves-Pereira, Mariottia Guiggi) (9 spp.). Type: C. fluminensis (Miq.) Backeb.
C. subg. Coleocephalocereus
1. C. fluminensis (Miq.) Backeb.
2. C. decumbens F.Ritter
3. C. pluricostatus Buin. & Brederoo
4. C. buxbaumianus Buin.
C. subg. Simplex N.P.Taylor. Type and only species:
5. C. goebelianus (Vaupel) Buin.
C. subg. Buiningia (F.Buxb.) P.J.Braun. Type: Buiningia brevicylindrica Buin. (=C. aureus F.Ritter)
6. C. aureus F.Ritter
7. C. purpureus (Buin. & Brederoo) F.Ritter.
Coleocephalocereus subg. Siccobaccatus (P.J.Braun & E.Esteves-Pereira) N.P.Taylor, comb. nov. Basionym: Siccobaccatus P.J.Braun & E.Esteves-Pereira, Succulenta (NL) 69: 7 (1990) (2 spp.). Type: S. dolichospermaticus (Buin. & Brederoo) P.J.Braun & E.Esteves-Pereira.
Differs from other subgenera of Coleocephalocereus in fruits which are dry and disintegrate at maturity; seeds elongate, testa ±smooth (wind dispersed).
8. Coleocephalocereus dolichospermaticus (Buin. & Brederoo) N.P.Taylor, comb. nov. Basionym: Austrocephalocereus dolichospermaticus Buin. & Brederoo, Kakt. and. Sukk. 25: 76–79 (1974).
9. Coleocephalocereus neoestevesii N.P.Taylor, nom. nov. Replaced synonym: Austrocephalocereus estevesii Buin. & Brederoo, Cact. Succ. J. (US) 47: 267 (1975), non Coleocephalocereus estevesii L.Diers (=C. buxbaumianus subsp. flavisetus (F.Ritter) N.P.Taylor & Zappi).
Pilosocereus Byles & G.Rowley (excl. P. subg. Gounellea Zappi and Caerulocereus Guiggi) (~60 spp.). Type: P. leucocephalus (Poselger) Byles & G.D.Rowley.
Incertae sedis within Cereinae:
1. Micranthocereus albicephalus (Buin. & Brederoo) F.Ritter
2. M. auriazureus Buin. & Brederoo.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Table S1. Sampling information of Cactaceae and Portulacaceae species used for the genomic dataset. Table S2. Sequence matrix of species and regions included in the gene-scale dataset. Figure S1. Heatmap indicating 591 orthologue recovery success per sample. Figure S2. Maximum-likelihood phylogenetic inference estimated by IQ-TREE2 using the gene-scale dataset. Figure S3. Phylogenetic tree reconstructions of coalescent-based inference (A) and maximum-likelihood (B) using the Cactaceae591 dataset. Figure S4. Phylogenetic tree reconstructions of coalescent-based inference (A) and maximum-likelihood (B) using the Angiosperm353 dataset. Figure S5. Gene-tree and species-tree conflicts estimated in PhyParts using coalescent-based phylogeny inferred with the Cactaceae591 dataset.
ACKNOWLEDGMENTS
We thank Gerardus Olsthoorn, Dr Lidyanne Y. S. Aona, Dr Diego R. Gonzaga and the Coleção de Cactos e Suculentas do Instituto de Pesquisas Jardim Botânico do Rio de Janeiro for contributing cactus samples used in this study. We thank Dr Juliana F. Martinez and Heidi S. M. Utsunomiya for technical support. We also thank Dr Matias Kohler and Dr Martin Lowry for sharing photos from representatives of tribe Cereeae and Dr Danilo T. Amaral and Dr Isabel A. S. Bonatelli for valuable comments and discussions that improved the manuscript. The authors declare that there is no conflict of interest regarding the publication of this article.
Contributor Information
Monique Romeiro-Brito, Departamento de Biologia, Centro de Ciências Humanas e Biológicas, Universidade Federal de São Carlos (UFSCar), Sorocaba, São Paulo, Brazil.
Nigel P Taylor, University of Gibraltar, Gibraltar Botanic Gardens Campus, Gibraltar.
Daniela C Zappi, Programa de Pós-Graduação em Botânica, Instituto de Ciências Biológicas Universidade de Brasília (UNB), Brasília, Distrito Federal, Brazil.
Milena C Telhe, Departamento de Biologia, Centro de Ciências Humanas e Biológicas, Universidade Federal de São Carlos (UFSCar), Sorocaba, São Paulo, Brazil.
Fernando F Franco, Departamento de Biologia, Centro de Ciências Humanas e Biológicas, Universidade Federal de São Carlos (UFSCar), Sorocaba, São Paulo, Brazil.
Evandro M Moraes, Departamento de Biologia, Centro de Ciências Humanas e Biológicas, Universidade Federal de São Carlos (UFSCar), Sorocaba, São Paulo, Brazil.
FUNDING
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP: 2018/06937-8 to M.R.B., 2019/11233-2 to M.C.T., 2020/15161-3 to F.F.F., 2018/03428-5 and 2019/03211-9 to E.M.M.] and Conselho Nacional de Desenvolvimento Científico e Tecnológico (04178/2021-7 to D.C.Z.).
LITERATURE CITED
- Acha S, Majure LC.. 2022. A new approach using targeted sequence capture for phylogenomic studies across Cactaceae. Genes 13: 350. doi: 10.3390/genes13020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DT, Bombonato JR, da Silva Andrade SC, Moraes EM, Franco FF.. 2021a. The genome of a thorny species: comparative genomic analysis among South and North American Cactaceae. Planta 254: 1–7. [DOI] [PubMed] [Google Scholar]
- Amaral DT, Bonatelli IA, Romeiro-Brito M, Moraes EM, Franco FF.. 2022. Spatial patterns of evolutionary diversity in Cactaceae show low ecological representation within protected areas. Biological Conservation 273: 109677. doi: 10.1016/j.biocon.2022.109677. [DOI] [Google Scholar]
- Amaral DT, Minhós‐Yano I, Oliveira JVM, et al. 2021b. Tracking the xeric biomes of South America: The spatiotemporal diversification of Mandacaru cactus. Journal of Biogeography 48: 3085–3103. [Google Scholar]
- Anderson EF. 2001.The cactus family. Portland, OR: Timber Press. [Google Scholar]
- Applequist WL, Wallace RS. 2002. Deletions in the plastid trnT–trnL intergenic spacer define clades within Cactaceae subfamily Cactoideae. Plant Systematics and Evolution 231: 153–162. doi: 10.jstor.org/stable/23644353. [DOI] [Google Scholar]
- Arakaki M, Christin PA, Nyffeler R, et al. 2011. Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proceedings of the National Academy of Sciences of the United States of America 108: 8379–8384. doi: 10.1073/pnas.1100628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki M, Speranza P, Soltis PS, Soltis DE.. 2021. Examination of reticulate evolution involving Haageocereus and Espostoa. Haseltonia 27: 102–112. [Google Scholar]
- Baker WJ, Bailey P, Barber V, et al. 2022. A comprehensive phylogenomic platform for exploring the angiosperm tree of life. Systematic Biology 71: 301–319. doi: 10.1093/sysbio/syab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcenas RT, Yesson C, Hawkins JA.. 2011. Molecular systematics of the Cactaceae. Cladistics 27: 470–489. doi: 10.1111/j.1096-0031.2011.00350.x. [DOI] [PubMed] [Google Scholar]
- Barthlott W, Hunt D.. 1993. Cactaceae. In: Kubitzki K, Rohwer JG, Bittrich V. eds. The families and genera of vascular plants, Vol. 2. Berlin: Springer, 161–197. [Google Scholar]
- Bombonato JR, Amaral DT, Silva GAR, et al. 2020. The potential of genome-wide RAD sequences for resolving rapid radiations: a case study in Cactaceae. Molecular Phylogenetics and Evolution 151: 106896. doi: 10.1016/j.ympev.2020.106896. [DOI] [PubMed] [Google Scholar]
- Borowiec ML. 2016. AMAS: A fast tool for alignment manipulation and computing of summary statistics. PeerJ 4: e1660–e1973. doi: 10.7717/peerj.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin PB, Wojciechowski MF, Majure LC.. 2021. Molecular phylogeny of the Mammilloid clade (Cactaceae) resolves the monophyly of Mammillaria. Taxon 70: 308–323. doi: 10.1002/tax.12451. [DOI] [Google Scholar]
- Brown JW, Walker JF, Smith SA.. 2017. Phyx: phylogenetic tools for unix. Bioinformatics 33: 1886–1888. doi: 10.1093/bioinformatics/btx063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Xi Z, Lemmon EM, et al. 2021. The perfect storm: gene tree estimation error, incomplete lineage sorting, and ancient gene flow explain the most recalcitrant ancient angiosperm clade, Malpighiales. Systematic Biology 70: 491–507. doi: 10.1093/sysbio/syaa083. [DOI] [PubMed] [Google Scholar]
- Calvente A, Moraes EM, Lavor P, et al. 2017. Phylogenetic analyses of Pilosocereus (Cactaceae) inferred from plastid and nuclear sequences. Botanical Journal of the Linnean Society 183: 25–38. [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li F, Xie F, et al. 2022. Pitaya genome and multiomics database (PGMD): a comprehensive and integrative resource of Selenicereus undatus. Genes 13: 745. doi: 10.3390/genes13050745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christe C, Boluda CG, Koubínová D, Gautier L, Naciri Y.. 2021. New genetic markers for Sapotaceae phylogenomics: More than 600 nuclear genes applicable from family to population levels. Molecular Phylogenetics and Evolution 160: 107123. doi: 10.1016/j.ympev.2021.107123. [DOI] [PubMed] [Google Scholar]
- Copetti D, Búrquez A, Bustamante E, et al. 2017. Extensive gene tree discordance and hemiplasy shaped the genomes of North American columnar cacti. Proceedings of the National Academy of Sciences of the United States of America 114: 12003–12008. doi: 10.1073/pnas.1706367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA.. 2006. Discordance of species trees with their most likely gene trees. PLoS Genetics 2: e68. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaio PH, Barfuss MH, Kiesling R, Till W, Chiapella JO.. 2011. Molecular phylogeny of Gymnocalycium (Cactaceae): assessment of alternative infrageneric systems, a new subgenus, and trends in the evolution of the genus. American Journal of Botany 98: 1841–1854. [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Nyffeler R, Donoghue MJ.. 2005. Basal cactus phylogeny: implications of Pereskia (Cactaceae) paraphyly for the transition to the cactus life form. American Journal of Botany 92: 1177–1188. doi: 10.3732/ajb.92.7.1177. [DOI] [PubMed] [Google Scholar]
- Eserman LA, Thomas SK, Coffey EE, Leebens‐Mack JH. 2021. Target sequence capture in orchids: developing a kit to sequence hundreds of single‐copy loci. Applications in Plant Sciences 9: e11416. doi: 10.1002/aps3.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantinati MR, Soffiatti P, Calvente A.. 2021. A new phylogenetic hypothesis for Cereinae (Cactaceae) points to a monophyletic subtribe. Systematic Botany 46: 689–699. doi: 10.1600/036364421x16312068417020. [DOI] [Google Scholar]
- Franck AR, Cochrane BJ, Garey JR.. 2013. Phylogeny, biogeography, and infrageneric classification of Harrisia (Cactaceae). Systematic Botany 38: 210–223. doi: 10.1600/036364413x662105. [DOI] [Google Scholar]
- Franco FF, Amaral DT, Bonatelli IA, Romeiro-Brito M, Telhe MC, Moraes EM.. 2022. Evolutionary genetics of cacti: research biases, advances and prospects. Genes 13: 452. doi: 10.3390/genes13030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco FF, Silva GAR, Moraes EM, et al. 2017. Plio-Pleistocene diversification of Cereus (Cactaceae, Cereeae) and closely allied genera. Botanical Journal of the Linnean Society 183: 199–210. doi: 10.1093/botlinnean/bow010. [DOI] [Google Scholar]
- Goettsch B, Hilton-Taylor C, Cruz-Piñón G, et al. 2015. High proportion of cactus species threatened with extinction. Nature Plants 1: 1–7. doi: 10.1038/nplants.2015.142. [DOI] [PubMed] [Google Scholar]
- Guerrero PC, Majure LC, Cornejo-Romero A, Hernández-Hernández T.. 2019. Phylogenetic relationships and evolutionary trends in the cactus family. The Journal of Heredity 110: 4–21. doi: 10.1093/jhered/esy064. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 30. Systematic Biology 59: 307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hernández‐Hernández T, Brown JW, Schlumpberger BO, Eguiarte LE, Magallón S.. 2014. Beyond aridification: multiple explanations for the elevated diversification of cacti in the New World Succulent Biome. New Phytologist 202: 1382–1397. doi: 10.1111/nph.12752. [DOI] [PubMed] [Google Scholar]
- Hernández‐Hernández T, Hernández HM, De‐Nova JA, Puente R, Eguiarte LE, Magallón S.. 2011. Phylogenetic relationships and evolution of growth form in Cactaceae (Caryophyllales, Eudicotyledoneae). American Journal of Botany 98: 44–61. [DOI] [PubMed] [Google Scholar]
- Hernández-Ledesma P, Berendsohn WG, Borsch T, et al. 2015. A taxonomic backbone for the global synthesis of species diversity in the angiosperm order Caryophyllales. Willdenowia 45: 281–383. doi: 10.3372/wi.45.45301. [DOI] [Google Scholar]
- Hibbins MS, Hahn MW.. 2022. Phylogenomic approaches to detecting and characterizing introgression. Genetics 220: iyab173. doi: 10.1093/genetics/iyab173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt D, Taylor NP, Charles G.. 2006. The new cactus lexicon, Vols 1 & 2. Milborne Port: dh Books. [Google Scholar]
- Hunt D, Taylor NP, Charles G.. 2013. The new cactus lexicon, Vol 2, illustrations. Milborne Port: dh Books. [Google Scholar]
- Inglis PW, Pappas MDCR, Resende LV, Grattapaglia D.. 2018. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS One 13: e0206085. doi: 10.1371/journal.pone.0206085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, et al. 2016. HybPiper: Extracting coding sequence and introns for phylogenetics from high‐throughput sequencing reads using target enrichment. Applications in Plant Sciences 4: 1600016. doi: 10.3732/apps.1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan G, Franco FF, Silva GA, et al. 2020. Maintaining genetic integrity with high promiscuity: Frequent hybridization with low introgression in multiple hybrid zones of Melocactus (Cactaceae). Molecular Phylogenetics and Evolution 142: 106642. [DOI] [PubMed] [Google Scholar]
- Korotkova N, Aquino D, Arias S, et al. 2021. Cactaceae at Caryophyllales org–a dynamic online species-level taxonomic backbone for the family. Willdenowia 51: 251–270. [Google Scholar]
- Lagomarsino LP, Frankel L, Uribe-Convers S, Antonelli A, Muchhala N.. 2022. Increased resolution in the face of conflict: phylogenomics of the Neotropical bellflowers (Campanulaceae: Lobelioideae), a rapid plant radiation. Annals of Botany 129: 723–736. doi: 10.1093/aob/mcac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavor P, Calvente A, Versieux LM, Sanmartin I. 2018. Bayesian spatio‐temporal reconstruction reveals rapid diversification and Pleistocene range expansion in the widespread columnar cactus Pilosocereus. Journal of Biogeography 46: 238–250. doi: 10.1111/jbi.13481. [DOI] [Google Scholar]
- Lendel A. 2013. South American Cacti in time and space: studies on the diversification of the tribe Cereeae, with particular focus on subtribe Trichocereinae (Cactaceae). PhD Thesis, University of Zurich, Switzerland. [Google Scholar]
- Machado MC. 2008. What is the role of hybridization in the evolution of the Cactaceae? Bradleya 26: 1–18. doi: 10.25223/brad.n26.2008.a1. [DOI] [Google Scholar]
- Majure LC, Baker MA, Cloud‐Hughes M, Salywon A, Neubig KM.. 2019. Phylogenomics in Cactaceae: A case study using the chollas sensu lato (Cylindropuntieae, Opuntioideae) reveals a common pattern out of the Chihuahuan and Sonoran deserts. American Journal of Botany 106: 1327–1345. doi: 10.1002/ajb2.1364. [DOI] [PubMed] [Google Scholar]
- Majure LC, Barrios D, Díaz E, Bacci LF, Piñeyro YE.. 2022. Phylogenomics of the Caribbean melocacti: Cryptic species and multiple invasions. Taxon 71: 993–1012. doi: 10.1002/tax.12791. [DOI] [Google Scholar]
- Majure LC, Barrios D, Díaz E, Zumwalde BA, Testo W, Negrón‐Ortíz V.. 2021. Pleistocene aridification underlies the evolutionary history of the Caribbean endemic, insular, giant Consolea (Opuntioideae). American Journal of Botany 108: 200–215. doi: 10.1002/ajb2.1610. [DOI] [PubMed] [Google Scholar]
- Martínez-González CR, Ramírez-Mendoza R, Jiménez-Ramírez J, Gallegos-Vázquez C, Luna-Vega I.. 2017. Improved method for genomic DNA extraction for Opuntia Mill (Cactaceae). Plant Methods 13: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merklinger FF, Böhnert T, Arakaki M, Weigend M, Quandt D, Luebert F.. 2021. Quaternary diversification of a columnar cactus in the driest place on earth. American Journal of Botany 108: 184–199. doi: 10.1002/ajb2.1608. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, et al. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiardino Koch N. 2021. Phylogenomic subsampling and the search for phylogenetically reliable loci. Molecular Biology and Evolution 38: 4025–4038. doi: 10.1093/molbev/msab151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Briones DF, Kadereit G, Tefarikis DT, et al. 2021. Disentangling sources of gene tree discordance in phylogenomic data sets: testing ancient hybridizations in Amaranthaceae sl. Systematic Biology 70: 219–235. doi: 10.1093/sysbio/syaa066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosti S, Bandara NL, Papini A.. 2011. Further insights and new combinations in Aylostera (Cactaceae) based on molecular and morphological data. Pakistan Journal of Botany 43: 2769–2785. [Google Scholar]
- Nyffeler R. 2002. Phylogenetic relationships in the cactus family (Cactaceae) based on evidence from trnK/matK and trnL‐trnF sequences. American Journal of Botany 89: 312–326. doi: 10.3732/ajb.89.2.312. [DOI] [PubMed] [Google Scholar]
- Nyffeler R, Eggli U.. 2010. A farewell to dated ideas and concepts: molecular phylogenetics and a revised suprageneric classification of the family Cactaceae. Schumannia 6: 109–149. [Google Scholar]
- Ogutcen E, Christe C, Nishii K, Salamin N, Möller M, Perret M.. 2021. Phylogenomics of Gesneriaceae using targeted capture of nuclear genes. Molecular Phylogenetics and Evolution 157: 107068. doi: 10.1016/j.ympev.2021.107068. [DOI] [PubMed] [Google Scholar]
- Pillet M, Goettsch B, Merow C, et al. 2022. Elevated extinction risk of cacti under climate change. Nature Plants 8: 366–372. doi: 10.1038/s41477-022-01130-0. [DOI] [PubMed] [Google Scholar]
- Portik DM, Wiens JJ.. 2020. SuperCRUNCH: A bioinformatics toolkit for creating and manipulating supermatrices and other large phylogenetic datasets. Methods in Ecology and Evolution 11: 763–772. doi: 10.1111/2041-210x.13392. [DOI] [Google Scholar]
- Ritz CM, Fickenscher K, Föller J, Herrmann K, Mecklenburg R, Wahl R.. 2016. Molecular phylogenetic relationships of the Andean genus Aylostera Speg (Cactaceae, Trichocereeae), a new classification and a morphological identification key. Plant Systematics and Evolution 302: 763–780. doi: 10.1007/s00606-016-1296-4. [DOI] [Google Scholar]
- Ritz CM, Martins L, Mecklenburg R, Goremykin V, Hellwig FH.. 2007. The molecular phylogeny of Rebutia (Cactaceae) and its allies demonstrates the influence of paleogeography on the evolution of South American mountain cacti. American Journal of Botany 94: 1321–1332. doi: 10.3732/ajb.94.8.1321. [DOI] [PubMed] [Google Scholar]
- Romeiro-Brito M, Telhe MC, Amaral DT, Franco FF, Moraes EM.. 2022. A target capture probe set useful for deep-and shallow-level phylogenetic studies in Cactaceae. Genes 13: 707. doi: 10.3390/genes13040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeiro‐Brito M, Khan G, Perez MF, et al. 2023. Revisiting phylogeny, systematics, and biogeography of a Pleistocene radiation. American Journal of Botany 110: e16134. doi: 10.1002/ajb2.16134. [DOI] [PubMed] [Google Scholar]
- Rowley GD. 1994. Spontaneous bigeneric hybrids in Cactaceae. Bradleya 12: 2–7. doi: 10.25223/brad.n12.1994.a2. [DOI] [Google Scholar]
- Santos MR. 2013. Filogenia Molecular, Taxonomia, Biogeografia e Conservação de Discocactus Pfeiff. PhD Thesis, Universidade Federal de Vicosa, Viçosa, Minas Gerais. [Google Scholar]
- Sayyari E, Mirarab S.. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Molecular Biology and Evolution 33: 1654–1668. doi: 10.1093/molbev/msw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schley RJ, Twyford AD, Pennington RT.. 2022. Hybridization: a ‘double-edged sword’ for Neotropical plant diversity. Botanical Journal of the Linnean Society 199: 331–356. doi: 10.1093/botlinnean/boab070. [DOI] [Google Scholar]
- Schlumpberger BO, Renner SS.. 2012. Molecular phylogenetics of Echinopsis (Cactaceae): Polyphyly at all levels and convergent evolution of pollination modes and growth forms. American Journal of Botany 99: 1335–1349. doi: 10.3732/ajb.1100288. [DOI] [PubMed] [Google Scholar]
- Schubert M, Lindgreen S, Orlando L.. 2016. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Research Notes 9: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GAR, Antonelli A, Lendel A, Moraes EM, Manfrin MH. 2018. The impact of early Quaternary climate change on the diversification and population dynamics of a South American cactus species. Journal of Biogeography 45: 76–88. [Google Scholar]
- Siniscalchi CM, Hidalgo O, Palazzesi L, et al. 2021. Lineage‐specific vs universal: a comparison of the Compositae1061 and Angiosperms353 enrichment panels in the sunflower family. Applications in Plant Sciences 9. doi: 10.1002/aps3.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Moore MJ, Brown JW, Yang Y.. 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evolutionary Biology 15: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffiatti P. 2003. Estudos Anatâmicos e Filogenéticos na Tribo Cereeae, Cactaceae. PhD Thesis, Universidade de São Paulo, São Paulo. [Google Scholar]
- Taylor NP, Albuquerque-Lima S.. 2020. Annotated checklist of Cactaceae in the Catimbau National Park, Pernambuco, Brazil. Bradleya 2020: 231–246. doi: 10.25223/brad.n38.2020.a22. [DOI] [Google Scholar]
- Taylor NP, Zappi DC.. 1989. An alternative view of generic delimitation and relationships in tribe Cereeae (Cactaceae). Bradleya 7: 13–40. doi: 10.25223/brad.n7.1989.a2. [DOI] [Google Scholar]
- Taylor NP, Zappi DC.. 2004. Cacti of Eastern Brazil. Kew: Royal Botanic Gardens. [Google Scholar]
- Ufimov R, Zeisek V, Píšová S, et al. 2021. Relative performance of customized and universal probe sets in target enrichment: A case study in subtribe Malinae. Applications in Plant Sciences 9: e11442. doi: 10.1002/aps3.11442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JF, Yang Y, Feng T, et al. 2018. From cacti to carnivores: Improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. American Journal of Botany 105: 446–462. doi: 10.1002/ajb2.1069. [DOI] [PubMed] [Google Scholar]
- Wang N, Yang Y, Moore MJ, et al. 2019. Evolution of Portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Molecular Biology and Evolution 36: 112–126. doi: 10.1093/molbev/msy200. [DOI] [PubMed] [Google Scholar]
- Yardeni G, Viruel J, Paris M, et al. 2022. Taxon‐specific or universal? Using target capture to study the evolutionary history of rapid radiations. Molecular Ecology Resources 22: 927–945. doi: 10.1111/1755-0998.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Mirarab S.. 2022. Weighting by gene tree uncertainty improves accuracy of quartet-based species trees. Molecular Biology and Evolution 39: msac215. doi: 10.1093/molbev/msac215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


