Abstract
Background
The molecular evolution of organellar genomes in angiosperms has been studied extensively, with some lineages, such as parasitic ones, displaying unique characteristics. Parasitism has emerged 12 times independently in angiosperm evolution. Holoparasitism is the most severe form of parasitism, and is found in ~10 % of parasitic angiosperms. Although a few holoparasitic species have been examined at the molecular level, most reports involve plastomes instead of mitogenomes. Parasitic plants establish vascular connections with their hosts through haustoria to obtain water and nutrients, which facilitates the exchange of genetic information, making them more susceptible to horizontal gene transfer (HGT). HGT is more prevalent in the mitochondria than in the chloroplast or nuclear compartments.
Scope
This review summarizes current knowledge on the plastid and mitochondrial genomes of holoparasitic angiosperms, compares the genomic features across the different lineages, and discusses their convergent evolutionary trajectories and distinctive features. We focused on Balanophoraceae (Santalales), which exhibits extraordinary traits in both their organelles.
Conclusions
Apart from morphological similarities, plastid genomes of holoparasitic plants also display other convergent features, such as rampant gene loss, biased nucleotide composition and accelerated evolutionary rates. In addition, the plastomes of Balanophoraceae have extremely low GC and gene content, and two unexpected changes in the genetic code. Limited data on the mitochondrial genomes of holoparasitic plants preclude thorough comparisons. Nonetheless, no obvious genomic features distinguish them from the mitochondria of free-living angiosperms, except for a higher incidence of HGT. HGT appears to be predominant in holoparasitic angiosperms with a long-lasting endophytic stage. Among the Balanophoraceae, mitochondrial genomes exhibit disparate evolutionary paths with notable levels of heteroplasmy in Rhopalocnemis and unprecedented levels of HGT in Lophophytum. Despite their differences, these Balanophoraceae share a multichromosomal mitogenome, a feature also found in a few free-living angiosperms.
Keywords: Balanophoraceae, Cuscuta, horizontal gene transfer, Lophophytum mirabile, mitochondria, non-photosynthetic plastid, Ombrophytum subterraneum, Rafflesiaceae, Rhopalocnemis phalloides, Santalales
INTRODUCTION
The genetic material within plant organelles contains genes necessary for photosynthesis and respiration, which are essential processes for plant survival. Examining the evolution of the plastid and mitochondrial genomes in parasitic plants helps to address key evolutionary questions, such as how parasitism developed independently many times in angiosperms and how holoparasitic plants have evolved to rely entirely on their host plants for survival. Furthermore, these studies improve our understanding of the cascading effects provoked by photosynthesis loss and the relaxation of plastid metabolism (Wicke and Naumann, 2018; Cai, 2023). Genomic and transcriptomic investigations provide information about the molecular and genomic changes that occurred during the shift from free-living to parasitic plants, such as those involving haustorium formation (Yoshida et al., 2016; Teixeira-Costa, 2021), cytonuclear interactions (Ceriotti et al., 2022), co-evolutionary dynamics between host and parasites, and the potential role of horizontal gene transfer (HGT) in driving molecular changes in both the nucleus and the organelles (Davis and Xi, 2015; Yang et al., 2019; Wickell and Li, 2020).
EXPLORING THE DIVERSITY OF HETEROTROPHIC PLANTS: MYCOHETEROTROPHS AND PARASITES
Heterotrophy has originated multiple times throughout the evolution of the Archaeplastida, including green algae, red algae and land plants. In streptophytes, the greatest diversity of heterotrophy occurs among angiosperms (Hadariová et al., 2018), where it exists in two different modes: mycoheterotrophy and parasitism. Mycoheterotrophs acquire nutrients directly from fungi or indirectly from another plant through a fungus (Graham et al., 2017). Fully mycoheterotrophic plants lost their ability to photosynthesize and associate with mycorrhizal fungi, which act as a bridge between the mycoheterotroph and an autotrophic plant (Merckx, 2013). Mycoheterotrophic species evolved independently in ten angiosperm families, namely Burmanniaceae, Corsiaceae, Ericaceae, Gentianaceae, Iridaceae, Petrosaviaceae, Polygalaceae, Thismiaceae, Triuridaceae and Orchidaceae, which have undergone at least 30 independent transitions to mycoheterotrophy (Merckx and Freudenstein, 2010; Nickrent, 2020). Parasitic plants, on the other hand, obtain part or all of their nutrients from the host plant through a modified root known as haustorium, which connects the parasite with the host’s vascular tissues (Kuijt, 1969; Nickrent, 2020). There are nearly 4750 species of parasitic angiosperms that feed on other plants, invading the roots or stems of their hosts (Heide-Jorgensen, 2008; Westwood et al., 2010; Nickrent, 2020). Parasites are distributed across 12 angiosperm lineages, namely Cassytha, Cynomoriaceae, Cytinaceae, Cuscuta, Hydnoraceae, Krameriaceae, Lennoaceae, Mitrastemonaceae, Orobanchaceae, Santalales, Apodanthaceae and Rafflesiaceae (Fig. 1). While parasitic angiosperms can have a positive role in ecosystems, some of them, such as the Orobanchaceae Striga and Orobanche or the dodder Cuscuta, can cause crop damage and have been studied more intensively (Press and Phoenix, 2005; Nickrent, 2020; Zagorchev et al., 2021; Albanova et al., 2023).
Fig. 1.
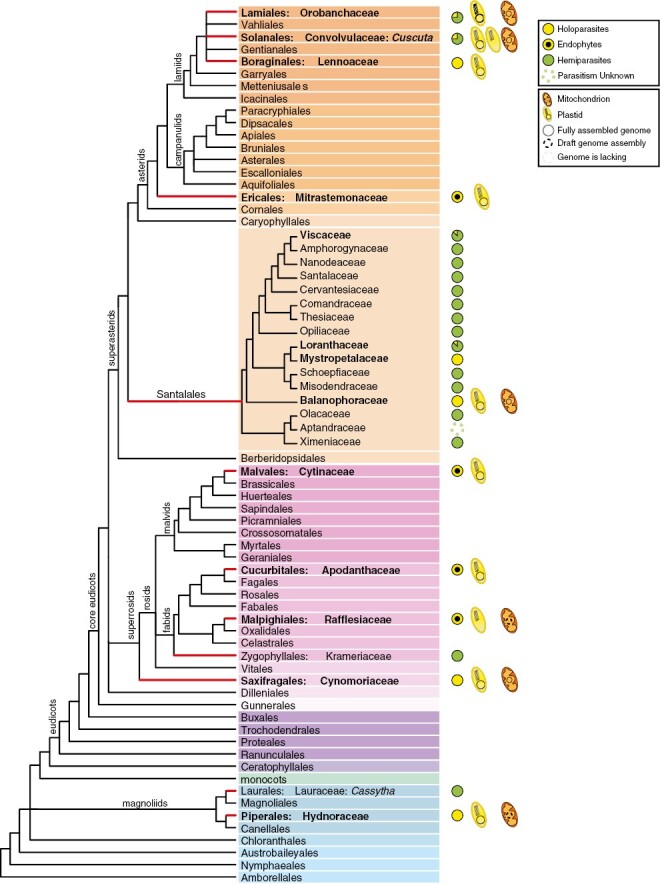
Phylogeny of angiosperms depicting parasitic lineages. Red branches depict lineages including parasitic plants. Families that contain holoparasitic plants are in boldface. The availability of organellar genomic resources of holoparasites is indicated by a schematic plastid or mitochondrion. A pie chart next to the parasitic lineages shows the proportion of hemi- and holoparasitic taxa. The phylogeny follows Angiosperm Phylogeny Group IV (2016) classification and the phylogeny of Santalales is based on Nickrent (2020).
The transition to a heterotrophic lifestyle is accompanied by numerous morphological and physiological changes that have occurred independently in distant angiosperm lineages, indicating the existence of highly convergent evolutionary trajectories. These changes include reductions in vegetative body, photosynthetic tissue and chlorophyll content. At the genetic level, convergent traits are also observed, particularly in the plastid genome. These convergent features are known as the parasitic reduction syndrome, and are shared by all heterotrophic plants, regardless of the feeding type (Wicke et al., 2013; Graham et al., 2017; Wicke and Naumann, 2018).
The phylogenetic affiliations of parasitic lineages have been a subject of conflict, but it is now widely accepted that the parasitic lifestyle originated 12 times independently in the evolution of angiosperms (Fig. 1). Parasitic lineages exhibit notable variability in terms of the number of species and range of nutritional modes, including hemiparasites (i.e. parasites able to photosynthesize) and holoparasites (i.e. completely non-photosynthetic parasites). For example, Orobanchaceae and Santalales have experienced remarkable expansion with more than 2000 described species, whereas others, such as Cynomoriaceae and Mitrastemonaceae, have only one or two described species (Nickrent, 2020). Among the 12 taxonomic lineages of parasitic plants, seven include exclusively holoparasites and two only contain hemiparasites (Krameriaceae and the genus Cassytha). The remaining three lineages (Santalales, Orobanchaceae and the genus Cuscuta) encompass both hemi- and holoparasites (Fig. 1).
The transition to a parasitic life form occurred at different times during the evolution of angiosperms, leading to the emergence of both older and more recent parasitic lineages. Santalales are the oldest (~109 Mya), along with Hydnoraceae (~101 Mya) and Cynomoriaceae (~100 Mya). These are followed by Rafflesiaceae (65–95 Mya), Cytinaceae (~72 Mya), Mitrastemonaceae (~78 Mya), Apodanthaceae (65-81 Mya), Krameriaceae (61.8 Mya), Cassytha (77 Mya) and Lennoaceae (40-67 Mya) (Naumann et al., 2013; Xi et al., 2013; Magallón et al., 2015; Bellot and Renner, 2016; Rose et al., 2018). The youngest parasitic lineages include Cuscuta and Orobanchaceae that originated independently 35–38 Mya (Naumann et al., 2013; Magallón et al., 2015; Xu et al., 2022).
HOLOPARASITIC ANGIOSPERMS
Holoparasitism is the most extreme form of parasitism in plants, whereby parasitic plants lack photosynthetic activity and are completely reliant on their host for their nutritional needs. Even though ten of the 12 parasitic angiosperm lineages include holoparasitic taxa (Fig. 1), holoparasites represent ~10 % of the parasitic species, and invade either the stem or the roots of the host plants. Some holoparasites have an endophytic stage that dominates their life cycle, and four parasitic families are exclusively endoparasitic, i.e. they only grow within their host for the majority of their lifespan (Teixeira-Costa and Davis, 2021). Holoparasitism has arisen about 15 times during the evolution of angiosperms, once in Apodanthaceae, Cynomoriaceae, Cytinaceae, Hydnoraceae, Lennoaceae, Mitrastemonaceae and Rafflesiaceae, twice in Cuscuta, three to five times within the Orobanchaceae, and twice within the Santalales leading to Balanophoraceae s.s. and Mystropetalaceae (Banerjee and Stefanović, 2019; Nickrent, 2020; Xu et al., 2022). Other Santalales approaching holoparasitism have also been described, such as the endoparasites Tristerix aphyllus (Loranthaceae) (Mauseth et al., 1984), Arceuthobium spp. (Viscaceae) (Nickrent and García, 2009) and Viscum minimum (Viscaceae) (Mauseth and Rezaei, 2013). The genus Cuscuta encompasses ~200 species, a number of which are considered functionally holoparasitic (van der Kooij et al., 2000; Banerjee and Stefanović, 2019).
Among the holoparasitic angiosperms, those in Balanophoraceae stand out for several reasons. First, it is one of the best studied holoparasitic lineages in terms of the number of characterized organellar genomes. Second, their plastomes exhibit unique features, even when considering all other living organisms: the highest AT content, the most biased codon usage and a novel genetic code change. Third, the mitochondrial genome of Lophophytum mirabile leads the ranking of functional horizontal transfers among those reported in the three domains of life.
The taxonomic complexity of the family Balanophoraceae: its position within the order Santalales and the phylogenetic relationships among its genera
The order Santalales is the largest lineage of parasitic plants comprising ~50 % of all parasitic angiosperms with 2428 species (Barkman et al., 2007; Su et al., 2015; Nickrent, 2020). It is the only order with more than one family of parasitic plants, consisting of ~15 families with worldwide distribution (Nickrent, 2020). The morphological losses and reductions that are characteristic of parasitic plants, especially in those that lost the ability to perform photosynthesis, led to remarkable convergent traits hindering an accurate phylogenetic classification. Therefore, despite strong support for the monophyly of the order (Soltis et al., 1999, 2003), the placement of Santalales within the overall angiosperm phylogeny has been uncertain (Leebens-Mack et al., 2019; Li et al., 2019), and several reviews of their complex taxonomic history have been published (Kuijt, 1969; Nickrent et al., 2010; Kuijt and Hansen, 2015; among others). The association of Balanophoraceae with Santalales dates back to the 19th century (Eichler, 1867; Van Tieghem, 1896), but it was not until the advent of molecular data that the relationships and position of Balanophoraceae began to be clarified (Nickrent and Duff, 1996; Su et al., 2012, 2015; Ceriotti et al., 2021). The analysis of several molecular markers from the nucleus, plastid and mitochondrial compartments supported the position of the family within Santalales, and the separation of Balanophoraceae s.l. into two clades, Balanophoraceae s.s. and Mystropetalaceae (Su et al., 2015). The split of Balanophoraceae s.l. indicates that holoparasitism evolved independently in these two clades within Santalales, although their exact phylogenetic positions within the order remain to be elucidated.
The family Balanophoraceae s.s. consists entirely of root holoparasites with 13 genera (Exorhopala has been placed within Helosis; Eberwein and Weber, 2004) and 53 species distributed in tropical and subtropical regions. Unlike cormophytes, species of this family lack typical structures such as roots, stems and leaves. Instead, they develop an underground vegetative body called tuber (Gonzalez and Mauseth, 2010; Sato and Gonzalez, 2016). During reproduction, the endotrophic tissue develops a fleshy inflorescence, often cylindrical, elongated, with unisexual and tiny flowers, observed in both monoecious and dioecious species (Kuijt and Hansen, 2015; Gonzalez and Sato, 2016; Sato and Gonzalez, 2016). Balanophora is the most species-rich genus within the family, whereas most other genera are monotypic or encompass a low number of species (Fig. 2).
Fig. 2.
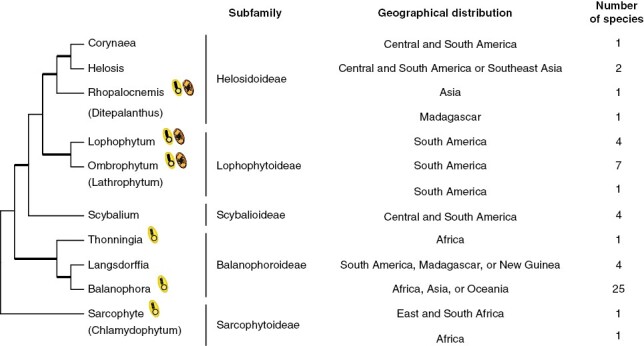
Phylogeny and features of Balanophoraceae. Maximum-likelihood phylogenetic tree based on a concatenated alignment of nuclear (rDNA operon) and mitochondrial (matR) sequences. No molecular data are available for Ditepalanthus, Lathrophytum or Chlamydophytum. Thick and thin branches indicate strongly or weakly supported relationships with bootstrap support values above or below 90 %, respectively. Details of the phylogenetic analysis are shown in Supplementary Data Figure S1. The availability of organellar genomic resources of holoparasites is indicated by a schematic plastid (in yellow) or mitochondrion (in orange). The geographical distribution and number of species was taken from https://powo.science.kew.org/.
Relationships among the genera of Balanophoraceae based on molecular data (Su et al., 2012, 2015; Ceriotti et al., 2021; Kim et al., 2023) are generally concordant with groups recognized in morphology-based classifications (Hansen, 1980; Takhtajan, 2009; Kuijt and Hansen, 2015), although a thorough analysis including all genera is still lacking. We performed a maximum-likelihood phylogenetic analysis (Fig. 2 and Supplementary Data Fig. S1) using a concatenated data set of nuclear (rDNA operon) and mitochondrial (matR) sequences, which represent all markers available in public databases encompassing most Balanophoraceae (Supplementary Data Table S1). The tree includes all genera except for Chlamydophytum, Ditepalanthus and Lathrophytum, for which no molecular data are available. The topology shown in Fig. 2 agrees with previous studies that included a smaller number of species or were based on smaller data sets (Su et al., 2012, 2015; Schelkunov et al., 2019; Ceriotti et al., 2021; Kim et al., 2023). The relationships among eight of the ten genera are well resolved, except for Sarcophyte and Scybalium, which show poorly supported affiliations. Overall, the tree shows that Lophophytum and Ombrophytum [and probably Lathrophytum based on morphological data (Hansen, 1980; Kuijt and Hansen, 2015)] represent a monophyletic group (named subfamily Lophophytoideae) sister to a clade formed by the subfamily Helosidoideae. On the other hand, Balanophora, Langsdorffia and Thonningia form a separate monophyletic group, recognized as the subfamily Balanophoroideae. Balanophora and Langsdorffia form a well-supported clade in the tree in contrast to previous classifications based on flower morphology that placed Langsdorffia as sister to Thonningia (Harms, 1935; Takhtajan, 2009). However, similarities in embryo sac development and the morphology of the pollen between Balanophora and Langsdorffia were recognized (Kuijt and Hansen, 2015) and agree with the molecular phylogeny.
Morphological analyses place Sarcophyte and Chlamydophytum as monophyletic due to their conspicuously branched inflorescences and sessile stigma (Hansen, 1980; Kuijt and Hansen, 2015). The phylogenetic tree shows the subfamily Sarcophytoideae as the earliest diverging branch, although with low bootstrap support. A recent phylogeny based on a large plastid gene data set also found Sarcophyte as sister to all other Balanophoraceae plastomes with strong support (Kim et al., 2023). The affiliation of Scybalium as sister to the subfamilies Helosidoideae and Lophophytoideae recovered in the phylogenetic analysis with strong support agrees with the presence of shared morphological characters, such as the unbranched inflorescences and the presence of flowers with two stylodia and starch in tubers (Hansen, 1980; Kuijt and Hansen, 2015). The advent of DNA sequences, especially from plastids, from all the described genera in the family will help to fully resolve the speciation events in the family Balanophoraceae.
ORGANELLE GENOMES OF HOLOPARASITES
Over 50 holoparasitic genera have been identified (Nickrent, 2020), but only a few of them have been studied at the genomic level (Table 1 and Supplementary Data Note S1). Complete mitochondrial genomes (mtDNAs) have been sequenced from representatives of nine holoparasitic genera from four different lineages, whereas plastid genomes (cpDNAs) have been reported from representatives of 28 genera from nine lineages (Table 1).
Table 1.
Organellar genomes in angiosperm holoparasitic lineages.
| Holoparasitic lineages and genera | Santalales, Balanophoraceae s.str.: 13 genera c |
Santalales, Mystropetalaceae: Dactylanthus, Hachettea, Mystropetalon | Piperales, Hydnoraceae: Hydnora, Prosopanche | Saxifragales, Cynomoriaceae: Cynomorium | Malpighiales, Rafflesiaceae s. str.: Rafflesia, Rhizanthes, Sapria | Malvales, Cytinaceae: Cytinus, Bdallophytum (including Sanguisuga) | Ericales, Mitrastemonaceae: Mitrastemon | Cucurbitales, Apodanthaceae: Apodanthes, Pilostyles | Boraginales, Lennoaceae: Lennoa, Pholisma |
Solanales, Convolvulaceae: Cuscuta (~200 spp) d | Lamiales, Orobanchaceae: 22 genera with holoparasitic species e |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type of parasitism | root parasite | root parasite | root parasite | root parasite | stem or root endophyte | root endophyte | root endophyte | stem endophyte | root parasite | stem parasite | root parasite |
| cpDNAs available in public databases? a | 10 species in 6 genera | none | 9 species in 2 genera | Cynomorium coccineum | Rafflesiaceae lacks a cpDNA | Cytinus hypocistis | Mitrastemon kanehirai, M. yamamotoi | Pilostyles aethiopica, P. hamiltonii | Lennoa madreporoides, Pholisma arenarium | 34 species of Cuscuta | 30 species in 13 genera |
| Main features of the cpDNAs | cpDNA size: 15,130 - 28,384 bp Gene content: 14 - 27 genes GC content: 11.56 - 21.2% Inverted repeats: absent trnE: present or absent Genetic code change: yes (TGA>Trp or TAG>Trp) or no |
NA | cpDNA size: 24,479 - 28,658 bp Gene content: 25 - 27 genes GC content: 20.4 - 24.1% Inverted repeats: present or absent (H. esculenta and Prosopanche spp.) trnE: present Genetic code change: no |
cpDNA size: 45,519 bp. Gene content: 27 genes GC content: 30% Inverted repeats: present trnE: present Genetic code change: no |
NA | cpDNA size: 19,400 bp. Gene content: 23 genes GC content: 29.9% Inverted repeats: absent trnE: present Genetic code change: no |
cpDNA size: 18,252 - 25,740 bp. Gene content: 19 - 24 genes GC content: 22.45 - 25.1% Inverted repeats: absent trnE: present Genetic code change: no |
cpDNA size: 11,348 - 15,167 bp Gene content: 5 - 6 genes GC content: 22.7 - 24.2% Inverted repeats: absent trnE: absent Genetic code change: no |
cpDNA size: 81,198 - 83,675 bp Gene content: 60 genes GC content: 37.1 - 38.1% Inverted repeats: present trnE: present Genetic code change: no |
cpDNA size: 60,959 - 125,373 bp. Gene content: 84 -103 genes GC content: 35 - 38.3% Inverted repeats: present or absent (in C. approximata and C. pedicellata) trnE: present Genetic code change: no |
cpDNA size: 45,673 - 150,504 bp Gene content: 21 - 48 genes GC content: 31.09 - 38.13% Inverted repeats: present or absent (in Conopholis americana and Phelipanche ramosa) trnE: present Genetic code change: No |
| Complete mtDNAs available in public databases? a | Lophophytum mirabile, Ombrophytum subterraneum, Rhopalocnemis phalloides | none | none | Cynomorium coccineum | none | none | none | none | none | C. australis, C. campestris, C. epilinum, C. europea, C. japonica | Aeginetia indica, Boschniakia himalaica, B. rossica, Christisonia kwangtungensis, Cistanche spp. |
| Main features of the mtDNAs b | mtDNA size: 170,713 - 821,919 bp Structure: multiple circular chromosomes GC content: 44.2 - 45% HGT: numerous genic and intergenic foreign regions from their hosts in Lophophytum spp. and O. subterraneum |
NA | NA (HGT: 2 chimeric genes in draft mtDNAs) |
mtDNA size: 1,106,389 bp Structure: multiple circular molecules GC content: 44.3% HGT: 5 foreign genes and intergenic DNA (MTPTs) |
NA (HGT: several mitochondrial genes and intergenic regions, e.g. MTPTs in draft mtDNAs) |
NA | NA | NA | NA | mtDNA size: 125,641 - 813,731 bp. Structure: circular GC content: 43.4 - 44.6% HGT: intergenic DNA |
mtDNA size: 401,628 - 3,978,341 bp Structure: one or more circular molecules GC content: 43.5 - 47.5% HGT: intergenic DNA. |
aDetailed information in Note S1.
bMTPTs refer to plastid derived DNA located within the mitochondrial genome.
c13 genera: Balanophora, Chlamydophytum, Corynaea, Ditepalanthus, Helosis, Lathrophytum, Langsdorffia, Lophophytum, Ombrophytum, Rhopalocnemis, Sarcophyte, Scybalium, Thonningia.
dSome species of Cuscuta are considered functionally holoparasitic
e22 genera: Aeginetia, Alectra (1sp; A. orobanchoides), Aphyllon, Boschniakia, Boulardia, Christisonia, Cistanche, Conopholis, Phelypaea, Epifagus, Eremitilla, Gleadovia, Harveya, Hyobanche, Kopsiopsis, Lathraea, Mannagettaea, Myzorrhiza, Orobanche, Phacellanthus, Phelipanche, Striga (some species).
Convergent traits in the plastomes of holoparasitic plants
The transition from an autotrophic to a heterotrophic lifestyle had a profound impact on plastid genomes, resulting in genomic features that differ significantly from those of free-living plants. This is particularly evident in holoparasitic plants, as the loss of photosynthetic capacity is associated with a relaxation of the evolutionary pressures on the plastome. These changes are essentially convergent and characterized by rampant gene loss, accelerations of molecular evolutionary rates and biased nucleotide compositions, in sharp contrast to the highly conserved plastid genomes of free-living plants (Wicke and Naumann, 2018). The reported plastomes from 28 holoparasitic genera share a reduction in gene content and genome size, primarily due to photosynthesis-related gene losses, and a decrease in overall GC content ranging between 11.56 % in Balanophoraceae and 38 % in Lennoaceae, Cuscuta and Orobanchaceae (Table 1). This contrasts with photosynthetic plants, which exhibit a genome size of 120–170 kb carrying 120–130 genes, and a GC content of 35–40 % (Ruhlman and Jansen, 2014). While the conserved large inverted repeats are believed to stabilize the plastome (Palmer and Thompson, 1982; Maréchal and Brisson, 2010), the typical quadripartite structure has been perturbed in species from seven holoparasitic lineages through the loss of the inverted repeats (Table 1). These changes often lead to genomic rearrangements (Wicke et al., 2013; Bellot and Renner, 2016; Roquet et al., 2016; Chen et al., 2019; Schelkunov et al., 2019; Su et al., 2019; Ceriotti et al., 2021; Jost et al., 2022). In addition, the complete loss of the plastid genome has been described in the Rafflesiaceae (Molina et al., 2014; Cai et al., 2021) and recently suggested for species in section Subulatae within the genus Cuscuta subgenus Grammica (Banerjee and Stefanović, 2023).
Plastid evolution in Balanophoraceae: extreme nucleotide composition, strong gene content reduction, and genetic code changes.
The complete loss of photosynthetic ability in Balanophoraceae led to the most extreme plastomes known to date. Initially, it was believed that Balanophoraceae had lost their plastids completely (Gedalovich-Shedletzky and Kuijt, 1990; Nickrent et al., 1997). However, recent reports of the plastid genome in Balanophora and ultrastructural observations of these organelles (Chen et al., 2019; Su et al., 2019), followed by the sequencing of plastomes from Rhopalocnemis (Schelkunov et al., 2019), Lophophytum and Ombrophytum (Ceriotti et al., 2021), and Sarcophyte and Thonningia (Kim et al., 2023), provide ample evidence to support the presence of reduced and aberrant plastids with even more odd plastomes (Fig. 3). Transmission electron micrographs of ovarian cells of Lophophytum pyramidale show the presence of non-photosynthetic plastids that accumulate starch (Fig. 3C), which are also observed with light microscopy of cells stained with iodine (Fig. 3D) (Sato and Gonzalez, 2016; Torres et al., 2021). Starch-containing plastids in Lophophytum contrast with the description of Balanophora plastids that exhibit only oil droplets (Su et al., 2019). Additionally, in the tissues of Balanophora and Lophophytum, large quantities of tannins were observed (Hsiao et al., 1995; Torres et al., 2021), which seem to have originated from plastids called tannosomes (Brillouet et al., 2013).
Fig. 3.

Lophophytum pyramidale. (A) Map of the plastome with the main features shown in the centre. (B) Inflorescences of L. pyramidale growing in the province of Misiones (Argentina). Yellow and whitish flowers are male and female, respectively. (C) Transmission electron micrograph of an ovarian cell. The arrow points to the non-photosynthetic plastid. (D) Light micrograph of a cell from the female flower stained with iodine. Arrowheads depict starch granules.
To date, six genera of the Balanophoraceae have their plastomes sequenced (Table 2). These plastomes have undergone significant reductions in size and gene content, and their nucleotide compositions are highly biased (Chen et al., 2019; Schelkunov et al., 2019; Su et al., 2019; Ceriotti et al., 2021; Kim et al., 2023). In fact, compared to hemiparasitic Santalales and free-living plants, more than 80 and 90 genes are missing, respectively. This severe gene loss probably took place in the common ancestor of Balanophoraceae, although additional gene losses occurred during species diversification (Ceriotti et al., 2021; Kim et al., 2023). It is worth noting that Sarcophyte, which diverged early in the evolution of the family (Kim et al., 2023), presents the largest gene content among the examined Balanophoraceae plastomes (Table 2). Interestingly, Balanophora spp. achieved an exceptional level of genome compaction, resulting from shrunken intergenic regions, genes that overlap and the outright loss of cis-introns in the retained genes (Chen et al., 2019; Su et al., 2019).
Table 2.
Plastid genomes of Balanophoraceae.
| Species | Lophophytum pyramidale | Ombrophytum subterraneum | Rhopalocnemis phalloides | Balanophora spp. a | Thonningia sanguinea b | Sarcophyte sanguinea b |
|---|---|---|---|---|---|---|
| cpDNA size | 20,892 bp | 17,313 bp | 18,622 bp | 15,130 - 16,056 bp | 18,560 - 19,013 bp | 28,372 - 28,384 bp |
| Gene content | 14 | 15 | 15 | 18-20 | 22 | 27 |
| GC (%) | 11.56 | 14.05 | 13.21 | 11.61 - 12.95 | 20.4 - 21.2 | 19.1 |
| Genetic code | UGA -> W | UGA -> W | standard | UAG -> W | UAG -> W | standard |
| Accession numbers | MT834848 | MT834847 | MK036331 | MN414176, MN414177, KX784265, KX784266, MT901289 | OQ810029-OQ810031 | OQ810027- OQ810028 |
| References | Ceriotti et al. 2021 | Ceriotti et al. 2021 | Schelkunov et al. 2019 | Chen et al. 2020; Su et al. 2019 | Kim et al. 2023 | Kim et al. 2023 |
aPlastid genomes of five species of Balanophora are available: B. fungosa, B. harlandii, B. laxiflora, B. reflexa, B. yakushimensis.
bThree and two individuals of Thonningia sanguinea and Sarcophyte sanguinea are available, respectively.
A major feature shared by all plastomes of the family is the low GC content (Schelkunov et al., 2019; Su et al., 2019; Ceriotti et al., 2021). The dramatic nucleotide bias probably occurred in the ancestor of the Balanophoraceae and became stronger after the divergence of Sarcophyte, as all other genera, except for Thonningia, exhibit GC contents < 14.05 % (Table 2) compared to the 36–38 % observed in other Santalales. This reduction in GC content has resulted in protein genes and proteins that are barely recognizable by comparative tools, and some Balanophoraceae exhibit the most biased codon usage and nucleotide compositions across the tree of life (Su et al., 2019). Only the plastomes of Apicomplexa (apicoplasts) and some Proteobacteria show comparable levels of nucleotide bias (Wilson et al., 1996; Su et al., 2019). The bias in the Balanophoraceae plastomes is likely to be the result of mutational forces, probably due to a relaxation and subsequent erosion of the maintenance machinery (Schelkunov et al., 2021; Ceriotti et al., 2022) and to genetic drift (Su et al., 2019; Ceriotti et al., 2021).
Another outstanding finding is that the only two genetic code changes found in plants occur in Balanophoraceae. Specifically, TAG and TGA, which are typically stop codons, code for tryptophan in the plastomes of Balanophora–Thonningia and Lophophytum–Ombrophytum, respectively. The reassignment of the stop codon UGA to a tryptophan coding codon is the most common genetic code change in eukaryotes, and is mainly found in mitochondrial genomes (Yokobori et al., 2010). In contrast, the Balanophora–Thonningia change has never been described before. The codon capture theory (Osawa and Jukes, 1989) has been favoured as the most plausible explanation for both genetic code changes (Su et al., 2019; Ceriotti et al., 2021). This hypothesis consists mainly of three steps: codon disappearance from the genome (probably due to gene loss and nucleotide bias), the stop codon is no longer recognized (which is supported by the loss of specific release factors in Lophophytum and Balanophora; Ceriotti, et al. 2021) and, finally, reassignment of the codons. For this last step, several unexplored complex scenarios involving tRNAs and related enzymes are possible, especially for Balanophora spp. and Thonningia (Su et al., 2019; Ceriotti et al., 2021). The family Balanophoraceae displays exceptional variation regarding plastid tRNA biology, which makes it a promising lineage to explore unknown aspects of tRNA metabolism in plastids and to capture the transitional steps that result in the functional replacement of plastid trnE and eventually lead to the loss of the entire cpDNA.
The minimal plastome for survival.
The characteristics of the minimum plastome needed for survival (if they exist) remain a matter of debate (Barbrook et al., 2006; Molina et al., 2014; Su et al., 2019). To date, the complete loss of the cpDNA has been described in only a few non-photosynthetic eukaryotes, namely the Rafflesiaceae and Cuscuta subgenus Grammica section Subulatae among angiosperms (Molina et al., 2014; Cai et al., 2021; Banerjee and Stefanović, 2023), and four distantly related protists and algae: Polytomella (Chlorophyta; Smith and Lee, 2014), Paraphysomonas (Chrysophyta; Dorrell et al., 2019), Perkinsus (Alveolata; Fernández Robledo et al., 2011) and Rhodelphis (sister to Rhodophyta; Gawryluk et al., 2019).
The plastid trnE gene is believed to be the raison d’être of maintaining a non-photosynthetic plastome due to its dual role in tetrapyrrole biosynthesis and translation (Barbrook et al., 2006). However, trnE is missing from the plastomes of Lophophytum, Ombrophytum and Rhopalocnemis (Schelkunov et al., 2019; Ceriotti et al., 2021), marking the second independent loss of this gene in angiosperms along with the holoparasitic Pilostyles spp. (Bellot and Renner, 2016). A functional substitution of the plastid trnE by the nuclear homologue has been proposed for these Balanophoraceae (Ceriotti et al., 2021), Polytomella (Smith and Lee, 2014) and Paraphysomonas (Dorrell et al., 2019). In Balanophora spp., however, a divergent anticodon-less trnE is present in all plastomes (Chen et al., 2019; Su et al., 2019), a feature only reported in the non-photosynthetic orchid Pogoniopsis schenckii (Klimpert et al., 2022). It has been suggested that the plastid trnE gene in Balanophora and Pogoniopsis functions in tetrapyrrole biosynthesis but not in translation (Su et al., 2019; Ceriotti et al., 2021; Klimpert et al., 2022), as evidenced by the loss of the anticodon and the conservation of most sequence determinants necessary for its recognition by tetrapyrrole synthesis enzymes (Ceriotti et al., 2021).
Evolutionary trends in the mitochondrial genomes of holoparasitic plants
The mitochondrial genome of holoparasitic plants is expected to be impacted to a lesser extent than plastomes by the loss of photosynthesis and a parasitic lifestyle. Angiosperm mitochondrial genomes are widely conserved in terms of gene content (Richardson et al., 2013), but their size and structure are dynamic, with genomes consisting of linear, circular or branched subgenomic molecules (Oldenburg and Bendich, 1996; Sloan, 2013; Kozik et al., 2019) varying in size up to 11.3 Mb (Sloan et al., 2012). Mitogenomes of parasitic plants have not been studied as extensively as plastomes. Complete mitogenomes are available from only nine genera that belong to four lineages of holoparasitic angiosperms, namely Cynomoriaceae, Balanophoraceae, Orobanchaceae and Cuscuta (Table 1). In addition, several partial mitochondrial assemblies have been reported (Table 1).
Comparisons of these mitogenomes show no evolutionary convergence in the structure, gene content, nucleotide composition, substitution rate or genome size as a result of their extreme lifestyle (Zervas et al., 2019; Petersen et al., 2020). Although initially, parasitic lineages were thought to exhibit accelerated substitution rates in all cell compartments (Bromham et al., 2013), recent analyses revealed that most parasites have ordinary or only slightly elevated rates in their mitochondrial genes, with a few exceptions (Skippington et al., 2015, 2017; Zervas et al., 2019; Ceriotti et al., 2022). Members of three holoparasitic lineages [Balanophoraceae, Boschniakia himalaica (Orobanchaceae) and Cynomorium coccineum (Cynomoriaceae)] exhibit a mitogenome assembled as multiple circular molecules (Bellot et al., 2016; Sanchez-Puerta et al., 2017; Roulet et al., 2020; Yu et al., 2022; Zhang et al., 2023), which has also been observed in some free-living angiosperms (Alverson et al., 2011; Sloan et al., 2012; Rice et al., 2013; Wu et al., 2015).
One aspect shared by the mitochondria of holoparasites is the presence of foreign DNA (Barkman et al., 2007; Xi et al., 2013; Davis and Xi, 2015; Bellot et al., 2016; Sanchez-Puerta et al., 2017; Petersen et al., 2020; Roulet et al., 2020). While many free-living angiosperms have occasionally acquired mtDNA from other plants (Richardson and Palmer, 2007; Bock, 2010; Wickell and Li, 2020), the impact of HGT is higher in parasitic plant mitochondria (Petersen et al., 2020). In particular, the Balanophoraceae Lophophytum mirabile takes foreign DNA content to new levels (see below).
Mitochondrial evolution in Balanophoraceae
Three mitogenomes of Balanophoraceae have been reported, revealing striking intrafamily differences (Table 3). In contrast to the radical features of their plastomes shared by all the Balanophoraceae analysed, the three mitogenomes investigated lack a nucleotide composition bias, have a similar gene content to that of free-living angiosperms and use the standard genetic code. Nevertheless, the mtDNAs exhibit other odd features that are either shared or variable within the family.
Table 3.
Mitochondrial genomes of Balanophoraceae.
| Species | Lophophytum mirabile | Ombrophytum subterraneum | Rhopalocnemis phalloides |
|---|---|---|---|
| mtDNA size | 821,906 bp | 713,777 bp | 130,713 bp |
| Protein-coding genes (including duplicates) | 35 (44) | 36 (51) | 36 |
| Mitogenome structure | 60 circular chromosomes | 54 circular chromosomes | 21 circular chromosomes |
| Non-coding chromosomes | 36 | 20 | 2 |
| GC (%) | 44.5 | 44.22 | 45.0 |
| Host-derived DNA (%) | from Fabaceae (59.1%), 29 foreign and 3 chimeric genes | from Fabaceae (15.4%); from Asteraceae (13.7%), 12 foreign genes | none reported |
| Heteroplasmy | no | no | yes |
| Accession numbers | KU992322-KU992380, KX792461 | MT076267-MT076320 | MZ269392-MZ269412 |
| References | Sanchez Puerta et al. 2017; 2019; Roulet et al. 2020 | Roulet et al. 2020 | Yu et al. 2022 |
The three genera share a particular genomic structure, i.e. a multichromosomal arrangement of their mitochondrial genome (Sanchez-Puerta et al., 2017; Roulet et al., 2020; Yu et al., 2022). The mitogenomes consist of 21–60 putatively autonomous circular chromosomes that range in size from 4.9 to 58 kb (Table 3). Similar to free-living species of Silene (Caryophyllaceae) (Wu et al., 2015), the mitogenomes of Balanophoraceae present 2–36 chromosomes that lack known mitochondrial genes (Table 3). These chromosomes may carry unidentified functional elements that are relevant for mitochondrial functions (Rice et al., 2013; Wu et al., 2015; Sanchez-Puerta et al., 2017). However, the variable presence of non-coding chromosomes across populations of Silene noctiflora (Caryophyllaceae) suggests that they are evolving by genetic drift (Wu et al., 2015). Comparable studies are necessary to test these hypotheses in Balanophoraceae.
Mitochondrial chromosomes in Rhopalocnemis are fewer and smaller (<8 kb) than those in Lophophytum and Ombrophytum (Table 3), with a much smaller mitogenome (131 kb vs. 714–822 kb). The difference in genome sizes is easily explained by the remarkable horizontal acquisition of mtDNA in both ancestral and independent events in Lophophytum and Ombrophytum (Sanchez-Puerta et al., 2017; Roulet et al., 2020). In addition, chromosomes in Rhopalocnemis mitochondria display a conserved arrangement (Fig. 4). The 21 minicircular chromosomes share an identical non-coding region, which carries a sequence capable of forming a stem loop and is believed to be the replication origin for the rolling circle replication mechanism (Yu et al., 2022). Minicircular organellar genomes with a similar structure, in which a replication origin is shared among chromosomes, were described in other eukaryotic lineages, such as the plastid of dinoflagellates (Zhang et al., 1999) or the mitochondria of lice (Shao et al., 2009) and nematodes (Gibson et al., 2007), among others. Such a particular structure is the result of convergent evolution in widely disparate eukaryotic lineages. In contrast, the mitochondrial chromosomes in Lophophytum and Ombrophytum do not have conserved repeats and the replication mechanism remains unknown. Another outstanding feature exclusive to Rhopalocnemis is the extreme level of heteroplasmy, mainly due to short indels, which are observed across the mitogenome, including frameshift mutations in the coding regions. This heteroplasmy is registered in the majority of the DNA sequence reads and in the RNA transcripts, indicating that the functional protein coding transcripts are the minority (Yu et al., 2022).
Fig. 4.
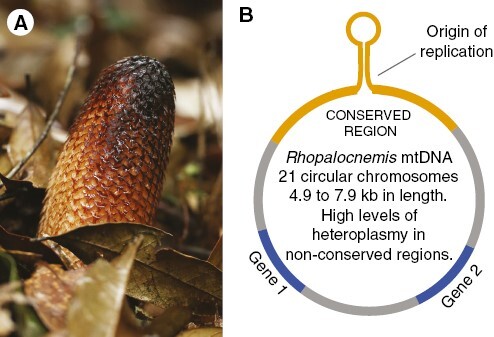
Rhopalocnemis phalloides. (A) Photo of an inflorescence of R. phalloides, courtesy of Runxian Yu. (B) Map of a mitochondrial chromosome of R. phalloides, depicting the conserved region (in orange) shared across chromosomes, which includes a region that can fold into a stem loop and is considered the origin of replication. Genes are shown in blue.
HGT IN HOLOPARASITIC PLANTS: A FREQUENT PROCESS IN MITOCHONDRIAL GENOMES
A hallmark of parasitic plants is the vascular connections established by haustoria, which allow for the transfer of water, nutrients and even nucleic acids, such as RNA and DNA, between parasitic plants and their hosts (Mower et al., 2010; Xi et al., 2013; Sanchez-Puerta, 2014; Davis and Xi, 2015; Góralski et al., 2021). Holoparasites, in particular, typically exhibit both xylem and phloem connections with their hosts (Gonzalez and Sato, 2016; Těšitel, 2016), establishing an intimate contact that facilitates the exchange of genetic information making them more susceptible to HGT (Mower et al., 2004, 2010; Davis et al., 2005; Barkman et al., 2007; Cusimano and Wicke, 2016; Yang et al., 2016). In addition to rare parasite-to-host transfer events (Mower et al., 2004, 2010; Davis et al., 2005), several reports of host-to-parasite HGT have accumulated over the years (Mower et al., 2004, 2010; Davis et al., 2005; Barkman et al., 2007; Xi et al., 2013; Cusimano and Wicke, 2016; Yang et al., 2016). The reasons for the larger impact of HGT on parasitic plants than on their hosts reside in the fact that parasites establish a connection with the host at an early stage of development and the acquired DNA may spread to germ cells and be transmitted to the mitochondria of offspring (Huang, 2012; Petersen et al., 2020). Petersen et al. (2020) compiled a comprehensive list of HGT events involving parasitic angiosperms.
Among the three DNA-containing compartments in plant cells, plant-to-plant HGT is far more common in the mitochondrial than in the chloroplast or nuclear genomes (Xi et al., 2013; Sanchez-Puerta, 2014; Davis and Xi, 2015). The lower frequency of HGT identified in the nuclear genome may be related to the limited availability of nuclear data. In fact, reports of HGT affecting nuclear genomes have increased in recent years (Dunning and Christin, 2020; Wickell and Li, 2020), while HGT in plastid genomes remains exceptional (Ma et al., 2015; Gandini and Sanchez-Puerta, 2017; Burke et al., 2018). Mitochondria from two free-living species, Amborella trichopoda (Rice et al., 2013) and Geranium brycei (Park et al., 2015), are particularly rich in HGT. Amborella, a flowering plant endemic to New Caledonia, stands out for having a 3900-kb mtDNA that carries foreign DNA equivalent to six mitochondrial genomes donated by green algae, mosses and other angiosperms, including parasitic Santalales (Rice et al., 2013).
The mobile cox1 intron
The most pervasive case of horizontal transfer among plant mitochondria involves an intron found in the mitochondrial cox1 gene (cytochrome c oxidase subunit 1) of many diverse and disparately related angiosperms (Cho et al., 1998). This group I intron was originally transferred to angiosperms from a fungal donor and subsequently spread among many diverse angiosperm lineages via hundreds (if not thousands) of plant-to-plant transfer events (Cho et al., 1998; Sanchez-Puerta et al., 2008, 2011). The cox1 intron is commonly found in parasitic plants (Barkman et al., 2007), including almost all holoparasites examined to date, with one exception in which an individual of Cynomorium songaricum has secondarily lost the intron (Sanchez-Puerta et al., 2011). However, it is uncertain whether these plants acquired the intron horizontally from their hosts or inherited it vertically from free-living ancestors (Barkman et al., 2007; Fan et al., 2016). Studies of the Orobanchaceae have shown that the cox1 intron was acquired by free-living ancestors (Fan et al., 2016), while family-wide analyses of the Hydnoraceae revealed independent acquisitions in the two holoparasitic genera Hydnora and Prosopanche, but the donor lineages remain unknown (Yu et al., 2023). Wider taxon sampling of free-living relatives is necessary to test these hypotheses across all plant parasitic lineages (Barkman et al., 2007; Fan et al., 2016).
Mitochondrial HGT in holoparasitic plants
Horizontal acquisition of mitochondrial genes from other plant mitochondria initially results in gene duplication in the recipient mtDNA. Foreign genes commonly coexist with native homologues leading to duplicative HGT, and the foreign copies generally become pseudogenes (Mower et al., 2010; Rice et al., 2013; Sanchez-Puerta, 2014). Occasionally, the foreign and native copies may undergo continuous or discontinuous gene conversion, generating chimeric gene copies, also known as chimeric or partial replacement HGT, which has been increasingly reported (Barkman et al., 2007; Hao et al., 2010; Mower et al., 2010; Sanchez-Puerta et al., 2019; Yu et al., 2021, 2023; Darshetkar et al., 2023). In rare cases, the native copy is lost and only the xenologue (foreign copy) remains, known as full replacement HGT. Despite the long list of horizontally acquired genes reported in angiosperm mitochondria, evidence of a functional role of these foreign genes is scarce (Bergthorsson et al., 2003; Hao et al., 2010; Garcia et al., 2021). Again, Lophophytum stands out as the angiosperm with the highest number of functional foreign mitochondrial genes (see below).
The impact of HGT across complete or draft mitogenomes of holoparasites has mainly been assessed in coding regions or in plastid-derived sequences (MTPTs) or not evaluated at all (Table 1). These mitogenomes exhibit a wide range of foreign DNA content, including coding and non-coding DNA, although intergenic DNA has rarely been examined (Bellot et al., 2016; Sanchez-Puerta et al., 2019; Roulet et al., 2020; Choi and Park, 2021; Lin et al., 2022a, b; Zhong et al., 2022). Among the holoparasites, the most striking examples are found within the Cynomoriaceae, the Rafflesiaceae and the Balanophoraceae exhibiting increasing levels of mitochondrial HGT. Foreign DNA might serve as a diagnostic of former host associations, as previously recognized in Boschniakia spp. (Zhang et al., 2023), Cynomorium coccineum (Cusimano and Renner, 2019) and Sapria himalayana (Cai et al., 2021).
Extent of mitochondrial HGT in holoparasitic lineages
The family Balanophoraceae exhibits divergent patterns of mitochondrial HGT, with no reported instances in Rhopalocnemis, and exceptional levels in the mitogenomes of Lophophytum and Ombrophytum (Sanchez-Puerta et al., 2017; Roulet et al., 2020). Extensive analyses of HGT across the entire mitogenomes of Lophophytum and Ombrophytum revealed that ~60 and 30 % of their mtDNA (including 29 and 12 foreign mitochondrial genes), respectively, was acquired from their hosts (Sanchez-Puerta et al., 2019; Roulet et al., 2020). However, these two species demonstrate disparate patterns in terms of the functional impact of HGT. While all foreign genes in Ombrophytum coexist with native homologues that presumably remain functional (Roulet et al., 2020), Lophophytum lost 20 native genes, which were functionally replaced by foreign homologues (Roulet et al., 2020; Garcia et al., 2021). In addition, Ombrophytum exhibits foreign DNA from Fabaceae and Asteraceae donors, both of which are known hosts of this holoparasite. Legume-derived DNA tracts are fragmented across the genome and seem to result from older horizontal transfer events. In contrast, more recently acquired DNA from Asteraceae is concentrated in nine mitochondrial chromosomes (Roulet et al., 2020).
Despite having only draft assemblies of their mitogenomes available, three species of Rafflesiaceae have been extensively studied. Foreign DNA in this lineage involves multiple mitochondrial genes, including five, five and 11 genes (28–41 %) acquired from their hosts by Rafflesia cantleyi, R. tuan-mudae and Sapria himalayana, respectively (Xi et al., 2013). Many of these foreign genes are transcribed and appear to have replaced the native homologues, but further analyses are needed to evaluate their functionality in these parasites (Garcia et al., 2021).
The mitogenome of the endophytic plant Cynomorium coccineum contains five foreign mitochondrial genes, as well as foreign plastid and nuclear-derived sequences obtained from two different plant host lineages (Bellot et al., 2016). While three foreign genes coexist with native homologues, atp1 and atp8 may have replaced the native copies (Bellot et al., 2016). However, the functionality of these genes was not analysed in depth. Evolutionary analyses of coding regions in the draft mitochondrial assemblies of the root holoparasites Hydnora and Prosopanche revealed a small incidence of HGT, with only short regions of the mitochondrial genes cox1 and atp8 replaced by foreign DNA (Yu et al., 2023).
The impact of mitochondrial HGT on holoparasites of the family Orobanchaceae is very small and limited to non-coding regions with no foreign mitochondrial genes reported (Choi and Park, 2021; Zhang et al., 2022, 2023; Zhong et al., 2022). Finally, the complete mitogenomes of five species of Cuscuta, C. australis, C. campestris (Anderson et al., 2021), C. apilinum, C. europea and C. japonica (Lin et al., 2022b), and several draft mitochondrial assemblies (Lin et al., 2022b) did not reveal any foreign genes. Nevertheless, mitochondrial intergenic regions of C. japonica were inferred as foreign based on similarity searches (Lin et al., 2022b). In a wide gene survey of the genus Cuscuta, atp1 was found to be chimeric in one of the 27 Cuscuta species examined (Lin et al., 2022a).
All these findings provide further evidence of the potential functional impact of mitochondrial HGT on holoparasitic plants and highlight the importance of further research to better understand the mechanisms and implications of HGT in parasitic plant evolution. To date, the numerous cases of horizontally transferred intergenic or non-functional genes reported in holoparasitic mitochondria indicate that functional HGT is extremely rare. However, elevated rates of mitochondrial horizontal transfer may occasionally result in exceptional cases of functional foreign genes, as observed in Lophophytum (Garcia et al., 2021).
Is mitochondrial HGT paralleled by nuclear HGT?
The large-scale horizontal transfer of mtDNA mainly from host plants to both Lophophytum and Ombrophytum (Sanchez-Puerta et al., 2017; Roulet et al., 2020) raises new questions that can be better addressed with the sequencing of nuclear genomes. Is the scale of nuclear HGT equivalent to that of mitochondrial HGT? Unfortunately, the nuclear genomes of species of Balanophoraceae have not been explored, and it does not seem to be an easy task since genome size estimations (e.g. 30 000 Mbp for Rhopalocnemis phalloides) indicate that their nuclear genomes are large (Schelkunov et al., 2019). Alternatively, the impact of HGT in the nuclear compartment can be evaluated based on transcriptomic data. Next, the role, if any, of the foreign nuclear genes in the holoparasites could be determined. Given the extent of mitochondrial HGT, we expect large-scale nuclear HGT in Lophophytum or Ombrophytum, but this aspect has not yet been analysed in depth. Likewise, the impact of HGT in the nuclear genomes of Cynomoriaceae or Hydnoraceae remains unknown.
Evolutionary analyses of nuclear HGT in parasitic plants of the Orobanchaceae, Rafflesiaceae and Cuscuta uncovered the expression of dozens of horizontally transferred nuclear genes (Xi et al., 2012; Yang et al., 2016, 2019; Vogel et al., 2018). Of those lineages, only Rafflesiaceae also exhibits massive mitochondrial HGT (Xi et al., 2013). A transcriptomic assay of Rafflesia cantleyi revealed more than 40 foreign nuclear genes (Xi et al., 2012), and ~1.2 % of the nuclear genome of S. himalayana was acquired from its hosts (Cai et al., 2021). Interestingly, while 84 nuclear genes were identified as foreign in the Orobanchaceae Aegenetia indica (Kado and Innan, 2018), its mitochondrial genome displays very limited evidence of host-to-parasite HGT and no mitochondrial genes are affected (Choi and Park, 2021; Zhong et al., 2022). Similarly, the mitochondrial genomes of some Cuscuta species were expected to be highly impacted by HGT, based on the identification of dozens of foreign genes in the nuclear genome of C. campestris, which were probably acquired from different donors (Vogel et al., 2018; Yang et al., 2019). However, the reported impact of mitochondrial HGT in Cuscuta spp. is minimal, as described above. This unexpected disparity between mitochondrial and nuclear HGT levels within a single species or lineage is intriguing. Uncommon features of their mitochondrial genomes, such as a small genome size or reduced rates of DNA incorporation, may contribute to lower levels of mitochondrial HGT (Anderson et al., 2021).
It has been proposed that many of the transferred genes in the nuclear genome may undergo neo-functionalization, such as roles related to invasive processes or to camouflage the invasion. Functional nuclear HGT events may represent an important evolutionary force, as they allow these organisms to expand their genetic tools and develop novel physiological capabilities (Yang et al., 2019). In three members of the family Orobanchaceae, six horizontally transferred genes related to defence responses against infection or insect toxins were detected (Yang et al., 2016). Cuscuta also has foreign genes that are related to enzymes that modify cell walls. These genes are expressed mainly in prehaustorial structures, suggesting a role in host invasion (Yang et al., 2019). Comparisons between foreign nuclear genes in Cuscuta spp. and Orobanchaceae revealed functional and transcriptional convergence. Approximately 18 foreign nuclear genes were independently acquired from their hosts by these two parasitic lineages (Yang et al., 2019). The extent of evolutionary convergence with other holoparasitic lineages remains to be elucidated.
THE CURIOUS CASE OF LOPHOPHYTUM MIRABILE
The mitogenome of Lophophytum mirabile (Balanophoraceae) stands out as being the first case of large-scale replacement HGT, in which two-thirds of the protein-coding genes have functionally supplanted the native homologues (Sanchez-Puerta et al., 2017; Garcia et al., 2021). Replacement HGT of the scale found in Lophophytum mtDNA is unprecedented for any genome or organism, including bacteria. A recipient cell faces potential barriers for the expression of foreign genes so that only rarely does a horizontally transferred gene become functional in a new genome. These barriers include recognizing a foreign promoter, accurately splicing foreign introns, and identifying and editing novel RNA editing sites. A total of 23 foreign protein-coding mitochondrial genes in Lophophytum have successfully overcome these challenges, leading to the functional replacement of native copies (Garcia et al., 2021). The use of host-derived genes may have a positive effect on the host–parasite relationship, but could also be the result of non-adaptive forces that led to neutral replacement despite a minor reduction in fitness, or even be deleterious by generating cytonuclear incompatibilities (Garcia et al., 2021; Gatica-Soria et al., 2022). These replacements raise questions concerning the factors that might enable or promote such a remarkably high level of foreign gene takeover, despite the low likelihood of overcoming the aforementioned expression barriers and the expected reluctance due to the risk of disrupting coevolved cytonuclear interactions.
Doubly chimeric OXPHOS complexes in Lophophytum
Several of the functional foreign genes in Lophophytum encode subunits of mitochondrial complexes, which are also formed by other subunits encoded either in the mitochondrial or nuclear genomes. For example, the oxidative phosphorylation system (OXPHOS) of plants, responsible for cellular respiration, requires the interaction of up to 20 mitochondrial-encoded proteins and 71 nuclear-encoded proteins (Meyer et al., 2019) generating multiple opportunities for mitonuclear coevolution. Thus, the replacement of coadapted subunits within OXPHOS is predicted to lead to cytonuclear incompatibilities. The complexity hypothesis establishes that genes that encode proteins that form part of multisubunit complexes are less likely to suffer functional replacement by HGT given the potential disruption of coevolved subunits (Jain et al., 1999; Cohen et al., 2011). In particular, proteins that have extensive molecular interactions are more recalcitrant to functional replacement. In contrast, other proteins present incredible tolerance to the functional replacement of native genes by foreign homologues, with no apparent negative effects (Woese et al., 2000; Adams and Palmer, 2003; Rice and Palmer, 2006; Sloan et al., 2022). That is, some systems are able to preserve the interchangeability of their components without generating incompatibilities (Sorek et al., 2007; Sloan et al., 2022). Four parameters that determine the incompatibility or interchangeability of systems have been proposed: the number of protein–protein interactions, the sensitivity to gene dosage, the evolutionary rate of the proteins and the relevance of the functional role (Sloan et al., 2022). The multisubunit structure, the fundamental functional role of OXPHOS and the elevated expression levels contribute to the prediction that subunits of these complexes are less likely to be interchangeable and thus less likely to suffer functional replacement by foreign homologues (Park and Zhang, 2012; Sloan et al., 2022). In fact, it has been shown that in hybridization events or in cytoplasmic hybrids (cybrids) produced in the laboratory, cytonuclear incompatibilities arise even between closely related species (Schmitz-Linneweber et al., 2005; Meiklejohn et al., 2013). A single nucleotide difference in mitochondrial genes could be responsible for strong phenotypic changes due to the disruption of OXPHOS complexes (Kim et al., 2009; Meiklejohn et al., 2013; Dahan et al., 2014; Qi et al., 2017).
Interestingly, the OXPHOS complexes are doubly chimeric in Lophophytum in terms of both the cell compartment in which the genes reside and the phylogenetic origin of the genes, as shown in Fig. 5B. Eleven OXPHOS subunits encoded in the mitochondrial genome of Lophophytum are partially or fully foreign, while eight mitochondrial-encoded subunits are native (Sanchez-Puerta et al., 2017; Garcia et al., 2021). Additionally, all but one (sdh3) of the nuclear-encoded subunits have a native origin (Gatica-Soria et al., 2022). Despite the expected tight coordination between nuclear and mitochondrial genes, Lophophytum does not display signs of cytonuclear incompatibilities in the OXPHOS that may have arisen from the functional replacement of many mitochondrial subunits (Ceriotti et al., 2022; Gatica-Soria et al., 2022). Physiological studies have revealed that Lophophytum exhibits levels of canonical and alternative respiration comparable to those of free-living angiosperms (L. M. Gatica-Soria et al., unpublished results), in contrast to hemiparasites of the genus Viscum (Santalales), which lack OXPHOS complex I and use alternative oxidases (Maclean et al., 2018; Senkler et al., 2018; Petersen et al., 2022). This unexpected replacement of mitochondrial subunits without apparent disruptive consequences has been explained by the high similarity of the mitochondrial-encoded OXPHOS subunits donated by the host and those native to Balanophoraceae, which minimizes the incompatibilities in the assembly and functioning of these multiprotein complexes (Gatica-Soria et al., 2022), as a low substitution rate has been proposed to mitigate the potential incompatibilities (Sloan et al., 2022).
Fig. 5.
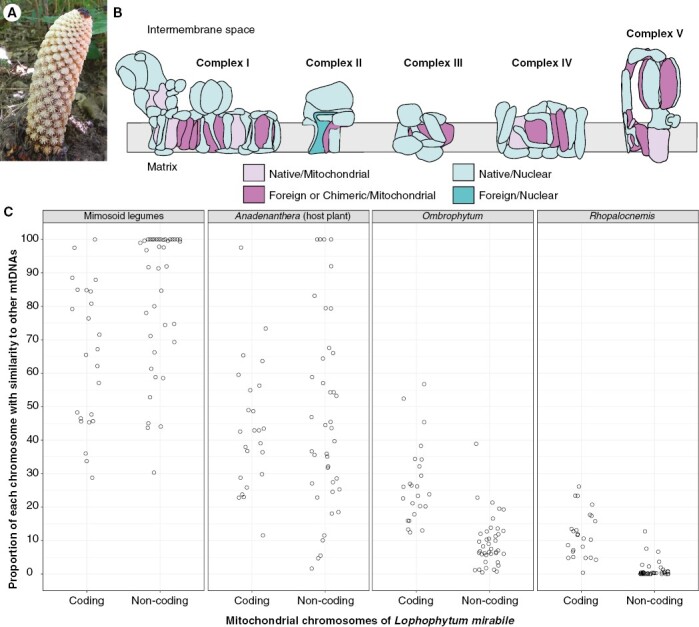
Lophophytum mirabile. (A) Inflorescence of L. mirabile growing in Jujuy (Argentina). Only the staminate flowers emerged above the soil level; female flowers are located below them. (B) Diagram of the internal membrane of the mitochondria with the five OXPHOS complexes of L. mirabile depicting the subunits coloured according to their phylogenetic origin and the genome in which the genes are located. Figure modified from Gatica-Soria et al. (2022). (C) Proportion of each of the mitochondrial chromosomes of L. mirabile with similarity (>80 %) to the mitogenomes of mimosoids, the actual host plant Anadenanthera colubrina, or to the close relatives Ombrophytum subterraneum and Rhopalocnemis phalloides.
In contrast, nuclear-encoded subunits of the OXPHOS are less conserved and thus less likely to be replaced. Nonetheless, the foreign nuclear-encoded subunit (SDH3) has functionally replaced the native subunit in Lophophytum (Garcia et al., 2021). Within OXPHOS complex II, SDH3 interacts extensively with SDH4, which is mitochondrial-encoded (SDH4) and foreign in Lophophytum. The cooccurrence of these two foreign genes acquired from mimosoid hosts could be the result of gene cooperation (Gatica-Soria et al., 2022), as the replacement of coevolved subunits often requires the parallel replacement of other interacting subunits to preserve compatible interactions (Waller et al., 2006; Monier et al., 2009).
Foreign DNA in non-coding regions of the Lophophytum mitogenome
Deciphering the entire mitochondrial genome of Lophophytum also revealed that it acquired over 60 % of the non-coding sequences from mimosoid hosts, even with a single mimosoid mitogenome available for comparison (Choi et al., 2019; Sanchez-Puerta et al., 2019). Today, DNA from six mimosoid mitogenomes has been sequenced, including the actual host of L. mirabile, Anadenanthera colubrina (M.E. Roulet et al., unpublished results). Similarity searches with each of the 60 mitochromosomes of Lophophytum against the mitogenomes of mimosoids, Anadenanthera, or the close relatives Ombrophytum and Rhopalocnemis uncovered a stunning picture (Fig. 5C). The proportion of mtDNA of Lophophytum with identity greater than 80 % when compared with their close relatives is strikingly low (4–10 % on average) in those chromosomes containing known genes and even lower in the non-coding chromosomes (0.7–6 % on average). These values are lower than expected for intrafamily comparisons of angiosperm mitochondrial genomes (Alverson et al., 2010; Choi et al., 2019; Gandini et al., 2019). However, what is particularly shocking is the large number of chromosomes with most of their sequences shared with legume mitochondrial genomes. In fact, there are at least 15 mitochondrial chromosomes of Lophophytum composed fully of mimosoid-like mtDNA, which is especially observed among non-coding chromosomes. Although genus-wide and population-level studies are still missing in Lophophytum, horizontal events may have occurred not only recently, but also in the past from ancestral legume hosts. This is supported by the relatively low number of chromosomes shared with the actual host Anadenanthera in comparison to those shared with the six mimosoids combined (Fig. 5C).
A ‘mitochondrial fusion-compatibility’ model has been proposed to explain the horizontal transfer of DNA between plant mitochondria from unrelated land plants. This model suggests that complete mitochondria enter a foreign plant cell and that the mitogenomes recombine with the native ones (Rice et al., 2013). In host–parasite relationships, entire organelles may move through the parasite haustoria, as observed in tissue grafts (Gurdon et al., 2016; Hertle et al., 2021). Lophophytum provides strong evidence for the fusion-compatibility model of mitochondrial HGT (Rice et al., 2013). However, the foreign DNA in Lophophytum mitochondria may not always or immediately recombine with the resident genome and may be perpetuated as separate molecules (M. E. Roulet et al., unpubl. data).
WHY IS THE EXTENT OF HGT DIFFERENT ACROSS HOLOPARASITES?
The intimate relationships of parasites and their hosts associated via vascular connections and the early establishment of this connection upon parasite germination provide ample opportunities for DNA transfer leading to expected high levels of HGT from host to parasite. Among parasitic plants, the degree of dependence on the host has been linked to a greater propensity for nuclear HGT (Yang et al., 2016; Yoshida and Kee, 2021; Ashapkin et al., 2023). Holoparasitic plants, which are in close contact with their host from an early stage of development and possess phloem connections, are more likely to experience HGT (Yang et al., 2016). Indeed, the highest levels of nuclear HGT were reported for the holoparasite Phelipanche in comparison with other Orobanchaceae ranging from free-living to obligate parasites (Yang et al., 2016; Kado and Innan, 2018; Yoshida and Kee, 2021). However, this correlation is not observed in the mitogenomes of the Orobanchaceae, which show very low levels of HGT, regardless of the degree of host dependency (Zhang et al., 2022; Zhong et al., 2022). A limited impact of HGT in the mitogenome has also been observed in other host–parasite relationships (Fan et al., 2016; Skippington et al., 2017; Zhang et al., 2022; Yu et al., 2023), while others have been strongly altered by HGT (Xi et al., 2013; Bellot et al., 2016; Sanchez-Puerta et al., 2017; Roulet et al., 2020).
Overall, the reported extent of mitochondrial HGT differs substantially across and within holoparasitic lineages. One possible explanation for this phenomenon is the type of parasitism, with endoparasites exhibiting higher levels of foreign DNA than exoparasites (Bellot et al., 2016). This hypothesis is supported by the observation that endoparasites in the Rafflesiaceae and Cynomoriaceae carry numerous foreign mitochondrial genes. It is expected that high levels of HGT will be discovered in unexplored endoparasitic lineages, such as Apodanthaceae and Mitrastemonaceae.
Interestingly, Lophophytum mirabile and Ombrophytum subterraneum are not endoparasites but contain large amounts of mitochondrial foreign DNA (Sanchez-Puerta et al., 2017; Roulet et al., 2020). In fact, Lophophytum ranks highest for mitochondrial HGT (Sanchez-Puerta et al., 2017, 2019). It is unclear why these taxa are particularly prone to HGT. Furthermore, the close relative Rhopalocnemis does not exhibit any evidence of foreign mtDNA (Yu et al., 2022). The variability in the degree of HGT observed in the mitogenomes of root holoparasites within the family Balanophoraceae is intriguing.
The unprecedented levels of HGT observed in Lophophytum may be explained by its unique reproductive dynamics. Specifically, parenchymatous cells of Lophophytum form small groups within the cambium area of the host root. This nodule generates new tubers over time, representing a form of vegetative reproduction. For this, the initial nodule may grow through the host phloem and periderm, emerge from the root, and produce a tuber outside of the host that will eventually flower (Gonzalez and Sato, 2016). This nodule, considered the endophytic stage, acts as a reservoir of parasitic cells within the host root and exhibits intimate contact with direct exchange of substances across intercellular communications between parasite and host cells, facilitating the incorporation and inheritance of host DNA, in agreement with the weak-link model (Huang, 2012). A similar anatomical structure has been observed in Ombrophytum growing within the host root of Asteraceae (Mauseth et al., 1992). However, the anatomy of Rhopalocnemis inside the host root remains unexplored to the best of our knowledge, preventing a further evaluation of the hypothesis for the differential mitochondrial HGT among Balanophoraceae.
We propose that parasitic plants with a stable endophytic phase, as described for Lophophytum and observed in those considered endophytes, may exhibit high levels of HGT, both nuclear and mitochondrial. Other factors that might contribute to increased levels of mitochondrial HGT include alteration of the DNA repair, replication and recombination processes, predisposition to undergo mitochondrial fusion with unrelated taxa, tolerance to fluctuations in mitochondrial genome size, phylogenetic distance between host and parasite, and occurrence of population bottlenecks. Within the mitogenome, foreign intergenic regions may be incorporated without negative consequences (except for the increase in genome size) and evolve neutrally under the whims of genetic drift. Thus, foreign non-coding regions are more likely to be found in parasitic plant mitochondria, making the examination of intergenic regions highly relevant to assess the degree of HGT.
CONCLUSIONS
In addition to their morphological similarities, plastid genomes of holoparasitic plants also display convergent features, such as biased nucleotide compositions, increased evolutionary rates and gene content reductions related to photosynthesis. In-depth comparisons among mitochondrial genomes of holoparasitic plants are limited by the amount of genomic data available. Based on the data at hand, there are no obvious genomic differences from mitogenomes of free-living angiosperms, except for a higher incidence of HGT, which is widely observed among holoparasites but is not universal.
The study of a greater diversity of parasitic lineages will increase our understanding of the evolutionary mechanisms and genomic changes experienced by parasites with variable degrees of dependence on the host. Furthermore, anatomical studies and detailed descriptions of the life cycles will be instrumental to understanding the evolutionary dynamics of HGT in holoparasitic species. Among the diversity of parasitic lineages, Balanophoraceae and endophytes, in particular, offer interesting evolutionary traits and intriguing features yet to be discovered.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Figure S1. Maximum-likelihood phylogenetic analysis of Balanophoraceae based on a concatenated alignment of nuclear and mitochondrial sequences. Table S1. Accession numbers or references of the nuclear and mitochondrial sequences used for the phylogenetic analysis of Balanophoraceae. Note S1. List of species and accession numbers of taxa mentioned in Table 1.
ACKNOWLEDGEMENTS
We thank Renchao Zhou and Runxian Yu for sharing the photo of Rhopalocnemis phalloides.
Contributor Information
M Virginia Sanchez-Puerta, IBAM, Universidad Nacional de Cuyo, CONICET, Facultad de Ciencias Agrarias, Almirante Brown 500, Chacras de Coria, M5528AHB, Mendoza, Argentina; Facultad de Ciencias Exactas y Naturales, Padre Jorge Contreras 1300, Universidad Nacional de Cuyo, M5502JMA, Mendoza, Argentina.
Luis F Ceriotti, IBAM, Universidad Nacional de Cuyo, CONICET, Facultad de Ciencias Agrarias, Almirante Brown 500, Chacras de Coria, M5528AHB, Mendoza, Argentina; Facultad de Ciencias Exactas y Naturales, Padre Jorge Contreras 1300, Universidad Nacional de Cuyo, M5502JMA, Mendoza, Argentina.
Leonardo M Gatica-Soria, IBAM, Universidad Nacional de Cuyo, CONICET, Facultad de Ciencias Agrarias, Almirante Brown 500, Chacras de Coria, M5528AHB, Mendoza, Argentina; Facultad de Ciencias Exactas y Naturales, Padre Jorge Contreras 1300, Universidad Nacional de Cuyo, M5502JMA, Mendoza, Argentina.
M Emilia Roulet, IBAM, Universidad Nacional de Cuyo, CONICET, Facultad de Ciencias Agrarias, Almirante Brown 500, Chacras de Coria, M5528AHB, Mendoza, Argentina.
Laura E Garcia, IBAM, Universidad Nacional de Cuyo, CONICET, Facultad de Ciencias Agrarias, Almirante Brown 500, Chacras de Coria, M5528AHB, Mendoza, Argentina; Facultad de Ciencias Exactas y Naturales, Padre Jorge Contreras 1300, Universidad Nacional de Cuyo, M5502JMA, Mendoza, Argentina.
Hector A Sato, Facultad de Ciencias Agrarias, Cátedra de Botánica General–Herbario JUA, Alberdi 47, Universidad Nacional de Jujuy, 4600 Jujuy, Argentina.
FUNDING
This work was supported by the Fondo para la Investigación Científica y Tecnológica (grant number PICT2020-01018) and Universidad Nacional de Cuyo (grant number 06/A092-T1).
LITERATURE CITED
- Adams KL, Palmer JD.. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Molecular Phylogenetics and Evolution 29: 380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Albanova IA, Zagorchev LI, Teofanova DR, Odjakova MK, Kutueva LI, Ashapkin VV.. 2023. Host resistance to parasitic plants: current knowledge and future perspectives. Plants 12: 1447. doi: 10.3390/plants12071447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD.. 2011. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell 23: 2499–2513. doi: 10.1105/tpc.111.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD.. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Molecular Biology and Evolution 27: 1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BM, Krause K, Petersen G.. 2021. Mitochondrial genomes of two parasitic Cuscuta species lack clear evidence of horizontal gene transfer and retain unusually fragmented ccmFC genes. BMC Genomics 22: 816. doi: 10.1186/s12864-021-08105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin VV, Kutueva LI, Aleksandrushkina NI, Vanyushin BF, Teofanova DR, Zagorchev LI.. 2023. Genomic and epigenomic mechanisms of the interaction between parasitic and host plants. International Journal of Molecular Sciences 24: 2647. doi: 10.3390/ijms24032647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Stefanović S.. 2019. Caught in action: fine-scale plastome evolution in the parasitic plants of Cuscuta section Ceratophorae (Convolvulaceae). Plant Molecular Biology 100: 621–634. doi: 10.1007/s11103-019-00884-0. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Stefanović S.. 2023. A comparative study across the parasitic plants of Cuscuta subgenus Grammica (Convolvulaceae) reveals a possible loss of the plastid genome in its section Subulatae. Planta 257: 66. doi: 10.1007/s00425-023-04099-y. [DOI] [PubMed] [Google Scholar]
- Barbrook A, Howe C, Purton S.. 2006. Why are plastid genomes retained in non-photosynthetic organisms? Trends in Plant Science 11: 101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Barkman TJ, McNeal JR, Lim SH, et al. 2007. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evolutionary Biology 7: 248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot S, Cusimano N, Luo S, et al. 2016. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Genome Biology and Evolution 8: 2214–2230. doi: 10.1093/gbe/evw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot S, Renner SS.. 2016. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biology and Evolution 8: 189–201. doi: 10.1093/gbe/evv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthorsson U, Adams KL, Thomason B, Palmer JD.. 2003. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424: 197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- Bock R. 2010. The give-and-take of DNA: horizontal gene transfer in plants. Trends in Plant Science 15: 11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Brillouet J-M, Romieu C, Schoefs B, et al. 2013. The Tannosome is an organelle forming condensed tannins in the Chlorophyllous organs of Tracheophyta. Annals of Botany 112: 1003–1014. doi: 10.1093/aob/mct168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L, Cowman PF, Lanfear R.. 2013. Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evolutionary Biology 13: 126. doi: 10.1186/1471-2148-13-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke SV, Ungerer MC, Duvall MR.. 2018. Investigation of mitochondrial-derived plastome sequences in the Paspalum lineage (Panicoideae; Poaceae). BMC Plant Biology 18: 152. doi: 10.1186/s12870-018-1379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L. 2023. Rethinking convergence in plant parasitism through the lens of molecular and population genetic processes. American Journal of Botany 110: e16174. doi: 10.1002/ajb2.16174. [DOI] [PubMed] [Google Scholar]
- Cai L, Arnold BJ, Xi Z, et al. 2021. Deeply altered genome architecture in the endoparasitic flowering plant Sapria himalayana Griff. (Rafflesiaceae). Current Biology 31: 1002–1011.e9. doi: 10.1016/j.cub.2020.12.045. [DOI] [PubMed] [Google Scholar]
- Ceriotti LF, Gatica-Soria L, Sanchez-Puerta MV.. 2022. Cytonuclear coevolution in a holoparasitic plant with highly disparate organellar genomes. Plant Molecular Biology 109: 673–688. doi: 10.1007/s11103-022-01266-9. [DOI] [PubMed] [Google Scholar]
- Ceriotti F, Roulet M, Sanchez-Puerta MV.. 2021. Plastomes in the holoparasitic family Balanophoraceae: extremely high AT content, severe gene content reduction, and two independent genetic code changes. Molecular Phylogenetics and Evolution 162: 107208. doi: 10.1016/j.ympev.2021.107208. [DOI] [PubMed] [Google Scholar]
- Chen X, Fang D, Wu C, et al. 2019. Comparative plastome analysis of root- and stem-feeding parasites of Santalales untangle the footprints of feeding mode and lifestyle transitions. Genome Biology and Evolution 12: 3663–3676. doi: 10.1093/gbe/evz271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Qiu YL, Kuhlman P, Palmer JD.. 1998. Explosive invasion of plant mitochondria by a group I intron. Proceedings of the National Academy of Sciences 95: 14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-S, Park S.. 2021. Complete plastid and mitochondrial genomes of Aeginetia indica reveal intracellular gene transfer (IGT), horizontal gene transfer (HGT), and cytoplasmic male sterility (CMS). International Journal of Molecular Sciences 22: 6143. doi: 10.3390/ijms22116143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I-S, Schwarz EN, Ruhlman TA, et al. 2019. Fluctuations in Fabaceae mitochondrial genome size and content are both ancient and recent. BMC Plant Biology 19: 448. doi: 10.1186/s12870-019-2064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O, Gophna U, Pupko T.. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Molecular Biology and Evolution 28: 1481–1489. doi: 10.1093/molbev/msq333. [DOI] [PubMed] [Google Scholar]
- Cusimano N, Renner SS.. 2019. Sequential horizontal gene transfers from different hosts in a widespread Eurasian parasitic plant, Cynomorium coccineum. American Journal of Botany 106: 679–689. doi: 10.1002/ajb2.1286. [DOI] [PubMed] [Google Scholar]
- Cusimano N, Wicke S.. 2016. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytologist 210: 680–693. doi: 10.1111/nph.13784. [DOI] [PubMed] [Google Scholar]
- Dahan J, Tcherkez G, Macherel D, et al. 2014. Disruption of the CYTOCHROME C OXIDASE DEFICIENT1 gene leads to cytochrome c oxidase depletion and reorchestrated respiratory metabolism in Arabidopsis. Plant Physiology 166: 1788–1802. doi: 10.1104/pp.114.248526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshetkar AM, Pable AA, Nadaf AB, Barvkar VT.. 2023. Understanding parasitism in Loranthaceae: insights from plastome and mitogenome of Helicanthes elastica. Gene 861: 147238. doi: 10.1016/j.gene.2023.147238. [DOI] [PubMed] [Google Scholar]
- Davis C, Andersen W, Wurdack K.. 2005. Gene transfer from a parasitic flowering plant to a fern. Proceedings of the Royal Society B-Biological Sciences 272: 2237–2242. doi: 10.1098/rspb.2005.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Xi Z.. 2015. Horizontal gene transfer in parasitic plants. Current Opinion in Plant Biology 26: 14–19. doi: 10.1016/j.pbi.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Dorrell RG, Azuma T, Nomura M, et al. 2019. Principles of plastid reductive evolution illuminated by nonphotosynthetic chrysophytes. Proceedings of the National Academy of Sciences 116: 6914–6923. doi: 10.1073/pnas.1819976116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning LT, Christin P-A.. 2020. Reticulate evolution, lateral gene transfer, and innovation in plants. American Journal of Botany 107: 541–544. doi: 10.1002/ajb2.1452. [DOI] [PubMed] [Google Scholar]
- Eberwein RK, Weber A.. 2004. Exorhopala ruficeps (Balanophoraceae): morphology and transfer to Helosis. Botanical Journal of the Linnean Society 146: 513–517. doi: 10.1111/j.1095-8339.2004.00321.x. [DOI] [Google Scholar]
- Eichler A. 1867. Sur la structure femelle de quelques Balanopho- racées. In: Fournier E, ed. Actes du Congrés international de botanique tenu a Paris en août 1867, sous les auspices de la Société botanique de France. Paris: G. Bailliére, 137–155. [Google Scholar]
- Fan W, Zhu A, Kozaczek M, et al. 2016. Limited mitogenomic degradation in response to a parasitic lifestyle in Orobanchaceae. Scientific Reports 6: 36285. doi: 10.1038/srep36285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández Robledo JA, Caler E, Matsuzaki M, et al. 2011. The search for the missing link: a relic plastid in Perkinsus? International Journal for Parasitology 41: 1217–1229. doi: 10.1016/j.ijpara.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandini CL, Garcia L, Abbona CC, Sanchez-Puerta MV.. 2019. The complete organelle genomes of Physochlaina orientalis: insights into short sequence repeats across seed plant mitochondrial genomes. Molecular Phylogenetics and Evolution 137: 274–284. doi: 10.1016/j.ympev.2019.05.012. [DOI] [PubMed] [Google Scholar]
- Garcia LE, Edera AA, Palmer JD, Sato H, Sanchez-Puerta MV.. 2021. Horizontal gene transfers dominate the functional mitochondrial gene space of a holoparasitic plant. New Phytologist 229: 1701–1714. doi: 10.1111/nph.16926. [DOI] [PubMed] [Google Scholar]
- Gandini CL, Sanchez-Puerta MV.. 2017. Foreign plastid sequences in plant mitochondria are frequently acquired via mitochondrion-to-mitochondrion horizontal transfer. Scientific Reports 7: 43402. doi: 10.1038/srep43402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica-Soria LM, Ceriotti LF, Garcia LE, Sanchez-Puerta MV.. 2022. Native and foreign mitochondrial and nuclear encoded proteins conform the OXPHOS complexes of a holoparasitic plant. Gene 817: 146176. doi: 10.1016/j.gene.2021.146176. [DOI] [PubMed] [Google Scholar]
- Gawryluk RMR, Tikhonenkov DV, Hehenberger E, Husnik F, Mylnikov AP, Keeling PJ.. 2019. Non-photosynthetic predators are sister to red algae. Nature 572: 240–243. doi: 10.1038/s41586-019-1398-6. [DOI] [PubMed] [Google Scholar]
- Gedalovich-Shedletzky E, Kuijt J.. 1990. An ultrastructural study of the tuber stands of Balanophora: Balanophoraceae. Canadian Journal of Botany 68: 1271–1279. doi: 10.1139/b90-162. [DOI] [Google Scholar]
- Gibson T, Blok VC, Phillips MS, et al. 2007. The mitochondrial subgenomes of the nematode Globodera pallida are mosaics: evidence of recombination in an animal mitochondrial genome. Journal of Molecular Evolution 64: 463–471. doi: 10.1007/s00239-006-0187-7. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Mauseth J.. 2010. Morphogenesis is highly aberrant in the vegetative body of the holoparasite Lophophytum leandrii (Balanophoraceae): all typical vegetative organs are absent and many tissues are highly modified. International Journal of Plant Science 171: 499–508. doi: 10.1086/651947. [DOI] [Google Scholar]
- Gonzalez A, Sato H.. 2016. Anatomía vegetativa de Lophophytum mirabile subsp. bolivianum (Balanophoraceae) y efecto de su parasitismo en la anatomía de las raíces de su hospedante Anadenanthera colubrina var. cebil. Anales Jardín Botánico Madrid 73: e039. doi: 10.3989/ajbm.2423. [DOI] [Google Scholar]
- Góralski G, Denysenko-Bennett M, Burda A, Staszecka-Moskal N, Kwolek D.. 2021. Achievements in horizontal gene transfer studies in parasitic plants. Acta Biologica Cracoviensia s. Botanica 63: 17–28. doi: 10.24425/abcsb.2021.136701. [DOI] [Google Scholar]
- Graham SW, Lam VKY, Merckx VSFT.. 2017. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytologist 214: 48–55. doi: 10.1111/nph.14398. [DOI] [PubMed] [Google Scholar]
- Gurdon C, Svab Z, Feng Y, Kumar D, Maliga P.. 2016. Cell-to-cell movement of mitochondria in plants. Proceedings of the National Academy of Sciences 113: 3395–3400. doi: 10.1073/pnas.1518644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadariová L, Vesteg M, Hampl V, Krajčovič J.. 2018. Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists. Current Genetics 64: 365–387. doi: 10.1007/s00294-017-0761-0. [DOI] [PubMed] [Google Scholar]
- Hansen B. 1980. Balanophoraceae. Flora Neotrópica 23: 1–80. [Google Scholar]
- Hao W, Richardson AO, Zheng Y, Palmer JD.. 2010. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proceedings of the National Academy of Sciences 107: 21576–21581. doi: 10.1073/pnas.1016295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms H. 1935. Balanophoraceae. In: Engler A, Prantl K, eds. Die natürlichen Planzenfamilien. Leipzig: Engelmann, 296–339. [Google Scholar]
- Heide-Jorgensen HS. 2008. Parasitic flowering plants. Leiden: Koninklijke Brill NV. [Google Scholar]
- Hertle AP, Haberl B, Bock R.. 2021. Horizontal genome transfer by cell-to-cell travel of whole organelles. Science Advances 7: eabd8215. doi: 10.1126/sciadv.abd8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao S-C, Mauseth J, Peng C-I.. 1995. Composite bundles, the host/parasite interface in the holoparasitic angiosperms Langsdorffia and Balanophora (Balanophoraceae). American Journal of Botany 82: 81–91. [Google Scholar]
- Huang J. 2012. Horizontal gene transfer in Eukaryotes: the weak-link model. Bioessays 35: 868–875. doi: 10.1002/bies.201300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Rivera MC, Lake JA.. 1999. Horizontal gene transfer among genomes: the complexity hypothesis. Proceedings of the National Academy of Sciences 96: 3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Naumann J, Bolin JF, et al. 2022. Structural plastome evolution in holoparasitic Hydnoraceae with special focus on inverted and direct repeats. Genome Biology and Evolution 14: evac077. doi: 10.1093/gbe/evac077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado T, Innan H.. 2018. Horizontal gene transfer in five parasite plant species in Orobanchaceae. Genome Biology and Evolution 10: 3196–3210. doi: 10.1093/gbe/evy219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Lautenschläger T, Bolin JF, et al. 2023. Extreme plastomes in holoparasitic Balanophoraceae are not the norm. BMC Genomics 24: 330. doi: 10.1186/s12864-023-09422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-R, Yang J-I, Moon S, et al. 2009. Rice OGR1 encodes a pentatricopeptide repeat–DYW protein and is essential for RNA editing in mitochondria. The Plant Journal 59: 738–749. doi: 10.1111/j.1365-313x.2009.03909.x. [DOI] [PubMed] [Google Scholar]
- Klimpert NJ, Mayer JLS, Sarzi DS, Prosdocimi F, Pinheiro F, Graham SW.. 2022. Phylogenomics and plastome evolution of a Brazilian mycoheterotrophic orchid, Pogoniopsis schenckii. American Journal of Botany 109: 2030–2050. doi: 10.1002/ajb2.16084. [DOI] [PubMed] [Google Scholar]
- van der Kooij TAW, Krause K, Dörr I, Krupinska K.. 2000. Molecular, functional and ultrastructural characterisation of plastids from six species of the parasitic flowering plant genus Cuscuta. Planta 210: 701–707. doi: 10.1007/s004250050670. [DOI] [PubMed] [Google Scholar]
- Kozik A, Rowan BA, Lavelle D, et al. 2019. The alternative reality of plant mitochondrial DNA: one ring does not rule them all. PLoS Genetics 15: e1008373. doi: 10.1371/journal.pgen.1008373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt J. 1969. The biology of parasitic flowering plants. Berkeley: University of California Press. [Google Scholar]
- Kuijt J, Hansen B.. 2015.Balanophoraceae. In: Kubitzki K, ed. Flowering plants. The families and genera of vascular plants. Flowering plants: Eudicots; Santalales, Balanophorales, Vol.12. Berlin: Springer, 193–208. doi: 10.1007/978-3-319-09296-6_23. [DOI] [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, et al. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H-T, Yi T-S, Gao L-M, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5: 461–470. doi: 10.1038/s41477-019-0421-0. [DOI] [PubMed] [Google Scholar]
- Lin Q, Banerjee A, Stefanović S.. 2022a. Mitochondrial phylogenomics of Cuscuta (Convolvulaceae) reveals a potentially functional horizontal gene transfer from the host. Genome Biology and Evolution 14: evac091. doi: 10.1093/gbe/evac091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Li P, Zhang Y, et al. 2022b. Unprecedented organelle genomic variations in morning glories reveal independent evolutionary scenarios of parasitic plants and the diversification of plant mitochondrial complexes. BMC Biology 20: 49. doi: 10.1186/s12915-022-01250-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P-F, Zhang Y-X, Guo Z-H, Li D-Z.. 2015. Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Scientific Reports 5: 11608. doi: 10.1038/srep11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean A, Hertle A, Ligas J, Bock R, Balk J, Meyer E.. 2018. Absence of complex I is associated with diminished respiratory chain function in European mistletoe. Current Biology 28: 1614–1619. doi: 10.1016/j.cub.2018.03.036 [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T.. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Brisson N.. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytologist 186: 299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- Mauseth JD, Hsiao S-C, Montenegro G.. 1992. Vegetative body of the parasitic angiosperm Ombrophytum subterraneum (Balanophoraceae). Bulletin of the Torrey Botanical Club 119: 407–417. doi: 10.2307/2996729. [DOI] [Google Scholar]
- Mauseth JD, Montenegro G, Walckowiak AM.. 1984. Studies of the holoparasite Tristerix aphyllus (Loranthaceae) infecting Trichocereus chilensis (Cactaceae). Canadian Journal of Botany 62: 847–857. doi: 10.1139/b84-124. [DOI] [Google Scholar]
- Mauseth JD, Rezaei K.. 2013. Morphogenesis in the parasitic plant Viscum minimum (Viscaceae) is highly altered, having apical meristems but lacking roots, stems, and leaves. International Journal of Plant Sciences 174: 791–801. doi: 10.1086/669926. [DOI] [Google Scholar]
- Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL.. 2013. An incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genetics 9: e1003238. doi: 10.1371/journal.pgen.1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merckx VSFT. 2013. Mycoheterotrophy: an introduction. In: Merckx V, ed. Mycoheterotrophy: the biology of plants living on Fungi. New York, NY: Springer, 1–17. [Google Scholar]
- Merckx V, Freudenstein JV.. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytologist 185: 605–609. doi: 10.1111/j.1469-8137.2009.03155.x. [DOI] [PubMed] [Google Scholar]
- Meyer EH, Welchen E, Carrie C.. 2019. Assembly of the complexes of the oxidative phosphorylation system in land plant mitochondria. Annual Review of Plant Biology 70: 23–50. doi: 10.1146/annurev-arplant-050718-100412. [DOI] [PubMed] [Google Scholar]
- Molina J, Hazzouri KM, Nickrent DL, et al. 2014. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Molecular Biology and Evolution 31: 793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Pagarete A, de Vargas C, et al. 2009. Horizontal gene transfer of an entire metabolic pathway between a eukaryotic alga and its DNA virus. Genome Research 19: 1441–1449. doi: 10.1101/gr.091686.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Hao W, et al. 2010. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biology 8: 150. doi: 10.1186/1741-7007-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Stefanovic S, Young GJ, Palmer JD.. 2004. Plant genetics: gene transfer from parasitic to host plants. Nature 432: 165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- Naumann J, Salomo K, Der JP, et al. 2013. Single-copy nuclear genes place haustorial Hydnoraceae within Piperales and reveal a cretaceous origin of multiple parasitic angiosperm lineages. PLoS One 8: e79204. doi: 10.1371/journal.pone.0079204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent D, Ouyang Y, Duff R, dePamphilis CW.. 1997. Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Molecular Biology 34: 717–729. doi: 10.1023/A:1005860632601. [DOI] [PubMed] [Google Scholar]
- Nickrent DL. 2020. Parasitic angiosperms: how often and how many? Taxon 69: 5–27. doi: 10.1002/tax.12195. [DOI] [Google Scholar]
- Nickrent DL, Duff RJ.. 1996. Molecular studies of parasitic plants using ribosomal RNA. In: Moreno MT, Cubero JI, Berner D, Joel D, Musselman LJ, Parker C, eds.. Advances in parasitic plant research. Cordoba, Spain: Junta de Andalucia, Dirección General de Investigación Agraria, 28–52. [Google Scholar]
- Nickrent DL, García MA.. 2009. On the brink of holoparasitism: plastome evolution in dwarf mistletoes (Arceuthobium, Viscaceae). Journal of Molecular Evolution 68: 603–615. doi: 10.1007/s00239-009-9224-7. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Malécot V, Vidal-Russell R, Der JP.. 2010. A revised classification of Santalales. Taxon 59: 538–558. doi: 10.1002/tax.592019. [DOI] [Google Scholar]
- Oldenburg D, Bendich A.. 1996. Size and structure of replicating mitochondrial DNA in cultured tobacco cells. Plant Cell 8: 447–461. doi: 10.1105/tpc.8.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S, Jukes TH.. 1989. Codon reassignment (codon capture) in evolution. Journal of Molecular Evolution 28: 271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF.. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29: 537–550. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- Park C, Zhang J.. 2012. High expression hampers horizontal gene transfer. Genome Biology and Evolution 4: 523–532. doi: 10.1093/gbe/evs030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Grewe F, Zhu A, et al. 2015. Dynamic evolution of Geranium mitochondrial genomes through multiple horizontal and intracellular gene transfers. New Phytologist 208: 570–583. [DOI] [PubMed] [Google Scholar]
- Petersen G, Anderson B, Braun H-P, Meyer EH, Møller IM.. 2020. Mitochondria in parasitic plants. Mitochondrion 52: 173–182. doi: 10.1016/j.mito.2020.03.008. [DOI] [PubMed] [Google Scholar]
- Petersen G, Shyama Prasad Rao R, Anderson B, et al. 2022. Genes from oxidative phosphorylation complexes II-V and two dual-function subunits of complex I are transcribed in Viscum album despite absence of the entire mitochondrial holo-complex I. Mitochondrion 62: 1–12. doi: 10.1016/j.mito.2021.10.006. [DOI] [PubMed] [Google Scholar]
- Press M, Phoenix G.. 2005. Impacts of parasitic plants on natural communities. New Phytologist 166: 737–751. [DOI] [PubMed] [Google Scholar]
- Qi W, Tian Z, Lu L, et al. 2017. Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics 205: 1489–1501. doi: 10.1534/genetics.116.199331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice D, Palmer JD.. 2006. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biology 4: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DW, Alverson AJ, Richardson AO, et al. 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342: 1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- Richardson AO, Palmer JD.. 2007. Horizontal gene transfer in plants. Journal of Experimental Botany 58: 1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD.. 2013. The ‘fossilized’ mitochondrial genome of Liriodendron tulipifera: an ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biology 11: 29. doi: 10.1186/1741-7007-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquet C, Coissac E, Cruaud C, et al. 2016. Understanding the evolution of holoparasitic plants: the complete plastid genome of the holoparasite Cytinus hypocistis (Cytinaceae). Annals of Botany 118: 885–896. doi: 10.1093/aob/mcw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JP, Kleist TJ, Löfstrand SD, Drew BT, Schönenberger J, Sytsma KJ.. 2018. Phylogeny, historical biogeography, and diversification of angiosperm order Ericales suggest ancient Neotropical and East Asian connections. Molecular Phylogenetics and Evolution 122: 59–79. doi: 10.1016/j.ympev.2018.01.014. [DOI] [PubMed] [Google Scholar]
- Roulet M, Garcia L, Gandini CL, Sato H, Ponce G, Sanchez-Puerta MV.. 2020. Multichromosomal structure and foreign tracts in the Ombrophytum subterraneum (Balanophoraceae) mitochondrial genome. Plant Molecular Biology 103: 623–638. doi: 10.1007/s11103-020-01014-x. [DOI] [PubMed] [Google Scholar]
- Ruhlman T, Jansen RK.. 2014. The plastid genomes of flowering plants. Methods in Molecular Biology 1132: 3–38. doi: 10.1007/978-1-62703-995-6_1 [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV. 2014. Involvement of plastid, mitochondrial and nuclear genomes in plant-to-plant horizontal gene transfer. Acta Societatis Botanicorum Poloniae 83: 317–323. doi: 10.5586/asbp.2014.041. [DOI] [Google Scholar]
- Sanchez-Puerta MV, Abbona CC, Zhuo S, et al. 2011. Multiple recent horizontal transfers of the cox1 intron in Solanaceae and extended coconversion of flanking exons. BMC Evolutionary Biology 11: 277. doi: 10.1186/1471-2148-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD.. 2008. Frequent, phylogenetically local horizontal transfer of the cox1 group I intron in flowering plant mitochondria. Molecular Biology and Evolution 25: 1762–1777. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Edera A, Gandini CL, et al. 2019. Genome-scale transfer of mitochondrial DNA from legume hosts to the holoparasite Lophophytum mirabile (Balanophoraceae). Molecular Phylogenetics and Evolution 132: 243–250. doi: 10.1016/j.ympev.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, García LE, Wohlfeiler J, Ceriotti F.. 2017. Unparalleled replacement of native mitochondrial genes by foreign homologs in a holoparasitic plant. New Phytologist 214: 376–387. doi: 10.1111/nph.14361. [DOI] [PubMed] [Google Scholar]
- Sato H, Gonzalez A.. 2016. Floral development and anatomy of pistillate flowers of Lophophytum (Balanophoraceae), with special reference to the embryo sac inversion. Flora 219: 35–47. [Google Scholar]
- Schelkunov M, Nuraliev M, Logacheva M.. 2019. Rhopalocnemis phalloides has one of the most reduced and mutated plastid genomes known. PeerJ 7: e7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelkunov M, Nuraliev M, Logacheva M.. 2021. Genomic comparison of non-photosynthetic plants from the family Balanophoraceae with their photosynthetic relatives. PeerJ 9: e12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Kushnir S, Babiychuk E, Poltnigg P, Herrmann RG, Maier RM.. 2005. Pigment deficiency in nightshade/tobacco cybrids is caused by the failure to edit the plastid ATPase ɑ-subunit mRNA. Plant Cell 17: 1815–1828. doi: 10.1105/tpc.105.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Rugen N, Eubel H, Hegermann J, Braun HP.. 2018. Absence of complex I implicates rearrangements of the respiratory chain in European mistletoe. Current Biology 28: 1606–1613.e4. doi: 10.1016/j.cub.2018.03.050. [DOI] [PubMed] [Google Scholar]
- Shao R, Kirkness E, Barker S.. 2009. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Research 19: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD.. 2015. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proceedings of the National Academy of Sciences 112: E3515–E3524. doi: 10.1073/pnas.1504491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD.. 2017. Comparative mitogenomics indicates respiratory competence in parasitic Viscum despite loss of complex I and extreme sequence divergence, and reveals horizontal gene transfer and remarkable variation in genome size. BMC Plant Biology 17: 49. doi: 10.1186/s12870-017-0992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan D. 2013. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytologist 200: 978–985. [DOI] [PubMed] [Google Scholar]
- Sloan D, Alverson AJ, Chuckalovcak J, et al. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biology 10: e1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Warren JM, Williams AM, Kuster SA, Forsythe ES.. 2022. Incompatibility and interchangeability in molecular evolution. Genome Biology and Evolution 15: evac184. doi: 10.1093/gbe/evac184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Lee RW.. 2014. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiology 164: 1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Senters AE, Zanis MJ, et al. 2003. Gunnerales are sister to other core eudicots: implications for the evolution of pentamery. American Journal of Botany 90: 461–470. doi: 10.3732/ajb.90.3.461. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE, Chase MW.. 1999. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402: 402–404. doi: 10.1038/46528. [DOI] [PubMed] [Google Scholar]
- Sorek R, Zhu Y, Creevey C, Francino M, Bork P, Rubin E.. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452. [DOI] [PubMed] [Google Scholar]
- Su H-J, Barkman TJ, Hao W, et al. 2019. Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proceedings of the National Academy of Sciences 116: 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H-J, Hu J-M, Anderson FE, Der JP, Nickrent DL.. 2015. Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon 64: 491–506. doi: 10.12705/643.2. [DOI] [Google Scholar]
- Su H-J, Murata J, Hu J-M.. 2012. Morphology and phylogenetics of two holoparasitic plants, Balanophora japonica and Balanophora yakushimensis (Balanophoraceae), and their hosts in Taiwan and Japan. Journal of Plant Research 125: 317–326. doi: 10.1007/s10265-011-0447-5. [DOI] [PubMed] [Google Scholar]
- Takhtajan A. 2009. Flowering plants. Berlin: Springer Science & Business Media. [Google Scholar]
- Teixeira-Costa L. 2021. A living bridge between two enemies: haustorium structure and evolution across parasitic flowering plants. Brazilian Journal of Botany 44: 165–178. doi: 10.1007/s40415-021-00704-0. [DOI] [Google Scholar]
- Teixeira-Costa L, Davis CC.. 2021. Life history, diversity, and distribution in parasitic flowering plants. Plant Physiology 187: 32–51. doi: 10.1093/plphys/kiab279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Těšitel J. 2016. Functional biology of parasitic plants: a review. Plant Ecology and Evolution 149: 5–20. doi: 10.5091/plecevo.2016.1097. [DOI] [Google Scholar]
- The Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. doi: 10.1111/boj.12385. [DOI] [Google Scholar]
- Torres CA, Pérez Zamora CM, Sato HA, Nuñez MB, Gonzalez AM.. 2021. Composición fitoquímica y cribado biológico de dos especies de Lophophytum. Boletín Latinoamericano y del Caribe de Plantas Medicinales y Aromáticas 20: 598–610. [Google Scholar]
- Van Tieghem MP. 1896. Sur l’organisation florale des Balanophoracées et sur la place de cette famille dans la sous-classe des dicotylédones inovulées ou Loranthinées. Bulletin de la Société Botanique de France 43: 295–310. doi: 10.1080/00378941.1896.10830679. [DOI] [Google Scholar]
- Vogel A, Schwacke R, Denton A, et al. 2018. Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nature Communications 9: 2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller R, Slamovits C, Keeling PJ.. 2006. Lateral gene transfer of a multi-gene region from cyanobacteria to dinoflagellates resulting in a novel plastid-targeted fusion protein. Molecular Biology and Evolution 23: 1437–1443. [DOI] [PubMed] [Google Scholar]
- Westwood JH, Yoder JA, Timko M, dePamphilis CW.. 2010. The evolution of parasitism in plants. Trends in Plant Science 15: 227–235. [DOI] [PubMed] [Google Scholar]
- Wicke S, Muller KF, de Pamphilis CW, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25: 3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Naumann J.. 2018. Molecular evolution of plastid genomes in parasitic flowering plants. In: Chaw S-M, Jansen RK, eds. Advances in botanical research. New York: Academic Press, 315–347. [Google Scholar]
- Wickell DA, Li FW.. 2020. On the evolutionary significance of horizontal gene transfers in plants. New Phytologist 225: 113–117. doi: 10.1111/nph.16022. [DOI] [PubMed] [Google Scholar]
- Wilson RJM, Denny PW, Preiser P, et al. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. Journal of Molecular Biology 261: 155–172. [DOI] [PubMed] [Google Scholar]
- Woese CR, Olsen GJ, Ibba M, Söll D.. 2000. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiology and Molecular Biology Reviews 64: 202–236. doi: 10.1128/MMBR.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Cuthbert JM, Taylor DR, Sloan DB.. 2015. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proceedings of the National Academy of Sciences 112: 10185–10191. doi: 10.1073/pnas.1421397112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Bradley R, Wurdack K, et al. 2012. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics 13: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Wang Y, Bradley R, et al. 2013. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genetics 9: e1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang J, Ma C, et al. 2022. Comparative genomics of orobanchaceous species with different parasitic lifestyles reveals the origin and stepwise evolution of plant parasitism. Molecular Plant 15: 1384–1399. doi: 10.1016/j.molp.2022.07.007. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wafula E, Kim G, et al. 2019. Convergent horizontal gene transfer and cross-talk of mobile nucleic acids in parasitic plants. Nature Plants 5: 991–1001. [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang Y, Wafula E, et al. 2016. Horizontal gene transfer is more frequent with increased heterotrophy and contributes to parasite adaptation. Proceedings of the National Academy of Sciences 113: E7010–E7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S-i, Ueda T, Watanabe K.. 2010.Egenetic code In: Encyclopedia of Life Sciences. Oxford: Wiley Blackwell. doi: 10.1002/9780470015902.a0000548.pub2. [DOI] [Google Scholar]
- Yoshida S, Cui S, Ichihashi Y, Shirasu K.. 2016. The haustorium, a specialized invasive organ in parasitic plants. Annual Review of Plant Biology 67: 643–667. doi: 10.1146/annurev-arplant-043015-111702. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kee YJ.. 2021. Large-scale sequencing paves the way for genomic and genetic analyses in parasitic plants. Current Opinion in Biotechnology 70: 248–254. doi: 10.1016/j.copbio.2021.06.011. [DOI] [PubMed] [Google Scholar]
- Yu R, Chen X, Long L, et al. 2023. De novo assembly and comparative analyses of mitochondrial genomes in Piperales. Genome Biology and Evolution 15: evad041. doi: 10.1093/gbe/evad041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Sun C, Liu Y, Zhou R.. 2021. Shifts from cis-to-trans splicing of five mitochondrial introns in Tolypanthus maclurei. PeerJ 9: e12260. doi: 10.7717/peerj.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Sun C, Zhong Y, et al. 2022. The minicircular and extremely heteroplasmic mitogenome of the holoparasitic plant Rhopalocnemis phalloides. Current Biology 32: 470–479.e5. doi: 10.1016/j.cub.2021.11.053. [DOI] [PubMed] [Google Scholar]
- Zagorchev L, Stöggl W, Teofanova D, Li J, Kranner I.. 2021. Plant parasites under pressure: effects of abiotic stress on the interactions between parasitic plants and their hosts. International Journal of Molecular Sciences 22: 7418. doi: 10.3390/ijms22147418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervas A, Petersen G, Seberg O.. 2019. Mitochondrial genome evolution in parasitic plants. BMC Evolutionary Biology 19: 87. doi: 10.1186/s12862-019-1401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Green BR, Cavalier-Smith T.. 1999. Single gene circles in dinoflagellate chloroplast genomes. Nature 400: 155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang Z, Fu W, et al. 2023. Host shift promotes divergent evolution between closely related holoparasitic species. Molecular Phylogenetics and Evolution 186: 107842. doi: 10.1016/j.ympev.2023.107842. [DOI] [PubMed] [Google Scholar]
- Zhang C, Lin Q, Zhang J, et al. 2022. Comparing complete organelle genomes of holoparasitic Christisonia kwangtungensis (Orabanchaceae) with its close relatives: how different are they? BMC Plant Biology 22: 444. doi: 10.1186/s12870-022-03814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Yu R, Chen J, Liu Y, Zhou R.. 2022. Highly active repeat-mediated recombination in the mitogenome of the holoparasitic plant Aeginetia indica. Frontiers in Plant Science 13: 988368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


