Abstract
Measuring protein thermostability provides valuable information on the biophysical rules that govern the structure-energy relationships of proteins. However, such measurements remain a challenge for membrane proteins. Here, we introduce a new experimental system to evaluate membrane protein thermostability. This system leverages a recently developed nonfluorescent membrane scaffold protein to reconstitute proteins into nanodiscs and is coupled with a nano-format of differential scanning fluorimetry (nanoDSF). This approach offers a label-free and direct measurement of the intrinsic tryptophan fluorescence of the membrane protein as it unfolds in solution without signal interference from the “dark” nanodisc. In this work, we demonstrate the application of this method using the disulfide bond formation protein B (DsbB) as a test membrane protein. NanoDSF measurements of DsbB reconstituted in dark nanodiscs loaded with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-dimyristoyl-sn-glycero-3-phosphorylglycerol (DMPG) lipids show a complex biphasic thermal unfolding pattern with a minor unfolding transition followed by a major transition. The inflection points of the thermal denaturation curve reveal two distinct unfolding midpoint melting temperatures (Tm) of 70.5°C and 77.5°C, consistent with a three-state unfolding model. Further, we show that the catalytically conserved disulfide bond between residues C41 and C130 drives the intermediate state of the unfolding pathway for DsbB in a DMPC and DMPG nanodisc. To extend the utility of this method, we evaluate and compare the thermostability of DsbB in different lipid environments. We introduce this method as a new tool that can be used to understand how compositionally and biophysically complex lipid environments drive membrane protein stability.
Significance
Lipids are essential for membrane protein stability and serve as determinants underlying the biophysical rules that regulate membrane protein folding, structural dynamics, and function. Experimental measurements of membrane protein thermostability provide thermodynamic parameters key to understanding these relationships, but such measurements are typically relegated to systems like detergent micelles. This work develops an experimental system to monitor the thermal unfolding of membrane proteins in spectroscopically silent “dark” nanodiscs by nanoDSF. We use DsbB to demonstrate the utility of this method and show that we can monitor the thermal unfolding of DsbB by nanoDSF without signal interference from the nanodisc. This dark nanodisc system provides a new tool to advance knowledge regarding the biophysical basis of lipid-mediated membrane protein thermal unfolding.
Introduction
Membrane proteins represent a class of proteins that are challenging targets for biophysical studies because they associate with unique asymmetric lipid environments. Evaluating the thermal stability of membrane proteins relies on spectroscopic or calorimetric assays that perform well and are robust for water-soluble proteins, such as circular dichroism (CD) (1), differential scanning calorimetry (DSC) (2), and differential scanning fluorimetry (DSF) (3). These tools often fall short for membrane proteins because they require a model membrane system for in vitro studies, which often convolute the signal and measurement. Detergent micelles are the most widely used model membrane systems for biophysical studies, despite their known effect on membrane protein structure, stability, and function (4,5). To date, few studies tabulated in the membrane protein thermodynamic database (MPTherm) report thermostability measurements for membrane proteins in the presence of lipids (6) despite the key role lipids play in thermal stability (7,8,9). This is largely due to technical hurdles associated with developing simple tools to investigate the effects of lipids on membrane protein stability.
Model membrane systems used for membrane protein stability measurements range from micelles and bicelles to more native-like systems such as membrane scaffold protein (MSP)-based nanodiscs (10,11), saposin-derived lipid nanoparticles (12), styrene-maleic acid copolymers (13), and proteoliposomes (14). Many sophisticated attempts have been made to evaluate how lipids affect membrane protein stability using a variety of approaches. For example, an ion mobility mass spectrometry approach showed that specific lipid types increased protein unfolding resistance for several membrane protein systems compared to micelle conditions alone (15). Using a green fluorescent protein (GFP)-based thermal shift assay, Nji et al. revealed that cardiolipin increased the thermal stability of detergent-solubilized sodium-proton antiporter (16). Treuheit et al. studied the thermal stability of a membrane-associated cytochrome P4503A4 demonstrating an increase in the thermal stabilization of cytochrome P4503A4 incorporated into nanodiscs by DSC, an approach that requires substantial sample consumption (17). Flayhan et al. tested the thermostability of three different membrane protein systems—DtpA, PepT, and the small-conductance mechanosensitive channel T2 in saposin lipid nanoparticles—using a nano-format of DSF (nanoDSF) (18). All three membrane protein test systems revealed an increase in thermal stabilization (i.e., melting temperature) in saposin lipid nanoparticles compared to n-dodecyl-β-D-maltoside (DDM) micelles but offer less control over lipid composition compared to MSPs. Although these methods have made valuable attempts to study the lipid regulation of membrane proteins, they often suffer from complexity, substantial sample requirements, reliance on the introduction of nonnative fluorescent tags, and lack of precise control over lipid composition, hindering quantitative comparisons. These attempts suggest we still lack robust tools for evaluating and quantitatively comparing how lipids regulate the structure and stability of membrane proteins.
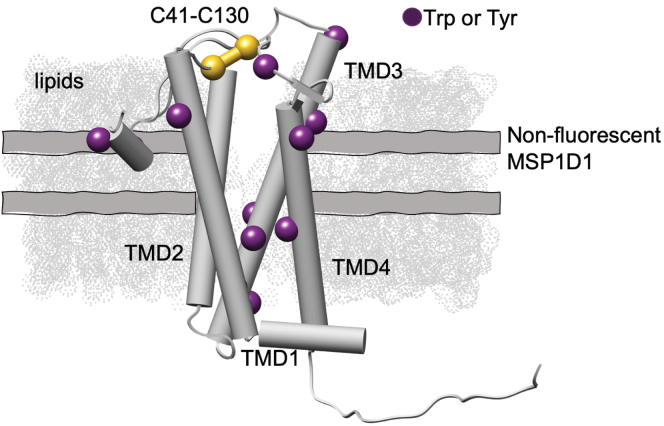
Recently, an MSP construct was engineered that replaces all tryptophan and tyrosine residues with phenylalanine for reconstituting membrane proteins in nanodiscs, making them fluorescently dark (19). Nanodiscs are attractive systems because the lipid composition can be precisely controlled and, therefore, can be used as a model membrane to systematically dissect the effect of lipid types on membrane protein thermal stability. In this work, we introduce a new method that couples the nonfluorescent MSP to form dark nanodiscs with nanoDSF measurements of intrinsic tryptophan fluorescence. This offers a facile measurement of the thermal unfolding of reconstituted membrane proteins without signal interference from the nanodisc. Further, thermostability measurements by nanoDSF offer many advantages in that it is a label-free approach that monitors the shift in intrinsic tryptophan fluorescence as the protein unfolds and does not require a high sample consumption. We test the utility of this approach using the Escherichia coli (E. coli) disulfide bond formation protein B (DsbB) as a model membrane protein system. The topology of DsbB is shown in Fig. 1 and consists of four transmembrane domains (TMD1–TMD4) with a catalytically conserved interloop disulfide bond that plays an important part in the reaction cycle (20). We use this as a model membrane protein because the thermodynamic behavior of DsbB has been extensively evaluated under chemical denaturation conditions, making it an ideal test system for comparison (21,22).
Figure 1.
The structural topology and location of the tryptophan and tyrosine residues mapped onto the NMR-derived model of DsbB reconstituted in a dark nanodisc (PDB: 2K73). DsbB is reconstituted in a dark nanodisc system loaded with DMPC and DMPG lipids at a 3:2 molar ratio and the nonfluorescent MSP. The four transmembrane domains are labeled TMD1–TMD4 and the interloop disulfide bond C41–C130 is shown in yellow. The C41–C130 interloop disulfide bond connects the two periplasmic loops. We also show the tryptophan and tyrosine residues as purple spheres for visualization.
Early studies by Otzen et al. combined fluorescence-based stopped-flow kinetic experiments and chemical denaturation in sodium dodecyl sulfate-DDM (SDS-DDM) mixed micelles to study the unfolding/refolding of DsbB. These experiments revealed a three-state unfolding model for DsbB consisting of the native state in DDM, an unfolding intermediate, and the SDS-denatured state (21). The authors also performed the same experiments under reducing conditions to investigate the role disulfide bonds play in the stability of DsbB. They showed that, under reducing conditions, unfolding rate constants increased and that the disulfide bonds contribute to the stability of the protein. Regardless, it remains unclear if the observed intermediate state in SDS-DDM mixed micelles is biophysically relevant.
In this work, we report nanoDSF thermostability data for DsbB reconstituted in dark nanodiscs to showcase the utility of our method. Our data suggest DsbB undergoes a three-state/biphasic unfolding process in dark nanodiscs loaded with 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC)/1,2-dimyristoyl-sn-glycero-3-phosphorylglycerol (DMPG) lipids. We compare these results to a panel of different detergent micelle conditions and under reducing conditions where the C41–C130 interloop disulfide bond is disrupted. Furthermore, we showcase that this method can be used to investigate how lipids with different physiochemical properties alter the thermostability profiles for DsbB.
Methods
Expression and purification of the nonfluorescent dark membrane scaffold protein and fluorescent MSP1D1 construct
The nonfluorescent and fluorescent membrane scaffold protein MSP1D1 genes were cloned in a pET28a vector containing a tobacco etch virus (TEV) protease-cleavable N-terminal His6 tag. BL21(DE3) competent E. coli cells were transformed with the target plasmid and grown on Luria-Bertani (LB) agar plates overnight at 37°C. A single colony was selected and used to inoculate a 150-mL LB starter culture and grown overnight at 37°C. Flasks containing 1 L of terrific broth medium were inoculated with 10 mL of the starter culture and grown at 37°C at 230 rpm in the presence of 100 ng/mL kanamycin. Protein expression was induced at an optical density 600 of 0.6–0.8 with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultured for another 24 h at 20°C. Cells were harvested by centrifugation at 6500 rpm for 20 min and cell pellets were stored at −80°C or immediately resuspended in lysis buffer for purification.
Cell pellets expressing the nonfluorescent or fluorescent MSP1D1 proteins were resuspended (5 mL/g of pellet) in buffer A (50 mM Tris, 300 mM NaCl, pH 8.0) with the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF), 1% Triton X-100, EDTA-free protease cocktail inhibitor tablet (Sigma-Aldrich) (1 tablet per 50 mL of buffer), and 5 mM Mg acetate. The cells were lysed by sonication for 10 min (60% amplitude with 5 s on/5 s off) on ice. The cell lysis solution was centrifuged at 50,000 × g for 20 min and the supernatant was loaded onto an Ni-NTA (nitriloacetic acid) gravity-flow column. The Ni-NTA resin was washed with 10 column volumes of the following in this order: buffer A and 1% Triton X-100, buffer A and 75 mM sodium cholate, buffer A, and buffer A plus 20 mM imidazole. Proteins were eluted with buffer A containing 500 mM imidazole. Isolated proteins were dialyzed against TEV cleavage buffer (20 mM Tris-HCl, 100 mM NaCl, 1 mM DTT, pH 7.5) at 4°C for 24 h. The TEV cleavage reaction was set up using TEV protease at a 1:50 ratio of protease to protein to remove the N-terminal His6 tag.
The cleaved sample was loaded onto a second Ni-NTA column to remove the TEV protease and N-terminal His6 tag. The flow-through was collected containing the target protein. The MSP1D1 samples were size excluded using a 120-mL HiLoad 16/600 Superdex S200 preparative column (Cytiva) equilibrated with reconstitution buffer (40 mM Tris-HCl, 200 mM NaCl, pH 7.5). Protein fractions were analyzed by SDS-PAGE. Pooled fractions were quantified using a Bradford assay and concentrated to 500 μM in an Amicon centrifugal filter with a molecular weight cutoff (MWCO) of 10,000 Daltons (Da) and analyzed by SDS-PAGE to confirm that the purity of the samples was greater than 95%. As a control, the Ni-NTA-bound TEV protease and His6 tag were eluted from the resin. The resin was washed with reconstitution buffer followed by a 10 mM imidazole wash. The TEV protease and cleaved His6 tag were eluted with reconstitution buffer and 500 mM imidazole. The final yield we obtained of the dark MSP1D1 was 0.55 mg per liter of culture.
Expression and purification of DsbB
The wild-type DsbB protein construct (20) was inserted into pET22b vector and transformed into BL21(DE3) E. coli cells. Cells were grown on LB agar plates containing 100 μg/mL ampicillin overnight at 37°C. A single colony was selected and used to inoculate a 150-mL starter culture grown overnight at 37°C. Flasks containing 1 L of minimal (M9) medium were inoculated with 10 mL of starter culture and grown in the presence of 100 μg/mL ampicillin. Cells were cultured at 37°C at 230 rpm until the optical density 600 reached 0.6–0.8. Cells were transferred to room temperature and protein expression was induced with 1 mM IPTG for 18 h. Cells were harvested by centrifugation at 6500 rpm for 15 min at 4°C. Cell pellets were stored at −80°C or immediately resuspended in lysis buffer for purification.
Each cell pellet was resuspended in 40 mL of buffer B (50 mM Tris-HCl, 300 mM NaCl, pH 8.0) with the addition of 1 mM PMSF, 5 mM Mg acetate, and 1 EDTA-free protease cocktail inhibitor tablet (Sigma-Aldrich) (1 tablet per 50 mL of buffer). The cell lysis solution was rotated at 4°C for 30 min and then lysed by sonication for 10 min (60% amplitude with 5 s on/5 s off). The membrane fraction was pelleted by ultracentrifugation at 100,000 × g for 1 h at 4°C and the supernatant was discarded. The membrane fraction was homogenized on ice with buffer B (20 mL/pellet) containing 1% dodecylphosphocholine (DPC, FOS-CHOLINE-12, Anatrace). Homogenized cells were ultracentrifuged at 100,000 × g for 30 min to remove the insoluble fraction. The remaining supernatant was loaded onto an Ni-NTA gravity-flow column equilibrated with buffer B and 0.05% DPC. The Ni-NTA column was washed with buffer B and 0.05% DPC followed by buffer B plus 0.05% DPC and 10 mM imidazole. DsbB was eluted with buffer B plus 0.05% DPC and 500 mM imidazole. The collected sample was concentrated to 500 μM and size excluded using a 120-mL HiLoad 16/600 Superdex S200 preparative column (Cytiva) equilibrated with fast protein liquid chromatography buffer (50 mM Tris-HCl, 300 mM NaCl, 0.15% DPC, pH 8.0). The pooled fractions were concentrated to 500 μM in an Amicon centrifugal filter (10,000 Da MWCO) and analyzed by SDS-PAGE to confirm the purity of the sample was greater than 95%. For the reduced DsbB sample, we followed the above protocol with the exception that each buffer in the purification protocol was supplemented with 2 mM tris (2-carboxyethyl) phosphine (TCEP).
Reconstitution reactions for DsbB in nanodiscs and empty nanodiscs
All purified proteins were concentrated to 500 μM as described above and reconstitutions were set up as previously described by Nasr. et al. (23). Lipids were prepared as an 80 mM stock of DMPC and DMPG lipids at a 3:2 molar ratio solubilized in 10% n-decyl-β-D-maltoside (DM). We also prepared 80 mM stocks of 100% 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) and E. coli polar lipid extract (EPL) solubilized in 10% DM. Bio-Beads SM2 resin (Bio-Rad) was prepared freshly before each reaction by washing the beads with 10 mL of methanol followed by excess deionized water. DsbB was assembled into nanodiscs by adding DsbB (160 μM), dark or fluorescent MSP1D1 (160 μM), and lipids (13.6 mM) to reconstitution buffer at a 1:1:85 ratio, respectively. Reaction components were added to the reconstitution buffer in the order of lipids, MSP1D1, and DsbB. The reaction was then equilibrated at room temperature for 1 h. Fresh Bio-Beads (0.5 g of wet beads/300 μL of total reaction volume) were added to the reaction and allowed to tumble overnight at room temperature to remove all detergents. The DsbB-containing nanodisc solution was collected to remove the Bio-Beads and loaded three times onto an Ni-NTA column equilibrated with reconstitution buffer (i.e., DsbB is His6 tagged). The Ni-NTA resin was washed with excess reconstitution buffer to remove empty nanodiscs and/or large vesicles. The assembled DsbB-nanodisc complex was then size excluded at room temperature using a 24 mL Superdex 200 Increase 10/300 GL column (Cytiva) equilibrated with reconstitution buffer. Selected fractions were analyzed by SDS-PAGE to confirm the purity of the DsbB-nanodisc samples. Fractions were pooled and concentrated at room temperature using a low spin speed (i.e., 2500 × g) in an Amicon centrifugal filter (30,000 Da MWCO) to 1 mg/mL. Empty nanodisc reconstitution reactions were performed using the same protocol described above with the exception that the volume of DsbB was replaced with reconstitution buffer (i.e., the reaction volumes were the same for each reconstitution). Reduced DsbB was assembled into nanodiscs as described above with the exception that the reconstitution buffer was supplemented with 2 mM TCEP.
NanoDSF measurements for detergent-solubilized DsbB under different micelle conditions
To prepare DsbB under different detergent micelle conditions, we followed the method outlined for a membrane protein stability detergent screen as described (24). Briefly, DPC-solubilized DsbB was diluted 10-fold into detergent buffers (LMNG, lauryl maltose neopentyl glycol; DDM, LMPG, lyso-myristoylphosphatidylglycerol; DM, and LDAO, lauryldimethylamine oxide) that were made with a 50-fold excess of the critical micelle concentration of the target detergent. The diluted DsbB samples were incubated at 4°C overnight to ensure detergents were fully exchanged. All detergent micelle samples were prepared to a final concentration of 1 mg/mL.
NanoDSF thermostability measurements
All samples were run on a Prometheus NT.48 nanoDSF instrument (NanoTemper Technologies). Samples were set up in triplicate at a final concentration of 1 mg/mL (10 μL/capillary). All nanodisc samples and detergent samples were measured at a scan rate of 0.1°C per minute over a temperature range of 25°C–95°C and 20°C–95°C, respectively. Samples were excited at 280 nm and the intrinsic tryptophan fluorescence at 350 and 330 nm were recorded as a function of temperature to monitor changes upon thermal unfolding.
CD spectroscopy
CD wavelength and temperature scans were performed using a Chirascan VX instrument (Applied Photophysics) to monitor changes to DsbB secondary structure upon thermal unfolding in nanodiscs. Each sample was buffer exchanged into 40 mM Tris-HCl and 100 mM NaCl at pH 7.5. The wavelength range covered 190–280 nm. All samples were measured at a scan rate of 1°C per minute over a thermal ramp of 25°C–95°C. Each sample was scanned in triplicate at a final concentration of 0.2 mg/mL in a 1-mm cuvette.
Quantification of free thiol content
The free thiol content for detergent-solubilized DsbB (DPC) and DsbB reconstituted in dark nanodiscs (DMPC/DMPG) was quantified using the Measure-IT Thiol Assay Kit (Invitrogen). Samples were measured in triplicate at a final DsbB concentration of 1 mg/mL and free thiol concentration was quantitated relative to a reduced glutathione standard. We calculated the average free thiol concentration and standard deviation and converted this into a percentage relative to the total number of cysteine residues. The dark MSP does not contribute to the thiol measurement because there is no cysteine content in the dark nanodisc construct.
Data analysis
All raw nanoDSF data were collected and then exported from the Prometheus Panta Control software for visualization in MoltenProt (24,25). The experimental derivatives were exported from MoltenProt and further analyzed in a Jupyter Notebook through Matplotlib (26). The F350/F330 ratios for each of the triplicate samples were averaged and normalized to the initial value with Matplotlib. The first derivatives were plotted with Matplotlib to determine the inflection points corresponding to the melting temperatures. The fraction unfolded curves were calculated and plotted by fitting the F350/F330 ratio data to a two-state equilibrium model. The rate constants of the unfolding transition and baseline transition as well as the baseline offset and noise were extrapolated. These parameters were utilized to calculate the baseline-corrected experimental curves from the equation utilized by MoltenProt (27). For each CD spectrum, the ellipticity value (Θ, mdeg) was converted to standard units (deg·cm2/dmol) to normalize molar ellipticity based on sample concentration, path length of the cuvette, and the number of amino acid residues (28,29). The molar ellipticity values were plotted as a function of wavelength in Matplotlib as an average of the triplicate scans (n = 3).
Results
The thermal unfolding curve for DsbB reconstituted in a dark nanodisc loaded with DMPC/DMPG lipids follows a biphasic unfolding pattern with a minor transition followed by a second major transition
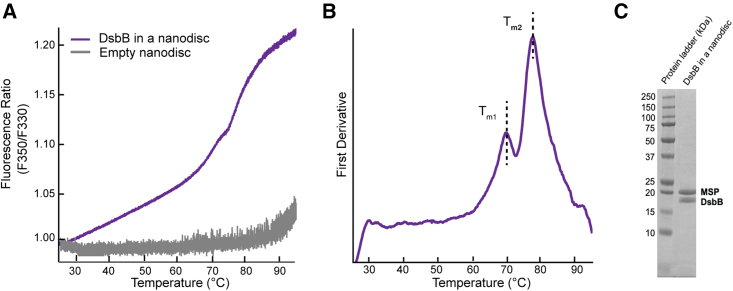
One of the most widely used proxies for overall protein stability is thermostability. We reconstituted DsbB in dark nanodiscs loaded with a 3:2 molar ratio of DMPC and DMPG lipids and measured the thermostability of the protein by nanoDSF. Fig. 2 A shows the intrinsic tryptophan fluorescence ratio at 350 and 330 nm for DsbB (F350/F330 ratio) over a thermal ramp from 25°C to 95°C. The inflection points of the unfolding curve determined from the experimental derivative correspond to the unfolding midpoint melting temperatures (Tm). Fig. 2 B shows the first derivative of the F350/F330 ratio as a function of temperature. Fig. 2 C shows a representative SDS-PAGE gel demonstrating sample purity after a reconstitution reaction for DsbB in a dark nanodisc. The experimental derivative of the unfolding curve reveals two distinct Tm values of 70.5 ± 0.3°C and 77.5 ± 0.1°C. These results suggest that DsbB in a nanodisc follows a three-state/biphasic unfolding model: native (N) → intermediate (I) → unfolded (U). The three-state/biphasic unfolding model of DsbB in a DMPC/DMPG dark nanodisc is consistent with prior observations of the folding/unfolding of DsbB in mixed micelles measured by stopped-flow kinetic experiments (21,30).
Figure 2.
NanoDSF thermal unfolding curve for DsbB in a dark nanodisc. (A) The F350/F330 thermal unfolding curve for DsbB reconstituted in a dark nanodisc model membrane system (violet). The thermal unfolding curve for empty dark nanodiscs is shown in gray. (B) The first derivative plot of the F350/F330 ratio with respect to temperature for DsbB in a nanodisc. The inflection points correspond to Tm values of 70.5 ± 0.3°C and 77.5 ± 0.1°C, respectively. All samples were run in triplicate for three independent preparations and the average is plotted. (C) Representative SDS-PAGE gel after size exclusion to showcase the successful reconstitution of DsbB in a dark nanodisc.
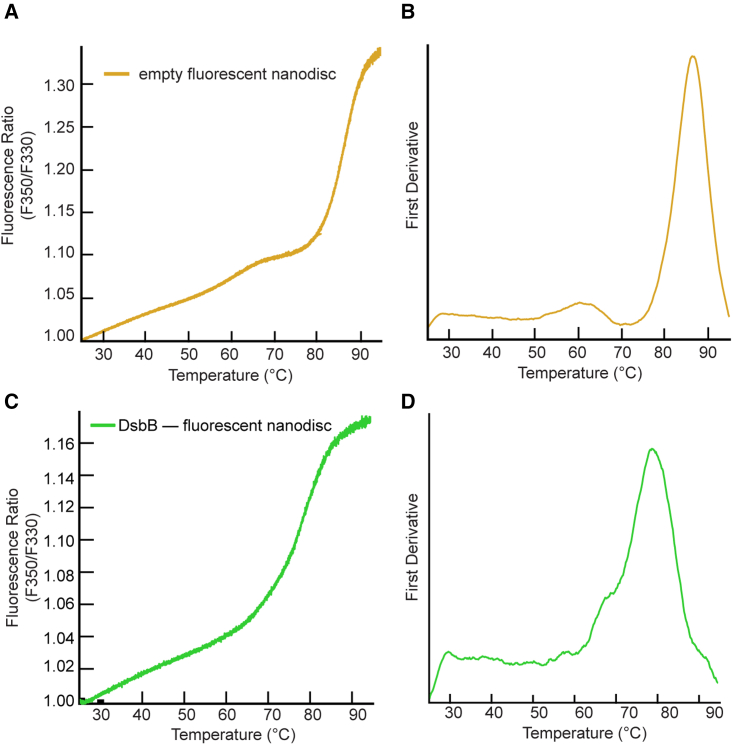
To confirm that the nanodisc does not contribute to the three-state/biphasic unfolding mechanism, we performed nanoDSF measurements on empty dark nanodiscs (i.e., in the absence of protein) loaded with the same lipid content. Indeed, the dark nanodisc itself is fluorescently undetectable over the temperature gradient (Fig. 2 A, gray line). This can be compared to the results for empty nanodiscs prepared using conventional (fluorescent) MSPs (Fig. 3 A). The near-baseline signal for the empty dark nanodisc sample up to roughly 85°C suggests that the engineered MSP used for reconstitutions does not interfere with the signal observed for the target DsbB protein. Furthermore, DMPC and DMPG lipids have melting temperatures below the range of the thermal gradient measured (i.e., below 25°C) and both are cylindrical lipids that lack the required geometry for a transition to a hexagonal or inverted hexagonal phase (31). Therefore, we do not expect that the phase transition of the lipids used in this work contributes to the unfolding transitions observed for DsbB.
Figure 3.
NanoDSF measurement for empty and DsbB reconstituted fluorescent MSP-based nanodiscs. (A) The F350/F330 thermal unfolding curve for the empty fluorescent MSP-based nanodisc (yellow). (B) The first derivative plot of the unfolding curve shows an inflection point at a Tm value of 86.6°C ± 0.6°C. All samples were run in triplicate and the average is plotted. (C) The F350/F330 thermal unfolding curves for DsbB reconstituted in a fluorescent MSP-based nanodisc (DMPC/DMPG). (D) The first derivative plot of the F350/F330 ratio with respect to temperature for DsbB in the fluorescent MSP-based nanodisc (green). The inflection point corresponds to a single Tm value of 79.0°C ± 0.4°C for DsbB. All samples were run in triplicate and the average is plotted.
For visualization of the Tm values at the inflection point, we also report the fraction unfolded curves (baseline corrected) in Fig. S1 A for DsbB in DMPC/DMPG dark nanodiscs. Furthermore, we report the dynamic light-scattering measurements collected in Fig. S2 A for DsbB reconstituted in a DMPC/DMPG dark nanodisc and Fig. S3 A for the empty dark DMPC/DMPG nanodisc as a cumulative radius plot to evaluate any contributions of aggregation to our unfolding curve data. In the case of DsbB in a dark DMPC/DMPG nanodisc, the cumulative radius plot shows that the system size increases moderately to 340 nm at a temperature of 76.5°C and 664 nm at a temperature of 83.9°C. These temperature-dependent changes in cumulative radius occur after the minor and major thermal unfolding states observed for DsbB. This information suggests that 1) the change in system size correlates with the unfolding of DsbB, and/or 2) protein unfolding at the midpoint melting temperatures is followed by protein aggregation.
The thermal unfolding curves for DsbB under a panel of detergent micelle conditions each show a major unfolding transition and decreased Tm values compared to nanodiscs
We selected a range of detergents with different physiochemical properties and hydrocarbon chain lengths to compare the thermostability of DsbB in dark nanodiscs to different micelle conditions. Fig. S4 A–E shows the F350/F330 ratio for the unfolding curves for detergent-solubilized DsbB in LMNG, DDM, LMPG, DM, and LDAO. Fig. S4 F–J shows the respective experimental derivative plots for the unfolding curves under each detergent micelle condition, revealing a single major inflection point corresponding to a single Tm value. Detergent-solubilized DsbB switches to a single major unfolding transition in the case of DDM and LMPG micelles. A second less-defined peak can be seen toward the end of the thermal gradient, roughly 40°C above the first transition, for DsbB under LMNG, DM, and LDAO conditions. The measured thermostability for DsbB in detergent micelles from most stable to least stable is LMNG > DDM > LMPG > DM > LDAO. The fraction unfolded curves (baseline corrected) for each detergent condition are reported in Fig. S5 for visualization of the Tm values at the inflection point as well as the dynamic light-scattering data collected as a cumulative radius plot in Fig. S4 K–O. The cumulative radius plots for DsbB in detergent micelles shows marked variability. DM, LMNG, and LDAO have similar readouts with a drastic increase in system size at the end of the thermal gradient. In contrast, DDM and LMPG both show a minimal increase in the cumulative radius at the end of the thermal ramp.
To deconvolute the second less-defined peak, which occurs 40°C post the first thermal transition observed peak for DsbB under LMNG, DM, and LDAO conditions, we evaluated the dynamic light-scattering traces collected in parallel with the fluorescence measurements (Fig. S4 K–O), which present several interesting observations. First, the cumulative radius plots for DsbB in DM, LMNG, and LDAO each show a significant increase in system size (i.e., >4000 nm) toward the end of the thermal gradient. This corresponds to the second less-defined peaks in the first derivative plots. This suggests that the delay in the onset between the first and second peaks is the result of protein aggregation and micelle dissociation from the unfolded protein state. Second, the cumulative radius plots for DsbB in detergent micelles show marked variability. LMNG, DM, and LDAO have similar readouts with a drastic increase in system size at the end of the thermal gradient. In contrast, DDM and LMPG both show a minimal increase in the cumulative radius at the end of the thermal ramp. We speculate the different patterns observed for the change in system size are attributed to the different chemical and physical properties of the detergent micelles. For example, DsbB has a +4 charge state at the pH of our measurements, and detergents such as LMPG are negatively charged (32). We propose that electrostatic interactions between LMNG and DsbB are strong enough to prevent micelle dissociation from the unfolded state and thus prevent aggregation.
The experimentally determined Tm value for the fluorescent MSP-based nanodisc is well outside the range of Tm values observed for DsbB
We tested the thermostability of a fluorescent MSP-based nanodisc to ensure that it was not contributing to the three-state/biphasic unfolding pattern observed for DsbB. Fig. 3 A shows the F350/F330 ratio for the unfolding curve for a fluorescent MSP-based nanodisc in the absence of DsbB. Fig. 3 B shows the respective experimental derivative plot for the unfolding curve. The inflection point corresponds to a Tm value of 86.6 ± 0.6°C, which is more than 10°C above the melting temperatures observed for DsbB in a dark nanodisc (see Fig. 2 B). The melting temperature we observe for the empty fluorescent nanodisc is consistent with previous studies that report a Tm value of roughly 85°C (33,34). Fig. S1 B shows the fraction unfolded curve (baseline corrected) for the fluorescent MSP-based nanodisc for visualization of the Tm value at the inflection point. These data illustrate that the baseline DSF trace from fluorescent nanodiscs would be problematic had these nanodiscs been used for this study of DsbB. The dynamic light-scattering data collected for the empty fluorescent nanodiscs are shown in Fig. S3 B (third row) as a cumulative radius plot indicating that aggregation of the empty fluorescent nanodiscs falls outside the range of Tm values observed for DsbB in the dark nanodisc. This suggests that the delay in the onset between the first and second peaks is the result of protein aggregation and micelle dissociation from the unfolded protein state.
The biphasic unfolding pattern for DsbB is broadened and not well defined in a fluorescent MSP1D1-based nanodisc (DMPC/DMPG)
We also reconstituted DsbB in a fluorescent MSP-based nanodisc (DMPC/DMPG) to determine how the fluorescent MSP affects the thermal unfolding curve of DsbB. Fig. 3 C shows the F350/F330 ratio for the unfolding curve for DsbB reconstituted in a fluorescent MSP-based nanodisc and Fig. 3 D shows the respective experimental derivative plot for the unfolding curve. These data reveal that the first inflection point (Tm1) is unresolvable, which convolutes the interpretation of the biphasic unfolding pattern compared to dark nanodiscs. We propose here that the unfolding of the fluorescent MSP masks the signal of the first inflection point through peak broadening of the unfolding curve. Fig. S1 C shows the fraction unfolded curve (baseline corrected) for DsbB reconstituted in the fluorescent MSP-based nanodisc for visualization of the Tm value at the inflection point and the respective dynamic light-scattering data collected are shown in Fig. S3 C (third row).
The thermal unfolding mechanism for DsbB measured under reducing conditions shifts the unfolding curve from three-state/biphasic to two-state/monophasic
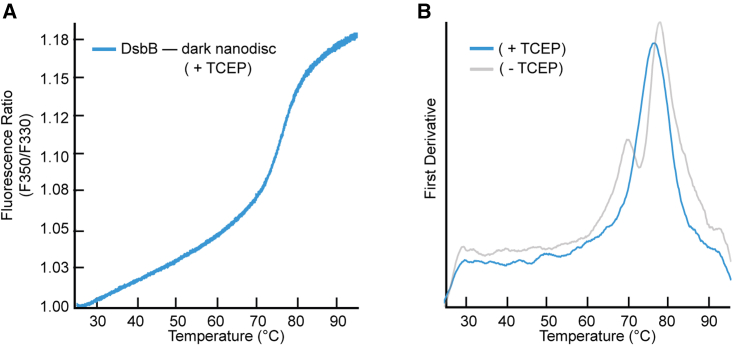
We purified wild-type DsbB under reducing conditions by adding 2 mM TCEP to test the hypothesis that the interloop disulfide bond between cysteine residues C41 and C130 plays a key role in the formation of the intermediate unfolding state. Fig. 4 A shows the F350/F330 ratio for the unfolding curve for C41–C130 reduced DsbB assembled in a dark nanodisc. Fig. 4 B shows the respective first derivative plot for the unfolding curve revealing a single inflection point corresponding to a Tm value of 76.6°C, which is within the range of the Tm2 value observed for DsbB under nonreducing conditions. This suggests that the intermediate unfolding state observed for DsbB is mediated by the C41–C130 disulfide bond. More interestingly, this distinct three-state/biphasic unfolding pattern is not observed under detergent conditions, further suggesting that this interloop disulfide bond intermediate state is mediated by the presence of DMPC/DMPG lipids. Fig. S1 D shows the fraction unfolded curve (baseline corrected) for C41–C130 reduced DsbB reconstituted in a nanodisc for visualization of the Tm value at the inflection point and Fig. S2 B (third row) shows dynamic light-scattering data collected for DsbB under reducing conditions in a dark nanodisc as a cumulative radius plot.
Figure 4.
NanoDSF measurements for DsbB reconstituted in dark nanodiscs under reducing conditions. (A) The F350/F330 thermal unfolding curve of DsbB reconstituted in the dark nanodisc in the presence of the reducing agent TCEP at 2 mM. (B) The first derivative plot shows a single inflection point corresponding to a Tm value of 76.6°C ± 0.5°C. The first derivative plot for DsbB reconstituted in the dark nanodisc in the absence of reducing conditions has been overlayed as a comparison. All samples were run in triplicate and the average is plotted.
We also quantified the free thiol content for DsbB reconstituted in dark nanodiscs (DMPC/DMPG) to determine the population of protein in the oxidized versus the reduced state post assembly into nanodiscs and compared this to detergent-solubilized DsbB in DPC micelles. The free thiol content determined for DsbB in micelles and DsbB reconstituted in nanodiscs was 2.25% and 1.59%, respectively, and is summarized in Table S1. This suggests that post reconstitution into nanodisc, DsbB retains its oxidized state and represents the major population, and the fraction of reduced cysteines is negligible in dark nanodiscs. These data support that the reconstitution of DsbB into dark nanodiscs does not result in a mixture of oxidized/reduced states and further confirms that loss of the minor transition observed here is indeed the result of the reduction of the C41–C130 disulfide bond.
CD measurements reveal that DsbB reconstituted in a dark nanodisc maintains residual α-helical content at 95°C
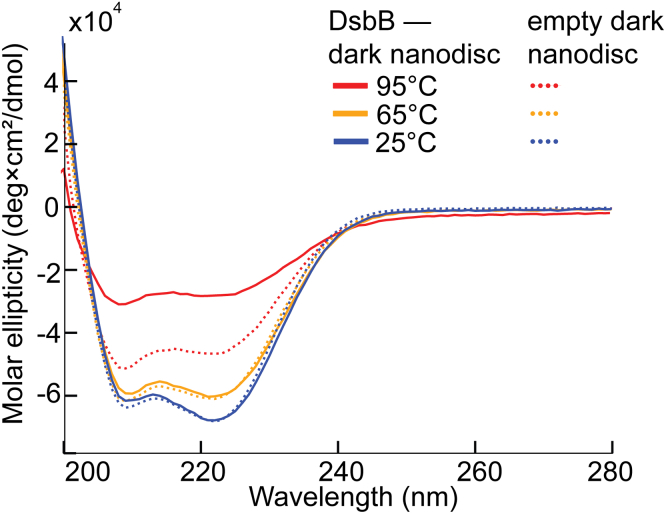
We reconstituted DsbB in a dark nanodisc using our standard DMPC/DMPG lipid composition and collected CD data to determine how the secondary structural content for DsbB changes along the thermal gradient. We also performed CD melting experiments for empty DMPC/DMPG dark nanodiscs as a control. Fig. 5 shows the CD signal for empty dark nanodiscs and DsbB reconstituted in dark nanodiscs plotted as molar ellipticity over the UV wavelength range from 190 to 280 nm. In the case of the empty dark nanodiscs, the molar ellipticity change is small over the thermal ramp from 25°C to 95°C (%Δ208 = 18.2% and %Δ222 = 31.8%). This is consistent with prior observations for empty MSP1D1 nanodiscs (35). For DsbB reconstituted in dark nanodiscs, the CD spectral signature for α-helix content is high at the beginning of the thermal ramp and dampens over the thermal melting experiments. This is reflected by the strong signal at 208 and 222 nm 25°C and the reduced signal at 208 and 222 nm 95°C (%Δ208 = 48.8% and %Δ222 = 58.8%). This suggests that the contribution of the dark MSP construct to the change in CD signal is dependent on DsbB. Therefore, in the case of DsbB-loaded nanodiscs, we interpret that the change in CD signal is largely related to the unfolding of DsbB. Furthermore, we would expect the CD signature to be flat at these two wavelengths if all α-helical content were lost after the thermal unfolding of DsbB, but the residual signature suggests that DsbB partially maintains its secondary structural content and is consistent with prior observations for DsbB (36,37,38).
Figure 5.
CD spectra for DsbB reconstituted in a dark nanodisc (DMPC/DMPG) and empty dark nanodiscs (DMPC/DMPG) in the absence of DsbB at different temperature points along a thermal gradient. CD spectra were collected over a thermal ramp ranging from 25°C to 95°C at a step of 1°C/min in the 190- to 280-nm UV region. The CD spectra for each sample are shown at 95°C (red), 65°C (orange), and 25°C (blue). DsbB reconstituted in a dark nanodisc is shown as a solid line and the empty nanodisc is shown as a dotted line at each respective temperature. All CD scans were done in triplicate and the average is plotted.
The thermostability profile for DsbB shifts in different lipid environments
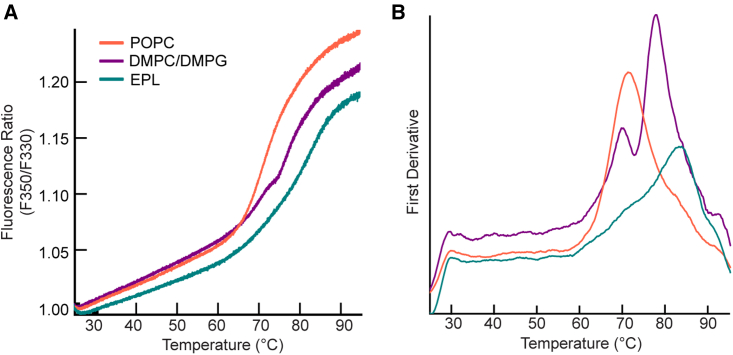
We reconstituted DsbB in dark nanodiscs using two different lipid compositions to compare to our standard DMPC/DMPG dark nanodiscs and showcase the utility of the presented method. We selected lipids based on the criteria that they 1) had different phase transition temperatures, 2) had a complex phospholipid content, and/or 3) represented a more native-like environment for DsbB. For these reasons, we tested POPC because the phase transition temperature is −2°C and well below that of DMPC (24°C) and DMPG (23°C). We also tested E. coli polar lipid extract (EPL) because the phospholipid content is compositionally diverse and consists of 67% phosphatidylethanolamine, 23.2% phosphatidylglycerol (PG), and 9.8% cardiolipin. Fig. 6 A shows an overlay of the F350/F330 ratios for the unfolding curves for each lipid composition tested and Fig. 6 B shows the respective first derivative plots. Surprisingly, compared to the standard DMPC/DMPG lipid composition used in our method development experiments, the unfolding pattern for both POPC and EPL is monophasic with Tm values of 71.2 ± 0.2°C and 82.9 ± 0.5°C, respectively. We attribute the reduced thermostability for DsbB in POPC-loaded dark nanodiscs (ΔTm = −6.3°C) to the lower phase transition temperature, resulting in a more fluid-like membrane environment where the lipids are disordered and have higher lateral mobility. Additionally, POPC contains a zwitterionic headgroup with a net charge of zero. We would expect negatively charged headgroups such as DMPG to enhance the stability of the cationic DsbB, which is consistent with our data. We observe the opposite effect for EPL (ΔTm = +5.4°C). This is expected as cardiolipin is well recognized for its stabilizing effects on membrane proteins (16) and because cardiolipin is negatively charged and may stabilize DsbB given that the protein carries a positive charge (+4) at physiological pH. Fig. S1 E and F show the fraction unfolded curve (baseline corrected) for DsbB reconstituted in POPC and EPL dark nanodiscs for visualization of the Tm value at the inflection point, and the respective dynamic light-scattering data collected are shown in Fig. S2 C and D (third row). Collectively, these data demonstrate the utility of our method and how lipids with different biophysical properties shift the thermostability profile of DsbB. For comparison, we summarize all nanoDSF recorded melting temperatures in Table 1.
Figure 6.
NanoDSF thermal unfolding curves for DsbB in dark nanodiscs loaded with different lipid compositions. (A) The F350/F330 thermal unfolding curves for DsbB reconstituted in a dark nanodisc loaded with 100% POPC (orange), DMPC and DMPG at a 3:2 molar ratio (purple), and 100% E. coli polar lipid extract (teal). (B) The first derivative plot of the F350/F330 ratio with respect to temperature for DsbB in a dark nanodisc for each lipid composition. The inflection points correspond to melting temperatures of Tm = 71.2 ± 0.2°C (POPC), Tm1 = 70.5 ± 0.3°C and Tm2 = 77.5 ± 0.1°C (DMPC/DMPG), and Tm = 82.9 ± 0.5°C (EPL). All samples were run in triplicate and the average is plotted.
Table 1.
Table summarizing the measured inflection points (Tm)
| Experimental condition | Tm1 (°C) | Tm2 (°C) |
|---|---|---|
| DsbB: dark nanodisc (DMPC/DMPG) | 70.5 ± 0.3 70.0 ± 0.1 70.3 ± 0.6 |
77.5 ± 0.1 79.1 ± 0.2 79.7 ± 0.5 |
| DsbB: dark nanodisc (POPC) | 71.2 ± 0.2 | – |
| DsbB: dark nanodisc (E. coli polar lipid extract) | 82.9 ± 0.5 | |
| Empty fluorescent nanodisc | 86.6 ± 0.6 | |
| DsbB: MSP1D1 nanodisc (DMPC/DMPG) | 79.0 ± 0.4 | |
| DsbB: dark nanodisc + TCEP | 76.6 ± 0.5 | |
| DsbB: LMNG | 56.4 ± 0.9 | |
| DsbB: DDM | 49.5 ± 1.0 | |
| DsbB: LMPG | 45.4 ± 0.1 | |
| DsbB: DM | 40.1 ± 0.9 | |
| DsbB: LDAO | 36.7 ± 0.2 |
Discussion
Tools to evaluate membrane protein stability in the presence of lipids or a lipid-based model membrane system remain a technical challenge and are often reverted to studies in detergent micelles. We describe in this work the development and evaluation of dark nanodiscs as a model membrane system for nanoDSF thermostability measurements. We present this as a new tool that can be used to evaluate how lipid-protein interactions regulate membrane protein stability. We show that the dark nanodiscs are fluorescently undetectable by nanoDSF, making them an ideal model membrane system to investigate the effect of lipids and specific lipid types on membrane protein thermostability.
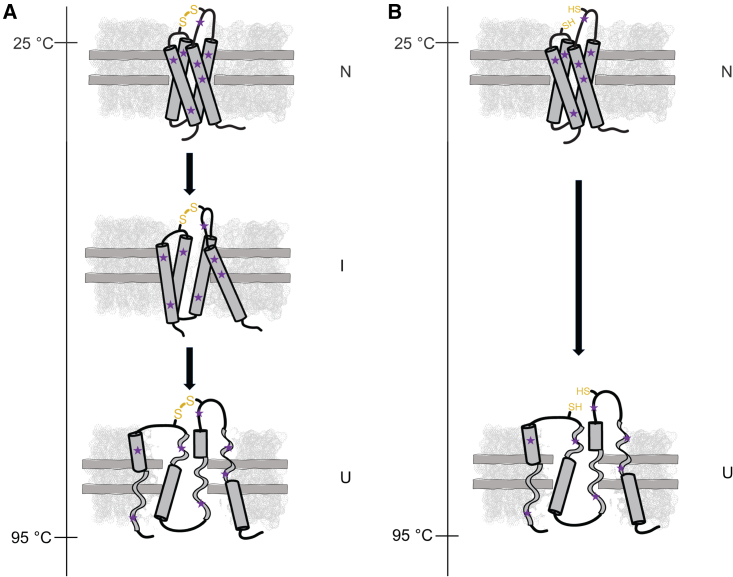
We observed that DsbB reconstituted in a dark nanodisc loaded with DMPC/DMPG lipids follows a three-state/biphasic thermal unfolding model that can be described in terms of a native (N) state, an intermediate (I) state, and an unfolded (U) state. Fig. 7 A depicts the assembled DsbB-dark nanodisc complex as it unfolds along the thermal gradient under nonreducing conditions where the interloop disulfide bond between residues C41 and C130 is in the oxidized state. In this model, DsbB retains part of its α-helical secondary structural content in the U state. This is supported by the observation that the nanoDSF thermal unfolding curve for DsbB does not reach a plateau (see Fig. 2 A) and is also reflected in the CD spectral fingerprint at the end of the thermal melt (see Fig. 5). For comparison, we also depict the unfolding pathway for DsbB in the case where the disulfide bond between residues C41 and C130 is in the reduced state in Fig. 7 B. The thermostability profile for DsbB in DMPC/DMPG dark nanodiscs compared to detergent micelles revealed two distinct differences: 1) the thermostability for DsbB in a nanodisc is significantly higher than under micelle conditions, and 2) the distinct three-state/biphasic unfolding model of DsbB in a nanodisc switches to a two-state/monophasic process under micelle conditions revealing a major thermal transition state. These observations further signify the impact and role lipids play in the thermal stability and thermal unfolding of membrane proteins.
Figure 7.
Schematic summarizing the proposed three-state unfolding model for DsbB in a dark nanodisc composed of DMPC/DMPG lipids. (A) DsbB reconstituted in a dark nanodisc loaded with DMPC/DMPG lipids under nonreducing conditions (i.e., the interloop C41–C130 disulfide bond is oxidized) as it thermally unfolds from the native state (N) to an intermediate state (I) and to the unfolded (U) state. We propose that the three-state/biphasic model is the result of a disulfide bond-mediated I state in the presence of DMPC/DMPG lipids. (B) DsbB reconstituted in a dark nanodisc under reducing conditions (i.e., the interloop C41–C130 disulfide bond is reduced) as it thermally unfolds from the N state to the U state. We do not observe the formation of an I state during thermal unfolding of DsbB in our dark nanodisc system under reducing conditions. DsbB is shown in dark gray, the dark nanodisc system is shown in light gray, the tryptophan and tyrosine content for DsbB is represented in purple, the loss in secondary structural content is reflected by gray ribbons in the U state, and the C41–C130 interloop disulfide bond in the oxidized state (left) or reduced state (right) is shown in yellow.
The decrease in thermal stability for detergent-solubilized DsbB in micelles compared to DsbB reconstituted in nanodiscs is not surprising. It is well recognized that the function and stability of membrane proteins are compromised during isolation from the membrane and delipidation through detergent solubilization and, even more, can be restored by reconstituting the protein back into a lipid-based model membrane system. For example, the light-harvesting chlorophyll a/b complex of photosystem II was reported to have a lower thermostability in DDM micelles (Tm = 59.4°C) and, when reconstituted in proteoliposomes, increased to Tm values as high as 74.9°C in the presence of monogalactosyl diacylglycerol lipids (39). Notably, the thermal stability for LHCllb was determined over a range of lipid compositions and revealed that specific lipid types can shift the Tm value as much as 15°C (i.e., phosphatidylglycerol vs. monogalactosyl diacylglycerol) (39). Thermograms for the Na,K-ATPase cation transporter reconstituted in dipalmitoyl phosphatidylcholine and dipalmitoyl phosphatidylethanolamine proteoliposomes revealed different thermostability profiles under varying concentrations of cholesterol, suggesting that the mechanism by which the protein thermally unfolds is dependent on cholesterol content (40).
We also showcase in the study here how different lipid compositions affect the thermostability of DsbB reconstituted in dark nanodiscs (see Fig. 6). Interestingly, the biphasic unfolding pattern observed for DsbB in DMPC/DMPG nanodiscs does not surface under POPC and EPL compositions. This further suggests that lipid-mediated interactions between the interloop disulfide bond and DMPC and/or DMPG lipid types specifically drive the three-state/biphasic thermal unfolding pattern. Prior studies have investigated disulfide bond interactions in the presence of lipids that contribute to the stability of proteins. For example, a Raman spectroscopy study of a lung surfactant peptide B co-solubilized in a lipid mixture of DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) and DOPG (1,2-dioleoyl-sn-glycero-3-[phosphor-rac-(1-glycerol)](sodium salt)) at a 4:1 lipid to protein ratio suggests that the presence of lipids “locks” the disulfide bond into a distinct conformation (41). Another study of antimicrobial peptides in lipid nanodiscs found that reduction of disulfide bonds can have a drastic impact on the ability of these peptides to interact with lipid bilayers (42). These studies show that different lipid types can affect disulfide bond conformations and interactions with disulfides. We propose here that the DMPC and/or DMPG lipids stabilize/interact with the C41–C130 disulfide bond, resulting in the formation of a metastable unfolding I state.
Time-resolved kinetic experiments showed that the stability of DsbB in mixed micelles was reduced by half upon the reduction of the two periplasmic disulfide bonds between residues C41–C44 and C104–C130 (21). Disulfide bonds have been shown to play key roles in membrane protein stability for a number of systems such as G protein-coupled receptors (43) and ATP-binding cassette (ABC) transporters (44). For example, a simulated thermal unfolding of the G protein-coupled receptor rhodopsin revealed that the C110–C187 disulfide bond is key to retinal binding and stabilizing the folded state of rhodopsin (43). In another example, the intramolecular disulfide bond C592–C608 located in the extracellular loop of the ABC transporter ABCG2 was shown to be critical for protein stability (i.e., plasma localization and expression levels), which was significantly reduced when this disulfide bond was mutated out with glycine residues (44). However, in the case of DsbB, we do not observe a significant reduction in the overall thermostability of DsbB in a dark nanodisc under reduced conditions. Rather, we do observe a change in the unfolding pathway and a shift from a three-state unfolding pathway for DsbB in the oxidized state to a two-state unfolding pathway and loss of the unfolding intermediate under reducing conditions. This suggests that the disulfide bond formed between the catalytically conserved cysteine residues contributes to the stabilization of a metastable I state during the thermal unfolding of DsbB but does not contribute to the overall thermostability of DsbB.
From these results, we conclude that the interloop disulfide bond 1) plays a critical role in the thermal unfolding of DsbB and 2) is specific to the presence of DMPC and/or DMPG lipids. Furthermore, the minor thermal transition state is not observed under the detergent conditions tested here or under POPC and EPL-based lipid compositions. In light of these findings, our methodology offers a promising and straightforward approach for future studies, enabling the exploration of crucial protein-lipid interactions for various membrane proteins of interest. Our quantitative measurements will find application in membrane protein-linked diseases such as cardiac long QT syndrome, cystic fibrosis, and Charcot-Marie-Tooth disease.
Conclusions
Overall, the method developed herein will be broadly applicable in membrane protein biophysics research. It provides a simple tool to dissect the effect of lipid-protein interactions on membrane protein thermostability, interactions that are typically missed or difficult to measure using existing methods. For example, genetic mutations lead to the loss of important intermolecular interactions that cause structural instability and misfolding of proteins. This leads to serious genetic diseases such as cystic fibrosis, long QT syndrome, and Charcot-Marie-Tooth disease. Specifically, these diseases are caused by the misfolding of an integral membrane protein including the cystic fibrosis transmembrane conductance regulator (45), the KCNQ1 ion channel (46), and the peripheral myelin protein 22 (47), respectively. In such cases, mutations compromise membrane protein stability, resulting in aberrant protein function. Experimental and high-throughput measurements are key for understanding 1) the effects of genetic mutations on the thermostability of membrane proteins and 2) how mutations reshape native lipid-protein interactions to drive disease states, which have been historically underappreciated (48,49). Furthermore, our method can also be used to test membrane protein design candidates in cases where the goal is to design a more thermostable protein to facilitate biochemical or structural studies (50,51).
Author contributions
Conceptualization, K.V.L.; data curation, J.A.S., E.F.M., P.D.N., and J.R.M.; formal analysis, J.A.S., E.F.M., J.R.M., and K.V.L.; funding acquisition, J.M.; investigation, J.A.S., E.F.M., P.D.N., J.R.M., and K.V.L.; methodology, K.V.L.; supervision, K.V.L. and J.M.; writing – original draft, J.A.S. and E.F.M.; writing – review & editing, E.F.M., J.M., and K.V.L. All authors have read and agreed to the published version of the manuscript.
Acknowledgments
This work was supported by NIH grants R01 GM080403, R01 HL122010, and R01 GM129261. The authors further acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG) through SFB1423, project number 421152132. J.M. is supported by a Humboldt Professorship of the Alexander von Humboldt Foundation. E.F.M. was supported by a predoctoral fellowship from the National Heart, Lung, and Blood Institute (F31 HL162483-01A1). We also thank Dr. Vadim Kotov for technical assistance and discussions regarding the use of MoltenProt for data visualization and interpretation. This work was conducted using the resources of the Center for Structural Biology at Vanderbilt University. We especially thank the Lab of Dr. Charles Sanders for technical assistance with the nanoDSF experiments and for insightful discussions. The nanoDSF instrument used in this work was acquired via an ANCORA grant to the Sanders lab from Deerfield Drug Discovery (3DC).
Declaration of interests
The authors declare no competing interests.
Editor: Daniel Huster.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.11.019.
Contributor Information
Jens Meiler, Email: jens@meilerlab.org.
Kaitlyn V. Ledwitch, Email: k.ledwitch@vanderbilt.edu.
Supporting material
References
- 1.Carra J.H., Murphy E.C., Privalov P.L. Thermodynamic effects of mutations on the denaturation of T4 lysozyme. Biophys. J. 1996;71:1994–2001. doi: 10.1016/S0006-3495(96)79397-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibarra-Molero B., Loladze V.V., et al. Sanchez-Ruiz J.M. Thermal versus guanidine-induced unfolding of ubiquitin. An analysis in terms of the contributions from charge− charge interactions to protein stability. Biochemistry. 1999;38:8138–8149. doi: 10.1021/bi9905819. [DOI] [PubMed] [Google Scholar]
- 3.Woods H., Schiano D.L., et al. Schoeder C.T. Computational modeling and prediction of deletion mutants. Structure. 2023 doi: 10.1016/j.str.2023.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurauskas V., Hessel A., et al. Schanda P. How Detergent Impacts Membrane Proteins: Atomic-Level Views of Mitochondrial Carriers in Dodecylphosphocholine. J. Phys. Chem. Lett. 2018;9:933–938. doi: 10.1021/acs.jpclett.8b00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bürck J., Wadhwani P., et al. Ulrich A.S. Oriented circular dichroism: a method to characterize membrane-active peptides in oriented lipid bilayers. Acc. Chem. Res. 2016;49:184–192. doi: 10.1021/acs.accounts.5b00346. [DOI] [PubMed] [Google Scholar]
- 6.Kulandaisamy A., Sakthivel R., Gromiha M.M. MPTherm: database for membrane protein thermodynamics for understanding folding and stability. Briefings Bioinf. 2021;22:2119–2125. doi: 10.1093/bib/bbaa064. [DOI] [PubMed] [Google Scholar]
- 7.González Flecha F.L. Kinetic stability of membrane proteins. Biophys. Rev. 2017;9:563–572. doi: 10.1007/s12551-017-0324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plasencia I., Survery S., et al. Mouritsen O.G. Structure and stability of the spinach aquaporin SoPIP2;1 in detergent micelles and lipid membranes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0014674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tol M.B., Deluz C., et al. Vogel H. Thermal unfolding of a mammalian pentameric ligand-gated ion channel proceeds at consecutive, distinct steps. J. Biol. Chem. 2013;288:5756–5769. doi: 10.1074/jbc.M112.422287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie T.K., Grinkova Y.V., et al. Sligar S.G. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denisov I.G., Grinkova Y.V., et al. Sligar S.G. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 12.Frauenfeld J., Löving R., et al. Nordlund P. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods. 2016;13:345–351. doi: 10.1038/nmeth.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y. Detergent-free systems for structural studies of membrane proteins. Biochem. Soc. Trans. 2021;49:1361–1374. doi: 10.1042/BST20201080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders M.R., Findlay H.E., Booth P.J. Lipid bilayer composition modulates the unfolding free energy of a knotted alpha-helical membrane protein. Proc. Natl. Acad. Sci. USA. 2018;115:E1799–E1808. doi: 10.1073/pnas.1714668115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laganowsky A., Reading E., et al. Robinson C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nji E., Chatzikyriakidou Y., et al. Drew D. An engineered thermal-shift screen reveals specific lipid preferences of eukaryotic and prokaryotic membrane proteins. Nat. Commun. 2018;9:4253. doi: 10.1038/s41467-018-06702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treuheit N.A., Redhair M., et al. Atkins W.M. Membrane Interactions, Ligand-Dependent Dynamics, and Stability of Cytochrome P4503A4 in Lipid Nanodiscs. Biochemistry. 2016;55:1058–1069. doi: 10.1021/acs.biochem.5b01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flayhan A., Mertens H.D.T., et al. Löw C. Saposin Lipid Nanoparticles: A Highly Versatile and Modular Tool for Membrane Protein Research. Structure. 2018;26:345–355.e5. doi: 10.1016/j.str.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLean M.A., Denisov I.G., et al. Sligar S.G. Ultra-Dark and Ultra-Bright Nanodiscs for membrane protein investigations. Anal. Biochem. 2020;607 doi: 10.1016/j.ab.2020.113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Cierpicki T., et al. Bushweller J.H. NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol. Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otzen D.E. Folding of DsbB in mixed micelles: a kinetic analysis of the stability of a bacterial membrane protein. J. Mol. Biol. 2003;330:641–649. doi: 10.1016/s0022-2836(03)00624-7. [DOI] [PubMed] [Google Scholar]
- 22.Sehgal P., Otzen D.E. Thermodynamics of unfolding of an integral membrane protein in mixed micelles. Protein Sci. 2006;15:890–899. doi: 10.1110/ps.052031306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nasr M.L., Baptista D., et al. Wagner G. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods. 2017;14:49–52. doi: 10.1038/nmeth.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotov V., Bartels K., et al. Garcia-Alai M.M. High-throughput stability screening for detergent-solubilized membrane proteins. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-46686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burastero O., Niebling S., et al. Garcia Alai M.M. eSPC: an online data-analysis platform for molecular biophysics. Acta Crystallogr. D Struct. Biol. 2021;77:1241–1250. doi: 10.1107/S2059798321008998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 27.Kotov V., Mlynek G., et al. Marlovits T.C. In-depth interrogation of protein thermal unfolding data with MoltenProt. Protein Sci. 2021;30:201–217. doi: 10.1002/pro.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akaike T., Sakurai Y., et al. Tsuruta T. Study on the interaction between plasma-proteins and polyion complex by circular-dichroism and ultraviolet spectroscopy. Kobunshi Ronbunshu. 1979;36:217–222. [Google Scholar]
- 29.Greenfield N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otzen D.E. Mapping the folding pathway of the transmembrane protein DsbB by protein engineering. Protein Eng. Des. Sel. 2011;24:139–149. doi: 10.1093/protein/gzq079. [DOI] [PubMed] [Google Scholar]
- 31.Lee A.G. Membrane lipids: it’s only a phase. Curr. Biol. 2000;10:R377–R380. doi: 10.1016/s0960-9822(00)00477-2. [DOI] [PubMed] [Google Scholar]
- 32.Lee H.J., Lee H.S., et al. Chae P.S. Affect of novel detergents on membrane protein studies. Chem. 2022;8:980–1013. [Google Scholar]
- 33.Hagn F., Etzkorn M., et al. Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miehling J., Goricanec D., Hagn F. A Split-Intein-Based Method for the Efficient Production of Circularized Nanodiscs for Structural Studies of Membrane Proteins. Chembiochem. 2018;19:1927–1933. doi: 10.1002/cbic.201800345. [DOI] [PubMed] [Google Scholar]
- 35.McClary W.D., Sumida J.P., et al. Atkins W.M. Membrane fluidity modulates thermal stability and ligand binding of cytochrome P4503A4 in lipid nanodiscs. Biochemistry. 2016;55:6258–6268. doi: 10.1021/acs.biochem.6b00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris N.J., Findlay H.E., et al. Booth P.J. Relative domain folding and stability of a membrane transport protein. J. Mol. Biol. 2014;426:1812–1825. doi: 10.1016/j.jmb.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Harris N.J., Reading E., et al. Booth P.J. Structure formation during translocon-unassisted co-translational membrane protein folding. Sci. Rep. 2017;7:8021. doi: 10.1038/s41598-017-08522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann J., Klein N., et al. Schneider D. Folding energetics and oligomerization of polytopic α-helical transmembrane proteins. Arch. Biochem. Biophys. 2014;564:281–296. doi: 10.1016/j.abb.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Yang C., Boggasch S., et al. Paulsen H. Thermal stability of trimeric light-harvesting chlorophyll a/b complex (LHCIIb) in liposomes of thylakoid lipids. Biochim. Biophys. Acta. 2006;1757:1642–1648. doi: 10.1016/j.bbabio.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 40.Yoneda J.S., Rigos C.F., et al. Ciancaglini P. K-ATPase reconstituted in ternary liposome: The presence of cholesterol affects protein activity and thermal stability. Arch. Biochem. Biophys. 2014;564:136–141. doi: 10.1016/j.abb.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Biswas N., Waring A.J., et al. Dluhy R.A. Structure and conformation of the disulfide bond in dimeric lung surfactant peptides SP-B1-25 and SP-B8-25. Biochim. Biophys. Acta. 2007;1768:1070–1082. doi: 10.1016/j.bbamem.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 42.Walker L.R., Marty M.T. Revealing the specificity of a range of antimicrobial peptides in lipid nanodiscs by native mass spectrometry. Biochemistry. 2020;59:2135–2142. doi: 10.1021/acs.biochem.0c00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rader A.J., Anderson G., et al. Klein-Seetharaman J. Identification of core amino acids stabilizing rhodopsin. Proc. Natl. Acad. Sci. USA. 2004;101:7246–7251. doi: 10.1073/pnas.0401429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakabayashi K., Nakagawa H., et al. Ishikawa T. Intramolecular disulfide bond is a critical check point determining degradative fates of ATP-binding cassette (ABC) transporter ABCG2 protein. J. Biol. Chem. 2007;282:27841–27846. doi: 10.1074/jbc.C700133200. [DOI] [PubMed] [Google Scholar]
- 45.McDonald E.F., Woods H., et al. Meiler J. Structural Comparative Modeling of Multi-Domain F508del CFTR. Biomolecules. 2022;12 doi: 10.3390/biom12030471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuenze G., Vanoye C.G., et al. Meiler J. Allosteric mechanism for KCNE1 modulation of KCNQ1 potassium channel activation. Elife. 2020;9 doi: 10.7554/eLife.57680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marinko J.T., Wright M.T., et al. Sanders C.R. Glycosylation limits forward trafficking of the tetraspan membrane protein PMP22. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levental I., Lyman E. Regulation of membrane protein structure and function by their lipid nano-environment. Nat. Rev. Mol. Cell Biol. 2022;24:107–122. doi: 10.1038/s41580-022-00524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marx D.C., Fleming K.G. Membrane proteins enter the fold. Curr. Opin. Struct. Biol. 2021;69:124–130. doi: 10.1016/j.sbi.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniilidis M., Brandl M.J., Hagn F. The advanced properties of circularized MSP nanodiscs facilitate high-resolution NMR studies of membrane proteins. J. Mol. Biol. 2022;434 doi: 10.1016/j.jmb.2022.167861. [DOI] [PubMed] [Google Scholar]
- 51.Zhang M., Gui M., et al. Wagner G. Cryo-EM structure of an activated GPCR–G protein complex in lipid nanodiscs. Nat. Struct. Mol. Biol. 2021;28:258–267. doi: 10.1038/s41594-020-00554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.