Abstract
The actomyosin cytoskeleton generates mechanical forces that power important cellular processes, such as cell migration, cell division, and mechanosensing. Actomyosin self-assembles into contractile networks and bundles that underlie force generation and transmission in cells. A central step is the assembly of the myosin II filament from myosin monomers, regulation of which has been extensively studied. However, myosin filaments are almost always found as clusters within the cell cortex. While recent studies characterized cluster nucleation dynamics at the cell periphery, how myosin clusters grow on stress fibers remains poorly characterized. Here, we utilize a U2OS osteosarcoma cell line with endogenously tagged myosin II to measure the myosin cluster size distribution in the lamella of adherent cells. We find that myosin clusters can grow with Rho-kinase (ROCK) activity alone in the absence of myosin motor activity. Time-lapse imaging reveals that myosin clusters grow via increased myosin association to existing clusters, which is potentiated by ROCK-dependent myosin filament assembly. Enabling myosin motor activity allows further myosin cluster growth through myosin association that is dependent on F-actin architecture. Using a toy model, we show that myosin self-affinity is sufficient to recapitulate the experimentally observed myosin cluster size distribution, and that myosin cluster sizes are determined by the pool of myosin available for cluster growth. Together, our findings provide new insights into the regulation of myosin cluster sizes within the lamellar actomyosin cytoskeleton.
Significance
Myosin II molecular motors assemble into higher-ordered clusters within the actin cytoskeleton and cortex. These clusters are thought to play important roles in controlling force transmission but their regulation is unknown. Here, we use quantitative fluorescence imaging of adherent cells with endogenously tagged myosin II to explore regulation of clusters in the actin cytoskeleton. We find the cluster size distribution is determined both by a limiting pool of myosin and by the underlying actin organization.
Introduction
Various cellular processes and dynamics in cells depend on mechanical forces generated by the actomyosin cytoskeleton, such as cell motility (1), adhesion (2,3), and mechanosensing (4). The central contractile molecular element of the actomyosin cytoskeleton is nonmuscle myosin II (NMII, hereafter referred to as myosin), which self-assembles into myosin filaments that contract actin filaments (F-actin) to generate contractile forces. In combination with cross-linkers and other actin binding proteins, actin and myosin build contractile actomyosin structures that underlie force production in cells. For example, in nonmuscle cells, F-actin bundles and myosin filaments form contractile bundles that span the cell, called stress fibers. While both F-actin and myosin must be regulated to build stress fibers, F-actin regulation has been well-studied (for example, reviewed in (5,6)) and myosin regulation is relatively less understood.
Most studies on myosin regulation have focused on the assembly of myosin filaments from myosin molecules. At the molecular scale, the ability of myosin to assemble into myosin filaments is determined by the transition between its assembly-incompetent autoinhibited state and its assembly-competent active state (7). This transition largely depends on phosphorylation on the regulatory light chain (RLC) of myosin. Several kinases, including the myosin light chain kinase (MLCK) and Rho-associated coiled-coil containing kinase (ROCK), can phosphorylate the Thr18 or Ser19 residues of the RLC to promote myosin filament assembly (1,8,9). Phosphorylation sites on the myosin heavy chain have also been shown to play a role (1,10). Aside from biochemical regulation, mechanical tension has also been proposed to impact the assembly of myosin filaments (11,12,13).
Despite the importance of myosin filament regulation, single myosin filaments are rarely observed in cells. While some individual myosin filaments may be observed near the cell periphery at the leading edge, most myosin filaments in cells form clusters, where multiple myosin filaments come into close contact with each other with their myosin head domains. Described as ribbons or stacks, myosin clusters were best visualized using electron microscopy (12,14,15,16). Myosin clusters at the leading edge of the cell usually contain fewer myosin filaments, while more myosin filaments cluster together toward the cell body as they associate with stress fibers. Myosin clusters can further register with each other to form superstructures on stress fibers (17). However, it remains largely unknown how myosin filaments self-associate to form clusters and how the process is regulated.
Recent studies have characterized the dynamics of myosin clusters to understand their formation at the leading cell edge. Using superresolution microscopy, myosin filaments were observed nucleating at the leading edge of the cell (18,19). After nucleation, more myosin filaments were recruited to the nucleated filament to form myosin clusters. Through rounds of amplification and splitting, myosin clusters amplify both in size and number before incorporating into stress fibers. The amplification and splitting dynamics have been shown to depend on myosin monomer availability, myosin motor activity, actin dynamics, and actin density (18,19). While myosin cluster dynamics at the leading edge and their regulation is thought to be general for all myosin clusters in cells, myosin cluster sizes on stress fibers have not been properly characterized.
In this work, we explore the regulation of myosin cluster sizes distribution in the lamella of adherent cells. To this end, we quantitatively characterized endogenously tagged nonmuscle myosin IIA (NMIIA) in adherent U2OS osteosarcoma cells. The size distribution of myosin clusters found within actin networks and bundles within the lamella is quite broad. ROCK activity is sufficient to grow myosin clusters without myosin motor activity by increasing the pool of assembled myosin that can associate with existing ROCK-independent myosin clusters. Myosin motor activity further enhances myosin cluster growth by myosin association, which is dependent on F-actin architecture. A toy model of myosin cluster growth with myosin self-affinity is sufficient to recapitulate the broad distribution of cluster sizes. Our results suggest that myosin cluster sizes within the lamella of adherent cells are set by a limiting pool of myosin available for cluster growth.
Materials and methods
Cell culture
U2OS cells were cultured in McCoy’s 5A Medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (Corning, Corning, NY) and 2 mM L-glutamine (Invitrogen, Waltham, MA). To visualize myosin, mScarlet was knocked in at the C-terminal locus of the MYH9 gene using CRISPR. The target/Cas9 plasmid pSpCas9(BB)-2A-Puro (PX459) V2.0 (http://www.addgene.org/62988/) with target sequence AGGTAGATGGCAAAGCGGAT was engineered according to established protocols (Ran et al., 2013). To create a donor plasmid, pUC57 was digested with EcoRI and StuI and purified. A four-piece Gibson assembly was then performed. Three gBlocks (5′ HDR, linker/mScarlet, and 3′ HDR) were obtained from IDT with overlapping extensions to mediate Gibson assembly. The 5′ HDR arm is 867 bp of genomic sequence terminating immediately before the endogenous STOP codon. Silent mutations (AaGTtGAcGGaAAAGCGGATGGt) were placed in the target sequence to prevent Cas9 binding and cleavage of the donor plasmid. The linker/mScarlet was placed in-frame with the coding sequence and consists of an 18-amino-acid GS-rich linker and mScarlet-I fluorophore. The 3′ HDR arm is 684 bp immediately downstream of the MYH9 coding sequence. U2OS cells were transfected with both Cas9 and donor plasmids. After ∼1 week in culture, a polyclonal mScarlet positive population was obtained using fluorescent-activated cell sorting (FACS). The resulting population shows only one single mScarlet-positive population (confirmed by flow cytometry). Coupled with our observation that the knockin efficiency is low (much less than 1%), our characterization suggests a homogeneous population of monoallelic-inserted cells.
For fluorescence recovery after photobleaching (FRAP) experiments, eGFP was knocked in at the N-terminal locus of the MYH9 gene instead. The following modifications were made for the CRISPR procedure. The target sequence for the guide RNA is AAACTTCATCAACAATCCGC. Donor plasmid was generated in pUCIDT-AmpR with internal mEGFP flanked by 5′ and 3′ HDR. The 5′ HDR is 498 bp upstream of endogenous start and 3′ HDR is 383 bp downstream of endogenous start. All cells were previously treated with BM cyclin (Roche, Basel, Switzerland) per manufacturer instructions to eliminate mycoplasma.
Drug perturbations
Cells were treated with 40 μM Y-27632 (EMD Millipore, Burlington, MA) or 50 μM blebbistatin (MilliporeSigma, Burlington, MA) for at least 30 min. For drug washout experiments, drugs were removed for 30 min before fixation and permeabilization. Latrunculin A (Sigma-Aldrich, St. Louis, MA) was used at a subsaturating concentration of 50 nM for 30 min.
Immunofluorescence
Cells were first washed with warm PBS. Then cells were fixed and permeabilized with 4% paraformaldehyde (EMS, Hatfield, PA) and 0.5% Triton X-100 (Fisher Scientific, Waltham, MA), diluted in 1.5% BSA (Fisher Scientific, Waltham, MA) and cytoskeletal buffer (0.01 M MES, 0.003 M MgCl2, 0.138 M KCl, 0.002 M EGTA [pH 6.8]). To visualize F-actin, cells were then washed with PBS then incubated with Alexa-647 phalloidin (Invitrogen, Waltham, MA) at 1:1000 dilution, 0.5% Triton X-100, and 1.5% BSA diluted in cytoskeletal buffer. Cells were then washed with PBS and mounted onto a coverslip with ProLong gold antifade reagent (Invitrogen, Waltham, MA).
Microscopy and live-cell imaging
Cells were imaged on an inverted Nikon Ti-E (Nikon, Melville, NY) with a Yokogawa CSU-X confocal scanning head (Yokogawa Electric, Tokyo, Japan) and laser merge model with 491, 561, and 642 nm laser lines (Spectral Applied Research, ON, Canada). Images were collected on a Zyla 4.2 sCMOS Camera (Andor, Belfast, UK). A 60× 1.2 Plan Apo water (Nikon) objective was used to collect images. MetaMorph Automation and Image Analysis Software (Molecular Devices, Sunnyvale, CA) controlled all hardware. For quantitative myosin cluster size analysis, all imaging conditions are controlled to be the same in each set of experiments, including laser intensity and exposure time. For FRAP experiments, Airyscan imaging was performed on a Zeiss LSM 980 microscope equipped with the Airyscan 2 detector. Images were acquired using the MPLX SR-4X mode and processed by Zen Blue 3.0 software using the Airyscan processing feature with default settings. For live-cell imaging, cells were mounted on an imaging chamber (Live Cell Instruments, Republic of Korea) and maintained at 37°C. For live-cell imaging, cell medium was replaced with Dulbecco’s modified Eagle medium without phenol red (Corning, Corning, NY) supplemented with 10% FBS, 2 mM L-glutamine, 1% penicillin-streptomycin (Corning, Corning, NY), 10 mM HEPES (Corning, Corning, NY), and 30 μL/mL Oxyrase (Oxyrase, Mansfield, OH).
Image processing
For the quantification of myosin cluster sizes, confocal images were captured ∼2 μm around the focal plane to capture all intensities and z projection ∼1 μm was performed around the focal plane where myosin is in focus. To remove diffuse myosin background, immunofluorescence images were first background subtracted using a top-hat filter with a circular element with a diameter of 50 pixels. Then myosin clusters were localized using a feature-finding code (20), where a bandpass filter is applied on the image to preserve circular features with a diameter of 7 pixels. The noise floor is identified as the maximum pixel intensity in a background region outside of the cell, and local maximum in the bandpassed space with an intensity higher than the noise floor is accepted as a feature candidate. The identified features were filtered based on the general criteria: maximum radius of gyration 4.75, maximum eccentricity 0.48, and intensity divided by radius of gyration. Features outside of the cell area and features closer than 2 pixels apart (0.2 μm) were also excluded. Myosin puncta intensities were then fitted to a lognormal distribution using maximum-likelihood estimation (MATLAB, The MathWorks, Natick, MA).
Live-cell imaging movies were first corrected for photobleaching. The photobleaching curve was generated by summing all pixel intensities within a cell. The photobleaching curve was then fitted empirically to a double exponential decay function of the form . Then intensities were corrected by dividing the imaging intensity to the curve . Afterward, movies were either denoised by applying a spatial Gaussian filter (σ = 1 pixel) or by Noise2Void (21).
To analyze FRAP experiments, both FRAP and control regions were identified. A double normalization scheme is used to normalize data and correct for photobleaching . The FRAP curve is fitted to to extract the recovery half time and the mobile fraction . All analysis codes are available upon request.
To track myosin clusters over time, we used the quantitative fluorescence speckle microscopy (QFSM) (22) package. The QFSM package was retrieved from GitHub (https://github.com/DanuserLab/QFSM). In brief, each image was first Gaussian filtered with a standard deviation of 0.36 pixels. Myosin clusters were localized using the speckle detection algorithm with the following parameters: standard deviation for Gaussian low-pass filtering was 0.36 pixels, α value for statistical selection was 0.5, with no iterative selection. The flow rate of speckles was determined using a template window size of 17 pixels, then particle tracking was conducted with a search radius of 4 pixels and correlation length of 33 pixels.
Simulation of myosin cluster growth
The simulation setup is summarized in the main text. To match the initial myosin cluster size distribution in the simulation with experimental conditions, each grid point is initialized with a cluster size that is randomly sampled from the experimental cumulative distribution of cluster sizes. After all grid points have been initialized, the initial total cluster size can be calculated, and Nmyo can be determined by percent increase of total myosin cluster sizes in experimental quantifications as described in the main text. The simulation iterates through the random binding of Nmyo myosin filaments to a grid. In each iteration, one myosin filament binds to a grid chosen by the default Python function choices, with the probability to choose each grid being as described in the main text. Code is available on GitHub (https://github.com/WHChou/Myosin-cluster-simulation).
Results
Myosin filaments form clusters of a broad range of sizes within lamellar actin networks and bundles
To study the regulation of myosin clusters, we chose the human osteosarcoma U2OS cell line because their lamellar actin architectures, including stress fibers, have been well characterized (23,24). Using a CRISPR knockin approach, we endogenously labeled nonmuscle myosin IIA by inserting an mScarlet gene at the C-terminal locus of MYH9 (see materials and methods). We chose to focus on nonmuscle myosin IIA because of its role in generating contractile forces in cells (25,26), and it is the dominant myosin isoform in U2OS cells. Since myosin assembles into a filament with their C-terminal tail overlapping at the bare zone, the fluorescence signal represents the central location of a myosin filament (Fig. 1 A). Using spinning disk confocal imaging, we see that most myosin appears as diffraction-limited puncta of similar physical dimensions but with varying intensities (Fig. 1 B). These myosin puncta often colocalize with F-actin bundles (Fig. 1 B). Since our imaging conditions are likely insufficient to confidently detect single myosin filaments, each punctum likely contains multiple myosin filaments. We therefore refer to these punctate structures as myosin clusters, which may represent single or multiple closely associated myosin stacks.
Figure 1.
Myosin filaments form clusters of a broad range of sizes on stress fibers. (A) Schematics of the experiment setup. Nonmuscle myosin IIA is endogenously tagged on the C-terminal tail so that myosin filaments will appear as diffraction-limited puncta. The intensity of each punctum is directly proportional with the amount of myosin filaments in each cluster. The number of myosin heads on a filament have been reduced for simplicity. (B) Representative images of nonmuscle myosin IIA (myosin) and F-actin in a U2OS cell. The right panels show zoom-in of myosin clusters from the two red boxes. Images are inverted. Scale bar, 10 μm. (C) Examples of myosin cluster localization. In the cluster identification column, each myosin intensity maxima is shown as a red dot. The brown circles indicate a typical area where the integrated intensity of a myosin punctum is calculated. The intensity profile column shows the intensity profile along the blue line in (B (i)) and (B (ii)). (D) Myosin clusters in cells color coded by their sizes. Nmin represents the minimal myosin cluster size that can be detected, as discussed in the text. (E) Histogram of myosin cluster sizes. Inset shows the cumulative distribution of myosin cluster sizes. To see this figure in color, go online.
To characterize myosin cluster dynamics, we conducted time-lapse live-cell imaging. While myosin clusters undergo retrograde motion and contract inward toward the cell center, they remain stable over time, persisting for at least 8 min (Fig. S1 A and Video S1). This suggests that myosin clusters are not multiple randomly overlapping structures but rather stable structures that move together as one unit. Despite their structural stability, the constituents of myosin clusters exchange rapidly. When we perform FRAP analysis on myosin clusters, we see that myosin fluorescence recovers with a half-time of ∼2 min and a mobile fraction of 80% (Fig. S1, B, D, and E), consistent with previous reports (23,25,27,28). We also observed FRAP recovery at the level of individual myosin clusters. Instead of assembling new myosin clusters or transporting clusters into the photobleached region, nearby myosin clusters undergo FRAP recovery at a similar rate (Fig. S1 C). This suggests that myosin is continually exchanging within myosin clusters, with constant association and dissociation of myosin. Since myosin clusters contain multiple myosin filaments (see characterization below), FRAP recovery may indicate the exchange of single myosin monomers, oligomers, or whole filaments.
Color is inverted. Time stamp indicates minutes and seconds.
Since myosin is endogenously labeled, myosin punctum intensity reflects the number of myosin filaments within each cluster. To quantify the intensities of myosin clusters bound to the lamellar actin network or bundles, we fixed and permeabilized the cells before imaging to remove unbound or weakly bound myosin. We noticed diffuse myosin intensity within the cell that is higher than background intensities even after fixation and permeabilization (Fig. S2, B–D); this may indicate myosin bound to the cellular cortex. To focus on quantifying the intensities of myosin puncta, we exclude the diffuse background by performing local background subtraction. Then, the centroid location of each myosin punctum is determined using previously developed methods (20). In brief, each image is passed through a bandpass filter to preserve circular features with a diameter of ∼700 nm, about the typical size of a myosin punctum. Local maxima with intensity higher than background noise are accepted as puncta candidates (Fig. 1 C). We further filter out puncta that are irregularly shaped or puncta whose intensity are too spread out (see materials and methods). The remaining puncta candidates are accepted as valid myosin clusters. Each myosin cluster’s intensity is then calculated as the integrated intensity of each punctum within a diameter of ∼700 nm.
While the absolute number of myosin molecules within each cluster requires careful calibration between fluorescence intensity and the number of fluorescent proteins (29), their relative sizes can be assessed without this calibration. To compare the sizes of myosin clusters across different cells and experimental conditions, we normalize all myosin cluster intensities within the same set of experiments against the minimum myosin intensity identified in each experiment. We posit that this minimum intensity (Imin) reflects the minimal number of myosin molecules in a cluster (Nmin) that we can detect above the noise floor. Nmin likely represents more than one myosin filament (see Quintanilla et al., 2023, who suggest that the vast majority of identifiable myosin structures at the leading edge of the cell contains more than one myosin filament). Since we keep the imaging conditions and analysis parameters constant in each experiment, we reasoned that Nmin will be the same but Imin can vary slightly across different experiments. Therefore, we normalize myosin cluster intensities in each experiment to Imin and present myosin cluster sizes as multiples of Nmin.
With this approach, we find a broad distribution of myosin cluster sizes across the cell, ranging up to 30-fold. Most myosin clusters contain three times more myosin than Nmin, but there is a sizable fraction of larger myosin clusters containing 20-fold more myosin than Nmin (Fig. 1, D and E). This is also demonstrated by the histogram of myosin cluster intensities, which shows a long-tailed distribution (Fig. 1 E). Myosin intensity is also spatially heterogeneous (Fig. 1 D). Myosin clusters tend to be smaller at the lamellar region or newly formed transverse arcs, containing only 2–4 times more myosin than Nmin (Fig. 1, C (i)). On the other hand, myosin clusters tend to be larger on mature transverse arcs in the cell body or on ventral stress fibers, with more than 15 times more myosin than Nmin (Fig. 1 C (ii)). In addition, the number of myosin clusters scale with cell area (Fig. S2 A), suggesting that there is a well-defined density of myosin clusters.
Myosin cluster sizes can be determined by their growth dynamics. Since we observed both de novo assembly and growth of existing clusters in our time-lapse imaging of myosin clusters (Video S1), we sought to determine which process dominates. We estimated their relative contributions by tracking myosin clusters in untreated cells over time using QFSM (22). To estimate de novo assembly rates, we looked at myosin clusters identified over a 5 min interval. We found that ∼70% of the myosin clusters were stable for at least 5 min while the remaining 30% are newly identified clusters. Of the new clusters, 45% are dim enough to be considered de novo assembly events since they have intensities smaller than three times the size of Nmin. The other 55% of the new clusters may be attributed to either cluster disassembly or tracking errors caused by particle-tracking analysis. Taken together, these values suggest that, over several-minute timescales, de novo assembly contributes a small fraction (∼13.5%) of myosin clusters while the vast majority (∼70%) of the myosin clusters are stable. This indicates that, during these timescales, the majority of myosin cluster growth occurs through the growth of existing clusters.
Rho-kinase activity drives growth through net myosin association to existing clusters
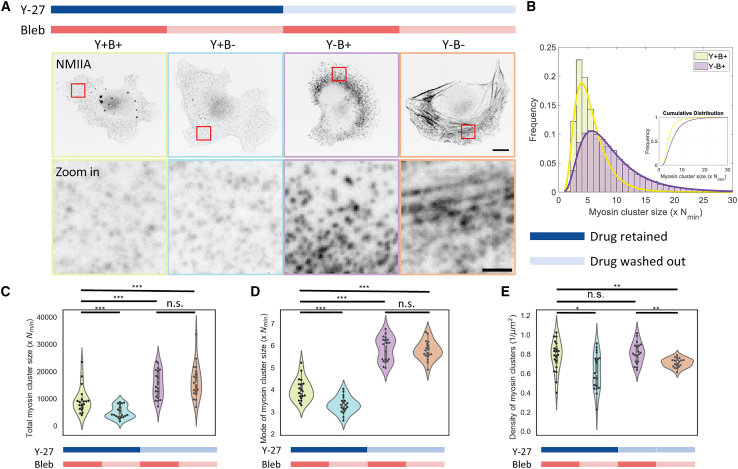
Recent literature showed that myosin clusters at the leading edge are regulated by myosin monomer availability and motor activity (18,19). We designed an experiment to test their respective contributions to the growth of myosin clusters within the lamellar actin. To promote myosin cluster disassembly, cells were treated with 40 μM ROCK inhibitor Y-27632 (Y-27, inhibits ROCK-dependent myosin light chain phosphorylation) and 50 μM blebbistatin (myosin II ATPase inhibitor, which locks myosin in a weak actin-binding state (30)) for 30 min. Even after treatment, residual myosin clusters with low fluorescence intensity remained (Fig. 2 A, Y+B+). The sizes of the remaining myosin clusters are smaller with less variation, averaging around two to three times the size of Nmin (Fig. 2 B, Y+B+). These myosin clusters are presumably formed by myosin filaments that are phosphorylated by other kinases, such as MLCK or MRCK, formed by increasing the total monomer pool above a critical threshold to enable spontaneous filament assembly (29), or as a result of incomplete ROCK inhibition.
Figure 2.
ROCK activity is sufficient for myosin clusters to grow in the absence of myosin motor activity. (A) Top row shows representative myosin images of cells with or without ROCK activity and myosin motor activity. In the left panel, myosin clusters were disassembled by both Y-27632 and blebbistatin (Y+B+). The middle panel shows cases where Y27 or blebbistatin are removed individually (Y+B– and Y–B+). The right panel shows the control condition where both inhibitors are removed (Y–B–). Zoomed in myosin images are shown in the bottom row. Images are inverted. Scale bars, 10 μm (top row) and 2 μm (bottom row). (B) Histogram of myosin cluster size between Y27/blebbistatin-treated and blebbistatin-treated cells. Inset shows the cumulative distribution of cluster sizes. (C) The sum of myosin cluster sizes. (D) Mode of myosin cluster size distribution. (E) Density of myosin clusters. n = 20 cells for each condition. Two-sample Kolmogorov-Smirnov tests were performed in (C–E) to test if the measured quantity differs between conditions. n.s. signifies p > 0.05, ∗ signifies p < 0.05, ∗∗ signifies p < 0.01, ∗∗∗ signifies p < 0.001. To see this figure in color, go online.
To decouple the role of ROCK and myosin motor activity on myosin cluster growth, we selectively retain one inhibitor during washout of the other. When we selectively washed out blebbistatin but retained Y-27, we saw no apparent increase in myosin cluster intensity (Fig. 2 A, Y+B–). The total amount of myosin contained within clusters marginally decreased compared with the cells treated with both inhibitors (Fig. 2 C). To quantify the size distribution of myosin clusters across different conditions, we fitted the myosin cluster intensity distribution to a lognormal distribution and used the mode of this distribution to represent the most probable myosin cluster size. The mode of myosin cluster intensity only marginally changed with blebbistatin washout (Fig. 2 D).
In contrast, when Y-27 is removed but blebbistatin is retained, the total amount of myosin contained within clusters across the lamella increased by ∼42% (Fig. 2 A and C, Y–B+). From the histogram of myosin cluster sizes, we estimate that myosin clusters grow up to 4–15 times the size of Nmin with ROCK activity alone (Fig. 2 B, Y–B+). Compared with myosin cluster size distributions in cells treated with both inhibitors, the peak of the distribution shifts toward larger sizes and the tail widens with ROCK activity. This indicates both that the most probable myosin cluster size and the fraction of larger myosin clusters increased. This is also shown by the increasing mode of myosin cluster intensity when Y-27 is selectively washed out (Fig. 2 D). Interestingly, myosin cluster density remained at similar levels in all drug conditions (Fig. 2 E). This suggests that ROCK activity is sufficient for myosin clusters to grow, predominantly through the accumulation of myosin to existing clusters but not through forming new clusters.
As a control, when we washed out both inhibitors, we observed the recovery of stress fibers, as demonstrated by the presence of linearly arranged myosin (Fig. 2 A, Y–B–). Interestingly, the total amount of myosin in clusters were not significantly different from cells where only Y-27 is washed out (Fig. 2 C, Y–B–). Similarly, the mode of myosin cluster sizes (Fig. 2 D) remains largely unchanged, while the myosin cluster density decreased slightly (Fig. 2 E). While one may expect this control condition to parallel untreated cells, this may suggest that the timescale of the washout was not sufficient for myosin motor activity to fully reestablish myosin cluster sizes. We will further investigate the role for myosin motor activity in the next section.
To visualize ROCK-mediated myosin cluster growth, we conducted live-cell imaging of fluorescently tagged myosin as Y-27 is selectively washed out in cells treated with both Y-27 and blebbistatin (Fig. 3 A). As Y-27 is washed out, the intensities of myosin clusters increased over 20 min (Fig. 3, A and B). We see a ∼6% increase in myosin density when we wash out the ROCK inhibitor (Fig. 3 B), which we suspect is at least partially caused by the reduced cytoplasmic background after ROCK washout that allows smaller myosin clusters to be resolved. Alternatively, this could reflect some myosin clusters assembling de novo. The dynamics of individual myosin clusters reveal that most clusters are very mobile and only appear transiently for a few frames after washing out Y-27 (Fig. 3 C and Video S2). We think that this does not represent rapid assembly and disassembly dynamics but instead reflects that myosin is weakly bound to F-actin due to blebbistatin treatment and, consequently, leaves the field of view. It is therefore challenging to track single myosin clusters over time. Nevertheless, we observed some myosin clusters growing in intensity without apparent interaction with other clusters (Fig. 3 D and Video S3). This suggests that myosin clusters grow via a net association of myosin monomers or filaments to existing myosin clusters, potentiated by the increase in assembly-competent myosin driven by ROCK activity (Fig. 3 E). Moreover, myosin clusters that appear at later times have higher intensities than myosin clusters that appear earlier in the washout (Fig. 3 C). This suggests that myosin clusters may grow while diffusing around the cytoplasm.
Figure 3.
Myosin clusters grow through net association of myosin to existing clusters without myosin motor activity. (A) Snapshots of myosin clusters after washing out Y-27632 from Y-27632/blebbistatin treated cells. Time after washout is shown on the top left corner of each image. Inset shows zoom-in of myosin clusters in the red squares. Images are inverted. Scale bars, 10 μm and 2 μm (inset). (B) The sum of intensities of all myosin clusters in the cell, normalized to their intensities before washout. n = 3 cells. (C) Kymograph of myosin clusters along the green line in the inset in (A). Yellow asterisks indicate representative myosin clusters that transiently appear in the field of view. Horizontal axis represents time and vertical axis represents spatial position. Timescale 5 min and spatial scale 1 μm. (D) Close-up snapshots of myosin cluster growth over time. Top left indicates time after washout. Arrowheads indicate trackable myosin clusters over time. (E) Schematic of myosin association to existing clusters. To see this figure in color, go online.
Color is inverted. Time stamp indicates minutes and seconds. Images are denoised with Noise2Void.
Arrowheads show transiently appearing myosin clusters that increase in intensity. Color is inverted. Time stamp indicates minutes and seconds. Images are denoised with Noise2Void.
Myosin motor activity allows myosin clusters to grow through F-actin-dependent myosin association
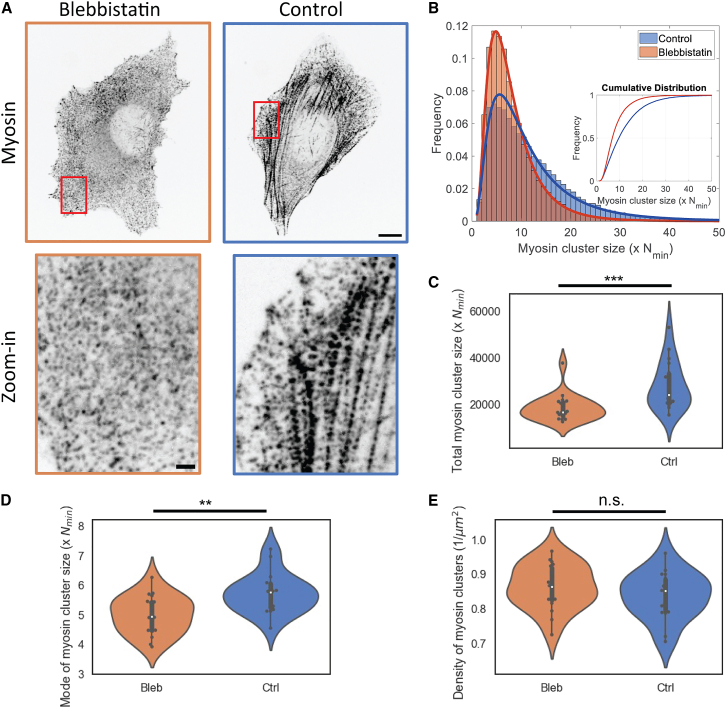
Our results so far show that ROCK activity is sufficient for myosin cluster growth without myosin motor activity. Next, we explored the role of myosin motor activity in myosin cluster growth. To this end, we compared myosin cluster sizes in cells treated with blebbistatin with untreated cells (Fig. 4 A). We see that the total myosin contained in clusters decreased by about 35% with blebbistatin treatment (Fig. 4 C). This is consistent with previous studies showing that myosin motor activity affects myosin filament assembly (12). At the level of single myosin clusters, most myosin clusters in blebbistatin-treated cells contain around 4 times more myosin than Nmin but can contain up to 18 times more than Nmin (Fig. 4 B), which is consistent with the case when Y-27 is washed out in cells treated with Y-27 and blebbistatin (Fig. 2 B, Y–B+). In contrast, while the most probable myosin cluster size only modestly increased to 5–6 times more than Nmin in control cells (Fig. 4, B and D), there is a larger fraction of myosin clusters that contain more than 15-fold of Nmin, reaching up to 28-fold of Nmin (Fig. 4 B). As in the case of ROCK-dependent myosin cluster growth, myosin cluster density remains the same with blebbistatin treatment (Fig. 4 E).
Figure 4.
Myosin motor activity allows for myosin cluster growth. (A) Top row shows representative myosin images in cells treated with or without blebbistatin. Bottom row shows zoomed-in myosin images in the red boxes in the top row. Images are inverted. Scale bars, 10 μm (top row) and 2 μm (bottom row). (B) Histogram of myosin cluster sizes between blebbistatin-treated and control cells. Inset shows the cumulative distribution of both conditions. (C) The sum of all myosin cluster sizes. (D) Mode of myosin cluster size distribution. (E) Density of myosin clusters. n = 20 cells for blebbistatin-treated cells and Nn= 18 cells for control cells. Two-sample Kolmogorov-Smirnov tests were performed in (C–E) to test if the measured quantity differs between conditions. n.s. signifies p > 0.05, ∗∗ signifies p < 0.01, ∗∗∗ signifies p < 0.001. To see this figure in color, go online.
To understand how myosin motor activity can further enhance myosin cluster size, we performed live-cell myosin imaging as we washed out blebbistatin (Fig. 5 A and Video S4). Consistent with fix and stain results, myosin cluster intensity increased as blebbistatin is washed out (Fig. 5 B). We also see a ∼10% change in myosin cluster density during our time-lapse imaging experiment (Fig. 5 B). During this process, myosin clusters are more stably bound, persisting for more than 10 min (Fig. 5 C). Some myosin clusters increase intensity over time without apparent local motion or interaction with neighboring clusters, suggesting an increased net myosin association to existing clusters (Fig. 5, C and D). We also observe that some myosin clusters undergo local motion toward each other and merge into a brighter cluster (Fig. 5, C and E), suggesting myosin cluster growth through actomyosin sliding. In addition, some de novo myosin cluster assembly events can be observed (Fig. 5 C).
Figure 5.
Myosin motor activity grows myosin clusters through net myosin association or cluster merging. (A) Snapshots of time-lapse imaging of myosin clusters during blebbistatin washout. Inset shows zoom-in of myosin clusters in the red squares. Images are inverted. Scale bars, 10 μm and 2 μm (inset). (B) Myosin cluster intensity and density over time. The left axis shows the mode of myosin cluster intensities in the cell, normalized to the mode intensity at the first frame of acquisition. The right axis shows the density of myosin clusters over time. (C) Kymograph of myosin clusters. Maroon asterisks indicate cluster growth events through myosin association and yellow asterisks indicate two clusters merging. (D) Close-up snapshots of myosin clusters increasing intensity over time through net myosin association. Arrowheads indicate example myosin clusters. (E) Schematic of myosin association with existing clusters. (F) Close-up snapshots of myosin clusters’ increasing intensity through merging with another cluster driven by actomyosin sliding. Arrowheads indicate example myosin clusters. (G) Schematic of myosin cluster growth via actomyosin sliding. To see this figure in color, go online.
Color is inverted. Time stamp indicates minutes and seconds.
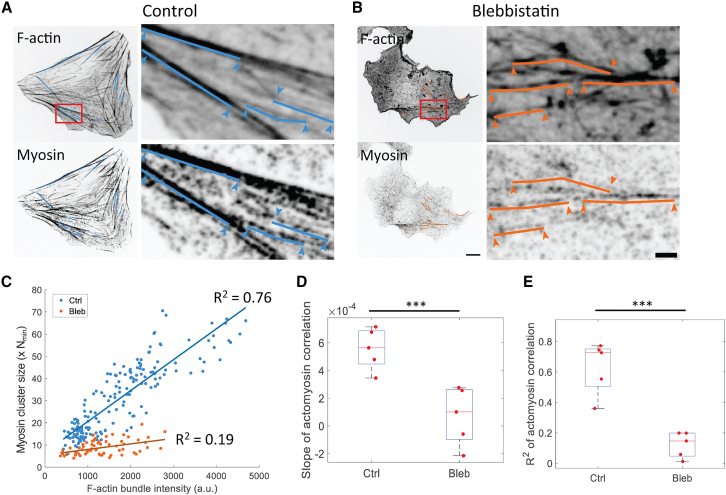
Since blebbistatin does not impact RLC phosphorylation, we reasoned that myosin motor activity can only indirectly affect myosin association. Since F-actin has been proposed to regulate myosin assembly and myosin cluster growth at the leading edge (18,19,31), we wondered if F-actin could play a role. Consistent with this idea, myosin cluster sizes across the cell positively correlate with the underlying F-actin intensity (Fig. S3, A–C). Since different stress fiber architecture contains variable amounts of myosin, we restricted our analysis to mature transverse arcs and ventral stress fibers. On these stress fibers, myosin cluster sizes correlate strongly with the intensity of F-actin bundles (Fig. 6, A and C). On the other hand, myosin cluster sizes across the cell showed much less positive correlation with the underlying F-actin intensity (Fig. S3 D). While there are few F-actin bundles in blebbistatin-treated cells, myosin cluster sizes showed much less positive correlation with the intensity of the remaining F-actin bundles (Fig. 6, B and C). Since both the slope and R2 of the correlation decreased with blebbistatin treatment (Fig. 6 D and E), this suggests that F-actin plays a role in myosin cluster growth only in the presence of myosin motor activity and/or strong binding.
Figure 6.
Myosin cluster sizes correlate with F-actin bundle intensity under myosin motor activity. (A) F-Actin and myosin on select F-actin bundles in control cells. Left column shows all segments of F-actin bundles that are selected for analysis. Right column shows zoomed-in view of a few F-actin bundles. Solid lines represent F-actin bundles where actomyosin correlation is examined. Arrowheads indicate the end points of the segment. (B) F-Actin and myosin on select F-actin bundles in blebbistatin-treated cells. Scale bars, 10 μm (whole cell image) and 2 μm (zoomed-in image). (C) Correlation between myosin cluster size and F-actin bundle intensity in control and blebbistatin-treated cells. Correlation is plotted for myosin clusters on every selected bundle in a single cell. (D) Slope of the correlation. n = 5 cells in each condition. The two-sample t-test is performed for statistical analysis. ∗∗∗ signifies p < 0.001. (E) R2 of the correlation. n = 5 cells in each condition. The two-sample t-test is performed for statistical analysis. ∗∗∗ signifies p < 0.001. To see this figure in color, go online.
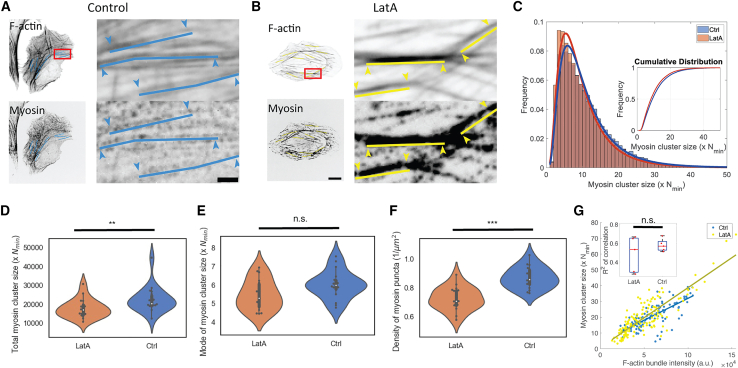
To further understand how F-actin architecture affects myosin cluster size, we perturb the stress fiber architecture and see how myosin clusters respond. With subsaturating concentrations of latrunculin A (LatA), we disrupted the smallest F-actin bundles and meshwork within the lamella. As a result, large F-actin bundles such as ventral stress fibers became the dominant F-actin architecture in cells (Fig. 7, A and B). While the overall F-actin intensity decreased, the intensity of remaining F-actin bundles remains largely unchanged (Fig. S4, A and B). Under LatA treatment, the sizes of myosin clusters did not change significantly (Fig. 7, C and E), but the total amount of myosin within clusters decreased (Fig. 7 D). Since LatA treatment has been reported to neither alter nor increase myosin light chain phosphorylation (32), we hypothesized that myosin cluster sizes are impacted by the change in F-actin architecture. Indeed, the reduction in the F-actin meshwork under LatA treatment is concurrent with the decrease in myosin cluster density (Fig. 7 F). With thin F-actin bundles depleted, myosin clusters accumulated on larger transverse arcs and ventral stress fibers. On these F-actin bundles, myosin cluster sizes exhibited a similar positive correlation with F-actin bundle intensity (Fig. 7 G), consistent with the correlation across the cell (Fig. S3 E). This suggests that myosin cluster sizes depend on the underlying F-actin architecture.
Figure 7.
Myosin association to F-actin bundles depends on F-actin architecture. (A) F-Actin and myosin on select F-actin bundles in control cells. (B) F-Actin and myosin on select F-actin bundles in LatA-treated cells. Scale bars 10 μm (whole cell image) and 2 μm (zoomed-in image). (C) Histogram of myosin cluster sizes between LatA-treated and control cells. Inset shows the cumulative distribution of both conditions. (D) The sum of all myosin cluster sizes. (E) Mode of myosin cluster size distribution. (F) Density of myosin clusters. n = 18 cells for LatA-treated cells and n = 18 cells for control cells. Two-sample Kolmogorov-Smirnov tests were performed in (D–F) to test if the measured quantity differs between conditions. n.s. signifies p > 0.05, ∗∗ signifies p < 0.01, ∗∗∗ signifies p < 0.001. (G) Correlation between myosin cluster intensity and F-actin bundle intensity in control and LatA-treated cells. Correlation is plotted for myosin clusters on every selected bundle in a single cell. Inset shows the R2 of the correlation between control and LatA-treated cells. n = 5 cells in each condition. The two-sample t-test is performed for statistical analysis. n.s., p > 0.05. To see this figure in color, go online.
Myosin cluster growth with a positive feedback is sufficient to recapitulate the broad range of myosin cluster sizes
With our experimental results showing myosin clusters growing via net association to existing clusters, we wanted to quantitatively describe myosin cluster growth. To this end, we constructed a toy model to simulate myosin cluster growth. We modeled myosin cluster growth as a Monte Carlo process, where a predetermined number of myosin filaments (Nmyo) iteratively bind randomly to a 1D grid of 10,000 points. The number of grid points is fixed because myosin cluster density remained similar across almost all experimental conditions (Figs. 2 E and 4 E). The simulation iterates through all myosin filaments, where one myosin filament binds randomly to a grid point in each iteration. (Fig. 8 A). The probability for the myosin filament on a given iteration to bind to grid point is given by , where is the number of myosin filaments at that grid point. The parameter is an affinity parameter that captures scenarios where myosin filaments preferentially bind to existing myosin clusters. This is motivated by recent literature that suggests myosin filaments preferentially bind to existing myosin clusters (18,19,29).
Figure 8.
Myosin cluster growth with positive feedback is sufficient to recapitulate the broad range of myosin cluster sizes. (A) Schematic of the simulation setup. (B) Simulation of myosin cluster growth without myosin motor activity. (i) Experimentally observed myosin cluster size distribution in cells before (Y+B+) and after (Y–B+) washing out Y-27 in cells that are treated with both Y-27 and blebbistatin. (ii) Simulation when myosin binding probability is uniform across all grid points (α = 0). Inset shows the cumulative distribution of the experimental and simulated results. (iii) Simulation when myosin has an affinity toward existing myosin clusters (α = 10). Inset shows the cumulative distribution of the experimental and simulated results. (iv) Average distance between cumulative distributions of the experimental and simulated cluster sizes across different α and Nmyo values. Stars show the Nmyo determined by experimental quantifications. (C) Simulation of myosin cluster growth with myosin motor activity. (i) Experimentally observed myosin cluster size distribution in cells treated with blebbistatin or control cells. (ii) Simulation when myosin binding probability is uniform across all grid points (α = 0). Inset shows the cumulative distribution of the experimental and simulated results. (iii) Simulation when myosin has an affinity toward existing myosin clusters (α = 10). Inset shows the cumulative distribution of the experimental and simulated results. (iv) Average distance between cumulative distributions of the experimental and simulated cluster sizes across different α and Nmyo values. Stars show the Nmyo values determined by experimental quantifications. To see this figure in color, go online.
First, we use our toy model to simulate myosin cluster growth without myosin motor activity, specifically when Y-27 is washed out in cells treated with both Y-27 and blebbistatin (Figs. 2 A and 8 B (i)). To make a meaningful comparison between the simulation and experiments, we initialized the simulation such that the size of myosin clusters on each grid matches the size distribution in cells treated with both Y-27 and blebbistatin (Fig. 8 B (i), Y+B+). This is done assuming that Nmin represents one myosin filament. As a result, the initial total amount of myosin within clusters in the simulation is approximately 47,000. Since our experimental results showed that ROCK-dependent myosin cluster growth increases assembled myosin by about 42% (Fig. 2 C), the simulation iterates through the binding of approximately 20,000 additional myosin filaments. If we assume that myosin filaments can bind to each grid point with equal probability (), corresponding to uniform growth rates across the cell, the myosin cluster sizes at the end of the simulation exhibit a Gaussian-like distribution (Fig. 8 B (ii)). However, this result contradicts the experimentally observed lognormal distribution when we selectively wash out Y-27 (Fig. 8 B (i), Y–B+). When the affinity parameter is nonzero (), the simulated cluster sizes start to qualitatively recapitulate the lognormal distribution of myosin cluster sizes in the absence of myosin motor activity (Fig. 8 B (iii)). We quantified the error between simulated and experimental cluster sizes by calculating the average distance between the cumulative distributions of experimental-measured and simulated myosin cluster sizes. The error decreased with increasing but started to converge around (Fig. S5). This suggests that the simulated results became insensitive to changes in , presumably due to the finite size of our simulation setup. On the other hand, the simulated results depend heavily on the amount of myosin iterated through the simulation. When we scan through a range of Nmyo, the simulation error is the lowest when Nmyo is 20,000, which coincides with experimentally quantified results (Fig. 8 B (iv))! This suggests that the limiting pool of available myosin is important for setting myosin cluster sizes.
We next explored if the same simulation framework and parameters can capture myosin cluster growth under myosin motor activity, specifically when blebbistatin is washed out from cells (Figs. 4 A and 8 C (i)). To make a meaningful comparison with experiments, we initialized the simulation such that myosin cluster size on each grid matches the myosin cluster size distribution in blebbistatin-treated cells (Fig. 8 C (i)). Using the same assumption as before, the initial total myosin contained in clusters in the simulation is approximately 81,000. Since untreated cells show 54% more assembled myosin than blebbistatin-treated cells in our experimental quantification (Fig. 4 C), the simulation iterates through the binding of about 44,000 additional myosin filaments. As in the case of myosin cluster growth without motor activity, uniform myosin binding probability () fails to capture the lognormal distribution of myosin cluster sizes in control cells (Fig. 8 C (ii)). When the affinity parameter is nonzero (), the simulated result once again qualitatively recapitulated the experimental cluster sizes in control cells (Fig. 8 C (iii)). The error between simulated and experimental results decreased with increasing but again converged around (Fig. S5). The optimal Nmyo that produces the least error between simulated and experimental results is around 37,500, which is about 15% less than the experimentally measured Nmyo (Fig. 8 C (iv)). This is likely due to the F-actin-dependent myosin cluster regulation not captured in this toy model. Taken together, our results suggest that myosin cluster sizes on stress fibers are set by a limiting pool of myosin available for cluster growth with myosin self-affinity.
Discussion
Regulation of myosin clusters in adherent cells occurs through spatiotemporal control of de novo assembly, growth, and disassembly. While these processes have been investigated thoroughly at the leading edge (18,19), we sought to explore growth mechanisms across the lamella. To this end, we designed experiments and perturbations over timescales to minimize effects of de novo assembly and disassembly. Within the experimental conditions of our work, at least 70% of myosin clusters in the lamellar network are stable for at least 5 min. Over this timescale, <15% of myosin clusters are estimated to undergo de novo assembly events. While de novo cluster formation dominates at the leading edge of the cell, we observed that growth of existing clusters occurs throughout the whole lamellar network over several minutes (Figs. 3 B and 5 B). Furthermore, myosin clusters that assemble de novo contain fewer myosin filaments. Compared with the increase in myosin cluster sizes enabled by ROCK and myosin motor activity, we reasoned that growth of existing clusters played a larger role than de novo assembly.
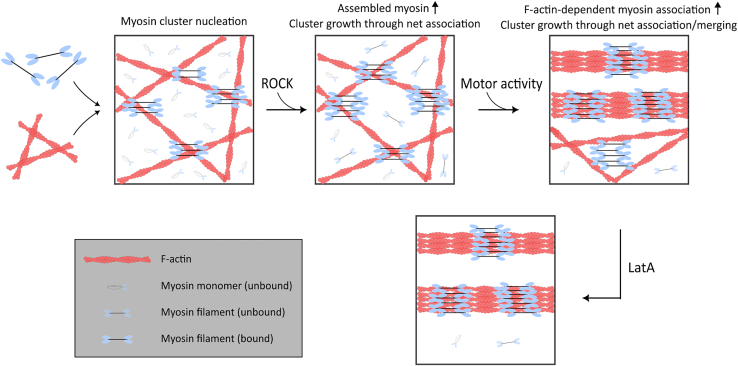
Our results are consistent with the picture that myosin clusters grow by myosin-myosin affinity. We found that the most important factor that determines myosin cluster size is the limited availability of cytoplasmic myosin available to grow clusters, regulated by both ROCK and myosin motor activity (Fig. 9). F-Actin architecture can further regulate myosin cluster sizes (Fig. 9). On the other hand, the nucleation of myosin clusters is independent of these regulations, as we found that the density of myosin clusters remains at similar levels. This insight is only possible through our combination of endogenous tagging, quantitative imaging, analysis, perturbation, and modeling.
Figure 9.
Myosin cluster sizes are set by a limiting pool of myosin that is available for cluster growth. Myosin filaments independent of ROCK activity nucleate myosin clusters. ROCK activity increases myosin assembly, which increases the pool of myosin available for cluster growth through increased net association. Myosin motor activity further enhances myosin cluster growth through F-actin-dependent myosin association. Myosin cluster sizes are perturbed when thin F-actin meshworks are depleted by LatA. To see this figure in color, go online.
A novel finding of this study is that myosin cluster growth can occur independently of myosin motor activity. ROCK activity alone allows myosin clusters to grow from 3 times to 4–15 times the size of Nmin. Since myosin self-affinity is required for our in silico model to recapitulate experimental results, our results suggest that there is biochemical affinity between myosin filaments. This may explain the stack or ribbon configuration of myosin clusters observed with platinum EM (14,15). However, to the best of our knowledge, there is no reported molecular or electrostatic interaction between pairs of assembled myosin filaments. We speculate that the affinity can arise from numerous weak intermolecular interactions between heads, light chains, and tails of myosin monomers in neighboring myosin filaments (33). These interactions can be amplified when multiple myosin filaments are involved. This is supported by a previous study that reported purified NMIIA can form large clusters in vitro (34). Alternatively, the affinity between myosin filaments can be indirectly mediated by other molecular components. For example, some proteins associate strongly with myosin filaments and have been proposed to regulate myosin structures, such as myosin-18B (35) or Tpm 3.1 (36). This will be an interesting topic for future research to identify the cause of myosin self-affinity, and the effect on myosin clusters when this interaction is disrupted.
Myosin cluster growth can also occur with myosin motor activity mainly through the addition of available myosin to existing myosin clusters. We think that this is due to the increase in myosin assembly from myosin motor activity. While myosin motor activity does not affect the phosphorylation of the RLC, previous studies have reported the reduction in myosin filament assembly under blebbistatin treatment (12). This has been attributed to the effect of mechanical tension on myosin assembly, specifically because of the catch-bond nature of myosin-actin interaction (11,12,37) or the mechanosensitive properties of myosin IIA (38). While recent studies suggest that only NMIIB exhibit catch-bond behavior while NMIIA is more like a slip-bond (28), NMIIA assembly can in principle be affected by NMIIB since NMIIA and NMIIB have been shown to copolymerize into filaments (16,39). Another possible molecular process that can result in myosin cluster growth under myosin motor activity is through the coalescence of myosin clusters resulting from their lateral motion promoted by actomyosin sliding. While coalescence occurs over a longer time scale than the association of free myosin, this can serve as a parallel mechanism for myosin cluster growth or as a means to organize the quasisarcomeric pattern (40).
The correlation between myosin cluster sizes and F-actin bundle size may be a result of nuanced feedback between F-actin and myosin clusters, with possible mutual regulation between myosin clusters and F-actin bundle size. On one hand, myosin can be regulated by F-actin. At the molecular level, it has been reported that myosin assembly is enhanced in the presence of F-actin (31,41,42). Some actin-binding protein can also regulate myosin motor activity, such as tropomyosin (43). On the other hand, myosin can remodel the F-actin network. For example, myosin filaments can contract and remodel F-actin networks into asters (44,45,46). The combined effect can effectively create a positive feedback loop to give rise to the correlation between myosin cluster sizes and F-actin bundle size.
Our understanding of myosin cluster regulation is consistent with the idea that the number and size of subcellular organelles are tightly controlled (47,48,49). One model of organelle size control is that a limited pool of cytoplasmic components sets the size of these structures (50). This concept has been demonstrated in several organelles, such as the flagella (51), centrosomes (52), and mitotic spindles (53). Similarly, structures that depend on the same building blocks can compete for these limited components, as observed with different F-actin architectures (5,54) and myosin clusters on various actomyosin structures (18,55). Our paper contributes to this idea by quantitatively demonstrating its applicability in stress fibers of nonmuscle cells. We believe that our approach, combined with advanced microscopy, molecular biology, and perturbation techniques, will lead to further insight into how cells balance and control organelles.
Author contributions
W.-H.C., M.M., P.O., J.B., and M.L.G. designed the research. W.-H.C. and M.M. performed the research and analyzed the data. W.-H.C., M.M., and H.W. provided the reagents. W.-H.C., M.M., P.O., J.B., and M.L.G. interpreted the results and wrote the paper.
Acknowledgments
The authors thank Melissa Quintanilla, Steven Redford, and Ed Munro for insightful discussions. This work is supported by NIH RO1 GM104032, NIH RO1 GM143792, and DMR 2215605 awards to M.L.G.
Declaration of interests
The authors declare no competing interests.
Editor: Dimitrios Vavylonis.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.12.004.
Supporting material
References
- 1.Vicente-Manzanares M., Ma X., et al. Horwitz A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oakes P.W., Beckham Y., et al. Gardel M.L. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 2012;196:363–374. doi: 10.1083/jcb.201107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stricker J., Beckham Y., et al. Gardel M.L. Myosin II-Mediated Focal Adhesion Maturation Is Tension Insensitive. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S., Kumar S. Actomyosin stress fiber mechanosensing in 2D and 3D. F1000Res. 2016;5 doi: 10.12688/f1000research.8800.1. F1000 Faculty Rev-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kadzik R.S., Homa K.E., Kovar D.R. F-Actin Cytoskeleton Network Self-Organization Through Competition and Cooperation. Annu. Rev. Cell Dev. Biol. 2020;36:35–60. doi: 10.1146/annurev-cellbio-032320-094706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard T.D. Regulation of Actin Filament Assembly by Arp2/3 Complex and Formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 7.Quintanilla M.A., Hammer J.A., Beach J.R. Non-muscle myosin 2 at a glance. J. Cell Sci. 2023;136:jcs260890. doi: 10.1242/jcs.260890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholey J.M., Taylor K.A., Kendrick-Jones J. Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature. 1980;287:233–235. doi: 10.1038/287233a0. [DOI] [PubMed] [Google Scholar]
- 9.Newell-Litwa K.A., Horwitz R., Lamers M.L. Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Dis. Model. Mech. 2015;8:1495–1515. doi: 10.1242/dmm.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulyaninova N.G., Bresnick A.R. The heavy chain has its day: Regulation of myosin-II assembly. BioArchitecture. 2013;3:77–85. doi: 10.4161/bioa.26133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo T., Mohan K., et al. Robinson D.N. Understanding the Cooperative Interaction between Myosin II and Actin Cross-Linkers Mediated by Actin Filaments during Mechanosensation. Biophys. J. 2012;102:238–247. doi: 10.1016/j.bpj.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shutova M., Yang C., et al. Svitkina T. Functions of Nonmuscle Myosin II in Assembly of the Cellular Contractile System. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grewe J., Schwarz U.S. Mechanosensitive self-assembly of myosin II minifilaments. Phys. Rev. E. 2020;101 doi: 10.1103/PhysRevE.101.022402. [DOI] [PubMed] [Google Scholar]
- 14.Verkhovsky A.B., Svitkina T.M., Borisy G.G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svitkina T.M., Verkhovsky A.B., et al. Borisy G.G. Analysis of the Actin–Myosin II System in Fish Epidermal Keratocytes: Mechanism of Cell Body Translocation. J. Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shutova M.S., Spessott W.A., et al. Svitkina T. Endogenous Species of Mammalian Nonmuscle Myosin IIA and IIB Include Activated Monomers and Heteropolymers. Curr. Biol. 2014;24:1958–1968. doi: 10.1016/j.cub.2014.07.070. –1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu S., Dasbiswas K., et al. Bershadsky A.D. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 2017;19:133–141. doi: 10.1038/ncb3466. [DOI] [PubMed] [Google Scholar]
- 18.Beach J.R., Bruun K.S., et al. Hammer J.A. Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat. Cell Biol. 2017;19:85–93. doi: 10.1038/ncb3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenix A.M., Taneja N., et al. Burnette D.T. Expansion and concatenation of nonmuscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol. Biol. Cell. 2016;27:1465–1478. doi: 10.1091/mbc.E15-10-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crocker J.C., Grier D.G. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 21.Krull A., Buchholz T.-O., Jug F. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) IEEE; 2019. Noise2Void - Learning Denoising From Single Noisy Images; pp. 2124–2132. [Google Scholar]
- 22.Ji L., Danuser G. Tracking quasi-stationary flow of weak fluorescent signals by adaptive multi-frame correlation. J. Microsc. 2005;220:150–167. doi: 10.1111/j.1365-2818.2005.01522.x. [DOI] [PubMed] [Google Scholar]
- 23.Hotulainen P., Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tojkander S., Gateva G., Lappalainen P. Actin stress fibers - assembly, dynamics and biological roles. J. Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- 25.Chang C.-W., Kumar S. Differential Contributions of Nonmuscle Myosin II Isoforms and Functional Domains to Stress Fiber Mechanics. Sci. Rep. 2015;5:13736. doi: 10.1038/srep13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weißenbruch K., Grewe J., et al. Bastmeyer M. Distinct roles of nonmuscle myosin II isoforms for establishing tension and elasticity during cell morphodynamics. Elife. 2021;10 doi: 10.7554/eLife.71888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breckenridge M.T., Dulyaninova N.G., Egelhoff T.T. Multiple Regulatory Steps Control Mammalian Nonmuscle Myosin II Assembly in Live Cells. Mol. Biol. Cell. 2009;20:338–347. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weißenbruch K., Fladung M., et al. Bastmeyer M. Nonmuscle myosin IIA dynamically guides regulatory light chain phosphorylation and assembly of nonmuscle myosin IIB. Eur. J. Cell Biol. 2022;101 doi: 10.1016/j.ejcb.2022.151213. [DOI] [PubMed] [Google Scholar]
- 29.Quintanilla M.A., Patel H., et al. Beach J.R. Local Monomer Levels and Established Filaments Potentiate Non-Muscle Myosin 2 Assembly. bioRxiv. 2023 doi: 10.1101/2023.04.26.538303. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovács M., Tóth J., et al. Sellers J.R. Mechanism of Blebbistatin Inhibition of Myosin II. J. Biol. Chem. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 31.Mahajan R.K., Vaughan K.T., et al. Pardee J.D. Actin filaments mediate Dictyostelium myosin assembly in vitro. Proc. Natl. Acad. Sci. USA. 1989;86:6161–6165. doi: 10.1073/pnas.86.16.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe T., Hosoya H., Yonemura S. Regulation of Myosin II Dynamics by Phosphorylation and Dephosphorylation of Its Light Chain in Epithelial Cells. Mol. Biol. Cell. 2007;18:605–616. doi: 10.1091/mbc.E06-07-0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S., Tiwari P., et al. Craig R. Cryo-EM structure of the inhibited (10S) form of myosin II. Nature. 2020;588:521–525. doi: 10.1038/s41586-020-3007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melli L., Billington N., et al. Sellers J.R. Bipolar filaments of human nonmuscle myosin 2-A and 2-B have distinct motile and mechanical properties. Elife. 2018;7:e32871. doi: 10.7554/eLife.32871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiu Y., Kumari R., et al. Lappalainen P. Myosin-18B Promotes the Assembly of Myosin II Stacks for Maturation of Contractile Actomyosin Bundles. Curr. Biol. 2019;29:81–92.e5. doi: 10.1016/j.cub.2018.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meiring J.C.M., Bryce N.S., et al. Gunning P.W. Colocation of Tpm3.1 and myosin IIa heads defines a discrete subdomain in stress fibres. J. Cell Sci. 2019;228916 doi: 10.1242/jcs.228916. [DOI] [PubMed] [Google Scholar]
- 37.Veigel C., Molloy J.E., et al. Kendrick-Jones J. Load-dependent kinetics of force production by smooth muscle myosin measured with optical tweezers. Nat. Cell Biol. 2003;5:980–986. doi: 10.1038/ncb1060. [DOI] [PubMed] [Google Scholar]
- 38.Schiffhauer E.S., Luo T., et al. Robinson D.N. Mechanoaccumulative Elements of the Mammalian Actin Cytoskeleton. Curr. Biol. 2016;26:1473–1479. doi: 10.1016/j.cub.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beach J.R., Shao L., et al. Hammer J.A. Nonmuscle Myosin II Isoforms Coassemble in Living Cells. Curr. Biol. 2014;24:1160–1166. doi: 10.1016/j.cub.2014.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thoresen T., Lenz M., Gardel M.L. Reconstitution of Contractile Actomyosin Bundles. Biophys. J. 2011;100:2698–2705. doi: 10.1016/j.bpj.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Applegate D., Pardee J.D. Actin-facilitated assembly of smooth muscle myosin induces formation of actomyosin fibrils. J. Cell Biol. 1992;117:1223–1230. doi: 10.1083/jcb.117.6.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahajan R.K., Pardee J.D. Assembly Mechanism of Dictyostelium Myosin II: Regulation by K+, Mg2+, and Actin Filaments. Biochemistry. 1996;35:15504–15514. doi: 10.1021/bi9618981. [DOI] [PubMed] [Google Scholar]
- 43.Barua B., Nagy A., et al. Hitchcock-DeGregori S.E. Regulation of Nonmuscle Myosin II by Tropomyosin. Biochemistry. 2014;53:4015–4024. doi: 10.1021/bi500162z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soares E Silva M., Depken M., et al. Koenderink G.H. Active multistage coarsening of actin networks driven by myosin motors. Proc. Natl. Acad. Sci. USA. 2011;108:9408–9413. doi: 10.1073/pnas.1016616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Köster D.V., Husain K., et al. Mayor S. Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc. Natl. Acad. Sci. USA. 2016;113:E1645–E1654. doi: 10.1073/pnas.1514030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stam S., Freedman S.L., et al. Gardel M.L. Filament rigidity and connectivity tune the deformation modes of active biopolymer networks. Proc. Natl. Acad. Sci. USA. 2017;114:E10037–E10045. doi: 10.1073/pnas.1708625114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall W.F. Cell Geometry: How Cells Count and Measure Size. Annu. Rev. Biophys. 2016;45:49–64. doi: 10.1146/annurev-biophys-062215-010905. [DOI] [PubMed] [Google Scholar]
- 48.Amiri K.P., Kalish A., Mukherji S. Robustness and Universality in Organelle Size Control. Phys. Rev. Lett. 2023;130 doi: 10.1103/PhysRevLett.130.018401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen P., Levy D.L. Regulation of organelle size and organization during development. Semin. Cell Dev. Biol. 2023;133:53–64. doi: 10.1016/j.semcdb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goehring N.W., Hyman A.A. Organelle Growth Control through Limiting Pools of Cytoplasmic Components. Curr. Biol. 2012;22:R330–R339. doi: 10.1016/j.cub.2012.03.046. [DOI] [PubMed] [Google Scholar]
- 51.Ludington W.B., Shi L.Z., et al. Marshall W.F. Organelle Size Equalization by a Constitutive Process. Curr. Biol. 2012;22:2173–2179. doi: 10.1016/j.cub.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Decker M., Jaensch S., et al. Hyman A.A. Limiting Amounts of Centrosome Material Set Centrosome Size in C. elegans Embryos. Curr. Biol. 2011;21:1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Mitchison T.J., Ishihara K., et al. Wühr M. Size Scaling of Microtubule Assemblies in Early Xenopus Embryos. Cold Spring Harbor Perspect. Biol. 2015;7:a019182. doi: 10.1101/cshperspect.a019182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suarez C., Kovar D.R. Internetwork competition for monomers governs actin cytoskeleton organization. Nat. Rev. Mol. Cell Biol. 2016;17:799–810. doi: 10.1038/nrm.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Najafabadi F.R., Leaver M., Grill S.W. Orchestrating Non-muscle myosin II filament assembly at the onset of cytokinesis. Mol. Biol. Cell. 2022;33:0599. doi: 10.1091/mbc.E21-12-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Color is inverted. Time stamp indicates minutes and seconds.
Color is inverted. Time stamp indicates minutes and seconds. Images are denoised with Noise2Void.
Arrowheads show transiently appearing myosin clusters that increase in intensity. Color is inverted. Time stamp indicates minutes and seconds. Images are denoised with Noise2Void.
Color is inverted. Time stamp indicates minutes and seconds.