Abstract
The body condition of dairy cows is a crucial health and welfare indicator that is widely acknowledged. Dairy herds with a well-management body condition tend to have more fertile and functional cows. Therefore, routine recording of high-quality body condition phenotypes is required. Automated prediction of body condition from 3D images can be a cost-effective approach to current manual recording by technicians. Using 3D-images, we aimed to build a reliable prediction model of body condition for Jersey cows. The dataset consisted of 808 individual Jersey cows with 2,253 phenotypes from three herds in Denmark. Body condition was scored on a 1 to 9 scale and transformed into a 1 to 5 scale with 0.5-unit differences. The cows’ back images were recorded using a 3D camera (Microsoft Xbox One Kinect v2). We used contour and back height features from 3D-images as predictors, together with class predictors (evaluator, herd, evaluation round, parity, lactation week). The performance of machine learning algorithms was assessed using H2O AutoML algorithm (h2o.ai). Based on outputs from AutoML, DeepLearning (DL; multi-layer feedforward artificial neural network) and Gradient Boosting Machine (GBM) algorithms were implemented for classification and regression tasks and compared on prediction accuracy. In addition, we compared the Partial Least Square (PLS) method for regression. The training and validation data were divided either through a random 7:3 split for 10 replicates or by allocating two herds for training and one herd for validation. The accuracy of classification models showed the DL algorithm performed better than the GBM algorithm. The DL model achieved a mean accuracy of 48.1% on the exact phenotype and 93.5% accuracy with a 0.5-unit deviation. The performances of PLS and DL regression methods were comparable, with mean coefficient of determination of 0.67 and 0.66, respectively. When we used data from two herds for training and the third herd as validation, we observed a slightly decreased prediction accuracy compared to the 7:3 split of the dataset. The accuracies for DL and PLS in the herd validation scenario were > 38% on the exact phenotype and > 87% accuracy with 0.5-unit deviation. This study demonstrates the feasibility of a reliable body condition prediction model in Jersey cows using 3D-images. The approach developed can be used for reliable and frequent prediction of cows’ body condition to improve dairy farm management and genetic evaluations.
Keywords: Body Condition, Herd Management, High Throughput Phenotyping, Jersey Dairy Cattle, Machine Learning, Precision Livestock Farming
This study demonstrates the feasibility of predicting body condition in Jersey dairy cows using 3D images. The developed model can be used for frequent prediction of cows’ body condition to improve dairy farm management.
Introduction
The body condition of a dairy cow is one of the important indicators of its welfare and health status (Roche et al., 2009; Welfare Quality®consortium, 2009). A good management practice on farms of the dairy cow’s body condition is associated with more functional cows (healthy, fertile, etc.; Roche et al., 2009). Continuously (weekly to monthly) scoring individual cow’s body condition adds value to dairy farm management, especially in early lactation, as it provides farmers an overview of how body condition changes through lactation (Bell et al., 2018). Furthermore, body condition phenotypes are important for genetic evaluations of feed efficiency, in order to properly account for the period of mobilization and deposition (Stephansen et al., 2021a).
The standard approach of assessing the body condition of dairy cows is through a body condition score (BCS) which is based on assessing the degree of apparent adiposity of dairy cows (Roche et al., 2009). Currently trained staffs (e.g., evaluators, veterinarians) visit farms to subjectively score body condition, typically on a 1 to 5 scale with 0.25-point differences, but other scales have also been used (Roche et al., 2009). Several countries use a 1 to 9 scale for scoring of BCS, following the ICAR recommendation (ICAR, 2022), which can be transformed to the 1 to 5 scale with 0.5-unit differences following Garnsworthy (2006).
A manual and visual scoring of BCS is time-consuming and costly, making it difficult to implement routinely on dairy farms. Therefore, frequent BCS scoring is typically limited in commercial farms, while mostly scored on research and nucleus farms. A low-cost automated system that can predict body condition traits often during lactation adds value to dairy farm management. In a review, Qiao et al. (2021) compared several studies on the predictability of BCS using 2D and 3D camera technologies. The authors concluded that automated techniques to predict BCS could improve cost efficiency and would play an important role in future dairy farm management. A recently developed system, Cattle Feed InTake (CFIT; Lassen et al., 2018), combines artificial intelligence (AI) and 3D images to identify cows and to predict their feed intake and body weight (BW) (Lassen and Borchersen, 2022; Gebreyesus et al., 2023; Lassen et al., 2023). Development of a model to predict BCS in real time using input data from CFIT system can potentially increase the management value of such a system. Furthermore, the predicted BCS phenotypes can be used to improve genetic selection indices for feed efficiency.
Several studies have investigated the possibility of using computer-vision techniques to predict phenotypes for genetic evaluation and farm management in dairy cattle (Qiao et al., 2021; Lassen and Borchersen, 2022). The field is rapidly advancing, offering potential for high-throughput phenotyping in on-farm settings. Nevertheless, assessing predictive capabilities across studies are challenging due to variations in modeling approaches (e.g., regression, classification), sample sizes, disparate employment of validation as well as evaluation procedures for predictive performance (Qiao et al., 2021). Furthermore, no studies have investigated the proportion of agreement between prediction BCS phenotypes from regression and classification models using the same algorithm.
Studies using 3D-images for prediction of BCS have typically been developed in specific time periods and most cases in a single research herd, or in few cases two herds (Qiao et al., 2021). Validating trained models of their predictability of BCS in different environments (herds) and longer time periods has not been reported so far. Thus, there is a gap in knowledge on the predictive performance of 3D-images for BCS in different environments (herds) and time periods, that is, of importance for the applicability of such systems in a commercial context.
Studies on the predictability of BCS have used depth images from 3D-cameras to train convolutional neural network (CNN) classification models (Rodríguez Alvarez et al., 2019; Yukun et al., 2019), or a similar type of neural network (PointNet++) (Shi et al., 2023). These studies had achieved high accuracies (>90%) with a human error judgement of 0.5-unit BCS, but these models were not validated in different farm environments. The neural network models are typically more complex models that requires more computational time than regression models but are more accurate. Fischer et al. (2015); Martins et al. (2020) and Zin et al. (2020) showed the feasibility of using regression models to predict BCS from 3D-images features, in small populations (<82 cows). Gebreyesus et al. (2023) showed that multi-breed prediction of BW with contour features as predictors, achieved a mean correlation coefficient of 0.94 between observed and predicted BW. However, there is a gap in knowledge on using contours from whole back 3D-images as predictors for BCS.
To our knowledge, no study has so far investigated the predictive ability of BCS of Jersey cows based on 3D-image generated contours. Therefore, the aim of this project was to set up a reliable prediction of BCS using machine learning techniques in Danish Jersey cows under commercial farm environments. Compared to previous studies (Qiao et al., 2021), we used one of the largest training data sets to develop prediction algorithm for BCS using contours as predictors and compared different machine learning models. In addition, we are the first to validate the model’s agreement between regression and classification models, as well as their predictive performance in different herds, where practical application of these models is expected.
Material and Methods
No Animal Care and Ethic Committee approval was required as data used in the study came from routine dairy herd management practice. No treatment or handling of animals was performed in this study.
Data collection of body condition phenotypes
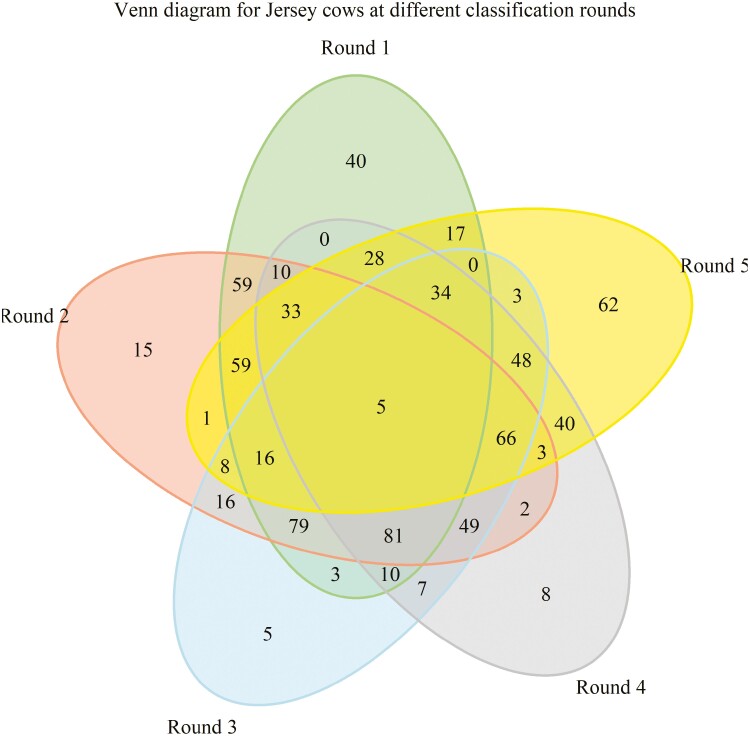
Three commercial Jersey cattle farms located on the island Fyn, Denmark participated in the project. The herd sizes were on average 150, 260, and 280 dairy cows per year and milked in either milking parlor (2 farms) or with automatic milking system (the largest herd). Scoring of BCS was done every other month from December 2021 until August 2022 by two trained evaluators from SEGES INNOVATION (Skejby, Denmark; https://www.seges.dk/), who took rotation to visit different herds during this recording period. Body condition scores were assessed for all cows in these project herds. Figure 1 shows the number of cows scored in the different rounds of evaluation, meaning most cows had repeated BCS measurements. Most cows had a minimum of two records, but a few were scored in all five rounds.
Figure 1.
Venn diagram showing the number of Jersey cows scored in the different rounds of evaluation, meaning most cows had repeated measurement.
In total 2,253 BCS phenotypes were recorded on 808 Jersey cows. The cows were scored on a scale from 1 to 9 according to ICAR (2022). As most studies (Qiao et al., 2021) and farms use the 1 to 5 scale, the score from the evaluators were transformed to the 1 to 5 scale following Garnsworthy (2006):
| (1) |
Basic information, such as calving date (December 2020 to August 2022) and lactation data (parity range 1-9, average parity 2.65 with the first and third quantiles of 1.0 and 4.0; days in milk in the range 10-401, average days in milk 142.4 d with the first and third quantiles of 76.0 and 205), were extracted from the Danish Cattle database and provided by SEGES Innovation (Skejby, Denmark; https://www.seges.dk/). The annual herd yields of the cows ranged between 10,900 and 11,750 kg energy corrected milk in the Danish test-day recording system (RYK, 2022).
Method for repeatability analysis of body condition score
A model to assess the repeatability of the scores was developed using the Proc MIXED procedure in SAS using the REML method and the following model:
| (2) |
where BCSijklm is the phenotype for body condition score for the ith cow on the jth week of lactation; µ is the intercept; is the fixed effect of the kth evaluator (2 levels) nested within the lth herd (3 levels); is the mth fixed regression on the jth week of lactation and nested within the lth herd; is the term of a 4th-order Legendre polynomial for the jth week of lactation; RD is the fixed effect for round of evaluation (5 rounds); animal is the random effect of animal; ɛ is the random residual from the model.
The order for Legendre Polynomials was tested by increasing the number of orders until the Akaike’s Information Criterion started to increase. The repeatability was defined as
where is the variance associated within animals and is the summed variance of the model.
Feature extraction from 3D images and quality control
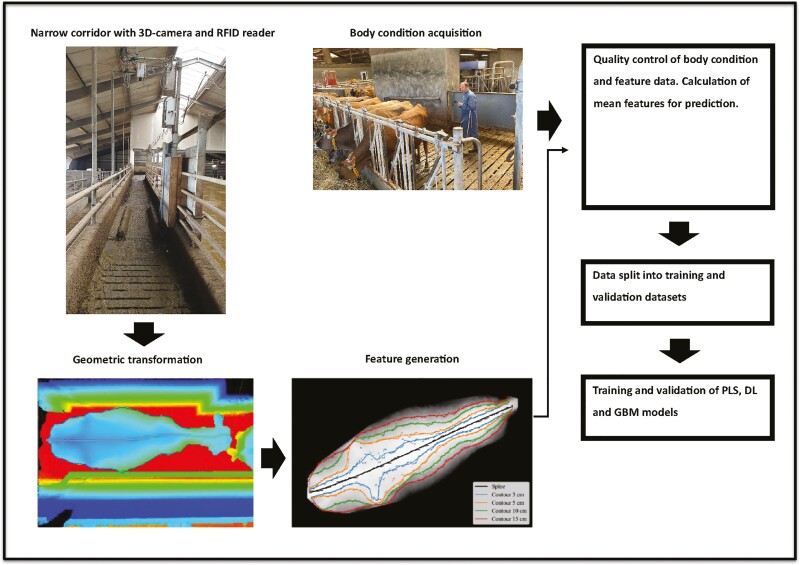
Feature data of the animals within +3 d from the day of BCS evaluation were provided by VikingGenetics (Randers, Denmark). A brief description of the system setup, data handling and feature extraction is described in Figure 2. Further details of the system can be found by Lassen and Borchersen (2022), Gebreyesus et al. (2023), and Lassen et al. (2023). The reference unit consists of a single 3D camera using Time of Flight technology (Microsoft Xbox One Kinect v2) to create a 3D image as well as a Radio Frequency Identification (RFID) reader (Agrident Sensor ASR550). A DELL T630 128 GB RAM server with 3090 RTX graphics card is used for the data analysis of the images recorded. The camera and ear tag reader were installed in a narrow corridor with a time-based trigger system that allocates all images taken within 3 s of reading an RFID to the associated ear tag. A maximum of 5 pictures are taken every 5 s. With this system, it was ensured that one reference image was obtained from each cow when they pass through the corridor. The corridor was narrower than a normal exit corridor to minimize that two cows were exiting together or cows turning around and exiting at strange angles. The 3D camera was placed at a height of 3.4 m above floor level, directly above the passing cows.
Figure 2.
Overview of data acquisition and building prediction algorithm. RFID = Radio Frequency Identification, PLS = Partial Least Square, DL = DeepLearning, GBM = Gradient Boosting Machine. (c) The highest point on an animal. Then contours 3, 5, 10, and 15 cm were found by how far left or right, respectively, should you go from the spine to drop 3, 5, 10, or 15 cm.
Before any cows enter the system, the fixed interior in the image of an empty corridor is annotated. This is done to avoid that anything enters, an image will be noticed as a change from the annotated picture and considered a cow. The first step in the image process is to estimate features from the geometric information in the 3D images, which are useful for separating the individuals. The second step is a calibration procedure that converts the region within the cow circumference to a point cloud, so each pixel in this region of the 3D image is transformed into the corresponding spatial 3D coordinates. The procedure is primarily done to remove distortions due to perspective. Furthermore, the calibration allows a combination of the point clouds information from two neighboring cameras if a cow is placed on the border between the camera’s field of view. In the third step, all images were standardized to have the same length and width. In the final step, a corrected depth image of the cow region is created by interpolating the point cloud back into a 2D depth image.
The feature extraction process started by finding the circumference and spine of the cow in the raw uncorrected 3D images. The circumference was defined as the last pixel before the image sees the annotated floor. Across the back of the cow the highest point was found and named the spine. This was simply the highest point across the whole corridor. The following step included finding the points on the corrected depth image lying 3, 5, 10, and 15 cm below the spine level of the cow. So how far left or right respectively should you go from the spine to drop 3, 5, 10, or 15 cm. This described the contour of the back of each cow. Based on the length standardization described above, 100 spots are placed for each of the 3, 5, 10, and 15 cm features. The features used to predict BCS were the spine (back height) and the distance between the 3, 5, 10, and 15 cm, respectively from left to right across the spine of the cow. In total, there were 900 spots for each image.
Quality control was undertaken on the feature data using the SAS software version 9.4, to remove outlier values. Features were set missing for values out of the range of mean ± 3SD. This outlier detection was done by cow and date of evaluation twice, to ensure very extreme outliers are removed in the first round of outlier detection. Hereafter features with a missing rate higher than 25% were discarded. Cows has on average 32.8 pictures (SD of 11.9) per round of evaluation. Animals with fewer than five pictures per evaluation round were deleted. This resulted in a total of 700 features used as predictors for training the models. We calculated a mean feature per round of scoring, for all 700 features, to give the most stable prediction of BCS. A mean feature was calculated by cow and evaluation date for the 700 individual features and weighted by
The weighting was used to put emphasis on features from the day of evaluation, assigning more weight to closer days apart between visual evaluation and image data capture. The variation among cows, when calculating mean features, was calculated as coefficient of variation (CV) by cow and date of scoring for five features, on two contours (3 and 10 cm) and back height. The features were located on the same position and equally distributed over both contours and back height.
Defining training and validation data
We predicted BCS phenotypes in Jersey cows from contour data generated from 3D cameras using both classification and regression machine learning (ML) models. Splitting training and validation datasets for model development is commonly done with a 7:3 random split of the data (Rodríguez Alvarez et al., 2019; Yukun et al., 2019). The 7:3 random split was performed using Proc Survey procedure in SAS version 9.4, and clustered by cow ID to ensure individual (cows) only appeared in either the training or validation dataset. Ten replicates of training and validation datasets were created to compare different ML algorithms. The two most extreme BCS classes (1.0 and 5.0) were grouped with the immediate next class due to very low observations (three in each) in training and validation sets. This was done to ensure adequate observations were available for the learning step and enabled class balances between the training and validation datasets across replicates.
We also tested the predictability of training the models on two herds and validating in the third herd, giving three different validation scenarios. When training the model, the class effect of herd was left out in this validation setup.
Learning algorithms
A general overview of the steps from image acquisition to the prediction model development is given in Figure 2. We used the AutoML algorithm from H2O package in R (LeDell et al., 2022) for testing best-performing classification and regression algorithms. We used the first training dataset from the random 7:3 split in the AutoML algorithm to test which ML algorithm performs best on that dataset. The non-default parameters in the AutoML algorithm were set to test maximum 2,000 models for classification or regression, and had seed set to 1 and nfolds to 10. Common class predictors including evaluator, parity number, round of evaluation and herd were considered across all the ML methods. Predictors were features from 3D images, which were standardized to a mean of 0 and SD of 1, and Legendre polynomials fitted on weeks of lactation up to 5th order. The Legendre polynomials were the same as used in the repeatability analysis (equation (2)). We also tested models in three scenarios where 1) only features were used as predictors (3D-image-based information), 2) only class predictors (cow-specific information) were used as predictors and 3) features together with class predictors (evaluator and round of evaluation) were used as predictors). This was tested to evaluate the predictability of the features. Results were reported in Supplementary Material.
Tuning parameters for the various classification and regression models in the AutoML algorithm were optimized based on cross-validation with logloss and mean squared error (MSE) as optimizing metrics for the classification and regression models, respectively.
Following models were implemented in the AutoML tuning fase (H2O.ai, 2023). DeepLearning (DL; classification and regression) is a multi-layer feedforward artificial neural network algorithm in H2O. XGBoost (classification) is an ensemble learning technique of many models that attempts to correct the deficiencies in the previous model. Gradient Boosting Machine (GBM; classification and regression) is a forward learning ensemble method and build regression trees on all features in the dataset. Distributed Random Forest (DRF; classification) is a classification method based on building uplift trees. Generalized Linear Model (GLM; regression) is a regression model. The non-default parameters from AutoML used to train the ML models can be found in Supplementary Material.
The implemented classification and regression approaches were compared for prediction accuracy amongst each other as well as with the Partial Least Square (PLS) model. The reason for testing the PLS algorithm were that the PLS algorithm works well on correlated predictors (James et al., 2013). The PLS model was tested in SAS with the Proc PLS procedure (SAS Institute Inc, 2013) fitting the same features and class variables as in all the classification and regression models. The first training and validation dataset from the random 7:3 split was used to fine-tune the PLS model and to define the optimum number of factors. The tunning process of PLS showed 20 factors were the optimum.
Evaluation of predictive performance
For classification models, four evaluation terms were reported for each model within a validation scenario. We grouped data into four individual classes based on confusion matrices between observed and predicted BCS: True Positives (TP), the number of correctly recognized phenotypes, True Negative (TN), the correctly predicted value that do not belong to the observed phenotype class, False Positives (FP) predicted phenotype assigned to the wrong observed class and False Negatives (FN) not recognized observed classes.
Accuracy of classification (AOC) was the effectiveness of a model to classify correctly and defined as (Rodríguez Alvarez et al., 2019):
Precision of prediction (POP) was the ability of the model not to label the predicted phenotype into the wrong observed phenotype and defined as
Recall of prediction (ROP), also called sensitivity is the ability of the model to assign observed phenotypes into the right predicted class, and defined as
F1-score is a measure that combined the trade-offs of precision and recall and defined as
All the evaluation parameters for classification models were evaluated for their ability to predict the exact phenotype, but also with a 0.50-unit deviation (DEV) to account for the human error judgement.
For regression models, the following terms were used to evaluate the predictive performance using an ANOVA analysis: R2 and root mean squared error (RMSE). These parameters were estimated with the Proc ANOVA procedure in SAS, using the predicted BCS as predictor of the observed BCS scored by trained evaluators. Another evaluation parameter for regression analysis methods was to evaluate the percentage of predicted BCS phenotypes that were equal to the observed phenotype and with a human error of judgment range at 0.5 BCS unit. This was implemented in such a way that the predicted BCS phenotype from a regression model was rounded to the nearest 0.5 unit. The percentage of correctly assigned phenotypes were then reported for each class of observed BCS, but also a weighted average based on frequency was reported.
The rounded BCS phenotypes from regression models were compared with predicted BCS phenotypes from classification models, within algorithm. The proportion of agreement between the predicted phenotypes from regression and classification models were reported on the exact and 0.5-unit DEV phenotypes.
Results
Body condition scores in Danish Jersey cattle
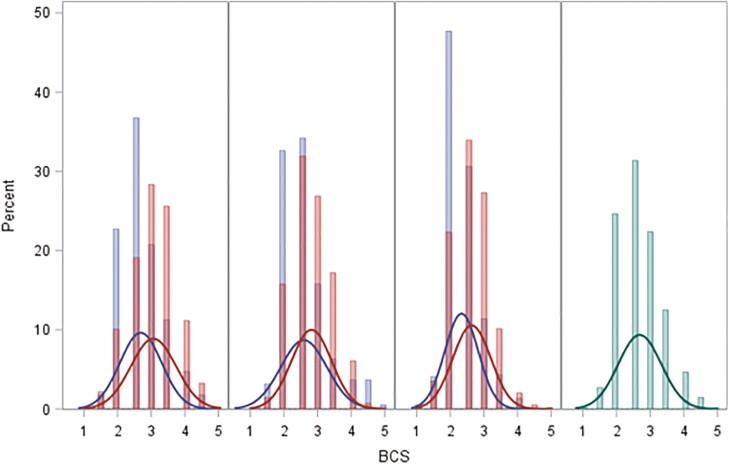
Figure 3 shows the distribution of BCS in three project herds by two evaluators. The density plots (red and blue) show difference between two evaluators. Overall, the evaluator represented by blue, scored cows to be leaner compared to the “red” evaluator. An ANOVA analysis with evaluator and herd as class predictors of BCS showed significant (P < 0.0001) effects of both evaluator and herd. The R2 of the ANOVA analysis was 0.097. The distribution of BCS in the training and the validation datasets were similar as observed for the whole dataset (Figure 3).
Figure 3.
Histogram and density plot of BCS in the different project herds (columns, last column combined herds) and evaluators (blue = evaluator 1, red = evaluator 2, green = both). BCS = Body Condition Score.
The repeatability (τ) measured from model (equation (2)) were 0.60, meaning 60% of the variation in BCS was related to the animal effect.
Variability in calculation of mean features
The highest variability for features were observed on contour at 3 cm and lowest variability on the features located on the back height. Furthermore, differences in variation were observed between rounds of evaluation for contour at 3 cm, though the variability in CV among cows were not extremely high (Figure 4). One possible reason can be that the features are not located on the exact same spot on consecutive (same day and across days) 3D-images as the cows are passing a narrow corridor when the pictures were captured. To show the calculation of mean features were minimally affected by location of features from 3D image to 3D image, we used heatmaps showing correlation structure between features.
Figure 4.
Box plots for coefficients of variance for individual cows at the different round of evaluation for five features on two contours and back height. Contour 1 represents the 3 cm contour and contour 3 represents 10 cm contour. The five features had the same position and were equally distributed at both contours and back height.
The adjacent features are highly correlated, as can be seen from the correlation heatmap of features (Figure 5). Distinct features for the two contours show lower correlations, varying from moderately high to zero. All features for back height show very high correlations, meaning the measure of height of the animal is very consistent across features (Figure 5).
Figure 5.
Heatmaps for the correlation structure among features on two contours and back height. Contour 1 represents the 3 cm contour and contour 3 represents 10 cm contour. On X- and Y-axis, the feature position on the contours and back height are indicated. Features are depth spots based on 3D images from the top of a cow.
Predictive performance of various machine learning techniques
The two best performing algorithms from the AutoML model for classification and regression models, were in both cases DL and GBM. We will in the following section present the validation results from the DL (classification & regression), GBM (classification & regression) and PLS (regression) models.
Predictive performance of classification models
On the exact and 0.5-unit DEV phenotypes, the DL classification model achieved the highest accuracy and the lowest range among the replicates (Table 1).
Table 1.
Accuracy in percentage of the classification models DL and GBM from the 7:3 random split
| DL, % | GBM, % | |||
|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| Mean | 48.1 | 93.5 | 46.0 | 90.9 |
| SD | 1.5 | 0.9 | 2.0 | 1.0 |
| Range | 45.9 to 50.7 | 92.7 to 95.3 | 42.6 to 49.6 | 89.7 to 92.7 |
DL = DeepLearning, GBM = Gradient Boosting Machine, Exact = exact score, DEV = deviation, SD = Standard deviation.
Validation results for sensitivity, precision, and F1-score of the DL classification model are presented in Table 2. The weighted average of sensitivity, precision and F1-score were on similar levels for both, the exact and 0.5-unit DEV. Highest sensitivity, precision and F1-score were achieved with a 0.5-unit DEV. The level of the sensitivity, precision and F1-score followed the pattern of the category frequency of BCS (Figure 5), meaning highest values were observed for BCS categories with highest frequency.
Table 2.
Validation results for sensitivity, precision, and F1-score in percentage for DL, using the 7:3 random split
| BCS | Sensitivity, % | Precision, % | F1-Score, % | |||
|---|---|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| 1.5 | 3 (0-11) | 88 (71-100) | 25 (0-100) | 40 (0-100) | 3 (0-14) | 39 (0-100) |
| 2.0 | 55 (45-62) | 99 (97-100) | 63(60-69) | 98 (96-99) | 59 (51-63) | 98 (97-99) |
| 2.5 | 67 (63-76) | 98 (96-100) | 47 (43-52) | 94 (93-97) | 55 (52-57) | 96 (95-97) |
| 3.0 | 33 (21-46) | 98 (96-99) | 40 (35-45) | 91 (88-97) | 36 (27-43) | 94 (93-97) |
| 3.5 | 44 (32-60) | 82 (77-93) | 41 (32-51) | 88 (82-93) | 42 (32-48) | 85 (80-92) |
| 4.0 | 7 (0-29) | 72 (61-83) | 30 (0-100) | 93 (71-100) | 9 (0-34) | 81 (73-91) |
| 4.5 | 3 (0-20) | 26 (0-55) | 13 (0-100) | 23 (0-100) | 4 (0-25) | 13 (0-57) |
| WAvg | 48 (46-51) | 94 (93-95) | 47 (44-52) | 91 (89-94) | 46 (44-49) | 91 (89-94) |
The parenthesis represents the range among replicates. DL = DeepLearning, BCS = Body Condition Score, Exact = exact score, DEV = deviation, WAvg = weighted average by frequency.
We observed the same patterns using the GBM model for classification of BCS as for the DL model (Table 3). However, in general the GBM model had lower predictive performance on the weighted average, sensitivity, precision and F1-score for both exact and 0.5-unit DEV phenotypes.
Table 3.
Validation results for sensitivity, precision, and F1-score in percentage for GBM, using the 7:3 random split
| BCS | Sensitivity, % | Precision, % | F1-Score, % | |||
|---|---|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| 1.5 | 2 (0-12) | 90 (80-100) | 25 (0-100) | 68 (0-100) | 4 (0-21) | 66 (0-98) |
| 2.0 | 65 (60-71) | 96 (94-98) | 58 (54-65) | 95 (92-98) | 61 (58-67) | 95 (94-97) |
| 2.5 | 54 (47-60) | 97 (95-99) | 47 (44-55) | 92 (88-94) | 50 (46-56) | 94 (93-96) |
| 3.0 | 36 (30-43) | 91 (88-96) | 38 (33-43) | 89 (86-94) | 37 (33-43) | 90 (88-93) |
| 3.5 | 32 (25-38) | 78 (71-83) | 37 (29-43) | 88 (75-94) | 34 (30-38) | 83 (78-87) |
| 4.0 | 13 (3-24) | 72 (57-89) | 26 (8-41) | 71 (44-91) | 17 (4-28) | 71 (55-84) |
| 4.5 | 10 (0-38) | 43 (20-73) | 32 (0-67) | 63 (33-100) | 13 (0-40) | 48 (32-84) |
| WAvg | 46 (43-50) | 91 (90-93) | 45 (40-50) | 89 (88-92) | 45 (41-48) | 90 (88-92) |
The parenthesis represents the range among replicates. GBM = Gradient Boosting Machine, BCS = Body Condition Score, Exact = exact score, DEV = deviation, WAvg = weighted average by frequency.
The DL model showed the highest validation accuracy in herd validation scenarios (Table 4). Across herds, both DL and GBM models showed variability in accuracy. The highest accuracies were seen with Herd 2 (lowest annual average number of cows) as the validation herd. The lowest accuracies were seen with Herd 3 (highest yield in kg energy corrected milk) as the validation herd (Table 4). Comparing the 7:3 random split with the herd validation results, DL showed similar levels of accuracies for Herd 1 and 2 as the validation herds. However, when Herd 3 was validation herd, the accuracies were lower compared to the random 7:3 split. The same pattern was seen for GBM on a lower level of accuracies. Tables with sensitivity, precision and F1-scores from the herd validation scenario can be found in Supplementary Material.
Table 4.
Accuracies from the herd validation scenario of classification models
| DL, % | GBM, % | |||
|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| Herd 1 | 46.8 | 91.2 | 43.6 | 86.8 |
| Herd 2 | 49.5 | 93.0 | 46.1 | 92.5 |
| Herd 3 | 38.3 | 87.4 | 35.9 | 81.4 |
DL = DeepLearning, GBM = Gradient Boosting Machine, Exact = exact score, DEV = deviation.
Predictive performance of regression models
Validation results from the regression models in a 7:3 random split of training and validation data, showed similar R2 and RMSE for PLS and DL models (Table 5). The GBM model performed poorer on R2 but achieved similar RMSE as PLS and DL. The accuracy for the rounded predicted BCS phenotype showed highest values from the DL model on the exact phenotype, but PLS performed best on the 0.5-unit DEV (Table 5).
Table 5.
Validation results from regression models using the 7:3 random split
| BCS | PLS | DL | GBM | |||
|---|---|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| 1.5 | 33 (19-52) | 91 (80-100) | 16 (6-29) | 89 (72-100) | 6 (0-18) | 84 (76-94) |
| 2.0 | 49 (46-52) | 97 (94-99) | 50 (45-54) | 97 (95-99) | 53 (47-58) | 97 (96-99) |
| 2.5 | 60 (56-69) | 98 (96-99) | 67 (61-73) | 98 (97-99) | 62 (57-66) | 98 (96-99) |
| 3.0 | 55 (51-58) | 98 (97-100) | 52 (44-60) | 98 (97-99) | 51 (41-57) | 97 (95-99) |
| 3.5 | 45 (36-51) | 94 (91-99) | 41 (34-48) | 91 (86-98) | 39 (33-45) | 90 (83-94) |
| 4.0 | 23 (10-35) | 86 (80-96) | 23 (7-29) | 78 (71-83) | 23 (10-31) | 75 (62-91) |
| 4.5 | 9 (0-20) | 65 (42-78) | 9 (0-20) | 64 (40-78) | 2 (0-20) | 47 (20-73) |
| WAvg | 51.2 | 96.1 | 52.0 | 95.5 | 50.4 | 94.3 |
| R 2 | 0.67 (0.65-0.68) | 0.66 (0.64-0.68) | 0.63 (0.61-0.66) | |||
| RMSE | 0.31 (0.29-0.33) | 0.29 (0.26-0.32) | 0.30 (0.28-0.32) | |||
Accuracy of the BCS categories is presented in percentage and parenthesis represents the range among replicates. PLS = Partial Least Square, DL = DeepLearning, GBM = Gradient Boosting Machine, BCS = Body Condition Score, R2 = R-square, RMSE = Root Mean Square Error, Exact = exact score, DEV = deviation, WAvg = weighted average by frequency.
The validation results for regression models across herd validation showed the DL model having the highest weighted accuracy across herds on the exact and 0.5-unit DEV phenotype (Table 6-8). For R2, the PLS model performed best in 2 out of 3 herds.
Table 6.
Validation results for the PLS model using the herd splitting method
| BCS | Herd 1 | Herd 2 | Herd 3 | |||
|---|---|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 5-unit DEV | Exact | 0.5-unit DEV | |
| 1.5 | 22 | 83 | 60 | 100 | 17 | 83 |
| 2.0 | 35 | 89 | 55 | 97 | 30 | 83 |
| 2.5 | 60 | 92 | 55 | 98 | 34 | 89 |
| 3.0 | 49 | 97 | 47 | 96 | 51 | 95 |
| 3.5 | 34 | 90 | 40 | 97 | 54 | 97 |
| 4.0 | 31 | 88 | 25 | 83 | 28 | 91 |
| 4.5 | 75 | 100 | 0 | 30 | 23 | 77 |
| WAvg | 45.7 | 91.5 | 48.4 | 95.1 | 39.6 | 90.3 |
| R 2 | 0.46 | 0.67 | 0.57 | |||
| RMSE | 0.39 | 0.33 | 0.34 | |||
Accuracy of the BCS categories is presented in percentage. PLS = Partial Least Square, BCS = Body Condition Score, R2 = R-square, RMSE = Root Mean Square Error, Exact = exact score, DEV = deviation, WAvg = weighted average by frequency.
Table 7.
Validation results for the DL model using the herd splitting method
| BCS | Herd 1 | Herd 2 | Herd 3 | |||
|---|---|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| 1.5 | 11 | 81 | 30 | 80 | 11 | 83 |
| 2.0 | 40 | 92 | 55 | 98 | 39 | 89 |
| 2.5 | 75 | 95 | 64 | 100 | 46 | 93 |
| 3.0 | 42 | 98 | 48 | 97 | 44 | 96 |
| 3.5 | 24 | 82 | 33 | 93 | 42 | 92 |
| 4.0 | 13 | 44 | 33 | 75 | 18 | 74 |
| 4.5 | 50 | 75 | 0 | 50 | 18 | 59 |
| WAvg | 49.3 | 92.3 | 50.7 | 95.3 | 40.4 | 90.6 |
| R 2 | 0.45 | 0.68 | 0.54 | |||
| RMSE | 0.33 | 0.31 | 0.35 | |||
Accuracy of the BCS categories is presented in percentage. DL = DeepLearning, BCS = Body Condition Score, R2 = R-square, RMSE = Root Mean Square Error, Exact = exact score, DEV = deviation, WAvg = weighted average by frequency.
Table 8.
Validation results for the GBM model using the herd splitting method
| BCS | Herd 1 | Herd 2 | Herd 3 | |||
|---|---|---|---|---|---|---|
| Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | Exact | 0.5-unit DEV | |
| 1.5 | 3 | 67 | 0 | 90 | 0 | 78 |
| 2.0 | 25 | 86 | 42 | 98 | 29 | 88 |
| 2.5 | 51 | 93 | 63 | 99 | 58 | 92 |
| 3.0 | 62 | 94 | 52 | 97 | 33 | 95 |
| 3.5 | 49 | 93 | 38 | 97 | 29 | 81 |
| 4.0 | 19 | 88 | 25 | 88 | 12 | 69 |
| 4.5 | 0 | 100 | 0 | 40 | 0 | 32 |
| WAvg | 42.2 | 89.8 | 48.0 | 95.9 | 35.9 | 86.6 |
| R 2 | 0.49 | 0.67 | 0.45 | |||
| RMSE | 0.32 | 0.33 | 0.35 | |||
Accuracy of the BCS categories is presented in percentage. GBM = Gradient Boosting Machine, BCS = Body Condition Score, R2 = R-square, RMSE = Root Mean Square Error, Exact = exact score, DEV = deviation, WAvg = weighted average by frequency.
Comparing predicted phenotypes from regression and classification models
The proportion of predicted BCS phenotypes from regression (predicted phenotypes rounded to closest 0.5 unit) and classification models, showed an accordance on the exact phenotype of 70.5% (range: 66.9 to 75.9) and 62.0% (range: 57.5 to 66.1) for DL and GBM, respectively. With a 0.5-unit DEV the values increased to 99.3% (range: 98.8 to 99.7) and 98.5% (range: 97.6 to 99.2) for DL and GBM, respectively. In the herd validation scenario, the accordance between models showed on the exact phenotype a range of 62.3% to 66.3% and 56.1% to 62.9% for DL and GBM, respectively. The proportion of accordance between models in the herd validation increased with a 0.5 unit DEV to 97.9% to 99.6% and 97.8% to 98.9% for DL and GBM, respectively.
Evaluating predictability of features
The test results using different predictors can be found in Supplementary Material. Using only features as predictors in DL and GBM classification models showed higher accuracies (40.6% to 41.1% for exact phenotype; 85.6% to 87.7% for 0.5-unit DEV) than models only using class predictors (evaluator, herd, etc.) (32.7% to 35.2% for exact phenotype; 78.5% to 80.1% for 0.5-unit DEV). Combining the class predictors evaluator and round of evaluation with features, showed similar validation results for accuracy as presented in Table 1. Similar patterns in results were obtained on F1-score for classification models, but also for metrics of validation from regression models (Supplementary Material). This shows that features from CFIT add additional information next to class predictors; however, models need information to correct for the effect of evaluator and round of evaluation.
Discussion
Performance of implemented learning techniques
In this study, we measured BCS on a discrete scale as previous studies, where both, classification (Rodríguez Alvarez et al., 2019; Shi et al., 2023) and regression models (Fischer et al., 2015; Zin et al., 2020) have been trained for prediction. We have trained both model types with the same algorithm to facilitate comparison. We implemented various evaluation parameters and strategies with the aim of ensuring comparability with other studies and for robust evaluation of our approaches. In addition, we applied a novel evaluation parameter for accuracy from regression models (rounded phenotypes), which approximates AOC in classification models. Moreover, we used R2 and RMSE from ANOVA analysis, as additional parameters to evaluate the regression models following Martins et al. (2020). For the classification models, we combined various evaluation parameters commonly used in assessing classification models (AOC, ROC, POC, F1-score) and with the exact and 0.5-unit DEV (Rodríguez Alvarez et al., 2019; Shi et al., 2023). Across the different ML approaches, we observed only minor differences in predictive performance, as measured by the various evaluation parameters. Notably, DL and GBM approaches demonstrated relatively higher performance when used in regression tasks compared to classification models across the different evaluation parameters. For all the classification approaches, DL outperformed GBM in all evaluation parameters employed.
In the regression analyses, differences between employed learning approaches (DL, GBM, PLS) varied according to employed evaluation parameters (R2, RMSE, AOC), the different human-error ranges allowed (exact vs 0.5-unit DEV) and validation strategies (within herd vs across herd). Accordingly, the PLS and DL models showed comparable overall prediction accuracy with DL outperforming PLS in the exact phenotype while PLS showed the highest accuracy when a human-error range of 0.5-unit DEV was assumed. Despite having the lowest prediction accuracy in terms of R2 values, the GBM approach was as predictive as the DL and PLS approaches in observed RMSE.
The PLS regression is a technique that reduces the dimensions of predictor variables into uncorrelated latent variables (James et al., 2013), while DL is a flexible nonparametric modeling approach that can adapt to associations beyond linearity and identify concealed patterns (Kononenko and Kukar, 2007). DL methods are becoming increasingly popular in various disciplines (Alom et al., 2019) but some limitations still linger including difficulty of interpretability and explainability (Alzubaidi et al., 2021). Our results suggest that, in the task of predicting BCS, PLS may be a promising alternative to DL, as it requires relatively fewer computational resources while achieving comparable accuracy levels.
A significant challenge hampering the comparability of results among studies investigating the feasibility of predicting BCS from image data is the inconsistent application of different modeling approaches (classification or regression). In this study, we aim to address this challenge by implementing both regression and classification approaches to predict BCS. Potential differences in prediction performance between regression and classification approaches could be due to the nature of the predicted or outcome variable (continuous versus categorical variables, respectively), model complexity, data distribution and evaluation metrics. We compared the accordance between proportion of BCS phenotypes predicted using regression and classification approaches. We did that to test the nature of predicted phenotypes and the associated choice of evaluation metrics in the relative performance of the two approaches using the same data, thus similar data distribution. Our results showed that DL and GBM regression and classification models had a moderately high level of accordance on the exact phenotype with minimum 56% and maximum 76%, having DL being superior to GBM. When allowing for a 0.5-unit DEV, both models showed a very high level of agreement (>97%) across 7:3 split validation and herd validation.
Comparison of different sets of predictors underscored the importance of including cow-specific effects of evaluator and round of evaluation in the present study. Unlike Shi et al. (2023) and Rodríguez Alvarez et al. (2019) who did not include cow-specific information in their algorithm development, this study diverges in its approach. While constructing the BCS reference data, Rodríguez Alvarez et al. (2019) and Shi et al. (2023) involved one or two trained evaluators scoring BCS simultaneous with 3D-image acquisition. Consequently, the effect of evaluator and round of evaluation (if repeated measures) became superfluous. However, we aimed to build a reference dataset representing a real-world situation on dairy cattle farms. Therefore, it was necessary to include cow-specific information (evaluator and round of evaluation) along with CFIT features, to produce similar predictability comparable to the relevant literature.
Predictive value of contour data from 3D-images for dairy cattle body condition
We used contour data extracted from 3D-images to predict BCS of Jersey dairy cows using different ML techniques. Studies on prediction of BCS using computer-vision investigated a variety of predictive features including angles, distances and curvatures at predefined anatomical points (Coffey et al., 2003; Ferguson et al., 2006; Bewley et al., 2008), the animal’s shape reconstructed from images captured with 2D (Azzaro et al., 2011) or thermal cameras (Halachmi et al., 2008, 2013) as well as contour or depth of an animal captured from top-down 3D images (Salau et al., 2014; Rodríguez Alvarez et al., 2019; Yukun et al., 2019; Liu et al., 2020; Shi et al., 2023).
In general, although studies based on anatomical points from 2D-images have achieved good results, it has been demonstrated that contour or depth-based features from 3D-images resulted in more robust prediction of BCS. The study of Liu et al. (2020) used 3D-shape features from six predefined regions which were used for prediction of BCS using ensemble models while that of Rodríguez Alvarez et al. (2019) fed pre-corrected depth images of cows directly into a CNN model, which learned to extract relevant features for BCS prediction through its layers of convolutions and pooling leading.
Rodríguez Alvarez et al. (2019) reported AOC of 41% and 97% on the exact and 0.5-unit DEV, for an ensemble model. The four models used for the ensemble model ranged in AOC from 30% to 40% and 89% to 97% for the exact and 0.5-unit DEV, respectively. Liu et al. (2020) showed also for an ensemble classification model an AOC of 56 and 94% for the exact and 0.5-unit DEV. However, both studies only validated (7:3 split or five-fold cross validation) their results within herd environment.
In our study, all contour features across the back of the animal were extracted from 3D-images and fed to robust models including DL to predict BCS. We showed one of the highest accuracies of up to 95.5% within 0.50 units indicating the potential of contour data for robust prediction of body condition score. The stability of the features used for prediction (Figure 4) showed back height were very stable, whereas the contours showed more variation from picture to picture. It is biologically meaningful that the back of a mature animal is rather stable. The higher variability in the contours comes from the changes in condition, but also from the annotation of the 3D-images and features extraction. That means the location of the different features on the contours can vary between pictures. However, the covariance between close related features within contour shows high covariance (Figure 5), showing that the features can be used for prediction.
Previous studies investigating the possibility of predicting BCS using image data in cattle have mainly been carried out in research farms. One of the challenges in the development and practical implementation of robust BCS prediction approaches using computer-vision might be the differences in efficiency of the developed systems and models in environments/farms than those used for training (O’Mahony et al., 2023). In this regard, our study conducts a sensitivity analysis of the developed system and prediction models, wherein a training dataset derived from various commercial dairy farms is utilized to predict cows’ BCS from a distinct, unobserved farm. We trained and validated both classification and regression models in a commercial context to assess how well the trained models can predict in commercial environments. The results indicate that prediction models performed well and without marking loss of accuracy when deployed for prediction in an unseen farm/dataset. It has been shown in various fields of application that the performance of various ML techniques, including DL, heavily relies on the amount and quality of data available for learning (Fan and Shi, 2022). One additional advantage in our study, compared to previous studies on prediction of BCS using computer-vision, is the use of relatively large amount of data, both in terms of number animals and number of records available for these animals.
Perspectives of using predicted body condition in management and breeding
The feasibility to predict BCS on dairy farms from 3D-images has been proposed in this study and in a review of Qiao et al. (2021). Predicted BCS phenotypes with satisfying accuracy, can help dairy farmers to improve management decisions. A good management tool to improve dairy farmers management decisions, could be daily or weekly phenotypes for energy balance calculated from frequent measurements of BCS along with BW (Thorup et al., 2018). Wathes et al. (2007) showed that severe negative energy balance in early lactation had adverse negative impact on dairy cows’ fertility performance, where fails to conceive resulted in culling. Randall et al. (2015) showed cows with a BCS below 2 where at greater risk of lameness compared to cows with a BCS higher than 2. Oltenacu and Broom (2010) found the genetic selection for improved milk production have increased the period and level of negative energy balance for dairy cows, which has adverse effects on metabolic health, fertility and productive health. Using the SimHerd simulation software, Anneberg et al. (2016) showed that reducing the risk of different diseases related to hoof, metabolic and reproductive performances had significant positive economic impact on dairy farms gross margin.
Veerkamp (2002) showed the importance of accounting for the variance in BW and BCS for feed efficiency evaluations. It is important to distinguish between muscle and adipose tissue because of the difference in energy density per kg tissue, where adipose tissue has the highest energy density. However, a few genetic evaluation centers have included BCS when modelling feed efficiency in genetic evaluations (Jamrozik et al., 2021; Parker Gaddis et al., 2021; Stephansen et al., 2021b). There is a big potential to include a predicted phenotype of BCS in feed efficiency models. That means feed efficiency models then could (to some extent) distinguish between mobilization or deposition of muscle and adipose tissue. Future research should focus on utilizing big data through precision livestock farming to improve genetic evaluation for feed efficiency.
Body condition score has been documented to describe the level of subcutaneous fat reserves with reasonable accuracies. Wright and Russel (1984) reported an R2 of 0.89 between BCS and subcutaneous fat in Friesian cows. However, in a review by Mann (2022) suggested that BCS is a poor proxy to describe the total body fat reserves, because BCS is inaccurate to describe visceral fat and differences in muscle mass. Ideally, we could measure the total body fat, but that is not feasible in large-scale recording and requires dissection of the animal. Future studies should focus on establishing a prediction of total body fat from various predictors (features from 3D-images, milk components, blood metabolites, etc.).
Conclusions
This study aimed to build a reliable prediction of BCS in Danish Jersey herds. We tested different methods for predicting BCS in commercial conditions, but also validated the results with a common 7:3 random split and a novel herd validation. In addition, we have tested for the first time the feasibility of using contour features from 3D-images in the prediction of BCS. The validation results show that predicting the exact phenotype can be done with an accuracy of ~50% for classification and regression models. Allowing a 0.5-unit deviation gives higher accuracies for all models. The best performing model across classification and regression models was DL. However, the results for regression models show that the PLS model have similar validation results as DL. In conclusion, it is possible to establish a reliable prediction of BCS with contour features from 3D-images in commercial Jersey Herds.
Supplementary Material
Acknowledgements
The research leading to the study results was funded by the CFIT project (9090-00083B), funded by the Innovation Fund Denmark. The authors thank the Danish evaluators Jørgen Knudsen and Villy Nicolajsen for an excellent job on scoring cows’ body conditions and Mogens H. Madsen for administrating the scoring of body condition. The authors also thank the farmers Palle Bjerggaard Hansen, Hans Ulvsbjerg Rasmussen and Viacheslav Kreitor for participating with their herds in the project.
Glossary
Abbreviations
- AOC
accuracy of classification
- AI
artificial intelligence
- BCS
body condition score
- BW
body weight
- CFIT
Cattle Feed InTake
- CNN
convolutional neural network
- DL
DeepLearning
- DEV
deviation
- DRF
distributed random forest
- FN
false negative
- FP
false positive
- GLM
generalized linear model
- GBM
gradient boosting machine
- ML
machine learning
- MSE
mean squared error
- PLS
partial least square
- POP
precision of prediction
- ROP
recall of prediction
- RMSE
root mean squared error
- TN
true negatives
- TP
true positives
Contributor Information
Rasmus B Stephansen, Center for Quantitative Genetics and Genomics, Aarhus University, 8000-Aarhus C, Denmark.
Coralia I V Manzanilla-Pech, Center for Quantitative Genetics and Genomics, Aarhus University, 8000-Aarhus C, Denmark.
Grum Gebreyesus, Center for Quantitative Genetics and Genomics, Aarhus University, 8000-Aarhus C, Denmark.
Goutam Sahana, Center for Quantitative Genetics and Genomics, Aarhus University, 8000-Aarhus C, Denmark.
Jan Lassen, Center for Quantitative Genetics and Genomics, Aarhus University, 8000-Aarhus C, Denmark; Viking Genetics, Assentoft, 8960-Randers, Denmark.
Author Contributions
Rasmus Bak Stephansen (Conceptualization, Methodology, Software, Validation, Formal analysis, Data curation, Writing—original draft, Visualization). Coralia I. V. Manzanilla-Pech (Conceptualization, Writing—Review & Editing, Supervision). Grum Gebreyesus (Conceptualization, Writing—Review & Editing). Goutam Sahana (Conceptualization, Writing—Review & Editing, Supervision, Project administration). Jan Lassen (Conceptualization, Data Curation, Writing—Review & Editing, Supervision, Funding acquisition).
Conflicit of Interest Statement
Part of the results has been presented at the annual ICAR conference from 21st to 26th May 2023 in Toledo, Spain. An extended abstract is the part of the conference proceedings. Jan Lassen was employed by the company VikingGenetics (Randers, Denmark).
References
- Alom, M. Z., Taha T. M., Yakopcic C., Westberg S., Sidike P., Nasrin M. S., Hasan M., Van Essen B. C., Awwal A. A., and Asari V. K... 2019. A state-of-the-art survey on deep learning theory and architectures. Electronics. 8:292. doi: 10.3390/electronics8030292 [DOI] [Google Scholar]
- Alzubaidi, L., Zhang J., Humaidi A. J., Al-Dujaili A., Duan Y., Al-Shamma O., Santamaría J., Fadhel M. A., Al-Amidie M., and Farhan L... 2021. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data. 8:1–74. doi: 10.1186/s40537-021-00444-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anneberg, I., Østergaard S., Ettema J. F., and Kudahl A. B... 2016. Economic figures in herd health programmes as motivation factors for farmers. Prev. Vet. Med. 134:170–178. doi: 10.1016/j.prevetmed.2016.10.007 [DOI] [PubMed] [Google Scholar]
- Azzaro, G., Caccamo M., Ferguson J. D., Battiato S., Farinella G. M., Guarnera G. C., Puglisi G., Petriglieri R., and Licitra G... 2011. Objective estimation of body condition score by modeling cow body shape from digital images. J. Dairy Sci. 94:2126–2137. doi: 10.3168/jds.2010-3467 [DOI] [PubMed] [Google Scholar]
- Bell, M. J., Maak M., Sorley M., and Proud R... 2018. Comparison of methods for monitoring the body condition of dairy cows. Front. Sustain. 2:80. doi: 10.3389/fsufs.2018.00080 [DOI] [Google Scholar]
- Bewley, J., Peacock A., Lewis O., Boyce R., Roberts D., Coffey M., Kenyon S., and Schutz M... 2008. Potential for estimation of body condition scores in dairy cattle from digital images. J. Dairy Sci. 91:3439–3453. doi: 10.3168/jds.2007-0836 [DOI] [PubMed] [Google Scholar]
- Coffey, M., Mottram T., and McFarlane N... 2003. A feasibility study on the automatic recording of condition score in dairy cows. Pages 131-131 in Proc. Proceedings of the BSAS. Cambridge University Press. Penicuik, Midlothian, United Kingdom. [Google Scholar]
- Fan, F. J., and Shi Y... 2022. Effects of data quality and quantity on deep learning for protein-ligand binding affinity prediction. Bioorg Med Chem. 72:117003. doi: 10.1016/j.bmc.2022.117003 [DOI] [PubMed] [Google Scholar]
- Ferguson, J., Azzaro G., and Licitra G... 2006. Body condition assessment using digital images. J. Dairy Sci. 89:3833–3841. doi: 10.3168/jds.s0022-0302(06)72425-0 [DOI] [PubMed] [Google Scholar]
- Fischer, A., Luginbühl T., Delattre L., Delouard J., and Faverdin P... 2015. Rear shape in 3 dimensions summarized by principal component analysis is a good predictor of body condition score in Holstein dairy cows. J. Dairy Sci. 98:4465–4476. doi: 10.3168/jds.2014-8969 [DOI] [PubMed] [Google Scholar]
- Garnsworthy, P. C. 2006. Body condition score in dairy cows: targets for production and fertility. Rec. Adv. An. 2006:61–86. doi: 10.5661/recadv-06-61 [DOI] [Google Scholar]
- Gebreyesus, G., Milkevych V., Lassen J., and Sahana G... 2023. Supervised learning techniques for dairy cattle body weight prediction from 3D digital images. Front. Genet. 13:947176. doi: 10.3389/fgene.2022.947176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- H2O.ai. 2023. Algorithms. [accessed 16 March 2023]. https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science.html#
- Halachmi, I., Polak P., Roberts D., and Klopcic M... 2008. Cow body shape and automation of condition scoring. J. Dairy Sci. 91:4444–4451. doi: 10.3168/jds.2007-0785 [DOI] [PubMed] [Google Scholar]
- Halachmi, I., Klopčič M., Polak P., Roberts D., and Bewley J... 2013. Automatic assessment of dairy cattle body condition score using thermal imaging. Comput. Electron. Agric. 99:35–40. doi: 10.1016/j.compag.2013.08.012 [DOI] [Google Scholar]
- ICAR. 2022. Section 5 - ICAR guidelines for conformation recording of dairy cattle, beef cattle, dual purpose cattle and dairy goats. [accessed January 16, 2023]. https://www.icar.org/Guidelines/05-Conformation-Recording.pdf
- James, G., Witten D., Hastie T., and Tibshirani R... 2013. An introduction to statistical learning. Vol. 112. New York, NY:Springer. [Google Scholar]
- Jamrozik, J., Kistemaker G., Sullivan P., Van Doormaal B., Chud T., Baes C., Schenkel F., and Miglior F... 2021. Genomic evaluation for feed efficiency in Canadian Holsteins. Pages 153-161 in Proc. Interbull Bulletin, Online.
- Kononenko, I. and Kukar M... 2007. Machine learning and data mining. Chichester, UK: Horwood Publishing. [Google Scholar]
- Lassen, J. and Borchersen S... 2022. Weight determination of an animal based on 3d imaging. [accessed April 25, 2023]. https://patents.google.com/patent/US20220221325A1/en
- Lassen, J., Thomasen J. R., Hansen R. H., Nielsen G. G. B., Olsen E., Stentebjerg P. R. B., Hansen N. W., and Borchersen S... 2018. Individual measure of feed intake on in-house commercial dairy cattle using 3D camera system. In: Proceedings of the 11th World Congress of Genetics Applied to Livestock Production, Auckland, New Zealand.
- Lassen, J., Thomasen J. R., and Borchersen S... 2023. Repeatabilities of individual measure of feed intake and body weight on in-house commercial dairy cattle using a 3D camera system. J. Dairy Sci. in press. doi: 10.3168/jds.2022-23177 [DOI] [PubMed] [Google Scholar]
- LeDell, E., Gill N., Aiello S., Fu A., Candel A., Click C., Kraljevic T., Nykodym T., Aboyoun P., Kurka M., and Malohlava M... 2022. R interface for the ‘H2O’ scalable machine learning platform. https://CRAN.R-project.org/package=h2o
- Liu, D., He D., and Norton T... 2020. Automatic estimation of dairy cattle body condition score from depth image using ensemble model. Biosyst. Eng. 194:16–27. doi: 10.1016/j.biosystemseng.2020.03.011 [DOI] [Google Scholar]
- Mann, S. 2022. Symposium review: The role of adipose tissue in transition dairy cows: Current knowledge and future opportunities. J. Dairy Sci. 105:3687–3701. doi: 10.3168/jds.2021-21215 [DOI] [PubMed] [Google Scholar]
- Martins, B., Mendes A., Silva L., Moreira T., Costa J., Rotta P., Chizzotti M., and Marcondes M... 2020. Estimating body weight, body condition score, and type traits in dairy cows using three dimensional cameras and manual body measurements. Livest Sci. 236:104054. doi: 10.1016/j.livsci.2020.104054 [DOI] [Google Scholar]
- O’Mahony, N., Krpalkova L., Sayers G., Krump L., Walsh J., and Riordan D... 2023. Two-and three-dimensional computer vision techniques for more reliable body condition scoring. Dairy. 4:1–25. doi: 10.3390/dairy4010001. doi: 10.3390/dairy4010001 [DOI] [Google Scholar]
- Oltenacu, P. A., and Broom D. M... 2010. The impact of genetic selection for increased milk yield on the welfare of dairy cows. Anim. Welf. 19:39–49. doi: 10.1017/s0962728600002220 [DOI] [Google Scholar]
- Parker Gaddis, K., VanRaden P., Tempelman R., Weigel K., White H., Peñagaricano F., Koltes J., Santos J., Baldwin R., Burchard J., and VandeHaar M. J... 2021. Implementation of Feed Saved evaluations in the US. Pages 147-152 in Proc. Interbull Bull., Online.
- Qiao, Y., Kong H., Clark C., Lomax S., Su D., Eiffert S., and Sukkarieh S... 2021. Intelligent perception for cattle monitoring: a review for cattle identification, body condition score evaluation, and weight estimation. Comput. Electron. Agric. 185:106143. doi: 10.1016/j.compag.2021.106143 [DOI] [Google Scholar]
- Randall, L., Green M., Chagunda M., Mason C., Archer S., Green L. E., and Huxley J... 2015. Low body condition predisposes cattle to lameness: an 8-year study of one dairy herd. J. Dairy Sci. 98:3766–3777. doi: 10.3168/jds.2014-8863 [DOI] [PubMed] [Google Scholar]
- Roche, J. R., Friggens N. C., Kay J. K., Fisher M. W., Stafford K. J., and Berry D. P... 2009. Invited review: body condition score and its association with dairy cow productivity, health, and welfare. J. Dairy Sci. 92:5769–5801. doi: 10.3168/jds.2009-2431 [DOI] [PubMed] [Google Scholar]
- Rodríguez Alvarez, J., Arroqui M., Mangudo P., Toloza J., Jatip D., Rodriguez J. M., Teyseyre A., Sanz C., Zunino A., and Machado C... 2019. Estimating body condition score in dairy cows from depth images using convolutional neural networks, transfer learning and model ensembling techniques. Agronomy. 9:90. doi: 10.3390/agronomy9020090 [DOI] [Google Scholar]
- RYK. 2022. Milkrecording. [accessed March 25, 2023]. https://www.ryk-fonden.dk/english
- Salau, J., Haas J. H., Junge W., Bauer U., Harms J., and Bieletzki S... 2014. Feasibility of automated body trait determination using the SR4K time-of-flight camera in cow barns. SpringerPlus. 3:1–16. doi: 10.1186/2193-1801-3-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc. 2013. The PLS procedure. [accessed February 25, 2023]. https://support.sas.com/documentation/onlinedoc/stat/131/pls.pdf
- Shi, W., Dai B., Shen W., Sun Y., Zhao K., and Zhang Y... 2023. Automatic estimation of dairy cow body condition score based on attention-guided 3D point cloud feature extraction. Comput. Electron. Agric. 206:107666. doi: 10.1016/j.compag.2023.107666 [DOI] [Google Scholar]
- Stephansen, R. B., Lassen J., Ettema J. F., Sørensen L. P., and Kargo M... 2021a. Economic value of residual feed intake in dairy cattle breeding goals. Livest Sci. 253:104696. doi: 10.1016/j.livsci.2021.104696 [DOI] [Google Scholar]
- Stephansen, R. S., Lidauer M. H., Nielsen U. S., Pösö J., Fikse F., Pech C. I. M., and Aamand G. P... 2021b. Genomic prediction of residual feed intake in the Nordic breeds using data from research herds and 3D cameras in commercial herds. Pages 162-166 in Proc. Interbull Bulletin, Online.
- Thorup, V. M., Chagunda M. G., Fischer A., Weisbjerg M. R., and Friggens N. C... 2018. Robustness and sensitivity of a blueprint for on-farm estimation of dairy cow energy balance. J. Dairy Sci. 101:6002–6018. doi: 10.3168/jds.2017-14290 [DOI] [PubMed] [Google Scholar]
- Veerkamp, R. 2002. Feed intake and energy balance in lactating animals. Pages 10-01 in Proc. Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, CD-ROM Session.
- Wathes, D., Fenwick M., Cheng Z., Bourne N., Llewellyn S., Morris D., Kenny D., Murphy J., and Fitzpatrick R... 2007. Influence of negative energy balance on cyclicity and fertility in the high producing dairy cow. Theriogenology. 68:S232–S241. doi: 10.1016/j.theriogenology.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Welfare Quality®consortium. 2009. Assessment protocol for cattle. Accessed 24. February, 2023. https://edepot.wur.nl/233467
- Wright, I., and Russel A... 1984. Estimation in vivo of the chemical composition of the bodies of mature cows. Anim. Sci. 38:33–44. doi: 10.1017/S0003356100041325 [DOI] [Google Scholar]
- Yukun, S., Pengju H., Yujie W., Ziqi C., Yang L., Baisheng D., Runze L., and Yonggen Z... 2019. Automatic monitoring system for individual dairy cows based on a deep learning framework that provides identification via body parts and estimation of body condition score. J. Dairy Sci. 102:10140–10151. doi: 10.3168/jds.2018-16164 [DOI] [PubMed] [Google Scholar]
- Zin, T. T., Seint P. T., Tin P., Horii Y., and Kobayashi I... 2020. Body condition score estimation based on regression analysis using a 3D camera. Sensors (Basel, Switzerland). 20:3705. doi: 10.3390/s20133705 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.