Abstract
Microbial cell factories (MCFs) convert low-cost carbon sources into valuable compounds. The CRISPR/Cas9 system has revolutionized MCF construction as a remarkable genome editing tool with unprecedented programmability. Recently, the CRISPR toolbox has been significantly expanded through the exploration of new CRISPR systems, the engineering of Cas effectors, and the incorporation of other effectors, enabling multi-level regulation and gene editing free of double-strand breaks. This expanded CRISPR toolbox powerfully promotes MCF construction by facilitating pathway construction, enzyme engineering, flux redistribution, and metabolic burden control. We summarize different CRISPR tool designs and their applications in MCF construction for gene editing, transcriptional regulation, and enzyme modulation. Finally, we also discuss future perspectives for the development and application of the CRISPR toolbox.
Keywords: CRISPR toolbox, Microbial cell factory, Metabolic engineering, Base editing, Tunable regulation, Synthetic metabolons
An expanded CRISPR toolbox expands microbial cell factory construction
Microbial cell factories (MCFs) are widely used to produce valuable compounds from low-cost carbon sources, offering a cost-effective and sustainable route for chemical production [1–5]. To achieve high performance, MCFs need to be robust and productive, which requires high strain stability, efficient biosynthesis pathways, optimized metabolic flux distribution and minimized metabolic burdens [6,7]. However, the existing cellular metabolism and regulation network of microbes have been evolved over numerous years to ensure growth and survival, rather than chemical production [8]. As a result, the desired high-performance MCFs often require complicated and laborious engineering, especially in microbial genomes.
The emergence of technologies derived from CRISPR-Cas (see Glossary) revolutionized the field of metabolic engineering by providing an unprecedentedly programmable, efficient, low-cost and precise genome editing tool [9–11]. The CRISPR toolbox has been robustly enriched in recent years. Advanced CRISPR gene editing systems, including CRISPR base editors, CRISPR prime editors, and EvolvR, have been developed, which can perform different kinds of in vivo mutagenesis without double-strand breaks (DSBs) [12–17]. Meanwhile, more CRISPR regulation toolkits have been established by sgRNA or Cas engineering, enabling more precise and tunable expression control. Additionally, CRISPR-mediated synthetic metabolons have been successfully created, expanding the CRISPR toolbox to protein-level regulation [18].
These CRISPR technologies can vastly improve complicated MCF engineering. Efficient CRISPR-mediated gene disruption or repression has accelerated the identification of new valuable genes. Applications of multiplexed gene editing have substantially contributed to pathway reprogramming and enzyme engineering. In addition, controllable and tunable regulation systems are particularly suitable to overcome the challenges in metabolic burden and metabolic flux rewiring for higher production. Here, we summarize the design of different CRISPR toolkits and comprehensively review and discuss their implementation in gene editing, transcriptional regulation, and enzyme modulation for the purpose of MCF construction (Figure 1, Key Figure). We also provide future perspectives on the development and application of the CRISPR toolbox.
Figure 1, Key Figure.
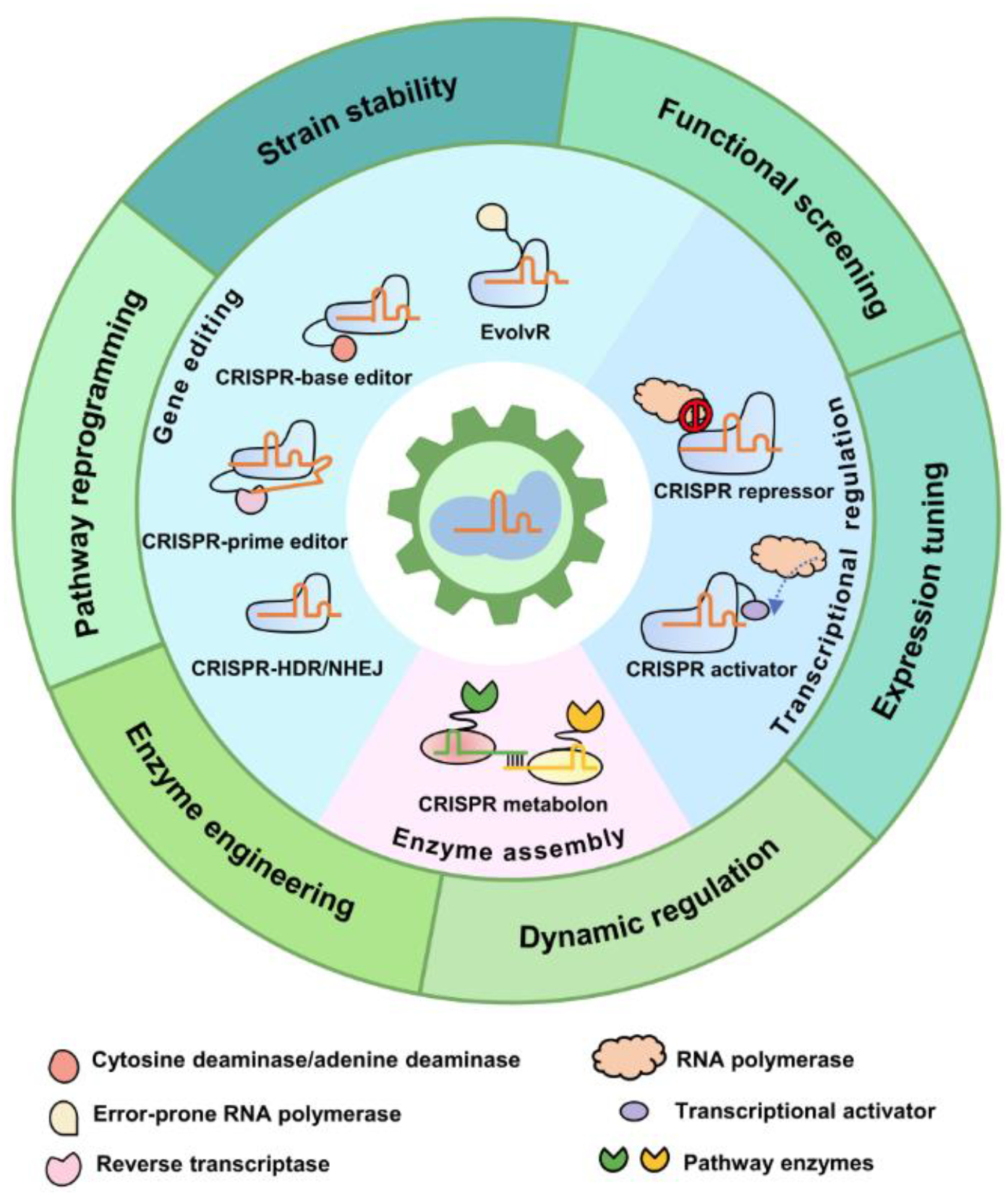
Overview of the CRISPR toolbox for MCF construction. The CRISPR toolbox consists of three types of tools respectively performing gene editing, transcriptional regulation, and enzyme assembly. CRISPR gene editing tools include CRISPR-mediated HDR and NHEJ, as well as the advanced CRISPR base editor, prime editor and EvolvR. Transcriptional regulation tools consist of CRISPRi and CRISPRa, which can be used to design more complex regulation circuits. Enzyme assembly has been achieved by a CRISPR mediated synthetic metabolon. These tools participate in MCF construction by facilitating stable overproducer development, functional screening, expression tuning, dynamic regulation, enzyme engineering and pathway reprogramming.
Gene editing
CRISPR-mediated homology directed repair (HDR) has been extensively utilized for efficient, markerless and multiplexed gene knock-in, knock-out and substitution, thereby facilitating gene discovery, pathway construction, and enzyme engineering. Recent development of CRISPR base editing, CRISPR prime editing, and EvolvR systems have further expanded the CRISPR gene editing toolbox. These advanced tools enable efficient in vivo mutagenesis without the need of DSBs and DNA donors and have started to showcase their applicability in MCF construction (Box 1). Each CRISPR editing system has unique advantages and limitations, and it is important to choose the most appropriate tool for the specific application (Table 1).
Box 1. Summary of CRISPR gene editing tools.
CRISPR-Cas9 can efficiently generate lethal DSBs in the target chromosome DNA loci with adoptable PAMs. In the pioneering work by Jiang and coworkers, phage lambda-red recombinase was harnessed to repair the DSBs with designed DNA donors as templates and introduce DNA mutations, including deletion, insertion or substitution in Escherichia coli [117]. Successful mutation causes PAM elimination or spacer mismatches, so repaired chromosome with desired DNA modification can escape from CRISPR cleavage while unmodified strains are killed, resulting in highly efficient gene editing [117,118]. Similar CRISPR HDR systems have been established in many other industrial microbes, with some relying on host native HDR or using recombinase T for higher efficiency [119–123].
Recently, the CRISPR gene editing toolbox has expanded by cooperating with other gene editing proteins. The new CRISPR tools avoid the lethal DSBs and require no DNA template for more efficient gene editing [124,125].
An EvolvR system was constructed by fusing nCas9 with an error-prone DNA polymerase. In this system, the fidelity-reduced DNA polymerase is guided by the CRISPR system to repair the nCas9-nicked strand and simultaneously introduce random mutations [17].
CRISPR base editors were designed by fusing an nCas9 (D10A) with a cytidine deaminase or adenosine deaminase, generating respectively CRISPR-guided CBE (cytosine base editors) or ABE (adenine base editors). Upon paring between sgRNA and the target DNA strand, the PAM-distal sequence on the non-target DNA strand becomes accessible to the deaminases, which convert cytosines (C) to uracils (U) or adenosines (A) to inosines (I) in this editing window. DNA replication and DNA repair recognizes U as thymine (T) and I as guanine (G), so nCas9 cutting the unmutated strand can stimulate the nick repairing using the mutated strand as template to promote the complete C:G to T:A or A:T to G:C conversion [13,14,126]. Fusing uracil glycosylase inhibitor proteins (UGIs) to CBEs efficiently improved its editing efficiency via obstructing the removal of U before complete conversion. More recently, C to A conversion in E. coli was demonstrated by introducing an uracil-DNA glycosylase into the CBE system, which excises the U after deamination and leaves an apurinic/apyrimidinic (AP) site. The AP site triggers DNA repair that preferably convert C to A in E. coli [127].
CRISPR guided prime editing is another advanced genome editing tool bypassing DSBs, which includes a nCas9 (H840A) fused with reverse transcriptase (RT) and a pegRNA consisting of a sgRNA of the CRISPR system, a primer binding site (PBS) to hybridize with the nicked DNA strand, and an RT template to introduce mutations [128]. Once bound to the target DNA, the active RuvC domain of nCas9 cleaves the non-target strand and then the 3’ end of the cleaved strand pairs with PBS, allowing the RT to use the designed RT template to extend the 3’ end and introduce desired mutations. For the unedited strand, using another simple sgRNA guiding the nCas9-RT complex to cleave it can promote its mutation by the DNA repair process using the edited strand as template. Such prime editing system was firstly established in human cells, and later developed for the E. coli [129]. More recently, researchers even demonstrated large size deletion and insertion by CRISPR guided prime editor, as well as the successful multiplex prime editing [108,110]. Additionally, it was revealed that the RT component is separable from nCas9, which overcame the challenge of expressing a large fusion protein and facilitated the rapid screening of more compact RTs [109].
Although the canonical Cas9 has been widely utilized in developing CRISPR gene editing toolkits, the use of CRISPR-Cas12a has also contributed to the expansion of the gene editing toolbox. With distinct PAM specificities and simpler gRNA processing, Cas12a has been employed to construct gene editing toolkits such as Cas12a-mediated HDR and base editor [130]. Moreover, Cas12a-mediated HDR even achieved better performance than the Cas9 system in some microbes such as Pichia pastoris and Methanococcus maripaludis [123,131,132].
Table 1.
Summary and comparison of different CRISPR gene editing tools.
| Gene editing tool | Application in MCFs | Representative achievements | Advantages | Limitations | Refs |
|---|---|---|---|---|---|
| CRISPR HDR | Gene knock-in | Chromosomal pathway integration resulted in 4.4-fold higher lycopene yield than the plasmid-based strain | High mutating efficiency Least limitation on editing length |
Lethal DSBs DNA donor required |
[22,25,39,47,117,118] |
| Gene knock-out | A 30-fold free fatty acid titer increase | ||||
| Pathway optimization | 3-fold higher xylose utilization rate | ||||
| Enzyme engineering | Engineered ERG12-encoded mevalonate kinase and ERG20-encoded farnesyl pyrophosphate synthase, synergistically leading to an 11-fold increase in carotenoids production. | ||||
| CRISPR NHEJ | Gene knock-out | Engineered heterothallic Kluyveromyces marxianus strains suitable for breeding and trait combination | No DNA donor required | Lethal DSBs Only efficient in some eukaryotic cells Unpredicted mutation |
[133–135] |
| CRISPR base editor | Gene knock-out | Identified two furfural tolerance-related genes, purU and serA, enabling an engineered strain with a 1.93-fold higher biomass under furfural stress. | No lethal DSBs No DNA donor required Precise mutagenesis High mutational density |
5–8 bp editing window Typically one base mutation pattern by each system |
[13,14,35,42,49,126,127] |
| Pathway optimization | 4.8-fold increase in lycopene production | ||||
| Enzyme engineering | A Sec-translocase mutant with 3.6-fold higher translocation efficiency | ||||
| EvolvR | Enzyme engineering | An ornithine aminotransferase mutant with a 2.85-fold increase in catalytic efficiency | No lethal DSBs High mutating rate |
Only applicable for enzyme engineering | [17,48] |
| CRISPR prime editor | To be demonstrated | To be demonstrated | Able to perform all types of editing (insertion, deletion, substitution) without DSBs | Low editing efficiency when used in multiplexing manner. | [110,128,129] |
Gene knock-in and knock-out
Although plasmid systems are commonly used in MCF construction to assemble and construct synthetic pathways for the ease of manipulation, chromosomal integration of the finalized pathways generates optimal high producers with less genetic variation, releases the limitation on DNA size, and reduces the metabolic burden for plasmid maintenance [19,20].
CRISPR-mediated HDR can efficiently insert a large DNA fragment into a precise genome locus by one single step and therefore has been broadly used to integrate large size biosynthesis pathways to generate stable MCFs (Figure 2A), exemplified by the chromosomal integration of 10 kb isobutanol synthesis pathway [21], and 12 kb lycopene synthetic pathway into Escherichia coli [22], with the latter achieved a 4.4-fold higher yield compared to the plasmid-based strain. Moreover, the multiplexing feature of CRISPR HDR enables simultaneous integration of multiple genes into different loci. As demonstrated in constructing a β-carotene producing Saccharomyces cerevisiae, three DNA donors with pathway genes of 6.6 kb, 5.8 kb, and 5.1 kb were integrated to their respective genome sites with 84% efficiency [23]. CRISPR knock-in has also been successfully implemented in non-model microorganisms, resulting in microbial chassis that are especially advantageous for producing complicated natural products. As a typical example, the integration of extra copies of the endogenous mevalonate pathway and other critical genes in Aspergillus oryzae created an optimized chassis for high production of pharmaceutically important terpenoids [24].
Figure 2.
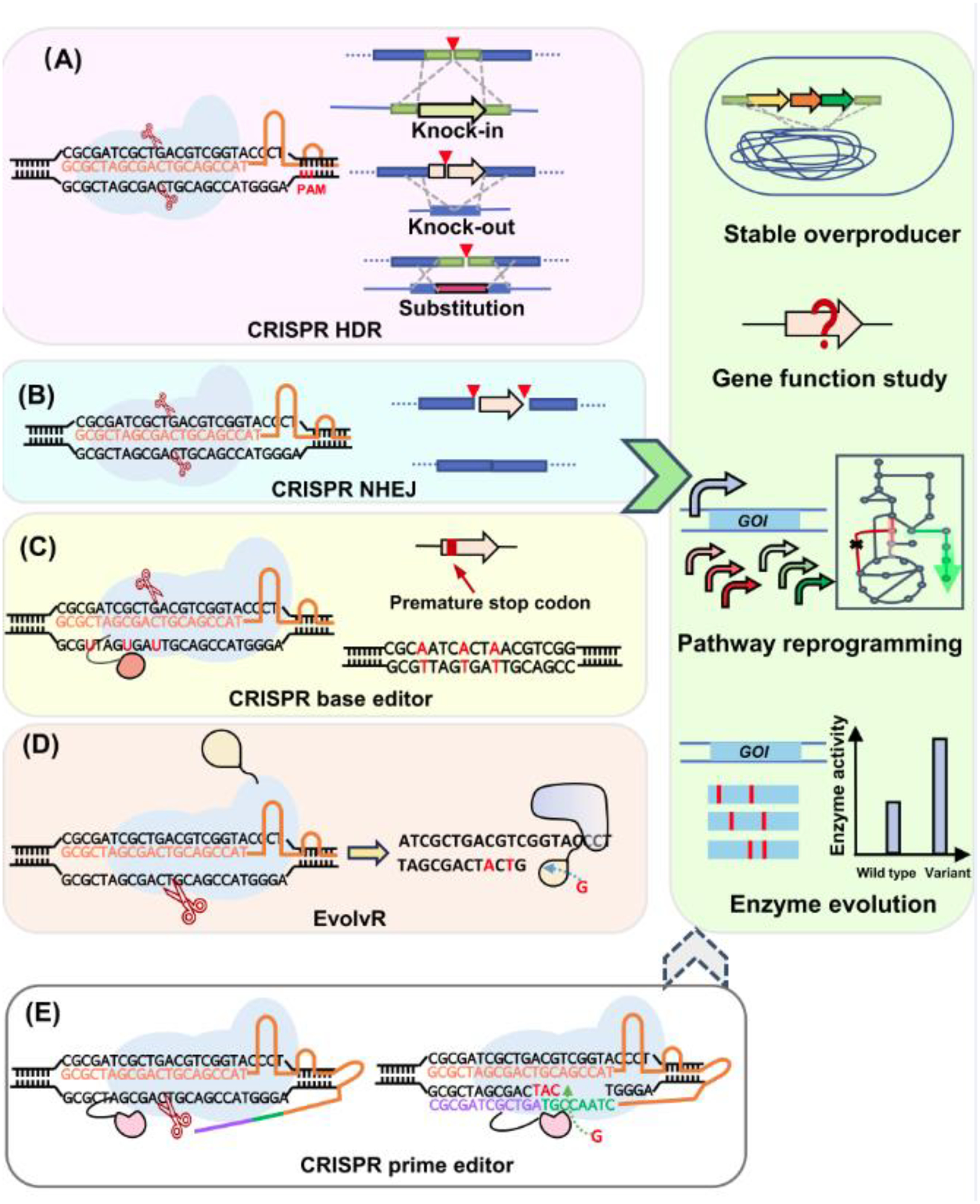
Mechanism and application of the CRISPR gene editing tools. (A) CRISPR-mediated HDR is triggered by the DSBs cleaved by Cas effectors. A DNA donor is required for the homology directed repair. By providing different DNA donors, CRISPR HDR can introduce gene knock-in, knock-out and substitution. It has been extensively used for sable overproducer construction, gene function study, pathway reprogramming and enzyme evolution. (B) CRISPR NHEJ directly ligases the two ends of DSBs and only performs gene knock-out. It has been used in some pathway reprogramming studies. (C) CRISPR base editor recruits cytosine deaminase or adenine deaminase to introduce point mutations on one strand in a limited editing window. The unmutated strand is usually nicked by nCas9 to promote complete mutation on both strands. CRISPR base editor can create premature stop codon to silent a gene and investigate its function. It can also diversify the multiple target sequences for pathway reprogramming and enzyme engineering. (D) EvolvR is designed by fusing an nCas9 with an error-prone RNA polymerase. After targeting and nicking, the error-prone RNA polymerase repairs the nicked strand and simultaneously introduces random mutations. The editing window is around 56 bp. Thus, it is mainly applied in enzyme engineering. (E) CRISPR prime editor is designed by fusing a reverse transcriptase (RT) with nCas9. The primer binding site (PBS, colored in purple) in the pegRNA can hybridize with the 3’ end of the nick, and the template sequence (colored in green) is used by the RT to repair the nick and introduce mutations.
Gene knockout can effectively redirect carbon flux towards the target product by deleting competing pathways and saving cellular resources [25]. Both CRISPR-mediated HDR and non-homologous end joining (NHEJ) can be recruited for gene deletion (Figure 2A and B). Reserachers utilized the multiplxing feature of CRISPR-enabled gene knockout to study the sequential and combinational deletion of 5 genes for high mevalonate production, resulting in a 41-fold titer increasement in S. cerevisiae [26]. In another study, Dong and coworkers deleted 33 native genes and optimized the butanal biosynthesis genes for higher butanol production in E. coli. The final strain produced 20 g/L butanol, which is 83% of the theoretical yield [27]. This simple and straight strategy has been extensively applied in metabolic engineering and contributed to the construction of overproducers for many valuable chemicals including polyhydroxyalkanoate [28], free fatty acid [25], β-carotene [29], and ergot alkaloids [30].
CRISPR-enabled gene knockout is not only useful for pathway programming, but also for studying gene functions and genotype-phenotype associations, providing important guidance for MCF construction. For example, Cas9-enabled gene deletion was used to elucidate the biosynthetic pathway of complicated nature products such as demethoxyviridin and talaromyolides [31–33]. CRISPR base editor can also be used to silent genes by introducing premature stop codons (Figure 2C). Without requiring DSBs and DNA donors, this approach introduces loss-of-function mutation with simpler process and therefore is more suitable for large scale screening [34]. A genome-scale screening by a nCas9-CBE (cytosine base editor) was performed in Corynebacterium glutamicum to identify genes related to stress tolerance. 98.1% of the total genes (3,041) were targeted for genetic perturbations and two genes, purU and serA, were identified to be related to the tolerance towards furfural, a toxic compound in pretreated lignocellulose inhibiting microbial growth. The engineered strain with purU deletion and serA mutation achieved 1.93-fold higher biomass under furfural stress [35]. Similar screening was performed across 16,452 perturbations in yeast to study regulators controlling protein abundance [36].
Pathway optimization
Modifying the regulation elements, such as promoters, ribosome binding sites (RBS) and 5′ untranslated regions (5’ UTR), is a prevalent approach to optimize metabolic pathways and rewire carbon flux for higher production [37,38]. In order to engineer complex cellular metabolism, it is often necessary to customize regulatory elements for multiple genes (Figure 2). The robust and multiplexing nature of the CRISPR system makes it an ideal tool for fulfilling these requirements and simplifying the strain optimization process.
The high efficiency and multiplexing capabilities of CRISPR HDR render it an invaluable asset for the multiplex automated genome engineering (MAGE) strategy to rapidly generate a large number of strain variants for overproducer screening using synthetic libraries of regulatory elements. For instance, Zhu and colleagues employed a regulator pool containing 6-bp randomized RBS to simultaneously engineer the expression levels of three genes in the xylose utilization pathway of E. coli. With 70% editing efficiency, the RBS optimization led to a 3-fold increase in xylose utilization [39]. In another study, four RBS sites in the isopropanol synthetic pathway of E. coli were optimized using an in silico designed RBS library. A total of 256 variants were constructed with 4 different levels of RBS available for each gene and the highest productivity was 2.8-fold higher than the control group [40]. This method can also be used to modulate competitive pathways to rewire more flux towards the final products. As an example, to redirect more flux from glycolysis towards aromatic amino acid synthesis, the promoters of PFK1, PFK2, and PYK1 in yeast glycolysis were simultaneously targeted for combinatorial engineering [41].
The use of CRISPR base editors eliminates the need for large combinatorial libraries as DNA donors and simplifies the optimization process for multigene expression (Figure 2C). As a proof-of-concept, Wang and coworkers constructed a “BRTTER” system by recruiting a nCas9-CBE to randomly mutate the multi-G/C region of RBS, 5’ untranslated regions, and promoters to engineer gene expression [42]. The system can target up to 10 genes simultaneously and generate sufficient variants for the screening. Applications included improvements of xylose catabolism and lycopene biosynthesis in C. glutamicum, as well as optimization of glycerol catabolism in Bacillus subtilis.
Protein evolution
Enzymes play a critical role in metabolic pathways, and their limited efficiency often hinders final production. Protein engineering can effectively solve this bottleneck problem, but rational engineering can be difficult or laborious considering the complexity of protein structure [43–45]. Directed evolution provides a powerful alternative strategy, which can be significantly assisted by the CRISPR editing technologies that directly and efficiently introduce protein mutants into the genome.
CRISPR HDR has been widely used in protein evolution to integrate mutagenesis libraries and generate protein variants. Enzymes like the folA encoded dihydrofolate reductase in E. coli and ERG12 and ERG21 in S. cerevisiae have been shown to acquire improved or enhanced properties using this approach, with the former leading to a 11-fold isoprenoid production increase [46,47]. In addition to the recombination method, nCas9 based gene editing tools avoiding DSBs were also applied for protein evolution. The EvolvR system, designed by Halperin and coworkers, can continuously and efficiently diversify DNA in an editing window of ~56 bp after the nick (Figure 2D), and has demonstrated multiplexed targeting by simultaneously engineering two proteins related to antibiotic resistances in E. coli [17]. Later, EvolvR was harnessed to engineer a heterologous ornithine aminotransferase in E. coli, and the best variant exhibited a 2.85-fold higher catalytic efficiency for L-proline synthesis [48]. CRISPR base editor also has been leveraged for protein engineering (Figure 2C). Hao and colleagues developed an optimized CBE system for protein evolution in B. subtilis, which they applied to improve two proteins related to bacitracin resistance including a Sec-translocase complex and a BceB encoding protein [49]. However, the limited single type of base transition by a CRISPR base editor might restrict their capability in generating diverse protein variants, which might be improved by the dual-base editor systems combining ABE and CBE [50]. Another limitation lies in the length of editing window. In practice, protein-encoding genes to be engineered are usually hundreds or thousands of base pairs in length. The multiplexity of CRISPR targeting and iterative editing are surely helpful to expand the mutating window. It is also crucial to narrow down the mutating ranges in long protein sequences by structure and mechanism analysis.
Transcriptional regulation
In addition to gene editing, CRISPR-Cas can be repurposed for transcriptional gene regulation. CRISPR interference (CRISPRi) and activation (CRISPRa) respectively down- and up-regulate gene expression by affecting RNA polymerase (RNAP) recruitment [51]. In bacteria, dCas9 itself can act as an efficient repressor blocking RNAP, while in eukaryotic microbes, an extra repressor domain is required to be fused with dCas9, such as Mxi1, UME6, MIG1, TUP1 or relevant variants [29,52,53]. On the other hand, bacterial CRISPRa suffers from low efficiency due to its position sensitivity, despite the attempts with multiple activators like SoxS and RNAP subunits, while eukaryotic microbial CRISPRa easily reaches high efficiency using activators such as VP64, VPR, p65AD and Rta [54–60].
Compared with genetically engineering the regulatory elements, transcriptional regulation through CRISPRi and CRISPRa causes no permanent DNA changes and is thus more reversible, controllable, and tunable. Given their exceptional features, CRISPRi and CRISPRa have become widely employed in designing dynamic regulation circuits to maximize the production of MCFs.
CRISPRi
With its ease-of-use and nearly knock-out level repression, CRISPRi has become a popular tool for functional screening in addition to gene knock-out [61]. Yao and colleagues designed a sgRNA library targeting all the genes in Cyanobacterium synechocystis and identified multiple repression targets benefiting L-lactate productivity [62]. Similar genome-scale screenings were performed to enhance free fatty acid production in E. coli and protein titer in C. glutamicum [63,64]. Liu and coworkers used CRISPRi to rapidly screen nearly 400 transporters in C. glutamicum and identified a novel L-proline exporter Cgl2622. They further overexpressed the exporter by chromosomally integrating an extra copy of it with a proper promoter via Cas9-mediated knock-in, resulting in a final strain with a high L-proline titer of 142.4 g/L [65].
In addition to the large-scale screening, rational analysis can help narrow down the testing range to identify effective repression targets. In a study, researchers focused on the important central metabolism was to screen the repression targets for higher flavonoid production in E. coli [66]. Combinatorial repression of the identified targets fabF, fumC, fabB, sucC and adhE resulted in a 7.2-fold increase of naringenin titer to 421.6 mg/L. Another study used a computational minimal cut set approach to predict the repression targets for indigoidine production in Pseudomonas putida [67]. Multiplexed CRISPRi targeting of all 14 genes led to a stable phenotype of 25.6 g/L titer, 0.22 g/l/h rate, and ~50% maximum theoretical yield. For higher isoprenol production, Wang and colleagues analyzed three limiting factors including phosphatase activity, precursor acetyl-COA accumulation and isoprenol toxicity, to screen for related genes [68]. A final titer of 3.63 g/L reaching 57% of the theoretical yield was achieved by the combinatorial repression on yggV and accA in E. coli.
In practical applications, precise tuning of high level CRISPRi repression is crucial, especially when essential genes for cell growth need to be regulated. Various CRISPRi regulation strategies have been developed including sgRNA positioning [51,69], expression control [70], and spacer sequence shortening or mismatching [51,71] (Figure 3A). More recently, a sgRNA scaffold engineering approach was reported, which regulates the binding affinity of sgRNA with dCas9 [72]. It has been applied in violacin and lycopene biosynthesis, with the latter showing a 2.7-fold improvement. Meanwhile, Cas9 engineering enabled a PAM-tuned CRISPRi approach for titratable gene control and led to a 2.6-fold increase in 4-hydroxycoumarin production in E. coli [73].
Figure 3.
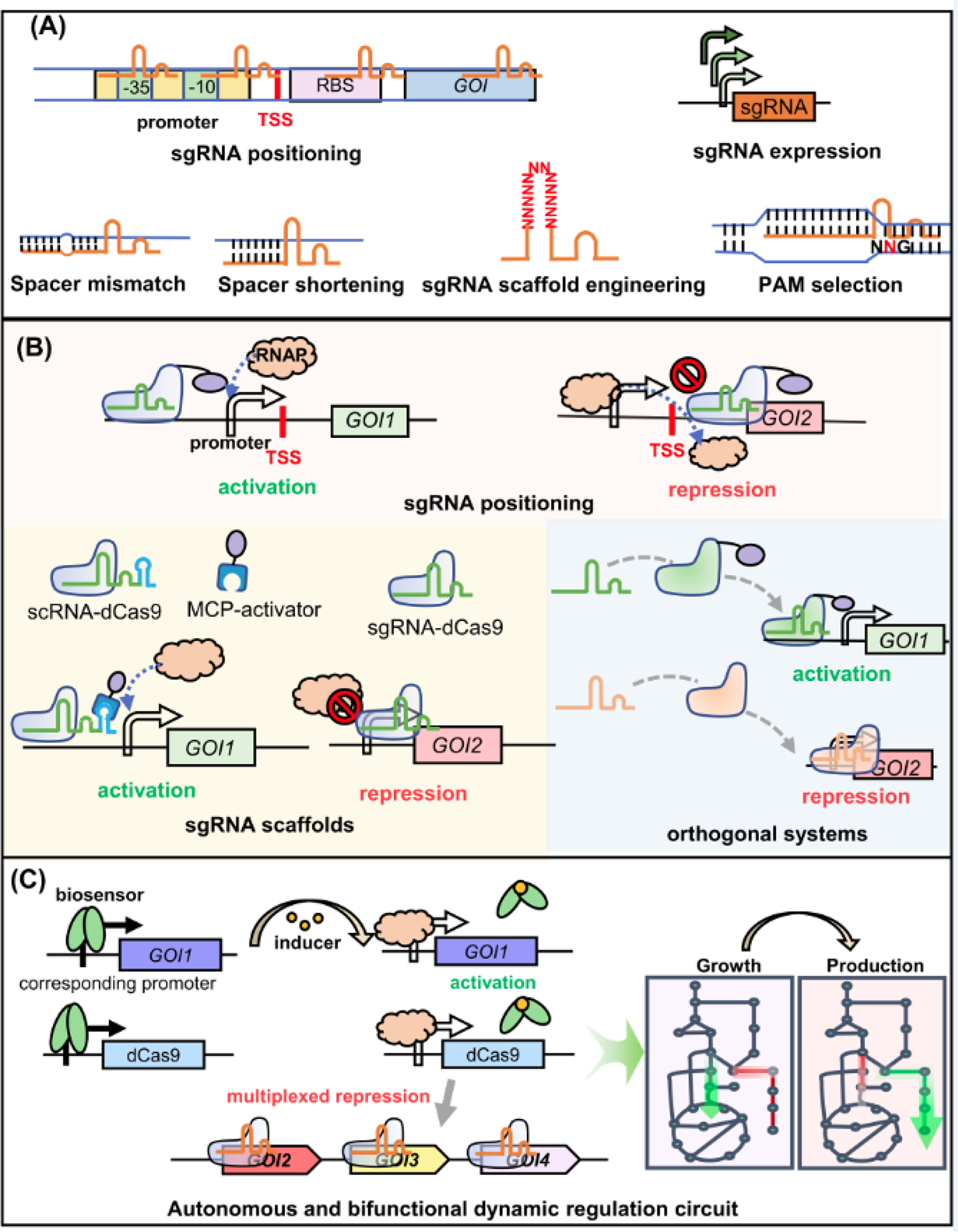
Schemes for the designs of different transcriptional regulation systems. (A) Approaches to tune CRISPRi level. High level CRISPRi repression can be tuned by designing sgRNAs to target TSS downstream regions with longer distances, decreasing the sgRNA expression level, creating spacer mismatches, shortening spacer length, engineering the sgRNA scaffold for less affinity against dCas9, and targeting the less preferred PAMs using the engineered Cas9 variants with expanded PAM range. (B) Different bifunctional regulation designs. Firstly, Cas9-activator fusion protein activates gene expression when targeted to the proper TSS upstream region, and represses gene expression when targeted to the TSS downstream region. Secondly, an engineered scRNA with an extra MS2 hairpin structure can guide Cas9 and simultaneously recruit the activator fused with MS2 coat protein (MCP), and therefore activate gene expression; on the contrary, the sgRNA-Cas9 complex represses gene expression. Thirdly, orthogonal CRISPR systems with no mutual interference can be recruited for simultaneous activation and repression. (C) A typical autonomous and bifunctional dynamic regulation circuit. Internal stimuli can cause conformational changes to the biosensor, releasing it from its corresponding promoter and activating downstream gene expression. The expression of dCas9 controlled by the same promoter will also be activated, which can be targeted to repress multiple genes. The activated GOI1 is usually a heterologous gene in biosynthesis, and repression targets GOI2–4 are endogenous genes essential for cell growth and competing fluxes with the biosynthetic pathway.
Furthermore, the tunability can cooperate with the multiplexity of CRISPRi to enable customized regulation of multiple genes. In a study aimed at redirecting the flux towards 4-hydroxybutyrate in E. coli, CRISPRi levels on multiple targets were tuned by their own sgRNA positions, resulting in different proportions of 4-hydroxybutyrate in the final product poly (3-hydroxybutyrate-co-4-hydroxybutyrate) [69]. Another study generated sgRNA libraries by a mismatching approach to fine tune the CRISPRi level for 20 genes at a full range [74]. With the assistance of biosensor screening, combinatorial knocking down two genes (pfkA and ptsI) at their respective optimal level reached over 40% increasement of the p-coumaric acid in E. coli to 1308.6 mg/L.
CRISPRa
While the highly efficient eukaryotic CRISPRa has been extensively applied for flux redirecting, bacterial CRISPRa has been limited by the sensitivity to the targeting position. To optimize the bacterial CRISPRa system, multiple activator domains have been explored, including the ω and α subunit of RNA polymerase, SoxS, PspF, AsiA, and related variants [55,56,75,76]. Among them, the SoxS is more frequently used and has enabled improved production of pinene and 4-hydroxyphenylacetic [77,78]. In addition to activator screening, expanding the PAM range of CRISPR is highly demanded so that the most propriate position can be targeted to reach the highest activation [73,79,80]. The Cas9 variant SpRY with the most expanded PAM range was used to construct a PAM-independent activation system [81]. Another Cas9 engineering approach was to connect its original N- and C- termini to create new termini that could be fused with the activator. This can locate the activator at different positions within the tertiary structure, resting in different position-dependent activation patterns and thus expand the targetable range [55].
Bifunctional regulation
Repression and activation are often both needed to respectively upregulate the pathway genes and downregulate the competitive genes to maximize the productivity and minimize metabolic burdens. Via proper design, CRISPR systems can be engineered to simultaneously implement CRISPRi and CRISPRa (Figure 3B). The simplest method is to design different sgRNAs to guide a Cas-activator fusion protein. When the sgRNA targets the appropriate site upstream of the TSS of a gene, the fusion protein functions as an activator, while targeting downstream of the TSS of a gene turns the protein into a repressor. Many effectors including dCas9-ω/α and dCas12a-SoxS have been applied for such bi-functional regulation in different microbes [81–83]. Another method for simultaneous CRISPRi and CRISPRa was designed by utilizing a MS2 hairpin and its interacting coat protein (MCP) [56]. The MS2 hairpin was fused with sgRNA, creating a scRNA that can recruit both dCas9 and MCP-SoxS at the target site for gene activation. Meanwhile, the original sgRNA only recruits dCas9 to repress the target gene expression [81,84,85]. The third method is to utilize orthogonal CRISPR systems. For instance, orthogonal dCas12a and dCas9 were used in combination for activation and repression, respectively. In cooperation with the efficiency expression of large DNA arrays, this system demonstrated a substantial improvement of succinic acid by 45-fold [86].
Autonomous dynamic regulation with CRISPR regulators
CRISPR has proven to be highly compatible with transcription factor-based biosensors. Researchers have designed many advanced dynamic regulation circuits by coupling CRISPR regulation with biosensors, endowing microbes with the intelligence to control their metabolic flux in response to changes of internal environment [4,87,88]. In these dynamic regulation circuits, biosensors detect various environmental changes acting as inputs, whereas CRISPR regulator contributes to the dynamic range, multiplexity, and versatility of outputs (Figure 3C). To control metabolic burdens, a dCas9-based negative feedback circuit was implemented in E. coli using a native heat stress-related promoter that responds to metabolic burdens [89]. Placing sgRNA under the control of this promoter allows the regulation on heterologous gene expression to automatically reduce the burdens. Another dCas9-based regulation circuit was developed to autonomously balance the flux between the flavonoid synthesis pathway and its competitive fatty acid synthesis pathway [90]. The sgRNA targeting fatty acid synthesis was placed under fatty acid-inducible promoters, enabling negative autoregulation to control the production of fatty acid byproducts and resulting in a 74.8% increase in naringenin production in Yarrowia lipolytica.
In addition to metabolite-responsive sensors, quorum sensing (QS) has been used to establish pathway-independent dynamic regulation that decouples cell growth and product synthesis [91]. By sensing cell density, it allows cell growth to take priority at the early phase for sufficient biomass accumulation, and then switches to production mode to maximize carbon flux towards the desired products. A QS-based CRISPRi circuit was reported to generated tunable and multiplexed repression on the competitive pathways of rapamycin synthesis, which increased the rapamycin titer to the highest reported level of 1836 mg/L in Streptomyces [92]. Similarly, a stationary phase promoter-controlled CRISPRi enabled such growth-to-production switch, contributing to high titer shikimic acid and glutaric acid production of respective 21 g/L and 26 g/L in 5-L bioreactor [93].
Furthermore, an autonomous dual-control biosensor-CRISPRi system was constructed in B. subtilis for N-acetylglucosamine production [94] (Figure 3C). The biosensor GamR responds to a pathway intermediate glucosamine 6-phosphate (GlcN6P). High level GlcN6P accumulation triggers GamR to switch on the downstream production pathways and the CRISPRi repressing the competitive pathways. The titer of N-acetylglucosamine increased from 59.9 g/L to 97.1 g/L in a 15-L fed-batch bioreactor.
Enzyme modulation via synthetic metabolons
Enzymes in metabolic pathways can be tethered together to form complexes termed as metabolons, which can increase pathway efficacy and control toxic intermediates by decreasing diffusion and transportation time of intermediates.
Lim and coworkers demonstrated CRISPR enabled in vitro programmable enzyme assembly to improve catalytic efficiency [95]. In this study, the dCas9s were respectively attached to five enzymes catalyzing L-tryptophan to violacein. Each protein complex was separately paired with a different sgRNA, which guided the complexes to a DNA scaffold after mixing. Both sgRNA and DNA scaffold are programmable, making the assembly highly modular. However, this strategy cannot be applied for microbial cell factory construction since the attachment between dCas9 and enzymes, and the pairing between the protein complexes and sgRNA were all non-specific and cannot be controlled in vivo. In another CRISPR dCas9 guided enzyme assembly system by Chen group, two Cas9 orthologs were directly fused with enzymes and could specifically interact with their own sgRNA [96]. Such specific interaction opens the opportunity for in vivo application. Notably, this system was also designed to be dynamically controlled by engineering the sgRNA with using a well-developed toehold-mediated strand displacement strategy (TMSD) [97]. In 2022, the same group reported the successful in vivo application of CRISPR mediated enzyme assembly in E. coli [18] (Figure 4). They further upgraded the system using smaller CRISPR-Cas6 and achieved dynamic assembly and disassembly. Different from Cas9, Cas6 is an endoribonuclease cutting the guide RNA and remains binding to it after cleavage. More importantly, Cas6 binds with the hairpin structure at the 3’ end of its guide RNA and leaves the 5’ end handle free. The complexes can be directly assembled by the 5’ end RNA hybridization, and the hybridization can be similarly interfered by the TMSD strategy to achieve dynamic turn-on and turn-off. The CRISPR enabled dynamic metabolons enhanced indole-3-acetic acid production by up to 9 folds and demonstrated multimeric enzymes cascading in malate production, leading to a 3-fold increase.
Figure 4.
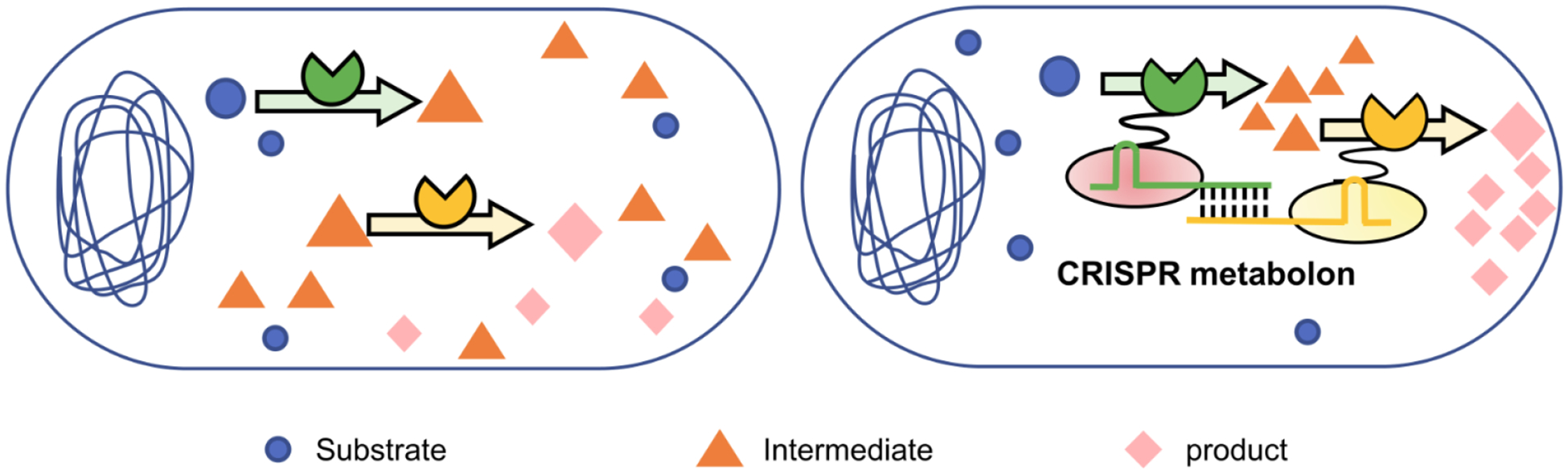
CRISPR-guided synthetic metabolons. Two enzymes catalyzing sequential reactions are connected to orthogonal Cas6s, each of which forms a complex with its specific guide RNA. Enzyme assembly can be achieved through RNA hybridization between the 5’ free handles of the guide RNAs. This CRISPR-driven enzyme cascading can improve sequential reaction efficiency, leading to less accumulation of the intermediate and more final products.
Concluding remarks and future perspectives
The CRISPR toolbox has undergone significant expansion with the development of DSB-free gene editing tools, tunable transcriptional regulation systems, and the application of CRISPR mediated synthetic metabolons for protein-level regulation. For MCF construction, the expanded CRISPR toolbox has enabled the engineering of different cellular processes, addressing the challenges regarding gene function, pathway reprogramming, and gene expression coordination.
However, the CRISPR toolbox still faces inherent limitations, including off-target effects, PAM limitations, and metabolic burdens. An attractive direction for further advancement is the exploration of new CRISPR systems (see Outstanding Questions). The emergence of miniature CRISPR systems with smaller nucleases provides an opportunity to develop improved CRISPR toolkits with higher fidelity, distinct PAM preferences, and reduced metabolic burdens. For example, CRISPR-Cas12f systems, with effectors ranging from 400 to 700 amino acids and a preference for T- or C-rich PAMs, have been utilized for gene editing and regulation, demonstrating advantages such as easier delivery and lower off-targeting effects [98–101]. Recent studies have revealed even smaller miniature systems, including TnpB and IscB, with sizes below 400 amino acids [102,103]. We anticipate more comprehensive investigation and exploitation of these systems in the future. Moreover, engineering the Cas effector has consistently proven to be a highly effective approach to relax the PAM requirement and minimize off-targeting. Notable examples include the engineered PAMless variant SpRY, high-fidelity Cas9 HF1, as well as the xCas9 and HiFi-Sc++ variants with both broader PAM ranges and enhanced fidelities [80,104–106].
Outstanding Questions Box.
What are the potential applications of recently discovered miniature CRISPR systems in developing new gene editing and regulation tools?
Is it possible to apply CRISPR prime editing in metabolic engineering?
Are there other gene editing proteins that can be combined with CRISPR systems to optimize current tools or explore new functions?
SpRY reached nearly PAMless with higher activity on NRN than NYN PAMs. How to develop more robust PAM-independent Cas effectors for the tools requiring precise targeting?
How to overcome the position sensitivity of bacterial CRISPRa and enhance its applicability?
Can CRISPR base editors, prime editors, or CRISPRi be successfully used in non-model microorganisms that are not amenable to CRISPR-HDR?
The CRISPR prime editor has not yet been implemented in MCF construction. This tool can perform more types of gene editing than EvolvR and base editor and has been used for introducing saturated mutagenesis for the herbicide resistance in plants [107]. We expect that the advancements of CRISPR prime editor reported in mammalian and plant cells, such as efficient multiplex editing and separable RTs, can be achieved in microbial systems to further promote MCF construction [108–110].
Furthermore, it is also necessary to promote the use of CRISPR tools in non-model microbes, which possess unique advantages for gene mining, substrate consumption or environmental tolerances [33,111–113]. One potential challenge lies in the unclear or unsupportive native DNA repair machinery, which requires thorough investigation or engineering [114]. Considering the wide distribution of CRISPR systems in bacteria and archaea, the identification and engineering of endogenous CRISPR systems are important to avoid any crosstalk pitfalls and ensure successful implementation of CRISPR technologies in non-model microorganisms [115]. Additionally, the existence and possible interference of anti-CRISPR proteins in some microbes should also be taken into consideration [116].
Highlights.
The development of CRISPR-base editors, CRISPR-prime editors, and EvolvR has enabled more efficient and precise gene editing without relying on double-stranded DNA breaks.
Transcriptional CRISPRi and CRISPRa systems have been developed with higher efficiency and tunability, enabling the design of more advanced regulation circuits.
The CRISPR toolbox has been expanded to protein-level regulation by the Cas6-enabled dynamic enzyme assembly.
The expanded CRISPR toolbox has comprehensively promoted the engineering of various components and cellular processes in microbes, driving the advancement of microbial cell factory (MCF) construction.
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM128620. We also acknowledge the support from the College of Engineering, The University of Georgia, Athens.
Glossary
- CRISPR-Cas
an adaptive immune system in bacteria using RNA-guided nucleases to target and cut foreign genetic elements
- CRISPR-Cas9
a Class 2, Type II CRISPR-Cas system. Cas9 is a RNA-guided endonuclease that cleave off the target DNA with an recognizable PAM and generate blunt DSBs. The guide RNA (gRNA) is a crRNA-tracrRNA duplex, with crRNA containing a sequence complementary to the target DNA and tracrRNA providing the binding scaffold for the endonuclease Cas 9 protein
- CRISPR-Cas12a
also known as Cpf1, a Class 2, Type V CRISPR-Cas system. Cas12a is an crRNA-guided endonuclease that cleaves off the target DNA with an recognizable PAM and generates staggered DNA DSBs
- CRISPR activation (CRISPRa)
equip the dCas effectors with trancriptional activators to enhace RNA polymerase recruiment and upregulate the expression of genes of interest
- CRISPR-mediated homology directed repair (HDR)
the repair of DSBs induced by CRISPR-Cas systems through homologous recombination using a DNA template
- CRISPR inteference (CRISPRi)
use dCas effectors or dCas-transcriptional repressor fusion protein to phsically repress RNA polymerase recruitment and downregulate the expressio of genes of interest
- CRISPR NHEJ
repairing the DSBs generated by CRISPR-Cas through non-homologous end joining, which directly ligases the break ends
- dCas9
endonuclease activity-deactivated Cas9
- Dynamic regulation
dynamically control gene expression as response to external signals and endogenous changes
- Metabolic burden
physiological stress on a cell due to the consumption of energy and resources caused by heterologous gene expression
- Metabolon
A complex formed by tethering enzymes that catalyze sequential metabolic pathways, allowing for efficient substrate channeling and coordinated regulation
- nCas9
endonuclease activity-partially deactivated Cas9 that only cleave one DNA strand
- Protospacer adjacent motif (PAM)
a short specific sequence following the target DNA sequence that is essential for cleavage by Cas nuclease
- sgRNA
a synthetic RNA molecure fusing the crRNA and the scaffold tracrRNA
- Spacer
the sequence in gRNA or sgRNA complementary to the target DNA
- Toehold-mediated strand displacement strategy (TMSD)
a molecular tool to exchange one strand of nucleic acid complex with another strand
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
No interests are declared.
References
- 1.Lu H et al. (2019) Modular Metabolic Engineering for Biobased Chemical Production. Trends Biotechnol 37, 152–166. 10.1016/j.tibtech.2018.07.003 [DOI] [PubMed] [Google Scholar]
- 2.Cho JS et al. (2022) Designing Microbial Cell Factories for the Production of Chemicals. JACS Au 2, 1781–1799. 10.1021/jacsau.2c00344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang T et al. (2020) Recent advances in improving metabolic robustness of microbial cell factories. Current Opinion in Biotechnology 66, 69–77. 10.1016/j.copbio.2020.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teng Y et al. (2022) Biosensor-enabled pathway optimization in metabolic engineering. Current Opinion in Biotechnology 75, 102696. 10.1016/j.copbio.2022.102696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C et al. (2022) Fine-tuning gene expression for improved biosynthesis of natural products: From transcriptional to post-translational regulation. Biotechnol Adv 54, 107853. 10.1016/j.biotechadv.2021.107853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu G et al. (2016) Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications. Trends Biotechnol 34, 652–664. 10.1016/j.tibtech.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 7.Ko YS et al. (2020) Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem Soc Rev 49, 4615–4636. 10.1039/d0cs00155d [DOI] [PubMed] [Google Scholar]
- 8.Nielsen J and Keasling JD (2016) Engineering Cellular Metabolism. Cell 164, 1185–1197. 10.1016/j.cell.2016.02.004 [DOI] [PubMed] [Google Scholar]
- 9.Cong L et al. (2013) Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M et al. (2012) A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakociunas T et al. (2016) CRISPR/Cas9 advances engineering of microbial cell factories. Metab Eng 34, 44–59. 10.1016/j.ymben.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 12.Chen L et al. (2021) Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nat Commun 12, 1384. 10.1038/s41467-021-21559-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudelli NM et al. (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komor AC et al. (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. 10.1038/nature17946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anzalone AV et al. (2022) Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol 40, 731–740. 10.1038/s41587-021-01133-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erwood S et al. (2022) Saturation variant interpretation using CRISPR prime editing. Nat Biotechnol 40, 885–895. 10.1038/s41587-021-01201-1 [DOI] [PubMed] [Google Scholar]
- 17.Halperin SO et al. (2018) CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252. 10.1038/s41586-018-0384-8 [DOI] [PubMed] [Google Scholar]
- 18.Mitkas AA et al. (2022) Dynamic modulation of enzyme activity by synthetic CRISPR-Cas6 endonucleases. Nat Chem Biol 18, 492–500. 10.1038/s41589-022-01005-7 [DOI] [PubMed] [Google Scholar]
- 19.Li L et al. (2019) Synthetic biology approaches for chromosomal integration of genes and pathways in industrial microbial systems. Biotechnol Adv 37, 730–745. 10.1016/j.biotechadv.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 20.Saleski TE et al. (2021) Optimized gene expression from bacterial chromosome by high-throughput integration and screening. Science Advances 7, eabe1767. doi: 10.1126/sciadv.abe1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassalo MC et al. (2016) Rapid and Efficient One-Step Metabolic Pathway Integration in E. coli. ACS Synth Biol 5, 561–568. 10.1021/acssynbio.5b00187 [DOI] [PubMed] [Google Scholar]
- 22.Su B et al. (2020) Homology-dependent recombination of large synthetic pathways into E. coli genome via lambda-Red and CRISPR/Cas9 dependent selection methodology. Microb Cell Fact 19, 108. 10.1186/s12934-020-01360-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronda C et al. (2015) CrEdit: CRISPR mediated multi-loci gene integration in Saccharomyces cerevisiae. Microb Cell Fact 14, 97. 10.1186/s12934-015-0288-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y et al. (2022) Efficient exploration of terpenoid biosynthetic gene clusters in filamentous fungi. Nature Catalysis 5, 277–287. 10.1038/s41929-022-00762-x [DOI] [Google Scholar]
- 25.Zhang Y et al. (2019) A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae. Nat Commun 10, 1053. 10.1038/s41467-019-09005-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakociunas T et al. (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng 28, 213–222. 10.1016/j.ymben.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 27.Dong H et al. (2017) A systematically chromosomally engineered Escherichia coli efficiently produces butanol. Metab Eng 44, 284–292. 10.1016/j.ymben.2017.10.014 [DOI] [PubMed] [Google Scholar]
- 28.Jung HR et al. (2019) Construction of Efficient Platform Escherichia coli Strains for Polyhydroxyalkanoate Production by Engineering Branched Pathway. Polymers (Basel) 11. 10.3390/polym11030509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lian J et al. (2017) Combinatorial metabolic engineering using an orthogonal trifunctional CRISPR system. Nat Commun 8, 1688. 10.1038/s41467-017-01695-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis KA et al. (2020) Genetic Reprogramming of the Ergot Alkaloid Pathway of Metarhizium brunneum. Appl Environ Microbiol 86. 10.1128/AEM.01251-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang GQ et al. (2018) Biosynthetic pathway for furanosteroid demethoxyviridin and identification of an unusual pregnane side-chain cleavage. Nat Commun 9, 1838. 10.1038/s41467-018-04298-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLorenzo DM et al. (2021) An Improved CRISPR Interference Tool to Engineer Rhodococcus opacus. ACS Synth Biol 10, 786–798. 10.1021/acssynbio.0c00591 [DOI] [PubMed] [Google Scholar]
- 33.Woodcraft C et al. (2023) The expanding CRISPR toolbox for natural product discovery and engineering in filamentous fungi. Nat Prod Rep 40, 158–173. 10.1039/d2np00055e [DOI] [PubMed] [Google Scholar]
- 34.Parrish PCR and Berger AH (2021) CRISPR base editor screens identify variant function at scale. Mol Cell 81, 647–648. 10.1016/j.molcel.2021.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y et al. (2022) Base editor enables rational genome-scale functional screening for enhanced industrial phenotypes in Corynebacterium glutamicum. Science Advances 8, eabq2157. doi: 10.1126/sciadv.abq2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert OT et al. (2022) Genome-wide base editor screen identifies regulators of protein abundance in yeast. Elife 11. 10.7554/eLife.79525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alper H et al. (2005) Tuning genetic control through promoter engineering. Proceedings of the National Academy of Sciences 102, 12678–12683. doi: 10.1073/pnas.0504604102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keasling JD (2012) Synthetic biology and the development of tools for metabolic engineering. Metabolic Engineering 14, 189–195. 10.1016/j.ymben.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 39.Zhu X et al. (2017) The CRISPR/Cas9-facilitated multiplex pathway optimization (CFPO) technique and its application to improve the Escherichia coli xylose utilization pathway. Metab Eng 43, 37–45. 10.1016/j.ymben.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 40.Liang L et al. (2017) CRISPR EnAbled Trackable genome Engineering for isopropanol production in Escherichia coli. Metab Eng 41, 1–10. 10.1016/j.ymben.2017.02.009 [DOI] [PubMed] [Google Scholar]
- 41.Liu Q et al. (2019) Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat Commun 10, 4976. 10.1038/s41467-019-12961-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y et al. (2021) In-situ generation of large numbers of genetic combinations for metabolic reprogramming via CRISPR-guided base editing. Nat Commun 12, 678. 10.1038/s41467-021-21003-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zha J et al. (2019) Pathway enzyme engineering for flavonoid production in recombinant microbes. Metabolic Engineering Communications 9. 10.1016/j.mec.2019.e00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang D et al. (2020) Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol 38, 745–765. 10.1016/j.tibtech.2019.11.007 [DOI] [PubMed] [Google Scholar]
- 45.Li C et al. (2020) Protein Engineering for Improving and Diversifying Natural Product Biosynthesis. Trends Biotechnol 38, 729–744. 10.1016/j.tibtech.2019.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garst AD et al. (2017) Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol 35, 48–55. 10.1038/nbt.3718 [DOI] [PubMed] [Google Scholar]
- 47.Jakociunas T et al. (2018) CasPER, a method for directed evolution in genomic contexts using mutagenesis and CRISPR/Cas9. Metab Eng 48, 288–296. 10.1016/j.ymben.2018.07.001 [DOI] [PubMed] [Google Scholar]
- 48.Long M et al. (2020) Directed Evolution of Ornithine Cyclodeaminase Using an EvolvR-Based Growth-Coupling Strategy for Efficient Biosynthesis of l-Proline. ACS Synth Biol 9, 1855–1863. 10.1021/acssynbio.0c00198 [DOI] [PubMed] [Google Scholar]
- 49.Hao W et al. (2021) Development of a base editor for protein evolution via in situ mutation in vivo. Nucleic Acids Res 49, 9594–9605. 10.1093/nar/gkab673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grunewald J et al. (2020) A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat Biotechnol 38, 861–864. 10.1038/s41587-020-0535-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi LS et al. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. 10.1016/j.cell.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gilbert LA et al. (2013) CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen MK (2018) Design principles for nuclease-deficient CRISPR-based transcriptional regulators. FEMS Yeast Res 18. 10.1093/femsyr/foy039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bikard D et al. (2013) Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res 41, 7429–7437. 10.1093/nar/gkt520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villegas Kcam MC et al. (2021) Rational engineering of a modular bacterial CRISPR-Cas activation platform with expanded target range. Nucleic Acids Res 49, 4793–4802. 10.1093/nar/gkab211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong C et al. (2018) Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat Commun 9, 2489. 10.1038/s41467-018-04901-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farzadfard F et al. (2013) Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol 2, 604–613. 10.1021/sb400081r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deaner M and Alper HS (2017) Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab Eng 40, 14–22. 10.1016/j.ymben.2017.01.012 [DOI] [PubMed] [Google Scholar]
- 59.Jensen ED et al. (2017) Transcriptional reprogramming in yeast using dCas9 and combinatorial gRNA strategies. Microb Cell Fact 16, 46. 10.1186/s12934-017-0664-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chavez A et al. (2015) Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Todor H et al. (2021) Bacterial CRISPR screens for gene function. Curr Opin Microbiol 59, 102–109. 10.1016/j.mib.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao L et al. (2020) Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nat Commun 11, 1666. 10.1038/s41467-020-15491-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu X et al. (2023) CRISPRi-microfluidics screening enables genome-scale target identification for high-titer protein production and secretion. Metab Eng 75, 192–204. 10.1016/j.ymben.2022.12.004 [DOI] [PubMed] [Google Scholar]
- 64.Fang L et al. (2021) Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids. Nature Communications 12. 10.1038/s41467-021-25243-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J et al. (2022) CRISPR-assisted rational flux-tuning and arrayed CRISPRi screening of an L-proline exporter for L-proline hyperproduction. Nat Commun 13, 891. 10.1038/s41467-022-28501-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J et al. (2015) Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci Rep 5, 13477. 10.1038/srep13477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Banerjee D et al. (2020) Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nat Commun 11, 5385. 10.1038/s41467-020-19171-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J et al. (2022) Improving isoprenol production via systematic CRISPRi screening in engineered Escherichia coli. Green Chemistry 24, 6955–6964. 10.1039/d2gc02255a [DOI] [Google Scholar]
- 69.Lv L et al. (2015) Application of CRISPRi for prokaryotic metabolic engineering involving multiple genes, a case study: Controllable P(3HB-co-4HB) biosynthesis. Metab Eng 29, 160–168. 10.1016/j.ymben.2015.03.013 [DOI] [PubMed] [Google Scholar]
- 70.Jang S et al. (2018) Toward tunable dynamic repression using CRISPRi. Biotechnol J 13, e1800152. 10.1002/biot.201800152 [DOI] [PubMed] [Google Scholar]
- 71.Jost M et al. (2020) Titrating gene expression using libraries of systematically attenuated CRISPR guide RNAs. Nat Biotechnol 38, 355–364. 10.1038/s41587-019-0387-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Byun G et al. (2023) CRISPRi-mediated tunable control of gene expression level with engineered single-guide RNA in Escherichia coli. Nucleic Acids Res. 10.1093/nar/gkad234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J et al. (2023) Exploring and engineering PAM-diverse Streptococci Cas9 for PAM-directed bifunctional and titratable gene control in bacteria. Metabolic Engineering 75, 68–77. 10.1016/j.ymben.2022.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang J et al. (2022) Biosensor-assisted titratable CRISPRi high-throughput (BATCH) screening for over-production phenotypes. Metab Eng 75, 58–67. 10.1016/j.ymben.2022.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho HI et al. (2020) Programmable CRISPR-Cas transcriptional activation in bacteria. Mol Syst Biol 16, e9427. 10.15252/msb.20199427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Y et al. (2019) Engineered CRISPRa enables programmable eukaryote-like gene activation in bacteria. Nat Commun 10, 3693. 10.1038/s41467-019-11479-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen YP et al. (2022) Biosensor-assisted evolution for high-level production of 4-hydroxyphenylacetic acid in Escherichia coli. Metab Eng 70, 1–11. 10.1016/j.ymben.2021.12.008 [DOI] [PubMed] [Google Scholar]
- 78.Niu FX et al. (2019) Genomic and transcriptional changes in response to pinene tolerance and overproduction in evolved Escherichia coli. Synth Syst Biotechnol 4, 113–119. 10.1016/j.synbio.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fontana J et al. (2020) Effective CRISPRa-mediated control of gene expression in bacteria must overcome strict target site requirements. Nat Commun 11, 1618. 10.1038/s41467-020-15454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu JH et al. (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. 10.1038/nature26155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klanschnig M et al. (2022) CRISPRactivation-SMS, a message for PAM sequence independent gene up-regulation in Escherichia coli. Nucleic Acids Res 50, 10772–10784. 10.1093/nar/gkac804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schilling C et al. (2020) Novel Prokaryotic CRISPR-Cas12a-Based Tool for Programmable Transcriptional Activation and Repression. ACS Synth Biol 9, 3353–3363. 10.1021/acssynbio.0c00424 [DOI] [PubMed] [Google Scholar]
- 83.Lu Z et al. (2019) CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis. Nucleic Acids Res 47, e40. 10.1093/nar/gkz072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kiattisewee C et al. (2021) Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metab Eng 66, 283–295. 10.1016/j.ymben.2021.04.002 [DOI] [PubMed] [Google Scholar]
- 85.Tickman BI et al. (2022) Multi-layer CRISPRa/i circuits for dynamic genetic programs in cell-free and bacterial systems. Cell Syst 13, 215–229 e218. 10.1016/j.cels.2021.10.008 [DOI] [PubMed] [Google Scholar]
- 86.Shaw WM et al. (2022) Inducible expression of large gRNA arrays for multiplexed CRISPRai applications. Nat Commun 13, 4984. 10.1038/s41467-022-32603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hartline CJ et al. (2021) Dynamic control in metabolic engineering: Theories, tools, and applications. Metab Eng 63, 126–140. 10.1016/j.ymben.2020.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang T et al. (2022) Establishing an Autonomous Cascaded Artificial Dynamic (AutoCAD) regulation system for improved pathway performance. Metabolic Engineering 74, 1–10. 10.1016/j.ymben.2022.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ceroni F et al. (2018) Burden-driven feedback control of gene expression. Nat Methods 15, 387–393. 10.1038/nmeth.4635 [DOI] [PubMed] [Google Scholar]
- 90.Lv Y et al. (2020) Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab Eng 61, 79–88. 10.1016/j.ymben.2020.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gupta A et al. (2017) Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol 35, 273–279. 10.1038/nbt.3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tian J et al. (2020) Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in Streptomyces. Nucleic Acids Res 48, 8188–8202. 10.1093/nar/gkaa602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao C et al. (2021) Engineering a CRISPRi Circuit for Autonomous Control of Metabolic Flux in Escherichia coli. ACS Synth Biol 10, 2661–2671. 10.1021/acssynbio.1c00294 [DOI] [PubMed] [Google Scholar]
- 94.Wu Y et al. (2020) Design of a programmable biosensor-CRISPRi genetic circuits for dynamic and autonomous dual-control of metabolic flux in Bacillus subtilis. Nucleic Acids Res 48, 996–1009. 10.1093/nar/gkz1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lim S et al. (2020) CRISPR/Cas-directed programmable assembly of multi-enzyme complexes. Chem Commun (Camb) 56, 4950–4953. 10.1039/d0cc01174f [DOI] [PubMed] [Google Scholar]
- 96.Berckman EA and Chen W (2019) Exploiting dCas9 fusion proteins for dynamic assembly of synthetic metabolons. Chem Commun (Camb) 55, 8219–8222. 10.1039/c9cc04002a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang DY and Winfree E (2009) Control of DNA Strand Displacement Kinetics Using Toehold Exchange. Journal of the American Chemical Society 131, 17303–17314. 10.1021/ja906987s [DOI] [PubMed] [Google Scholar]
- 98.Harrington LB et al. (2018) Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842. doi: 10.1126/science.aav4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karvelis T et al. (2020) PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res 48, 5016–5023. 10.1093/nar/gkaa208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xin C et al. (2022) Comprehensive assessment of miniature CRISPR-Cas12f nucleases for gene disruption. Nat Commun 13, 5623. 10.1038/s41467-022-33346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xu X et al. (2021) Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell 81, 4333–4345 e4334. 10.1016/j.molcel.2021.08.008 [DOI] [PubMed] [Google Scholar]
- 102.Altae-Tran H et al. (2021) The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65. doi: 10.1126/science.abj6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Karvelis T et al. (2021) Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696. 10.1038/s41586-021-04058-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Walton RT et al. (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. doi: 10.1126/science.aba8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kleinstiver BP et al. (2016) High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. 10.1038/nature16526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chatterjee P et al. (2020) An engineered ScCas9 with broad PAM range and high specificity and activity. Nat Biotechnol 38, 1154–1158. 10.1038/s41587-020-0517-0 [DOI] [PubMed] [Google Scholar]
- 107.Xu R et al. (2021) Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nat Plants 7, 888–892. 10.1038/s41477-021-00942-w [DOI] [PubMed] [Google Scholar]
- 108.Yuan Q and Gao X (2022) Multiplex base- and prime-editing with drive-and-process CRISPR arrays. Nat Commun 13, 2771. 10.1038/s41467-022-30514-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grunewald J et al. (2023) Engineered CRISPR prime editors with compact, untethered reverse transcriptases. Nat Biotechnol 41, 337–343. 10.1038/s41587-022-01473-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang J et al. (2022) Efficient targeted insertion of large DNA fragments without DNA donors. Nat Methods 19, 331–340. 10.1038/s41592-022-01399-1 [DOI] [PubMed] [Google Scholar]
- 111.Cai P et al. (2019) CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb Cell Fact 18, 63. 10.1186/s12934-019-1112-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu L et al. (2022) CRISPR-based metabolic engineering in non-model microorganisms. Curr Opin Biotechnol 75, 102698. 10.1016/j.copbio.2022.102698 [DOI] [PubMed] [Google Scholar]
- 113.Fatma Z et al. (2020) Recent advances in domesticating non-model microorganisms. Biotechnol Prog 36, e3008. 10.1002/btpr.3008 [DOI] [PubMed] [Google Scholar]
- 114.Cai P et al. (2021) Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res 49, 7791–7805. 10.1093/nar/gkab535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goh YJ and Barrangou R (2019) Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli. Curr Opin Biotechnol 56, 163–171. 10.1016/j.copbio.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 116.Mahendra C et al. (2020) Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol 5, 620–629. 10.1038/s41564-020-0692-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang W et al. (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31, 233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Selle K and Barrangou R (2015) Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol 23, 225–232. 10.1016/j.tim.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 119.Stovicek V et al. (2017) CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res 17. 10.1093/femsyr/fox030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oh JH and van Pijkeren JP (2014) CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42, e131. 10.1093/nar/gku623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang H et al. (2019) Development of a RecE/T-Assisted CRISPR-Cas9 Toolbox for Lactobacillus. Biotechnol J 14, e1800690. 10.1002/biot.201800690 [DOI] [PubMed] [Google Scholar]
- 122.Wang B et al. (2018) A RecET-assisted CRISPR-Cas9 genome editing in Corynebacterium glutamicum. Microb Cell Fact 17, 63. 10.1186/s12934-018-0910-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Behler J et al. (2018) CRISPR-Based Technologies for Metabolic Engineering in Cyanobacteria. Trends Biotechnol 36, 996–1010. 10.1016/j.tibtech.2018.05.011 [DOI] [PubMed] [Google Scholar]
- 124.Arroyo-Olarte RD et al. (2021) Genome Editing in Bacteria: CRISPR-Cas and Beyond. Microorganisms 9. 10.3390/microorganisms9040844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anzalone AV et al. (2020) Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38, 824–844. 10.1038/s41587-020-0561-9 [DOI] [PubMed] [Google Scholar]
- 126.Nishida K et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353. 10.1126/science.aaf8729 [DOI] [PubMed] [Google Scholar]
- 127.Zhao D et al. (2021) Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol 39, 35–40. 10.1038/s41587-020-0592-2 [DOI] [PubMed] [Google Scholar]
- 128.Anzalone AV et al. (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tong Y et al. (2021) A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat Commun 12, 5206. 10.1038/s41467-021-25541-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meliawati M et al. (2021) Recent advances of Cas12a applications in bacteria. Appl Microbiol Biotechnol 105, 2981–2990. 10.1007/s00253-021-11243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bao J et al. (2022) Efficient CRISPR/Cas12a-Based Genome-Editing Toolbox for Metabolic Engineering in Methanococcus maripaludis. ACS Synth Biol 11, 2496–2503. 10.1021/acssynbio.2c00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang X et al. (2021) A Novel and Efficient Genome Editing Tool Assisted by CRISPR-Cas12a/Cpf1 for Pichia pastoris. ACS Synth Biol 10, 2927–2937. 10.1021/acssynbio.1c00172 [DOI] [PubMed] [Google Scholar]
- 133.Nishida K and Kondo A (2021) CRISPR-derived genome editing technologies for metabolic engineering. Metab Eng 63, 141–147. 10.1016/j.ymben.2020.12.002 [DOI] [PubMed] [Google Scholar]
- 134.Zhao D et al. (2021) CRISPR-based metabolic pathway engineering. Metab Eng 63, 148–159. 10.1016/j.ymben.2020.10.004 [DOI] [PubMed] [Google Scholar]
- 135.Cernak P et al. (2018) Engineering Kluyveromyces marxianus as a Robust Synthetic Biology Platform Host. mBio 9. 10.1128/mBio.01410-18 [DOI] [PMC free article] [PubMed] [Google Scholar]


