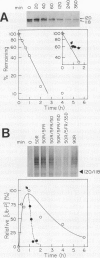
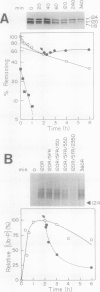
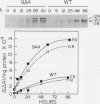
Abstract
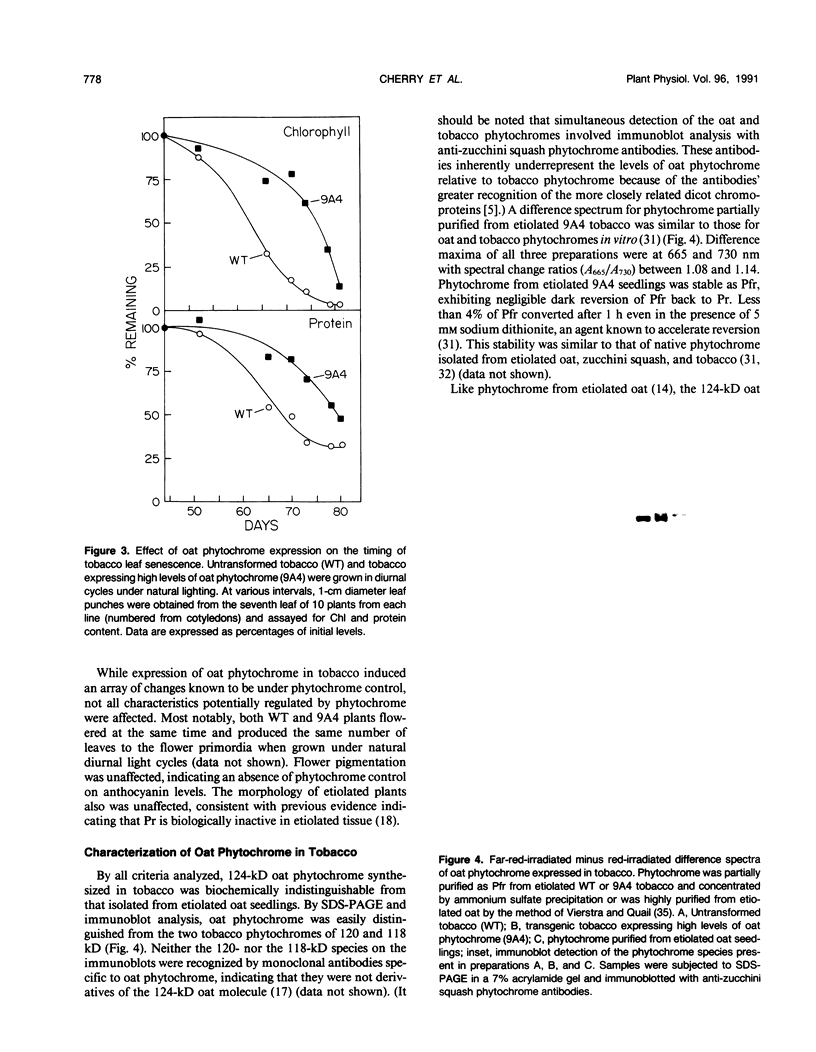
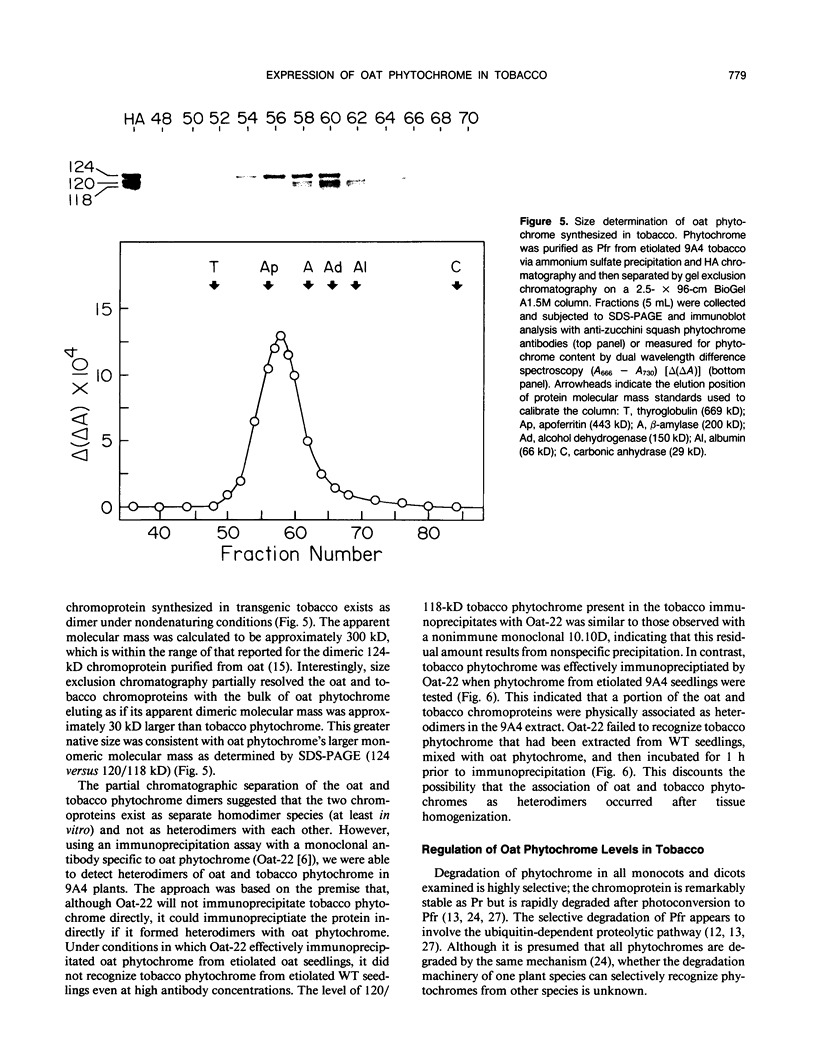
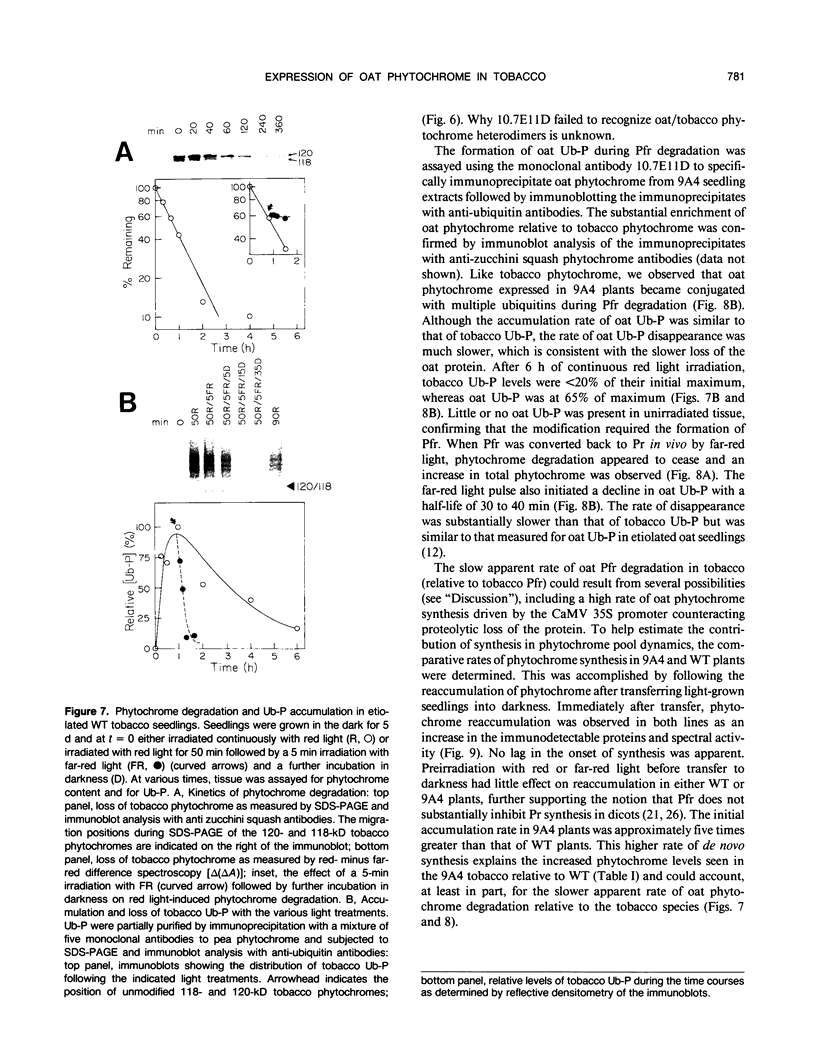
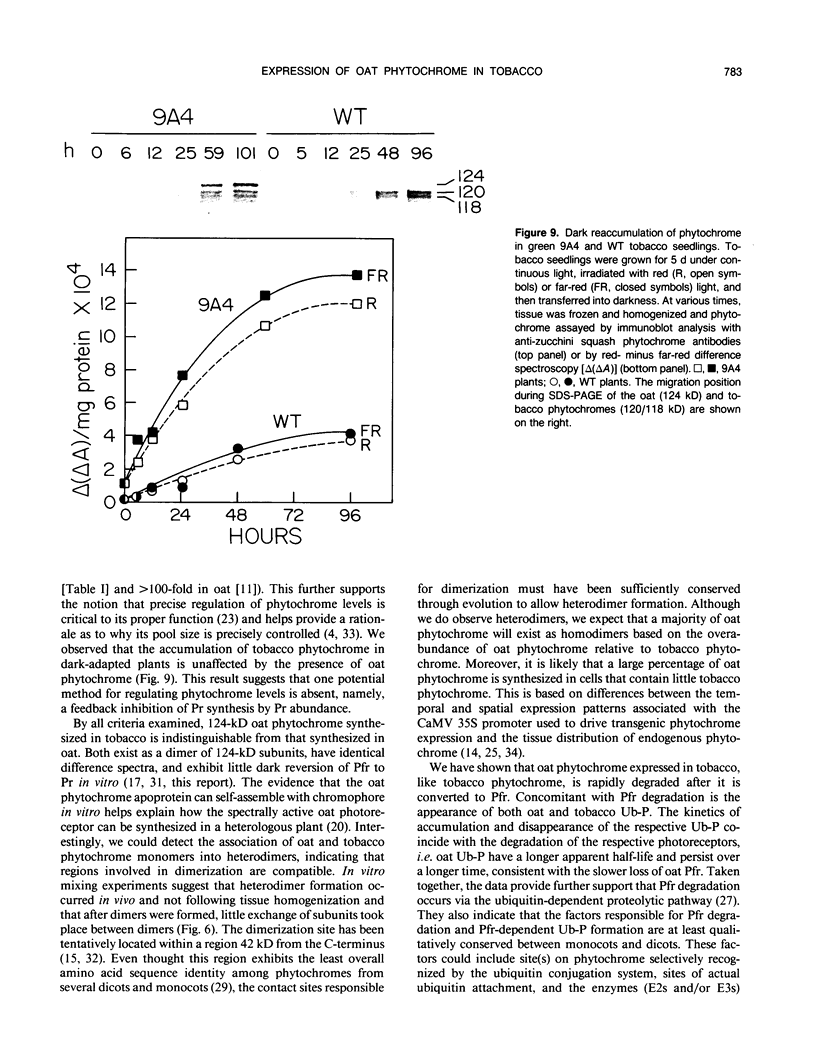
Constitutive expression of a chimeric oat phytochrome gene in tobacco (Nicotiana tabacum) results in the accumulation of a functional 124-kilodalton photoreceptor that markedly alters the phenotype of light-grown tobacco (Keller et al. [1989] EMBO J 8: 1005-1012). Here, we provide a detailed phenotypic and biochemical characterization of homozygous tobacco expressing high levels of oat phytochrome. Phenotypic changes include a substantial inhibition of stem elongation, decreased apical dominance, increased leaf chlorophyll content, and delayed leaf senescence. Oat phytochrome synthesized in tobacco is indistinguishable from that present in etiolated oats, having photoreversible difference spectrum maxima at 665 and 730 nanometers, exhibiting negligible dark reversion of phytochrome—far red-absorbing form (Pfr) to phytochrome—red-absorbing form (Pr), and existing as a dimer with an apparent size of approximately 300 kilodaltons. Heterodimers between the oat and tobacco chromoproteins were detected. Endogenous tobacco phytochrome and transgenically expressed oat phytochrome are rapidly degraded in vivo upon photoconversion of Pr to Pfr. Breakdown of both oat and tobacco Pfr is associated with the accumulation of ubiquitin-phytochrome conjugates, suggesting that degradation occurs via the ubiquitin-dependent proteolytic pathway. This result indicates that the factors responsible for selective recognition of Pfr by the ubiquitin pathway are conserved between monocot and dicot phytochromes. More broadly, it demonstrates that the domain(s) within a plant protein responsible for its selective breakdown can be recognized by the degradation machinery of heterologous species.
Full text
PDF

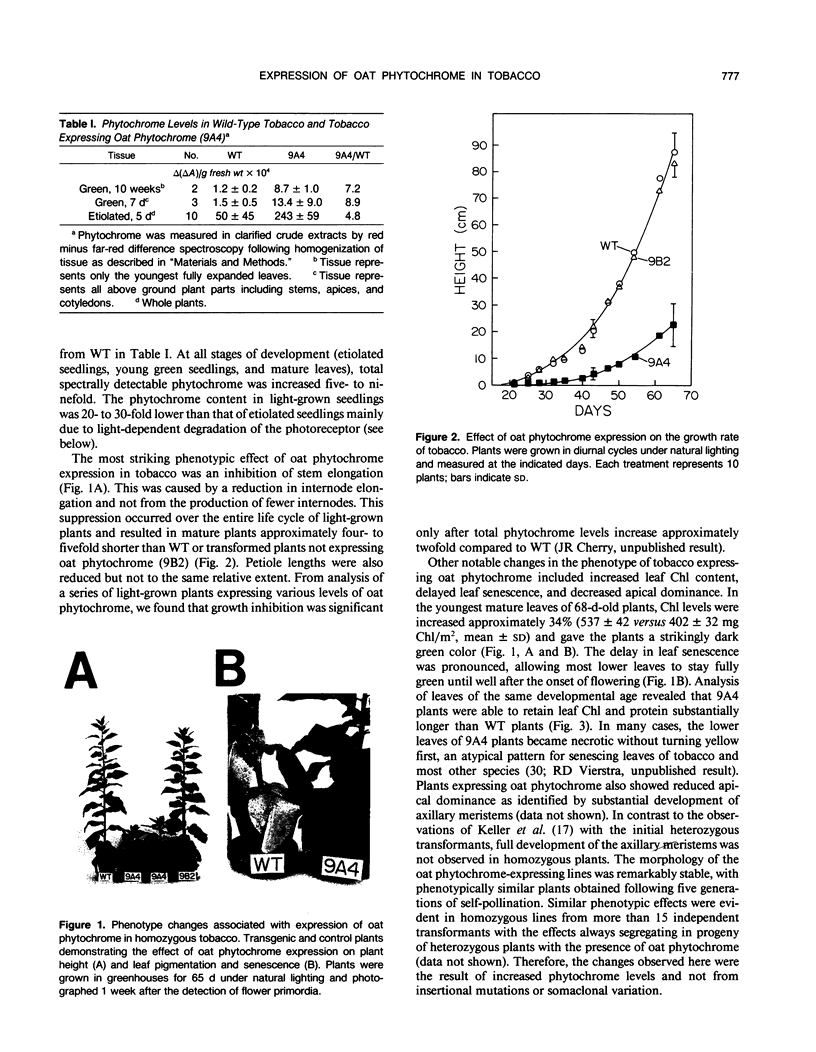




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boylan M. T., Quail P. H. Oat Phytochrome Is Biologically Active in Transgenic Tomatoes. Plant Cell. 1989 Aug;1(8):765–773. doi: 10.1105/tpc.1.8.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briggs W. R., Mösinger E., Schäfer E. Phytochrome regulation of greening in barley-effects on chlorophyll accumulation. Plant Physiol. 1988 Feb;86(2):435–440. doi: 10.1104/pp.86.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordonnier M. M., Greppin H., Pratt L. H. Characterization by enzyme-linked immunosorbent assay of monoclonal antibodies to pisum and Avena phytochrome. Plant Physiol. 1984 Jan;74(1):123–127. doi: 10.1104/pp.74.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordonnier M. M., Pratt L. H. Comparative Phytochrome Immunochemistry as Assayed by Antisera against Both Monocotyledonous and Dicotyledonous Phytochrome. Plant Physiol. 1982 Sep;70(3):912–916. doi: 10.1104/pp.70.3.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. M., Quail P. H. Monoclonal antibodies to three separate domains on 124 kilodalton phytochrome from Avena. Plant Physiol. 1984 Nov;76(3):622–626. doi: 10.1104/pp.76.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin P. M., Sarokin L., Memelink J., Chua N. H. Molecular light switches for plant genes. Plant Cell. 1990 May;2(5):369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey H. P., Barker R. F., Idler K. B., Lissemore J. L., Quail P. H. Analysis of cloned cDNA and genomic sequences for phytochrome: complete amino acid sequences for two gene products expressed in etiolated Avena. Nucleic Acids Res. 1985 Dec 9;13(23):8543–8559. doi: 10.1093/nar/13.23.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. Ubiquitin-mediated protein degradation. J Biol Chem. 1988 Oct 25;263(30):15237–15240. [PubMed] [Google Scholar]
- Hunt R. E., Pratt L. H. Phytochrome radioimmunoassay. Plant Physiol. 1979 Aug;64(2):327–331. doi: 10.1104/pp.64.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabben M., Shanklin J., Vierstra R. D. Red light-induced accumulation of ubiquitin-phytochrome conjugates in both monocots and dicots. Plant Physiol. 1989 Jun;90(2):380–384. doi: 10.1104/pp.90.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabben M., Shanklin J., Vierstra R. D. Ubiquitin-phytochrome conjugates. Pool dynamics during in vivo phytochrome degradation. J Biol Chem. 1989 Mar 25;264(9):4998–5005. [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A., Bevan M. W. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987 Dec 20;6(13):3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay S. A., Nagatani A., Keith B., Deak M., Furuya M., Chua N. H. Rice Phytochrome Is Biologically Active in Transgenic Tobacco. Plant Cell. 1989 Aug;1(8):775–782. doi: 10.1105/tpc.1.8.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller J. M., Shanklin J., Vierstra R. D., Hershey H. P. Expression of a functional monocotyledonous phytochrome in transgenic tobacco. EMBO J. 1989 Apr;8(4):1005–1012. doi: 10.1002/j.1460-2075.1989.tb03467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagarias J. C., Lagarias D. M. Self-assembly of synthetic phytochrome holoprotein in vitro. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5778–5780. doi: 10.1073/pnas.86.15.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P. H., Schäfer E., Marmé D. Turnover of phytochrome in pumpkin cotyledons. Plant Physiol. 1973 Aug;52(2):128–131. doi: 10.1104/pp.52.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J., Jabben M., Vierstra R. D. Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc Natl Acad Sci U S A. 1987 Jan;84(2):359–363. doi: 10.1073/pnas.84.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R. A., Lissemore J. L., Quail P. H. Nucleotide and amino acid sequence of a Cucurbita phytochrome cDNA clone: identification of conserved features by comparison with Avena phytochrome. Gene. 1986;47(2-3):287–295. doi: 10.1016/0378-1119(86)90072-7. [DOI] [PubMed] [Google Scholar]
- Vierstra R. D., Quail P. H. Spectral Characterization and Proteolytic Mapping of Native 120-Kilodalton Phytochrome from Cucurbita pepo L. Plant Physiol. 1985 Apr;77(4):990–998. doi: 10.1104/pp.77.4.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson J. D., Hirsch-Wyncott M. E., Larkins B. A., Gelvin S. B. Differential Accumulation of a Transcript Driven by the CaMV 35S Promoter in Transgenic Tobacco. Plant Physiol. 1989 Aug;90(4):1570–1576. doi: 10.1104/pp.90.4.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans J. F., de Mots A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim Biophys Acta. 1965 Nov 29;109(2):448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]