Abstract
Background
The taxonomic composition of the gut microbiome undergoes rapid development during the first 2–3 y of life. Poor diet during complementary feeding has been associated with alterations in infant growth and compromised bone, immune system, and neurodevelopment, but how it may affect gut microbial composition is unknown.
Objectives
This cross-sectional study aimed to examine the associations between early-life nutrition and the developing infant gut microbiota at 6 mo of age.
Methods
Latino mother–infant pairs from the Mother’s Milk Study (n = 105) were included. Infant gut microbiota and dietary intake were analyzed at 6 mo of age using 16S ribosomal RNA amplicon sequencing and 24-h dietary recalls, respectively. Poisson generalized linear regression analysis was performed to examine associations between dietary nutrients and microbial community abundance while adjusting for infants’ mode of delivery, antibiotics, infant feeding type, time of introduction of solid foods, energy intake, and body weight. A P value of <0.05 was used to determine the statistical significance in the study.
Results
Infants with higher consumption of total sugar exhibited a lower relative abundance of the genera Bacteroides (β = −0.01; 95% CI: −0.02, −0.00; P = 0.03) and genus Clostridium belonging to the Lachnospiraceae family (β = −0.02; 95% CI: −0.03, −0.00; P = 0.01). In addition, a higher intake of free sugar (which excludes sugar from milk, dairy, and whole fruit) was associated with several bacteria at the genus level, including Parabacteroides genus (β = 0.03; 95% CI: 0.01, 0.05; P = 0.001). Total insoluble fiber intake was associated with favorable bacteria at the genus level such as Faecalibacterium (β = 0.28; 95% CI: 0.03, 0.52; P = 0.02) and Coprococcus (β = 0.28; 95% CI: 0.02, 0.52; P = 0.03).
Conclusion
These findings demonstrate that early-life dietary intake at 6 mo impacts the developing gut microbiome associated with the presence of both unfavorable gut microbes and dietary fiber-associated commensal microbes.
Keywords: infant diet, gut microbiome, early-life nutrition, complementary diet, Latino
Introduction
The first 3 y of life are important for the establishment of the gut microbiome. The neonatal gastrointestinal tract becomes rapidly colonized with an assembly of microorganisms reaching from 108 to up to 1011–1012 microorganisms in a matter of months [1,2]. Early-life patterns of microbial colonization have been shown to influence the development of metabolic, endocrine, neural, and immune functions, and are related to future health outcomes [3]. Thus, disrupted optimal microbial succession may lead to lifelong and intergenerational growth and developmental deficits [4]. The infant gut microbiota is less stable and resilient to external perturbation compared with adult gut microbiota [5]. The development of the infant gut microbiota is a dynamic and nonrandom process [6], developing and stabilizing over the first years of life and resembling an adult-like composition by the age of 3–5 y [2,7,8]. Numerous factors, including delivery mode, gestational age, antibiotic exposure, supplement use, maternal diet, infant feeding (breastmilk compared with formula feeding), geographic location, ethnicity, and the introduction of solid foods, are known to affect infant gut bacterial diversity in the first 1–3 y of life [9,10]. Although all these factors have an impact on the developing gut microbiome, infant diet composition is considered a major modifiable contributor to gut bacterial development in infants but is relatively understudied other than the general effect of breastfeeding compared with formula feeding [11,12].
Although the impact of the early-life diet, including formula compared with breast milk feeding, has been well studied, the influence of complementary feeding on gut microbial composition has not been widely explored [1,13,14]. There is a lack of consensus on how the introduction of specific nutrients affects gut microbial taxonomy, diversity, and richness in early life [9,15]. In addition, limited studies have been conducted on Latinos, which is one of the fastest growing demographic segments of the United States population [16] and comprise ∼40% of the population in the state of California [17]. Latinos are a group known to harbor a distinct gut microbiome composition because of variations in diet and exposure to new natural and built environment influencers [18]. Moreover, higher rates of obesity have been reported in Latino as compared with non-Latino children, and feeding practices related to the selection of solid foods at the beginning of infancy may contribute to this disparity [19]. One study examined dietary data to compare the food consumption and introduction of specific foods in Latino infants with non-Latino infants living in the United States [19]. The study findings indicated notable differences in the early food experiences of Latino infants in comparison with non-Latino infants, whereby a greater proportion of Latino infants were fed fresh fruits, fruit-flavored drinks, infant cookies, and traditional foods such as soups, rice, and beans [19]. In the current study, we aimed to examine the effects of early-life dietary sugars and fiber during the introductory solid food feeding phase at 6 mo on the gut microbiome composition in a cohort of healthy Latino infants.
Methods
Study participants and design
This study utilized data collected at 6 mo of age from the Southern California Mother’s Milk Study, which is a longitudinal cohort of Latino mother–infant pairs that were enrolled at 1 mo of infant age and assessed throughout the first 24 mo of infancy [[20], [21], [22]]. Participants were recruited from various maternity clinics affiliated with the University of Southern California in Los Angeles County. We excluded participants with the following criteria: 1) physician diagnosis of inflammatory or infectious diseases and fatal abnormalities, 2) preterm/low birth weight infants, 3) mothers aged <18 y at delivery (to prevent the confounding effect of adolescent growth), and 4) pregnancy complications (e.g., hypertensive disorders and severe anemia). Infants who used antibiotics or probiotics were not excluded but were rigorously documented and included in all models as potential confounders. At baseline, participants underwent a comprehensive assessment that included recording demographic data, such as age and sex, as well as a detailed family history and physical examination. Antibiotic exposures including the exact medication and dosage were collected at each visit to control for potential covariates. The participant flowchart is included in Supplemental Figure 1. This study was reviewed and approved by the Children’s Hospital Los Angeles Institutional Review Board. Written consent forms were obtained from each mother at recruitment. In this analysis, we included data from 105 infants at 6 mo who had complete data related to gut microbiota and dietary intake.
Assessment of dietary intake
As part of the study protocol, mothers were asked to complete an infant diet questionnaire at baseline to record their infant’s feeding practices. In addition, at 6 mo, mothers completed 24-h dietary recalls on behalf of their infants in a structured interview with a trained bilingual dietician, which involved describing the food and drinks consumed by their child during the preceding 24-h period, including the assessment of breastfeeding and infant formula use. These assessments were conducted to track changes in the infants’ diet over time. Infant dietary intake at 6 mo of age was measured on the basis of the average from multiple 24-h dietary recalls per infant, consisting of 2 weekday recalls and 1 weekend recall. Mothers were interviewed to complete detailed dietary recalls including portion sizes, brand names, and preparation methods on behalf of their infants. They were asked to describe what they fed their child during the previous 24-h period. Before completing the recalls, the research team provided instructions on how to estimate food and beverage portion sizes, and mothers were provided with portion size information booklets by Nutrition Data System for Research software (NDSR). We analyzed dietary intake using the NDSR (2014–2019), which has been extensively used for studies on the Latino population [23,24]. For this study, we explored the associations between gram intakes of dietary sugar (total, free, and added sugar) and fiber (soluble and insoluble fiber) from all sources at 6 mo of age.
Description of sugar categories in infant diet recalls
As part of the dietary assessment, sugars were classified into 3 categories on the basis of the guidelines from the WHO and the nutrient analysis software NDSR [25,26]. The 3 categories of sugars include free sugars, added sugars, and total sugars. Free sugars include sugars naturally present in honey, syrups, and fruit juices, as well as those added to food and drinks. Added sugars refer to all sugars added to foods and drinks during manufacturing, cooking, or processing, and do not include naturally occurring sugars including sugars in juice. Total sugars include all sugars found in a food or drink, regardless of whether they are naturally occurring or added. Total sugars include free sugars, intrinsic sugars, and milk sugars. On the basis of the data obtained from the 24-h dietary recalls, NDSR was used to analyze infant dietary data to calculate the total and added sugar intake. To estimate the free sugar intake, the research team calculated the free sugars from beverages and nonbeverages separately. For beverages, lactose was subtracted from the total sugar content, whereas for nonbeverages, only added sugars were considered free sugars. These values were then summed to obtain the total free sugar intake for each participant.
Stool collection and microbiome analysis
We used OMNIgene·GUT kits to collect a sample of infant stool at 6 mo postpartum during the mother/infant study visit. In cases where sample collection during the study visit was not feasible, the participant was provided with a collection kit to take home, and the samples were retrieved by the study staff. On receipt in the laboratory, the collected stool samples were homogenized, aliquoted into the cryotubes with O-ring, and stored in the laboratory freezer at −80°C until DNA extraction. A detailed description of the 16S rRNA amplicon sequencing method has been previously published [20,27]. Briefly, microbial DNA was extracted from stool samples using the MoBio PowerSoil kit for 16S rRNA amplicon sequencing. The bacterial 16S rRNA gene was sequenced using the 515/806 barcoded primer pair (515 F [Parada]): GTGYCAGC MGCCGCGGTAA, 806 R: GGACTACN VGGGTWTCTAAT) and then standardized comparatively to the protocols in the Earth Microbiome Project [28]. We performed paired-end, 2 × 150 bp, next-generation DNA sequencing on the existing Illumina MiSeq platform in the Institute for Genomic Medicine at the University of California San Diego [29]. Demultiplexed files were processed using Qiita (https://qiita.ucsd.edu) [30]. Sequences were trimmed to 150-bp, suspected error sequences were removed, and amplicon sequence variants called suboperational taxonomic units (sOTUs) were assigned using Deblur, a reference-free method [31]. In subsequent steps, a feature table containing counts for each sOTU for each sample was generated. To generate a phylogeny, we inserted Deblur tag sequences into the GreenGenes 13_8 backbone phylogeny using Sample Adaptive Treatment Effect-enabled phylogenic placement (SATE-SEPP) [32]. Any sOTUs not placed during SEPP were excluded from the analysis. sOTUs were assigned taxonomy using the GreenGenes 13_8 database and the q2-feature-classifier classify-sklearn from QIIME2 [33].
Statistical analysis
Initial descriptive and exploratory analyses were performed to assess variable distributions. We performed Spearman rank correlation analysis to quantify the direction and strength of the linear association between the relative abundances of bacteria at the genus level with infants’ dietary intake at 6 mo of age. The correlation heatmaps were used as visualization without adjusting for confounders. The Poisson regression model, which is a type of generalized linear model, was employed to investigate how intake of dietary sugar and fiber at 6 mo was associated with bacterial relative abundances at the genus level at 6 mo [34]. For each type of dietary sugar or fiber, we analyzed separate models, allowing us to assess their individual associations with bacterial abundance. The Poisson regression model is appropriate for count data and assumes that the dependent variable follows a Poisson distribution. Therefore, the assumptions for Poisson regression focus on the model's linearity, independence of observations, and absence of overdispersion [35]. We performed a complete case analysis to handle missing covariate data, where only participants with complete data for all covariates were included in the analysis.
The analysis of infant gut microbiota involved several key steps. First, the total reads (counts) for each taxonomic group were calculated by summing the counts across all samples. This provided an overall measure of abundance for each taxonomic group. Next, bacterial relative abundances were log-transformed (Equation 1) to better satisfy the regression modeling assumptions [36]. To focus on more abundant taxa, taxonomic groups with low total reads (below a certain threshold, for example, 10) were filtered out. This step helps to reduce noise and ensures that the analysis focuses on taxa that are more consistently observed across samples. In addition, sparse taxa that were observed in fewer than 50% of the samples were filtered out. This further reduces noise and allows for the analysis to focus on taxa that are more prevalent in the dataset [37,38].
Regression models were adjusted for infants’ mode of delivery, current antibiotics (antibiotics within the past 30 d), infant feeding type (refer to primarily breastfed or formula-fed at 6 mo), time of introduction of solid foods (age of infant when started eating solid foods), energy intake, and body weight. Descriptive statistics are presented as mean ± SD for continuous variables and as frequency (percentage) for categorical variables. We used the Benjamini–Hochberg method for false discovery rate correction at a 5% significance level to address multiple comparisons in analyzing microbial abundance and diet correlations. The statistical significance threshold was set at a False Discovery Rate corrected p-value (pFDR) of less than 0.05. All statistical analyses were carried out in QIIME2 v.2020.11 and R (version 4.1.1).
Equation 1. General Equation for Relative Abundance Log Transformation
Results
Study characteristics
Maternal and infant characteristics and dietary intake variables of interest at 6 mo of infant age are presented in TABLE 1, TABLE 2, respectively. In the subsample of 105 infants, 54.3% were women and 45.7% were men, with an average age of 185.2 ± 8.5 d (and an average age at stool sample collection of 186.2 ± 9.5). In addition, 82% of infants were still breastfed at 6 mo, with 46.6% being exclusively breastfed at 6 mo. At 6 mo, the infants had an average weight of 8.0 ± 0.8 kg and an average length of 66.6 ± 2.1 cm. The average age of introduction to complementary foods was 5.59 ± 1.28 mo (range: 4–12 mo). Dietary intake of carbohydrates, protein, fat, fiber, and sugar at 6 mo are presented in Table 2. The mean energy intake was 668 ± 150.2 kcal. The infants’ diet consisted of 44.7% carbohydrates, 7.6% protein, and 47.5% fat. The mean intake of added sugar was 14.3 ± 16.3 g, which accounted for 8.6% of the total energy intake. The mean total sugar intake was 61.8 ± 18 g, which accounted for 37.1% of the total energy intake. The infants’ diet also included 2.1 g of total fiber, 1 g of insoluble fiber, and 1.1 g of soluble fiber. Our observation of infant diets revealed interesting patterns in the sources of total sugar and dietary fiber (Supplemental Table 1). The majority of total sugar content was found to originate from homemade fresh fruit purees, such as apples, bananas, and pears. In addition, commercially available infant foods and infant formula contributed to the total sugar intake. In contrast, added sugars were predominantly derived from sources such as infant formula and commercialized desserts, including fruit juices and sweet treats. When examining dietary fiber, we observed that it primarily came from vegetables, including beans, such as black beans and pinto beans cooked from dried, as well as avocado. Furthermore, fiber-rich foods, such as cooked vegetables, fresh fruits, and homemade fruit puree, played a significant role in contributing to the dietary fiber content in infants’ diets.
TABLE 1.
Characteristics of infants from the Southern California Mother’s Milk Study at 6 mo of infant age
| Variable | Subsample (n = 105) |
|---|---|
| Sex | |
| Female | 57 (54.3%) |
| Male | 48 (45.7%) |
| Age | |
| Days | 185.2 (8.5) |
| Age (stool sample collection) | |
| Days | 186.2 (9.5) |
| SES index | 27.1 (11.5) |
| Mode of delivery | |
| Vaginal | 82 (78%) |
| Cesarean | 23 (22%) |
| Breastfeeding | |
| Yes | 86 (82%) |
| No | 19 (18%) |
| Exclusive breastfeeding | |
| Yes | 49 (46.6%) |
| No | 36 (34.3%) |
| N/A | 20 (19.1%) |
| Antibiotics | |
| Yes | 3 (2.9%) |
| No | 93 (88.6%) |
| N/A | 9 (8.6%) |
| Infant anthropometrics | |
| Weight (kg) | 8 (0.8) |
| Length (cm) | 66.6 (2.1) |
| Weight Z-score | 0.4 (0.8) |
| Length Z-score | −0.1 (0.9) |
| Weight for length Z-score | −0.3 (1.3) |
| Maternal characteristics | |
| Age (y) | 29.7 (6.4) |
| BMI (kg/m2) | 30.2 (5.5) |
| Prepregnancy BMI (kg/m2) | 28 (5.6) |
Six-month characteristics of 105 Latino mother–infant dyads from the Southern California Mother’s Milk Study. Participants who had complete data for the microbiome and dietary intake data were included in the analysis. Unless otherwise indicated, values are presented as n (%) or mean ± SDs. The age in days for each infant was calculated using their date of birth and the date of the visit. This was done by subtracting the date of birth from the date of visit and converting the result to days. The calculated age was then compared with the expected age on the basis of the dates of birth and visits to ensure accuracy. Age is reported in days and includes infant age at the time of the visit (6 mo) and age at the time of stool sample collection. Breastfeeding refers to any amount of breastfeeding or pumping for the infant, regardless of whether it was exclusive or partial, at the time of stool sample collection. Exclusive breastfeeding was defined as the infant receiving only breast milk and no other foods or liquids, including water. The percentage of infants exclusively breastfed at the time of stool sample collection is reported for the subset of participants with available data. The antibiotics variable indicates whether the infant was currently taking antibiotics within 30 d of the stool sample collection date. N/A in the antibiotics list indicates missing or unavailable data on antibiotic use for the corresponding sample.
TABLE 2.
Nutrient intake at 6 mo of infant age (n = 105)
| Variable | 6 mo Mean ± SD | % of calories |
|---|---|---|
| Age of introduction of solid foods (mo) | 5.6 (1.3) | |
| Energy (kcal) | 668 (150.2) | |
| Carbohydrate (g) | 76 (17.3) | 44.7 |
| Protein (g) | 12.3 (3.8) | 7.6 |
| Fat (g) | 35.8 (9.6) | 47.5 |
| Added sugar (g) | 14.3 (16.3) | 8.6 |
| Total sugar (g) | 61.8 (18) | 37.1 |
| Free sugar (g) | 5 (8) | 3 |
| Total fiber (g) | 2.1 (1.4) | — |
| Insoluble fiber (g) | 1 (0.8) | — |
| Soluble fiber (g) | 1.1 (1) | — |
The data presented represent the average nutrient intake over a 3-d food recall period. Grams of nutrient intake are reported as daily intake, and the “% of calories” column indicates the percentage of total caloric intake that is contributed by each nutrient. It is important to note that added sugar, free sugar, and total sugar are subcategories of carbohydrates, and as such, the percentage of calories from each of these subcategories should be added up to get the total percentage of calories from carbohydrates. However, it is incorrect to add all the percentages of calories from each nutrient, as this will exceed 100%. Rather, it is important to interpret the percentages of calories from each nutrient in the context of the total calorie intake, and the total percentage of calories should add ≤100%.
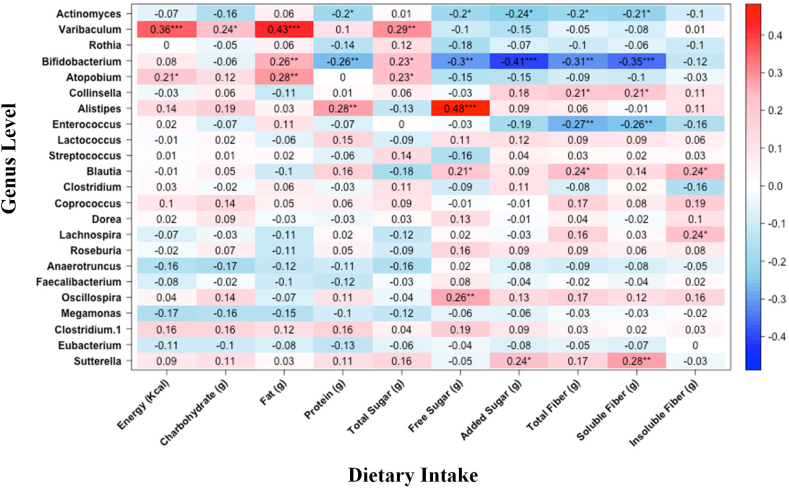
Association between dietary intake and bacterial abundance
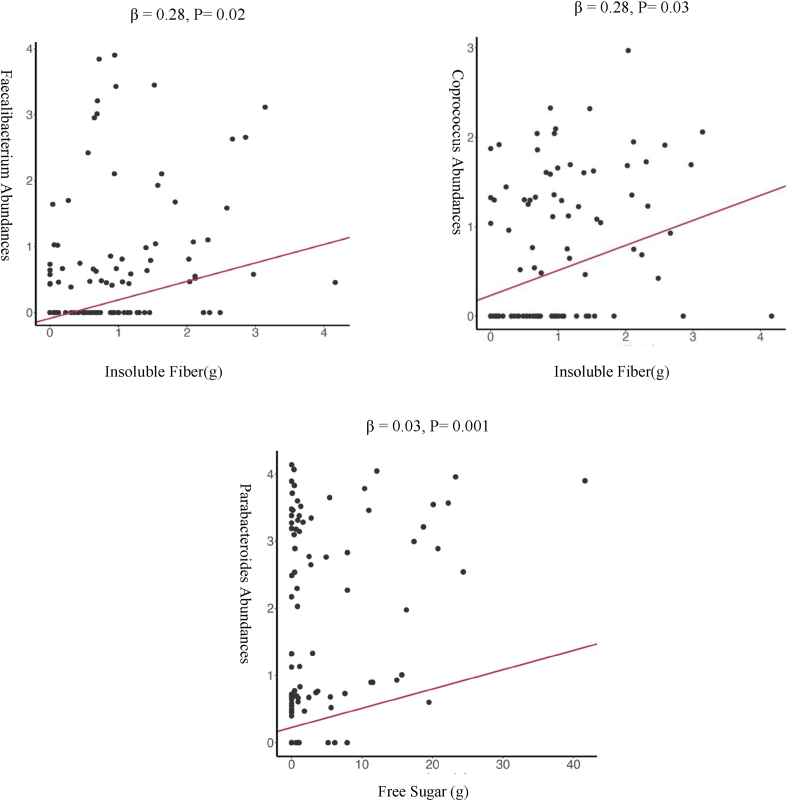
Figure 1 displays the Spearman correlations of nutrient intakes of interest with bacterial relative abundance. The dietary intake correlation matrix is included in Supplemental Figure 2 to investigate associations between dietary components. We further performed a Poisson generalized linear regression analysis to identify the associations between dietary intake with microbial composition. TABLE 3, TABLE 4 present the complete set of significant associations identified. Multiple bacteria at the genus level were associated with sugar and fiber intake. Figure 2 illustrates the scatter plots of dietary sugar and fiber with the relative abundance of several bacteria. The associations highlighted in scatter plots between sugar and fiber intake and the abundance of specific bacterial taxa were chosen based on their previous linkage to metabolic disorders and inflammation [39,40]. Associations that had been previously supported by research in similar populations were also prioritized. Although there were additional significant associations that were not illustrated in plots, we chose to focus on the associations that were most relevant and informative to our research questions. The relative abundance of Blautia was lower in infants who consumed a greater amount of total sugar (β = −0.02; 95% CI: −0.03, −0.00; P = 0.01). Conversely, we observed a higher relative abundance of the Blautia (β = 0.29; 95% CI: 0.14, 0.43; P ≤ 0.001) with increasing intake of total fiber consumption. In addition, a higher intake of free sugar was associated with a higher abundance of Parabacteroides genus (β = 0.03; 95% CI: 0.01, 0.05; P = 0.001) and a lower abundance of genus Clostridium (β = −0.04; 95% CI: −0.07, −0.0; P = 0.02).
FIGURE 1.
Spearman correlation analysis heatmap of bacterial genera and dietary factors. Bacterial taxa were associated with dietary variable: negative associations are shown in blue and positive associations are shown in red. P values with asterisks represent statistical significance. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
TABLE 3.
Summary of regression analyses for gut microbiome associations with infant sugar intake at 6 mo (n = 105)
| Variable | Total sugar intake (g) |
Added sugar intake (g) |
Free sugar intake (g) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | P | PFDR | β (95% CI) | P | PFDR | Β (95% CI) | P | PFDR | |
| Bifidobacterium | 0.01 (−0.00, 0.02) | 0.15 | 0.26 | −0.01 (−0.01, 0.00) | 0.24 | 0.52 | −0.01 (−0.03, 0.00) | 0.14 | 0.24 |
| Bacteroides | −0.01 (−0.02, 0.00) | 0.031 | 0.12 | 0.00 (−0.01, 0.01) | 0.43 | 0.73 | 0.01 (−0.00, 0.02) | 0.14 | 0.25 |
| Streptococcus | 0.02 (0.00, 0.04) | 0.041 | 0.13 | −0.00 (−0.02, 0.01) | 0.56 | 0.76 | −0.03 (−0.06, 0.00) | 0.07 | 0.16 |
| Lachnospiraceae;g__Clostridium | −0.02 (−0.03, −0.00) | 0.011 | 0.12 | 0.00 (−0.00, 0.01) | 0.52 | 0.76 | 0.03 (0.01, 0.05) | 0.001 | 0.011 |
| f_Clostridiaceae;g_Clostridium | 0.02 (0.00, 0.03) | 0.031 | 0.12 | 0.00 (−0.01, 0.01) | 0.89 | 0.93 | −0.04 (−0.07, −0.01) | 0.021 | 0.05 |
| f_Erysipelotrichaceae;g_Clostridium | −0.00 (−0.02, 0.02) | 0.71 | 0.82 | 0.02 (0.01, 0.03) | 0.001 | 0.041 | 0.01 (−0.02, 0.03) | 0.55 | 0.61 |
| Veillonella | 0.01 (−0.00, 0.02) | 0.07 | 0.17 | −0.00 (−0.01, 0.01) | 0.60 | 0.85 | −0.02 (−0.04,0.00) | 0.06 | 0.16 |
| Blautia | −0.02 (-0.03, −0.00) | 0.011 | 0.12 | 0.02 (0.01, −0.03) | 0.001 | 0.031 | 0.03 (0.01, −0.05) | 0.0021 | 0.011 |
| Actinomyces | 0.03 (−0.00, 0.07) | 0.08 | 0.17 | −0.03 (−0.06, −0.00) | 0.05 | 0.19 | −0.15 (−0.31, −0.05) | 0.021 | 0.06 |
| Dorea | −0.02 (−0.04, −0.00) | 0.021 | 0.12 | 0.02 (0.01, 0.04) | 0.001 | 0.041 | 0.04 (0.02, 0.07) | 0.001 | 0.001 |
| Oscillospira | −0.01 (−0.03, 0.00) | 0.05 | 0.13 | 0.01 (0.00, 0.03) | 0.011 | 0.05 | 0.03 (0.01, 0.05) | 0.001 | 0.011 |
| Parabacteroides | −0.01 (−0.03, −0.00) | 0.041 | 0.12 | 0.01 (-0.00, 0.02) | 0.21 | 0.48 | 0.03 (0.01, 0.05) | 0.0011 | 0.011 |
| Lactobacillus | 0.02 (−0.01, 0.05) | 0.14 | 0.26 | −0.04 (−0.06, −0.01) | 0.011 | 0.05 | −0.04 (−0.09, 0.01) | 0.13 | 0.25 |
| Roseburia | −0.02 (−0.04,0.00) | 0.041 | 0.12 | −0.00 (−0.02, 0.01) | 0.78 | 0.86 | 0.01 (−0.02,0.04) | 0.35 | 0.45 |
β-coefficients and 95% confidence intervals (CIs) from Poisson generalized linear regression analysis were used to examine the associations between the relative abundance of genera with gram total sugar, added sugar, and free sugar intake in infants at 6 mo of age. Sugar intake was measured using 24-h dietary recalls and the abundance of genera was computed using 16S rRNA sequencing. Models adjusted for infants’ mode of delivery, antibiotics, infant feeding type, time of introduction of solid foods, energy intake, and body weight.
TABLE 4.
Summary of regression analyses for variables predicting infant fiber intake at 6 mo (n =105)
| Variable | Total fiber (g) |
Soluble fiber (g) |
Insoluble fiber (g) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | P | PFDR | β (95% CI) | P | PFDR | β (95% CI) | P | PFDR | |
| Blautia | 0.39 (0.14, 0.43) | <0.0011 | <0.0011 | 0.36 (0.17, 0.55) | <0.0011 | 0.001 | 0.22 (0.00, 0.42) | 0.031 | 0.15 |
| Dorea | 0.32 (0.14, 0.49) | <0.0011 | 0.001 | 0.35 (0.11, 0.57) | 0.001 | 0.021 | 0.32 (0.06, 0.56) | 0.011 | 0.15 |
| Oscillospira | 0.22 (0.19, 0.35) | 0.001 | 0.001 | 0.27 (0.19, 0.44) | 0.001 | 0.021 | 0.20 (0.00, 0.39) | 0.041 | 0.15 |
| Faecalibacterium | 0.14 (−0.04, 0.31) | 0.12 | 0.25 | 0.03 (−0.25, 0.29) | 0.14 | 0.45 | 0.28 (0.03, 0.52) | 0.021 | 0.15 |
| f__Erysipelotrichaceae;g__Clostridium | 0.10 (−0.06, 0.27) | 0.21 | 0.36 | 0.27 (0.04, 0.50) | 0.021 | 0.10 | −0.06 (−0.34, 0.20) | 0.68 | 0.74 |
| Lachnospiraceae;g__Clostridium | 0.14 (0.01, 0.26) | 0.031 | 0.14 | 0.08 (−0.10, 0.26) | 0.37 | 0.63 | 0.23 (0.04, 0.41) | 0.011 | 0.15 |
| Coprococcus | 0.21 (0.03, 0.38) | 0.011 | 0.11 | 0.19 (−0.07, 0.44) | 0.14 | 0.45 | 0.28 (0.02, 0.52) | 0.031 | 0.15 |
| Bacteroides | 0.19 (−0.01, 0.17) | 0.06 | 0.21 | 0.10 (−0.06, 0.20) | 0.28 | 0.52 | 0.13 (−0.02, 0.26) | 0.08 | 0.25 |
| Lactobacillus | −0.18 (−0.40, 0.02) | 0.09 | 0.22 | −0.47 (−0.91, −0.19) | 0.021 | 0.10 | −0.11 (−0.44, 0.22) | 0.56 | 0.70 |
1The P values indicate statistical significance at 0.05.
β-coefficients and 95% confidence intervals (CIs) from Poisson generalized linear analysis were used to examine the associations between the relative abundance of genera with gram total fiber, soluble fiber, and insoluble fiber intake in infants at 6 mo of age. Fiber intake was measured using 24-h dietary recalls and the abundance of genera was computed using 16S rRNA sequencing. Models adjusted for infants’ mode of delivery, antibiotics, infant feeding type, time of introduction of solid foods, energy intake, and body weight.
FIGURE 2.
Log-normalized relative abundances of bacteria at the genus level that are significantly associated with gram sugar and fiber consumption. Scatter plots present the Poisson generalized linear regression for the Log-normalized relative abundances of bacteria taxa at genus level in relation to dietary intake (g) at 6 mo of age.
The relative abundance of Faecalibacterium was positively associated with insoluble fiber consumption (β = 0.28; 95% CI: 0.03, 0.52; P = 0.02) but no significant association was observed between Faecalibacterium and sugar consumption. Furthermore, the genus Clostridium of family Lachnospiraceae exhibited a negative association with total sugar consumption (β = −0.02; 95% CI: −0.03, −0.00; P = 0.01) and a positive association with total fiber intake (β = 0.14; 95% CI: 0.01, 0.26; P = 0.03). Infants with higher total fiber consumption exhibited an increased relative abundance of Coprococcus (β = 0.21; 95% CI: 0.03, 0.38; P = 0.01), Dorea (β = 0.3; 95% CI: 0.14, 0.49; P ≤ 0.001), and Oscillospira (β = 0.22; 95% CI: 0.09, 0.35; P ≤ 0.01) genera.
Discussion
In this study, we examined the association between dietary carbohydrate intake at 6 mo of age from all sources and the gut microbiome. We identified that dietary sugar and fiber intake at 6 mo of age were associated with specific bacteria at the genus level. Gut microbiome composition has been indicated as a potentially important factor in long-term disease development [41]. It has been hypothesized that alterations in the composition and colonization of the gut microbiome can be altered by increased sugar consumption, which may play a causal role in the development of numerous prevalent westernized diseases, such as obesity, diabetes, cardiovascular disease, liver disease, as well as negative cognitive outcomes [41].
Carbohydrate utilization by gut microbiota affects the stability and diversity of the gut ecosystem and has been linked to glycolipid metabolism diseases [[42], [43], [44]]. High-sugar consumption can alter the carbohydrate pools and create an environment that favors pathogenic microbes [41]. Various carbohydrates regulate the intestinal microbiota, playing a role in alleviating glycolipid metabolism diseases [44]. Short-chain fatty acids (SCFAs) are end products of dietary fiber metabolism [45] and mediate microbiota–host interactions during complementary feeding [20]. Complementary solid foods contribute to the transition of gut microbiota from a Bifidobacterium-rich community toward a more diverse community composition [1].
The variation in the gut microbiota composition is reliant on the variation in sugars existing along the gut. Accumulation of a single sugar can sway microbial abundance [41]. Our results indicate that dietary sugar and fiber intake can be positively or negatively associated with the abundance of multiple bacteria at the genus level. Here, we show the relative abundance of the Bifidobacterium genus significantly increased with a higher intake of total sugar (includes all sources of sugar, including from milk and formula) at 6 mo of age [46]. On the contrary, the consumption of free sugar and added sugar at 6 mo of age is negatively associated with microbes belonging to the Bifidobacterium genus.
We found that a higher consumption of total sugar was associated with the genus Bifidobacterium, a major lactic acid producer, likely because of the presence of lactose in milk and formula. A longitudinal study conducted on Norwegian infants from birth to 3 y of age revealed that the gut microbiome composition transition from infancy to adulthood was partially driven by the disappearance of Bifidobacteria, supporting previous research suggesting that Bifidobacteria act as biological gatekeepers in the succession process of the gut microbiome [47]. In addition, certain genomes of infant gut commensals, particularly those of Bifidobacterial species, are genetically adapted to utilize human milk oligosaccharides found in human milk, leading to a beneficial coevolutionary relationship between the host and microbes [48,49]. A gene-association study of 1126 twins in the U.K. provides mechanistic support for the positive association between Bifidobacteria and lactose via the LCT gene locus that encodes lactase to hydrolyze lactose in the mammalian upper gastrointestinal tract [50]. This genetic association elucidates higher levels of Bifidobacterium in infants with an absence of the enzyme lactase, promoting the growth of lactose-utilizing Bifidobacteria [50].
Our study also found a positive correlation between free sugar intake at 6 mo of age and the relative abundance of Parabacteroides, consistent with previous studies in a rodent model. Noble et al. [51] reported that rats fed excessive sugar had a significantly elevated relative abundance of Parabacteroides, which was negatively correlated with hippocampal-dependent memory function and behavioral outcomes. Furthermore, the administration of Parabacteroides to rodents induced disruption in the performance of hippocampal- and perirhinal cortex-dependent memory resulting in impairment of neurocognitive outcome [51]. Taken together, these findings suggest a mechanistic role of altered gut microbiota induced by early introduction of added sugar on neurocognitive impairments [51]. Parabacteroides have been reported as an opportunistic pathogen in multiple infectious diseases in previous studies [52].
In addition, we observed a trend indicating that a higher intake of free sugar was inversely associated with the abundance of Streptococcus; however, this association did not reach statistical significance. Our study is consistent with previous research on Latino teenagers with obesity, which found a negative association between fructose intake and nonpathogenic Streptococcus bacteria [53]. The underlying mechanism of this association is not well understood, although Streptococcus populations are commonly found and highly active in utilizing and fermenting dietary carbohydrates. Streptococcus populations play a significant role in the primary carbohydrate metabolism of the small intestinal ecosystem, suggesting their ability to metabolize carbohydrates [[53], [54], [55]]. Our findings add to the growing body of literature suggesting that the consumption of free sugars, such as those found in fruit juice, may contribute to an unfavorable gut microbiome composition starting in infancy. High-fructose consumption can alter the gut microbiome through gut barrier disruption, alterations in microbial profile and diversity, and influencing gut microbial metabolites [56].
One possible mechanism linking excess sugars to the gut microbiome in early life is via fructose malabsorption. In early life, infants lack the GLUT-5 transporter, which is specific for fructose absorption [57], and therefore, it is likely that in infants any fructose that is consumed would be metabolized by the gut microbiota and alter its composition [58]. This fructose could be consumed as part of any added sugar or in free form (for example, in fruit juices). Hence, a greater amount of fructose reaches the colon, which may alter gut microbiome composition contributing to the development of obesity and metabolic dysfunction [59]. Given the role of Streptococcus in fructose metabolism, the result of the current study and previous studies explain the lower abundances of the commensal bacteria belonging to the Streptococcus genus in the increased consumption of free sugar. It is essential to recognize that the role of Streptococcus in fructose metabolism may vary between species and strains within the genus. Some strains may have a greater capacity for fructose utilization, whereas others may have a limited or nonexistent capacity for fructose metabolism. The metabolic activities of Streptococcus and their interactions with fructose in the gut environment are complex and influenced by multiple factors, including the gut microbiota's overall composition, dietary factors, and host-specific characteristics [55,60].
In our study, we observed a higher abundance of Faecalibacterium and Coprococcus in relation to increased consumption of insoluble fiber intake, which is consistent with previous research. One study investigated the influence of prenatal, perinatal, and postnatal factors, with a specific emphasis on nutrition on the composition of gut microbiota in 1-y-old children [61]. They have shown that high abundances of Faecalibacterium have been linked to infants introduced to solid foods from 4 mo of age. Faecalibacterium and Coprococcus are known for their role in butyrate production in the human gut, particularly through the fermentation of nondigestible dietary fibers [39,61]. These fibers, found in foods, such as whole wheat flour, brown rice, nuts, beans, and vegetables, undergo high fermentation by the gut microbiota, resulting in the production of SCFAs, such as butyrate [62]. Furthermore, SCFAs, including butyrate, are important for epithelial barrier function. Butyrate has a bidirectional effect on epithelial barrier function, promoting the production of tight junction proteins and enhancing mucosal integrity [63]. These findings highlight the potential role of insoluble fiber in shaping the abundance of specific bacterial genera, such as Faecalibacterium and Coprococcus, and their production of beneficial SCFAs, ultimately influencing gut health and epithelial barrier function.
During the introduction of solid foods, some genera that were not previously detected at high abundances in the gut microbiota expand, suggesting that changes in dietary behaviors or nutritional components could potentially facilitate the establishment of new microbiome community compositions. This observation is in agreement with the concept that microbial communities are dynamic and subject to changes, particularly during periods of intense disturbances, such as the transition to solid foods [64,65]. It highlights the importance of studying the development of the infant gut microbiota during critical periods of growth and development to better understand the factors that shape the microbial composition and potential long-term health outcomes [65]. To our knowledge, this study presents a novel contribution to the field, as it is the first to investigate the impact of dietary intake at 6 mo in conjunction with fecal sampling during a crucial dietary transition in infants among Latinos, a segment of the population with increased susceptibility to chronic disease development.
There are several limitations to this study that we are hoping to address in future work. The measurement of true SCFA production rates is urgently needed, as well as an understanding of how specific carbohydrates and bacteria influence SCFA composition. In addition, conducting rigorous studies with the ability to evaluate longitudinal changes in infant gut microbiome at species-level resolution and metagenomic functionality using whole-genome sequencing techniques is warranted. The reliance on dietary data reported by caregivers may be subject to recall bias and measurement errors. In addition, the study did not directly measure the nutrient composition of breast milk. Although the NDSR provides a comprehensive database for estimating nutrient values, it is important to acknowledge that the composition of breast milk can vary among individuals and over time. However, our previous investigations [66,67] have revealed a negligible presence of free sugars, beyond lactose, in breast milk. Furthermore, the study focused on a specific population of Latino infants and its findings may not be generalizable to other demographic groups.
Despite these limitations, this study provides valuable insights into the association between dietary factors and gut microbiota composition in the studied population at a critical period of nutritional transition at 6 mo of age. Future research should consider more precise measurements of breast milk nutrients and include larger and more diverse populations to enhance the generalizability of the findings. Further longitudinal studies with the possibility of tracking these infants would be beneficial in providing insight into microbiome development and maturation. It would be interesting to observe how the dietary intake at different time points impacts the gut microbiome diversity and composition over time. In addition, tracking infants beyond the 6-mo time point would allow for the observation of how the gut microbiome continues to develop and mature through infancy and into early childhood. These types of longitudinal studies would provide valuable information for understanding the complex interplay between early-life dietary patterns, gut microbiome development, and long-term health outcomes.
In conclusion, our study found that the introduction of complementary foods at 6 mo of age had significant impacts on the developing microbiome, which may have significant implications for future growth and development. The results from the sample of Latino infants showed associations between dietary sugar and specific genera, including opportunistic or sugar-digesting bacteria as well as Parabacteroides, which has been linked to alterations in hippocampal and memory development in rodent studies. Conversely, the early introduction of dietary fiber has beneficial effects, including increases in favorable SCFA-producing bacteria. These findings support recommendations for avoiding added and free sugars in early life and promoting higher fiber intake while introducing solid foods. Although these initial findings are limited to a cross-sectional analysis at 6 mo, they provide a foundation for future longitudinal analysis to explore how early-life dietary factors impact the developing microbiome, potentially affecting future outcomes related to obesity, diabetes, other chronic diseases, and cognitive development in children.
Author contributions
The authors’ responsibilities were as follows – PM, MIG, TLA: designed the research; MIG: provided essential materials; PM, EAH, BNC, CJM: analyzed the data; PM, TLA, KAS, MB, SA: interpreted the results; PM: wrote the paper; MIG, PM: had primary responsibility for the final content; and all authors: read and approved the final manuscript.
Conflict of interest
MIG is a scientific advisor for Yumi Foods and Else Nutrition and receives book royalties from Penguin Random House. All other authors have no conflicts of interest to disclose.
Funding
This research was funded by the National Institute of Diabetes and Digestive Kidney Diseases (R01DK110793), the Gerber Foundation (15PN-013), The Southern California Center for Latino Health (P50 MD017344), and the Atkins Foundation.
Data availability
Data described in the manuscript, code book, and analytic code will be made available upon request.
Acknowledgments
We would like to thank the following individuals for their expertise and assistance throughout all aspects of our study: Carla Flores, Elizabeth Campbell, Yessica Corona, Yareli, Rosalba Cain, Estella Duran, Claudia Rios, and Jennifer Fogel. We would also like to acknowledge The Saban Research Institute for providing additional funding for this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tjnut.2023.09.009.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Laursen M.F. Gut microbiota development: influence of diet from infancy to toddlerhood. Ann. Nutr. Metab. 2021;77(3):1–14. doi: 10.1159/000517912. [DOI] [PubMed] [Google Scholar]
- 2.Bittinger K., Zhao C., Li Y., Ford E., Friedman E.S., Ni J., et al. Bacterial colonization reprograms the neonatal gut metabolome. Nat. Microbiol. 2020;5(6):838–847. doi: 10.1038/s41564-020-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamburini S., Shen N., Wu H.C., Clemente J.C. The microbiome in early life: implications for health outcomes. Nat. Med. 2016;22(7):713–722. doi: 10.1038/nm.4142. [DOI] [PubMed] [Google Scholar]
- 4.Robertson R.C., Manges A.R., Finlay B.B., Prendergast A.J. The human microbiome and child growth–first 1000 days and beyond. Trends Microbiol. 2019;27(2):131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Y., Cai X., Ye Y., Wang F., Chen F., Zheng C. The role of microbiota in infant health: from early life to adulthood. Front. Immunol. 2021;12:708472. doi: 10.3389/fimmu.2021.708472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charbonneau M.R., Blanton L.V., DiGiulio D.B., Relman D.A., Lebrilla C.B., Mills D.A., et al. A microbial perspective of human developmental biology. Nature. 2016;535(7610):48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Homann C.M., Rossel C.A.J., Dizzell S., Bervoets L., Simioni J., Li J., et al. Infants’ first solid foods: impact on gut microbiota development in two intercontinental cohorts. Nutrients. 2021;13(8):2639. doi: 10.3390/nu13082639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maher S.E., O’Brien E.C., Moore R.L., Byrne D.F., Geraghty A.A., Saldova R., et al. The association between the maternal diet and the maternal and infant gut microbiome: a systematic review. Br. J. Nutr. 2020:1–29. doi: 10.1017/S0007114520000847. [DOI] [PubMed] [Google Scholar]
- 11.Stewart C.J., Ajami N.J., O’Brien J.L., Hutchinson D.S., Smith D.P., Wong M.C., et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galazzo G., van Best N., Bervoets L., Dapaah I.O., Savelkoul P.H., Hornef M.W., et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. 2020;158(6):1584–1596. doi: 10.1053/j.gastro.2020.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Fragkou P.C., Karaviti D., Zemlin M., Skevaki C. Impact of early life nutrition on children’s immune system and noncommunicable diseases through its effects on the bacterial microbiome, virome and mycobiome. Front. Immunol. 2021;12:644269. doi: 10.3389/fimmu.2021.644269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laursen M.F., Bahl M.I., Michaelsen K.F., Licht T.R. First foods and gut microbes. Front. Microbiol. 2017;8:356. doi: 10.3389/fmicb.2017.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fallani M., Amarri S., Uusijarvi A., Adam R., Khanna S., Aguilera M., et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology (Reading) 2011;157(5):1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 16.Bureau U.S.C. United States Census Bureau; Washington, DC, USA: 2021. 2020 Census Statistics Highlight Local Population Changes and Nation’s Racial and Ethnic Diversity. [Google Scholar]
- 17.Riley A.R., et al. Excess Mortality Among Latino People in California During the COVID-19 Pandemic. SSM Popul Health. 2021;15:100860. doi: 10.1016/j.ssmph.2021.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan R.C., Wang Z., Usyk M., Sotres-Alvarez D., Daviglus M.L., Schneiderman N., et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019;20(1):219. doi: 10.1186/s13059-019-1831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck A.L., Hoeft K.S., Takayama J.I., Barker J.C. Beliefs and practices regarding solid food introduction among Latino parents in Northern California. Appetite. 2018;120:381–387. doi: 10.1016/j.appet.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alderete T.L., Jones R.B., Shaffer J.P., Holzhausen E.A., Patterson W.B., Kazemian E., et al. Early life gut microbiota is associated with rapid infant growth in Hispanics from Southern California. Gut Microbes. 2021;13(1):1961203. doi: 10.1080/19490976.2021.1961203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alderete T.L., Wild L.E., Mierau S.M., Bailey M.J., Patterson W.B., Berger P.K., et al. Added sugar and sugar-sweetened beverages are associated with increased postpartum weight gain and soluble fiber intake is associated with postpartum weight loss in Hispanic women from Southern California. Am. J. Clin. Nutr. 2020;112(3):519–526. doi: 10.1093/ajcn/nqaa156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger P.K., Plows J.F., Jones R.B., Pollock N.K., Alderete T.L., Ryoo J.H., et al. Maternal blood pressure mediates the association between maternal obesity and infant weight gain in early postpartum. Pediatr. Obes. 2019;14(11) doi: 10.1111/ijpo.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook L.T., O'Reilly G.A., Goran M.I., Weigensberg M.J., Spruijt-Metz D., Davis J.N. Vegetable consumption is linked to decreased visceral and liver fat and improved insulin resistance in overweight Latino youth. J. Acad. Nutr. Diet. 2014;114:1776–1783. doi: 10.1016/j.jand.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis J.N., Le K.A., Walker R.W., Vikman S., Spruijt-Metz D., Weigensberg M.J., et al. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am. J. Clin. Nutr. 2010;92(6):1522–1527. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization . Guideline. World Health Organization; Geneva, Switzerland: 2015. Sugars intake for adults and children. [Google Scholar]
- 26.MINNESOTA U.O. NDSR); 2021. Nutrition Data System for Research. [Google Scholar]
- 27.Bailey M.J., Holzhausen E.A., Morgan Z.E.M., Naik N., Shaffer J.P., Liang D., et al. Postnatal exposure to ambient air pollutants is associated with the composition of the infant gut microbiota at 6-months of age. Gut Microbes. 2022;14(1):2105096. doi: 10.1080/19490976.2022.2105096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.16S Illumina Amplicon Protocol: earthmicrobiome. 2022. https://earthmicrobiome.org/protocols-and-standards/16s/ [Internet] [cited October 12. Available from: [Google Scholar]
- 29.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez A., Navas-Molina J.A., Kosciolek T., McDonald D., Vázquez-Baeza Y., Ackermann G., et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods. 2018;15(10):796–798. doi: 10.1038/s41592-018-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., et al. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2017;2(2):e00191. doi: 10.1128/mSystems.00191-16. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen S., McDonald D., Gonzalez A., Navas-Molina J.A., Jiang L., Xu Z.Z., et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3(3):e00021. doi: 10.1128/mSystems.00021-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen .E., Knight R., et al. Optimizing taxonomic classification of marker gene amplicon sequences. PeerJ. 2018;6 doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aldirawi H., Morales F.G. Univariate and multivariate statistical analysis of microbiome data: an overview. Appl. Microbiol. 2023;3(2):322–338. [Google Scholar]
- 35.Schober P., Vetter T.R. Count Data in Medical Research: Poisson Regression and Negative Binomial Regression. Anesth Analg. 2021;132(5):1378–1379. doi: 10.1213/ANE.0000000000005398. [DOI] [PubMed] [Google Scholar]
- 36.Kim H.Y. Statistical notes for clinical researchers: simple linear regression 3–residual analysis. Restor. Dent. Endod. 2019;44(1):e11. doi: 10.5395/rde.2019.44.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morelan I.A., Gaulke C.A., Sharpton T.J., Vega Thurber R., Denver D.R. Microbiome variation in an intertidal sea anemone across latitudes and symbiotic states. Front. Mar. Sci. 2019;6:7. [Google Scholar]
- 38.Kartal E., Schmidt T.S., Molina-Montes E., Rodríguez-Perales S., Wirbel J., Maistrenko O.M., et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut. 2022;71(7):1359–1372. doi: 10.1136/gutjnl-2021-324755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vijay A., Valdes A.M. Role of the gut microbiome in chronic diseases: a narrative review. Eur. J. Clin. Nutr. 2022;76(4):489–501. doi: 10.1038/s41430-021-00991-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Cui Y., Zhang L., Wang X., Yi Y., Shan Y., Liu B., et al. Roles of intestinal Parabacteroides in human health and diseases. FEMS Microbiol. Lett. 2022;369(1) doi: 10.1093/femsle/fnac072. fnac072. [DOI] [PubMed] [Google Scholar]
- 41.Di Rienzi S.C., Britton R.A. Adaptation of the gut microbiota to modern dietary sugars and sweeteners. Adv. Nutr. 2020;11(3):616–629. doi: 10.1093/advances/nmz118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonnenburg E.D., Smits S.A., Tikhonov M., Higginbottom S.K., Wingreen N.S., Sonnenburg J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529(7585):212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng X., Zheng J., Lin A., Xia H., Zhang Z., Gao Q., et al. A review: roles of carbohydrates in human diseases through regulation of imbalanced intestinal microbiota. J. Funct. Foods. 2020;74:104197. [Google Scholar]
- 44.Jardon K.M., Canfora E.E., Goossens G.H., Blaak E.E. Dietary macronutrients and the gut microbiome: a precision nutrition approach to improve cardiometabolic health. Gut. 2022;71(6):1214–1226. doi: 10.1136/gutjnl-2020-323715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott K.P., Duncan S.H., Flint H.J. Dietary fibre and the gut microbiota. Nutr. Bull. 2008;33(3):201–211. [Google Scholar]
- 46.Mela D.J., Woolner E.M. Perspective: total, added, or free? What kind of sugars should we be talking about? Adv. Nutr. 2018;9(2):63–69. doi: 10.1093/advances/nmx020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Avershina E., Lundgård K., Sekelja M., Dotterud C., Storrø O., Øien T., et al. Transition from infant- to adult-like gut microbiota. Environ. Microbiol. 2016;18(7):2226–2236. doi: 10.1111/1462-2920.13248. [DOI] [PubMed] [Google Scholar]
- 48.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017;81(4):e00036. doi: 10.1128/MMBR.00036-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walsh C., Lane J.A., van Sinderen D., Hickey R.M. Human milk oligosaccharides: shaping the infant gut microbiota and supporting health. J. Funct. Foods. 2020;72:104074. doi: 10.1016/j.jff.2020.104074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodrich J.K., Davenport E.R., Beaumont M., Jackson M.A., Knight R., Ober C., et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19(5):731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noble E.E., Olson C.A., Davis E., Tsan L., Chen Y.W., Schade R., et al. Gut microbial taxa elevated by dietary sugar disrupt memory function. Transl. Psychiatry. 2021;11(1):194. doi: 10.1038/s41398-021-01309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boente R.F., Ferreira L.Q., Falcão L.S., Miranda K.R., Guimarães P.L., Santos-Filho J., et al. Detection of resistance genes and susceptibility patterns in Bacteroides and Parabacteroides strains. Anaerobe. 2010;16(3):190–194. doi: 10.1016/j.anaerobe.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Jones R.B., Alderete T.L., Kim J.S., Millstein J., Gilliland F.D., Goran M.I. High intake of dietary fructose in overweight/obese teenagers associated with depletion of Eubacterium and Streptococcus in gut microbiome. Gut Microbes. 2019;10(6):712–719. doi: 10.1080/19490976.2019.1592420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hutkins R.W., Morris H.A. Carbohydrate metabolism by Streptococcus thermophilus: a review. J. Food Prot. 1987;50(10):876–884. doi: 10.4315/0362-028X-50.10.876. [DOI] [PubMed] [Google Scholar]
- 55.van den Bogert B., Erkus O., Boekhorst J., Goffau M.d., Smid E.J., Zoetendal E.G., et al. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol. Ecol. 2013;85(2):376–388. doi: 10.1111/1574-6941.12127. [DOI] [PubMed] [Google Scholar]
- 56.Hsu C.N., Yu H.R., Chan J.Y.H., Wu K.L.H., Lee W.C., Tain Y.L. The impact of gut microbiome on maternal fructose intake-induced developmental programming of adult disease. Nutrients. 2022;14(5):1031. doi: 10.3390/nu14051031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferraris R.P., Choe J.Y., Patel C.R. Intestinal absorption of fructose. Annu. Rev. Nutr. 2018;38:41–67. doi: 10.1146/annurev-nutr-082117-051707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramne S., Brunkwall L., Ericson U., Gray N., Kuhnle G.G.C., Nilsson P.M., et al. Gut microbiota composition in relation to intake of added sugar, sugar-sweetened beverages and artificially sweetened beverages in the Malmö Offspring Study. Eur. J. Nutr. 2021;60(4):2087–2097. doi: 10.1007/s00394-020-02392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Payne A.N., Chassard C., Lacroix C. Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols: implications for host–microbe interactions contributing to obesity. Obes. Rev. 2012;13(9):799–809. doi: 10.1111/j.1467-789X.2012.01009.x. [DOI] [PubMed] [Google Scholar]
- 60.Vernocchi P., Del Chierico F., Putignani L. Gut microbiota metabolism and interaction with food components. Int. J. Mol. Sci. 2020;21(10):3688. doi: 10.3390/ijms21103688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vacca M., Raspini B., Calabrese F.M., Porri D., De Giuseppe R., Chieppa M., et al. The establishment of the gut microbiota in 1-year-aged infants: from birth to family food. Eur. J. Nutr. 2022;61(5):2517–2530. doi: 10.1007/s00394-022-02822-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Portincasa P., Bonfrate L., Vacca M., De Angelis M., Farella I., Lanza E., et al. Gut microbiota and short chain fatty acids: implications in glucose homeostasis. Int. J. Mol. Sci. 2022;23(3):1105. doi: 10.3390/ijms23031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Silva Y.P., Bernardi A., Frozza R. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. (Lausanne). 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boldison J., Wong F.S. Immune and pancreatic β cell interactions in type 1 diabetes, Trends Endocrinol. Metab. 2016;27(12):856–867. doi: 10.1016/j.tem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Ronan V., Yeasin R., Claud E.C. Childhood development and the microbiome-the intestinal microbiota in maintenance of health and development of disease during childhood development. Gastroenterology. 2021;160(2):495–506. doi: 10.1053/j.gastro.2020.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berger P.K., Plows J.F., Demerath E.W., Fields D.A. Carbohydrate composition in breast milk and its effect on infant health. Curr. Opin. Clin. Nutr. Metab. Care. 2020;23(4):277–281. doi: 10.1097/mco.0000000000000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goran M.I., Martin A.A., Alderete T.L., Fujiwara H., Fields D.A. Fructose in breast milk is positively associated with infant body composition at 6 months of age. Nutrients. 2017;9(2) doi: 10.3390/nu9020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript, code book, and analytic code will be made available upon request.