Abstract
Compound-protein interactions (CPIs) are critical in drug discovery for identifying therapeutic targets, drug side effects, and repurposing existing drugs. Machine learning (ML) algorithms have emerged as powerful tools for CPI prediction, offering notable advantages in cost-effectiveness and efficiency. This review provides an overview of recent advances in both structure-based and non-structure-based CPI prediction ML models, highlighting their performance and achievements. It also offers insights into CPI prediction-related datasets and evaluation benchmarks. Lastly, the article presents a comprehensive assessment of the current landscape of CPI prediction, elucidating the challenges faced and outlining emerging trends to advance the field.
Keywords: compound-protein interaction prediction, drug discovery, artificial intelligence, scoring function, chemogenomics
Background
The affinity between a ligand molecule and a target reflects how tightly the ligand binds to a particular target. The identification of compound-protein interactions (CPI) plays a decisive role in drug discovery as it provides insights into therapeutic targets [1], drug side effects [2], and the new use of old drugs [3]. However, the experimental determination of compound-protein interactions (CPIs), quantified by measures such as the dissociation constant (Kd), inhibition constant (Ki), half maximal inhibitory concentration (IC50), etc., is often time-consuming and laborious. Furthermore, experimental methods remain limited both in coverage and throughput [4]. To systematically prioritize and speed up experimental work, researchers have developed many computational methods to predict CPI.
Recent developments in the field of artificial intelligence (AI) have brought new opportunities for drug discovery. AI provides promising tools for several areas of pharmacology, including prediction of protein-ligand interactions (PLI), drug-target interactions (DTI), and protein-protein interactions (PPI). CPI prediction is one of the highlights [5]. Various ligand- and/or structure-based machine learning (ML) models have been developed to study the relationships between compounds and their potential target space, such as Support Vector Machines (SVM) [6], Random Forests (RF) [7], Gaussian Processes [8], and Boosting [9]. In recent years, with the increasing publication of large-scale CPI datasets, such as PDBbind [10], CrossDocked [11], BindingDB [12], and DrugBank [13], the applications of AI have enhanced chemogenomics approaches, quantitative structure activity relationships (QSARs), and molecular docking [14], [15], [16], [17], showing improved accuracy and efficiency on CPI prediction [18].
The decision to establish a CPI prediction model greatly depends on whether the crystal structure of the target protein is present or not. The protein’s crystal structure provides more interaction information, which helps establish a model with better predictive performance. However, most proteins’ crystal structures have not been resolved, and determining the three-dimensional structure of a protein experimentally by X-ray crystallography and nuclear magnetic resonance (NMR) can be resource-intensive and time-consuming. Therefore, many CPI prediction models based on non-structural information have been proposed.
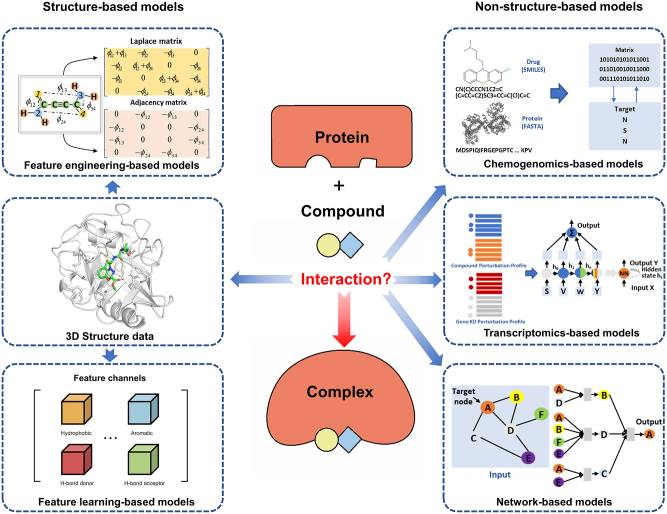
In this review, we categorize CPI prediction models into two types: structure-based and non-structure-based models, depending on whether three-dimensional (3D) structure data of the protein is utilized (Figure 1). Protein structure-based CPI prediction introduces representative structure-based CPI prediction models, including scoring functions and conformational prediction models. Non-structure-based CPI prediction introduces recent advances in non-structure-based CPI prediction models, including chemogenomics-based models, transcriptomics-based models, and network-based models. Additionally, we describe commonly used datasets and evaluation benchmarks for CPI prediction. In Discussion we summarize the current status of CPI prediction models, identify trends in the field, and propose strategies for developing better CPI prediction models.
Figure 1:
Categorization of compound-protein interaction (CPI) prediction models. FASTA, FAST-All; SMILES, Simplified molecular input line entry system.
Protein structure-based CPI prediction
Compound-protein interactions occur in three-dimensional space. The 3D structural data of protein-compound interactions can provide visualization of the important interaction motifs between them. This information can guide the structural modification of compounds to improve their binding affinity and selectivity, as well as help to explain the causes of activity cliffs, which are drastic changes in biological activity caused by small changes in compound structure. Therefore, using the 3D structural data of compound-protein complexes can lead to more accurate prediction of their interactions and guide the rational design of compounds, improving the efficiency of the drug discovery process.
While free energy perturbation (FEP) [19] and thermodynamic integration (TI) [20] can predict binding free energy more accurately, their applications are limited due to low computational efficiency. As an alternative, the two-end-state free energy calculation approaches, molecular mechanics energies combined with the Poisson-Boltzmann or generalized Born and surface area continuum solvation (MM/PBSA and MM/GBSA) methods, provide a balance between speed and accuracy but still rely on time-consuming molecular dynamics simulations for conformational sampling [21].
To address the need for rapid binding affinity predictions between molecules and targets in large-scale compound libraries, molecular docking has been developed to obtain compound-protein interactions and affinities accurately and rapidly. The reliability of molecular docking largely depends on the accuracy of the adopted scoring function (SF), which is used to determine the binding mode and site of a ligand and predict its binding affinity for a given protein target. A classical SF assumes a predetermined theory-inspired functional form for the relationship between the features characterizing the structure of the protein–ligand complex and its predicted binding affinity, while machine learning-based scoring functions (MLSFs) utilize machine learning algorithms to capture the nonlinear relationship between features and binding strength rather than relying on linear regression methods. More recently, geometric deep learning models enable direct-shot prediction of the optimal ligand conformation within the protein pocket to complete the docking process [22].
Overview of scoring functions
Scoring functions can be methodologically divided into four categories: physics-based, empirical, knowledge-based, and MLSFs [23]. The initial three classical scoring functions primarily employ the linear regression method, albeit with variations in the types of feature items they incorporate. In contrast, MLSFs integrate a nonlinear regression machine-learning approach (Table 1).
Table 1:
Summary of four types of scoring functions.
| Scoring function | Description | Representative methods |
|---|---|---|
| Physics-based | Calculate the interactions between protein and ligand atoms by summing the energy terms obtained from a molecular mechanics force field | GoldScore [27]; UCSF DOCK [28] |
| Empirical | A linear function of predetermined energetic factors, whose weights are determined through regression analysis using experimental data | ChemScore [31]; PLP [32]; X-score [33]; GlideScore [34] |
| Knowledge-based | The pairwise sum of statistical potentials between atom pairs from protein-ligand complexes | PMF [36]; ASP [37]; DrugScore [38] |
| Machine learning-based | Utilize non-linear machine learning algorithms to capture the nonlinear relationship between features and binding strength | NNscore [42]; RF-score [43]; SFCscoreRF [44]; ID-score [45] |
Physics-based SFs use linear additive energy terms derived from a molecular mechanics force-field to directly compute the interactions between the atoms of protein and ligand. The non-bond interaction energy is usually expressed as the sum of van der Waals and electrostatic interaction terms. Additional shorter-range terms are added to account for hydrogen bond. Considering entropy and solvent effect by incorporating the torsion entropy of ligand and the solvation/desolvation effect can further improve the predictive accuracy of Physics-based SFs. Recent research has also introduced SFs based on quantum mechanics (QM) and hybrid quantum mechanical/molecular mechanics (QM/MM) approach to address challenges related to covalent interactions, polarization, and charge transfer. However, MM or QM models of physics-based SFs are computationally expensive [24], [25], [26]. Representative methods within physics-based SFs include GoldScore [27] and UCSF DOCK [28]. Their general functional form is as follows:
| (1) |
The empirical SFs decompose protein–ligand binding affinities into several individual energetic factors, such as hydrogen bonding, hydrophobic effects, steric clashes, etc. The functional form of them is similar to physics-based scoring functions, but the weights of their energetic factors are optimized by linear regression analysis employing a training set with known binding affinities [25]. The flexible and intuitive functional form for these simple energy terms allows the development for customized empirical SFs to enhance performance for specific molecular systems [29]. However, they often face challenges related to double-counting issues [30]. The majority of widely-used docking software applications, including AutoDock, GOLD, and Glide, rely on empirical SFs. Notably, these include ChemScore [31], PLP [32], X-Score [33], and GlideScore [34]. For example, ChemScore was trained by regression against binding affinity data for 82 complexes. It takes account rewarding scores (“S”) for hydrogen bonding, coordination bonds with metal ions, and lipophilic contacts. It also assigns penalties (“P”) for frozen rotatable bonds, the internal strain energy of the ligand, and steric clashes between the protein and ligand, resulting in the following formula:
| (2) |
Knowledge-based SFs are based on the pairwise sum of statistical potentials between interacting atom pairs from protein-ligand complexes by applying an inverse Boltzmann analysis in a large data set of protein-ligand complexes. The primary advantage of knowledge-based scoring functions lies in their conceptual simplicity and computational efficiency [23]. However, their drawback arises from the fact that the assessment of atom pair distributions and frequencies does not fully align with actual scenarios [35]. Representative methods within empirical SFs include PMF [36], ASP [37], DrugScore [38]. Its general functional form is as follows:
| (3) |
| (4) |
Here, ρ ij (r) is the number density of pairs of type i-j at distance r while is the same quantity for a reference state where there is no interaction between types i and j. Then, with the relative number density of atom pairwise i-j at distance r, denoted as g ij (r), Boltzmann constant k B , and the absolute temperature T, the distance-dependent potential between atom pair i-j ω ij (r) can be obtained. Finally, the pairwise statistical potentials between protein and ligand are summed up to represent binding strength.
The introduction of ML techniques has been a breakthrough for structure-based drug design (SBDD), and many papers have reported outstanding performance of ML-based compound-protein binding prediction, especially MLSFs in the last decade [39], [40], [41]. Unlike classical SFs, MLSFs utilize machine learning algorithms to capture the nonlinear relationship between features and binding strength rather than relying on linear regression methods. This allows MLSFs to accurately describe complicated relationships in many systems. Additionally, they offer better flexibility as they do not require a predefined function form.
The construction of ML-based compound-protein binding prediction model can be divided into four steps: (1) preparing protein-ligand complex datasets for training and testing, (2) characterizing the protein-ligand interaction, (3) using ML algorithms to learn the relationship between features and binding strength, and (4) using various evaluation metrics for evaluation. According to the Comparative Assessment of Scoring Functions-2016 (CASF) [46], an SF should be assessed for four aspects: (1) Scoring power, which refers to the ability of SFs to reproduce the linear correlation between predicted and experimental values. (2) Ranking power, which refers to the ability of SFs to accurately rank the molecules of a specific target. (3) Docking power, which refers to the ability of SFs to identify the natural binding conformation from a set of decoy conformations. (4) Screening power, which refers to the ability of SFs to distinguish active molecules from inactive ones in virtual screening (VS).
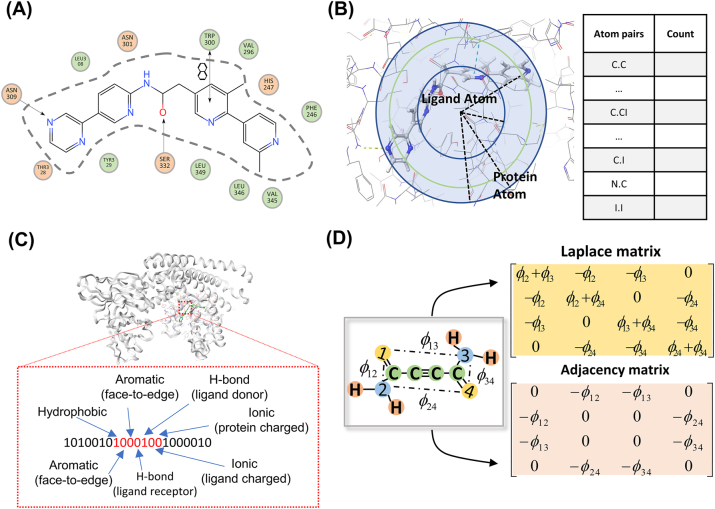
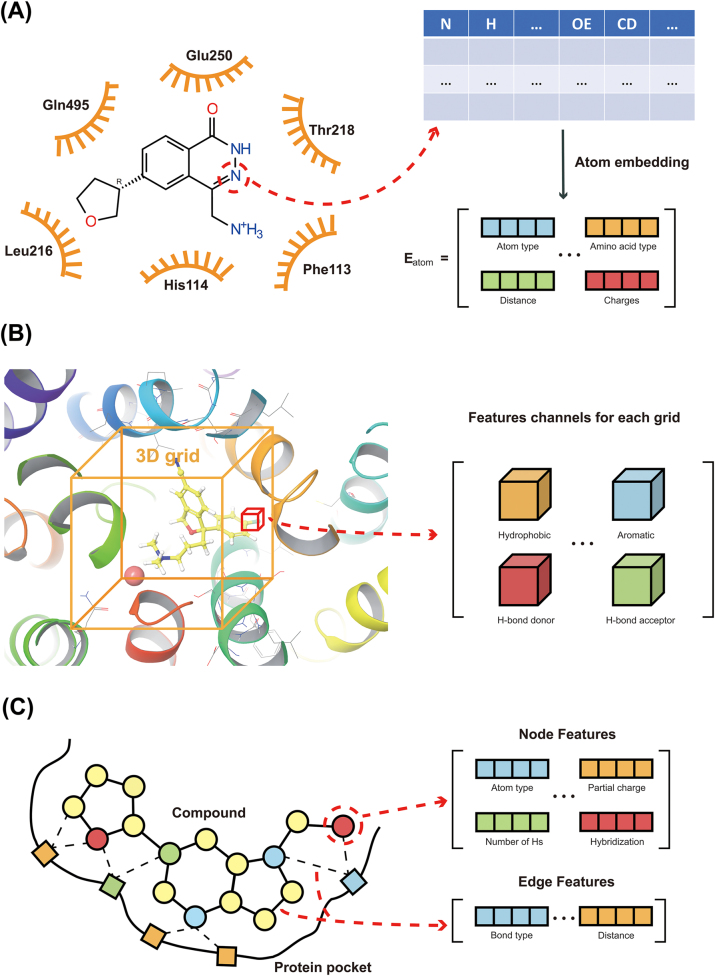
The protein-ligand complex featurization strategy plays the key role on the capacity of MLSFs. Featurization strategies can be classified into two categories based on the way of feature extraction: feature engineering-based strategies and feature learning-based strategies [47]. Feature engineering-based strategies require manual design and selection of feature, including specific energy features, pairwise counts and potentials of protein-ligand atoms, protein-ligand interaction fingerprints, and mathematical features (Figure 2 and Table 2). Feature learning-based strategies, on the other hand, automatically extract features using end-to-end deep learning algorithms. This method can obtain diverse, nonlinear, and latent feature sets to achieve better performance on sufficient data. The features extracted by such strategies can be further classified as atom context-based features, grid-based features, and graph-based features (Figure 3 and Table 3). Furthermore, the SE(3)-equivariant and invariant-based graph models expanded feature learning-based MLSFs, enabling the completion of the entire docking process. In the next section, we will introduce these featurization strategies and representative recent work in the field of ML-based compound-protein binding prediction.
Figure 2:
Feature engineering-based strategies. (A) Specific energy terms. (B) Protein-ligand atom pairwise counts. (C) Protein-ligand interaction fingerprints. (D) Mathematical features.
Table 2:
Feature engineering-based MLSFs.
| Model | Backbone | Main application |
|---|---|---|
| Specific energy terms | ||
| ID-score [45] | SVM | Scoring |
| SFCscoreRF [44] | RF | Scoring |
| ΔvinaRF20 [48] | RF | Scoring, ranking, pose prediction, VS |
| ΔvinaXGB [50] | XGBoost | Scoring, ranking, docking, VS |
| ΔLin_F9XGB [51] | XGBoost | Scoring, ranking, docking, VS |
| EAT-score [52] | RF, XGBoost | Scoring, ranking, docking, VS |
| Protein-ligand atom pairwise counts | ||
| NNscore [42] | ANN | VS |
| RF-score [43] | RF | Scoring |
| RF-score-v2 [53] | RF | Scoring |
| RF-score-v3 [54] | RF | Scoring |
| HydraMap [55, 56] | RF | Scoring |
| Protein-ligand interaction fingerprints | ||
| Pharm-IF [59] | SVM, NBC, RF, ANN | VS |
| PLEC-FP [60] | LR, RF, ANN | Scoring |
| Ding et al. [63] | SVM | VS |
| MIEC-GBDT [64] | GBDT | VS |
| LUNA [62] | DNN | Scoring, ranking, docking, VS |
| Mathematical features | ||
| T-bind [65] | GBDT | Scoring |
| TopologyNet [66] | CNN | Scoring |
| TopBP, TopVS [67] | GBDT, CNN | Scoring, VS |
| AGL-score [68] | GBDT | Scoring |
| Wee et al. [69] | GBT | Scoring |
SVM, support vector machine; RF, random forest; VS, virtual screening; XGBoost, eXtreme gradient boosting; ANN, artificial neural network; NBC, naive bayes classifier; LR, logistic regression; GBT, gradient boosting tree; DNN, deep neural networks; GBDT, gradient boosting decision tree; CNN, convolutional neural networks.
Figure 3:
Feature learning-based representation strategies. (A) Atom context-based features. (B) Grid-based features. (C) Graph-based features.
Table 3:
Feature learning-based MLSFs.
| Model | Backbone | Main application |
|---|---|---|
| Atom context-based features | ||
| DeepVS [71] | CNN | VS |
| DeepDock [72] | GNN | VS |
| RTMScore [73] | Graph transformer | Scoring, ranking |
| Grid-based features | ||
| Pafnucy [74] | CNN | Scoring |
| Kdeep [75] | CNN | Scoring |
| GNINA [76] | CNN | Docking |
| Graph-based features | ||
| PotentialNet [77] | GNN | Scoring |
| InteractionGraphNet [78] | GNN | Scoring, VS |
| PIGNet [79] | GNN | Docking, VS, ranking |
| SE(3)-equivariant and invariant-based features | ||
| EquiBind [22] | IEGMN | Binding pose prediction |
| TankBind [83] | GNN | Binding pose prediction |
| DiffDock [84] | Diffusion | Binding pose prediction |
| Uni-mol [85] | Transformer | Binding pose prediction |
CNN, convolutional neural networks; VS, virtual screening; GNN, graph neural networks; IEGMN, independent E(3)-equivariant graph matching network.
Feature engineering-based MLSFs
Featurization based on energy terms
Empirical SFs provide energy terms such as van der Waals and electrostatic potentials that are commonly used as features when developing MLSFs. These energy terms encompass all physicochemical factors related to molecular recognition, including enthalpy, desolvation, entropy, and pure ligand or receptor information. Representative SFs include ΔvinaRF20 [48], SFCscoreRF [44], and ID-Score [45].
SFCscoreRF shares most of the descriptors of SFCscore, including rotatable bonds, hydrogen bonds, and hydrophobic area, among others. The previous regression algorithm used for training has been replaced with the machine learning algorithm RF, using data from PDBbind. The IDscore is similar to SFCscoreRF. ΔvinaRF20 predicts the correction term of Autodock Vina SFs using RF instead of predicting the binding affinity, resulting in improved accuracy. The data set includes natural conformations from the decoys set of CASR and weak affinity conformations from the General Set of PDBbind-v2014. Feature extraction involves selecting 10 energy terms from AutoDock Vina and 10 feature terms related to solvent accessible surface area from the MSMS program [49]. The model outperformed other SFs on the CASF-2007 and CASF-2016 benchmarks. In 2019, Zhang et al. optimized ΔvinaRF20 to obtain ΔvinaXGB, which uses eXtreme Gradient Boosting (XGBoost) instead of RF for improved scoring accuracy and stability [50]. The extra consideration of ligand-bound water molecules as features improves the scoring performance. In 2022, Zhang et al. further developed ΔvinaXGB to obtain ΔLin_F9XGB, which performs even better on the CASF-2016 [51]. These results suggest that optimizing correction terms using AI algorithms is a promising way to improve SFs’ performance effectively.
Ye et al. proposed a novel SF called EAT-Score, which directly utilizes the energy auxiliary terms (EAT) from molecular docking scoring through XGBoost [52]. The model combines various energy terms from several classical SFs and protein-ligand interaction fingerprint information for prediction. The performance of EAT-Score in discriminating actives from decoys was validated on DUD-E using different performance metrics. The results showed that EAT-Score outperformed classical SFs in virtual screening, with ROC (area under Receiver Operating Characteristic curve) values exhibiting an improvement of around 0.3.
Featurization based on protein-ligand atom pairwise counts
The featurization strategy based on protein-ligand atom pairwise counts is similar to classical knowledge-based SFs. Atom pairwise counts or potentials between protein and ligand atoms are used to represent protein-ligand interactions, based on the assumption that the frequency of atom pairwise counts is correlated with their importance. This strategy is more computationally efficient and can provide more structural information about the complexes, as represented by SFs such as NNscore [42] and RF-score [43].
NNscore is a binary classifier that distinguishes well-docked and poorly docked complexes. It uses five types of counts and potentials of atom pairs from classical knowledge-based SFs to represent protein-ligand proximity contact, medium distance contact, electrostatic interaction energy, number of ligand atom types, and number of ligand rotatable bonds. The 194 features obtained are then fed into a fully connected network to predict the probability of whether the molecule is well-docked or not.
RF-score is a representative MLSF that was proposed in 2010 and has since been developed to the fourth generation. It uses the frequency of protein-ligand atom pair occurrences within a certain distance as its feature, defining 9 common atomic element types (C, N, O, S, P, F, Cl, Br, and I) for proteins and ligands. Since F, P, Cl, Br, and I atoms are not present in proteins, each protein-ligand complex can be represented as a vector containing (4*9) features. The model is then combined with RF algorithms and trained on PDBbind-v2007. The results showed that the best Rp (Pearson’s correlation coefficient) value of the model could reach 0.776, which is significantly better than most classical SFs. RF-Score-v2 optimized parameters such as atom pairwise type, featurization strategy, and model selection, and Rp can reach 0.803 [53]. RF-Score-v3 introduced 6 additional empirical energy terms of AutoDock Vina based on the features of the first version, improving the generalization ability of the model [54].
Li et al. and Qu et al. developed HydraMap to predict the favorable hydration sites in the binding pocket of a protein molecule [55, 56]. This method uses statistical potentials to quantify the interactions between protein atoms and water molecules. Such statistical potentials were derived from 10,987 crystal structures selected from the Protein Data Bank (PDB), and then the model calculates the frequency of occurrence of atom pairwise formed by water and protein-ligand atoms. Finally, authors incorporated the features extracted from protein-bound waters obtained in this way together with the amino acid fingerprinting of the bound water site, buried solvent availability surface area (bSASA), into three ML-based SFs (RF-score, ECIF, and PLEC). The result tested on CASF-2016 showed that the introduction of HydraMap significantly improved the performance of SFs.
Featurization based on protein-ligand interaction fingerprints
The concept of protein-ligand interaction fingerprints (IFPs) was originally introduced for docking scenarios. Over the past few decades, several IFPs have been proposed, the pioneer of which is SIFt, a model proposed by Deng et al. in 2004 [57]. SIFt generates fingerprints that convert complex 3D structural information into one-dimensional binary strings, making data visualization, analysis, and organization easier. When applied to the field of MLSFs, IFPs can be subdivided into classical IFPs based on geometrical and pharmacophore information, and energy-based IFPs based on energy information. Representative IFPs include SPLIF [58] and Pharm-IF [59].
Pharm-IF, a pharmacophore-based IFP, calculates residue-based IFPs to detect 12 types of protein-ligand interactions. The pharmacophoric features of ligand atoms and their distances are then used to characterize the interaction pairs. The feature vector is created by adding all the values of interaction pairs together. Da et al. developed SPLIF, a method similar to SIFt, but SPLIF maps more types of interactions into the fingerprint (e.g., π-π stacking, polarization interactions, etc.) [58]. Another team proposed PLEC-FP, a method based on Extended Connectivity Fingerprinting (ECFP) that identifies each pair of interacting atoms within a distance of 4.5 Å between the ligand and protein [60]. Then, these pairs of atoms are processed through a hashing function to produce a PLEC fingerprint that represents protein-ligand interactions. The PLEC fingerprint outperforms the other two IFPs, SILIRID [61] and SPLIF [58], for binding affinity prediction.
Fassio et al. proposed LUNA, a model that integrates three IPFs: EIEP, FIFP, and HIFP [62]. EIEP records the atomic invariant features in the complex, such as the number of heavy atoms covalently bound to the atom, the chemical valence minus the number of adjacent hydrogens, the atomic number, the number of atomic isotopes, the atomic charge, the number of bound hydrogens, and whether the atom belongs to a ring. FIFP encodes features of the atom and the atomic group to which it belongs, e.g., assigning chemical features of aromatic rings to each atom that belongs to them. FIFP represents a more abstract role-based substructural feature. HIFP is a mixture of encoded atomic invariants and physicochemical features (for atomic groups). After characterizing all the IFPs, the three fingerprint information codes are integrated using a hashing function to obtain a 32-bit integer identifier. The results showed that LUNA has good interpretability and can distinguish ligands by identifying their pose similarities.
As a subclass of IPFs, energy-based IFPs can be regarded as differential forms of specific energy features. That is, global energy terms are decomposed into the contributions of individual important amino acid residues in the binding site. Ding et al. constructed an SVM classifier for screening HIV-1 protease inhibitors [63]. The model calculates molecular interaction energy components (MIEC) of each protein residue, including van der Waals forces, electrostatic forces, hydrogen bonding, solvation energy, and geometric constraints, to depict protein-ligand interactions. When trained on a small dataset with only 50 known inhibitors, it could achieve improved enrichment of actives in the top 20 candidates. Chen et al. developed a sieve sequence model based on MIEC and gradient boosting decision tree (GBDT) algorithms to identify potential luciferase inhibitors [64]. It performs MM/GBSA free energy calculations and energy decomposition of the complexes. The residue-ligand interactions are represented by van der Waals interactions, electrostatic interactions, and two solvation energy terms. Then, these four decomposed terms of residues and the corresponding total energy terms were calculated to generate the MIEC matrix. The prediction accuracy of the best model for inhibitors and non-inhibitors reached 87.20 % and 90.30 %.
Featurization based on mathematical features
These strategies utilize mathematical ideas and methods, such as algebraic topology, differential geometry, and graph theory, to extract features of protein-ligand interactions. These related models have several applications, including virtual screening, binding affinity prediction, and toxicity prediction.
Wei et al. proposed several combined affinity prediction models, such as T-bind [65], TopologyNet [66], TopBP [67], and TopVS [67], based on the idea of persistent homology (PH) in algebraic topology. Instead of extracting traditional interaction features, such as hydrogen bonds and van der Waals forces, these models extract the three-dimensional information of the complexes as topological invariants, including independent components, rings, and cavities, and later use these invariants as features.
However, extracting topological features can lead to the loss of a large amount of biological and chemical information since the functions of biomolecules are closely related to their structures. To overcome this difficulty, they proposed element-specific PH, multi-component PH, and multi-level PH. For example, they performed PH calculations for four protein heavy atoms (C/N/O/S) and nine ligand heavy atoms (C/N/P/S/P/F/Cl/Br/I) within a certain distance, and characterized the interaction information of the complexes by atom pair networks. Using PH features of carbon atom pairs to indicate hydrophobic interactions, PH features of N and O to indicate hydrogen bonding interactions, etc. The results demonstrated that PH achieved a synthetic featurization of complex biomolecules and retained indispensable biological information while reducing ML dimensionality.
Nguyen et al. proposed AGL-Score based on multiscale weighted colored subgraphs [68]. The main task undertaken by the algebraic graph was to convert high-dimensional interaction information into low-dimensional representations. Three kinds of algebraic graphs, including Laplacian matrix, pseudo-inverse of Laplacian matrix, and adjacency matrices, were used in this study to depict protein-ligand interactions in different ways. Wee et al. used Ollivier persistent ricci curves (OPRCs) to characterize the complexes based on the idea of differential geometry [69]. The process mainly involves modeling the molecular structure and interactions as a graph, calculating Ollivier-ricci curves on the graph to obtain geometric descriptors, and finally combining them with the gradient boosting tree (GBT) algorithm for prediction of binding affinity. Overall, these mathematical features provide a new way of developing SFs, but the complexity and abstraction of such methods can also limit their application.
Feature learning-based MLSFs
Traditional feature engineering-based MLSFs involve pre-defined feature calculations and selection for both the compound and protein. These approaches heavily rely on domain knowledge to extract useful features from the compound and protein data [70]. In contrast, feature learning-based compound-protein binding prediction using end-to-end deep learning frameworks to extract structural information from the compound and protein automatically. The representation strategies can be categorized into atom context-based, grid-based, and graph-based approaches, as well as graph-based approaches with SE(3)-equivariant features [47]. These strategies aim to map the input data to a high-dimensional space to capture essential information for learning compound-protein interactions, encompassing not only scoring but also binding poses. Figure 3 depicts three kinds of feature learning-based representation strategies. Table 3 lists various feature learning-based compound-protein binding prediction methods introduced in this paper, along with their categories, main applications, and the backbone they are based on.
Atom context-based features
Atom context-based features refer to the extraction of features from the local environment of each compound atom. This includes information such as atom type, coordinates, distances, and amino acid type. By utilizing this approach, it is possible to convert the complex interactions between protein and compound into a fixed number of dimensional vectors.
One example of this approach is DeepVS, which extracts a set of atom contexts and then feeds them into a convolutional layer for further analysis [71]. DeepDock uses geometric deep learning for molecular docking and virtual screens [72]. In this approach, compounds are represented as graphs, and the intramolecular context of each atom is extracted using a graph convolutional neural network. The protein target surrounding the ligand is represented as a 3D mesh with four properties (electrostatics, hydropathy, hydrogen-bond donor/acceptor, and shape index) at each point, which is employed in a graph convolutional neural network to extract features. Pairwise-concatenated features are then used as input for a mixture density network (MDN) to obtain a probability density function for each complex, which determines the potential of the ligand fitting into the protein.
RTMScore improves over DeepDock by representing proteins as undirected graphs at the residue level and using a graph transformer for protein and ligand feature extraction [73]. The protein and ligand are converted into 3D residue graphs and 2D molecular graphs, respectively. Two sets of independent graph transformer layers are used to learn the node representations of protein and ligand. The node representations of protein and ligand are then concatenated pairwise and fed into an MDN to calculate the necessary parameters for a mixture density model. This model can generate a probability distribution for the minimum distance between each residue and each ligand atom, which is used to create a statistical potential by summing all independent negative log-likelihood values.
Grid-based features
Grid-based feature extraction is a common approach in deep learning-based prediction of CPI. The binding sites of protein-ligand complexes are transformed into a set of 3D lattice dots, where each dot is linked to a set of channels to capture various structural characteristics. This allows for the representation of protein-ligand interactions in a structured and organized manner, which is suitable for deep learning algorithms.
Pafnucy [74] and Kdeep [75] are two typical examples which use a 3D grid-based representation strategy to segment the binding sites of protein-ligand complexes into multiple 3D grid points. By using the grid points as the fundamental unit, atomic basic information is fed into a 3D convolutional neural network to predict the binding affinity of protein-ligand pairs. GNINA 1.0 is a molecular docking software that uses an ensemble of convolutional neural networks (CNNs) as a scoring function [76]. The built-in CNN models include Default2017, Default2018, and Dense, all of which rely on grid-based features. For each type of atom present in the ligand, a precomputed grid is generated. Each grid point is then assigned a value calculated using a single atom of the corresponding type. To score the entire ligand, the values for each of its atoms are interpolated from the grid and added together. GNINA also uses a hybrid docking approach that combines the search efficiency of a rigid-body docking algorithm with the accuracy of a flexible docking algorithm. The rigid-body docking step uses precomputed grids to speed up the search for the optimal pose of the ligand, while the flexible docking step uses a Monte Carlo algorithm to sample the conformational space of the protein-ligand complex.
Graph-based features
Compared to the traditional flat 2D graph, structure-based CPI prediction converts protein-ligand complexes into a 3D graph format. In this format, atoms are represented as nodes that hold information about their properties. The connections (edges) between atoms can be either covalent or non-covalent interactions, and the distance between the atoms is also considered.
PotentialNet is a typical graph neural network (GNN) architecture that employs graph-based features for protein-ligand binding affinity prediction [77]. It uses adjacency from the atomic distance matrix to encompass a wider range of neighbor interactions and includes three stages: covalent-only propagation, dual noncovalent and covalent propagation, and ligand-based graph gather.
InteractionGraphNet (IGN) is a deep graph representation learning framework that uses the 3D structures of compound-protein complexes to learn the interactions between them [78]. A protein-ligand complex is represented as three graphs with 3D structural and chemical information: a ligand graph, a protein graph, and a bipartite protein-ligand graph. One graph convolution module is shared between the ligand graph and the protein pocket graph for intramolecular representation extraction. Then, the intermolecular graph convolution module is sequentially stacked to extract edge representations (which learn about atom-pair interactions) in the bipartite protein-ligand graph. Finally, the edge representations are fed into a downstream fully connected neural network (FCNN) for downstream decision-making.
Structure-based deep learning models, such as the ones described above, demonstrate high accuracy due to the use of 3D information between ligands and proteins. However, the deficiency in 3D structural data of the protein–ligand complexes could drive the models to be over-fitted to the training data, might fail to generalize in a broader context. PIGNet is a physics-informed graph neural network for the prediction of binding affinity of a protein–ligand complex [79]. To learn the specific pattern of the interaction, binding affinity is defined as a sum of atom–atom pairwise interactions, which are the combinations of the four energy components – van der Waals (vdW) interaction, hydrogen bond, metal–ligand interaction, and hydrophobic interaction. It predicts CPI by incorporating a priori physical knowledge into deep learning to make the model more interpretable.
SE(3)-equivariant and invariant-based features
Recently, there has been emerging research utilizing SE(3)-equivariant or invariant features. The concepts of SE(3)-equivariance and invariance are critical physical characteristics in science, from classical and quantum physics to computational biology [80, 81]. SE(3)-equivariance refers to the property where the output undergoes equivalent rotations and translations when the input data is subjected to rotations and translations while invariance implies that the output remains unchanged despite rotations and translations applied to the input. Taking the example of a molecule, when the molecule undergoes rotations and translations in three-dimensional space, causing a change in the coordinates of its atoms, its atomic dipoles or forces (vector quantities) exhibit equivariance, while the bond energies and radial distances (scalar quantities) remain invariant. These properties ensure the ability to predict the same binding complex regardless of the initial positioning and orientation of the molecules in space, which is especially needed for data-scarce problems such as compound-protein binding prediction [22, 82].
EquiBind is an SE(3)-equivariant geometric deep learning model that enables direct-shot prediction of both the receptor binding location and the ligand’s bound pose and orientation [22]. It takes as input a ligand molecular graph with a random associated unbound 3D conformer, and receptor-bound structure, both represented as spatial k-nearest neighbor (k-NN) graphs. In the ligand graph, atoms are represented as nodes, using their respective 3D coordinates from the unbound conformer and initial features such as atom type. Edges are established between all pairs of atoms within a distance cutoff of 4 Å. The receptor graph consists of residues as nodes, where the α-carbon locations determine their 3D coordinates. Each node in the graph is connected to its ten closest neighboring nodes within a distance of less than 30 Å. To process these graphs, EquiBind utilizes an Independent E(3)-Equivariant Graph Matching Network (IEGMN) to transform both the features and 3D coordinates. This transformation facilitates intra and inter neural graph message passing, allowing for the extraction of coordinate E(3)-equivariant transformations and feature embeddings. The coordinate transformations are used to identify the rigid body transformation through ligand and receptor binding keypoints, as well as to model ligand flexibility. The binding keypoints are trained to match the ground truth binding pocket points using an optimal transport loss, which recovers their alignment. Ligand flexibility is modeled by predicting an atomic point cloud of the deformed molecule and subsequently utilizing a fast algorithm to extract internal changes in rotatable bonds’ torsion angles that best align with the point cloud.
TankBind improves on EquiBind by predicting a docking pose for each possible pocket independently [83]. This pose is represented as an interatomic distance matrix. The predicted poses are then ranked to determine the optimal conformation of a ligand in the protein pocket. This method is more effective than EquiBind as it allows for independent prediction of docking poses, which results in a more accurate ranking of the ligand conformations. EquiBind and TankBind treat docking as a regression problem, eliminating the need for extensive sampling of possible binding locations and poses employed by previous methods. These approach enables the attainment of the optimal ligand conformation within the protein pocket to complete the docking process, and achieves significant enhancements in both speed and prediction quality when compared to previous search-based binding that use one or more conformational predictions.
Unlike these two approaches, DiffDock is a diffusion generative model that treats molecular docking as a generative problem [84]. DiffDock involves a diffusion process on the degrees of freedom related to ligand poses, which include the position of the ligand relative to the protein pocket, its orientation within the pocket, and the torsional angles that describe its conformation. In the denoising process, it searches for the optimal conformation of a ligand within the protein pocket.
To enhance the performance of tasks involving protein-ligand structures and interactions, Uni-mol employs a Molecular Representation Learning (MRL) approach, which involves pre-training on extensive 3D structure data of organic molecules and candidate protein pockets [85]. To predict the optimal conformation of a ligand in a specific protein pocket, Uni-mol first obtains representations of the protein pocket and the ligand from the two pre-trained SE(3) Transformer models and concatenates them as input to the 4-layer Uni-mol architecture decoder. During the fine-tuning process for binding pose prediction, a scoring function is constructed based on the difference between the distance matrices of the predicted and true atom pairs, enabling the optimization of input coordinates. This approach enhances the representation ability for 3D spatial tasks, and shows excellent performance in complex conformation prediction, as well as molecular property prediction, molecular conformation generation, and pocket property prediction.
Data for protein structure-based CPI prediction
Structure-based prediction of CPI requires the co-crystallization of compound-protein complexes or protein folding structure. Obtaining high-quality three-dimensional structures of proteins has long been a bottleneck that limits the performance of constructed scoring functions. However, with the development of technology and instrumentation, especially cryo-electron microscopy and the advent of AlphaFold, 3D structural data has been supplemented and many databases containing 3D structures have been developed. This section introduces some datasets widely used for the construction of structure-based CPI models, and all dataset summaries are listed in Table 4.
Table 4:
Databases for protein structure-based CPI prediction.
| Databases | Main content | Links |
|---|---|---|
| PDBbind | Binding affinities and 3D structure for the protein ligand complexes. | http://www.pdbbind.org.cn/ |
| CrossDocked 2020 | Cross-docked protein ligand complexes from PDBbind. | https://github.com/gnina/models/ |
| Binding MOAD | Well resolved protein crystal structures biological relevant ligands with binding data. | http://www.bindingmoad.org/ |
| DUD-E | Benchmark with decoys. | https://dude.docking.org/ |
The PDBbind v.2020 database provides 19,443 experimentally determined protein-ligand binding affinity data (in the form of Kd, Ki, or IC50 values) collected from the RCSB Protein Data Bank. Besides protein-ligand interaction data, it also provides structural information about the protein-ligand complexes, i.e., the specific binding sites. These two types of information are useful for studying the relationship between binding affinity and complex structure using computational and statistical methods in drug discovery. PDBbind is regularly updated with the growth of PDB.
The Binding MOAD is another comprehensive collection of experimentally determined protein-ligand structures and their binding affinities from PDB or extracted from literature [86]. It contains >40,000 protein-ligand complexes with high-quality experimental data on binding affinity and well-resolved protein crystal structures. It is freely available and widely used in many studies and applications. The Binding MOAD database is continuously updated and curated to ensure high quality and relevance for current research needs.
The DUD-E dataset is a widely-used benchmark dataset for evaluating virtual screening methods by providing decoys in drug discovery [87]. It consists of two parts: the “decoy” set and the “active” set. The “active” set includes active compounds that are known to bind to protein targets with specific binding affinities, while the “decoy” set contains 50 molecules for each active having similar physicochemical properties but dissimilar 2D topology that are not known to bind to the target. This dataset challenges virtual screening methods to identify the active molecules from the decoys. However, there are still biases in this dataset leading to an unfair test [88, 89]. This indicates an urgent need for the construction of high-quality benchmark datasets.
Most methods for predicting protein-ligand interactions focus on replicating the structure of known complexes (redocking). However, in a realistic application scenario, our main goal is to determine how a novel ligand binds to a given protein pocket structure. Since deep learning methods prefer large amounts of data for training to get excellent results, currently, there is not enough 3D structural data available.
To address this challenge, CrossDocked expanded the available 3D structural data for CPI prediction [11]. Similar ligand binding sites were downloaded from PDB and grouped as the original data source of CrossDocked. Then, smina was used to augment docked poses by docking ligands to a cognate receptor and by intentionally docking to non-cognate receptors to generate counterexamples [90]. In total, the CrossDocked2020 set contains 13,780 unique ligands, 41.9 % of which have binding affinity data aligned to the PDBbind database, 2,922 pockets, 18,450 complexes, and 22,584,102 binding poses. However, the noise present in this dataset should be noted because some assumptions made in the construction of the dataset, such as that a given ligand has the same binding affinity for all receptors of a given pocket.
Non-structure-based CPI prediction
Conventional methods for predicting CPI can be classified into two types: structure-based and ligand-based. The structure-based method includes docking simulation, and it relies on the knowledge of 3D structure of the target protein [91]. The ligand-based method predicts CPI by comparing candidate ligands to known ligands of the target protein. However, it is not applicable when the target protein has very few known ligands [92].
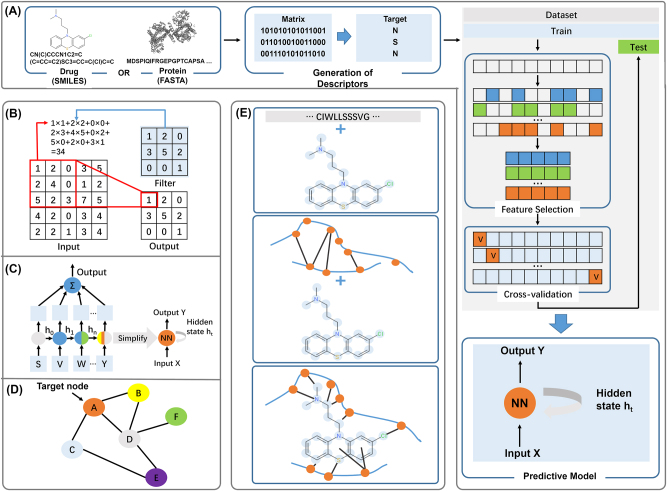
Bredel and Jacoby introduced a chemogenomics approach to predict CPI without using the 3D structure of the target protein [93]. Chemogenomics approaches consider multiple types of information simultaneously, including drug-related information (e.g., chemical information), target-related information (e.g., protein sequence), and known interaction information. Genomic features can also be used for functional annotation of small molecules and genes. When integrating multiple sources of biological information such as drug-target interactions (DTI), drug-drug interactions, and protein-protein interactions, network-based or knowledge graph-based approaches can be established for CPI prediction. Non-structure-based CPI prediction can be further divided into chemogenomics-based methods, transcriptomics-based methods, and network-based methods (Table 5). These methods share the same primary procedures: (1) Data collection; (2) Mathematical descriptor generation; (3) Search for the best subset of variables; (4) Model training; and (5) Model validation. Among them, the core step of model training is to represent small molecules or biomacromolecules with descriptors that can capture both molecular properties and structural characteristics well (Figure 4A).
Table 5:
Non-structure-based CPI prediction models.
| Model | Description |
|---|---|
| Chemogenomics-based CPI prediction | |
| DeepDTA [98] | DeepDTA applies CNN to extract low-dimensional real-value characteristics of compounds and proteins, then concatenates the two vectors and computes the final output via a fully-connected layers. |
| GraphDTA [102] | Based on DeepDTA, GraphDTA changed part of the CNN layers to the GNN layers. GraphDTA’s performance has been improved compared to DeepDTA on the same dataset. |
| TransformerCPI [99] | TransformerCPI is a transformer-based CPI prediction model, applying a gated convolutional network and a GCN to learn protein sequence representations and molecular graphs of compounds, respectively. |
| DGraphDTA [100] | DGraphDTA utilizes contact maps predicted from protein sequences as the input for the protein encoder. The inclusion of structural features through contact maps further enhances the performance of DTA predictions. |
| DrugVQA [101] | DrugVQA is an end-to-end deep learning framework for predicting interactions. In this framework, proteins are represented using a two-dimensional distance map and learned through a dynamic attentive convolutional neural network. |
| MGraphDTA [103] | MGraphDTA proposes a multiscale graph neural network with a novel visual explanation method named gradient-weighted affinity activation mapping for DTA prediction and interpretation, creating a probability map that highlights significant atoms contributing the most to the DTA. |
| Transcriptomics-based CPI prediction | |
| CMap [106] | CMap created the first reference sets of gene expression profiles from cultured human cells treated with bioactive small molecules, providing indirect clues for CPI. |
| ProTINA [108] | ProTINA creates a cell type-specific protein-gene regulatory network based on differential gene expression profiles, and applies a dynamic model to infer drug targets. |
| SSGCN [107] | SSGCN is a Siamese spectral-based graph convolutional network model for inferring the protein targets of chemical compounds from gene transcriptional profiles. SSGCN applies two parallel GCN to extract features from differential gene expression profiles between CP-signatures and KD-signatures. |
| Network-based CPI prediction | |
| DTINet [110] | DTINet applies an unsupervised approach to learn the low-dimensional characteristic representation of drugs and target proteins from heterogeneous data, and completed the prediction of DTI via an inductive matrix. |
| NeoDTI [111] | NeoDTI can integrate various information from heterogeneous network data and automatically learn topologically retained representations of drugs and targets to further facilitate DTI prediction |
| EEG-DTI [115] | EEG-DTI builds biological networks that connect biological entities including drug, protein, disease, and side effect based on two types of edges: Relative interaction edge and similarity edge. |
| CPI-IGAE [116] | CPI-IGAE converts the heterogeneous graph into a homograph with directed edges and weighted edges, and adjusts the induction aggregator of GraphSAGE to fit the CPI prediction task. |
| HampDTI [117] | HampDTI is a novel heterogeneous network-based method that automatically extract meta-paths through a learnable attention mechanism instead of a pre-defined one. |
CNN, convolutional neural networks; GNN, graph neural networks; GCN, graph convolutional networks; DTI, drug-target interaction; CPI, compound-protein interaction; CP-signatures, compound-induced signatures; KD-signatures, gene knock-down-induced signatures.
Figure 4:
Non-structure-based CPI prediction methodology. (A) CPI prediction model pipeline; (B) architecture of convolutional neural network; (C) architecture of recurrent neural network; (D) architecture of graph neural network; (E) three common input types of models: proteins are input as sequence and small molecules are constructed as graphs; protein pockets and small molecules are constructed into different graphs; protein pockets and small molecules are converted into the same graphs.
Chemogenomics-based CPI prediction model
Several chemogenomics methods have been proposed in the last decade, differing mainly in how proteins, ligands, and similarities between them are depicted. Playe et al. have comprehensively evaluated machine learning methods for chemogenomics-based CPI prediction [14]. Early chemogenomics-based CPI prediction methods mainly used predefined molecular fingerprints and protein descriptors to measure similarities between objects or as input features. With the continuous development of deep learning, more end-to-end frameworks have been applied to predict CPI, automatically extracting features of protein and compound to contain more information than predefined features. When proteins and compounds are represented in text form (sequence) or images (3D grids), CNN (Figure 4B) or recursive neural networks (RNN, Figure 4C) can handle protein sequence and molecular representation such as Simplified Molecular Input Line Entry System (SMILES) [94, 95]. When the molecular graph (represented by atomic characteristics, bond characteristics, and adjacency matrix) is used as input, the GNN [96] can be adopted to complete the CPI prediction task [97]. Unlike sequence-based methods, GNN captures non-Euclidean information and directly learns the representation of molecular structure (Figure 4D).
DeepDTA applies CNN to extract low-dimensional real-value characteristics of compounds and proteins, then concatenates the two vectors and computes the final output via a fully-connected layer [98]. Inspired by the transformer architecture’s powerful ability to capture features from sequences, TranformerCPI treats compound and protein sequences as input sequences, applying a gated convolutional network to learn representations of proteins and a graph convolutional network (GCN) to learn molecular graphs of compounds [99]. A label reversal experiment was proposed to test whether a model learns true interaction features other than hidden ligand bias introduced in model training. Compared with other models, TransformerCPI achieved significantly improved performance on the new experiments, suggesting it can learn desired interaction features and decrease the risk of hidden ligand bias.
To overcome the limitation of protein sequences in accurately representing interactions in three-dimensional space, several methods have utilized structural-related features of proteins as inputs to improve drug-target affinity (DTA) prediction. Jiang et al. proposed DGraphDTA that utilizes contact maps predicted from protein sequences as the input for the protein encoder, which helps improve the performance of DTA predictions [100]. Another approach is presented by Zheng et al. who developed an end-to-end deep learning framework for predicting interactions [101]. In this framework, proteins are represented using a two-dimensional distance map, and a dynamic attentive convolutional neural network is employed to learn fixed-size representations from the variable-length distance maps.
On the other hand, to overcome the limitation of losing structural information when representing drugs as sequences like SMILES, several models based on GNN have been developed for CPI prediction, which represent drugs as graphs. GraphDTA adapted part of the CNN layers to GNN layers without changing the remaining part of the model, demonstrating that GNN may be more suitable for feature extraction from chemical structures than CNN [102]. GraphDTA’s performance has been improved compared to DeepDTA on the same dataset. However, GNNs with a small number of layers may not capture the global structure of compounds effectively. To overcome this limitation, MGraphDTA proposed a multiscale graph neural network (MGNN) for DTA prediction and interpretation [103]. MGraphDTA introduces dense connections into the GNN, enabling all layers to have direct access to the gradients of the loss function with respect to each weight. This approach mitigates the vanishing gradient problem and allows for training very deep GNNs, thereby capturing multiscale features of proteins and drugs. Additionally, MGraphDTA introduces a novel visual explanation method called Gradient-Weighted Affinity Activation Mapping (Grad-AAM). Grad-AAM creates a probability map that highlights significant atoms contributing the most to the DTA.
Transcriptomics-based CPI prediction model
Since Hughes et al.’s landmark study demonstrated that gene expression data profiles can be used for functional annotation of small molecules and genes in yeast [104], various databases of expression profiles have been developed to identify potential mechanisms of action of chemicals [105]. CMap created the first reference sets of gene expression profiles from cultured human cells treated with bioactive small molecules [106]. These profiles can be used to find connections between small molecules and physiological processes, diseases, and drugs with the same mechanism, providing clues for CPI analyses.
Comparing differential expression patterns induced by chemical perturbation with those induced by genetic perturbation may reveal potential information about compound target interactions [107]. ProTINA creates a cell type-specific protein-gene regulatory network based on differential gene expression profiles and applies a dynamic model to infer drug targets [108]. Pabon et al. implemented an RF model to explore the correlations between compound-induced signatures (CP-signatures) and gene knock-down-induced signatures (KD-signatures) from CMap to predict drug targets [109]. Zhong et al. designed a Siamese spectral-based graph convolutional network (SSGCN) model for inferring the protein targets of chemical compounds from gene transcriptional profiles [107]. SSGCN applies two parallel GCN to extract features from differential gene expression profiles between CP-signatures and KD-signatures. By this way, SSGCN introduces fewer assumptions and can learn the underlying relationships between compound perturbations and gene perturbations. A compound-centric target inference pipeline was established to identify the potential host targets of nelfinavir (NFV), and a target-centric prediction pipeline was established to find novel small molecule inhibitors of ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1) by screening 22,425 compound perturbation profiles. These highlight SSGCN as a useful tool to infer the interacting targets of active compounds, or reversely, to find novel inhibitors of a given target of interest.
Network-based CPI prediction model
Most network-based methods assume that similar drugs may have similar targets, and vice versa. These approaches establish drug-target networks that incorporate multiple data sources, such as DTI, drug-drug interactions, and protein-protein interactions. Computational methods and known Compound-Protein Interactions) are then used to predict new interactions (Figure 4E). Additionally, heterogeneous data, including drug side effects, drug-disease associations, and genomics data, are employed to enhance CPI predictions.
DTINet applied an unsupervised approach to learn the low-dimensional characteristic representation of drugs and target proteins from heterogeneous data and completed the prediction of DTI via an inductive matrix [110]. NeoDTI, developed by Wan et al. is an end-to-end approach that integrates various information from heterogeneous network data and automatically learns topologically retained representations of drugs and targets to further facilitate DTI prediction [111]. Both DTINet and NeoDTI are based on various curated public drug-related databases, including DrugBank [13], Comparative Toxicogenomics Database (CTD) [112], Human Protein Reference Database (HPRD) [113], and Side Effect Resources (SIDER) [114].
EEG-DTI is a novel, end-to-end heterogeneous graph model, which builds biological networks that connect biological entities, including drug, protein, disease, and side effect based on two types of edges: relative interaction edge and similarity edge [115]. The heterogeneous graph convolutional neural network is applied to obtain low-dimensional representations of the drugs and targets, and the inner product of the representation of a drug and a target is considered as the interaction score.
Although heterogeneous graphs can integrate many types of entities and interactions in a single network, it is still challenging to aggregate the heterogeneous properties of different types of nodes or edges to obtain a graph representation. CPI-IGAE converts the heterogeneous graph into a homograph with directed and weighted edges, and adjusts the induction aggregator of GraphSAGE to fit the CPI prediction task. The edges in the graph are constructed by Dice similarity coefficient (DSCs) [116].
HampDTI is a novel heterogeneous network-based method, which automatically extracts meta-paths through a learnable attention mechanism instead of a pre-defined one [117]. Such meta-path graph implicitly measures the importance of every possible meta-path between drugs and targets. The experiments on benchmark datasets show both the superiority of HampDTI in DTI prediction over several baseline methods and the effectiveness of the model in discovering important meta-paths.
Data for non-structure-based CPI prediction
Compared to structure-based CPI prediction models, non-structure-based CPI prediction models do not require 3D structure information, and a large amount of experimental data is available. This section introduces some commonly used databases for constructing non-structure-based CPI models. The descriptions of these datasets are summarized in Table 6.
Table 6:
Databases for non-structure-based CPI prediction.
| Databases | Main content | Links |
|---|---|---|
| BindingDB | Binding affinity entries between small drug-like molecules and potential target proteins | https://www.bindingdb.org/ |
| STITCH | Experimentally determined interactions and predicted interactions between chemical compounds and proteins | http://stitch.embl.de/ |
| DrugBank | Clinical level information and molecular level data about drugs | https://go.drugbank.com/ |
| ChEMBL | Chemical, bioactivity and genomic data of bioactive molecules | https://www.ebi.ac.uk/chembl/ |
| PubChem | Chemical structures, identifiers, chemical and physical properties, biological activities, patents, health, safety, toxicity data, and many others | https://pubchem.ncbi.nlm.nih.gov/ |
| Davis | 72 kinase inhibitors with 442 kinases of human catalytic protein kinome | Refer to the supplementary materials of the original article [122] |
| KIBA | 52,498 chemical compounds and 467 kinase targets with various bioactivity | Refer to the supplementary materials of the original article [121] |
| KEGG | Genomic, chemical and systemic functional information | https://www.genome.jp/kegg/ |
| CLUE | Microarray data of treatment of 81,979 perturbagens including 33,609 small molecule drugs in 240 cell contexts. | https://clue.io/ |
BindingDB [12] and STITCH [118] are two large CPI databases of experimentally determined binding affinities between small molecules, such as drugs and other biologically active compounds, and their target proteins, such as enzymes, receptors, and transporters. PubChem [119] and ChEMBL [120] are the broadest databases, including molecular properties and bioactivity from patents or scientific literature. DrugBank is an online database containing clinical information on drugs and drug targets [13]. Alongside these comprehensive CPI databases, there are two widely utilized smaller databases named KIBA [121] and Davis [122], which contain measured bioactivity specifically for kinase inhibitors.
KEGG is a database that contains genomic information at the molecular level, using large-scale molecular databases generated by high-throughput technologies to understand high-level functions and utilities of cells, organisms, and ecosystems [123]. CMap collects gene-expression profiles from cultured human cells treated with bioactive small molecules, together with pattern-matching software to mine this data [106]. Leveraging a new, low-cost, high throughput reduced representation expression profiling method called L1000, a 1,000-fold scale-up of the CMap, named CLUE, has been accomplished [124]. The latest release is an expansion upon the 2017 data, encompassing 3.02 M profiles from over 80,000 perturbagens, including compounds, mechanisms of action (MoAs) and unique genes. These gene-expression profiles can be used to find connections among small molecules sharing a mechanism of action, chemicals and physiological processes, and diseases and drugs.
Discussion
In drug design, understanding the relationship between protein structure, conformation, and function is of great importance [125]. When the 3D structure of the target protein is available, structure-based CPI prediction can provide more reliable results by leveraging the structural information of the protein, which enables the extraction of interaction patterns that can be generalized to novel ligands [126]. Nevertheless, most of these methods necessitate the prior identification of the binding pocket for the ligand within the protein to determine 3D protein-ligand complex structures. While traditional computational methodologies for pocket identification, such as SiteMap [127], serve as viable alternatives to costly experimental techniques, they still exhibit limitations and constraints [128]. The emergence of SE(3)-equivariant geometric deep learning model and diffusion generative model that enables direct-shot prediction of both the receptor binding location and the ligand’s bound pose and orientation, eliminating the need for extensive sampling of possible binding locations [22]. On the other hand, when the knowledge of the 3D structure is inaccessible, non-structure-based CPI prediction models are an alternative. These models can be built based on large datasets that incorporate more diverse data types. However, non-structure-based CPI prediction models cannot account for complex interactions well [129], especially when these interactions are susceptible to conformational changes or the chain flexibility of the protein [130].
The integration of machine learning has significantly enhanced the predictive capability of both structure- and non-structure-based methods. However, it has also brought some challenges.
Model interpretability
Interpretability remains a significant concern for improving the performance of CPI prediction models. The three-dimensional structure of proteins can provide more information about the interaction characteristics between proteins and ligands, and more than that, researchers can also use deep learning models that combine methods such as attention mechanisms to give correlation between structural information and binding affinity of protein-ligand complexes, which can be further used for structural modification of compounds in drug design [131]. Recently, the progress in conversational AI represented by ChatGPT has had a profound impact on various fields. As a powerful language processing model, ChatGPT is designed to generate human-like conversations by understanding the context of a conversation and generating appropriate responses. We can use similar methods to interpret complex CPI prediction results, identify potential shortcomings, and guide future ligand structural optimization.
Data problem
Despite the availability of many large databases, they still cannot fully meet all requirements. For structure-based CPI prediction models, one of the most challenging problems is the lack of 3D protein-ligand complex structures for training, which results in a decline in the model’s generalization ability. For example, PDBbind, a widely used database for building structure-based CPI models, contains less than 20,000 data entries. Although AlphaFold2 has significantly contributed to the expansion of 3D structural data, they are still imperfect for accurate structure-based CPI modeling [132, 133]. Moreover, many CPI prediction models suffer from a high false positive rate problem that arises from imbalanced data in training [134, 135]. DUD-E creates more negative samples to address this issue, but also introduces hidden biases [88]. Such biases could be easily learned by neural network architectures, providing the wrong “guidance” to distinguish active molecules from negative ones.
Feature extraction
Optimizing feature extraction is an important direction for exploration. In the realm of structure-based CPI prediction models, conventional feature engineering-based MLSFs rely on expert domain knowledge for extracting valuable features, such as energy terms or non-covalent interactions. Nonetheless, this method may inadvertently introduce irrelevant features due to artificial bias [40]. Conversely, feature learning-based MLSFs generally adopt a data-driven approach, requiring straightforward inputs like atom type, distance, and charge. While this end-to-end approach can minimize the introduction of artificial errors, it often sacrifices interpretability [40]. As the utilization of 3D features in deep learning techniques becomes more widespread, incorporating them into feature learning-based MLSFs can lead to the automatic capture of vital interaction information. This enhancement has the potential to improve both the accuracy and interpretability of structure-based CPI prediction models [136, 137].
Non-structure-based CPI prediction models typically use protein residues as the basic unit of interaction with ligand atoms. While this simplifies representation, it can lead to the problem of information loss. To address this issue, prior knowledge of protein structural or functional characteristics is required, such as solvent accessible surface (SAS), secondary structure (SS), backbone torsion angle (BTA), etc. Additionally, combining non-structure-based CPI prediction models with protein structure folding algorithms may be helpful. For instance, AlphaFold2 can deduce the three-dimensional structure of proteins from protein sequences [138, 139]. Integrating the learned features of AlphaFold2 into chemogenomics-based CPI prediction models may compensate for the information loss of non-structure-based models.
In general, protein-ligand interactions should be represented from a multi-perspective, complementary, and comprehensive way, as information captured by different featurization strategies can be vastly different. While many attempts have been made to study feature combinations, increasing feature complexity does not guarantee improved model performance and may instead introduce the risk of overfitting [140]. Therefore, it is important to carefully consider the selection and prioritization of features. Combining multimodality to improve CPI prediction performance is also a feasible direction. Techniques such as data fusion, multitask learning, and transfer learning can integrate data from different sources into CPI prediction models, and are worthy of further investigation to improve the prediction performance of these models.
Model evaluation
Currently, there are common issues with data distribution bias and data leakage in CPI datasets. These problems make it difficult to obtain objective model evaluation results. Recently, the Critical Assessment of Computational Hit-Finding Experiments (CACHE) project was initiated to assess the effectiveness of computational CPI methods through experimental validation, which is the most rigorous way to evaluate models [141]. For CACHE challenges, rapid and high-quality testing of predicted outcomes is provided, and all predictions, including false negative samples, will be published. This is valuable as it can alleviate the problem of insufficient negative data in the database and enable further refinement of the model construction. Therefore, this project is expected to serve as a paradigm for integrating computational methods with experimental approaches in the field of building CPI prediction models.
Summary
This review explores recent progress in CPI prediction. The main part details recent advancements and innovative models that have emerged in this domain. The article delves into a detailed analysis of representative CPI models, highlighting their unique features and contributions. Furthermore, the review provides an overview of pertinent datasets employed in CPI modeling. The review concludes with a critical evaluation of the strengths and weaknesses exhibited by different types of CPI models, summarizing common challenges encountered in this field. Notably, feasible recommendations are presented, aiming to guide and facilitate the development of high-performance CPI models in future endeavors.
Footnotes
Ethical approval: Not applicable.
Informed consent: Not applicable.
Author contributions: All authors have accepted responsibility for the entire content of this manuscript and approved its submission.
Competing interests: Authors state no conflict of interest.
Research funding: This work was supported by National Natural Science Foundation of China (T2225002 and 82273855 to M.Y.Z., 82204278 to X.T.L.), Lingang Laboratory (LG202102-01-02 to M.Y.Z.), National Key Research and Development Program of China (2022YFC3400504 to M.Y.Z.), SIMM-SHUTCM Traditional Chinese Medicine Innovation Joint Research Program (E2G805H to M.Y.Z.), Shanghai Municipal Science and Technology Major Project and China Postdoctoral Science Foundation (2022M720153 to X.T.L.).
References
- 1.Zhang X, Wu F, Yang N, Zhan X, Liao J, Mai S, et al. In silico methods for identification of potential therapeutic targets. Interdiscip Sci. 2022;14:285–310. doi: 10.1007/s12539-021-00491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, Jenkins JL, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–7. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–81. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pahikkala T, Airola A, Pietilä S, Shakyawar S, Szwajda A, Tang J, et al. Toward more realistic drug–target interaction predictions. Briefings Bioinf. 2015;16:325–37. doi: 10.1093/bib/bbu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayatunga MK, Xie W, Ruder L, Schulze U, Meier C. AI in small-molecule drug discovery: a coming wave. Nat Rev Drug Discov. 2022;21:175–6. doi: 10.1038/d41573-022-00025-1. [DOI] [PubMed] [Google Scholar]
- 6.Keum J, Nam H. SELF-BLM: prediction of drug-target interactions via self-training SVM. PLoS One. 2017;12:e0171839. doi: 10.1371/journal.pone.0171839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H, Liu S, Chen J, Li X, Ma Q, Yu B. Predicting drug-target interactions using Lasso with random forest based on evolutionary information and chemical structure. Genomics. 2019;111:1839–52. doi: 10.1016/j.ygeno.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 8.van Laarhoven T, Nabuurs SB, Marchiori E. Gaussian interaction profile kernels for predicting drug–target interaction. Bioinformatics. 2011;27:3036–43. doi: 10.1093/bioinformatics/btr500. [DOI] [PubMed] [Google Scholar]
- 9.He T, Heidemeyer M, Ban F, Cherkasov A, Ester M. SimBoost: a read-across approach for predicting drug–target binding affinities using gradient boosting machines. J Cheminf. 2017;9:24. doi: 10.1186/s13321-017-0209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Su M, Han L, Liu J, Yang Q, Li Y, et al. Forging the basis for developing protein–ligand interaction scoring functions. Acc Chem Res. 2017;50:302–9. doi: 10.1021/acs.accounts.6b00491. [DOI] [PubMed] [Google Scholar]
- 11.Francoeur PG, Masuda T, Sunseri J, Jia A, Iovanisci RB, Snyder I, et al. Three-dimensional convolutional neural networks and a cross-docked data set for structure-based drug design. J Chem Inf Model. 2020;60:4200–15. doi: 10.1021/acs.jcim.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–53. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–82. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Playe B, Stoven V. Evaluation of deep and shallow learning methods in chemogenomics for the prediction of drugs specificity. J Cheminf. 2020;12:11. doi: 10.1186/s13321-020-0413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Yu J, Chen L, Li X, Jiang H, Chen K, et al. A hybrid framework for improving uncertainty quantification in deep learning-based QSAR regression modeling. J Cheminf. 2021;13:69. doi: 10.1186/s13321-021-00551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koutsoukas A, Monaghan KJ, Li X, Huan J. Deep-learning: investigating deep neural networks hyper-parameters and comparison of performance to shallow methods for modeling bioactivity data. J Cheminf. 2017;9:1–13. doi: 10.1186/s13321-017-0226-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentile F, Yaacoub JC, Gleave J, Fernandez M, Ton A-T, Ban F, et al. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nat Protoc. 2022;17:672–97. doi: 10.1038/s41596-021-00659-2. [DOI] [PubMed] [Google Scholar]
- 18.Tsubaki M, Tomii K, Sese J. Compound–protein interaction prediction with end-to-end learning of neural networks for graphs and sequences. Bioinformatics. 2019;35:309–18. doi: 10.1093/bioinformatics/bty535. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Chambers J, Abel R. Protein-ligand binding free energy calculations with FEP. Methods Mol Biol. 2019;2022:201–32. doi: 10.1007/978-1-4939-9608-7_9. [DOI] [PubMed] [Google Scholar]
- 20.Garbett NC, Chaires JB. Thermodynamic studies for drug design and screening. Expet Opin Drug Discov. 2012;7:299–314. doi: 10.1517/17460441.2012.666235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin Drug Discov. 2015;10:449–61. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark H, Ganea OE, Pattanaik L, Barzilay R, Jaakkola T, editors. EquiBind: geometric deep learning for drug binding structure prediction. 2022. pp. 20503–21. [Google Scholar]
- 23.Liu J, Wang R. Classification of current scoring functions. J Chem Inf Model. 2015;55:475–82. doi: 10.1021/ci500731a. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Peng J, Sidorov P, Leung Y, Leung K-S, Wong M-H, et al. Classical scoring functions for docking are unable to exploit large volumes of structural and interaction data. Bioinformatics. 2019;35:3989–95. doi: 10.1093/bioinformatics/btz183. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Fu A, Zhang L. An overview of scoring functions used for protein–ligand interactions in molecular docking. Interdiscip Sci. 2019;11:320–8. doi: 10.1007/s12539-019-00327-w. [DOI] [PubMed] [Google Scholar]
- 26.Meli R, Morris GM, Biggin PC. Scoring functions for protein-ligand binding affinity prediction using structure-based deep learning: a review. Front Neuroinf. 2022;2:57. doi: 10.3389/fbinf.2022.885983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking11Edited by F. E. Cohen. J Mol Biol. 1997;267:727–48. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 28.Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, et al. Dock 6: impact of new features and current docking performance. J Comput Chem. 2015;36:1132–56. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catana C, Stouten PFW. Novel, customizable scoring functions, parameterized using N-PLS, for structure-based drug discovery. J Chem Inf Model. 2007;47:85–91. doi: 10.1021/ci600357t. [DOI] [PubMed] [Google Scholar]
- 30.Thornton BF, Wik M, Crill PM. Double-counting challenges the accuracy of high-latitude methane inventories. Geophys Res Lett. 2016;43:12569–1277. doi: 10.1002/2016gl071772. [DOI] [Google Scholar]
- 31.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997;11:425–45. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 32.Verkhivker G, Appelt K, Freer ST, Villafranca JE. Empirical free energy calculations of ligand-protein crystallographic complexes. I. Knowledge-based ligand-protein interaction potentials applied to the prediction of human immunodeficiency virus 1 protease binding affinity. Protein Eng Des Sel. 1995;8:677–91. doi: 10.1093/protein/8.7.677. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 34.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 35.Ben-Naim A. Statistical potentials extracted from protein structures: are these meaningful potentials? J Chem Phys. 1997;107:3698–706. doi: 10.1063/1.474725. [DOI] [Google Scholar]
- 36.Muegge I, Martin YC. A general and fast scoring function for Protein−Ligand interactions: a simplified potential approach. J Med Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 37.Mooij WTM, Verdonk ML. General and targeted statistical potentials for protein–ligand interactions. Proteins. 2005;61:272–87. doi: 10.1002/prot.20588. [DOI] [PubMed] [Google Scholar]
- 38.Velec HFG, Gohlke H, Klebe G. DrugScoreCSDKnowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J Med Chem. 2005;48:6296–303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- 39.Ain QU, Aleksandrova A, Roessler FD, Ballester PJ. Machine‐learning scoring functions to improve structure‐based binding affinity prediction and virtual screening. Wires Comput Mol Sci. 2015;5:405–24. doi: 10.1002/wcms.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen C, Ding J, Wang Z, Cao D, Ding X, Hou T. From machine learning to deep learning: advances in scoring functions for protein–ligand docking. Wires Comput Mol Sci. 2020;10:e1429. doi: 10.1002/wcms.1429. [DOI] [Google Scholar]
- 41.Li H, Sze KH, Lu G, Ballester PJ. Machine‐learning scoring functions for structure‐based virtual screening. Wires Comput Mol Sci. 2021;11:e1478. doi: 10.1002/wcms.1478. [DOI] [Google Scholar]
- 42.Durrant JD, McCammon JA. NNScore: a neural-network-based scoring function for the characterization of protein−ligand complexes. J Chem Inf Model. 2010;50:1865–71. doi: 10.1021/ci100244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballester PJ, Mitchell JB. A machine learning approach to predicting protein–ligand binding affinity with applications to molecular docking. Bioinformatics. 2010;26:1169–75. doi: 10.1093/bioinformatics/btq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zilian D, Sotriffer CA. Sfcscore rf: a random forest-based scoring function for improved affinity prediction of protein–ligand complexes. J Chem Inf Model. 2013;53:1923–33. doi: 10.1021/ci400120b. [DOI] [PubMed] [Google Scholar]
- 45.Li G-B, Yang L-L, Wang W-J, Li L-L, Yang S-Y. ID-Score: a new empirical scoring function based on a comprehensive set of descriptors related to protein–ligand interactions. J Chem Inf Model. 2013;53:592–600. doi: 10.1021/ci300493w. [DOI] [PubMed] [Google Scholar]
- 46.Su M, Yang Q, Du Y, Feng G, Liu Z, Li Y, et al. Comparative assessment of scoring functions: the CASF-2016 update. J Chem Inf Model. 2018;59:895–913. doi: 10.1021/acs.jcim.8b00545. [DOI] [PubMed] [Google Scholar]
- 47.Xiong G, Shen C, Yang Z, Jiang D, Liu S, Lu A, et al. Featurization strategies for protein–ligand interactions and their applications in scoring function development. Wires Comput Mol Sci. 2022;12:e1567. doi: 10.1002/wcms.1567. [DOI] [Google Scholar]
- 48.Wang C, Zhang Y. Improving scoring‐docking‐screening powers of protein–ligand scoring functions using random forest. J Comput Chem. 2017;38:169–77. doi: 10.1002/jcc.24667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge‐based desolvation. J Comput Chem. 2007;28:1145–52. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 50.Lu J, Hou X, Wang C, Zhang Y. Incorporating explicit water molecules and ligand conformation stability in machine-learning scoring functions. J Chem Inf Model. 2019;59:4540–9. doi: 10.1021/acs.jcim.9b00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C, Zhang Y. Delta machine learning to improve scoring-ranking-screening performances of protein–ligand scoring functions. J Chem Inf Model. 2022;62:2696–712. doi: 10.1021/acs.jcim.2c00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye W-L, Shen C, Xiong G-L, Ding J-J, Lu A-P, Hou T-J, et al. Improving docking-based virtual screening ability by integrating multiple energy auxiliary terms from molecular docking scoring. J Chem Inf Model. 2020;60:4216–30. doi: 10.1021/acs.jcim.9b00977. [DOI] [PubMed] [Google Scholar]
- 53.Ballester PJ, Schreyer A, Blundell TL. Does a more precise chemical description of protein–ligand complexes lead to more accurate prediction of binding affinity? J Chem Inf Model. 2014;54:944–55. doi: 10.1021/ci500091r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Leung KS, Wong MH, Ballester PJ. Improving AutoDock Vina using random forest: the growing accuracy of binding affinity prediction by the effective exploitation of larger data sets. Mol Inform. 2015;34:115–26. doi: 10.1002/minf.201400132. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Gao Y, Holloway MK, Wang R. Prediction of the favorable hydration sites in a protein binding pocket and its application to scoring function formulation. J Chem Inf Model. 2020;60:4359–75. doi: 10.1021/acs.jcim.9b00619. [DOI] [PubMed] [Google Scholar]
- 56.Qu X, Dong L, Zhang J, Si Y, Wang B. Systematic improvement of the performance of machine learning scoring functions by incorporating features of protein-bound water molecules. J Chem Inf Model. 2022;62:4369–79. doi: 10.1021/acs.jcim.2c00916. [DOI] [PubMed] [Google Scholar]
- 57.Deng Z, Chuaqui C, Singh J. Structural interaction fingerprint (SIFt): a novel method for analyzing three-dimensional protein− ligand binding interactions. J Med Chem. 2004;47:337–44. doi: 10.1021/jm030331x. [DOI] [PubMed] [Google Scholar]
- 58.Da C, Kireev D. Structural protein–ligand interaction fingerprints (SPLIF) for structure-based virtual screening: method and benchmark study. J Chem Inf Model. 2014;54:2555–61. doi: 10.1021/ci500319f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato T, Honma T, Yokoyama S. Combining machine learning and pharmacophore-based interaction fingerprint for in silico screening. J Chem Inf Model. 2010;50:170–85. doi: 10.1021/ci900382e. [DOI] [PubMed] [Google Scholar]
- 60.Wójcikowski M, Kukiełka M, Stepniewska-Dziubinska MM, Siedlecki P. Development of a protein–ligand extended connectivity (PLEC) fingerprint and its application for binding affinity predictions. Bioinformatics. 2018;35:1334–41. doi: 10.1093/bioinformatics/bty757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chupakhin V, Marcou G, Gaspar H, Varnek A. Simple ligand–receptor interaction descriptor (SILIRID) for alignment-free binding site comparison. Comput Struct Biotec. 2014;10:33–7. doi: 10.1016/j.csbj.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fassio AV, Shub L, Ponzoni L, McKinley J, O’Meara MJ, Ferreira RS, et al. Prioritizing virtual screening with interpretable interaction fingerprints. J Chem Inf Model. 2022;62:4300–18. doi: 10.1021/acs.jcim.2c00695. [DOI] [PubMed] [Google Scholar]
- 63.Ding B, Wang J, Li N, Wang W. Characterization of small molecule binding. I. Accurate identification of strong inhibitors in virtual screening. J Chem Inf Model. 2013;53:114–22. doi: 10.1021/ci300508m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen F, Sun H, Liu H, Li D, Li Y, Hou T. Prediction of luciferase inhibitors by the high-performance MIEC-GBDT approach based on interaction energetic patterns. Phys Chem Chem Phys. 2017;19:10163–76. doi: 10.1039/c6cp08232g. [DOI] [PubMed] [Google Scholar]
- 65.Cang Z, Wei GW. Integration of element specific persistent homology and machine learning for protein‐ligand binding affinity prediction. Int J Numer Method Biomed Eng. 2018;34:e2914. doi: 10.1002/cnm.2914. [DOI] [PubMed] [Google Scholar]
- 66.Cang Z, Wei GW. TopologyNet: topology based deep convolutional and multi-task neural networks for biomolecular property predictions. PLoS Comput Biol. 2017;13:e1005690. doi: 10.1371/journal.pcbi.1005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cang Z, Mu L, Wei G-W. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput Biol. 2018;14:e1005929. doi: 10.1371/journal.pcbi.1005929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen DD, Wei G-W. AGL-score: algebraic graph learning score for protein–ligand binding scoring, ranking, docking, and screening. J Chem Inf Model. 2019;59:3291–304. doi: 10.1021/acs.jcim.9b00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wee J, Xia K. Ollivier persistent Ricci curvature-based machine learning for the protein–ligand binding affinity prediction. J Chem Inf Model. 2021;61:1617–26. doi: 10.1021/acs.jcim.0c01415. [DOI] [PubMed] [Google Scholar]
- 70.Du B-X, Qin Y, Jiang Y-F, Xu Y, Yiu S-M, Yu H, et al. Compound-protein interaction prediction by deep learning: databases, descriptors and models. Drug Discov Today. 2022;27:1350–66. doi: 10.1016/j.drudis.2022.02.023. [DOI] [PubMed] [Google Scholar]
- 71.Pereira JC, Caffarena ER, Dos Santos CN. Boosting docking-based virtual screening with deep learning. J Chem Inf Model. 2016;56:2495–506. doi: 10.1021/acs.jcim.6b00355. [DOI] [PubMed] [Google Scholar]
- 72.Méndez-Lucio O, Ahmad M, del Rio-Chanona EA, Wegner JK. A geometric deep learning approach to predict binding conformations of bioactive molecules. Nat Mach Intell. 2021;3:1033–9. doi: 10.1038/s42256-021-00409-9. [DOI] [Google Scholar]
- 73.Shen C, Zhang X, Deng Y, Gao J, Wang D, Xu L, et al. Boosting protein-ligand binding pose prediction and virtual screening based on residue-atom distance likelihood potential and graph transformer. J Med Chem. 2022;65:10691–706. doi: 10.1021/acs.jmedchem.2c00991. [DOI] [PubMed] [Google Scholar]
- 74.Stepniewska-Dziubinska MM, Zielenkiewicz P, Siedlecki P. Development and evaluation of a deep learning model for protein–ligand binding affinity prediction. Bioinformatics. 2018;34:3666–74. doi: 10.1093/bioinformatics/bty374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiménez J, Škalič M, Martínez-Rosell G, De Fabritiis G. KDEEP: protein-ligand absolute binding affinity prediction via 3D-convolutional neural networks. J Chem Inf Model. 2018;58:287–96. doi: 10.1021/acs.jcim.7b00650. [DOI] [PubMed] [Google Scholar]
- 76.McNutt AT, Francoeur P, Aggarwal R, Masuda T, Meli R, Ragoza M, et al. GNINA 1.0: molecular docking with deep learning. J Cheminf. 2021;13:1–20. doi: 10.1186/s13321-021-00522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feinberg EN, Sur D, Wu Z, Husic BE, Mai H, Li Y, et al. PotentialNet for molecular property prediction. ACS Cent Sci. 2018;4:1520–30. doi: 10.1021/acscentsci.8b00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang D, Hsieh C-Y, Wu Z, Kang Y, Wang J, Wang E, et al. InteractionGraphNet: a novel and efficient deep graph representation learning framework for accurate protein-ligand interaction predictions. J Med Chem. 2021;64:18209–32. doi: 10.1021/acs.jmedchem.1c01830. [DOI] [PubMed] [Google Scholar]
- 79.Moon S, Zhung W, Yang S, Lim J, Kim WY. PIGNet: a physics-informed deep learning model toward generalized drug-target interaction predictions. Chem Sci. 2022;13:3661–73. doi: 10.1039/d1sc06946b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grisafi A, Wilkins DM, Willatt MJ, Ceriotti M. Atomic-scale representation and statistical learning of tensorial properties. Machine learning in chemistry: data-driven algorithms, learning systems, and predictions. ACS Symposium Series. 2019;1326:1–21. [Google Scholar]
- 81.Du W, Zhang H, Du Y, Meng Q, Chen W, Zheng N, et al. SE(3) equivariant graph neural networks with complete local frames. Vol. 162. PMLR; 2022. pp. 5583–608. Proceedings of the 39th international conference on Machine Learning. Baltimore, Maryland, USA. [Google Scholar]
- 82.Ganea O-E, Huang X, Bunne C, Bian Y, Barzilay R, Jaakkola T, et al. Independent SE(3)-equivariant models for end-to-end rigid protein docking. . 2021 arXiv preprint arXiv:2111.07786. [Google Scholar]
- 83.Lu W, Wu Q, Zhang J, Rao J, Li C, Zheng S. Tankbind: trigonometry-aware neural networks for drug-protein binding structure prediction. Adv Neural Inf Process Syst. 2022;35:7236–49. [Google Scholar]
- 84.Corso G, Stärk H, Jing B, Barzilay R, Jaakkola T. Diffdock: diffusion steps, twists, and turns for molecular docking. . 2022 arXiv preprint arXiv:2210.01776. [Google Scholar]
- 85.Zhou G, Gao Z, Ding Q, Zheng H, Xu H, Wei Z, et al., editors. Uni-Mol: a universal 3D molecular representation learning framework. In: The eleventh international conference on Learning Representations. Kigali, Rwanda.
- 86.Smith RD, Clark JJ, Ahmed A, Orban ZJ, Dunbar JB, Carlson HA. Updates to binding MOAD (mother of all databases): polypharmacology tools and their utility in drug repurposing. J Mol Biol. 2019;431:2423–33. doi: 10.1016/j.jmb.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem. 2012;55:6582–94. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L, Cruz A, Ramsey S, Dickson CJ, Duca JS, Hornak V, et al. Hidden bias in the DUD-E dataset leads to misleading performance of deep learning in structure-based virtual screening. PLoS One. 2019;14:e0220113. doi: 10.1371/journal.pone.0220113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia J, Tilahun EL, Reid TE, Zhang L, Wang XS. Benchmarking methods and data sets for ligand enrichment assessment in virtual screening. Methods. 2015;71:146–57. doi: 10.1016/j.ymeth.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koes DR, Baumgartner MP, Camacho CJ. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J Chem Inf Model. 2013;53:1893–904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, Caffrey DR, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol. 2007;25:71–5. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 92.Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol Inform. 2010;29:476–88. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 93.Bredel M, Jacoby E. Chemogenomics: an emerging strategy for rapid target and drug discovery. Nat Rev Genet. 2004;5:262–75. doi: 10.1038/nrg1317. [DOI] [PubMed] [Google Scholar]
- 94.Ucak UV, Ashyrmamatov I, Lee J. Reconstruction of lossless molecular representations from fingerprints. J Cheminf. 2023;15:1–11. doi: 10.1186/s13321-023-00693-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee I, Keum J, Nam H. DeepConv-DTI. Prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Comput Biol. 2019;15:e1007129. doi: 10.1371/journal.pcbi.1007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ryu S, Kwon Y, Kim WY. A Bayesian graph convolutional network for reliable prediction of molecular properties with uncertainty quantification. Chem Sci. 2019;10:8438–46. doi: 10.1039/c9sc01992h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li P, Li Y, Hsieh C-Y, Zhang S, Liu X, Liu H, et al. TrimNet: learning molecular representation from triplet messages for biomedicine. Briefings Bioinf. 2021;22:bbaa266. doi: 10.1093/bib/bbaa266. [DOI] [PubMed] [Google Scholar]
- 98.Öztürk H, Özgür A, Ozkirimli E. DeepDTA: deep drug–target binding affinity prediction. Bioinformatics. 2018;34:i821–9. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen L, Tan X, Wang D, Zhong F, Liu X, Yang T, et al. TransformerCPI: improving compound–protein interaction prediction by sequence-based deep learning with self-attention mechanism and label reversal experiments. Bioinformatics. 2020;36:4406–14. doi: 10.1093/bioinformatics/btaa524. [DOI] [PubMed] [Google Scholar]
- 100.Jiang M, Li Z, Zhang S, Wang S, Wang X, Yuan Q, et al. Drug–target affinity prediction using graph neural network and contact maps. RSC Adv. 2020;10:20701–12. doi: 10.1039/d0ra02297g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng S, Li Y, Chen S, Xu J, Yang Y. Predicting drug–protein interaction using quasi-visual questionanswering system. Nat Mach Intell. 2020;2:134–40. doi: 10.1038/s42256-020-0152-y. [DOI] [Google Scholar]
- 102.Nguyen T, Le H, Quinn TP, Nguyen T, Le TD, Venkatesh S. GraphDTA: predicting drug–target binding affinity with graph neural networks. Bioinformatics. 2020;37:1140–7. doi: 10.1093/bioinformatics/btaa921. [DOI] [PubMed] [Google Scholar]
- 103.Yang Z, Zhong W, Zhao L, Yu-Chian Chen C. MGraphDTA: deep multiscale graph neural network for explainable drug-target binding affinity prediction. Chem Sci. 2022;13:816–33. doi: 10.1039/d1sc05180f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–26. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 105.Fielden MR, Eynon BP, Natsoulis G, Jarnagin K, Banas D, Kolaja KL. A gene expression signature that predicts the future onset of drug-induced renal tubular toxicity. Toxicol Pathol. 2005;33:675–83. doi: 10.1080/01926230500321213. [DOI] [PubMed] [Google Scholar]
- 106.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 107.Zhong F, Wu X, Yang R, Li X, Wang D, Fu Z, et al. Drug target inference by mining transcriptional data using a novel graph convolutional network framework. Protein Cell. 2022;13:281–301. doi: 10.1007/s13238-021-00885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Noh H, Shoemaker JE, Gunawan R. Network perturbation analysis of gene transcriptional profiles reveals protein targets and mechanism of action of drugs and influenza A viral infection. Nucleic Acids Res. 2018;46:e34. doi: 10.1093/nar/gkx1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pabon NA, Xia Y, Estabrooks SK, Ye Z, Herbrand AK, Süß E, et al. Predicting protein targets for drug-like compounds using transcriptomics. PLoS Comput Biol. 2018;14:e1006651. doi: 10.1371/journal.pcbi.1006651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo Y, Zhao X, Zhou J, Yang J, Zhang Y, Kuang W, et al. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat Commun. 2017;8:573. doi: 10.1038/s41467-017-00680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wan F, Hong L, Xiao A, Jiang T, Zeng J. NeoDTI: neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics. 2018;35:104–11. doi: 10.1093/bioinformatics/bty543. [DOI] [PubMed] [Google Scholar]
- 112.Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, et al. The comparative Toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47:D948–54. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human protein reference database--2009 update. Nucleic Acids Res. 2009;37:D767–2. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peng J, Wang Y, Guan J, Li J, Han R, Hao J, et al. An end-to-end heterogeneous graph representation learning-based framework for drug–target interaction prediction. Briefings Bioinf. 2021;22:bbaa430. doi: 10.1093/bib/bbaa430. [DOI] [PubMed] [Google Scholar]
- 116.Wan X, Wu X, Wang D, Tan X, Liu X, Fu Z, et al. An inductive graph neural network model for compound–protein interaction prediction based on a homogeneous graph. Briefings Bioinf. 2022;23:bbac073. doi: 10.1093/bib/bbac073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang H, Huang F, Xiong Z, Zhang W. A heterogeneous network-based method with attentive meta-path extraction for predicting drug–target interactions. Briefings Bioinf. 2022;23:bbac184. doi: 10.1093/bib/bbac184. [DOI] [PubMed] [Google Scholar]
- 118.Szklarczyk D, Santos A, Von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–4. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2023 update. Nucleic Acids Res. 2023;51:D1373–80. doi: 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–7. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tang J, Szwajda A, Shakyawar S, Xu T, Hintsanen P, Wennerberg K, et al. Making sense of large-scale kinase inhibitor bioactivity data sets: a comparative and integrative analysis. J Chem Inf Model. 2014;54:735–43. doi: 10.1021/ci400709d. [DOI] [PubMed] [Google Scholar]
- 122.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 123.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51:D587–92. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–52. doi: 10.1016/j.cell.2017.10.049. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang D, Zhou Q, Labroska V, Qin S, Darbalaei S, Wu Y, et al. G protein-coupled receptors: structure- and function-based drug discovery. Int J Software Tool Technol Tran. 2021;6:7. doi: 10.1038/s41392-020-00435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kimani SW, Owen J, Green SR, Li F, Li Y, Dong A, et al. Discovery of a novel DCAF1 ligand using a drug–target interaction prediction model: generalizing machine learning to new drug targets. J Chem Inf Model. 2023;63:4070–8. doi: 10.1021/acs.jcim.3c00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Halgren TA. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model. 2009;49:377–89. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- 128.Broomhead NK, Soliman ME. Can we rely on computational predictions to correctly identify ligand binding sites on novel protein drug targets? Assessment of binding site prediction methods and a protocol for validation of predicted binding sites. Cell Biochem Biophys. 2017;75:15–23. doi: 10.1007/s12013-016-0769-y. [DOI] [PubMed] [Google Scholar]
- 129.Li S, Wan F, Shu H, Jiang T, Zhao D, Zeng J. MONN: a multi-objective neural network for predicting compound-protein interactions and affinities. Cell Syst. 2020;10:308–22.e11. doi: 10.1016/j.cels.2020.03.002. [DOI] [Google Scholar]
- 130.Lima AN, Philot EA, Trossini GHG, Scott LPB, Maltarollo VG, Honorio KM. Use of machine learning approaches for novel drug discovery. Expet Opin Drug Discov. 2016;11:225–39. doi: 10.1517/17460441.2016.1146250. [DOI] [PubMed] [Google Scholar]
- 131.Krishnan SR, Bung N, Vangala SR, Srinivasan R, Bulusu G, Roy A. De novo structure-based drug design using deep learning. J Chem Inf Model. 2022;62:5100–9. doi: 10.1021/acs.jcim.1c01319. [DOI] [PubMed] [Google Scholar]
- 132.Scardino V, Di Filippo JI, Cavasotto CN. How good are AlphaFold models for docking-based virtual screening? iScience. 2023;26:105920. doi: 10.1016/j.isci.2022.105920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.He X-h, You C-z, Jiang H-l, Jiang Y, Xu HE, Cheng X. AlphaFold2 versus experimental structures: evaluation on G protein-coupled receptors. Acta Pharmacol Sin. 2023;44:1–7. doi: 10.1038/s41401-022-00938-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stumpfe D, Bajorath J. Current trends, overlooked issues, and unmet challenges in virtual screening. J Chem Inf Model. 2020;60:4112–5. doi: 10.1021/acs.jcim.9b01101. [DOI] [PubMed] [Google Scholar]
- 135.Lyu J, Irwin JJ, Shoichet BK. Modeling the expansion of virtual screening libraries. Nat Chem Biol. 2023;19:712–8. doi: 10.1038/s41589-022-01234-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Preuer K, Klambauer G, Rippmann F, Hochreiter S, Unterthiner T. Interpretable deep learning in drug discovery. Samek W, Montavon G, Vedaldi A, Hansen L, Müller KR, editors. Explainable AI: interpreting, explaining and visualizing deep learning. Cham: Springer. 2019;11700:331–45. doi: 10.1007/978-3-030-28954-6_18. [DOI] [Google Scholar]
- 137.Brown BP, Mendenhall J, Geanes AR, Meiler J. General purpose structure-based drug discovery neural network score functions with human-interpretable pharmacophore maps. J Chem Inf Model. 2021;61:603–20. doi: 10.1021/acs.jcim.0c01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–9. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–4. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang S, Yang K, Liu Z, Lai X, Yang Z, Zeng J, et al. DrugAI: a multi-view deep learning model for predicting drug–target activating/inhibiting mechanisms. Briefings Bioinf. 2023;24:bbac526. doi: 10.1093/bib/bbac526. [DOI] [PubMed] [Google Scholar]
- 141.Ackloo S, Al-awar R, Amaro RE, Arrowsmith CH, Azevedo H, Batey RA, et al. CACHE (Critical Assessment of Computational Hit-finding Experiments): a public–private partnership benchmarking initiative to enable the development of computational methods for hit-finding. Nat Rev Chem. 2022;6:287–95. doi: 10.1038/s41570-022-00363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]