Abstract
RNA-binding proteins (RBPs) are key post-transcriptional regulators, and the malfunctions of RBP-RNA binding lead to diverse human diseases. However, prediction of RBP binding sites is largely based on RNA sequence features, whereas in vivo RNA structural features based on high-throughput sequencing are rarely incorporated. Here, we designed a deep bimodal information fusion network called DeepFusion for unraveling protein-RNA interactions by incorporating structural features derived from DMS-seq data. DeepFusion integrates two sub-models to extract local motif-like information and long-term context information. We show that DeepFusion performs best compared with other cutting-edge methods with only sequence inputs on two datasets. DeepFusion’s performance is further improved with bimodal input after adding in vivo DMS-seq structural features. Furthermore, DeepFusion can be used for analyzing RNA degradation, demonstrating significantly different RBP-binding scores in genes with slow degradation rates versus those with rapid degradation rates. DeepFusion thus provides enhanced abilities for further analysis of functional RNAs. DeepFusion’s code and data are available at http://bioinfo.org/deepfusion/.
Keywords: Information fusion, Deep learning, RNA-binding proteins, In vivo RNA structures, Motifs
Graphical Abstract
1. Introduction
RNA-binding proteins (RBPs) have emerged as key regulators that play important roles in post-transcriptional regulation. These different RBPs bound their RNA targets competitively, constituting dynamic complexes that regulate numerous cellular processes in diverse cell types, tissues, and physiopathologic states. Such processes include splicing, degradation, and translation [1]. For example, RBPs like OTUD3, TTP, and HNRNPK affect mRNA degradation and regulate the abundance of diverse transcripts. Dysregulation of these RBPs is associated with esophageal cancer, systemic inflammation, and progenitor cell differentiation [2], [3], [4], [5].
The high-throughput technique of CLIP-seq became widely used to discover the mechanisms by which RBPs bind to RNA targets. CLIP-seq experiments can comprehensively identify the landscape of RNA binding sites for each RBP in single-nucleotide resolution in vivo by using UV radiation to guide cross-linking and immunoprecipitation (CLIP) of a particular RBP and its binding RNAs followed by reverse transcription and high-throughput sequencing of these RNA segments. Diverse CLIP-seq protocols have been developed, such as HITS-CLIP, iCLIP, eCLIP, etc. [6]. As a result, publicly available CLIP-seq datasets have been accumulated by different laboratories [7], [8], [9]. The largest such dataset has been produced by the ENCODE consortium using the eCLIP protocol, covering 133 different human RBPs in the human cell lines [10]. These resources allow us to better understand the binding preferences of RBPs [11].
Using machine-learning or deep-learning techniques to identify RBP binding sites, the sequence specificity of each RBP can be determined from experimental data. Traditional machine-learning-based algorithms require sophisticated feature engineering [12]. Comparatively, deep-learning-based methods automatically extract features from the sequences derived from the CLIP-seq data. The first of such methods, DeepBind, applied convolutional neural networks (CNNs) to the problem of RBP-RNA and TF-DNA interactions to automatically extract patterns of short sequence segments using the motif detector based on a convolution kernel [13]. Since then, there have been several advanced methods to improve this prediction by refining the model structure. For example, Pan et al. built several predictive models for RBP-RNA interactions on several datasets [14], including iDeep [15], iDeepV [16], and iDeepE [9]. iDeepE provides the best performance by merging two types of CNN sequence features, one for the global view and the other for the local view. In addition, DeepCLIP enhances the model capacity by combining a CNN architecture with a bi-directional long short-term memory layer (BLSTM) for capturing the contextual dependencies [17]. Finally, EDCNN improves the predictions of a CNN by introducing an additional evolutionary algorithm for optimizing the hyper-parameters [18].
The binding preferences of RBPs depend not only on the RNA sequences but also on the structural features of these RBP binding sites [19], [20], [21], [22], [23], [24], [25]. For example, A G-rich internal loop in the lncRNA Braveheart is critical for binding to a zinc-finger protein CNBP, which is required for the cardiac specification in mice [26]. Therefore, incorporating RNA structure-related features into the predictive models can improve their capacity to reveal the mechanisms of RNA-protein interactions [27], [28]. For example, GraphProt used the predicted secondary structures derived from RNAshapes [29] and a graph kernel to learn both sequence and structural features of the binding preference for 24 RBPs [30]. In addition, RPI-Net introduced the predicted RNA secondary structures as a base pairing probability matrix using RNAplfold [31] and takes this adjacency matrix into a graph neural network [32]. Moreover, deepnet-rbp utilized the secondary and tertiary RNA structures predicted by RNAshapes [29] and JAR3D [33], [34] to generate a multi-modal deep-learning framework for predicting RBP binding sites [35]. However, the RNA structures used in these studies are prediction-based, and their accuracy is limited by factors such as thermodynamic parameters and kinetic barrier errors [36]. Moreover, these in vitro predictions differ significantly from the in vivo RNA structure states.
Fortunately, in vivo RNA structures have been probed in human cell lines by diverse experimental protocols, the most representative of which are icSHAPE [37] and DMS-seq [38]. icSHAPE measures the nucleotide flexibility at base resolution by treating cells with a structure-sensitive molecule NAI-N3 and then enriching and sequencing the NAI-N3-modified RNAs to generate flexibility scores for every base with signals. In contrast, DMS-seq uses dimethyl sulphate (DMS), which is small enough to penetrate cells and react with solvent-accessible unpaired adenine and cytosine residues. As a result, the in vivo pairing states of these A/C bases can be revealed by using the DMS-seq technique. These data allow researchers to model both in vivo RNA structures and in vivo RNA-protein interactions, leading to better information fusion. A previous study called PrismNet showed that incorporating icSHAPE data can better predict RBP binding sites [39]. However, in vivo DMS-seq data have never been systematically evaluated in this context.
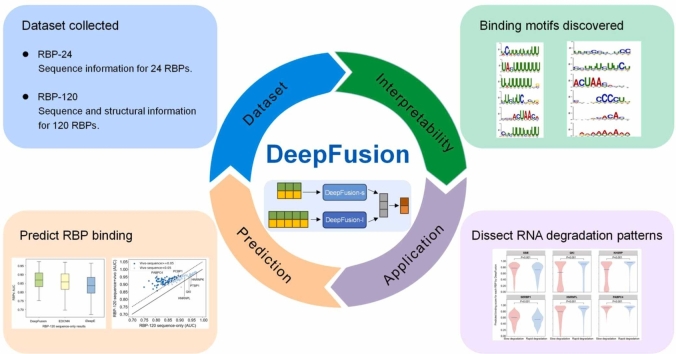
In this work, we introduce the in vivo human DMS-seq data and design a deep bimodal information fusion network called DeepFusion for unraveling protein-RNA interactions by incorporating both RNA sequence and structural features. DeepFusion integrates two sub-models based on convolutional neural networks and long short-term memory networks to extract local motif-like and long-term context information, respectively. DeepFusion can accept single- or bimodal data as input. We first show that DeepFusion with single-modal sequence input improves the prediction accuracy over a series of cutting-edge methods. Second, we show that DeepFusion with bimodal input, which adds in vivo DMS-seq structural features to sequence features, substantially improves the model performance and interpretability for resolving the RBP binding motifs. Third, we extend the application of DeepFusion in the context of RNA degradation. DeepFusion’s prediction scores for degradation-related RBPs differ statistically in genes with slow and rapid degradation rates, demonstrating that DeepFusion can capture biologically meaningful associations for further analysis of functional RNAs.
2. Materials and methods
2.1. Data resource
To evaluate DeepFusion and other tools, we used two characteristic datasets. The first dataset was downloaded from the GraphProt paper [40] called “RBP-24″. The second dataset was derived from the ENCODE eCLIP-seq data called “RBP-120″. We used the eCLIP-seq data for 120 RBPs in the human K562 cell line because the matched DMS-seq data in the same cell line was also available [10].
2.1.1. The RBP-24 dataset
We downloaded the RBP-24 dataset from the GraphProt website (http://www.bioinf.uni-freiburg.de/Software/GraphProt/). The RBP-24 dataset is a standard benchmark dataset frequently used in the field. These CLIP-seq data for 24 RBPs are produced using HITS-CLIP, PAR-CLIP, and iCLIP protocols in diverse cell types. The dataset provides sequences for binding sites (positive set) and nonbinding sites (negative set) for these 24 RBPs. Each sequence contains three parts: one motif sequence with a length of up to 75 nucleotides (nt) and two surrounding sequences, including 150 nt upstream and 150 nt downstream. We also utilized cd-hit-est in the CD-HIT tool [41] for each RBP to eliminate redundant RNA sequences in the test set with greater than 80% sequence similarity to any of the sequences in the training and validation sets, thus creating a non-redundant RBP-24 dataset. The detailed distribution of RBP-24 is shown in Supplementary Table S1.
2.1.2. The RBP-120 dataset
We downloaded alignments in BAM files and peaks in BED files of the eCLIP-seq data for the human K562 cell line from the ENCODE website (https://www.encodeproject.org). We merged the peaks from two biological replicates of eCLIP-seq data for each RBP. To deal with inconsistency, we first obtained the intersected region supported by both replicates using BEDtools [42] and then expanded the intersected region by 15 upstream and downstream nucleotides [40], [43]. We re-calculated the eCLIP-seq signals for these refined peak regions using SAMtools [44]. Then, we normalized CLIP BAM over input BAM using the ENCODE script “overlap_peakfi_with_bam.pl” [10]. The averaged signals across the two replicates were used as representatives.
We selected robust positive sets by requiring that the peaks have an averaged log2(CLIP/input) signal ≥ 1, which means that the normalized read count in the CLIP sample is at least twice that in the mock input). We further required the peaks with a max length of 75 nt following the criteria of the GraphProt paper. We then created paired negative datasets by shuffling unbound sites of similar length and genomic distribution as binding sites. We used the human genome version hg19 and annotation version GENCODE v38lift37. We partitioned the genome into five different types, including CDS, 3′UTR, 5′UTR, intron, and intergenic, in a hierarchical way using the UCSC table browser. We ensured that each pair of positive and negative samples distributed in the same genomic type. We also ensured the non-binding sites in the negative sets did not intersect with any peak in either biological replicate of eCLIP-seq data. In addition, we created non-redundant RBP-120 datasets by excluding the RNA sequences in test set with sequence similarity greater than 80% to any sequences in training and validation set for each RBP by using the cd-hit-est in CD-HIT tool [41].
As a result, sequences for the raw and de-redundant RBP-120 set were released (http://bioinfo.org/deepfusion/). The peak regions for both positive and negative sets are padded to 75 nt with flanking Ns for missing bases, and the extended sequences covering peak, upstream, and downstream regions are padded to 375 nt, like GraphProt. The structural signals for the RBP-120 were derived from DMS-seq data, which are introduced in the following section. The detailed distribution of RBP-120 is shown in Supplementary Table S1.
2.1.3. The DMS-seq data
We used the DMS-seq data probed in the human K562 cell line [38], which has been aligned to the human genome (hg19 assembly). The reverse transcription (RT) stop counts are considered as the raw DMS-seq signal C(i) for each nucleotide i and processed in the bigwig format in three conditions: in vivo, in vitro, and control [45]. Then we normalized the raw signal C(i) for each nucleotide across the whole transcriptome in each condition using the same formula in the original paper [36].
where N is the length of the extended sequences for the peak, upstream, and downstream regions surrounding nucleotide i. We then removed the background by subtracting the normalized control count C′control(i) from the normalized in vivo count C′vivo(i) or in vitro count C′vitro(i). As a result, we obtained the DMS-reactivity scores for each nucleotide i for in vivo and in vitro conditions, respectively. We released the structural files for both positive and negative sets of 120 RBPs (http://bioinfo.org/deepfusion/). Similarly, the DMS reactivity is padded to 75 nt and 375 nt for the peak and extended regions, respectively.
2.2. The DeepFusion model
2.2.1. Bimodal inputs of DeepFusion
DeepFusion accepts both RNA sequences and structural signals as input, with the latter optional. RNA sequences encoded by one-hot matrices of dimension 4 ×N convert linear biological sequences into image-like matrices that apply directly to convolutional neural networks. The first dimension, "4", refers to four types of bases, A, C, G, and U. The second dimension, "N," is the length of the RNA fragments in the input dataset, which are set to be 75 for local peak regions and 375 for extended long sequences, respectively. When ambiguous Ns are provided, they are encoded in equal-weight mode [0.25, 0.25, 0.25, 0.25]. For structural input, we preprocessed the DMS-seq read counts into normalized DMS-reactivity scores for each base, with higher scores representing higher probabilities of the given base being single-stranded. We encoded these scores as a 1 ×N vector. The fusion of the bimodal inputs is done by concatenating the 4 ×N sequence matrix with the 1 ×N structural vector to form a 5 ×N heterogeneous input matrix.
2.2.2. Bimodal architecture of DeepFusion
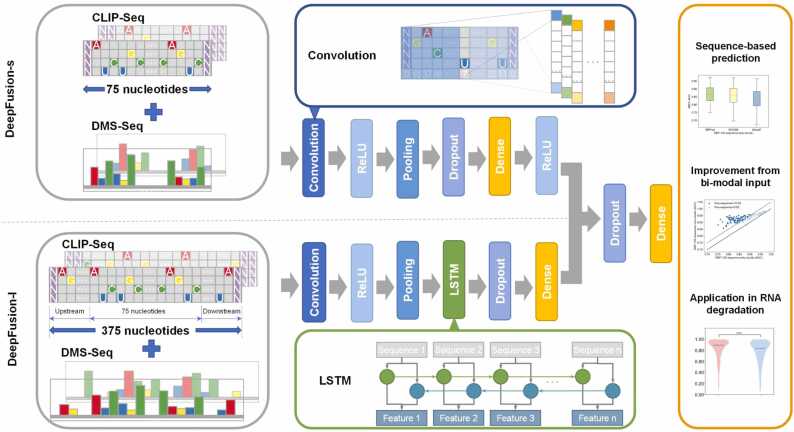
DeepFusion is a deep-learning network designed in a bimodal architecture, with a first sub-model used to extract motif-like information around the local peak regions and a second sub-model revealing the wider range interaction from the extended regions, as shown in Fig. 1. We named the sub-models as DeepFusion-s and DeepFusion-l, respectively.
Fig. 1.
DeepFusion workflow. DeepFusion consists of two parts: The upper part is called DeepFusion-s, and the lower part is called DeepFusion-l. The two boxes on the left indicate the bimodal input data, including the RNA sequences and the DMS-seq structural signals. The boxes in the middle indicate the model architecture, with convolution and LSTM architectures in detail. The box on the right shows three applications of DeepFusion.
The sub-model DeepFusion-s extracts motif-like information from the local peak regions. It takes a 4 × 75 or 5 × 75 data matrix as input, depending on only sequences or sequences and structures inputs. It then feeds the input matrix into a convolution layer for local feature extraction. A ReLU activation layer is used to increase the nonlinear representation of the whole model, and a max-pooling operation is used for sampling the most important features and reducing the dimensions of extracted features. Subsequently, a dropout layer is added to avoid over-fitting, and a fully connected layer plus a second ReLU function is used for extracting the final features from the peak region with properly reduced dimensions. The major operation of DeepFusion-s is based on convolution, with a kernel length set to 10 according to the average length of RBP binding sites [46].
The sub-model DeepFusion-l was introduced to extract wider range interaction information from the extended regions, including peak regions and the genomic context upstream and downstream. A 4 × 375 or 5 × 375 data matrix is required as single-modal sequence input or bimodal sequence and structure input. DeepFusion-l first uses the convolution with a kernel length of five, ReLU, and the max-pooling for the initial feature extraction, just like DeepFusion-s. It then adds another layer called a bidirectional long short-term memory network (BLSTM), a variant of a recurrent neural network (RNN) suitable for learning long-range associations. The RNN extracts information progressively by sequentially connecting hidden layer units in a sequential manner and sharing parameters between the different base inputs. LSTM preserves long-range inter-base dependencies by using input, forget, and output gates across the LSTM memory cells. Bidirectional LSTM combines forward and backward memory to learn how upstream and downstream sequences affect the binding between RBPs and their recognized RNA segments. DeepFusion-l thus learns abstract information about the extended regions and remembers valuable features between wider range interactions. Next, a dropout operation prevents over-fitting, and a fully connected layer is used to obtain the final feature description. In this way, DeepFusion-l is not limited to the local peak regions but explores the impact of complicated long-term contextual information.
We combined DeepFusion-s and DeepFusion-l into the whole DeepFusion model, which we used to analyze bimodal inputs to leverage the dual effect of the local motifs and the wider range protein-RNA interactions. The fusion of the two sub-models occurs on the feature-vector level, i.e., concatenates the features of DeepFusion-s and DeepFusion-l to form a uniform feature vector. After that, a dropout operation was used to avoid overfitting, and a fully connected layer is used to predict binding or non-binding.
2.2.3. Bi-dataset evaluation of DeepFusion
We evaluated the performance of DeepFusion and other tools when applied to both the RBP-24 and RBP-120 datasets based only on sequence input. There are two reasons for making a sequence-based comparison: First, collecting matching DMS-seq data is difficult for the RBP-24 dataset because the CLIP-seq data for these 24 RBPs are in multiple cell lines. Second, most cutting-edge methods for comparing with DeepFusion cannot accept in vivo RNA structural features as input. For fairness, we thus evaluated DeepFusion with single-modal input.
For the RBP-24 dataset, we compared DeepFusion with a series of tools (GraphProt [40], iDeepE [9], DeepCLIP [17], EDCNN [18], RPI-Net [32], deepnet-rbp [47], and PrismNet [39]) that were also previously evaluated in the same dataset. These tools mostly accept only sequence input, except for GraphProt and the latter three methods using the RNA structural features predicted from sequences or derived from icSHAPE. For the RBP-120 dataset, we selected iDeepE and EDCNN for sequence-based comparisons. The training strategies and hyperparameters of the compared methods were set in the same way as in the original papers (Supplementary Table S1).
We next evaluated the improvement of the DeepFusion model upon supplying it with in vivo RNA structures. This evaluation is based only on the RBP-120 dataset, which contains matching eCLIP-seq data for 120 RBPs and DMS-seq data from the human K562 cell line. We re-processed the DMS-seq signals to DMS-reactivity scores for both in vivo and in vitro experimental conditions. As a result, when we use DeepFusion with bimodal inputs, we can input either sequence and in vivo structural scores or sequence and in vitro structural scores. We thus compared the three modes of DeepFusion: sequence-only, sequence+vivo, and sequence+vitro.
In all these evaluations, we train one model per RBP. We separated the training, test, and validation sets independently to ensure the generalizability of DeepFusion. For the original RBP-24 dataset, the training and test data were already separately provided. We further divided its training set into a secondary training and validation set at a ratio of 85: 15. For the newly generated RBP-120 dataset, we divided the training, test, and validation sets at the ratios 76.5: 10: 13.5. The parameters were trained by an Adam optimizer using the cross-entropy loss function for measuring the distance between the predicted results and the actual labels in the training set. Then, the model with the lowest loss function on the validation set was selected as the best model. All performances were measured by the area under the receiver operating characteristic curve (AUC), Matthews correlation coefficient (MCC), and F1 score based on the test set.
2.2.4. Interpretation of DeepFusion results
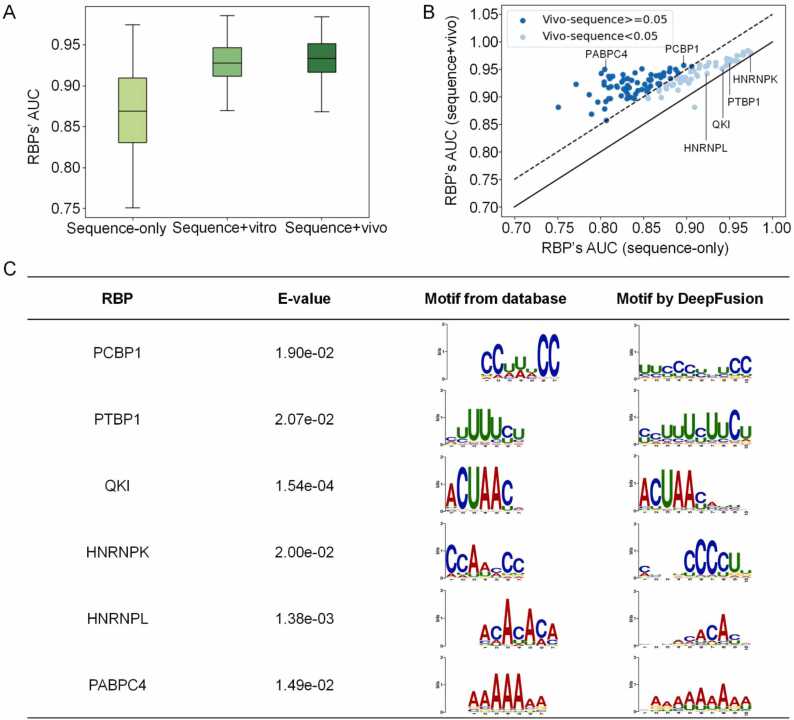
To interpret the results of DeepFusion, we extracted the sequence motifs around the peak regions and compared them with those recruited from existing databases. To this end, we extracted the 2000 largest feature values across the feature map generated by the first convolution layer in the sub-model DeepFusion-s, as done in a previous study [9]. The base composition of these 2000 sequences can be represented by a position weight matrix for depicting the preferences of bases in each position, i.e., a ten-mer sequence motif. We used the TOMTOM platform [48] to visualize the DeepFusion-derived position weight matrices as sequence logos and compared them against a database of known motifs [46]. We compared the motifs using the E-value scores that measured the expected number of false positives after adjusting the P-values. TOMTOM gave E-values based on Bonferroni correction of empirical motif P-values from searching shuffled query motifs against shuffled target motifs. As a result, a small E-value indicates a tiny probability of observing the alignment.
2.3. Application of DeepFusion to RNA degradation
We further extended the application of DeepFusion to RNA degradation. We used the SLAM-seq data that measured the endogenous mRNA decay in the K562 cell line [49], from which the matching eCLIP-seq data were also available. In this work, the K562 cells were fed with S4U to block transcription, following performed time-course SLAM-seq, and then calculated the decay rate for 8861 genes. We thus divided these genes into two groups: the first group was the genes with rapid degradation (i.e., decay rates ranked in the top 25%), and the second group was the genes with slow degradation (i.e., decay rates ranked in the bottom 25%).
We then set out to see whether these two gene groups differ in their RBP binding profiles according to DeepFusion’s prediction scores. We selected the mRNA with the longest 3′UTR as the representative of each gene and required the minimum length of the 3′UTR region to exceed 75 nt. We predicted each mRNA’s RBP binding profiles using DeepFusion in a sliding window mode, with a widow size of 75 nucleotides and a step size of 50. In this way, the model can predict binding probability for each window sliding over the putative binding site. We then took the maximum of DeepFusion’s prediction scores across all 3′UTR windows on this mRNA as the final binding score. Then, we evaluated whether the two groups of genes with rapid degradation or slow degradation differ in their RBP binding profiles. Moreover, we re-analyzed this issue in an opposite way, named Degradation-opposite. To this end, we first divided two new groups of genes according to their binding scores predicted by DeepFusion, one group with scores of the top 25% and the other with the bottom 25%, and compared the differences in degradation rates between the two new groups.
3. Results
3.1. DeepFusion performs best when evaluated in RBP-24 dataset
We first evaluated DeepFusion and other representative methods in the RBP-24 dataset based on single-modal sequence inputs. The detailed AUC scores for each RBP for these methods are listed in Table 1, with the best performance for each row shown in bold. DeepFusion performs the best with an average AUC of 0.952, which is significantly higher than that for EDCNN (0.944), DeepCLIP (0.935), iDeepE (0.931), PrismNet (0.827), and other methods. The differences between DeepFusion and other methods are all statistically significant under the two-tailed paired T-test. For example, the P-value of DeepFusion vs. EDCNN (with the second highest AUCavg) comparison is 0.022, and the P-value of DeepFusion vs. DeepCLIP (with the third highest AUCavg) comparison is 0.0006. We found that DeepFusion still performs best when using other metrics. Take the second-rank EDCNN as an example. The differences between DeepFusion and EDCNN are still significant, with a P-value of 0.044 for MCC (Matthews correlation coefficient) and a P-value = 0.042 for F1 score. Moreover, DeepFusion’s performance is higher than its two sub-models, DeepFusion-s and DeepFusion-l in the RBP-24 dataset. In addition, we evaluated DeepFusion on the stringent non-redundant RBP-24 dataset, in which CD-HIT was used to exclude any sequence in the test set that had greater than 80% sequence similarity to the training and validation sets. We found that the fraction of redundant sequences between the training and the test set is rather low (8.1%). And DeepFusion still performs the best after de-redundancy. All these results can be found in the Supplementary Table S2.
Table 1.
Comparison of DeepFusion performance with that of seven representative methods using the RBP-24 dataset.
| RBP | GraphProt | iDeepE | DeepCLIP | EDCNN | RPI-Net (GNN) (debiased) | deepnet-rbp (mDBN+) | PrismNet | DeepFusion |
|---|---|---|---|---|---|---|---|---|
| AGO1-4 | 0.895 | 0.915 | 0.918 | 0.934 | 0.927 | 0.881 | 0.800 | 0.951 |
| AGO2 | 0.765 | 0.884 | 0.859 | 0.895 | 0.877 | 0.809 | 0.795 | 0.916 |
| ALKBH5 | 0.680 | 0.758 | 0.716 | 0.768 | 0.724 | 0.714 | 0.761 | 0.770 |
| C17ORF85 | 0.800 | 0.830 | 0.898 | 0.880 | 0.844 | 0.820 | 0.757 | 0.892 |
| C22ORF28 | 0.751 | 0.837 | 0.838 | 0.869 | 0.849 | 0.792 | 0.786 | 0.904 |
| CAPRIN1 | 0.855 | 0.893 | 0.948 | 0.912 | 0.869 | 0.834 | 0.755 | 0.956 |
| ELAVL1 | 0.955 | 0.979 | 0.981 | 0.981 | 0.971 | 0.966 | 0.849 | 0.984 |
| ELAVL1 (A) | 0.959 | 0.964 | 0.982 | 0.977 | 0.968 | 0.966 | 0.853 | 0.979 |
| ELAVL1 (B) | 0.935 | 0.971 | 0.982 | 0.982 | 0.964 | 0.961 | 0.884 | 0.984 |
| ELAVL1(C)/HuR | 0.991 | 0.988 | 0.995 | 0.993 | 0.995 | 0.994 | 0.929 | 0.997 |
| EWSR1 | 0.935 | 0.969 | 0.973 | 0.976 | 0.967 | 0.966 | 0.799 | 0.985 |
| FUS | 0.968 | 0.985 | 0.986 | 0.988 | 0.980 | 0.980 | 0.827 | 0.992 |
| HNRNPC | 0.952 | 0.976 | 0.983 | 0.983 | 0.986 | 0.962 | 0.860 | 0.984 |
| IGF2BP1-3 | 0.889 | 0.947 | 0.898 | 0.969 | 0.912 | 0.879 | 0.758 | 0.926 |
| MOV10 | 0.863 | 0.916 | 0.940 | 0.940 | 0.875 | 0.854 | 0.818 | 0.948 |
| PTBP1/PTB | 0.937 | 0.944 | 0.927 | 0.954 | 0.958 | 0.983 | 0.893 | 0.959 |
| PUM2 | 0.954 | 0.967 | 0.969 | 0.974 | 0.972 | 0.971 | 0.906 | 0.982 |
| QKI | 0.957 | 0.970 | 0.975 | 0.973 | 0.977 | 0.983 | 0.929 | 0.978 |
| SFRS1 | 0.898 | 0.946 | 0.955 | 0.957 | 0.941 | 0.931 | 0.825 | 0.965 |
| TAF15 | 0.970 | 0.976 | 0.982 | 0.982 | 0.981 | 0.983 | 0.806 | 0.987 |
| TDP-43 | 0.874 | 0.945 | 0.905 | 0.955 | 0.959 | 0.876 | 0.901 | 0.958 |
| TIA1 | 0.861 | 0.937 | 0.945 | 0.950 | 0.963 | 0.891 | 0.823 | 0.950 |
| TIAL1 | 0.833 | 0.934 | 0.943 | 0.944 | 0.959 | 0.870 | 0.805 | 0.951 |
| ZC3H7B | 0.820 | 0.907 | 0.933 | 0.920 | 0.838 | 0.796 | 0.728 | 0.942 |
| Average | 0.887 | 0.931 | 0.935 | 0.944 | 0.927 | 0.903 | 0.827 | 0.952 |
Note: the bolded font indicates that the model performed best on this RBP among all models.
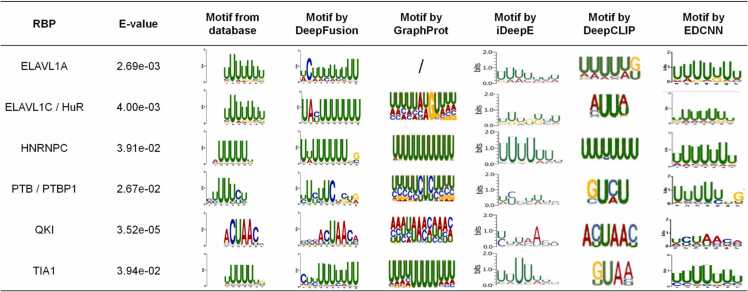
We then analyzed why DeepFusion provides such an improvement by visualizing the sequence motifs identified by DeepFusion. To this end, we extract the 10-mer sequences with the largest 2000 feature values across the feature map generated by the first convolution layer in the sub-model DeepFusion-s. We compare the extracted motif of DeepFusion with the known motif from the TOMTOM database and with that derived from other models, as shown in Fig. 2. It indicates that DeepFusion extracted U-rich motifs for ELAVL1A, ELAVL1C, HNRNPC, and TIA1 consistent with their reference motifs [50], [51], [52]. DeepFusion also obtains other informative motifs for QKI and PTB, for example, the “ACUAAC” motif for QKI [46]. The consistency between the sequence motif extracted by the model and that restored in the known database can reflect the effectiveness of the feature extraction methods designed in the DeepFusion framework.
Fig. 2.
Visualization of motifs extracted by DeepFusion on the RBP-24 dataset The motifs from existing databases and extracted by representative methods are shown. We showed the E-values that quantify the statistical significance of the similarity between DeepFusion’s predicted motifs and known motifs by using TOMTOM in the RBP-24 dataset.
3.2. DeepFusion performs best when evaluated in RBP-120 dataset
Subsequently, we generated a larger dataset consisting of 120 RBPs following the same procedure of RBP-24 for consistency. Firstly, we performed model comparisons based on only sequence input. We selected representative methods, EDCNN and iDeepE, for comparison with DeepFusion, as they perform well in the RBP-24 dataset, and all of them are built under the same deep-learning framework PyTorch [53].
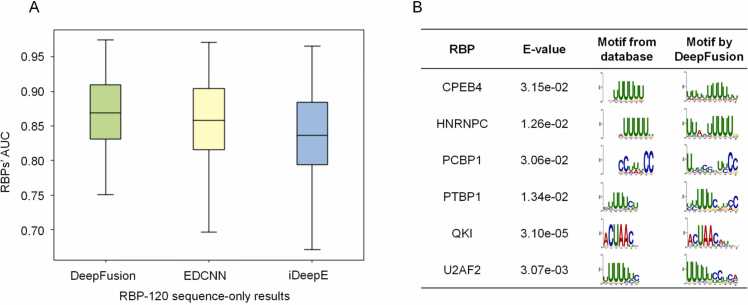
Fig. 3A shows their performances via box plots, which show the upper quantile, median, and lower quantile of the AUC performances for all 120 RBPs for the three methods. DeepFusion performs best, with a mean AUC of 0.874 for the 120 RBPs. Comparatively, EDCNN and iDeepE reported mean AUCs of 0.860 and 0.839, respectively, significantly lower than that for DeepFusion under the two-tailed paired t-test (1.24 ×10−9 and 3.50 ×10−30). As some RBPs cannot be predicted by EDCNN, we also compared the average performance across the remaining 99 RBPs, and DeepFusion still performs best. Moreover, DeepFusion’s performance is higher than its two submodels, DeepFusion-s and DeepFusion-l in the RBP-120 dataset. In addition, DeepFusion's performance superiority is maintained using the MCC and F1 score metrics. Again, we also created a non-redundant RBP-120 dataset and found that DeepFusion still performs best after de-redundancy. All these results can be found in the Supplementary Table S3.
Fig. 3.
Evaluation of DeepFusion’s performance in the RBP-120 dataset Performance of DeepFusion, iDeepE, and EDCNN using sequence-only input measured by AUC. B. Visualization of motifs extracted by DeepFusion and comparison with known motifs using TOMTOM in the RBP-120 dataset.
We also show in Fig. 3B the sequence motifs extracted by DeepFusion in the RBP-120 dataset. Some RBPs appear in the RBP-24 and RBP-120 datasets and have known motifs in the TOMTOM database, such as HNRNPC, PTBP1 (PTB), and QKI. DeepFusion extracts similar sequence motifs for them. Other new RBPs introduced in the RBP-120 dataset (e.g., CPEB4, PCBP1, and U2AF2) also have known motifs. DeepFusion extracts the U-rich motifs for CPEB4 and U2AF2 with E-values of 3.15 × 10−2 and 3.07 × 10−3, respectively, and the C-rich motifs for PCBP1 with an E-value of 3.06 × 10−2. These results demonstrate that DeepFusion can identify the intrinsic sequence features recognized by diverse RNA-binding proteins.
3.3. DeepFusion improves performance with the aid of in vivo RNA structures
We further evaluate DeepFusion on RBP-120 with bimodal inputs to fully exploit its bimodal architecture. We compare the performance of DeepFusion in three modes: The first mode accepts only single-modal sequence input, called sequence-only, and the latter two modes accept bimodal inputs with both sequence and structural information derived from the DMS-seq data. When pre-processing the DMS-seq data, we obtain the DMS reactivities for both in vivo and in vitro conditions. 69.2%/66.1% of the sequences in the RBP-120 dataset have in vivo/in vitro DMS-seq structural signals, which reflecting the single-stranded states of probed bases. As a result, the DeepFusion evaluation with bimodal inputs can be further classified into sequence+vitro or sequence+vivo modes.
Fig. 4A shows DeepFusion’s AUC scores for the three evaluations. The box plots indicate that DeepFusion improves performance with DMS-seq-derived structural information, especially in vivo RNA structures. The mean AUC goes from 0.874 in the sequence-only model to 0.927 in the sequence+vitro model and 0.933 in the sequence+vivo mode. In addition, the elevation is statistically significant, with a P-value of 3.08 × 10−30 between adding the in vitro signals or not and 6.72 × 10−35 for in vivo under the paired T-test, two-tailed. The detailed AUC score for each RBP for the three modes is provided in Supplementary Table S4. More performance improvement from the in vivo DMS-seq inputs is consistent with the biological rationale. CLIP-seq experiments are also performed under the in vivo conditions, making the bimodal inputs of sequence+vivo more coherent and compatible.
Fig. 4.
Performance improvement of DeepFusion when using DMS-seq structural input Performance of DeepFusion as measured by AUC on the RBP-120 dataset with different inputs, including sequence-only, sequence+vitro, and sequence+vivo. B. Scatter plot of each RBP’s performance as measured by AUC in the sequence+vivo mode compared with the sequence-only mode. C. Visualization of motifs extracted by DeepFusion after the addition of in vivo structural signal and comparison with known motifs using TOMTOM.
The scatter plot in Fig. 4B shows the performance difference between sequence-only and the sequence+vivo mode. Almost all RBPs get higher AUC scores after adding the in vivo structural signals and 71 of them improve performance by over 5%. We show that the better the sequence-only prediction, the less improvement from adding structural information. We also compared the AUC of sequence+vivo over sequence+vitro in a scatter plot in Supplementary Fig. 1. We further investigate how adding in vivo structural features to DeepFusion affects its sequence motif extraction, as shown in Fig. 4C. For RBPs whose motifs can be revealed only by sequence features (Fig. 3B), the sequence motifs can still be dissected after incorporating in vivo RNA structural features, such as PCBP1, PTBP1, and QKI. For other RBPs, adding structural signals clarifies their sequence motifs. For example, both DeepFusion and the previous database demonstrate an ACACAC motif for HNRNPL [46], and a poly-C motif for HNRNPK [54]. This motif analysis demonstrates that DeepFusion can use different modal data to improve the predictive power and interpretability for resolving the binding patterns of RBPs.
3.4. DeepFusion helps to dissect RNA degradation patterns
We further extend the application of DeepFusion in RNA degradation because RNA degradation is essential in the post-transcriptional regulation of RNAs regulated by RBPs. We use third-party data to distinguish two groups of genes: one group consists of 2181 genes with a low rate of degradation, and the other group consists of 2194 genes with a rapid rate of degradation. The degradation rate is measured by SLAM-seq in the same K562 cell line as the eCLIP-seq data [49].
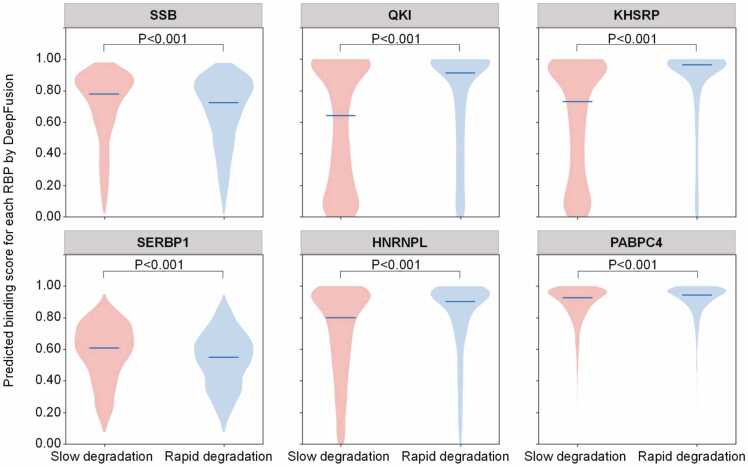
For the two gene groups, we evaluated whether they differ in their RBP binding profiles, as shown in Fig. 5. We selected six representative RBPs known to be involved in RNA stability. For example, QKI affects endolysosome-dependent degradation in glioma stem cells [55], and SERBP1 is known to regulate Serpine1 mRNA stability in human skeletal muscle [56]. Our analysis shows that the DeepFusion predictions for these RBPs in genes with slow degradation differ from those with rapid degradation. For instance, SSB and SERBP1 tend to concentrate more on genes with slow degradation than genes with rapid degradation. The median binding scores of the two RBPs with slow (rapid) degradation are 0.777 and 0.607 (0.724 and 0.547), respectively. In contrast, QKI, KHSRP, HNRNPL, and PABPC4 concentrate more on genes with rapid degradation than genes with slow degradation, with statistically higher binding scores in the rapid-degradation group. This suggests that DeepFusion’s prediction about RBP binding can give further insights about RNA degradation. In addition, we also conducted an opposite analysis and got significantly different degradation rates in genes with high DeepFusion predicted scores versus those with low scores. We showed that these analyses are statistically significant by providing detailed P-values in Supplementary Table S5. These results demonstrate the extended utility of DeepFusion.
Fig. 5.
Predicted RBP-binding scores by DeepFusion for genes with different RNA degradation rates. In each panel, genes that degrade slowly appear on the left and are colored in red, and genes that degrade rapidly appear on the right and are colored in blue, according to their decay rates as measured by SLAM-seq. For the two groups of genes, DeepFusion’s prediction scores of whether they are bound by six degradation-related RBPs are shown on the Y axis. The blue lines in the middle of each violin show the median scores. A two-tailed independent t-test measured the statistical significance of the differences between the two groups.
4. Conclusions
Analytical algorithms can determine the binding preferences of RBPs from experimental data. However, in vivo RNA structures based on high-throughput sequencing are rarely incorporated in algorithms for dissecting protein-RNA interactions. In this work, we introduce DeepFusion, a deep bimodal information fusion network containing RNA sequences and in vivo DMS-seq structural features to predict RBP bindings on RNAs. We evaluated DeepFusion and compared it with other cutting-edge methods based on two sets of CLIP-seq data: one is a widely used dataset called RBP-24, and the other is a comprehensive set called RBP-120 based on the ENCODE project. The results show that DeepFusion performs best on two independent datasets with only sequence inputs, and performs better when using bimodal input after adding in vivo DMS-seq structural features. The alignment of DeepFusion-predicted motifs with the motifs in existing databases can provide interpretations for DeepFusion's prediction results. In addition, DeepFusion can be used to analyze RNA degradation, expanding its potential application. Taken together, DeepFusion offers enhanced abilities to dissect the patterns of RNA-protein interactions and provides more support for further analysis of therapeutic RNAs.
Funding
This work was supported by the National Key R&D Program of China (2022YFC3500105); the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16021400); the National Natural Science Foundation of China (32070670); the Innovation Fund from Institute of Computing Technology, CAS (E161080) and the Zhejiang Provincial Natural Science Foundation of China (LY21C060003); Beijing Natural Science Foundation Haidian Origination and Innovation Joint Fund (L222007); National Natural Science Foundation of China (82172864). Funding for open access charge: the National Key R&D Program of China.
CRediT authorship contribution statement
Yixuan Qiao: Conceptualization, Methodology, Validation, Data analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization. Rui Yang: Methodology. Yang Liu: Methodology. Jiaxin Chen: Investigation. Lianhe Zhao: Writing – review & editing. Peipei Huo: Website. Zhihao Wang: Website. Dechao Bu: Website. Yang Wu: Conceptualization, Methodology, Validation, Data analysis, Data curation, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition. Yi Zhao: Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Thanks to Chunlong Luo, Yufan Luo, Jianwei Hong and Dr. Lianhe Zhao from the institute of computing technology, Chinese academy of sciences for their helpful discussions.
Code and data availability statement
We released the DeepFusion code as well as the entire RBP-120 dataset at http://bioinfo.org/deepfusion/. The DeepFusion code were also available at https://github.com/Qiaoyx97/DeepFusion.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.12.040.
Contributor Information
Yang Wu, Email: wuyang@ict.ac.cn.
Yi Zhao, Email: biozy@ict.ac.cn.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Turner M., Diaz-Munoz M.D. RNA-binding proteins control gene expression and cell fate in the immune system. Nat Immunol. 2018;19(2):120–129. doi: 10.1038/s41590-017-0028-4. [DOI] [PubMed] [Google Scholar]
- 2.Wang M., et al. Nicotine-mediated OTUD3 downregulation inhibits VEGF-C mRNA decay to promote lymphatic metastasis of human esophageal cancer. Nat Commun. 2021;12(1) doi: 10.1038/s41467-021-27348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akira S., Maeda K. Control of RNA Stability in Immunity. Annu Rev Immunol. 2021;39:481–509. doi: 10.1146/annurev-immunol-101819-075147. [DOI] [PubMed] [Google Scholar]
- 4.Akiyama T., Suzuki T., Yamamoto T. RNA decay machinery safeguards immune cell development and immunological responses. Trends Immunol. 2021;42(5):447–460. doi: 10.1016/j.it.2021.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Li J.T., et al. HNRNPK maintains epidermal progenitor function through transcription of proliferation genes and degrading differentiation promoting mRNAs. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-12238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramanathan M., Porter D.F., Khavari P.A. Methods to study RNA-protein interactions. Nat Methods. 2019;16(3):225–234. doi: 10.1038/s41592-019-0330-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strazar M., et al. Orthogonal matrix factorization enables integrative analysis of multiple RNA binding proteins. Bioinformatics. 2016;32(10):1527–1535. doi: 10.1093/bioinformatics/btw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corrado G., et al. RNAcommender: genome-wide recommendation of RNA-protein interactions. Bioinformatics. 2016;32(23):3627–3634. doi: 10.1093/bioinformatics/btw517. [DOI] [PubMed] [Google Scholar]
- 9.Pan X.Y., Shen H.B. Predicting RNA-protein binding sites and motifs through combining local and global deep convolutional neural networks. Bioinformatics. 2018;34(20):3427–3436. doi: 10.1093/bioinformatics/bty364. [DOI] [PubMed] [Google Scholar]
- 10.Van Nostrand E.L., et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13(6):508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hentze M.W., et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19(5):327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 12.Li X.T., Zhang S.X., Wong K.C. Multiobjective genome-wide RNA-binding event identification from CLIP-seq data. IEEE Trans Cybern. 2021;51(12):5811–5824. doi: 10.1109/TCYB.2019.2960515. [DOI] [PubMed] [Google Scholar]
- 13.Alipanahi B., et al. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33(8):831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 14.Pan X.Y., et al. Recent methodology progress of deep learning for RNA-protein interaction prediction. Wiley Interdiscip Rev-Rna. 2019;10(6) doi: 10.1002/wrna.1544. [DOI] [PubMed] [Google Scholar]
- 15.Pan X., Shen H.B. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinforma. 2017;18(1) doi: 10.1186/s12859-017-1561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan X.Y., Shen H.B. Learning distributed representations of RNA sequences and its application for predicting RNA-protein binding sites with a convolutional neural network. Neurocomputing. 2018;305:51–58. [Google Scholar]
- 17.Gronning A.G.B., et al. DeepCLIP: predicting the effect of mutations on protein-RNA binding with deep learning. Nucleic Acids Res. 2020;48(13):7099–7118. doi: 10.1093/nar/gkaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y.W., et al. EDCNN: identification of genome-wide RNA-binding proteins using evolutionary deep convolutional neural network. Bioinformatics. 2022;38(3):678–686. doi: 10.1093/bioinformatics/btab739. [DOI] [PubMed] [Google Scholar]
- 19.Ghanbari M., Ohler U. Deep neural networks for interpreting RNA-binding protein target preferences. Genome Res. 2020;30(2):214–226. doi: 10.1101/gr.247494.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J.X., et al. Genome-wide RNA structure changes during human neurogenesis modulate gene regulatory networks. Mol Cell. 2021;81(23) doi: 10.1016/j.molcel.2021.09.027. 4942-+ [DOI] [PubMed] [Google Scholar]
- 21.Dominguez D., et al. Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell. 2018;70(5) doi: 10.1016/j.molcel.2018.05.001. 854-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laverty K.U., et al. PRIESSTESS: interpretable, high-performing models of the sequence and structure preferences of RNA-binding proteins. Nucleic Acids Res. 2022;50(19) doi: 10.1093/nar/gkac694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L., et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell. 2021;184(7):1865–1883. doi: 10.1016/j.cell.2021.02.008. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., et al. Genome-wide RNA structure changes during human neurogenesis modulate gene regulatory networks. Mol Cell. 2021;81(23):4942–4953. doi: 10.1016/j.molcel.2021.09.027. e8. [DOI] [PubMed] [Google Scholar]
- 25.Yu B., et al. Differential analysis of RNA structure probing experiments at nucleotide resolution: uncovering regulatory functions of RNA structure. Nat Commun. 2022;13(1) doi: 10.1038/s41467-022-31875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Z., et al. A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell. 2016;64(1):37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orenstein Y., Wang Y.H., Berger B. RCK: accurate and efficient inference of sequence- and structure-based protein-RNA binding models from RNAcompete data. Bioinformatics. 2016;32(12):351–359. doi: 10.1093/bioinformatics/btw259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ben-Bassat I., Chor B., Orenstein Y. A deep neural network approach for learning intrinsic protein-RNA binding preferences. Bioinformatics. 2018;34(17):638–646. doi: 10.1093/bioinformatics/bty600. [DOI] [PubMed] [Google Scholar]
- 29.Steffen P., et al. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22(4):500–503. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- 30.Maticzka D., et al. GraphProt: modeling binding preferences of RNA-binding proteins. Genome Biol. 2014;15(1):R17. doi: 10.1186/gb-2014-15-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernhart S.H., Hofacker I.L., Stadler P.F. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22(5):614–615. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- 32.Yan Z.C., Hamilton W.L., Blanchette M. Graph neural representational learning of RNA secondary structures for predicting RNA-protein interactions. Bioinformatics. 2020;36:276–284. doi: 10.1093/bioinformatics/btaa456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarver M., et al. FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J Math Biol. 2008;56(1-2):215–252. doi: 10.1007/s00285-007-0110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahrig R.R., Leontis N.B., Zirbel C.L. R3D Align: global pairwise alignment of RNA 3D structures using local superpositions. Bioinformatics. 2010;26(21):2689–2697. doi: 10.1093/bioinformatics/btq506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S., et al. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2016;44(4) doi: 10.1093/nar/gkv1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y., et al. Improved prediction of RNA secondary structure by integrating the free energy model with restraints derived from experimental probing data. Nucleic Acids Res. 2015;43(15):7247–7259. doi: 10.1093/nar/gkv706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spitale R.C., et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519(7544) doi: 10.1038/nature14263. 486-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rouskin S., et al. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505(7485) doi: 10.1038/nature12894. 701-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun L., et al. Predicting dynamic cellular protein-RNA interactions by deep learning using in vivo RNA structures. Cell Res. 2021;31(5):495–516. doi: 10.1038/s41422-021-00476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maticzka D., et al. GraphProt: modeling binding preferences of RNA-binding proteins. Genome Biol. 2014;15(1) doi: 10.1186/gb-2014-15-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y., et al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakrabarti A.M., Haberman N., Praznik A., Luscombe N.M., Ule J. Data science issues in studying protein–RNA interactions with CLIP technologies. Annu Rev Biomed Data Sci. 2018;1:235–261. doi: 10.1146/annurev-biodatasci-080917-013525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Danecek P., et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10(2) doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y., et al. RNAex: an RNA secondary structure prediction server enhanced by high-throughput structure-probing data. Nucleic Acids Res. 2016;44(W1):W294–W301. doi: 10.1093/nar/gkw362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ray D., et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S., et al. A deep learning framework for modeling structural features of RNA-binding protein targets. Nucleic Acids Res. 2016;44(4) doi: 10.1093/nar/gkv1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S., et al. Quantifying similarity between motifs. Genome Biol. 2007;8(2) doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu Q.S., et al. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife. 2019:8. doi: 10.7554/eLife.45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao F.B., et al. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci USA. 1994;91(23):11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konig J., et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–U166. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dember L.M., et al. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J Biol Chem. 1996;271(5):2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 53.Paszke A., et al. Pytorch: an imperative style, high-performance deep learning library. Adv Neural Inf Process Syst. 2019:32. [Google Scholar]
- 54.Nakamoto M.Y., et al. hnRNPK recognition of the B motif of Xist and other biological RNAs. Nucleic Acids Res. 2020;48(16):9320–9335. doi: 10.1093/nar/gkaa677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shingu T., et al. Qki deficiency maintains stemness of glioma stem cells in suboptimal environment by downregulating endolysosomal degradation. Nat Genet. 2017;49(1):75–86. doi: 10.1038/ng.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Needham E.J., et al. Phosphoproteomics of acute cell stressors targeting exercise signaling networks reveal drug interactions regulating protein secretion. Cell Rep. 2019;29(6):1524–1538. doi: 10.1016/j.celrep.2019.10.001. e6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material