Abstract
Gastrointestinal Stromal Tumors (GIST) are the most frequent mesenchymal neoplasia of the digestive tract. Genomic alterations in KIT, PDFGRA, SDH, and BRAF genes are essential in GIST oncogenesis. Therefore, the mutations in these genes have demonstrated clinical implications. Tumors with deletions in KIT-exon 11 or duplications in exon 9 are associated with a worse prognosis. In contrast, KIT-exon 11 substitutions and duplications are associated with a better clinical outcome. Moreover, mutations in Kit exon 9 and 11 are actionable, due to their response to imatinib, while mutations in PDGFRA respond to sunitinib and/or avapritinib. Although, molecular testing on tissue samples is effective; it is invasive, requires adequate amounts of tissue, and a long experimental process is needed for results. In contrast, liquid biopsy has been proposed as a simple and non-invasive method to test biomarkers in cancer. The most common molecule analyzed by liquid biopsy is circulating tumor DNA (ctDNA). GISTs ctDNA testing has been demonstrated to be effective in identifying known and novel KIT mutations that were not detected using traditional tissue DNA testing and have been useful in determining progression risk and response to TKI therapy. This allows the clinician to have an accurate picture of the genetic changes of the tumor over time. In this work, we aimed to discuss the implications of mutational testing in clinical outcomes, the methods to test ctDNA and the future challenges in the establishment of alternatives of personalized medicine.
Keywords: GIST, Genomic profiling, Mutations, Treatment, KIT, PDGFRA
Synopsis.
Molecular testing is fundamental to assessing therapeutic decisions in GIST patients. Liquid biopsy as a feasible clinical biomarker with clinical applications in improving diagnosis, clinical management, and outcomes in GIST patients.
Alt-text: Unlabelled box
Introduction
Gastrointestinal stromal tumors (GIST) are the most frequent mesenchymal cancer of the gastrointestinal tract, worwilde incidence of these tumors is estimated at two cases per 100,000 and the prevalence is estimated at 13 cases per 100,000 [1]. Most GISTs are derived from interstitial cells of Cajal (ICC), meanwhile, a less proportion is developed from telocytes, and smooth muscle cells [2]. The main location of primary GISTs is the stomach (60 %), followed by the small intestine (20 %); while the rectum, colon, and esophagus are less frequent sites reported [3,4]. The gold standard technique for diagnosis is the fine needle aspiration, that had demonstrated clinical advantages compared to traditional biopsies [5]. Additionally, the analysis of immunohistochemistry (IHC) and immunophenotype markers as CD-117, DOG1, CD-34, and KIT are essential to a proper tumor classification [3]. After diagnosis, the establishment of risk stratification through the evaluation of tumor size, anatomical site, and mitotic index is fundamental to assessing treatment response, progression, and overall survival [2,6]. Furthermore, molecular profiling is essential for GIST prognosis [7], the most frequent mutations are located in KIT (80 %), platelet-derived growth factor receptor alpha (PDGFRA) (10 %), and BRAF, KRAS, PIK3CA, NF-1 [6] (altogether 10 %) genes [8]. Mutation analysis is often carried out in a tumor tissue sample, notwithstanding, it has some limitations including the quality of samples, the amount of tissue required, and the time to perform the test [9]. On the other hand, a recently proposed method called liquid biopsy allows the detection, analysis and monitoring of tumor mutations through a peripheral blood sample [10], [11], [12]. Using this method, the measurement of circulating tumoral cells (CTCs), free circulating nucleic acids (DNA, mRNA, non-coding RNA), "tumor-educated platelets" (TEPs) or vesicles such as exosomes can be tested [13]. Along these lines, the main of this review is to provide a comprehensive analysis of the oncogenic drivers in GIST, outstanding the launching of liquid biopsy as an alternative method for molecular testing that improves the stratification, clinical management and outcome of GIST.
Landscape of genomic and epigenomic alterations in GIST
Neoplastic transformation and progression of GIST involve several molecular modifications such as genetic mutations, chromosomal alterations, and epigenetic abnormalities. For an appropriated clinical management of GISTs the genomic profiling and molecular classification are fundamental. Approximately 80 % of GISTs harbor activating mutations in KIT gene, [14,15]. KIT encodes the 145 kDa receptor tyrosine kinase c-KIT that is member of the type III tyrosine kinase receptor family [16]. Its cytoplasmic domain is constituted by a Juxta-membrane domain (JM) and tyrosine kinase domains 1 and 2 (TK1 and TK2). TK domains contain a binding site for ATP and a phosphotransferase region separated by a kinase insert. In the absence of the KIT ligand Stem cell factor (SCF), the JM domain inhibits the tyrosine kinase activity of KIT [17]. SCF-KIT pathway activates downstream MAP kinase, PI3K/AKT, and STAT3 signaling pathways, signals leading to cell growth [16]. The most prevalent mutations in the KIT gene are located in exons 8, 9, 11, 13, and 17, among them, mutations in exon 11 are the most frequent and affect the juxta-membrane (JM) domain of the protein [18]. The JM domain normally functions to stabilize the KIT receptor in an inactive conformation and inhibits dimerization. Mutations that cause loss of function of the JM domain induce dimerization and autophosphorylation [7], leading to a constant autonomous activation, uncontrolled proliferation, and apoptosis inhibition. Mutations in the JM domain are mostly caused by in-frame deletions in codon Gln550 and Glu560, known as a hot spot region [18]. Besides, codons 557-558 are also known as hot spots, deletions of W557 and/or K558 have been reported in 28 % of all GISTs and are associated with high-risk tumors due to their clinicopathological features such as higher mitotic index (>50 HPF), larger tumor size, the high incidence in young populations (<60 years), metastatic phenotype, and poor recurrence-free survival [7,16,18]. On the other hand, mutations in exon 9 are found in approximately 10 % of cases and are characterized by tandem duplication of six nucleotides at codons 502-503 (p. A502_Y503dup) and are reflected in extracellular domains of protein [18]. These types of GISTs arise commonly in the small bowel and are often associated with more aggressive characteristics such as larger tumor size, advanced age (>60 years), female sex, and spindle cell morphology [16,18]. Other less frequent KIT spots are in exon 13, 17, and 8, and occur in approximately 1 %–2 % of KIT-GISTs. Exon 13 mutations (TK[I]: ATP-binding pocket), result in p.K642E, and therefore suppressing auto-inhibition of the JM domain [7]. These tumors are most often found in the small intestine, usually have a spindle cell morphology, are slightly larger, and are more aggressive tumors than other types of GIST. Regarding exon 17, the 70 % of mutations are c.2487T>A substitution mutation, affecting codon 822 (TK[II]: kinase activation loop). These tumors arise frequently in the small intestine and usually present a spindle cell morphology. Furthermore, mutations in exon 8 occur rarely in GIST. These tumors are associated with a malignant phenotype, and multiple peritoneum metastasis [16].
As we mentioned before mutations in the PDGFRA gene are the second most common molecular subtype of GIST. The mutations are described in exon 12 (1 %) which codifies a portion of the JM domain of the protein, in exon 14 (<1 %) coding ATP binding domain, and in exon 18 (5 %) which comprises the activation loop. All these mutations cause constitutive activation of PDGFRA in the absence of ligand binding [19]. Similar to KIT mutations, PDGFRA mutations can activate a number of signal transduction molecules, including MAPK, AKT, STAT1, and STAT3 [16]. GISTs with PDGFRA mutations originated mainly in the stomach (90 %-93 %), are histologically characterized by epithelioid or mixed epithelioid and spindle cell tissue, and are often with myxoid stromal change [20,21]. The mutation in p. D842V originated in exon 18 (accounts for 60-65 % of all PDGFRA mutations in GIST), a region that encodes the second kinase domain and is associated with extremely favorable disease-free survival compared to other types of mutations [16]. Also, exon 12 PDGFRA mutation is account for 1-2 % of overall GISTs. This mutation is more frequently detected as a deletion than a duplication, and the most frequent site is 1821T>A, resulting in a Val561Asp substitution at the protein level. Otherwise, exon 14 mutation induces N659K substitution in protein, this mutation is relatively rare compared to others and is associated with a better clinical outcome [21].
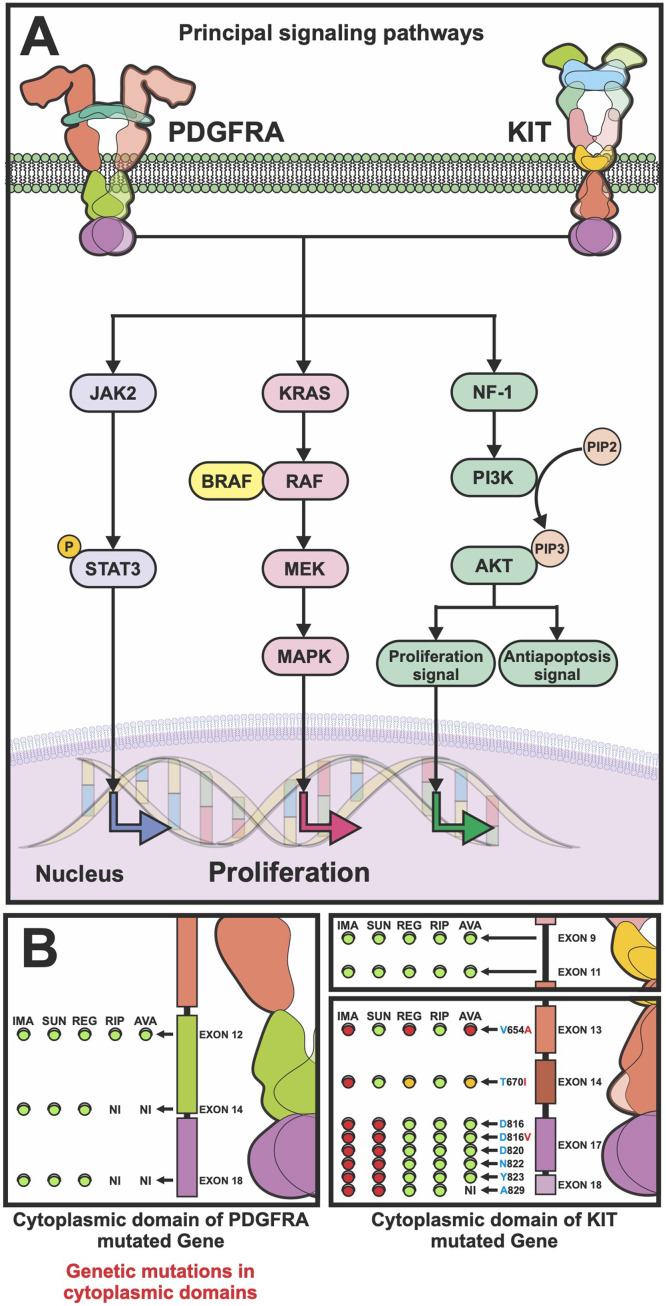
Other GIST mutations (BRAF, KRAS, and PIK3CA) are detected in less than 10 % of cases. BRAF and KRAS genes play a crucial role in tumorigenesis, known as the most deregulated genes among different types of cancer [22]. The BRAF gene codes for a serine/threonine protein kinase controls proliferation and differentiation through the Ras-Raf-MAPK kinase pathway [23]. BRAF is a member of the RAS-RAF-MEK-ERK pathway that is involved in cell cycle regulation and oncogenic modulation of cellular responses to growth signals by activating the mitogen-activated protein kinase (MAPK) pathway [24]. Most GIST-BRAF mutations cluster in a hot spot at nucleotide 1799 of exon 15 leading to the substitution of valine for glutamic acid at codon 600 (V600E) [24]. Besides, it has been reported that patients with BRAF mutations were also detected wild type for the BRAF gene, confirming the exclusivity of these 2 gene mutations [24]. KRAS gene is frequently mutated at codon 12 (G12D) or 13 (G13D and G12A/G13D). The tumors carrying the G12D and G12A/G13D substitutions also show deletions in exon 11 of KIT (Δ570-576 and Δ579), while the tumor with the G13D substitution had additional mutations in exon 18 (D842V) of PDGFRA gene [24]. BRAF-KRAS mutated GISTs are often found in small GISTs (4 mm long) thus this mutation is described to appear on the onset of GIST development [25]. In addition to mutations, up to 70 % of GISTs have been associated with alterations in chromosome 14, including loss of 14q and monosomy 14. The loss of 14q is associated with gastric localization, predominantly stable karyotypes, and favorable clinical outcomes [26]. Moreover, almost half of GISTs show loss of 22q, while losses of 1p, 9p, 10q, 11p, 13q, 15q and 17p are less frequent [26]. On the other hand, epigenetic changes such as significant hypomethylation in LINE-1 have been observed in high-risk GISTs, especially in those with metastases, worse prognosis, and resistance to treatment due to rapid cell proliferation, high mitotic index, and large tumor size [26]. Another molecular subtype of GIST is succinate dehydrogenase (SDH) deficient, these types of GISTs have specific clinical features, morphological and immunohistochemical characteristics including the absence of KIT/PDGFRA, but positive staining for cKIT, DOG1 [26], and defects in energy metabolism as a key oncogenic mechanism. Another molecular marker is the insulin-like growth factor 1 receptor (IGF1R), which is overexpressed in GISTs with KIT/PDGFR and is particularly elevated in SDH-deficient GISTs [14]. The IGF family consists of two ligands (IGF1 and IGF2), two receptors (IGFR1 and IGFR2) and 6 IGF-binding proteins (IGFBPs). The IGF and IGFR binding activates downstream signaling, including MAPK and PI3K/AKT pathways [14,27]. Inhibition of IGF1R induces apoptosis and represses AKT and MAPK signaling of SDH-deficient GISTs [26,18]. SDH-deficient GISTs diagnosis is considered mostly in gastric GIST and is particularly common in childhood and adolescence [21]. This kind of tumor is commonly multifocal and/or show a lobulated and multinodular growth pattern, are often associated with metastatic disease, and resistance to treatment [21]. A syndrome associated in 7 % of GISTs is neurofibromatosis type 1 (NF1), these patients show somatic mutations or loss of the remaining neurofibromin 1 (NF1) allele, leading to increased signal transduction through MAP-kinase pathway [27,28]. These GISTs are often located in small intestine and metastatic, nevertheless, have low mitotic rate and are associated to a good prognosis [29,30]. In Fig. 1A the principal signaling pathways activated in GIST development and progression, by consequence of genomic alterations described above, are schematized.
Fig. 1.
Molecular landscape in GIST is related to treatment response. (A) principal signaling pathways involved in GIST development and progression. The hyperactivity of tyrosine kinase receptors triggers Jack/Stat, K-ras/MAPK and NF-1 signaling pathways to induce cell proliferation. (B) Drug response of GIST tumors is related to KIT or PDGFRA gene mutations. Tyrosine kinase inhibitors are directed to alterations in cytoplasmic domain of the codified-receptors. Color circles represent sensitivity (green) or resistance (red) to the respective pharmacologic agent.
Therapeutic outline: challenges in primary and second-line resistance to treatment
Tyrosine kinase inhibitors (TKI) are the gold standard treatment for advanced GISTs. The first line treatment approved worldwide is imatinib, followed by sunitinib, regorafenib, and ripretinib approved as the second, third, and fourth line respectively [31]. Resistance to treatment represents a challenge for therapeutic approaches, compromising the quality of life and overall survival rates for GISTs patients. On behalf of this, progression has been studied to underline primary and secondary resistance. Both can be explained by mutations that carry out variant conformational changes in the KIT and PDFGRA kinase domain triggering a constitutively activated state [18]. Primary resistance (PR) is defined as the tumor progression within the first 6 months of treatment [32]. Due that imatinib binds only to the inactive conformation of KIT and PDGFRA receptors, patients with mutations in PDGFRA D842V and KIT exon 9-mutated GISTs (codify active forms of protein receptors) are reported to have primary resistance to treatment [18,32]. On the other hand, secondary resistance (SR) appears to be after 12–36 months of first-line treatment and has been reported in up to two-thirds of GISTs [32]. This resistance is driven by secondary mutations or genomic amplifications of KIT/PDGFRA genes that lead to alterations in the kinase domain of the receptor to trigger the oncogenes activation. KIT secondary mutations are non-randomly distributed single nucleotide substitutions in 2 regions: (1) The encoded by exons 13 and 14 that codify ATP-interaction pocket and interfere with the TKI binding; (2) Mutations in exon 17 induce the stabilization of KIT in the active conformation and thus preclude TKI interaction [19]. These SR-KIT mutations are found in 50–67 % of patients with secondary imatinib-resistant. Therefore, the monitoring of them during treatment is essential to make timely decisions about therapeutic options in GIST [19]. Fig. 1B, summarizes the therapy response to the five tyrosine kinase inhibitors currently approved for GIST treatment depending on the molecular profiling. The drugs resistance is represented in red light and the sensitivity is highlighted in green. In this context, the molecular landscape determination in GIST is relevant not only at the time of diagnosis to the establishment of prognostic assessment, but also in monitoring the diseases during pharmacological treatment. The gold-standard method of profiling tumors historically involves obtaining tumor samples from surgery. Nevertheless, the limitations of these invasive procedures include the difficulty of obtaining the quality and quantity of tumor samples required. In addition, in case of high burden or metastatic disease, multiple biopsies may be needed. Due to the challenges described before with traditional biopsies, oncology research has focused on analyzing other biological fluids, also known as liquid biopsy, for tumor-derived components [33]. This method has been demonstrated to be feasible and non-invasive to screening resistant mutations and therefore enhance progression-free survival and overall survival for GIST patients [18].
Liquid biopsy: the future of GIST diagnosis and prognosis
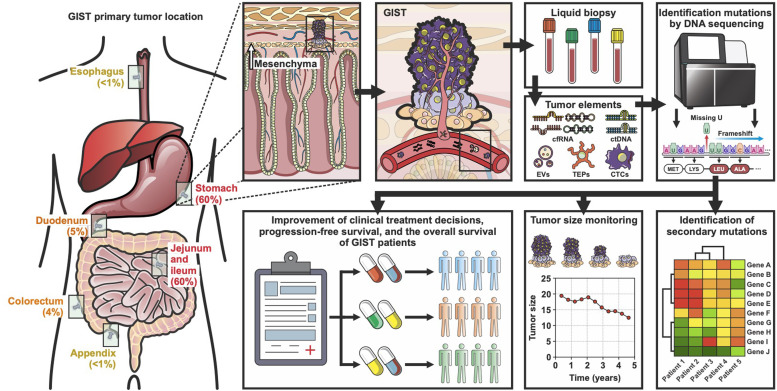
Precision medicine has led to monumental breakthroughs in oncology. The aim of precision oncology is to individualize treatment based on molecular profiling, identifying the main drivers of progression and metastasis. Even though immunohistochemistry is useful to assess GIST diagnosis, it is not enough to determine specific harbor mutations. Consequently, molecular profiling analysis is fundamental to guiding treatment and assessing the prognosis of GIST patients [34]. Nevertheless, mutational testing from tissue biopsy has some limitations, including large time to response and accessibility. Thus, novel, fast, accurate, and affordable techniques remain fundamental to include mutational testing in gist diagnosis [35,36]. Contrary to tumor tissue biopsy, liquid biopsy is a novel technique characterized for being noninvasive, safe, and rapid to assess for genetic mutations at the time of diagnosis, during treatment, and progression. This technique can be used to determine genomic and proteomic tumor-derived moieties, including circulating tumor DNA (ctDNA), circulating tumor cells (CTCs); tumor-derived extracellular vesicles (EVs); tumor-educated platelets (TEPs), and circulating cell-free RNA (cfRNA), composed of small RNAs/miRNAs in liquid tissues as peripheral blood. For instance, ctDNA assessments have been demonstrated to be effective in non-small cell lung cancer (NSCLC) [37], colon [38], and GISTs [39]. Regarding GIST, Arshad et al, compared ctDNA and solid tissue with next generation sequencing (NGS) technique, demonstrating a positive predictive value (PPV) of 100 %, a specificity concordance of 100 %, and a sensitivity of 56 % in metastatic patients or high burden tumor [39]. Besides, other clinical characteristics have been correlated with the positivity rate of CTCs in liquid biopsies, such as Ki 67 expression, mitotic count, and tumor diameter, all three characteristics used to evaluate progression risk in GIST tumors. Furthermore, ctDNA analysis has also been demonstrated to be effective in monitoring tumor burden and the response to treatment in cKIT and PDGFRA-mutated GISTs [40]. Additional to NGS, ctDNA can be analyzed by other techniques as qPCR, BEAMing, and droplet digital PCR (ddPCR) [41,42]. Recently, ddPCR has been proposed as a reliable and affordable method to the detection of GIST mutations that mediate TKI-response showing high analytical sensitivity [42], [43], [44], [45], [46], [47]. All this evidence supports the idea that ctDNA analysis, either by NGS or ddPCR, can complement early diagnosis, predict response evaluation, and detection of progression, representing a master key in the era of personalized medicine [42]. Fig. 2, represents the most frequent tumor locations, workflow and clinical applications of liquid biopsy in GIST tumors. Even though, some limitations challenge the use of liquid biopsy as a feasible clinical biomarker, including experimental design, variety of methodology, and preanalytical handling of samples the use of this technique has demonstrated to have great potential to improve clinical management and outcome in GIST patients.
Fig. 2.
Liquid biopsy implications in genomic profiling of GIST. The utility of liquid biopsy in diagnosis and monitoring of GIST is schematized. Liquid biopsy is one of the most useful techniques to determine molecular profiling, define the optimal choice of therapy, and predict response to treatment based on resistance mutations, representing a master key in the era of personalized medicine.
Conclusions and perspectives
Mutational testing is fundamental for clinical management and monitoring responses to the treatment of GIST patients. Nevertheless, genomic profiling is not a mandatory procedure in clinical guidelines worldwide, due to the limitations involved in tumor tissue biopsies such as high costs, not accessible tumors, and the tissue complex quality standards for its proper analysis. Besides, in patients with non-resectable tumors, the determination of the genomic landscape is crucial to pharmacological treatment decisions. However, GIST diagnosis in the metastatic scenario is performed through a tru-cut needle biopsy accessing to a low quantity and quality of the sample which precludes the analysis. On the other hand, liquid biopsy is an accessible, feasible, noninvasive, novel method that can be an alternative to making a proper diagnosis. In addition, the ctDNA detection rates in metastatic disease, particularly in primary and secondary resistance landscapes, have increased ctDNA shedding with higher mutated allele fractions. Consequently, liquid biopsy is a feasible alternative not only in the detection of primary detections, but also in monitoring resistance to treatment and therefore to enhancing the quality of life and progression-free survival of patients with metastatic disease. For the reasons mentioned above, the implementation of genomic profiles using liquid biopsy can improve clinical treatment decisions, progression-free survival, and the overall survival of GIST patients. Moreover, ctDNA analysis should be studied prospectively in real-world clinical-genomic trials to propose the routine clinical use of liquid biopsy.
Grant support
The authors did not receive support from any organization for the submitted work.
CRediT authorship contribution statement
German Calderillo-Ruíz: Conceptualization, Methodology. Eloy Andrés Pérez-Yepez: Conceptualization, Methodology, Writing – original draft. María Alejandra García-Gámez: Writing – original draft. Oliver Millan-Catalan: Writing – original draft. Consuelo Díaz-Romero: Writing – review & editing. Paul Ugalde-Silva: Writing – review & editing. Rodrigo Salas-Benavides: Writing – review & editing. Carlos Pérez-Plasencia: Conceptualization, Methodology. Berenice Carbajal-López: Conceptualization, Methodology, Writing – original draft.
Declaration of competing interest
The authors declare no conflict of interest. This manuscript has not been submitted in other journal.
Acknowledgments
Fundación GIST México ABP, Laboratorio de Genómica, Instituto Nacional de Cancerología.
Contributor Information
Carlos Pérez-Plasencia, Email: carlos.pplas@unam.mx.
Berenice Carbajal-López, Email: eb.carbajalopez@gmail.com.
References
- 1.Mandahl, N.; Mertens, F. Soft tissue tumors; 2010; ISBN 9780470181799.
- 2.Parab T.M., DeRogatis M.J., Boaz A.M., Grasso S.A., Issack P.S., Duarte D.A., Urayeneza O., Vahdat S., Qiao J.H., Hinika G.S. Gastrointestinal stromal tumors: a comprehensive review. J. gastrointest. Oncol. 2019;10:144–154. doi: 10.21037/JGO.2018.08.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Share B., Alloghbi A., Al Hallak M.N., Uddin H., Azmi A., Mohammad R.M., Kim S.H., Shields A.F., Philip P.A. Gastrointestinal stromal tumor: a review of current and emerging therapies. Cancer Metastasis Rev. 2021;40:625–641. doi: 10.1007/S10555-021-09961-7. [DOI] [PubMed] [Google Scholar]
- 4.Peng F., Liu Y. Gastrointestinal stromal tumors of the small intestine: progress in diagnosis and treatment research. Cancer Manag. Res. 2020;12:3877–3889. doi: 10.2147/CMAR.S238227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Renberg S., Papakonstantinou A., Haglund de Flon F. Diagnosing gastrointestinal stromal tumors: the utility of fine-needle aspiration cytology versus biopsy. Cancer Med. 2022;11:2729–2734. doi: 10.1002/CAM4.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casali P.G., Abecassis N., Bauer S., Biagini R., Bielack S., Bonvalot S., Boukovinas I., Bovee J.V.M.G., Brodowicz T., Broto J.M., et al. Gastrointestinal stromal tumours: ESMO-EURACAN clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018;29:iv68–iv78. doi: 10.1093/annonc/mdy095. [DOI] [PubMed] [Google Scholar]
- 7.Blay, J. Gastrointestinal stromal tumours 1 ✉ 4. 2021, 0123456789, 1–22. [DOI] [PubMed]
- 8.Go, D.; Garcı, A.; Pilco-janeta, D. Liquid biopsy in gastrointestinal stromal tumors: ready for prime time? 2021, doi:10.1007/s11864-021-00832-5. [DOI] [PubMed]
- 9.Ou S.-H.I., Nagasaka M., Zhu V.W. Vol. 38. American Society of Clinical Oncology; 2018. Liquid biopsy to identify actionable genomic alterations; p. 978. (American Society of Clinical Oncology Educational Book). Annual Meeting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tay T.K.Y., Tan P.H. Liquid biopsy in breast cancer: a focused review. Arch. Pathol. Lab. Med. 2021;145:678–686. doi: 10.5858/ARPA.2019-0559-RA. [DOI] [PubMed] [Google Scholar]
- 11.Vacante, M.; Ciuni, R.; Basile, F.; Biondi, A. Biomedicines the liquid biopsy in the management of colorectal cancer: an overview., doi:10.3390/biomedicines8090308. [DOI] [PMC free article] [PubMed]
- 12.Nagasaka M., Uddin M.H., Al-Hallak M.N., Rahman S., Balasubramanian S., Sukari A., Azmi A.S. Liquid biopsy for therapy monitoring in early-stage non-small cell lung cancer. Mol. Cancer. 2021;20:1–16. doi: 10.1186/S12943-021-01371-1. 2021 20:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulet G., Massias J., Taly V. Liquid biopsy: general concepts. Acta Cytol. 2019;63:449–455. doi: 10.1159/000499337. [DOI] [PubMed] [Google Scholar]
- 14.Wu C.E., Tzen C.Y., Wang S.Y., Yeh C.N. Clinical diagnosis of Gastrointestinal Stromal Tumor (GIST): from the molecular genetic point of view. Cancers. 2019:11. doi: 10.3390/CANCERS11050679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi J.H., Ro J.Y. The recent advances in molecular diagnosis of soft tissue tumors. Int. J. Mol. Sci. 2023;24 doi: 10.3390/IJMS24065934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niinuma T., Suzuki H., Sugai T. Molecular characterization and pathogenesis of gastrointestinal stromal tumor. Transl. Gastroenterol. Hepatol. 2018;2018:1–15. doi: 10.21037/tgh.2018.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buleje J., Yábar A., Guevara-Fujita M., Fujita R., De Revisión A. Características genético-moleculares de los tumores estromales Gastrointestinales (GIST) Rev. Gastroenterol. Perú. 2012;32:394–399. [PubMed] [Google Scholar]
- 18.Brčić I., Argyropoulos A., Liegl-Atzwanger B. Update on molecular genetics of gastrointestinal stromal tumors. Diagnostics (Basel, Switzerland) 2021;11 doi: 10.3390/DIAGNOSTICS11020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masucci M.T., Motti M.L., Minopoli M., Di Carluccio G., Carriero M.V. Emerging targeted therapeutic strategies to overcome imatinib resistance of gastrointestinal stromal tumors. Int. J. Mol. Sci. 2023;24 doi: 10.3390/IJMS24076026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H., Liu Q. Prognostic indicators for gastrointestinal stromal tumors: a review. Transl. Oncol. 2020;13 doi: 10.1016/J.TRANON.2020.100812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bannon A.E., Klug L.R., Corless C.L., Heinrich M.C. Using molecular diagnostic testing to personalize the treatment of patients with gastrointestinal stromal tumors. Expert Rev. Mol. Diagn. 2017;17:445–457. doi: 10.1080/14737159.2017.1308826. [DOI] [PubMed] [Google Scholar]
- 22.Mei L., Smith S.C., Faber A.C., Trent J., Grossman S.R., Stratakis C.A., Boikos S.A. Gastrointestinal stromal tumors: the GIST of precision medicine. Trends Cancer. 2018;4:74–91. doi: 10.1016/J.TRECAN.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Szucs Z., Thway K., Fisher C., Bulusu R., Constantinidou A., Benson C., Van Der Graaf W.T.A., Jones R.L. Molecular subtypes of gastrointestinal stromal tumors and their prognostic and therapeutic implications. Future Oncol. 2017;13:93–107. doi: 10.2217/FON-2016-0192. (London, England) [DOI] [PubMed] [Google Scholar]
- 24.Huss S., Pasternack H., Ihle M.A., Merkelbach-Bruse S., Heitkötter B., Hartmann W., Trautmann M., Gevensleben H., Büttner R., Schildhaus H.U., et al. Clinicopathological and molecular features of a large cohort of Gastrointestinal Stromal Tumors (GISTs) and review of the literature: BRAF mutations in KIT/PDGFRA wild-type GISTs are rare events. Hum. Pathol. 2017;62:206–214. doi: 10.1016/J.HUMPATH.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Avery T.Y., Köhler N., Zeiser R., Brummer T., Ruess D.A. Onco-immunomodulatory properties of pharmacological interference with RAS-RAF-MEK-ERK pathway hyperactivation. Front. Oncol. 2022;12 doi: 10.3389/FONC.2022.931774/PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niinuma T., Suzuki H., Sugai T. Molecular characterization and pathogenesis of gastrointestinal stromal tumor. Transl. Gastroenterol. Hepatol. 2018;3 doi: 10.21037/TGH.2018.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeRoith D., Holly J.M.P., Forbes B.E. Insulin-like growth factors: ligands, binding proteins, and receptors. Mol. Metab. 2021;52 doi: 10.1016/J.MOLMET.2021.101245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bharath B.G., Rastogi S., Ahmed S., Barwad A. Challenges in the management of metastatic gastrointestinal stromal tumor in a patient with neurofibromatosis type 1: a case report. J. Med. Case Rep. 2022;16:1–7. doi: 10.1186/S13256-022-03382-Y/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arshad J., Ahmed J., Subhawong T., Trent J.C. Progress in determining response to treatment in gastrointestinal stromal tumor. Expert Rev. Anticancer Ther. 2020;20:279. doi: 10.1080/14737140.2020.1745068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segawa K., Sugita S., Sugawara T., Ito Y., Tsujiwaki M., Fujita H., Ono Y., Kobayashi K., Hirobe M., Yoshida M., et al. Multiple gastrointestinal stromal tumors involving extragastrointestinal sites in neurofibromatosis type 1. Pathol. Int. 2018;68:142–144. doi: 10.1111/PIN.12620. [DOI] [PubMed] [Google Scholar]
- 31.Li Y., Gao Z.D., Yang X.D., Ye Y.J., Wang S., Jiang K.W. Gastrointestinal stromal tumors with KIT mutation coexisting with wild-type gastrointestinal stromal tumors in a patient with neurofibromatosis type 1. Chin. Med. J. (Engl.) 2018;131:2244. doi: 10.4103/0366-6999.240814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallilas C., Sarantis P., Kyriazoglou A., Koustas E., Theocharis S., Papavassiliou A.G., Karamouzis M.V. Gastrointestinal Stromal Tumors (GISTS): novel therapeutic strategies with immunotherapy and small molecules. Int. J. Mol. Sci. 2021;22:1–13. doi: 10.3390/IJMS22020493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Renberg S., Papakonstantinou A., Haglund de Flon F. Diagnosing gastrointestinal stromal tumors: the utility of fine-needle aspiration cytology versus biopsy. Cancer Med. 2022;11:2729. doi: 10.1002/CAM4.4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arshad J., Roberts A., Ahmed J., Cotta J., Pico B.A., Kwon D., Trent J.C. Utility of circulating tumor DNA in the management of patients with GI stromal tumor: analysis of 243 patients. JCO Precis. Oncol. 2020;4:66–73. doi: 10.1200/PO.19.00253. [DOI] [PubMed] [Google Scholar]
- 35.Lone S.N., Nisar S., Masoodi T., Singh M., Rizwan A., Hashem S., El-Rifai W., Bedognetti D., Batra S.K., Haris M., et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer. 2022;21:1–22. doi: 10.1186/S12943-022-01543-7. 2022 21:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravarty D., Gao J., Phillips S., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H., et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017;2017:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartzberg L., Kim E.S., Liu D., Schrag D. Vol. 37. American Society of Clinical Oncology; 2017. Precision oncology: who, how, what, when, and when not? pp. 160–169. (American Society of Clinical Oncology Educational Book). Annual Meeting. [DOI] [PubMed] [Google Scholar]
- 38.Dienstmann R., Tabernero J. A precision approach to tumour treatment. Nature. 2017;548:40–41. doi: 10.1038/nature23101. 2017 548:7665. [DOI] [PubMed] [Google Scholar]
- 39.Schøler L.V., Reinert T., Ørntoft M.B.W., Kassentoft C.G., Arnadøttir S.S., Vang S., Nordentoft I., Knudsen M., Lamy P., Andreasen D., et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin. Cancer Res. 2017;23:5437–5445. doi: 10.1158/1078-0432.CCR-17-0510. an official journal of the American Association for Cancer Research. [DOI] [PubMed] [Google Scholar]
- 40.Remon J., García-Campelo R., de Álava E., Vera R., Rodríguez-Peralto J.L., Rodríguez-Lescure, Bellosillo B., Garrido P., Rojo F., Álvarez-Alegret R. Liquid biopsy in oncology: a consensus statement of the Spanish society of pathology and the Spanish society of medical oncology. Clin. Transl. Oncol. 2020;22:823–834. doi: 10.1007/S12094-019-02211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gómez-Peregrina D., García-Valverde A., Pilco-Janeta D., Serrano C. Liquid biopsy in gastrointestinal stromal tumors: ready for prime time? Curr. Treat. Opt. Oncol. 2021;22:1–19. doi: 10.1007/S11864-021-00832-5/METRICS. [DOI] [PubMed] [Google Scholar]
- 42.Rassner M., Waldeck S., Follo M., Jilg S., Philipp U., Jolic M., Wehrle J., Jost P.J., Peschel C., Illert A.L., et al. Development of highly sensitive digital droplet PCR for detection of CKIT mutations in circulating free DNA that mediate resistance to TKI treatment for Gastrointestinal Stromal Tumor (GIST) Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24065411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sci-Hub An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2023;20(5):548–554. doi: 10.1038/Nm.3519. Available online: (accessed on 10 October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schøler L.V., Reinert T., Ørntoft M.B.W., Kassentoft C.G., Arnadøttir S.S., Vang S., Nordentoft I., Knudsen M., Lamy P., Andreasen D., et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin. Cancer Res. 2017;23:5437–5445. doi: 10.1158/1078-0432.CCR-17-0510. [DOI] [PubMed] [Google Scholar]
- 45.Yoo C., Ryu M.H., Na Y.S., Ryoo B.Y., Park S.R., Kang Y.K. Analysis of serum protein biomarkers, circulating tumor DNA, and dovitinib activity in patients with tyrosine kinase inhibitor-refractory gastrointestinal stromal tumors. Ann. Oncol. 2014;25:2272–2277. doi: 10.1093/annonc/mdu386. [DOI] [PubMed] [Google Scholar]
- 46.Maier J., Lange T., Kerle I., Specht K., Bruegel M., Wickenhauser C., Jost P., Niederwieser D., Peschel C., Duyster J, et al. Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin. Cancer Res. 2013;19(17):4854–4867. doi: 10.1158/1078-0432.CCR-13-0765. [DOI] [PubMed] [Google Scholar]
- 47.Serrano C., Leal A., Kuang Y., Morgan J.A., Barysauskas C.M., Phallen J, Triplett O., Mariño-Enríquez A., Wagner A.J., Demetri G.D., et al. Clinical trials: targeted therapy phase I study of rapid alternation of sunitinib and regorafenib for the treatment of tyrosine kinase inhibitor refractory gastrointestinal stromal tumors. Clin. Cancer Res. 2019;25(24):7287–7293. doi: 10.1158/1078-0432.CCR-19-2150. [DOI] [PubMed] [Google Scholar]