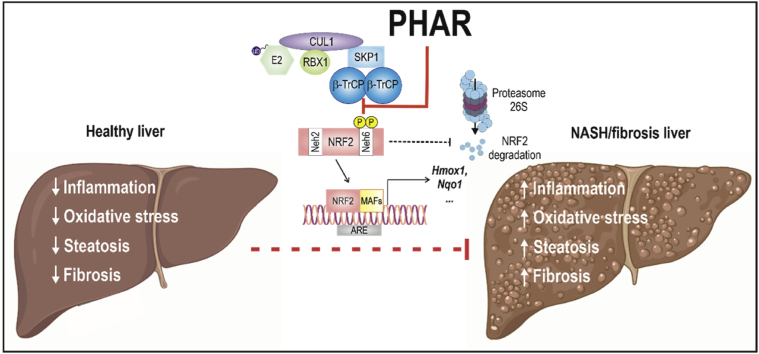
Abstract
Non-alcoholic steatohepatitis (NASH) is a common chronic liver disease that compromises liver function, for which there is not a specifically approved medicine. Recent research has identified transcription factor NRF2 as a potential therapeutic target. However, current NRF2 activators, designed to inhibit its repressor KEAP1, exhibit unwanted side effects. Alternatively, we previously introduced PHAR, a protein-protein interaction inhibitor of NRF2/β-TrCP, which induces a mild NRF2 activation and selectively activates NRF2 in the liver, close to normal physiological levels. Herein, we assessed the effect of PHAR in protection against NASH and its progression to fibrosis. We conducted experiments to demonstrate that PHAR effectively activated NRF2 in hepatocytes, Kupffer cells, and stellate cells. Then, we used the STAM mouse model of NASH, based on partial damage of endocrine pancreas and insulin secretion impairment, followed by a high fat diet. Non-invasive analysis using MRI revealed that PHAR protects against liver fat accumulation. Moreover, PHAR attenuated key markers of NASH progression, including liver steatosis, hepatocellular ballooning, inflammation, and fibrosis. Notably, transcriptomic data indicate that PHAR led to upregulation of 3 anti-fibrotic genes (Plg, Serpina1a, and Bmp7) and downregulation of 6 pro-fibrotic (including Acta2 and Col3a1), 11 extracellular matrix remodeling, and 8 inflammatory genes. Overall, our study suggests that the mild activation of NRF2 via the protein-protein interaction inhibitor PHAR holds promise as a strategy for addressing NASH and its progression to liver fibrosis.
Keywords: NRF2, β-TrCP, NASH, Fibrosis
Graphical abstract
Highlights
-
•
PHAR triggers NRF2 activation in hepatocytes, Kupffer cells, and stellate cells.
-
•
Extended use of PHAR in mice demonstrates its safety.
-
•
PHAR protects against intrahepatic lipid accumulation, oxidative stress, inflammation, and fibrosis.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most prevalent liver disorder and affects approximately 25 % of the world's population [1,2]. NAFLD is characterized by excessive accumulation of fat in the absence of a history of alcohol use or other liver diseases [2]. When liver fat exceeds 5 % and is accompanied by inflammation and swelling of hepatocytes (ballooning), a very serious condition termed non-alcoholic steatohepatitis (NASH) develops. NASH leads to the progressive death of hepatocytes, progressing to fibrosis, cirrhosis, and even hepatocellular carcinoma [3,4].
The underlying mechanisms for the development and progression of NASH are complex and multifactorial and include hepatic steatosis, mainly characterized by an excess of triacyl glycerides and hepatocellular injury, mitochondrial dysfunction, exacerbated production of reactive oxygen species (ROS) exciding homeostatic capacity, and the presence of an inflammatory infiltrate and pro-inflammatory signaling [5,6]. NASH leads to liver fibrosis, which is the main driver of mortality in NASH [7]. Probably due to the difficulty to reverse all these pathological hallmarks with a single drug, the pharmaceutical industry has been unsuccessful at tackling this medical problem.
Proof-of-concept studies suggest that it may be possible to combat several underlying mechanisms in NASH with a single hit at transcription factor NRF2 (Nuclear factor erythroid 2-related factor 2; gene name NFE2L2). Transcriptomic analyses indicate that NRF2 controls the expression of around 250 genes that participate in various cytoprotective functions, and it is nowadays considered a master regulator of cellular homeostasis [8,9]. Consistently, NASH progression in NRF2-deficient mice is exacerbated in comparison with wild type mice, and display enhanced steatohepatitis, oxidative stress, and inflammation [[10], [11], [12], [13], [14]]. Thus, in the high-fat diet NASH model, the susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis was associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes [12], and a proteomic analysis revealed that cellular defence and lipid metabolism are primary NRF2-associated pathways in the liver [14]. As a complamentary approach, pharmacological activation of NRF2 with TBE-31, reduced the halmarks of experimental NASH and liver fibrosis [15].
Pharmacological activation of NRF2 has been achieved through inhibition of its main repressor Kelch Like ECH Associated Protein 1 (KEAP1), which targets the transcription factor for ubiquitin/proteasome degradation under non-stress conditions [9]. Briefly, under basal conditions NRF2 is quickly degraded due to binding to the E3 ubiquitin ligase adapter KEAP1 that leads to its ubiquitinoylation and subsequent proteasomal degradation. KEAP1 contains several redox sensitive cysteines that can be modified by oxidants or electrophilic compounds. Such modifications disrupt the KEAP1/NRF2 interaction in a way that bound NRF2 cannot be degraded, making a stable non-enzymatically active complex. Overall, this limits the availability of free KEAP1 for binding newly-synthesized NRF2, which leads to accumulation of the transcription factor, followed by nuclear translocation and enhanced transcription of its target genes [16]. Electrophilic KEAP1 inhibitors such as TBE-31 and omaveloxolone ameliorate NASH progression in high-fat high-fructose diet-fed mice [15] and STAM mice [17]. However, electrophiles also react with several other cysteines in off-target proteins, leading to unwanted effects. Thus, a new generation of KEAP1 inhibitors is being developed based on the disruption of the KEAP1/NRF2 complex with protein-protein interaction inhibitors [18]. The rational is to develop non-covalent KEAP1 binders that limit the availability of free KEAP1 to interact with NRF2 [19]. One of these compounds, termed S217879, recently demonstrated a positive effect on NASH resolution and reduction of liver fibrosis in mice fed a methionine and choline-deficient diet and diet-induced obesity [18]. However, this potent drug, as well as NRF2 increase in Keap1-knockout mice, triggers hepatomegaly [20,21]. Moreover, strong NRF2 activation due to somatic point mutations in the interface between NRF2 and KEAP1 are found in a high fraction of tumors including 10 % of hepatocellular carcinomas [22]. Besides, clinical data on multiple sclerosis patients that were treated with the electrophile dimethyl fumarate indicate that almost half of them had an alteration of hepatic parameters which in 1 % led to treatment discontinuation [23]. Together these observations suggest caution with hyper-physiological NRF2 activation resulting from KEAP1 inhibition.
We have previously described an alternative, less potent mechanism for proteasomal degradation of NRF2 [24,25]. Under basal conditions, glycogen synthase kinase 3 (GSK-3) is active and phosphorylates NRF2, thus creating a recognition motif for the E3 ligase adapter Beta-Transducin Repeat Containing E3 Ubiquitin Protein Ligase (β-TrCP) and resulting in its degradation [24,25]. Disrupting the β-TrCP/NRF2 complex as a means to modestly activate the NRF2 pathway, is possible as we have previously reported the first protein-protein interaction inhibitor of β-TrCP/NRF2, known as PHAR [26]. Disruption of the β-TrCP/NRF2 interaction is pharmacologically attractive for at least four reasons: 1) the β-TrCP/NRF2 interaction is weaker than that between β-TrCP and other substrates, thus enabling the development of protein/protein interaction inhibitors that might displace NRF2, but not other β-TrCP substrates; 2) the activation of NRF2 following β-TrCP inhibition is weaker compared to KEAP1 inhibition [26], thus remaining close to homeostatic variations; 3) somatic oncogenic mutations in the interface of interaction between NRF2 and β-TrCP have not been reported; 4) in vivo pharmacokinetic studies have demonstrated selective localization of the drug and NRF2 activation in the liver with PHAR [26].
In the present study, we tested whether pharmacologic activation of NRF2 with the NRF2/β-TrCP protein-protein inhibitor PHAR could suppress NASH progression including fibrosis. We chose the STAM model, based on single dose of streptozotocin in neonatal mice, leading to partial damage of endocrine pancreas and insulin secretion impairment, followed by a high fat diet [[27], [28], [29]]. The STAM model recapitulates the full spectrum of human NAFLD from steatosis to NASH and hepatic fibrosis [28]. The histological phenotypes observed in this model are like those seen in human clinical simples [30]. Due to partial damage of the endocrine pancreas, the model does not allow reversion of insulin resistance. The STAM model is similar to human NASH with a background of diabetes mellitus and resembles the lean-type NASH that is more prevalent in Asians [31]. Our results demonstrate a highly protective effect of PHAR in the reduction of hepatocellular lipid accumulation, ballooning, and inflammation. Very importantly, our results also show an anti-fibrotic effect of PHAR and, therefore, provide a new therapeutic strategy to tackle this fatal disease.
2. Materials and methods
Cell culture and reagents. Immortalized mouse hepatocytes [32] were cultured in DMEM supplemented with 10 % fetal bovine serum (FBS), 2 mM glutamine (Gibco/Life Technologies), 1 mM sodium pyruvate (Gibco/Life Technologies), 5 mM HEPES pH 7.4, and 80 μg/ml gentamicin (Gibco/Life Technologies). Human LX-2 stellate cells were cultured in DMEM supplemented with 10 % FBS, 2 mM glutamine, 5 mM HEPES, pH 7.4 and 80 μg/ml gentamicin. Mouse kupffer cells were cultured in RPMI1640 medium (Sigma-Aldrich) with 10 % FBS and 80 μg/ml gentamicin. PHAR (synthesized by Enamine, https://enamine.net/) was dissolved in dimethyl sulfoxide (DMSO). The final concentration of DMSO in cell culture was less than 0.2 %. Lipopolysaccharide from Escherichia coli O111:B4 (LPS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT), and streptozotocin (STZ) were purchased from Sigma-Aldrich. TGF-β was purchased from PeproTech.
Animals and treatments. All experimental procedures were performed following the Guide for the Care and Use of Laboratory Animals and were previously approved by the Autonomous Community of Madrid (PROEX 105/18). All efforts were made to minimize animal suffering and to reduce the number of animals used. Animals were housed under controlled conditions (22 ± 1 °C, 55–65 % humidity, 12 h light-dark cycle) with free access to water and standard laboratory chow. NASH-STAM mice were generated as described previously [28]. Although the reason is not clear, this NASH model addressed NASH only in males as females seem to be somehow protected [28]. Briefly, newborn (2–5 days) C57BL/6 male mice were subjected to a single subcutaneous (s.c.) injection of vehicle (citrate buffer, pH 4.5) or 200 μg streptozotocin (STZ) (Cat. S0130, Sigma-Aldrich), to partially damage pancreatic islets of Langerhans, impair insulin secretion, and induce insulin resistance and oxidative stress [28,33]. Starting from week 4 after the injection and up to the end of the experiment, the STAM group was fed a high fat diet (60 % kcal from fat) (TD.06414, Envigo) or standard diet according to the classification of control of STAM groups. Hyperglycemia along with a high-fat diet gradually led to different stages of progressive liver damage: steatosis (week 6 post-STZ injection); inflammation (week 8) and fibrosis (week 10). We treated STAM or control mice in the NASH inflammatory stage (week 8) with an i.p dose of vehicle (Tween-80/PBS, 1:13) or PHAR (50 mg/kg) for 5 days/week for 2 weeks in order to test progression to fibrosis. Mice were sacrificed 2 h after the last administration, and liver and blood were collected for biochemical analyses.
Immunoblotting. This protocol was essentially performed as previously reported [34]. Briefly, cells were homogenized in lysis buffer (50 mM TRIS pH 7.6, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1 % SDS), denaturalized at 95 °C for 15 min, sonicated, and pre-cleared by centrifugation. Twenty mg of total proteins lysate were resolved in SDS-PAGE, transferred to Immobilon-P (Millipore) membranes and proteins of interest were detected with the following primary antibodies: NRF2 (made in-house, validated in Ref. [24], HO-1 (made in-house, validated in Ref. [35], NQO1 (ab2346, abcam), β-Catenin (610153, BD Transduction laboratories), GAPDH (CB1001, Merck Millipore), LAMINB (sc-6217, Santa Cruz Biotechnology), β-actin (sc-1616, Santa Cruz Biotechnology), Vinculin (E1E9V, Cell Signaling Technology), pre-IL1β (AF-401-NA, RD Systems), α-SMA (a2647, Sigma Aldrich), COL1A1 (sc-293112, Santa Cruz Biotechnology), YAP/TAZ (D24E4, Cell Signaling Technology), GLI2 (sc-271786, Santa Cruz Biotechnology), IκBα (sc-1643, Santa Cruz Biotechnology). Proper peroxidase-conjugated secondary antibodies were used for detection by enhanced chemiluminescence (GE Healthcare).
Analysis of mRNA levels. Total RNA extraction, reverse transcription, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) were done as detailed in Ref. [36]. Mouse and human primer sequences are shown in Supplemental Tables 1 and 2, respectively. Normalization of the reaction was done following amplification of Actb, Gapdh, and Tbp housekeeping transcripts. Data analysis was based on the ΔΔCT method, with normalization of the raw data by the geometric mean of the housekeeping genes Actb, Gapdh, and Tbp (Applied Biosystems). All PCRs were performed from at least triplicate samples.
Cell viability assessed by MTT reduction. In live cells but not in dead ones, the tetrazolium ring of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) can be reduced by active dehydrogenases to produce a formazan precipitate. At the end of the experiments, cells were washed three times with phosphate-buffered saline (PBS) followed by the addition of MTT (0.125 mg/ml) and incubation for 1 h at 37 °C. Thereafter, the media was removed and DMSO was added to each well to dissolve the formazan precipitate for 30 min, thereby determining the relative number of alive cells. An aliquot (100 μl) of the supernatants were analyzed in 96-well multiwell plates at 550 nm in a VERSAmax microplate reader (Molecular Devices).
Magnetic resonance imaging (MRI). Mice were anesthetized with 1–2% isoflurane mixed in 1 L of oxygen and maintained during the experiment employing a mask and the flow of anesthetic gas was constantly regulated to maintain a breathing rate of 60 ± 20 bpm. The animal temperature was maintained at approximately at 37 °C by passing warm water through a heat exchanger machine into the animal platform. The physiological state of the mouse was monitored using an MRI-compatible small animal gating system (SA Instruments; Stony Brook, NY) that controlled the respiratory rate and body temperature. All data acquisition was performed in a 7.0T 16 cm bore Bruker Biospect MRI system (Bruker Medical Gmbh, Ettlingen, Germany) in the High Field Magnetic Resonance Spectroscopy and Imaging Facility (SIERMAC) of the Institute of Biomedical Research “Alberto Sols” CSIC/UAM, Madrid (Spain), equipped with a quadrature 40 mm coil and a 90 mm Bruker gradient coil insert (maximum intensity 360 G/cm). Axial and coronal T2-weighted images were acquired with a rapid acquisition with relaxation enhancement (RARE) sequence with the following parameters: TR:1000 ms; TE:20 ms, rare factor, 4; number of acquisitions, 4; field of view: 35 × 35 mm; matrix 256 × 256; slice thickness 0.75 mm without gap. Single voxel in vivo 1H MRS was carried out with a PRESS method without suppression pulse water and the following parameters: voxel size, 3 mm 3; TR, 1000 ms; TE, 20 ms; averages 128 and 8 dummy scan. Spectroscopic data were acquired using 2018 points and 7.5 KHz spectral width. The Mnova software was used to estimate the intensity of the fat and water signal.
Kupffer cells isolation. We followed the protocol reported in Ref. [37] with minor modifications. The latest supernatant was centrifuged at 300 g for 5 min at 4 °C and the pellet containing the Kupffer cells was resuspended in attachment media. Kupffer cells were separated by two-step Percoll (Sigma Aldrich) gradient method. After centrifugation at 800g for 20 min (with break off), Kupffer cells were enriched between 25 % and 50 % Percoll. Finally, the Kupffer cells pellet was washed with RPMI1640 medium, centrifuged at 500 g for 10 min at 4 °C to wash out the residual Percoll solution, and cells were resuspended in RPMI1640 supplemented with 10 % FBS and 80 μg/ml gentamicin.
Reduced and oxidized glutathione levels. Liver samples were homogenized in phosphate buffer and submitted to the protocol described in Ref. [38]. Liver tissue samples were homogenized in 1 ml of EDTA (0.5 M)-phosphate buffer saline (pH 8.0) containing 300 μl of HPO3 (25 %) and then centrifuged at 3000 g for 10 min. Subsequently, 500 μl of the resulting supernatant was mixed with 100 μl of o-phthalaldehyde (5 mg/ml) for derivatization. The mixtures were incubated at 25 °C for 15 min, and the amount of glutathione (GSH) was determined by measuring the fluorescent signal using a Glomax Discover (Promega) at excitation and emission wavelengths of 350 nm and 420 nm, respectively. To evaluate the total glutathione, a reduction step of oxidized glutathione (GSSG) was performed. Briefly, 500 μl of the sample was combined with 100 μl of 25 mM dithiothreitol and incubated at 4 °C for 30 min. After centrifugation (5000 g, 10 min), the supernatants were used for derivatization. The concentration of GSSG was determined by subtracting the experimental GSH values from the whole GSH values obtained after the reduction step. The obtained data were adjusted based on the protein content.
Protein carbonyl content. As previously described in Ref. [25], the liver homogenates were treated with 1 % streptomycin sulfate to eliminate nucleic acids and then centrifuged at 6000 g (4 °C, 10 min). The resulting supernatants were subjected to a 1-h incubation at 25 °C with 10 mM 2,4-dinitrophenylhydrazine (DNPH) dissolved in 2.5 M HCl. Following the addition of 10 % trichloroacetic acid, the samples were centrifuged at 3000 g (4 °C) for 10 min. The resulting pellets were dissolved using 6 M guanidine hydrochloride in phosphate buffer saline (pH 8.0) and centrifuged at 5000 g (4 °C) for 5 min. The absorbance of the samples was measured at 370 nm using a spectrometer. The protein carbonyl content was expressed as nmol DNPH per milligram of protein.
Malondialdehyde determination. Lipid peroxidation was determined as the formation of thiobarbituric acid-reactive substances (TBA-RS), according to a previous report [25]. Briefly, 200 μl of liver homogenates were mixed with 400 μl of TBA reagent, which consisted of 0.375 g of TBA, 15 g of trichloroacetic acid (TCA), 2.5 ml of 37 % HCl plus phosphate buffer saline until 100 ml. The mixture was then incubated at 95 °C for 15 min, resulting in the production of a pink chromophore directly proportional to the amount of peroxidized products present. After incubation, the samples were placed on ice for 5 min and subsequently centrifuged at 3000 g for 15 min. The optical density of the resulting supernatants was measured using a spectrometer at a wavelength of 490 nm. The quantity of TBA-reactive substances (mostly malondialdehyde [MDA]) was determined by interpolating the values on a constructed MDA standard curve utilizing 1,1,3,3-tetrametoxypropane. The results were expressed as nanomoles of MDA per milligram of protein.
NAFLD activity scores. We assessed the NAFLD activity scores (NAS) in H&E stained sections, according to the method of Kleiner et al. [39]. NAS scores were double-blind evaluated as a composite parameter based on separate scores for steatosis (0–3), hepatocellular ballooning (0–2), and lobular inflammation (0–3). The total NAS score is the sum of these separate scores, and values ≥ 5 are correlated with a diagnosis of NASH in humans.
Serum analytics. Blood samples were left at room temperature for 30 min and then centrifuged at 400 g for 15 min at 4 °C. The supernatant was collected and stored at −80 °C until use. Serum analysis was outsourced to IDEXX BioAnalytics, which used a Beckman Coulter AU480.
RT2Profiler PCR Array of mouse fibrosis. RNA samples were extracted using TRIzol (Invitrogen, ID 15596026, Waltham, MA, USA), according to the manufacturer's instructions. Five hundred ng of each mRNA sample were retrotranscribed using the RT2 First Strand kit (QIAGEN, ID 330401, Hilden, Germany). Transcript levels for 84 different fibrosis-related genes were analyzed on an RT2 Profiler™ PCR Array Mouse Fibrosis, from QIAGEN (ID 330231, GeneGlobe ID PAMM-120ZA; https://geneglobe.qiagen.com/us/product-groups/rt2-profiler-pcr-arrays, accessed on February 24, 2023), according to the manufacturer's instructions, using RT2 SYBR Green/ROX qPCR Master Mix (QIAGEN, ID 330522) for the PCR reaction. The PCR reaction was performed on a 7900 HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). Reverse transcription quantitative PCR (RT-qPCR) data were analyzed using the R programming language. Ct values for each gene (Acta2; Agt; Ccl11; Ccl12; Ccl3; Ccn2; Grem1; Il13; Il13ra2; Il4; Il5; Snai1; Bmp7; Hgf; Ifng; Il10; Col1a2; Col3a1; Lox; Mmp1a; Mmp13; Mmp14; Mmp2; Mmp3; Mmp8; Mmp9: Plat; Plau; Plg; Serpina1a; Serpine1; Serpinh1; Timp1; Timp2; Timp3; Timp4; Itga1; Itga2; Itga3; Itgav; Itgb1; Itgb3; Itgb5; Itgb6; Itgb8; Ccr2; Cxcr4; Il1a; Il1b; Ilk; Tnf; Edn1; Egf; Pdgfa; Pdgfb; Vegfa; Cav1; Dcn; Eng; Inhbe; Ltbp1; Smad2; Smad3; Smad4; Smad6; Smad7; Tgfb1; Tgfb2; Tgfb3; Tgfbr1; Tgfbr2; Tgif1; Thbs1; Thbs2; Cebpb; Jun; Myc; Nfkb1; Sp1; Stat1; Stat6; Akt1; Bcl2; Fasl) were normalized to the geometric mean of the housekeeping genes (Actb; B2m; Gapdh; Gusb; Hsp90ab1) using the ΔCt method. ΔCt values for each gene were further normalized by subtracting the mean of the ΔCt values for all animals, yielding ΔΔCt values. The negative of the ΔΔCt values were used for further analysis as log2 of the fold of change and are presented as a heatmap. To determine the effects of treatment (vehicle or PHAR) and NASH status (control vs. STAM) on gene expression, a linear model was fitted to the log2 of the fold of change for each gene using the lm() function. The model considered the interaction between treatment and NASH status variables. The emmeans package (v1.8.6) was used to test for statistically significant differences in the mean log2 fold of change values among the different groups. To perform contrast tests, the emmeans() function was used for each linear model of each gene across conditions, and Bonferroni's correction was applied for multiple testing. The contrasts tested were limited to control-vehicle vs. STAM-Vehicle and STAM-Vehicle vs. STAM-PHAR groups. Significance was determined by comparing the adjusted p-values against a Bonferroni's adjusted threshold of p < 0.05. The heatmap was made with Morpheus (https://software.broadinstitute.org/morpheus). For volcano plot visualization, log2 of the fold of change for each gene of the STAM-PHAR group was normalized by subtracting the mean of the log2 of the fold of change for each gene of the STAM-VEH group and –log10 of the Bonferroni's adjusted p-value for STAM-VEH vs. STAM-PHAR comparison was calculated.
Statistical analyses. Unless otherwise indicated, all experiments were performed at least 3 times and all data presented in the graphs are the mean of at least 3 independent samples. Data are presented as mean ± S.D (standard deviation). Statistical differences between groups were assessed using GraphPad Prism 8 software by the unpaired Student's t-test. One and two-way analyses of variance with post-Bonferroni's test were used for multiple comparisons. Statistically significant differences are indicated in the figures (*** indicate p values < 0.001, ** <0.01 and * <0.05).
3. Results
3.1. PHAR activates NRF2 in hepatocytes, kupffer and stellate cells
Murine hepatocytes, murine Kupffer and human stellate cells were pre-incubated in a low-serum medium (less than 0.5 %) for 16 h before PHAR treatment. This treatment is necessary to create the GSK-3-dependent phosphorylation motif in NRF2 and promote its interaction with the E3 ligase β-TrCP [26].
First, we confirmed that several liver cell types express NRF2 and are responsive to PHAR. Low-serum growing immortalized hepatocytes were treated with 10 μM PHAR for 2, 4, 8, 16 and 24 h (Suppl. Fig. 1). The results showed that PHAR increased NRF2 protein levels from 2 h until 16 h. At these time points NRF2 activation was further reflected by an increase in the protein and transcript levels of two well-established targets, heme oxygenase-1 (HO-1) and NADPH quinone oxidoreductase (NQO1), encoded by Hmox1 and Nqo1 genes, respectively. Furthermore, PHAR significantly increased the expression of other weaker NRF2-targets, such as gamma-glutamyl cysteine ligase catalytic (Gclc) and modulator (Gclm) genes. Additionally, we noted that PHAR affects the expression of several lipid metabolism genes, as evidenced by the increased expression of lipid translocators to the cell (Cd36) and to the mitochondria (Cpt1a), and fatty acid beta-oxidation (Acox1) (Suppl. Fig. 1D).
Low-serum growing Kupffer cells isolated from wild-type mice were pre-treated with PHAR (10 μM, 8 h) or vehicle (DMSO) and then stimulated with LPS (100 ng/ml, 4 h) to trigger an acute inflammatory response. PHAR increased NRF2 and HO-1 protein levels in these liver macrophages (Suppl. Figs. 2A–B), and, importantly, PHAR attenuated the LPS-induced inflammatory response, measured by the analysis of pre-IL1β protein and Il1b, Il6, and Tnf transcript levels (Suppl. Fig. 2C). Taken together, we conclude that PHAR activates NRF2 and attenuates LPS-induced inflammation in hepatic resident macrophages.
TGF-β is considered the most relevant molecule in the pathogenesis of fibrosis and hepatic stellate cells (HSCs) activation [40,41]. We challenged low-serum growing LX-2 human HSCs with PHAR and/or TGF-β for 16 h. As shown in Suppl. Fig. 3, this compound increased NRF2 and HO-1 at protein and mRNA levels. TGF-β led to the expected increase in two fibrotic markers, alpha Smooth Muscle Actin (α-SMA) and Collagen Type I alpha 1 Chain (COL1A1) at protein and mRNA levels. Importantly, this increase was significantly lower in cells treated with PHAR (Suppl. Figs. 3A–C). The analysis of cell viability in the same cells, evaluated with an MTT assay, indicated that PHAR, TGF-β and/or the co-stimulation are not toxic at any condition (Suppl. Fig. 3D). These results suggest that PHAR prevents TGF-β-induced fibrosis signaling elicited by HSCs.
3.2. Prolonged administration of PHAR in mice is safe
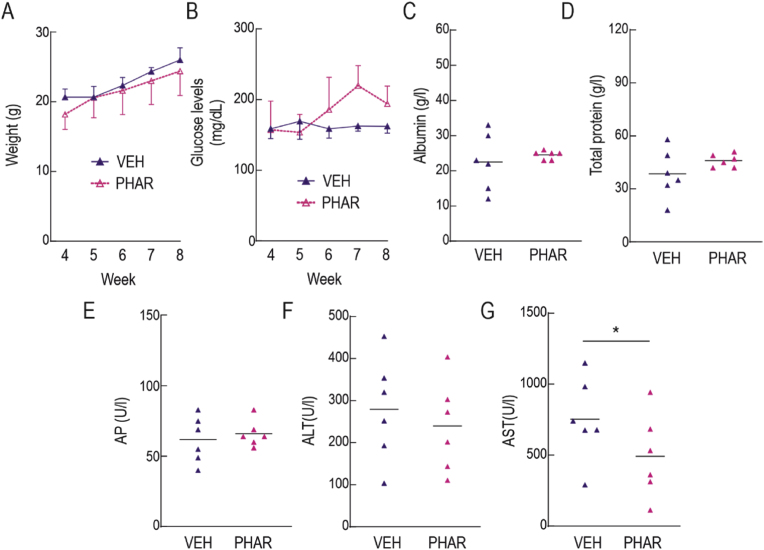
We conducted safety studies in eight-week-old C57BL/6 mice to evaluate the potential risks associated with prolonged administration of PHAR. The mice were subjected to an intraperitoneal (i.p.) treatment of vehicle (Tween-80/PBS; 1/13) or 50 mg/kg PHAR, 5 days/week for 4 weeks. During the treatment, the mice did not experience any weight loss, which is a highly sensitive indicator of disease in these animals (Fig. 1A). Additionally, glucose levels remained stable throughout the study (Fig. 1B). We also analyzed total protein serum levels (TP), as well as several hepatic parameters, including albumin (Alb), alkaline phosphatase (AP), aspartate aminotransferase (AST), and alanine aminotransferase (ALT) (Fig. 1C-G). These parameters were not statistically different between PHAR and vehicle-treated mice except AST levels which were slightly lower in the presence of PHAR. These results suggest that prolonged administration of PHAR is safe and non-toxic, least for a 4-week exposure to this treatment.
Fig. 1.
Safety of prolonged administration of PHAR in mice. A, evolution of weight during the weeks of treatment. B, blood glucose levels, under fed conditions, throughout the weeks of treatment. C, serum albumin, D, total serum protein. E, alkaline phosphatase (AP). F, alanine aminotransferase (ALT). G, aspartate aminotransferase (AST). Data are mean ± S.D. (n = 6). *p < 0.05; according to a Student's t-test. Animals in this experiment were 11-month-old at the beginning of a 4-week treatment.
3.3. PHAR activates the NRF2 signature in the liver and protects against oxidative stress
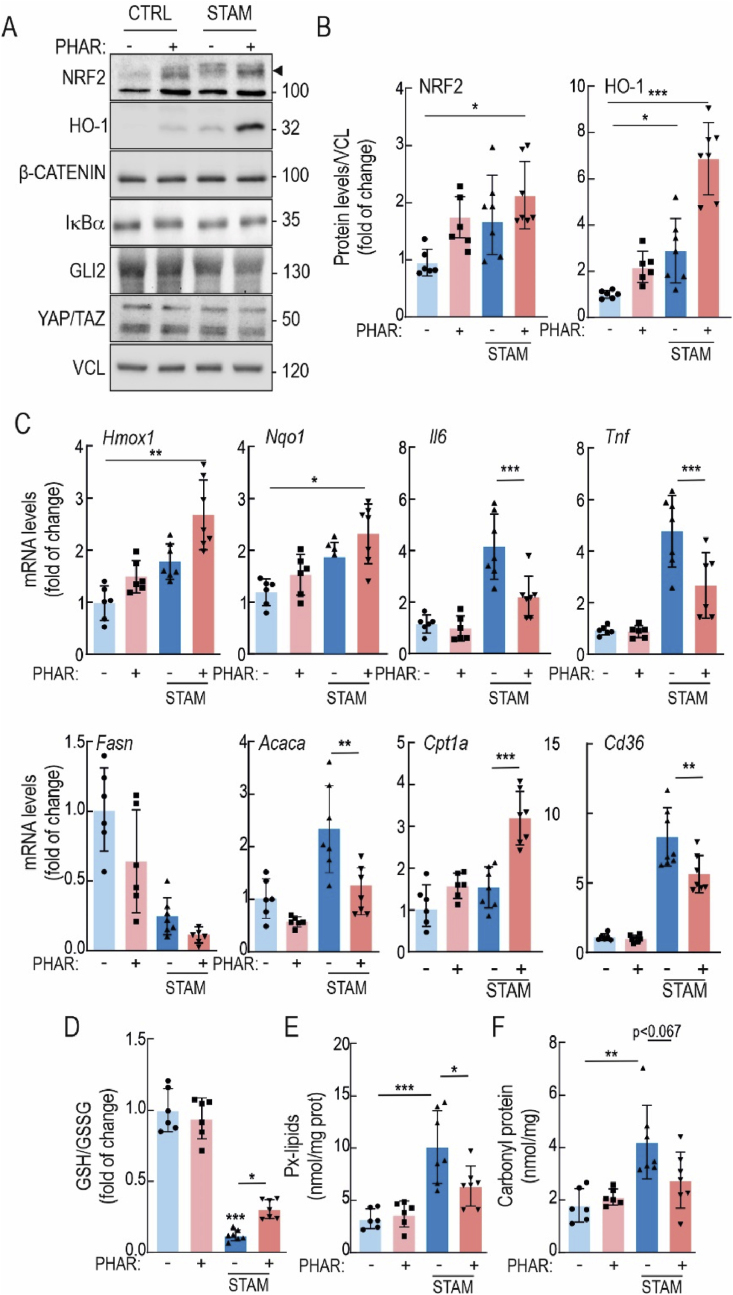
Eight-week-old control and STAM mice received a daily i.p. injection of vehicle or PHAR (50 mg/kg), 5 days/week for 2 weeks. As observed in Fig. 2A–C, PHAR induced the protein expression of NRF2 and HO-1, as well as Hmox1 and Nqo1 transcripts, used as readouts of NRF2 transcriptional activity. We have previously reported an in silico analysis of the energy of binding of several substrates to beta-TrCP and suggested that NRF2 is among the substrates with weakest affinity, therefore providing selectivity for PHAR-mediated displacement of NRF2 [24]. Here, we further analyzed in liver the effect of PHAR on several known key substrates of β-TrCP: β-CATENIN, YAP/TAZ and GLI2 (Fig. 2A). PHAR did not modify the levels of any of them, indicating that this compound shows preference for inhibition of the NRF2/β-TrCP interaction. The transcript levels of the inflammatory cytokines Il6 and Tnf were also significantly reduced by PHAR in the STAM mice. (Fig. 2C). Regarding lipid metabolism, transcript levels of Fasn, Acaca and Cd36 exhibited a tendency to decrease while Cpt1a was increased in the presence of PHAR (Fig. 2C). A major hallmark of NASH is the presence of oxidative stress [11,42]. Consistent with the well-established role of NRF2 as a master regulator of redox homeostasis, PHAR partially rescued STAM livers from oxidative stress as determined by partial normalization of the GSH/GSSG ratio, and the levels of lipid peroxides (malondialdehyde, MDA) and carbonylated proteins (adducts of 2,4-dinitrophenylhydrazine, DNPH) (Fig. 2D–F). Together, these results show target engagement (NRF2) by PHAR in the liver and reduction of inflammatory and oxidative markers as well as modification of lipid metabolism markers.
Fig. 2.
PHAR activates NRF2 in liver and protects against oxidative stress and inflammation in the STAM model of NASH. During weeks 8–10, mice in either the control or STAM groups were administered PHAR at a dose of 50 mg/kg/day. A, representative immunoblots of NRF2, HO-1, β-CATENIN, IĸBα, GLI2, YAP/TAZ, and VCL as a loading control. Black arrow indicates NRF2 specific band. B, densitometric analysis of NRF2 and HO-1 protein levels from representative immunoblots from A, expressed as a ratio of VCL. Data are mean ± S.D. (n = 6–7). *p < 0.05; ***p < 0,001 vs. CTRL-VEH according to a two-way ANOVA test followed by Bonferroni post-hoc test. C, mRNA levels of NRF2 targets Hmox1, and Nqo1, inflammatory markers Il6 and Tnf, lipid metabolic markers Fasn, Acaca, Cpt1a and Cd36, were determined by qRT-PCR and normalized by the geometric mean of Gapdh, Tbp, and Actb levels. Data are mean ± S.D. (n = 6–7). *p < 0,5; **p < 0,01; ***p < 0.001 vs. CTRL-VEH or STAM-VEH according to a two-way ANOVA followed by Bonferroni post-hoc test. D, levels of reduced glutathione (GSH) normalized with total oxidized glutathione (GSSG). E, Peroxidized lipids (Px-lipids) representing mostly MDA. F, protein carbonyl content as determined by DNPH levels. Data are mean ± S.D. (n = 6–7). *p < 0.05; **p < 0.01; ***p < 0.001 vs. CTRL-VEH or STAM-VEH according to a two-way ANOVA followed by Bonferroni post-hoc test.
3.4. PHAR decreases intrahepatic lipid accumulation
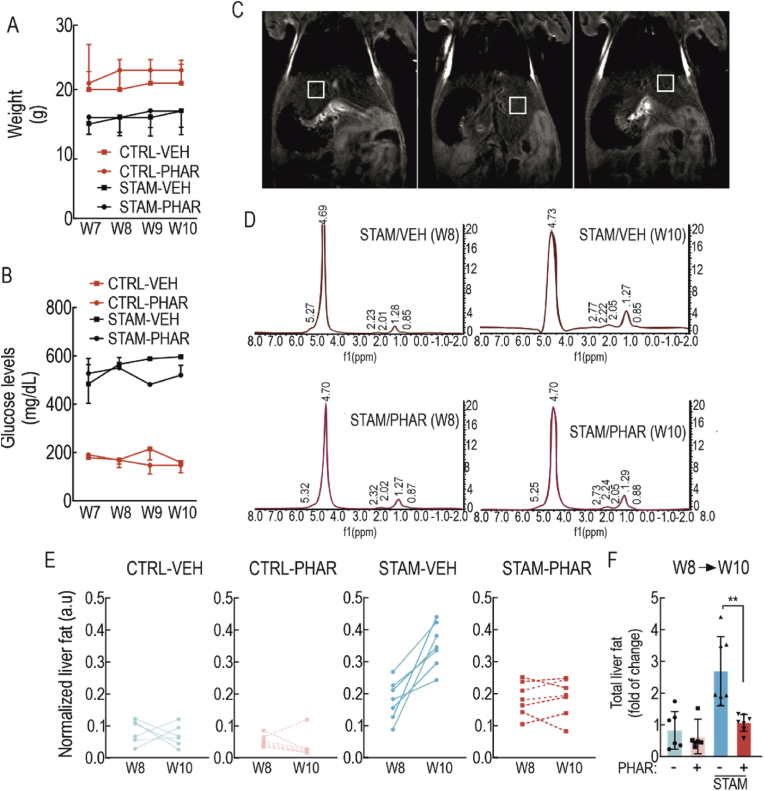
From the point of view of its therapeutic use, we analyzed the effect of PHAR in eight-week-old control and STAM mice, a moment that correlates with incipient NASH in humans at the time of clinical diagnosis. As previously described, these mice received i.p. administration of vehicle or 50 mg/kg of PHAR, 5 days/week until 10 weeks of age, a period at which these mice develop fibrosis [28]. As previously reported [28], we observed a significant decrease in the body weight of the STAM mice compared to control mice, but there was no change in weight in either group due to PHAR treatment (Fig. 3A). Also as expected, STAM mice exhibited hyperglycemia (Fig. 3B), and PHAR did not modify glucose levels.
Fig. 3.
PHAR decreases intrahepatic lipid accumulation. During weeks 8–10, mice in either the control or STAM model groups were administered PHAR at a dose of 50 mg/kg/day. This treatment was conducted during the transition from NASH stage (week 8, W8) to fibrosis stage (week 10, W10). A, weight changes during the last 4 weeks. B, blood glucose levels during the last four weeks. C, representative magnetic resonance images of a mouse per group. For each mouse, the hepatic fat of 3 different regions of the liver was measured with a 3 mm3 voxel by MRI. D, representative MRI spectra derived from STAM-VEH or STAM-PHAR. The peak that appears at an approximate frequency of 4.7 ppm corresponds to the water molecule (majority), while the rest of the peaks give information about the amount of liver fat. The ratio between the sum of the fat peaks vs. water peaks indicates the total liver fat content. E, evolution profiles of the normalized liver fat per mouse measured by MRI at week 8 (NASH) and at week 10 (fibrosis) of each experimental group. Each lane corresponds to the same mouse at the beginning and end of the treatment. F, normalized quantification of the amount of liver fat per mouse. Data are mean ± S.D. (n = 6–7). **p < 0.01 vs. STAM-VEH according to a two-way ANOVA followed by Bonferroni post-hoc test.
To monitor the effect of PHAR on hepatic fat, we used magnetic resonance imaging (MRI) at the beginning and end points of each experimental condition. As liver fat is diffuse, we took three measurements with a 3 mm3 voxel from three different liver regions in each mouse (Fig. 3C), as previously published [43]. In agreement with former reports [28], STAM mice accumulated fat in the liver, despite the animals were not obese. However, PHAR-treated mice showed significant protection against liver steatosis (Fig. 3D–F). Therefore, this integrative non-invasive strategy indicated that PHAR exerts global protection against lipid accumulation during NASH progression.
3.5. PHAR preserves liver integrity
We analyzed serum levels of albumin, ALT, AST, AP, and total protein. In the STAM mice, ALT levels were greatly increased, and PHAR significantly protected against this effect. Serum AST and AP levels also exhibited a slight non-significant increase and, again, PHAR tended to attenuate this effect (Table 1).
Table 1.
Effect of PHAR on liver parameters analyzed in serum of STAM mice. Serum albumin (Alb), alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (AP), and total protein levels (TP) were analyzed for the different experimental groups using a Beckman Coulter AU480. Data are mean ± S.D. (n = 6–7). ***p < 0.001 vs. CTRL; ###p < 0.001 vs. STAM-VEH according to two-way ANOVA followed by Bonferroni post-hoc test. Animals in this experiment were 8-week-old at the beginning of a 2-weeks treatment.
| Groups | Alb (g/L) | ALT (U/L) | AST (U/L) | AP (U/L) | TP (g/L) |
|---|---|---|---|---|---|
| CTRL | 22 ± 7.6 | 37.4 ± 10.3 | 86.7 ± 14.1 | 64.4 ± 13.9 | 35.6 ± 14.9 |
| CTRL-PHAR | 25 ± 1.2 | 46.5 ± 18.3 | 79.4 ± 14.8 | 59.5 ± 15.2 | 46.6 ± 3.3 |
| STAM-VEH | 18.5 ± 1.0 | 324.7 ± 70.8*** | 101.0 ± 20.8 | 125.2 ± 72.3 | 35.2 ± 6.7 |
| STAM-PHAR | 21.5 ± 2.6 | 185.3 ± 64.1### | 95.3 ± 7.8 | 103.5 ± 11.5 | 49.2 ± 10.4 |
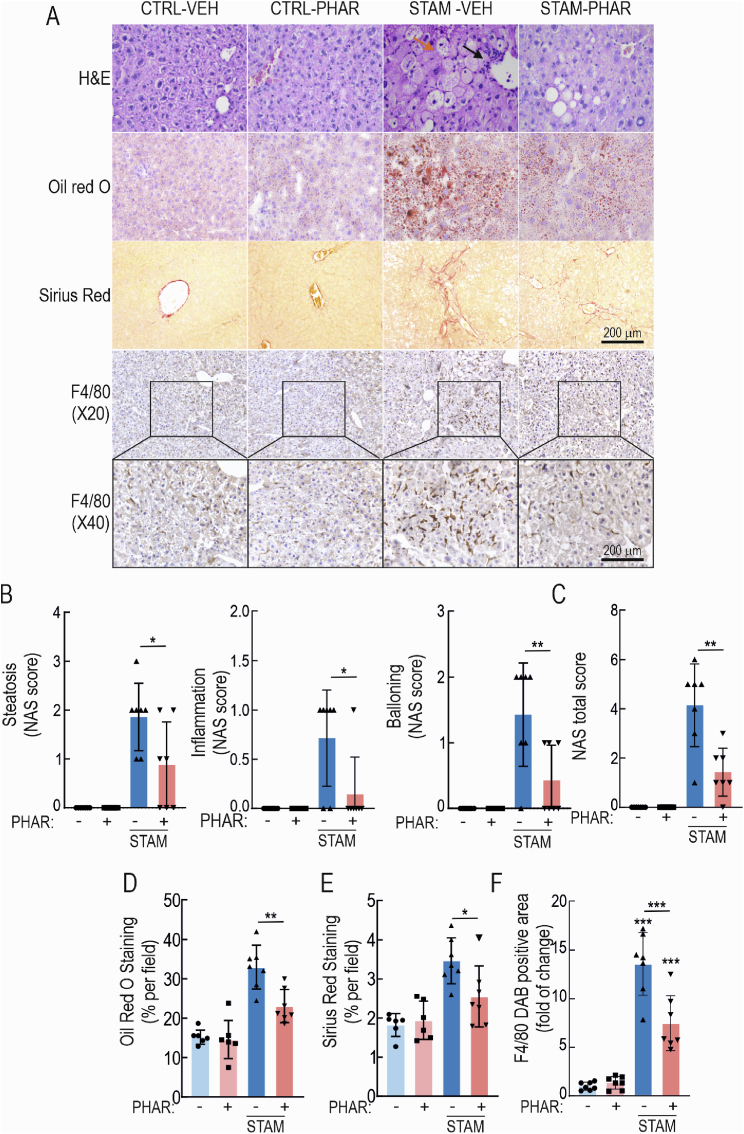
Hematoxylin and eosin (H&E)-stained sections confirmed the normal liver histology in mice receiving PHAR (Fig. 4A). Furthermore, double-blind analysis of the levels of steatosis, inflammatory cells, and ballooning (NAS score) revealed a high NAS score in STAM mice that was significantly lower in PHAR-treated mice, consistent with liver protection against NASH (Fig. 4B and C).
Fig. 4.
PHAR prevents the development of NASH and fibrosis. A, histochemical analysis of H&E, Oil red O (neutral fat), Sirius red. (collagen I and III), and immunohistochemistry for F4/80. Black arrow in H&E photographs points an area with an inflammatory focus; orange arrow indicates an area of ballooning. B–C, steatosis, inflammation, and ballooning, and integration of the three parameters in the NAFLD activity score (NAS). Data are mean ± S.D. (n = 6–7). *p < 0.01; **p < 0.01 vs. STAM vehicle according to a two-way ANOVA followed by Bonferroni post-hoc test. D, quantification of Oil red O positive area. Data are mean ± S.D. (n = 6–7). **p < 0.01 vs STAM vehicle according to a two-way ANOVA test. E, quantification of Sirius red positive area. Data are mean ± S.D. (n = 6–7). *p < 0.05 vs. STAM-VEH according to a two-way ANOVA followed by Bonferroni post-hoc test. F, Quantification of DAB-staining positive area of F4/80. Data are mean ± S.D. (n = 6–7). ***p < 0.001 vs. CTRL-VEH or STAM-VEH according to a two-way ANOVA test. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Liver lipid content was further analyzed by Oil red O staining (Fig. 4A). In agreement with the MRI data of Fig. 3, ten-week-old STAM mice developed strong steatosis, and PHAR greatly attenuated this effect (Fig. 4D). Importantly, collagens I and III content, analyzed by Sirius red staining, was significantly decreased by PHAR (Fig. 4A and E), therefore anticipating an anti-fibrotic effect. Immunohistochemical staining with anti-F4/80 of hepatic macrophages (Kupffer cells) revealed that STAM model significantly increases staining of this cell type, as expected, and that this increase is greatly diminished in STAM PHAR-treated mice (Fig. 4A and F). Therefore, these results confirm that PHAR favors an inflammation-protective environment in mouse liver in STAM model.
3.6. The expression profile of fibrotic genes is attenuated by PHAR
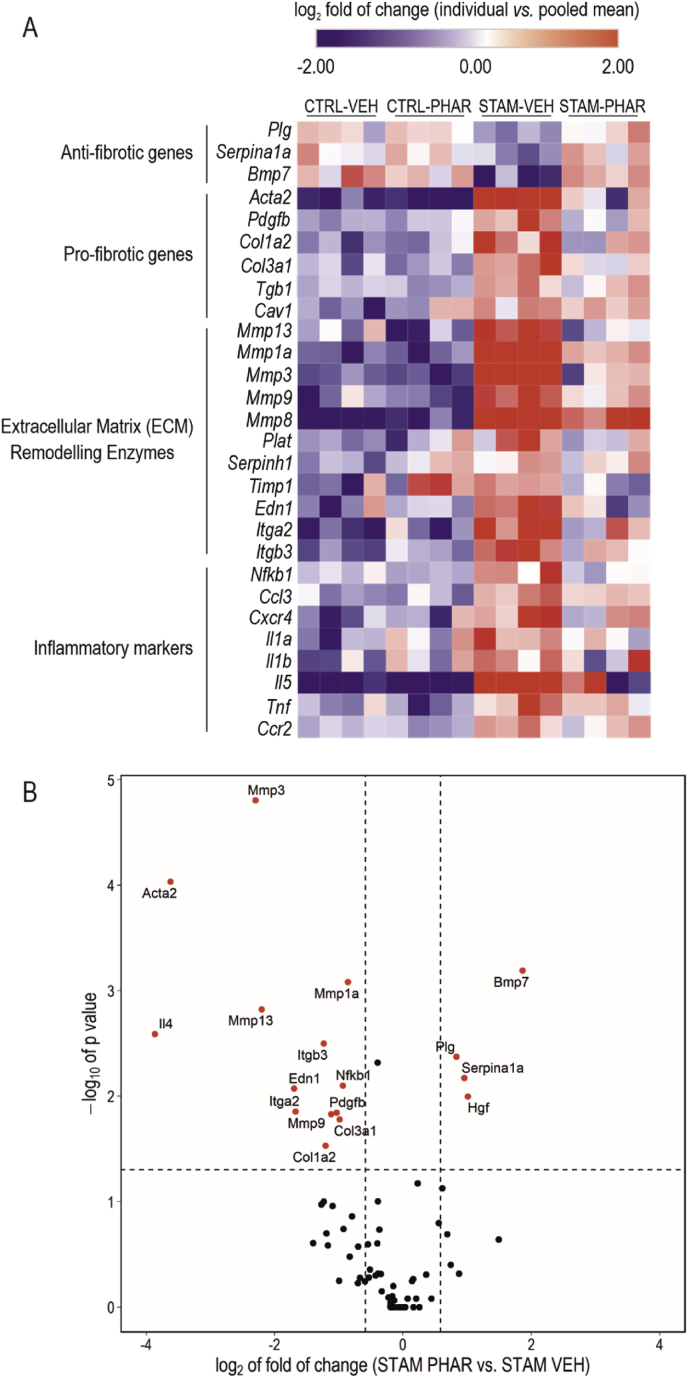
We analyzed the expression of a battery of 84 genes involved in liver fibrosis using RT2 Profiler™ PCR Array Mouse Fibrosis (see Materials and Methods). We grouped all genes with significant expression changes (CTRL-VEH vs. STAM-VEH; p-value ≤0.05) by their biological function in a heatmap (Fig. 5A) (anti-fibrotic, profibrotic, extracellular matrix remodeling, and inflammatory genes). We further analyzed transcript levels of these genes in a volcano plot (Fig. 5B). In STAM mice, we found a reduction of several anti-fibrotic genes (Plg, Serpina1a, and Bmp7) and, importantly, PHAR shifted their levels towards normal physiological values. On the other hand, the expression of several pro-fibrotic genes (Acta2, Pdgfb, Col1a2, Col3a1, Tgb1, and Cav1), extracellular matrix remodeling genes (Mmp13, Mmp1a, Mmp3, Mmp9, MmP8, Plat, Serpinh1, Timp1, Edn1, Itga2, and Itgb3), and inflammation markers (Nfkb1, Ccl3, Cxcr4, Il1a, Il1b, Il5, Tnf, and Ccr2) were increased. Of note, PHAR strongly attenuated the expression of these genes. Altogether, these results provide additional information about the effect of PHAR in attenuating liver fibrosis.
Fig. 5.
PHAR counteracts the fibrotic transcriptional signature. A, Changes in expression of 28 out of 84 genes involved in liver fibrosis were analyzed through RT2 Profiler™ PCR Array Mouse Fibrosis Qiagen for CTRL-VEH, CTRL-PHAR, STAM-VEH and STAM-PHAR livers. The 28 genes showed significant expression changes between CTRL-VEH vs. STAM-VEH (n = 4 per experimental group; p-value ≤0.05; two-way ANOVA, Bonferroni's post-hoc test). These genes were grouped by their biological function in a heatmap, representing the individual log2 of the fold of change. The color scale is capped at −2 and 2 of log2 fold of change. B, Volcano plot representing -log10 of p-value (calculated by a two way-ANOVA, Bonferroni's post-hoc test) and log2 of the fold of change for differences in gene expression between STAM-VEH and STAM-PHAR. The threshold p-value (dashed horizontal line) was established to <0.05, and the threshold for log2-fold change (dashed vertical lines) was established at -log2(1.5) and log2(1.5). Genes surpassing both thresholds are shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Despite huge efforts in clinical research, no licensed drugs for NASH treatment have been approved so far. In general, it seems that an approach based only on the use of metabolically active drugs that have been previously used to control obesity or hyperglycemia may not be sufficient to combat NASH [[44], [45], [46], [47], [48]]. Similarly, several anti-fibrotic drugs have failed to resolve NASH in phase 2 and 3 clinical trials [49] demonstrating the need for new pharmacological approaches. Probably, this failure reflects the complexity of NASH, which might be viewed as a syndrome resulting from environmental factors and lifestyle, biochemical causes, and interindividual genetic differences [11]. For this reason, we have pursued the preclinical validation of a recently proposed new target, the pleiotropic transcription factor NRF2, which is a master regulator of multiple homeostatic functions including lipid metabolism, inflammation, and oxidative stress [8].
The identification of NRF2 as a new therapeutic target for NASH has been suggested by several studies [11]. For example, NRF2-deficient mice are more susceptible to NASH development than their wild-type littermates [50]. Moreover, in recent years several NRF2 activators have demonstrated a beneficial effect in several murine NASH models in proof-of-concept studies. Just as a few examples, constitutive NRF2 activation due to genetic Keap1 knockdown leads to reduced liver lipogenesis, lipid accumulation and hepatic steatosis [51,52]. More in connection with NASH, Nrf2-knockout mice submitted to several protocols of a high fat diet administration exhibit increased oxidative stress and hyperlipidemia [12,53,54]. Pharmacological activation of NRF2 also elicits several protective effects against hyperlipidemia, inflammation and fibrosis [15]. It seems however surprising that there is not yet a clear pipeline to NRF2 activators for NASH therapy. Our view is that the previous studies have focused on inhibiting the potent NRF2 regulator KEAP1, therefore, raising concerns about the long-term side effects of chronic treatment. By contrast, our study is the first to target NRF2 with a completely new approach based on the inhibition of its interaction with the mild regulator β-TrCP [26]. Thus, regarding the STAM model, our study is in line with a recent report with a the KEAP1/NRF2 interaction inhibitor S217879 [18] which protected against similar hallmarks of NASH, including liver fat accumulation, inflammation and fibrosis. However, S217879, led to increased liver weight and hepatomegaly, which was not observed with our β-TrCP/NRF2 interaction inhibitor.
Although NRF2 is a ubiquitous transcription factor, there is a high variability in its cell and tissue expression, probably related to its epigenetic regulation. Thus, under basal conditions, stellate and Kupffer cells appear to have about 64 % higher NRF2 transcript levels than hepatocytes, according to a single cell RNA analysis of the Liver Cell Atlas Database [55]. Therefore, we analyzed the NRF2 signature in these specific liver cells in response to PHAR. We found that the three cell types were responsive to PHAR by upregulating NRF2 and modulating LPS and TGF-β-responses in Kupffer and HSCs, respectively. These results are in agreement with previous studies showing that a cell-permeable PROTAC degrader against KEAP1 suppresses HSC activation through the antioxidant and anti-inflammatory pathway [56]. Therefore, while this study supports a new strategy to combat NASH, further work is still needed to determine PHAR selectivity of the beta-TrCP/NRF2 axis and if its affects are hepatocyte-dependent or involve other liver cell types including dendritic and stellate cells. By comparison with other stem cells, for instance in the adult brain, it is plausible that NRF2 favors the maintenance of liver progenitor niches, which are otherwise exhausted in the progression towards cirrhosis, in a similar way as it does in the neurogenic niches [57].
Consistent with the cell culture data, we found that PHAR induces NRF2 expression and activates the NRF2 transcriptional response in the liver as determined by the increases in transcript levels of the bona fide NRF2 targets Hmox1 and Nqo1, and the widely reported effect of NRF2 in modulation of oxidative stress (GSH/GSG ratio and reduced levels of lipid peroxides and carbonylated proteins) and inflammation (Il6 and Tnf). A prediction of the pharmacokinetics profile with the DataWarrior and ADMETsar program [58,59] suggested that PHAR is not toxic and, consistently, mice submitted to daily i.p. injections of PHAR for 4 weeks (5 doses/week) did not show any evidence of toxicity. Further studies will be required to analyze in more detail the safety of PHAR administration, but based on these preliminary observations we went ahead to further evaluate the efficacy in NASH.
Considering interindividual variations and to better mimic the situation of recently diagnosed NASH patients, we tracked NASH progression by MRI. We found that from week 8–10 the ratio lipid/water was increased in vehicle-treated STAM mice as expected [28]. By contrast, PHAR greatly protected STAM mice from lipid accumulation during this time frame. Encouraged by this finding, we explored independent parameters of NASH progression. Although PHAR protected the liver from steatosis, oxidative stress and inflammation, the current challenge is to protect the liver against fibrosis since the clinical course of NASH is significantly dependent on the progression rate of fibrosis [60]. Importantly, Sirius red staining, a general marker of liver fibrosis, evidenced perivascular collagen accumulation after 10 weeks in control STAM mice, most likely contributing to portal hypertension. By contrast, the deposition of collagen was markedly reduced in PHAR-treated mice. Consistent with an antifibrotic role, the TGF-β-dependent induction of α-SMA and COL1A1 was greatly attenuated in HSCs which are the main cell type contributing to fibrosis (Supplem. Fig. 2). We analyzed the expression of 84 genes highly connected with liver fibrosis by using a customized array. In agreement with the results shown in Supplem. Fig. 2, Fig. 3 and Fig.5, the expression of inflammatory markers Il6 and Tnf, and profibrotic markers, Acta2 and Col1a1, was highly attenuated in the presence of PHAR. The array data further extended these observations to show increased levels of anti-fibrotic genes and reduced levels of pro-fibrotic genes. Among the anti-fibrotic genes, Bmp7 showed the highest statistically significant increase in response to PHAR. This protein is an antagonist of TGF-β [60]. Among the profibrotic genes, Acta2, encoding α-SMA, a well-established marker for myofibroblast formation and progression towards hepatic cirrhosis, Mmp3 and Mmp13 were highly downregulated. Of relevance, the cluster of genes that were downregulated by PHAR also includes those involved in extracellular matrix remodeling, probably connected with persistent activation of interstitial myofibroblasts [61].
Further studies are required to determine how NRF2 reduces fibrosis. Direct repression of pro-fibrotic or activation of anti-fibrotic genes is unlikely because after analysis of ChIP-Atlas database for NRF2 ChIP peaks [62] we could not retrieve any putative NRF2-regulated Antioxidant Response Elements (AREs) in the regulatory regions of those genes. Considering the widely accepted role of NRF2 in the protection against oxidative and inflammatory stress, it is more plausible that NRF2 controls indirectly the fibrotic response. One plausible possibility that will be explored in future studies is that NRF2 might regulate the expression of antifibrotic microRNAs. Additionally, the normalization of hepatic metabolism and attenuation of oxidative and inflammatory stress by PHAR may be the driver for its protection against fibrosis.
In conclusion, this study presents an innovative approach to therapeutically activate NRF2 by using a disrupter of its interaction with β-TrCP. Our findings suggest that PHAR may hold promise for the treatment of chronic liver diseases, such as NASH.
Funding
This research was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) (grants PID2019-110061RB-I00, PID-2021-122766OB-100 and PDC2021-121421-I00, PDC2022-133765-I00, MCIN/AEI/10.13039/501100011033 and “ERDF A way of making Europe” by the European Union.), CIBERdem and CIBERned (ISCIII), and The Autonomous Community of Madrid (grant P2022/BMD-7230). RFG enjoyed a FPI contract of MINECO (FPI-2017). DCS is a holder of a FPI contract of MICINN (Ministry of Science and Innovation, FPI-2020, PRE2020-091886). JJV is holder of a FPU contract of MIU (Ministry of Universities, FPU2020, FPU20/03326).
CRediT authorship contribution statement
Raquel Fernández-Ginés: Conceptualization, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. José Antonio Encinar: Writing – review & editing. Maribel Escoll: Formal analysis, Validation, Visualization, Writing – review & editing. Daniel Carnicero-Senabre: Formal analysis, Validation, Visualization, Writing – review & editing. José Jiménez-Villegas: Formal analysis, Validation, Visualization, Writing – review & editing. Ángel J. García-Yagüe: Formal analysis, Validation, Visualization, Writing – review & editing. Águeda González-Rodríguez: Formal analysis, Validation, Visualization, Writing – review & editing. Irma Garcia-Martinez: Formal analysis, Validation, Visualization, Writing – review & editing. Ángela M Valverde: Formal analysis, Validation, Visualization, Writing – review & editing. Ana I. Rojo: Conceptualization, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. Antonio Cuadrado: Conceptualization, Formal analysis, Funding acquisition, Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Antonio Cuadrado is funder of the biopharmaceutical company Servatrix Biomed S.L. The other authors declare no known competing financial interest.
Acknowledgments
This article is based upon work from COST Action CA20121, supported by COST (European Cooperation in Science and Technology) (www.cost.eu) (https://benbedphar.org/about-benbedphar/). PHAR is patented with WO 2022/152800 A1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103027.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Maurice J., Manousou P. Non-alcoholic fatty liver disease. Clin. Med. 2018;18:245–250. doi: 10.7861/clinmedicine.18-3-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pouwels S., Sakran N., Graham Y., Leal A., Pintar T., Yang W., Kassir R., Singhal R., Mahawar K., Ramnarain D. Non-alcoholic fatty liver disease (NAFLD): a review of pathophysiology, clinical management and effects of weight loss. BMC Endocr. Disord. 2022;22:63. doi: 10.1186/s12902-022-00980-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massoud O., Charlton M. Nonalcoholic fatty liver disease/nonalcoholic steatohepatitis and hepatocellular carcinoma. Clin. Liver Dis. 2018;22:201–211. doi: 10.1016/J.CLD.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Manne V., Handa P., Kowdley K.V. Pathophysiology of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. Clin. Liver Dis. 2018;22:23–37. doi: 10.1016/J.CLD.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Noureddin M., Sanyal A.J. Pathogenesis of NASH: the impact of multiple pathways. Curr. Hepatol. Reports. 2018;17:350–360. doi: 10.1007/s11901-018-0425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng C., Stewart A.G., Woodman O.L., Ritchie R.H., Qin C.X. Non-alcoholic steatohepatitis: a review of its mechanism, models and medical treatments. 2020. [DOI] [PMC free article] [PubMed]
- 7.Schwabe R.F., Tabas I., Pajvani U.B. Mechanisms of fibrosis development in nonalcoholic steatohepatitis. Gastroenterology. 2020;158:1913–1928. doi: 10.1053/j.gastro.2019.11.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi E.H., Suzuki T., Funayama R., Nagashima T., Hayashi M., Sekine H., Tanaka N., Moriguchi T., Motohashi H., Nakayama K., Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016;7 doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bathish B., Robertson H., Dillon J.F., Dinkova-Kostova A.T., Hayes J.D. Nonalcoholic steatohepatitis and mechanisms by which it is ameliorated by activation of the CNC-bZIP transcription factor Nrf2. Free Radic. Biol. Med. 2022;188:221–261. doi: 10.1016/j.freeradbiomed.2022.06.226. [DOI] [PubMed] [Google Scholar]
- 12.Meakin P.J., Chowdhry S., Sharma R.S., Ashford F.B., V Walsh S., McCrimmon R.J., Dinkova-Kostova A.T., Dillon J.F., Hayes J.D., Ashford M.L.J. Susceptibility of Nrf2-null mice to steatohepatitis and cirrhosis upon consumption of a high-fat diet is associated with oxidative stress, perturbation of the unfolded protein response, and disturbance in the expression of metabolic enzymes but not with. Mol. Cell Biol. 2014;34:3305–3320. doi: 10.1128/MCB.00677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.V Knatko E., Tatham M.H., Zhang Y., Castro C., Higgins M., Dayalan Naidu S., Leonardi C., de la Vega L., Honda T., Griffin J.L., Hay R.T., Dinkova-Kostova A.T. Downregulation of Keap1 confers features of a fasted metabolic state. iScience. 2020;23 doi: 10.1016/j.isci.2020.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitteringham N.R., Abdullah A., Walsh J., Randle L., Jenkins R.E., Sison R., Goldring C.E.P., Powell H., Sanderson C., Williams S., Higgins L., Yamamoto M., Hayes J., Park B.K. Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J. Proteonomics. 2010;73:1612–1631. doi: 10.1016/j.jprot.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma R.S., Harrison D.J., Kisielewski D., Cassidy D.M., McNeilly A.D., Gallagher J.R., V Walsh S., Honda T., McCrimmon R.J., Dinkova-Kostova A.T., Ashford M.L.J., Dillon J.F., Hayes J.D. Experimental nonalcoholic steatohepatitis and liver fibrosis are ameliorated by pharmacologic activation of Nrf2 (NF-E2 p45-related factor 2) Cell. Mol. Gastroenterol. Hepatol. 2018;5:367–398. doi: 10.1016/j.jcmgh.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horie Y., Suzuki T., Inoue J., Iso T., Wells G., Moore T.W., Mizushima T., Dinkova-Kostova A.T., Kasai T., Kamei T., Koshiba S., Yamamoto M. Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism. Commun. Biol. 2021;4:576. doi: 10.1038/s42003-021-02100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reisman S.A., Ferguson D.A., Lee C.-Y.I., Proksch J.W. Omaveloxolone and TX63682 are hepatoprotective in the STAM mouse model of nonalcoholic steatohepatitis. J. Biochem. Mol. Toxicol. 2020 doi: 10.1002/jbt.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seedorf K., Weber C., Vinson C., Berger S., Vuillard L.-M., Kiss A., Creusot S., Broux O., Geant A., Ilic C., Lemaitre K., Richard J., Hammoutene A., Mahieux J., Martiny V., Durand D., Melchiore F., Nyerges M., Paradis V., Provost N., Duvivier V., Delerive P. Selective disruption of NRF2-KEAP1 interaction leads to NASH resolution and reduction of liver fibrosis in mice. JHEP Reports Innov. Hepatol. 2023;5 doi: 10.1016/j.jhepr.2022.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barreca M., Qin Y., Cadot M.E.H., Barraja P., Bach A. Advances in developing noncovalent small molecules targeting Keap1. Drug Discov. Today. 2023;28 doi: 10.1016/j.drudis.2023.103800. [DOI] [PubMed] [Google Scholar]
- 20.He F., Antonucci L., Yamachika S., Zhang Z., Taniguchi K., Umemura A., Hatzivassiliou G., Roose-Girma M., Reina-Campos M., Duran A., Diaz-Meco M.T., Moscat J., Sun B., Karin M. NRF2 activates growth factor genes and downstream AKT signaling to induce mouse and human hepatomegaly. J. Hepatol. 2020;72:1182–1195. doi: 10.1016/j.jhep.2020.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler U.A., Kurinna S., Schwitter D., Marti A., Schäfer M., Hellerbrand C., Speicher T., Werner S. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology. 2014;60:670–678. doi: 10.1002/hep.26964. [DOI] [PubMed] [Google Scholar]
- 22.Taguchi K., Yamamoto M. The KEAP1-NRF2 system as a molecular target of cancer treatment. Cancers. 2020;13:46. doi: 10.3390/cancers13010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biolato M., Bianco A., Lucchini M., Gasbarrini A., Mirabella M., Grieco A. The disease-modifying therapies of relapsing-remitting multiple sclerosis and liver injury: a narrative review. CNS Drugs. 2021;35:861–880. doi: 10.1007/s40263-021-00842-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a keap1-independent manner. Mol. Cell Biol. 2011;31:1121–1133. doi: 10.1128/mcb.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rada P., Rojo A.I., Evrard-Todeschi N., Innamorato N.G., Cotte A., Jaworski T., Tobon-Velasco J.C., Devijver H., Garcia-Mayoral M.F., Van Leuven F., Hayes J.D., Bertho G., Cuadrado A. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/-TrCP Axis. Mol. Cell Biol. 2012;32:3486–3499. doi: 10.1128/mcb.00180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández-Ginés R., Encinar J.A., Hayes J.D., Oliva B., Rodríguez-Franco M.I., Rojo A.I., Cuadrado A. An inhibitor of interaction between the transcription factor NRF2 and the E3 ubiquitin ligase adapter β-TrCP delivers anti-inflammatory responses in mouse liver. Redox Biol. 2022;55 doi: 10.1016/j.redox.2022.102396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eslam M., El-Serag H.B., Francque S., Sarin S.K., Wei L., Bugianesi E., George J. Metabolic (dysfunction)-associated fatty liver disease in individuals of normal weight. Nat. Rev. Gastroenterol. Hepatol. 2022;19:638–651. doi: 10.1038/s41575-022-00635-5. [DOI] [PubMed] [Google Scholar]
- 28.Fujii M., Shibazaki Y., Wakamatsu K., Honda Y., Kawauchi Y., Suzuki K., Arumugam S., Watanabe K., Ichida T., Asakura H., Yoneyama H. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med. Mol. Morphol. 2013;46:141–152. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 29.Van der Schueren B., Vangoitsenhoven R., Geeraert B., De Keyzer D., Hulsmans M., Lannoo M., Huber H.J., Mathieu C., Holvoet P. Low cytochrome oxidase 4I1 links mitochondrial dysfunction to obesity and type 2 diabetes in humans and mice. Int. J. Obes. 2015;39:1254–1263. doi: 10.1038/ijo.2015.58. [DOI] [PubMed] [Google Scholar]
- 30.Saito K., Uebanso T., Maekawa K., Ishikawa M., Taguchi R., Nammo T., Nishimaki-Mogami T., Udagawa H., Fujii M., Shibazaki Y., Yoneyama H., Yasuda K., Saito Y. Characterization of hepatic lipid profiles in a mouse model with nonalcoholic steatohepatitis and subsequent fibrosis. Sci. Rep. 2015;5 doi: 10.1038/srep12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seto W.-K., Yuen M.-F. Nonalcoholic fatty liver disease in Asia: emerging perspectives. J. Gastroenterol. 2017;52:164–174. doi: 10.1007/s00535-016-1264-3. [DOI] [PubMed] [Google Scholar]
- 32.Pardo V., González-Rodríguez Á., Guijas C., Balsinde J., Valverde Á.M. Opposite cross-talk by oleate and palmitate on insulin signaling in hepatocytes through macrophage activation. J. Biol. Chem. 2015;290:11663–11677. doi: 10.1074/jbc.M115.649483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiura M., Ohshima M., Ogawa K., Yano M. Chronic administration of Satsuma Mandarin fruit (Citrus unshiu MARC.) improves oxidative stress in streptozotocin-induced diabetic rat liver. Biol. Pharm. Bull. 2006;29:588–591. doi: 10.1248/bpb.29.588. [DOI] [PubMed] [Google Scholar]
- 34.Pajares M., Jiménez-Moreno N., García-Yagüe Á.J., Escoll M., de Ceballos M.L., Van Leuven F., Rábano A., Yamamoto M., Rojo A.I., Cuadrado A., Jim enez-Moreno N., Garc ıa-Yag A.J., ıa de Ceballos M.L., Abano A.R. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12:1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojo A.I., Rada P., Mendiola M., Ortega-Molina A., Wojdyla K., Rogowska-Wrzesinska A., Hardisson D., Serrano M., Cuadrado A. The PTEN/NRF2 axis promotes human carcinogenesis. Antioxidants Redox Signal. 2014;21:2498–2514. doi: 10.1089/ars.2014.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rojo A.I., Innamorato N.G., Moreno A.M. Mart In, De Ceballos I.L., Yamamoto M., Cuadrado A. Nrf2 regulates microglial dynamics and neuroinflammation in experimental Parkinson's disease. Glia. 2010;8:588–598. doi: 10.1002/glia.20947. [DOI] [PubMed] [Google Scholar]
- 37.García-Ruiz I., Blanes Ruiz N., Rada P., Pardo V., Ruiz L., Blas-García A., Valdecantos M.P., Grau Sanz M., Solís Herruzo J.A., Valverde Á.M. Protein tyrosine phosphatase 1b deficiency protects against hepatic fibrosis by modulating nadph oxidases. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rojo A.I., Pajares M., Rada P., Nuñez A., Nevado-Holgado A.J., Killik R., Van Leuven F., Ribe E., Lovestone S., Yamamoto M., Cuadrado A. NRF2 deficiency replicates transcriptomic changes in Alzheimer's patients and worsens APP and TAU pathology. Redox Biol. 2017;13:444–451. doi: 10.1016/j.redox.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W., Ferrell L.D., Liu Y.-C., Torbenson M.S., Unalp-Arida A., Yeh M., Mccullough A.J., Sanyal A.J. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. 2005. [DOI] [PubMed]
- 40.Dewidar B., Meyer C., Dooley S., Meindl-Beinker A.N. TGF-β in hepatic stellate cell activation and liver fibrogenesis-updated 2019. Cells. 2019;8 doi: 10.3390/cells8111419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C.-Y., Yuan W.-G., He P., Lei J.-H., Wang C.-X. Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J. Gastroenterol. 2016;22:10512–10522. doi: 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z., Tian R., She Z., Cai J., Li H. Role of oxidative stress in the pathogenesis of nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2020;152:116–141. doi: 10.1016/j.freeradbiomed.2020.02.025. [DOI] [PubMed] [Google Scholar]
- 43.Metelo A.M., Pérez-Carro R., Castro M.M.C.A., López-Larrubia P. VO(dmpp)2 normalizes pre-diabetic parameters as assessed by in vivo magnetic resonance imaging and spectroscopy. J. Inorg. Biochem. 2012;115:44–49. doi: 10.1016/j.jinorgbio.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 44.O'Neil P.M., Birkenfeld A.L., McGowan B., Mosenzon O., Pedersen S.D., Wharton S., Carson C.G., Jepsen C.H., Kabisch M., Wilding J.P.H. Efficacy and safety of semaglutide compared with liraglutide and placebo for weight loss in patients with obesity: a randomised, double-blind, placebo and active controlled, dose-ranging, phase 2 trial. Lancet (London, England) 2018;392:637–649. doi: 10.1016/S0140-6736(18)31773-2. [DOI] [PubMed] [Google Scholar]
- 45.Newsome P., Francque S., Harrison S., Ratziu V., Van Gaal L., Calanna S., Hansen M., Linder M., Sanyal A. Effect of semaglutide on liver enzymes and markers of inflammation in subjects with type 2 diabetes and/or obesity. Aliment. Pharmacol. Ther. 2019;50:193–203. doi: 10.1111/apt.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Younes R., Bugianesi E. NASH in lean individuals. Semin. Liver Dis. 2019;39:86–95. doi: 10.1055/s-0038-1677517. [DOI] [PubMed] [Google Scholar]
- 47.Wang W., Ren J., Zhou W., Huang J., Wu G., Yang F., Yuan S., Fang J., Liu J., Jin Y., Qi H., Miao Y., Le Y., Ge C., Qiu X., Wang J., Huang P., Liu Z., Wang S. Lean non-alcoholic fatty liver disease (Lean-NAFLD) and the development of metabolic syndrome: a retrospective study. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-14701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenstock J., Wysham C., Frías J.P., Kaneko S., Lee C.J., Fernández Landó L., Mao H., Cui X., Karanikas C.A., Thieu V.T. Efficacy and safety of a novel dual GIP and GLP-1 receptor agonist tirzepatide in patients with type 2 diabetes (SURPASS-1): a double-blind, randomised, phase 3 trial. Lancet (London, England) 2021;398:143–155. doi: 10.1016/S0140-6736(21)01324-6. [DOI] [PubMed] [Google Scholar]
- 49.Tacke F., Weiskirchen R. Non-alcoholic fatty liver disease (NAFLD)/non-alcoholic steatohepatitis (NASH)-related liver fibrosis: mechanisms, treatment and prevention. Ann. Transl. Med. 2021;9:729. doi: 10.21037/atm-20-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akiyama K., Warabi E., Okada K., Yanagawa T., Ishii T., Kose K., Tokushige K., Ishige K., Mizokami Y., Yamagata K., Onizawa K., Ariizumi S.-I., Yamamoto M., Shoda J. Deletion of both p62 and Nrf2 spontaneously results in the development of nonalcoholic steatohepatitis. Exp. Anim. 2018;67:201–218. doi: 10.1538/expanim.17-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J., Donepudi A.C., Moscovitz J.E., Slitt A.L. Keap1-knockdown decreases fasting-induced fatty liver via altered lipid metabolism and decreased fatty acid mobilization from adipose tissue. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulkarni S.R., Armstrong L.E., Slitt A.L. Caloric restriction-mediated induction of lipid metabolism gene expression in liver is enhanced by Keap1-knockdown. Pharm. Res. (N. Y.) 2013;30:2221–2231. doi: 10.1007/s11095-013-1138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka Y., Aleksunes L.M., Yeager R.L., Gyamfi M.A., Esterly N., Guo G.L., Klaassen C.D. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J. Pharmacol. Exp. Therapeut. 2008;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 54.Shin S., Wakabayashi J., Yates M.S., Wakabayashi N., Dolan P.M., Aja S., Liby K.T., Sporn M.B., Yamamoto M., Kensler T.W. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur. J. Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brancale J., Vilarinho S. A single cell gene expression atlas of 28 human livers. J. Hepatol. 2021;75:219–220. doi: 10.1016/j.jhep.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F., Zhan Y., Li M., Wang L., Zheng A., Liu C., Wang H., Wang T. Cell-permeable PROTAC degraders against KEAP1 efficiently suppress hepatic stellate cell activation through the antioxidant and anti-inflammatory pathway. ACS Pharmacol. Transl. Sci. 2023;6:76–87. doi: 10.1021/acsptsci.2c00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robledinos-Antón N., Rojo A.I., Ferreiro E., Núñez Á., Krause K.-H., Jaquet V., Cuadrado A. Transcription factor NRF2 controls the fate of neural stem cells in the subgranular zone of the hippocampus. Redox Biol. 2017;13:393–401. doi: 10.1016/j.redox.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sander T., Freyss J., Korff V., Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. 2015. [DOI] [PubMed]
- 59.Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. AdmetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/bioinformatics/bty707. [DOI] [PubMed] [Google Scholar]
- 60.Gressner O.A., Gao C. Monitoring fibrogenic progression in the liver. Clin. Chim. Acta. 2014;433:111–122. doi: 10.1016/j.cca.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 61.Flevaris P., Vaughan D. The role of plasminogen activator inhibitor type-1 in fibrosis. Semin. Thromb. Hemost. 2017;43:169–177. doi: 10.1055/s-0036-1586228. [DOI] [PubMed] [Google Scholar]
- 62.Zou Z., Ohta T., Miura F., Oki S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 2022;50:W175–W182. doi: 10.1093/nar/gkac199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.