Abstract
Background
A general view in the study of pollination syndromes is that floral traits usually represent convergent floral adaptations to specific functional pollinator groups. However, the definition of convergence is elusive and contradictory in the literature. Is convergence the independent evolution of either the same trait or similar traits with the same function? A review of the concept of convergence in developmental biology and phylogenetic systematics may shed new light in studies of pollination syndromes.
Scope
The aims of this article are (1) to explore the notion of convergence and other concepts (analogy, homoplasy and parallelism) within the theory and practice of developmental evolution and phylogenetic systematics; (2) to modify the definitions of syndromes in order to embrace the concepts of analogy and convergence; (3) to revisit the bat pollination syndrome in the context of angiosperm phylogeny, with focus on the showy ‘petaloid’ organs associated with the syndrome; (4) to revisit the genetic-developmental basis of flower colour; (5) to raise evolutionary hypotheses of floral evolution associated with the bat pollination syndrome; and (6) to highlight some of the current frontiers of research on the origin and evolution of flowers and its impact on pollination syndrome studies in the 21st century.
Conclusions
The inclusion of the concepts of analogy and convergence within the concept of syndromes will constitute a new agenda of inquiry that integrates floral biology, phylogenetic systematics and developmental biology. Phyllostomid and pteropodid bat pollination syndrome traits in eudicots and monocots represent cases of analogous and convergent evolution. Pollination syndromes are a multivariate concept intrinsically related to the understanding of flower organogenesis and evolution. The formulation of hypotheses of pollination syndromes must consider the phylogenetic levels of universality for both plant and animal taxa, flower development, genetics, homology and evolution, and a clear definition of evolutionary concepts, including analogy, convergence, homoplasy and parallelism.
Keywords: Analogy, convergence, developmental biology, flower colour, flower development, flower evolution, homoplasy, parallelism, phylogenetic systematics
How does one ‘test’ the pollination syndromes? This is not obvious, which is probably one reason for the paucity of tests! As we see it, there are three steps. First, one must define what one means by the syndromes. Secondly, one must make the syndromes operational in order to test them quantitatively. Finally, one must decide what properties or predictions of the syndromes are the most important one to scrutinize.
(Ollerton et al., 2009, p. 1471).
What is the ancestral pollination syndrome? How often have shifts in pollination syndrome occurred? Is there a recurring shift directionality? Are there any reversals? Are some syndromes more likely to shift than others?
(Dellinger, 2020, p. 1205)
INTRODUCTION
Pollination syndromes are suites of floral traits related to the attraction and utilization of a particular group of animal agents as pollinators, so that a correlation among multiples traits across independent evolutionary events is expected (Fenster et al., 2004). Commonly recorded floral traits include flower colour and scent, flower orientation, flower size and symmetry, overall corolla shape, position of sexual organs, timing of anthesis, reward type, floral exposure from foliage, sturdiness, subtler quantitative colour differences (e.g. UV patterning), scent bouquets, and nectar composition (Raguso et al., 2003; Ollerton et al., 2009; Reynolds et al., 2009; Rosas-Guerrero et al., 2014; Dellinger, 2020). Among them, colour, reward, scent and floral shape have been used most often to justify syndrome expectations (Dellinger, 2020). The floral traits are likely to determine pollinator-mediated selection and capture differences in activity patterns, sensory attributes, dietary preferences, morphology and the behaviour of the pollinator (Fenster et al., 2004; Dellinger, 2020). Current studies on pollination syndromes have identified 11 functional groups: bat, bee, beetle, bird, butterfly, carrion fly, fly, long-tongued fly, moth, non-flying mammal and wasp (Rosas-Guerrero et al., 2014; Dellinger, 2020). In this way, a pollination syndrome is intrinsically associated with a functional pollinator group, which is defined as pollinators who select for an equivalent combination of floral traits, while distinct functional groups will select for distinct trait combinations (Fenster et al., 2004).
A global test of the pollination syndrome hypothesis showed that the hypothesis as classically articulated does not successfully represent the diversity of floral phenotypes or predict the pollinators of most studied plant species (Ollerton et al., 2009). In contrast, Dellinger (2020), in her review of pollination syndrome studies in the 21st century, endorsed the view that pollination syndromes constitute a robust concept to accurately delimit plant–pollinator relationships at large and small scales.
Sinnott-Armstrong et al. (2022) reviewed the concept of syndromes, recognizing it in two primary arenas: (1) macroevolution: the independent evolution of traits onto a phylogenetic hypothesis; and (2) community ecology: species classification into distinct syndromes based on their trait combinations. In order to avoid theoretical and practical confusion, they contrasted two definitions of syndromes: (1) Adaptive syndrome, which has three features: (a) convergent evolution of (b) multiple traits (c) adapted to a particular driver; and (2) Trait syndrome, which has two of these three criteria, namely (a) convergent evolution of (b) multiple traits (Sinnott-Armstrong et al., 2022, p. 4).
In the case of pollination syndromes, a general view is that floral traits usually represent convergent floral adaptations to specific functional pollinator groups (Fenster et al., 2004; Schiestl and Johnson, 2013; Sobel and Streisfeld, 2013; Rosas-Guerrero et al., 2014; Dellinger, 2020). Yet the definition of convergence or convergent evolution is elusive and contradictory in the literature. For instance, Fenster et al. (2004, p. 376) asked: ‘How, then, can we reconcile this apparent paradox of diverse visitors at flowers with our observations of widespread convergence in floral traits?’ According to Ollerton et al. (2009, p. 1471), ‘Convergent evolution is a ubiquitous feature of the biosphere, as indicated by correlations between phenotype and ecology across distantly related taxa’. For Schiestl and Johnson (2013, p. 310): ‘Unrelated plants pollinated by the same pollinators tend to exhibit convergence in their floral traits, including advertizing signals’. Dellinger (2020, p. 1194) said: ‘Generally, pollination syndromes represent convergent floral adaptations to specific functional pollinator groups’. Although evoking floral convergence, these authors did not define categorically what convergence is. Differently, Sobel and Streisfeld (2013, p. 1; italics added) clearly stated: ‘The independent evolution of the same trait among populations or species is known as phenotypic convergence’. By contrast, Ng and Smith (2016, p. 407; italics added) wrote: ‘Phenotypic convergence, whereby distantly related species evolve similar traits, provides a unique opportunity to study naturally occurring evolutionary replicates’. Finally, for Sinnott-Armstrong et al. (2022, p. 1; italics added): ‘One of the most striking and commonly studied phenomena in biology is that of convergent evolution, whereby distantly related species evolve similar phenotypes as adaptations to similar selective pressures’.
These elusive and contradictory definitions invite us to ask: is convergence the independent evolution of either the same trait or similar traits with the same function? The widespread use of the concept of convergence in the literature of floral biology may result in confusing comparative and evolutionary understanding. So, what is convergence indeed? The concept has been extensively revisited within the scope of both developmental evolution and phylogenetic systematics, resulting in competing and constructive interpretations (e.g. Haas and Simpson, 1946; Hennig, 1966; Patterson, 1982; Hall, 2007; Arendt and Reznick, 2008; Leander, 2008; Scotland, 2011), which will potentially shed new light on studies of pollination syndromes. Importantly, two or more structures are considered to be the same trait if they are phenotypically and genetically correspondent (= equivalent), whereas similar traits are structures that are not phenotypically correspondent, although they might be or not be genetically.
The main aim of this article is to provide a successful link among floral biology, developmental evolution and phylogenetic systematics in an attempt to guide studies of pollination syndromes. I will attempt to exemplify it through the integration of three subjects: (1) bat pollination and angiosperm phylogeny; (2) the origin and evolution of the perianth in angiosperms; and (3) flower colour as a model system for studies of plant evo-devo. In the first section, I explore the notion of convergent evolution and other concepts (analogy, homoplasy and parallelism) within the theory and practice of developmental evolution and phylogenetic systematics. Modified definitions of ‘syndrome’ are proposed, which embrace both analogy and convergence as distinct genetic-developmental and evolutionary phenomena. In the second section, I revisit the bat pollination syndrome in the context of angiosperm phylogeny, with focus on bat flowers. More specifically, I concentrate on the features of the showy ‘petaloid’ organs associated with the bat pollination syndrome. Since perianth corolla morphology is a trait traditionally considered to be associated with pollination syndromes, the third section deals with the origin and evolution of the perianth in angiosperms, showing the progress and challenges in understanding its developmental and evolutionary nature. Following this understanding, I criticize the naïve and widespread use of the terms ‘corolla shape’, ‘corolla tube’, ‘open corolla’, ‘corolla colour’ and ‘petals’ in studies of pollination syndromes, irrespective of theories of homology, analogy and convergence. In the fourth section, I revisit the genetic-developmental basis of flower colour and its potential impact on the discovery of analogous and convergent traits. The fifth section is a synthesis and application of the ideas presented in the previous ones, in which I raise evolutionary hypotheses of floral evolution associated with the bat pollination syndrome. I conclude by highlighting some of the current frontiers of research on the origin and evolution of flowers and its impact in pollination syndrome studies in the 21st century.
CONVERGENCE AND ANALOGY, WITH MODIFIED DEFINITIONS OF ‘SYNDROME’
As shown above, there are elusive and contradictory definitions of convergence in pollination syndrome studies. I suppose that such a confusion is due to the long and dynamic history of the concept itself. It is beyond the scope of this section to present an extensive review of the concept. The argumentation I will present here is based on a critical analysis provided by Scotland (2011), who recognized the problem in the definition of two important concepts in evolutionary biology: convergence and parallelism. Both concepts have been interpreted as examples of homoplasy, although in several distinct ways. The author (Scotland, 2011, p. 217) reviewed the following interpretations:
(1) homoplastic phenotypes are structurally correspondent in parallelism but non-correspondent in convergence (Patterson, 1982);
(2) homoplastic phenotypes in closely related taxa are parallelism but in distantly related taxa are convergence (Arendt and Reznick, 2008);
(3) homoplastic phenotypes have the same ancestral character states for parallelism but different ancestral character states for convergence (Leander, 2008); and
(4) parallelism comprises homoplastic phenotypes caused by the same underlying genetics resulting from an ancestral predisposition to evolve the same character states, whereas convergent phenotypes are caused by dissimilar genetics (Haas and Simpson, 1946).
By contrast, Arendt and Reznick (2008) also proposed to abandon the concept of parallelism and only adopt convergence to the independent evolution of a given phenotype. Subsequently, Scotland (2011, p. 219) proposed a new way:
The criteria listed … to distinguish parallelism from convergence may be unsuccessful because they treat these two ideas as mutually exclusive alternatives. But what if parallelism and convergence are not regarded as alternatives, but rather that the parallel evolution of genetic traits represents one of several possible types of explanation of phenotypic convergence. Under this model the parallel evolution of the same genetic traits can underpin and explain some – although not all – instances of phenotypic convergence.
In this way, Scotland (2011, Fig. 3, p. 218) originally linked convergence and parallelism with phenotype and genotype, respectively. Following his proposition, convergence means the independent (convergent) evolution of the same phenotypic trait controlled, in part, by the independent (parallel) evolution of the same genetic basis. Differently, the independent evolution of similar phenotypic traits controlled by the same or distinct genes represents analogy (Scotland, 2011). It is expected that analogous traits are regulated by distinct genes or genetic-regulatory pathways. But it has been reported that analogous phenotypes can be underpinned by the same pre-existing genetic regulatory circuits – a phenomenon called ‘deep homology’ (Shubin et al., 2009, p. 818). Therefore, both convergence (sensuScotland, 2011) and analogy can also be explained by deep homology (= latent and process homology; Ballego-Campos et al., 2023; see also de Beer, 1971; Gilbert and Bolker, 2001).
Based on this brief review, there are several ways to define convergence, a central concept that is part of the concept of ‘syndrome’. I sympathize with the definitions proposed by Haas and Simpson (1946) and Scotland (2011), as they link evolution, development and genetics. More precisely, I will follow Scotland (2011) and use hereafter the following comparative concepts and their respective definitions (Fig. 1):
Fig. 1.
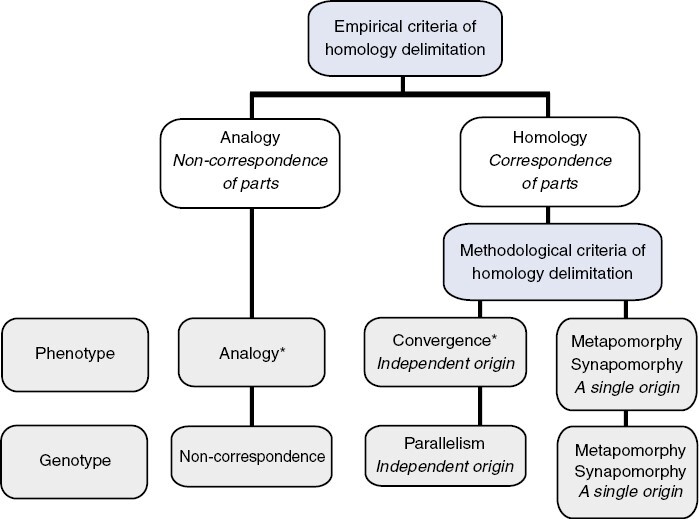
The formulation of hypotheses of analogy, homoplasy (convergence and parallelism) and phylogenetic homology (synapomorphy and metapomorphy) based on empirical and methodological criteria of homology delimitation (modified from Scotland, 2011, Fig. 3, p. 218; and Ballego-Campos et al., 2023, Fig. 2, p. 411). Note that the delimitation of analogy (= non-correspondence of parts) and homology (= correspondence of parts) depends on empirical criteria (genetic developmental causes, epigenetic causes, ontogeny timing, and position or topology), whereas the delimitation of phylogenetic homologies (metapomorphy and synapomorphy) and homoplasies (convergence and parallelism) depends on methodological criteria (parsimony, Bayesian and maximum likelihood analysis). Convergence and parallelism are associated with the phenotypic and genotypic levels, respectively. *Multiple hypotheses of analogy and convergence may constitute syndrome pollination traits grounded in a particular phylogenetic hypothesis.
(1) analogy: the phenomenon of independent evolution of similar phenotypes with the same function (Darwin, 1859) controlled by the same or distinct genetic regulatory basis (Shubin et al., 2009; Scotland, 2011);
(2) homoplasy: the phenomenon of independent evolution of the same phenotype (convergence) and genotype (parallelism) (Scotland, 2011);
(3) convergence: the phenomenon of independent evolution of the same phenotype controlled by the same genetic regulatory basis (Scotland, 2011); and
(4) parallelism: the phenomenon of independent evolution of the same genotype (Scotland, 2011).
Since analogy and convergence constitute distinct phenomena, i.e. the independent origin of similar and the same traits, respectively, to say that all events of independent evolution are convergence (e.g. Arendt and Reznick, 2008) is inconsistent with these two distinct patterns of phenotypic development and evolution. In line with this, I propose that the definitions of syndromes be modified in order to embrace analogy and convergence:
(1) adaptive syndrome, which has three features: (a) analogous and/or convergent evolution of (b) multiple traits (c) adapted to a particular driver (modified from Sinnott-Armstrong et al., 2022, p. 4).
(2) trait syndrome, which has two of these three criteria, namely that there has been (a) analogous and/or convergent evolution of (b) multiple traits (modified from Sinnott-Armstrong et al., 2022, p. 4).
In addition to the concepts of ‘syndrome’, there is discussion in the literature about the concept of ‘trait’. A recent review provided by Dawson et al. (2021) recognized distinct definitions of ‘trait’ and ‘functional trait’ across distinct biological areas and study systems (e.g. individual organism, taxa and biome). Yet Dawson et al. (2021, p. 16 441) proposed a return to the basics, endorsing a simplified version that could potentially satisfy all areas of biology, as follows:
A trait is a measurable characteristic (morphological, phenological, physiological, behavioural, or cultural) of an individual organism that is measured at either the individual or other relevant level of organization.
Such a definition contemplates the traits assessed in studies of pollination syndromes (see Dellinger, 2020) and it is adopted here.
Sinnott-Armstrong et al. (2022, Fig. 1, p. 5) proposed a schematic illustration of the three main features of syndromes and several approaches to deal with syndromes inquiry, including: (1) convergent evolution of traits: adequate character and taxa samples across phylogeny, an explicit ‘test’ for convergence, and identification of independent origin of trait combination; (2) multiple traits: sample trait space across the community and phylogeny, data collection of relevant traits, and data collection at the appropriate scale; and (3) adaptation: conducting ecological observations, experimental research, manipulation of the system, and assessment of correlated evolution with a phylogenetic hypothesis. As the pollination syndrome hypothesis depends on phylogenetic analysis, I propose that the delimitation of syndrome traits follows the logical delimitation of characters in phylogenetic systematics, including neomorphic versus transformational characters (Sereno, 2007; Assis, 2019), characters versus character states (Wagner, 2007) and organs versus attributes (DiFrisco et al., 2022). In line with this, I stimulate researchers to: (1) indicate the taxonomic lineages (i.e. species and monophyletic groups) of plants and animals involved in the syndrome; (2) use and compare competing phylogenetic hypotheses and relative evidence; and (3) present clear definitions of both comparative (e.g. analogy, convergence, homoplasy and parallelism) and syndrome concepts they will adopt in their studies.
REVISITING THE BAT POLLINATION SYNDROME
The bat pollination syndrome classically includes ‘nocturnal anthesis, drab coloration (i.e. white and green), musty smell, flowers often located on branches or tree trunks (cauliflory) or suspended on long stalks (flagelliflory), and tubular or radially symmetrical flowers, often of the “shaving brush” type, that produce relatively large amounts of hexose-rich nectar’ (Fleming et al., 2009, p. 1020). In more detail, Fleming et al. (2009, p. 1023) divided the flowers into three categories based on shape: (1) shaving brush or stamen ball with many projecting stamens (Fig. 2A); (2) bell-shaped with the corolla forming a tube (Fig. 2B); and (3) cup-shaped with an ‘open corolla’ (Fig. 2C). Two phylogenetically distant clades of bats are pollinators: (1) the New World phyllostomids, which are specialized nectar-feeders that usually visit tubular flowers produced by epiphytes and shrubs; and (2) the Old World pteropodids that visit edna shaving brush flowers produced by trees (Fleming et al., 2009). According to these authors, bat pollination has been reported for about 250 genera, 67 families and 28 orders of angiosperms. Using a hypothetical phylogeny of the angiosperms (Soltis et al., 2005), they estimated that bat pollination evolved independently in about 85 % of the angiosperm families, and that of the 28 orders pollinated by bats, only eight (29 %) contain taxa visited by species of the two clades of bats.
Fig. 2.

Three categories of bat-pollinated flowers. (A) Shaving brush or stamen ball with many projecting stamens; Abutilon regnellii (Malvaceae) visited by Anoura caudifer (Phyllostomidae). (B) Bell-shaped with the corolla forming a tube; Chelonanthus alatus (Gentianaceae) visited by Glossophaga soricina (Phyllostomidae). (C) Cup-shaped with an open corolla; Passiflora ovalis (Passifloraceae) visited by Anoura caudifer (Phyllostomidae). Photograph credits: Marlies Sazima and Ivan Sazima.
Despite enumerating different floral traits relative to phyllostomid and pteropodid pollination, Fleming et al. (2009) endorsed the heuristic value of the concept of bat pollination syndrome. Considering the distinct flower categories associated with these two distinct pollinator clades, it seems that rather than being restricted to a single and general concept of bat pollination syndrome (e.g. Faegri and van der Pijl, 1979), it would be more rewarding to consider two bat pollination syndromes: phyllostomid and pteropodid. However, before accepting this, it is necessary to assess if the floral traits associated with these two putative syndromes are analogous and/or convergent and adapted to a particular driver. The two bat clades seem to represent specific drivers. But what about the floral traits? The expressive independent evolution of bat pollination with an angiosperm phylogenetic hypothesis (Soltis et al., 2005) may suggest that the associated floral traits also evolved independently (Fleming et al., 2009). But are they analogous and/or convergent traits? Strikingly, could some of the traditional bat pollination floral traits be plesiomorphic traits, which do not satisfy the concepts of adaptive and trait syndrome? In order to infer this, it is necessary to assess the genetic-developmental and evolutionary nature of these traits. In the next sections, I will focus on the nature of the perianth and its colours.
ORIGIN AND EVOLUTION OF THE ANGIOSPERM PERIANTH
A flower may be interpreted from multiple points of view for artists, ecologists, developmental biologists and plant systematists, so leading to distinct scenarios (Assis, 2018). Usually, the most attractive flower component is the corolla. In her review of pollination syndrome, Dellinger (2020) showed that, among 244 articles recording floral traits, 52 (21.3 %) were based on the corolla. Traditionally, the corolla is defined by the set of petals, and the floral calyx by the set of sepals. The concept of petals means floral organs that are coloured or attractive, whereas sepals are generally green and dull. In addition, the corolla and the calyx constitute the inner and outer whorls of the perianth, respectively. However, these morphological characterizations are not rules, as noted by Ronse De Craene and Brockington (2013, p. 5):
the recognition of petals becomes problematic in cases where no clear distinction can be made between greenish sepals and showy petals, such as when both whorls are sepaloid (e.g. Juncaceae) or when the outer perianth whorl is petaloid and similar in appearance to the inner whorl (e.g. the ‘monocotyledonous’ perianth of Passifloraceae).
Furthermore, Ronse De Craene (2008, p. 302) highlighted:
The differentiation of the perianth in the core eudicots is obviously highly complex and statements about homology of petals appear to be at least ‘shaky’. Petaloidy (viz. the possession of petal features such as pigmentation) is often confused with the presence of petals, as any organ such as staminodes, petals proper, sepals, or bracts can be brightly coloured and look like petals.
In terms of genetic control, there are four classes of homoeotic genes that, alone or in combination, underlie floral organ development: (1) A-function genes alone control the identity of sepals; (2) A- and B-function genes control the development of petals; (3) B- and C-function genes control the development of stamens; (4) C-function genes alone control the identity of carpels; and (5) E acts in combination with all these genes to determine organ identity (Coen and Meyerowitz, 1991; Honma and Goto, 2001). Accordingly, the genes can be expressed in different regions of the floral meristem, resulting in two petaloid floral whorls in monocotyledons and magnoliids. In terms of position, the petaloid outer whorl in monocotyledons is potentially homologous to the sepaloid outer whorl of others angiosperms, although in terms of genetic control they are not homologous. Therefore, the use of position and genetic control as criteria of homology delimitation may lead to distinct comparative hypotheses (De-Paula et al., 2018; Ballego-Campos et al., 2023).
In terms of evolutionary or historical derivation, the origin of the petals has been viewed from two scenarios: (2) bracteopetals and (2) andropetals (Endress, 1994; Friis et al., 2006; Ronse De Craene, 2007, 2008; Ronse De Craene and Brockington, 2013). Yet a third and poorly discussed scenario is de novo origin (Albert et al., 2002; Ronse De Craene, 2008). Traditionally, the bracteopetals occur usually in the spiral perianth and present bract-like features, including the breadth of primordia, chloroplast, three vascular traces and cellular structure (Ronse De Craene, 2008; Ronse De Craene and Brockington, 2013). Andropetals are organs with a narrow base, a single vascular trace, pigmentation of leucoplasts and chromoplasts, conical and elongated cells at the adaxial surface, and the presence of volatile oils (Ronse De Craene and Brockington, 2013). Despite these potential differences, there are practical challenges in the differentiation of bracteo- and andropetals (Ronse De Craene, 2007, 2008). With respect to the large angiosperm groups, it is hypothesized that magnoliids, monocots, Amborellales, Austrobayleiales, Nymphaeales and Illiciales present bracteopetals, whereas eudicots present andropetals (Endress, 1994; Friis et al., 2006, Fig. 20, p. 283). However, Ronse De Craene (2007, 2008) and Ronse De Craene and Brockington (2013) have challenged the traditional view that most eudicots have andropetals (e.g. Endress, 1994; Friis et al., 2006). According to these authors, petals in core eudicots present the same morphological features and structure as both sepals and bracts. Furthermore, changes of function in the eudicot perianth are associated with shifts in petaloidy to the other perianth whorl, or losses of the inner and outer whorls that can be secondarily compensated by the incorporation of bracts in the floral axes (Ronse De Craene, 2008). Evidence for andropetals in core eudicots include in a few taxa: (1) fusion with the staminal tube (e.g. Molluginaceae or Caryophyllaceae); (2) the impossibility of differentiating petals from staminodes (e.g. Ranunculaceae, Dichapetalaceae, Theophrastaceae); and (3) a common stamen–petal primordium (e.g. Primulaceae, Plumbaginaceae, Rhamnaceae, Caryophyllales). Within the clade Caryophyllales, both bracteo- and andropetals may have evolved. In the case of the Phytolacca clade and Portulaca clade, the outer perianth whorl (i.e. the calyx) is coloured like petals (Ronse De Craene, 2007, 2008).
In sum, the problem of homologizing the perianth organs was clearly recognized by Ronse De Craene and Brockington (2013, p. 7; see also De-Paula et al., 2018; Ballego-Campos et al., 2023) as follows:
we have explored the definition and homology of the angiosperm petal from the perspective of various criteria: topology, historical derivation, morphology, and developmental genetics. It is evident that none of these criteria, either alone or in combination, provide a unifying principle with which to homologise the petal. This is perhaps an unsurprising conclusion, because at the level of angiosperms, petals are probably non-homologous structures that have attained an analogous showy petaloid condition in order to perform a common function in the attraction of pollinators.
These theoretical and practical considerations stimulate us to raise some fundamental questions about studies of pollination syndromes. (1) How will floral homology assessment change our view of the evolution of syndromes? (2) How should we deal with the formulation of hypotheses concerning the origin and evolution of showy petaloid organs based on competing criteria of homology delimitation (e.g. topology, historical derivation, morphology and developmental genetics)? (3) Which showy petaloid organs are analogous? (4) Which ones are convergent? (5) Which phenotypic components make the showy petaloid organs? (6) Which phenotypic components are analogous syndrome traits? (7) Which ones are convergent?
THE EVO-DEVO OF FLOWER COLOR
Colour, shape, nectar guides, scent and microscopic structures are phenotypic components of the distinct showy petaloid organs of angiosperms involved in pollination syndromes (Moyroud and Glover, 2017; Nadot and Carrive, 2021). In relation to colour, three classes of flower pigments are known: (1) anthocyanins; (2) betalains; and (3) carotenoids. Anthocyanins, broadly distributed in angiosperms, are hydrophilic pigments that belong to the well-known flavonoid biosynthetic pathway, and give rise to blue, purple and red (Grotewold, 2006; Tanaka et al., 2008). They are synthesized in the cytosol and accumulate in the vacuoles (Grotewold, 2006; Tanaka et al., 2008). Intermolecular copigmentation results from the interaction between anthocyanins with other non-coloured flavonoids (e.g. flavonols), so that the latter provide ‘depth’ to several white or cream organs (Grotewold, 2006). Biosynthetic pathways, gene expression, cellular location and vacuolar pH are responsible for colouring anthocyanin pigments (Grotewold, 2006). Betalains are also hydrophilic and nitrogen-containing compounds that accumulate in the vacuoles as glycosides, and give rise to yellow, orange-red to violet (Grotewold, 2006; Tanaka et al., 2008). In plants, they are exclusively found in the order Caryophyllales, with reversion to anthocyanins in the families Caryophyllaceae and Molluginaceae (Brockington et al., 2011, 2015). Betalains and anthocyanins share part of their biosynthetic pathways and are mutually exclusive, although they may combine with carotenoids (Grotewold, 2006; Nadot and Carrive, 2021). Carotenoids are lipophilic molecules synthesized in the chromoplasts (Grotewold, 2006; Tanaka et al., 2008). They are yellow, orange and orange-red, and combining with red or purple anthocyanin results in brown and bronze hues that any pigment cannot provide by itself (Grotewold, 2006). Importantly, in terms of biosynthesis, the colours or the loss of pigmentation (i.e. white and pale) resulting from these three distinct pathways are analogous to each other, even though they are visually undistinguishable (e.g. white, yellow and red). Such an understanding is a cue to the inference of colour analogy and convergence in pollination syndrome studies.
Although the biosynthetic pathways of flower pigments are well known (mostly for anthocyanins), an emerging and fascinating area is the evo-devo of flower pigmentation, which represents the genetic and molecular basis of the evolutionary transitions in flower colour (Rausher, 2008; Sobel and Streisfeld, 2013). Importantly, independent changes in the same gene, involving the same substitutions (i.e. parallelism), result in convergent phenotypes, whereas changes in distinct genes (i.e. non-correspondence) result in analogous phenotypes. Furthermore, deep homology also explains analogy (Shubin et al., 2009). Flower colour changes within populations, species and clades also result from phenotypic plasticity, genetic drift, indirect selection, or as a direct target of selection via pollinator visitation, physiological features and pleiotropic consequences (Sobel and Streisfeld, 2013; Nadot and Carrive, 2021). Hence, the evo-devo of flower colour is a fundamental area of research to delimit analogous and convergent traits in pollination syndrome inquiry.
Sobel and Streisfeld (2013), for instance, substantially interested in the independent evolution of mutations (parallelism) underlying the independent evolution (convergence) of gains and losses of anthocyanin pigmentation, presented a set of studies that mapped the genetic basis involved in these evolutionary changes. They showed that different genes control the white colour of the flowers of Petunia axillaris (Solanaceae) (R2R3-MYB genes) (Quattrocchio et al., 1999), Ipomoea purpurea (Convolvulaceae) (R2R3-MYB and CHS genes) (Habu et al., 1998; Chang et al., 2005) and Parrya nudicaulis (Brassicaceae) (CHS gene) and Mimulus lewisii (Phrymaceae) (DFR gene) (Wu et al., 2013). Hence, the white colour is an analogy shared by these species. In line with these findings, Sobel and Streisfeld (2013) highlighted the need to continue dissecting the genetic basis of independent evolution across phylogenetic scales, including the further development and evolution of flower colour as an essential model trait in evo-devo.
Another constructive study was conducted by Ng and Smith (2016), who hypothesized the independent evolution of colour in Solanaceae via alternative biochemical pathways. The authors investigated the evolutionary patterns of red flowers underpinned by anthocyanin and carotenoid pathways using large-scale data mining and new sequence data to hypothetically reconstruct the phylogeny of 1341 species Solanaceae. For them, all independent evolution is ‘Phenotypic convergence, whereby distantly related species evolve similar traits’ (Ng and Smith, 2016, p. 407; italics added). In this way, Ng and Smith (2016) found that red-flowered species used distinct pathways to make red as follows: (1) 12 species used exclusively anthocyanin; (2) two species exclusively carotenoids; and (3) 13 species both pigments. The character evolution reconstruction supported the utilization of anthocyanins to make red flowers at least 11 times, the utilization of carotenoids twice, and the dual use of both pigments 12 times. In turn, they delimit three levels of convergence: (1) red-anthocyanin flowers; (2) red-carotenoid flowers; and (3) red-anthocyanin-carotenoid flowers.
I see the Solanaceae problem from a different perspective. First, as previously presented, there are two scenarios of independent evolution of phenotypes: analogy and convergence. Second, in order to avoid terminological confusion, it is critical to note that what Ng and Smith (2016) called ‘convergence’ I call ‘analogy’ in the present article. In line with this, the independent evolution of red flowers in Solanaceae may represent cases of analogy (i.e. similar phenotypes controlled by the same or different genotypes) and convergence (the same phenotypes controlled by the same genotypes). In other words, red flowers exclusively originating from distinct (anthocyanin or carotenoid) pathways are phenotypically and genetically analogous to each other, whereas red flowers resulting from the same biosynthetic pathway may be analogous or convergent depending on the genetic regulatory basis (e.g. Sobel and Streisfeld, 2013). Because of the lack of information at the genetic-molecular level, the hypothesis of analogous and convergent evolution of red-anthocyanin flowers in Solanaceae cannot be effectively supported. The same is true for red-carotenoid flowers. Indeed, Ng and Smith (2016) concluded that increasing knowledge of the biochemistry and function of plant pigment pathways will provide a great opportunity for identifying the genetic-molecular changes underpinning red flower evolution across Solanaceae and testing for pleiotropic effects of colour transitions.
BAT POLLINATION SYNDROME AND ANGIOSPERM PHYLOGENY
Based on the points previously discussed and on a list of plant families pollinated by bats (Fleming et al., 2009, pp. 1026–1027), I present in Table 1 the showy petaloid organs relative to bat pollination and their putative evolutionary derivations, i.e. from either bracts or stamens. Some theoretical and practical points are important. First, showy petaloid organs means they are coloured and attractive. Hence, it is necessary to note that both organ and colour constitute distinct phenotypic elements of character statements. Organ is a neomorphic character with two states (absent versus present), whereas colour is the variable (or an attribute) of a transformational character including two or more states (Sereno, 2007; Wagner, 2007; Di-Frisco et al., 2022). Variables or attributes are essentially dependent on the organ or body part – ‘there is no colored phenotype without a body part to have that color’ (DiFrisco et al., 2022, p. 8). Yet the same organ or body part in two or more organisms may have distinct states (e.g. blue, red, white) of a variable or attribute. By contrast, neomorphic characters (e.g. organs) are separable from each other, and they can be or not be developmentally individualized (DiFrisco et al., 2022). In this way, analogous, homologous and convergent organs might present analogous, homologous or convergent colours (or other attributes), depending on the evolution of the genetic-developmental regulatory systems and biosynthetic pathways (carotenoid, anthocyanin and betalain) underlying flower colour, and the angiospermic phylogenetic levels of analysis as well. Second, despite the practical challenges of distinguishing bracteo- and andropetals, as well as establishing homology relationships of the perianth organs (Ronse De Craene and Brockington, 2013; De-Paula et al., 2018; Ballego-Campos et al., 2023), I endorse that all these fundamentally comparative problems are constructive in formulating and refreshing pollination syndrome hypotheses. Hence, the origins of the floral organs presented in Table 1 must be continuously assessed through new evidence. Third, distinct organs have been usually called ‘corolla’ (or ‘petals’), and such an essentialist interpretation of the corolla as a pollination syndrome trait, irrespective of theories of homology, analogy and convergence, is challenged here. Moreover, in order to satisfy the concepts of syndrome (modified from Sinnott-Armstrong et al., 2022), pollination syndrome studies need to be critically supported by comparative and phylogenetic analyses in terms of competing evolutionary hypotheses of relationship and floral character evolution (cf. Assis, 2018), as well as underlying criteria of homology delimitation (cf. Scotland, 2011; Ronse De Craene and Brockington, 2013; De-Paula et al., 2018; Ballego-Campos et al., 2023).
Table 1.
Putative evolutionary origins (i.e. historical derivations) of the showy petaloid organs in bat-pollinated plant families. Bat-pollinated taxa extracted from Fleming et al. (2009, p. 1026–1027)
| Lineage | Order | Family | Showy petaloid organs or whorls of organs | Origin of outer whorl of perianth organs | Origin of inner whorl(s) of perianth organs | References |
|---|---|---|---|---|---|---|
| Magnoliids | Laurales | Lauraceae | Outer and inner perianth whorls (united with each other and the staminal whorls, forming a hypanthium) | Bracteal Androecial |
Bracteal Androecial |
Ronse de Craene and Brockington (2013)
Chanderbali et al. (2006) |
| Magnoliales | Annonaceae | Two inner perianth whorls (free from each other; and organs free from each other) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | |
| Monocots | Arecales | Arecaceae | Outer and inner perianth whorls (free from each other; and organs of each whorl free from each other) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) |
| Asparagales | Agavaceae | Outer and inner perianth whorls (united with each other and the staminal whorls, forming a hypanthium) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | |
| Amaryllidaceae | Outer and inner perianth whorls (united with each other and the staminal whorls, forming a hypanthium) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Asphodelaceae | Outer and inner perianth whorls (forming or not a hypanthium) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Asteliaceae | Outer and inner perianth whorls (united with each other and the staminal whorls, forming a hypanthium) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Xanthorrhoeaceae | Outer and inner perianth whorls (free from each other; and organs of each whorl free from each other) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Pandanales | Pandanaceae | Bracts | N.A. | N.A. | N.A. | |
| Velloziaceae | Outer and inner perianth whorls (united or not with each other and the staminal whorls, forming a hypanthium) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Poales | Bromeliaceae | Outer and inner perianth whorls (united or not with each other and the staminal whorls, forming a hypanthium) | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | |
| Zingiberales | Cannaceae | Staminal whorls | N.A. | N.A. | N.A. | |
| Heliconiaceae | Bracts; outer and inner perianth whorls | Bracteal | Bracteal | |||
| Musaceae | Bracts; outer and inner perianth whorls | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Strelitziaceae | Outer perianth whorl | Bracteal | Bracteal | Endress (1994); Friis et al. (2006, Fig. 20, p. 283) | ||
| Basal eudicots | Caryophyllales | Cactaceae | Outer perianth (organs free from each other) | Androecial Bracteal |
N.A. |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283) Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
| Nyctaginaceae | Bracts; outer perianth (organs usually united with each other) | Androecial Bracteal |
N.A. |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283) Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Proteales | Proteaceae | Outer perianth whorl (united with the staminal whorl, forming a hypanthium) | Bracteal | N.A. | Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) | |
| Santalales | Loranthaceae | Inner perianth whorl (united with the staminal whorl, forming a hypanthium) | Androecial Bracteal |
Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Rosids | Brassicales | Capparaceae | Inner perianth whorl | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008) Ronse de Craene and Brockington (2013) |
| Salvandoraceae | Inner perianth whorl | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Celastrales | Celastraceae | Inflorescence; inner perianth whorl | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Cucurbitales | Cucurbitaceae | Inner perianth whorl (organs usually united with each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Fabales | Fabaceae | Inner perianth whorl (organs free from or united with each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Geraniales | Geraniaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Malpighiales | Caryocaraceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Chrysobalanaceae | Inflorescence; inner perianth whorl (organs free from or united with each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Clusiaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Euphorbiaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Passifloraceae | Inner perianth whorl (organs usually free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Rhizophoraceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Salicaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Malvales | Malvaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Myrtales | Combretaceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Lythraceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Melastomataceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Myrtaceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Onagraceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Vochysiaceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Oxalydales | Elaeocarpaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Rosales | Moraceae | Outer perianth whorl (organs free from each other) | Bracteal | N.A. | Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) | |
| Rhamnaceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial | Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) | ||
| Rosaceae | Inner perianth whorl (united with both outer perianth whorl and staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Sapindales | Anacardiaceae | Inner perianth whorl (usually united with the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Meliaceae | Inner perianth whorl (usually united with the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Sapindaceae | Inner perianth whorl (organs free from each other) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Asterids | Apiales | Araliaceae | Inner perianth whorl (organs usually free from each other at maturity) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
| Asterales | Asteraceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Campanulaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Ericales | Ericaceae | Inner perianth whorl (organs united with each other and usually with the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Marcgraviaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Polemoniaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Sapotaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Gentianales | Apocynaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Gentianaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Loganiaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Rubiaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Lamiales | Acanthaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Bignoniaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Gesneriaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Lamiaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Scrophulariaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Verbenaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Solanales | Boraginaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
|
| Convolvulaceae | Inner perianth whorl (organs united with each other and the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
||
| Solanaceae | Inner perianth whorl (organs united with each other and usually with the staminal whorl, forming a hypanthium) | Bracteal | Androecial Bracteal |
Endress (1994); Friis et al. (2006, Fig. 20, p. 283); Ronse de Craene (2007, 2008); Ronse de Craene and Brockington (2013) |
N.A., not applicable.
With respect to bat pollination, the organs constituting what the authors (e.g. Fenster et al., 2004, p. 376; Fleming et al., 2009, p. 1023; Moyroud and Glover, 2017, p. 350; Dellinger, 2020, p. 1201) have usually called ‘corolla’ correspond to analogous and convergent organs, depending on the phylogenetic level of inquiry. In monocots (Fleming et al., 2009, Fig. 3, p. 1021), they correspond to both outer and inner perianth whorls putatively derived from bracts, although in Cannaceae and Zingiberaceae they are elements of the androecium itself. In eudicots (Fleming et al., 2009, Fig. 4, p. 1022), they represent the inner perianth whorl putatively derived from bracts and stamens, although in Nyctaginaceae and Proteaceae they are the outer bract-derived perianth whorl. In Cactaceae, they are putatively bract-derived and spirally arranged. Additionally, the floral tube in some monocots and asterids usually constitutes the hypanthium (i.e. the tube formed by the fusion of distinct floral whorls; Ronse De Craene, 2010), which is developmentally analogous in both clades (Ronse De Craene and Brockington, 2013) (Table 1). Therefore, considering the distinct historical, topological and developmental origins of both showy petaloid organs and floral tubes, hypotheses of bat pollination syndrome are associated with the analogous evolution of organs in monocots (Fig. 3A, B) and eudicots (Fig. 3C, D). Yet it is still necessary to assess if the variables or attributes of attraction (e.g. colour and scent) found in these organs evolved through analogous or convergent evolution (see below).
Fig. 3.

Showy petaloid organs in monocots and eudicots. (A) Yellow inner perianth whorl; Vriesea sazimae (Bromeliaceae, monocot) visited by Anoura caudifer (Phyllostomidae). (B) Greenish outer and inner perianth whorls; Dyckia subsecunda (Bromeliaceae) visited by Lonchophylla bokermanni (Phyllostomidae). (C) Yellow-greenish inner perianth whorl; Siphocampylus sulfureus (Campanulaceae, eudicot) visited by Anoura caudifer (Phyllostomidae). (D) Greenish inner perianth whorl; Dyssochroma viridiflorum (Solanaceae, eudicot) visited by Glossophaga soricina (Phyllostomidae). Photograph credits: Marlies Sazima and Ivan Sazima.
Within specific lineages, where the perianth parts are potentially the same (homologous), it is necessary to hypothesize if they evolved once (synapomorphies or metapomorphies; Assis, 2017) or multiple times (convergences). For instance, it has been hypothesized that the tubular inner perianth whorl (i.e. ‘corolla tube’; Fleming et al., 2009, p. 1023) (Figs 2B and 3C, D) in eudicots is an apomorphic state, whereas the open inner perianth (i.e. ‘open corolla’; Fleming et al., 2009, p. 1023) whorl (Fig. 2A) is a plesiomorphic one or reversion (Ronse De Craene, 2010). In line with this, we cannot regard the open inner perianth whorl as a convergent trait (unless it is a reversion). In other words, although Fleming et al. (2009) relates open flowers to pteropodid pollination, such a feature cannot be considered a syndrome trait, as it is a symplesiomorphic state within the eudicots (cf. Ronse De Craene, 2010) also associated with phyllostomid and other modes of pollination (cf. Sazima et al., 1999; Queiroz et al., 2020). At the same time the floral tube represents an apomorphic state for the asterids (Ronse De Craene, 2010); it represents a plesiomorphic state within the clade, which is also associated with distinct modes of pollination. Indeed, bat pollination has evolved from other modes (e.g. bee, butterfly, moth and hummingbird pollination) (Fleming et al., 2009 and references therein), and all of them share the floral tube as a symplesiomorphic feature within asterids. Hence, it is more appropriate to say that what evolved multiple times along the phylogeny of asterids were both flower tube shapes and colours (e.g. Acanthaceae: Tripp and Manos, 2008; Gesneriaceae: Martén-Rodríguez et al., 2009; Lobeliaceae: Knox et al., 2008). With respect to shaving brush flowers, they are not exclusively associated with bat pollination. In the tribe Pachycereeae (Cactaceae), for instance, bat pollination is a hypothetical ancestral mode that may have been derived from bee pollination (Wallace, 2002; Fleming et al., 2009). In this case, shaving brush flowers represent a symplesiomorphic state in Cactaceae shared by bat and bee pollination. In other angiosperm groups, however, they might be analogously or convergently associated with bat pollination syndrome, but further investigation is necessary.
The occurrence of white, pale and greenish flowers in 67 families and 28 orders of angiosperms pollinated by bats (Fleming et al., 2009) leads us to a constructive question: are these colours analogous or convergent syndrome traits within and among bat-pollinated angiosperm lineages? To answer this question it is necessary to investigate the biosynthetic pathways and genetic-molecular basis of the colour. However, the understanding of syndromes at the genomic level is still incipient. The vast majority of biological systems and plant–pollinator interactions have not been addressed within the scope of developmental evolution. In her review of pollination syndrome, Dellinger (2020) showed that, among 346 articles on the subject, only 20 (5.8 %) addressed syndromes in the genomic dimension (e.g. evolution of colour genes). With respect to the bat pollination syndrome, some few inferences can be made.
Tripp and Manos (2008) studied the evolution of pollination systems in the large genus Ruellia (Acanthaceae) on the basis of phylogenetic analyses, morphological ordinations, ancestral state reconstruction and character mapping reconstruction. They revealed key patterns in the direction and lability of floral characters associated with pollination. The evolutionary hypothesis is that a hummingbird ancestral syndrome switched to new pollination modes, with repeated evolution of bee- and insect-adapted species through distinct developmental means and morphological histories. In addition, bats and hawkmoths presented as specialized evolutionary dead ends. The authors (p. 1727) strikingly highlighted that ‘Species that have dramatically reduced or have lost their floral pigmentation are probably evolutionary dead-ends with respect to new pollination systems. … Once a character is lost, here, red or purple floral pigmentation, it cannot be regained’. An explanation for this pattern in Ruellia and other angiosperms (e.g. Ipomoea sect. Mina; Zufall and Rausher, 2004) is the irreversibility of changes in the underlying anthocyanin pathway. Possibly in line with these findings, Fleming et al. (2009) cited 14 examples of the evolution of bat pollination from other modes in 12 angiosperm families – Acanthaceae (Tripp and Manos, 2008), Agavaceae (Good-Avila et al., 2006), Bromeliaceae (Endress 1994; Benzing, 2000), Cactaceae (Wallace, 2002), Fabaceae (Luckow and Hopkins, 1995), Gesneriaceae (Perret et al., 2007; Martén-Rodríguez et al., 2009), Lecythidaceae (Mori and Boeke, 1987; Mori et al., 2007), Lobeliaceae (Knox et al., 2008), Malvaceae (Baum et al., 1998; Nyffeler and Baum, 2001), Passifloraceae (Hansen et al., 2006), Polemoniaceae (Prather, 1999) and Strelitziaceae (Kress et al., 1994) – but not the other way around. Yet eight of these studies were based on phylogenetic analyses, whereas the others are equivocal pending species-level phylogenetic analyses (Fleming et al., 2009). By contrast, a recent study (Karimi et al., 2022) in the African baobab (Adansonia digitata) (Malvaceae) showed hawkmoth pollination as a derived mode from bat pollination. Assessment of whether the white and pale colours are analogies and convergences within and among all these families depends on further biosynthetic and genetic-molecular analyses.
Differently across Solanaceae, rare red flowers have been identified as an evolutionary dead end derived independently from blue, purple and white, and a potential explanation is ‘nonequilibrium dynamics, with selection for red flowers, or the ability to produce red pigments, arising only recently, leaving the number of taxa with this state far lower than expected’ (Ng and Smith, 2016, p. 414). Interestingly, in Acanthaceae, white colour has been, otherwise, hypothesized as a dead end, explained by the irreversibility of changes in the underlying anthocyanin pathway (Tripp and Manos, 2008). The contrasting colour dead-end patterns in Acanthaceae and Solanaceae represent distinct evolutionary and genetic-developmental changes within the same anthocyanin biosynthetic pathway. Furthermore, singular pollinator mediations and pleiotropic and selective causes may explain this mosaic evolution (i.e. heterobathmy; Hennig, 1966; Rieppel, 2023) of flower colour in asterids.
Considering that betalain biosynthesis is a metapomorphy for the order Caryophyllales, in which the families Cactaceae and Nyctaginaceae are included, it is expected that the derived white and pale floral colours found in these families are analogous in relation to the white and pale colours found in the other bat-pollinated families. Because of the deep time of phylogenetic divergence between eudicots and monocots, it would be expected that they share non-correspondent genotypes controlling analogous flower colours. This expectation is grounded on the analysis of distinct genes controlling the white flower colour in eudicot species (Sobel and Streisfeld, 2013), including Petunia axillaris (Solanaceae) (R2R3-MYB genes) (Quattrocchio et al., 1999), Ipomoea purpurea (Convolvulaceae) (R2R3-MYB and CHS genes) (Habu et al., 1998; Chang et al., 2005), Parrya nudicaulis (Brassicaceae) (CHS gene) and Mimulus lewisii (Phrymaceae) (DFR gene) (Wu et al., 2013). Differently, Arendt and Reznick (2008) showed examples of closely related species and populations evolving similar phenotypes via distinct genes, whereas distantly related species with similar phenotypes share the same genes. Indeed, R2R3-MYB genes have also been identified in monocots and some magnoliids (e.g. Muñoz-Gómez et al., 2021), constituting a widespread ancestral genetic regulatory circuit across angiosperm lineages.
The bat pollination syndrome involves both analogous and convergent traits. Considering historical derivation, development and genetics it is expected that the phenotypic traits are analogous between monocots and eudicots. Yet cases of analogy and convergence can be found within these large groups. Even though ‘nocturnal anthesis, drab coloration (i.e. white and green), musty smell, flowers often located on branches or tree trunks (cauliflory) or suspended on long stalks (flagelliflory), and tubular or radially symmetrical flowers, often of the “shaving brush” type, that produce relatively large amount of hexose-rich nectar’ (Fleming et al., 2009, p. 1020) have been traditionally classified as traits linked with the bat pollination syndrome, it is necessary to make a developmental and evolutionary analysis to hypothesize if they evolved analogously or convergently, or if they are, in some cases, symplesiomorphies shared by distinct pollination modes (e.g. the flower tube in asterids and shaving brush flowers in Cactaceae). In this article, I consider that the analogous and/or convergent traits associated with the phyllostomid syndrome include the long floral tube (Figs 2B and 3) and white, pale, greenish and yellow-greenish organs (Figs 2 and 3). The long floral tube in eudicots and monocots is analogous. Although white, pale, greenish and yellow-greenish colours evolved multiple times along angiosperm phylogeny, it is still unknown if they represent analogy or convergence, or both. Other great candidates to be bat pollination syndrome traits include nocturnal anthesis and flower placement away from foliage, which are universal or nearly so for all bat flowers (Fleming et al., 2009), and sulphur-containing perfumes, commonly associated with bat pollination as well (Karimi et al., 2022). Even though these features evolved multiple times, it is unknown if they are analogies or convergences. Genetic-developmental and phylogenetic analyses are necessary to clarify this. Floral traits associated with pteropodid syndrome need to be further investigated.
These considerations about bat pollination show us that generalizations about pollination syndrome traits are problematic, leading to spurious scenarios. They are more idealistic than evolutionary. On the one hand, criticisms of the theory of pollination syndrome rest on the fact that certain flowers considered especially linked to a pollination syndrome are visited by multiple functional groups. Based on their global analysis, Ollerton et al. (2009) stressed that they do not take their results as evidence against the independent origin of floral syndrome traits resulting from pollinator-mediated natural selection. Yet the authors proposed that thinking only in terms of selection by a single ‘most effective pollinator’ fails to capture the range of possibilities. In addition, these authors highlighted that the study of pollination syndromes must consider the idiosyncrasies of both geographic distribution and plant lineages. Another problem is pleiotropy and genetic linkage, which can result in the appearance of several unrelated traits evolving correlatedly (Sinnott-Armstrong et al., 2022). On the other hand, some examples have reinforced the theory of pollination syndromes, so that certain authors have strongly endorsed its heuristic value (e.g. Fenster et al., 2004; Fleming et al., 2009; Schiestl and Johnson, 2013; Rosas-Guerrero et al., 2014; Dellinger, 2020). Dellinger (2020), in her review, demonstrated that some problems, including predictive inaccuracy and visitation by multiple functional groups, have been solved by refining techniques, while others prevail. In addition, she proposed the use of sophisticated statistical tools plus the quantification of the contribution of distinct flower visitors to remedy typological–essentialist classifications. In order to avoid generalizations and oversimplifications, I propose that the formulation of hypotheses of pollination syndromes must consider (1) the phylogenetic levels of universality at both plant and animal taxonomic lineages; (2) a critical comparative analysis of character development, homology and evolution in these lineages; and (3) a clear definition of evolutionary concepts, including analogy, convergence and parallelism.
CONCLUSIONS
On the one hand, some people working with syndromes might say ‘so what?’ – it does not matter to a pollinator if the traits have evolved convergently or analogously. On the other hand, the distinction between analogy and convergence is a paramount issue in evolutionary and comparative biology. The inclusion of both concepts within the concept of syndrome will constitute a new agenda of inquiry that integrates floral biology, phylogenetic systematics and developmental biology. Thus, rather than solely saying that multiple traits evolved independently onto a phylogenetic scaffold, or that all independent evolution is convergence, it is necessary to assess if they evolved to the same (convergent) or similar (analogous) phenotypes. At a large phylogenetic scale, phyllostomid and pteropodid bat pollination syndrome traits in eudicots and monocots represent cases of analogous and convergent evolution. In line with this, some exciting and deep questions come to mind: (1) Are there more convergences than analogies in pollination syndromes, or vice versa? (2) Which plant lineages evolved analogies? (3) Which ones evolved convergences? (4) Did traits of a particular syndrome evolve either de novo or from pre-existing features? (5) Is the independent evolution of colours, shapes, nectar guides, scents and microscopic structures convergent or analogous? (6) In the case of analogous traits, do they share the same (i.e. deep homology) or distinct genetic regulatory systems? (7) Which floral organ attributes or variables evolved from phenotypic plasticity? (8) Which ones evolved from genetic changes?
The classical idea of pollination syndromes only links floral features with functional groups. I endorse the idea that the singular evolutionary and ecological interactions between plant and animal lineages cannot be generalized or oversimplified in this way. In order to achieve a refined evolutionary delimitation of pollination syndrome, it is important to indicate analogous and/or convergent traits, as well as specifying the taxonomic plant and animal lineages involved. Organs and their attributes or variables constitute distinct phenotypic elements. Both plant and animal lineages may present a set of known and still unknown attributes and variables that may confirm a pollination syndrome. Depending on the phylogenetic levels of universality, certain features might be syndrome traits specific to some lineages but possibly not to others. Indeed, some morphological character states may represent symplesiomorphies across certain lineages. Because of the mosaic evolution of characters across the phylogeny, pollination mode and pollination syndrome cannot be synonymous concepts. The former is a broader concept that embraces plesiomorphic and apomorphic floral characters, whereas the latter includes floral analogous and/or convergent specializations associated with specific plant–animal lineages.
Pollination syndromes are a multivariate concept (Dellinger, 2020) that relies on multi- and interdisciplinary approaches. Its understanding is intrinsically related to the understanding of flower organogenesis and evolution. Therefore, current frontiers of research on the origin and evolution of flowers will have a definite impact on pollination syndrome studies in the 21st century. Homology, analogy and convergence assessment of floral organs and their attributes continues to be a theoretical and practical challenge, regarding the distinct criteria of homology delimitation (Ronse De Craene and Brockington, 2013; Ballego-Campos et al., 2023). The historical origins of the perianth from bracts or stamens have led to competing and constructive hypotheses (Ronse De Craene, 2007, 2008; Ronse De Craene and Brockington, 2013), although a de novo origin of the angiosperm flower has also been raised (Albert et al., 2002; Ronse De Craene, 2008). The discovery of genetic-developmental programs regulating the development of bracteo- and andropetals is needed (Ronse De Craene, 2007, 2008). The understanding of serial homologies (Di-Frisco et al., 2022) may explain de novo origins of multiple floral organs and whorls. Time of initiation, pressure of organs, alterations of the size of the floral meristems, and auxin mediation constitute critical mechanisms to understand floral organogenesis and evolution in parallel with genetics (Ronse De Craene, 2008). These physico-dynamic mechanisms associated with 3-D morphometrics (e.g. Dellinger, 2020) are critical to a realistic understanding of pollination syndromes in terms of flower ontogeny and natural selection. The analysis of the large diversity of flowers will potentially reveal new biosynthetic pathways, novel structures and genetic regulatory systems of flower colour (Tanaka et al., 2008). How can fossil information contribute to pollination syndrome studies (e.g. Friis et al., 2006)? The use of evolutionary pathways and cryptic markers (i.e. ‘fossil fingerprints’; Sanders et al., 2007, p. 719) may reveal new hypotheses of plant–pollinator adaptation and exaptation at large and small scales of geological time (e.g. colour as a plesiomorphic thermoregulatory component; and the presence of petaloid organs in the extinct Bennettitales, a potential sister group of angiosperms; Rudall, 2020). Finally, I encourage the assessment and comparison of competing hypotheses and evidence of angiosperm phylogenetic relationships (Assis, 2009, 2013, 2018), which may result in distinct hypotheses of floral evolution and pollination syndromes.
ACKNOWLEDGEMENTS
I thank Agnes Dellinger, two anonymous referees and the Handling Editor Gitte Petersen for valuable comments on an early version of this article, and Marlies Sazima and Ivan Sazima for sharing the pictures of flowers and bats.
LITERATURE CITED
- Albert VA, Oppenheimer GG, Lindqvist V.. 2002. Pleiotropy, redundancy and the evolution of flowers. Trends in Plant Science 7: 297–301. [DOI] [PubMed] [Google Scholar]
- Arendt J, Reznick D.. 2008. Convergence and parallelism reconsidered: what have we learned about the genetics of adaptation? Trends in Ecology and Evolution 23: 26–32. doi: 10.1016/j.tree.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Assis LCS. 2009. Coherence, correspondence, and the renaissance of morphology in phylogenetic systematics. Cladistics 25: 528–544. doi: 10.1111/j.1096-0031.2009.00261.x. [DOI] [PubMed] [Google Scholar]
- Assis LCS. 2013. Testing evolutionary hypotheses: from Willi Hennig to the Angiosperm Phylogeny Group. Cladistics 30: 240–242. doi: 10.1111/cla.12048. [DOI] [PubMed] [Google Scholar]
- Assis LCS. 2017. Patterns of character evolution in phylogenies. Journal of Systematics and Evolution 55: 225–230. doi: 10.1111/jse.12241. [DOI] [Google Scholar]
- Assis LCS. 2018. Revisiting the Darwinian shortfall in biodiversity conservation. Biodiversity and Conservation 27: 2859–2875. doi: 10.1007/s10531-018-1573-3. [DOI] [Google Scholar]
- Assis LCS. 2019. The criterion of conjunction in plant systematics and evolution. Plant Systematics and Evolution 305: 925–931. doi: 10.1007/s00606-019-01612-3. [DOI] [Google Scholar]
- Ballego-Campos I, Bonifácio SKV, Assis LCS.. 2023. A unified view of homology. Cladistics 39: 398–417. doi: 10.1111/cla.12541. [DOI] [PubMed] [Google Scholar]
- Baum DA, Small RL, Wendel JF.. 1998. Biogeography and floral evolution of baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Systematic Biology 47: 181–207. doi: 10.1080/106351598260879. [DOI] [PubMed] [Google Scholar]
- de Beer G. 1971. Homology, an unsolved problem. Oxford: Oxford University Press. [Google Scholar]
- Benzing DH. 2000. Bromeliaceae: profile of an adaptive radiation. Cambridge: Cambridge University Press. [Google Scholar]
- Brockington SF, Walker RH, Glover BJ, Soltis PS, Soltis DE.. 2011. Complex pigment evolution in the Caryophyllales: research review. New Phytologist 190: 854–864. doi: 10.1111/j.1469-8137.2011.03687.x. [DOI] [PubMed] [Google Scholar]
- Brockington SF, Yang Y, Gandia-Herrero F, et al. 2015. Lineage-specific gene radiations underlie the evolution of novel betalain pigmentation in Caryophyllales. New Phytologist 207: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanderbali AS, Kim S, Buzgo M, et al. 2006. Genetic footprints of stamen ancestors guide perianth evolution in Persea (Lauraceae). International Journal of Plant Science 167: 1075–1089. [Google Scholar]
- Chang SM, Lu YQ, Rausher MD.. 2005. Neutral evolution of the nonbinding region of the anthocyanin regulatory gene Ipmyb1 in Ipomoea. Genetics 170: 1967–1978. doi: 10.1534/genetics.104.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM.. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Dawson SK, Carmona CP, González-Suárez M, et al. 2021. The traits of ‘trait ecologists’: an analysis of the use of trait and functional trait terminology. Ecology and Evolution 11: 16434–16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger AS. 2020. Pollination syndromes in the 21st century: where do we stand and where may we go? New Phytologist 228: 1193–1213. doi: 10.1111/nph.16793. [DOI] [PubMed] [Google Scholar]
- De-Paula OC, Assis LCS, Ronse De Craene LP.. 2018. Unbuttoning the ancestral flower of angiosperms. Trends in Plant Science 23: 551–554. [DOI] [PubMed] [Google Scholar]
- DiFrisco J, Love AC, Wagner GP.. 2022. The hierarchical basis of serial homology and evolutionary novelty. Journal of Morphology 284: e21531. doi: 10.1002/jmor.21531. [DOI] [PubMed] [Google Scholar]
- Endress PK. 1994. Diversity and evolutionary biology of tropical flowers. Cambridge: Cambridge University Press. [Google Scholar]
- Faegri K, van der Pijl L.. 1979. The principles of pollination ecology, 3rd edn. Oxford: Pergamon Press. [Google Scholar]
- Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD.. 2004. Pollination syndromes and floral specialization. Annual Review of Ecology, Evolution, and Systematics 35: 375–403. doi: 10.1146/annurev.ecolsys.34.011802.132347. [DOI] [Google Scholar]
- Fleming TH, Geiselman C, Kress WJ.. 2009. The evolution of bat pollination: a phylogenetic perspective. Annals of Botany 104: 1017–1043. doi: 10.1093/aob/mcp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis EM, Pedersen KR, Crane PR.. 2006. Cretaceous angiosperm flowers: innovation and evolution in plant reproduction. Palaeogeography, Palaeoclimatology, Palaeoecology 232: 251–293. doi: 10.1016/j.palaeo.2005.07.006. [DOI] [Google Scholar]
- Gilbert SF, Bolker JA.. 2001. Homologies of process and modular elements of embryonic construction. Journal of Experimental Zoology 291: 1–12. [DOI] [PubMed] [Google Scholar]
- Good-Avila SV, Souza V, Gaut BS, Eguiarte LE.. 2006. Timing and rate of speciation in Agave (Agavaceae). Proceedings of the National Academy of Sciences of the USA 103: 9124–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. 2006. The genetics and biochemistry of floral pigments. Annual Review of Plant Biology 57: 761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- Haas O, Simpson GG.. 1946. Analysis of some phylogenetic terms, with attempts at redefinition. Proceedings of the American Philosophical Society 90: 319–349. [PubMed] [Google Scholar]
- Habu Y, Hisatomi Y, Lida S.. 1998. Molecular characterization of the mutable flaked allele for flower variegation in the common morning glory. Plant Journal 16: 371–376. [DOI] [PubMed] [Google Scholar]
- Hall BK. 2007. Homology and homoplasy: dichotomy or continuum? Journal of Human Evolution 52: 473–479. doi: 10.1016/j.jhevol.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK.. 2006. Phylogenetic relationships and chromosome number evolution in Passiflora. Systematic Botany 31: 138–150. doi: 10.1600/036364406775971769. [DOI] [Google Scholar]
- Hennig W. 1966. Phylogenetic systematics. Urbana: University of Illinois Press. [Google Scholar]
- Honma T, Goto K.. 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- Karimi N, Saghafi S, Keefover-Ring K, Venter SM, Ané C, Baum DA.. 2022. Evidence for hawkmoth pollination in the chiropterophilous African baobab (Adansonia digitata). Biotropica 54: 113–124. [Google Scholar]
- Knox EB, Muasya AM, Muchhala N.. 2008. The predominantly South American clade of Lobeliaceae. Systematic Botany 33: 462–468. doi: 10.1600/036364408784571590. [DOI] [Google Scholar]
- Kress WJ, Schatz GE, Adrianifahanana M, Morland HS.. 1994. Pollination of Ravenala madagascariensis (Strelitziaceae) by lemurs in Madagascar: evidence of an archaic coevolutionary system? American Journal of Botany 81: 542–551. [Google Scholar]
- Leander BS. 2008. Different modes of convergent evolution reflect phylogenetic distances: a reply to Arendt and Reznick. Trends in Ecology and Evolution 23: 481–482. [DOI] [PubMed] [Google Scholar]
- Luckow M, Hopkins HCF.. 1995. A cladistic analysis of Parkia (Leguminosae: Mimosoideae). American Journal of Botany 82: 1300–1320. doi: 10.1002/j.1537-2197.1995.tb12664.x. [DOI] [Google Scholar]
- Martén-Rodríguez S, Almarales-Castro A, Fenster CB.. 2009. Evaluation of pollination syndromes in Antillean Gesneriaceae: evidence for bat, hummingbird and generalized flowers. Journal of Ecology 97: 348–359. doi: 10.1111/j.1365-2745.2008.01465.x. [DOI] [Google Scholar]
- Mori S, Boeke JD.. 1987. Pollination. Memoirs of the New York Botanical Garden 44: 137–155. [Google Scholar]
- Mori S, Tsou C-H, Wu C-C, Cronholm B, Anderberg AA.. 2007. Evolution of Lecythidaceae with an emphasis on the circumscription of neotropical genera: information from combined ndhF and trnL-F sequence data. American Journal of Botany 94: 289–301. [DOI] [PubMed] [Google Scholar]
- Moyroud E, Glover BJ.. 2017. The physics of pollinator attraction. New Phytologist 216: 350–354. [DOI] [PubMed] [Google Scholar]
- Muñoz-Gómez S, Suárez-Báron H, Alzate JF, Gonzáles F, Pabón-Mora N.. 2021. Evolution of the subgroup 6 R2R3-MYB genes and their contribution to floral color in the perianth-bearing Piperales. Frontiers in Plant Science 12: 633227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadot S, Carrive L.. 2021. The colourful life of flowers. Botany Letters 168: 120–130. [Google Scholar]
- Ng J, Smith SD.. 2016. Widespread flower color convergence in Solanaceae via alternate biochemical pathways. New Phytologist 209: 407–417. [DOI] [PubMed] [Google Scholar]
- Nyffeler R, Baum DA.. 2001. Systematics and character evolution in Durio s. lat. (Malvaceae/Helicteroideae/Durioneae or Bombacaceae-Durioneae). Organisms Diversity and Evolution 1: 165–178. [Google Scholar]
- Ollerton J, Alarcón R, Waser NM, et al. 2009. A global test of the pollination syndrome hypothesis. Annals of Botany 103: 1471–1480. doi: 10.1093/aob/mcp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C. 1982. Morphological characters and homology. In: Joysey KA, Friday AE, eds. Problems of phylogenetic reconstruction. London: Academic Press, 21–74. [Google Scholar]
- Perret M, Chautems A, Spichiger R, Barraclough TG, Savolainen V.. 2007. The geographical pattern of speciation and floral diversification in the neotropics: the tribe Sinningieae (Gesneriaceae) as a case study. Evolution 61: 1641–1660. doi: 10.1111/j.1558-5646.2007.00136.x. [DOI] [PubMed] [Google Scholar]
- Prather LA. 1999. The relative lability of floral vs. non-floral characters and a morphological phylogenetic analysis of Cobaea (Polemoniaceae). Botanical Journal of the Linnean Society 131: 433–450. doi: 10.1111/j.1095-8339.1999.tb01525.x. [DOI] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, et al. 1999. Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11: 14330–11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz JA, Diniz UM, Vásquez DP, et al. 2020. Bats and hawkmoths form mixed modules with flowering plants in a nocturnal interaction network. Biotropica 53: 596–607. [Google Scholar]
- Raguso RA, Levin RA, Foose SE, Holmberg MW, McDade LA.. 2003. Fragrance chemistry, nocturnal rhythms and pollination ‘syndromes’ in Nicotiana. Phytochemistry 63: 265–284. doi: 10.1016/s0031-9422(03)00113-4. [DOI] [PubMed] [Google Scholar]
- Rausher MD. 2008. Evolutionary transitions in floral color. International Journal of Plant Science 169: 7–21. [Google Scholar]
- Reynolds RJ, Westbrook MJ, Rohde AS, Cridland JM, Fenster CB, Dudash MR.. 2009. Pollinator specialization and pollination syndromes of three related North American Silene. Ecology 90: 2077–2087. doi: 10.1890/08-1141.1. [DOI] [PubMed] [Google Scholar]
- Rieppel O. 2023. ‘Living fossil’ and the mosaic evolution of characters. Frontiers in Ecology and Evolution 11: 1119418. [Google Scholar]
- Ronse De Craene LP. 2007. Are petals sterile stamens or bracts? The origin and evolution of petals in the core eudicots. Annals of Botany 100: 621–630. doi: 10.1093/aob/mcm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronse De Craene LP. 2008. Homology and evolution of petals in the core eudicot. Systematic Botany 33: 301–325. doi: 10.1600/036364408784571680. [DOI] [Google Scholar]
- Ronse De Craene LP. 2010. Floral diagrams: an aid to understanding flower morphology and evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Ronse De Craene LP, Brockington SF.. 2013. Origin and evolution of petals in angiosperms. Plant Ecology and Evolution 146: 5–25. [Google Scholar]
- Rosas-Guerrero V, Aguilar R, Martén-Rodríguez S, et al. 2014. A quantitative review of pollination syndromes: do floral traits predict effective pollinators? Ecology Letters 17: 388–400. doi: 10.1111/ele.12224. [DOI] [PubMed] [Google Scholar]
- Rudall PJ. 2020. Colourful cones: how did flower colour first evolve? Journal of Experimental Botany 71: 759–767. [DOI] [PubMed] [Google Scholar]
- Sanders H, Rothwell GW, Wyatt S.. 2007. Paleontological context for the developmental mechanisms of evolution. International Journal of Plant Science 168: 719–728. [Google Scholar]
- Sazima M, Buzato S, Sazima I.. 1999. Bat-pollinated flower assemblages and bat visitors at two Atlantic forest sites in Brazil. Annals of Botany 83: 705–712. [Google Scholar]
- Schiestl FP, Johnson SD.. 2013. Pollinator-mediated evolution of floral signals. Trends in Plant Science 28: 307–315. [DOI] [PubMed] [Google Scholar]
- Scotland RW. 2011. What is parallelism? Evolution and Development 13: 214–227. doi: 10.1111/j.1525-142x.2011.00471.x. [DOI] [PubMed] [Google Scholar]
- Sereno P. 2007. Logical basis for morphological characters in phylogenetics. Cladistics 23: 565–587. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S.. 2009. Deep homology and the origins of evolutionary novelty. Nature 457: 818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- Sinnott-Armstrong MA, Deanna R, Pretz C, et al. 2022. How to apply the study of syndromes in macroevolution and ecology. Ecology and Evolution 12: e8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JM, Streisfeld MA.. 2013. Flower color as a model system for studies of plant evo-devo. Frontiers in Plant Science 4: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Endress PK, Chase MW.. 2005. Phylogeny and evolution of angiosperms. Sunderland: Sinauer Associates. [Google Scholar]
- Tanaka Y, Sasaki N, Ohmiya A.. 2008. Biosynthesis of plant pigments: anthocyanins, betalains, and carotenoids. Plant Journal 54: 733–749. doi: 10.1111/j.1365-313x.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- Tripp EA, Manos PS.. 2008. Is floral specialization an evolutionary dead-end? Pollination system transitions in Ruellia (Acanthaceae). Evolution 62: 1712–1737. doi: 10.1111/j.1558-5646.2008.00398.x. [DOI] [PubMed] [Google Scholar]
- Wagner GP. 2007. The developmental genetics of homology. Nature Reviews Genetics 8: 473–479. doi: 10.1038/nrg2099. [DOI] [PubMed] [Google Scholar]
- Wallace RS. 2002. The phylogeny and systematics of columnar cacti: an overview. In: Fleming TH, Valiente-Banuet A, eds. Columnar cacti and their mutualists: evolution, ecology, and conservation. Tucson: University of Arizona Press, 42–65. [Google Scholar]
- Wu CA, Streisfeld MA, Nutter LI, Cross KA.. 2013. The genetic basis of a rare flower color polymorphism in Mimulus lewisii provides insight into the repeatability of evolution. PLoS One 8: e81173. doi: 10.1371/journal.pone.0081173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall RA, Rausher MD.. 2004. Genetic changes associated with floral adaptation restrict future evolutionary potential. Nature 428: 847–850. doi: 10.1038/nature02489. [DOI] [PubMed] [Google Scholar]


