Abstract
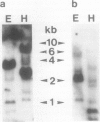
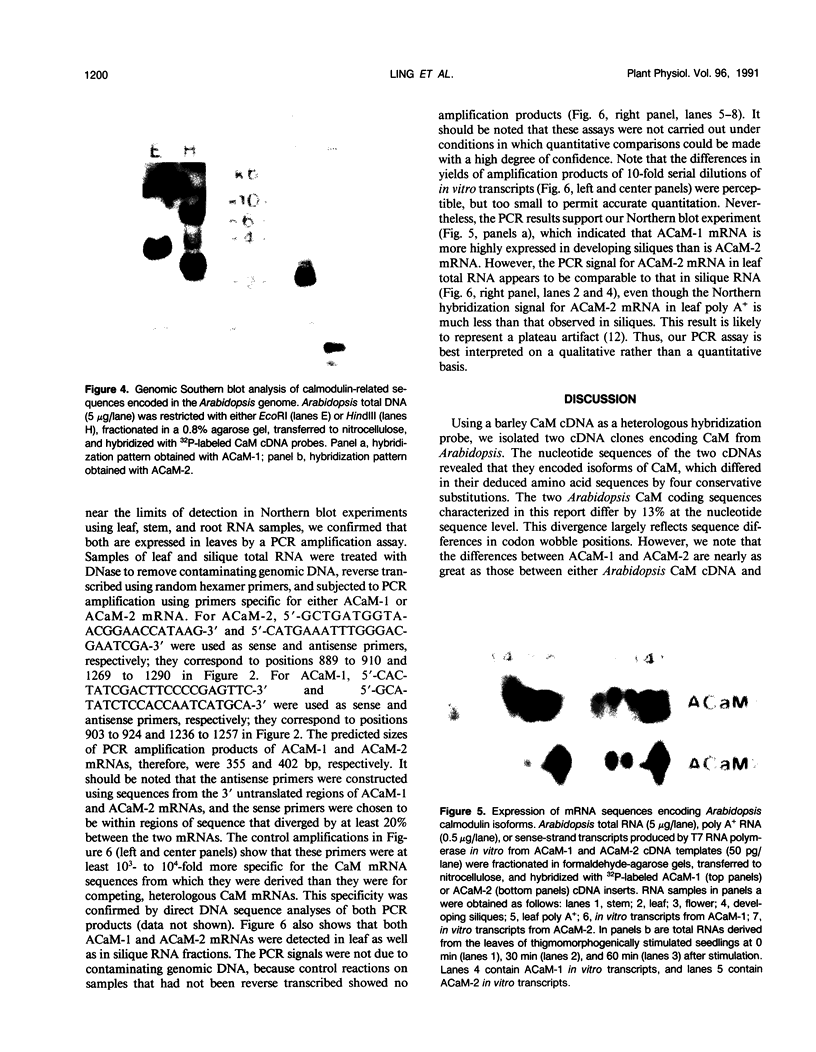
Complementary DNA (cDNA) clones encoding calmodulin isoforms were isolated from an Arabidopsis leaf λgt10 library by screening with cloned barley calmodulin cDNA probes. Two cDNAs, one a 626-base pair partial-length clone (ACaM-1) and one a 1400-base pair full-length clone (ACaM-2), encode calmodulin polypeptides that differ by four conservative amino acid substitutions. None of the amino acid sequence differences occur within the four Ca2+-binding domains of the proteins. Whereas the deduced amino acid sequences of the two Arabidopsis calmodulin isoforms share 97% identity, the nucleotide sequences encoding the two isoforms share 87% sequence identity. Most of these nucleotide sequence differences (80%) occur in codon wobble positions. ACaM-1 and ACaM-2 both hybridize with a distinct set of restriction fragments of Arabidopsis total DNA, indicating that they were derived from transcripts of separate genes; these genes are single- or very low-copy in the Arabidopsis genome. Both cDNAs hybridize to messenger RNA (mRNA) species of 0.8 kilobases that are expressed to a greater extent in developing siliques compared with leaves, flowers, and stems. Northern blot and polymerase chain reaction assays both indicate that ACaM-1 mRNA is more highly expressed than ACaM-2 mRNA in developing siliques. The steady-state levels of both isoform mRNAs increase as a result of touch stimulation; the kinetics and extent of increase are comparable for the two mRNAs.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barnett M. J., Long S. R. Nucleotide sequence of an alfalfa calmodulin cDNA. Nucleic Acids Res. 1990 Jun 11;18(11):3395–3395. doi: 10.1093/nar/18.11.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz G. A., Jacobs K. A., Eickbush T. H., Cherbas P. T., Kafatos F. C. Isolation of multigene families and determination of homologies by filter hybridization methods. Methods Enzymol. 1983;100:266–285. doi: 10.1016/0076-6879(83)00061-0. [DOI] [PubMed] [Google Scholar]
- Braam J., Davis R. W. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell. 1990 Feb 9;60(3):357–364. doi: 10.1016/0092-8674(90)90587-5. [DOI] [PubMed] [Google Scholar]
- Burgess W. H. Characterization of calmodulin and calmodulin isotypes from sea urchin gametes. J Biol Chem. 1982 Feb 25;257(4):1800–1804. [PubMed] [Google Scholar]
- Chien Y. H., Dawid I. B. Isolation and characterization of calmodulin genes from Xenopus laevis. Mol Cell Biol. 1984 Mar;4(3):507–513. doi: 10.1128/mcb.4.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R., Koller M., Flura M., Mathews S., Strehler-Page M. A., Krebs J., Penniston J. T., Carafoli E., Strehler E. E. Multiple divergent mRNAs code for a single human calmodulin. J Biol Chem. 1988 Nov 15;263(32):17055–17062. [PubMed] [Google Scholar]
- Hall T. C., Ma Y., Buchbinder B. U., Pyne J. W., Sun S. M., Bliss F. A. Messenger RNA for G1 protein of French bean seeds: Cell-free translation and product characterization. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3196–3200. doi: 10.1073/pnas.75.7.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy D. O., Bender P. K., Kretsinger R. H. Two calmodulin genes are expressed in Arbacia punctulata. An ancient gene duplication is indicated. J Mol Biol. 1988 Jan 5;199(1):223–227. doi: 10.1016/0022-2836(88)90392-0. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hodgson C. P., Fisk R. Z. Hybridization probe size control: optimized 'oligolabelling'. Nucleic Acids Res. 1987 Aug 11;15(15):6295–6295. doi: 10.1093/nar/15.15.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena P. K., Reddy A. S., Poovaiah B. W. Molecular cloning and sequencing of a cDNA for plant calmodulin: signal-induced changes in the expression of calmodulin. Proc Natl Acad Sci U S A. 1989 May;86(10):3644–3648. doi: 10.1073/pnas.86.10.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling V., Zielinski R. E. Cloning of cDNA Sequences Encoding the Calcium-Binding Protein, Calmodulin, from Barley (Hordeum vulgare L.). Plant Physiol. 1989 Jun;90(2):714–719. doi: 10.1104/pp.90.2.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas T. J., Iverson D. B., Schleicher M., Watterson D. M. Structural characterization of a higher plant calmodulin : spinacia oleracea. Plant Physiol. 1984 Jul;75(3):788–795. doi: 10.1104/pp.75.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas T. J., Wiggins M. E., Watterson D. M. Amino Acid sequence of a novel calmodulin from the unicellular alga chlamydomonas. Plant Physiol. 1985 Jul;78(3):477–483. doi: 10.1104/pp.78.3.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak D. R., Clarke M., Roberts D. M., Watterson D. M. Structural and functional properties of calmodulin from the eukaryotic microorganism Dictyostelium discoideum. Biochemistry. 1984 Jun 19;23(13):2891–2899. doi: 10.1021/bi00308a007. [DOI] [PubMed] [Google Scholar]
- Nojima H., Kishi K., Sokabe H. Multiple calmodulin mRNA species are derived from two distinct genes. Mol Cell Biol. 1987 May;7(5):1873–1880. doi: 10.1128/mcb.7.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovaiah B. W., Reddy A. S. Calcium messenger system in plants. CRC Crit Rev Plant Sci. 1987;6(1):47–103. doi: 10.1080/07352688709382247. [DOI] [PubMed] [Google Scholar]
- Roberts D. M., Burgess W. H., Watterson D. M. Comparison of the NAD Kinase and Myosin Light Chain Kinase Activator Properties of Vertebrate, Higher Plant, and Algal Calmodulins. Plant Physiol. 1984 Jul;75(3):796–798. doi: 10.1104/pp.75.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. M., Crea R., Malecha M., Alvarado-Urbina G., Chiarello R. H., Watterson D. M. Chemical synthesis and expression of a calmodulin gene designed for site-specific mutagenesis. Biochemistry. 1985 Sep 10;24(19):5090–5098. doi: 10.1021/bi00340a020. [DOI] [PubMed] [Google Scholar]
- Roux S. J., Wayne R. O., Datta N. Role of calcium ions in phytochrome responses: an update. Physiol Plant. 1986;66:344–348. doi: 10.1111/j.1399-3054.1986.tb02430.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter G. R., Means A. R. Analysis of expression of multiple genes encoding calmodulin during spermatogenesis. Mol Endocrinol. 1989 Oct;3(10):1569–1578. doi: 10.1210/mend-3-10-1569. [DOI] [PubMed] [Google Scholar]
- Tschudi C., Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988 Feb;7(2):455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschudi C., Young A. S., Ruben L., Patton C. L., Richards F. F. Calmodulin genes in trypanosomes are tandemly repeated and produce multiple mRNAs with a common 5' leader sequence. Proc Natl Acad Sci U S A. 1985 Jun;82(12):3998–4002. doi: 10.1073/pnas.82.12.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Lukas T. J., Craig T. A., Wilson E., King M. M., Kwiatkowski A. P., Watterson D. M. Computational and site-specific mutagenesis analyses of the asymmetric charge distribution on calmodulin. Proteins. 1989;6(1):70–85. doi: 10.1002/prot.340060107. [DOI] [PubMed] [Google Scholar]
- Zielinski R. E. Calmodulin mRNA in Barley (Hordeum vulgare L.) : Apparent Regulation by Cell Proliferation and Light. Plant Physiol. 1987 Jul;84(3):937–943. doi: 10.1104/pp.84.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]